Abstract
Objective
More than 80% of autoimmune disease is female dominant, but the mechanism for this female bias is poorly understood. We suspected an X chromosome dose effect and hypothesized that trisomy X (47,XXX , 1 in ~1,000 live female births) would be increased in female predominant diseases (e.g. systemic lupus erythematosus [SLE], primary Sjögren’s syndrome [SS], primary biliary cirrhosis [PBC] and rheumatoid arthritis [RA]) compared to diseases without female predominance (sarcoidosis) and controls.
Methods
We identified 47,XXX subjects using aggregate data from single nucleotide polymorphism (SNP) arrays and confirmed, when possible, by fluorescent in situ hybridization (FISH) or quantitative polymerase chain reaction (q-PCR).
Results
We found 47,XXX in seven of 2,826 SLE and three of 1,033 SS female patients, but only in two of the 7,074 female controls (p=0.003, OR=8.78, 95% CI: 1.67-86.79 and p=0.02, OR=10.29, 95% CI: 1.18-123.47; respectively). One 47,XXX subject was present for ~404 SLE women and ~344 SS women. 47,XXX was present in excess among SLE and SS subjects.
Conclusion
The estimated prevalence of SLE and SS in women with 47,XXX was respectively ~2.5 and ~2.9 times higher than in 46,XX women and ~25 and ~41 times higher than in 46,XY men. No statistically significant increase of 47,XXX was observed in other female-biased diseases (PBC or RA), supporting the idea of multiple pathways to sex bias in autoimmunity.
Introduction
Autoimmune disease affects 5-10% of the global population. Female preponderance is highlighted by the fact that almost 80% of autoimmune patients are female(1). Systemic lupus erythematosus (SLE), primary Sjögren's syndrome (SS), primary biliary cirrhosis (PBC), rheumatoid arthritis (RA) all have a female sex bias, though the ratios vary from fourteen women to one man in SS to only two to six women to one man in RA(2-5).
Several hypotheses have been proposed to explain the female preponderance in autoimmunity, such as sex hormones(6) and X monosomy(7, 8), but origins remain unknown. We have previously shown that Klinelfelter’s syndrome (47,XXY) is found in excess among men with SLE, and the calculated prevalence of SLE among Klinefelter’ syndrome is similar to the prevalence in women (46,XX)(9). Data for SLE in four core genotype mice show that the disease is associated with the number of X chromosomes, and not phenotypic sex (10, 11). In aggregate these data are consistent with the gene dose of chromosome X being a major determining factor for the female predominance of SLE in man and mouse.
In order to further test this X chromosome dose hypothesis, we asked whether having a third X chromosome increases the risk of developing SLE or other autoimmune diseases. Trisomy X (47,XXX) is present in approximately 1 out of every 1000 live-born girls(12, 13). There are no consistently recognized abnormalities in sex hormone levels, sexual development, fertility, or pregnancy(14), though premature ovarian failure has been associated with carriage of 47,XXX(15-17). One suggested phenotype difference is that 47,XXX women may have more anxiety and shyness and lower self-esteem(18). In fact, 47,XXX is usually unrecognized, with over 90% of the patients undiagnosed unless patients are screened for chromosomal abnormalities for other reasons(14). Herein, we show that 47,XXX is enriched among patients with SLE and SS, but not enriched in patients with PBC, RA or the non-autoimmune disease sarcoidosis.
Materials and Methods
Subjects and Genotyping Methods
We used genotyping records of subjects enrolled in multiple different genetic studies (some published) for this work. We excluded all male subjects from our study. To be included as a case in this study, subjects need to have a confirmed diagnosis of a disease of interest: SLE, SS, PBC, RA or sarcoidosis. For controls, we used non-affected SLE family controls, non-auto-inflammatory subjects, population controls and non-sarcoidosis controls. A summary of recruitment and inclusion information for each cohort used in this study is available in the supplementary information (Supplementary table 1a, 1b). Institutional review boards (IRB) at each site provided approval for this study, and individual informed consent was obtained from all participants.
All patients with SLE met the American College of Rheumatology (ACR) classification criteria for systemic lupus erythematosus (19). Subjects with SS met the 2002 American-European Consensus Group (AECG) classification criteria for primary Sjögren's syndrome (20). PBC patients met the American Association for the Study of Liver Diseases Diagnostic Criteria for PBC(21). Participants with RA met 1987 American College of Rheumatology criteria for the classification of RA (22). Sarcoidosis patients had a) tissue confirmation of granuloma, or b) chest radiographic evidence of bilateral symmetrical hilar adenopathy with either a history of erythema nodosum or at least two years observation during which time no other disease was found to explain radiographic abnormalities(23-26) (Supplementary table 1a).
Controls include first degree family members (sisters and mothers) of SLE patients, all of whom were shown not to have SLE (27). Other controls are healthy individual with no auto-inflammatory diseases from three cohorts (28-30). Population controls of European decedents recruited globally were also added (31). Another population control group consisted of self-identified African Americans receiving care at Mt. Sinai Hospital in New York. The last control cohorts were recruited as part of the sarcoidosis studies, and were without sarcoidosis (23-26) (Supplementary table 1b).
Genotyping records are from various platforms: ImmunoChip [196,524 single nucleotide polymorphisms (SNPs), Illumina], HumanHap370 Beadchip (300,000 SNPs, Illumina). Omni1 chip (1,134,514 SNPs, Illumina), OmniExpress chip (741,000 SNPs, Illumina) or Omni5-Quad chip (4,301,331 SNPs, Illumina). A summary of genotyping platforms for each cohort used in this study is available in the supplementary information (Supplementary table 1a, 1b).
47,XXX identification
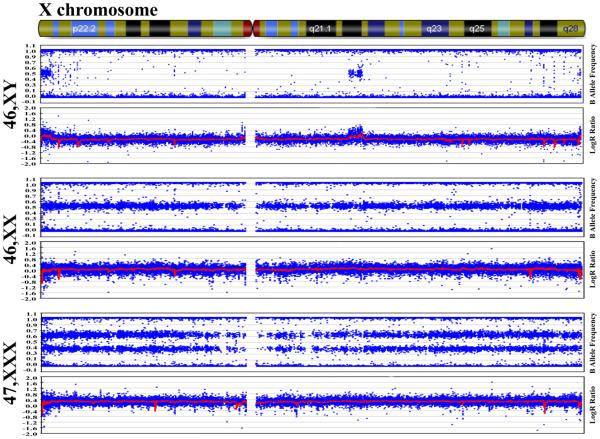
X chromosome copy number variations were found by visually inspecting B allele frequency and LogR ratio (LRR) plot of each subject's X chromosome for abnormalities using Genome Studio (Illumina), then mean logR ratio was calculated to confirm gain of copy number. In the B allele frequency plot, fluorescence intensity of the B allele of any SNP is plotted over the total fluorescence for that SNP in a given individual. Therefore, at a SNP with BB homozygosity the result is 100%, with AA homozygosity the result is 0%, and with AB heterozygosity the result is 50%. In the LRR plot, the fluorescence intensity of total allele A and B of the given sample is normalized by the reference genome to differentiate between copy number gains and losses.
Normal females (46,XX) were identified by the heterozygous "three band" pattern in the B allele frequency plot, which corresponds to the 0%, 50%, and 100% SNP frequencies (i.e. AA, AB, BB). The mean logR ratio, shown by a red line at 0.0 in the LRR plot, indicates there are two copies of the X chromosome (Figure 1).
Fig. 1. 47,XXX identification.
B allele frequency and logR ratio plot of 46,XY from a representative normal man (first panel, for comparison purposes only as men were not included in the present study), 46,XX from a normal woman (second panel) and 47,XXX from a trisomy X syndrome patient (third panel).
Trisomy X (47,XXX) subjects were identified by the "four band" pattern in the B allele frequency plot, which corresponds to 0%, 33%, 66%, and 100% SNP frequencies (i.e. AAA, AAB, ABB, BBB) with the accompanying mean LRR near 0.2 (Figure 1).
Fluorescence in situ hybridization (FISH)
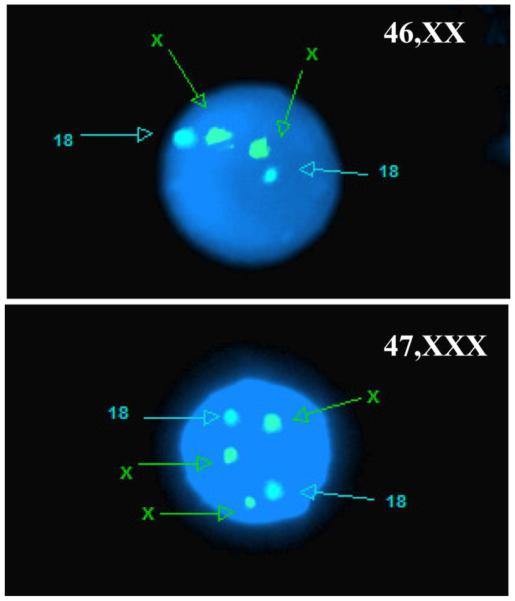
When cells were available, samples with chromosome abnormalities identified by B allele frequency and LRR plots were validated by FISH, as previously reported(32). For subjects that had frozen peripheral blood mononuclear cells (PBMCs), we used commercial FISH probes that recognize the centromeres of chromosomes X and Y to confirm aneuploidies of chromosome X (alpha-satellite repeats DXZ1 & DYZ3, PID# KI-20030, Veridex). Fifty nuclei were scored for each subject. For subjects that only had Epstein-Barr virus transformed B lymphocyte cell lines (LCLs or lymphoblastoid cell lines) available, we used commercial FISH probes that recognize the X centromeres, Yp and chromosome 18 (DXZ1, DYZ3 and D18Z1, Vysis) to confirm aneuploidies of chromosome X. Chromosome 18 probe signals served as controls to establish that the cell lines tested had a normal diploid genome. Two hundred nuclei were scored for each subject. Possible mosaicism was assessed according to the percentage of 45,X, 46,XX, or 47,XXX cells enumerated (Figure 2).
Fig. 2. 47,XXX validation by fluorescence in situ hybridization assay (FISH).
Validation of 46,XX from a normal female (upper) and 47,XXX from a trisomy X syndrome patient (bottom). The images are showing a single representative nucleus. 200 nuclei were counted.
Polymerase Chain Reaction (PCR)
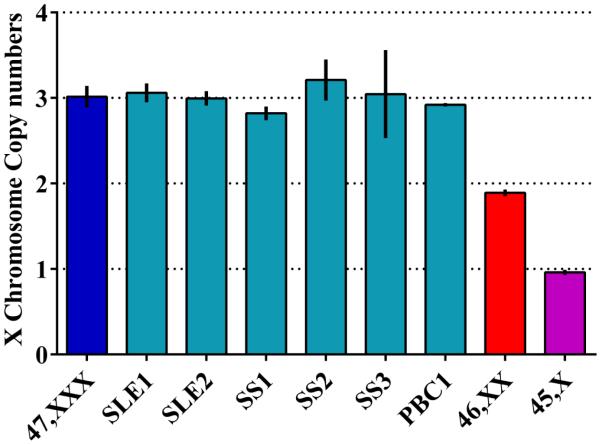
47,XXX samples having only DNA available were confirmed by PCR using TaqMan® copy variation assays (Life technology). Copy number of a gene on the X chromosome (YIPF6) was normalized to TaqMan® copy number reference assay, RNase P, on chromosome 14. Real time PCR was performed on an Applied Biosystems 7500 thermocycler. Data were analyzed using CopyCaller v2.0 software (Figure 3).
Fig. 3. 47,XXX validation by PCR.
DNA amplification validation of 47,XXX for two systemic lupus erythematosus patients (SLE1, SLE2), two primary Sjögren's syndrome patients (SS1, SS2) and one primary biliary cirrhosis patient (PBC1). As calibrators, we used known 47,XXX (shown in blue), 46,XX (shown in red) and 45X (shown in purple) samples. Calibrators were all validated by FISH and used to determine copy number in the experimental samples.
Quality control and statistical analysis
Subjects that had a call frequency lower than 0.90, genetically XY individuals self-reported as females, subjects whose samples indicated mixing of samples from multiple individuals or duplicated samples were removed.
The enrichment of 47,XXX in SLE, SS, PBC, RA subjects versus control subjects were tested by two-tailed Fisher’s exact test using R (Version 3.2.2) (Table 1).
Table 1.
47,XXX incidence. 47,XXX in systemic lupus erythematosus (SLE), primary Sjögren's syndrome (SS), primary biliary cirrhosis (PBC), rheumatoid arthritis (RA), sarcoidosis and controls. Each disease state is compared to the control group. P-value, OR with 95% CI were calculated by Fisher exact test.
| Disease | F/M ratio |
Sample Size |
47,XXX | Odds Ratio (95%CI) |
p value | Incidence |
|---|---|---|---|---|---|---|
| Systemic lupus erythematosus |
10:1 | 2826 | 7 | 8.78 (1.67-86.79) |
0.003 | 1/404 |
| Sjögren’s syndrome | 14:1 | 1033 | 3 | 10.29 (1.18-123.47) |
0.02 | 1/344 |
| Primary Biliary Cirrhosis |
10:1 | 1118 | 1 | 3.16 (0.05-60.89) |
NS | 1/1118 |
| Rheumatoid Arthritis | 2~3:1 | 1710 | 1 | 2.07 (0.03-39.75) |
NS | 1/1710 |
| Sarcoidosis | - | 939 | 0 | - | - | - |
| Controls | - | 7074 | 2 | - | - | 1/3537 |
The prevalence of SLE and SS among women with 47,XXX was estimated using Bayes’ theorem. Below are the Bayes’ theorem (P [B/A] = (P [A/B]*P [B])/P [A]) calculations to estimate the prevalence of SLE and Sjögren’s syndrome in females with 47,XXX.
A = the frequency of trisomy X, B = the frequency of SLE or SS in females, P [A/B] represents frequency of individuals with 47,XXX in SLE or SS, and P [B/A] would indicate the probability of SLE or SS within the group with trisomy X.
| Condition | Prevalences used in the Bayes’ theorem calculations |
|---|---|
| 1) SLE | = 1 in 1,000 women in the USA(2) |
| 2) SS | = 1 in 200 women in the USA(33) |
| 3) 47,XXX | = 1 in 1,000 women(34) |
| 4) 47,XXX with SLE | = 1 in 404 SLE women (data from this study) |
| 5) 47,XXX with SS | = 1 in 344 SS women (data from this study) |
| The Bayes’ theorem calculations: | |
| Prevalence of SLE in females with 47, XXX | |
| = [Freq (47,XXX in SLE) *Freq (SLE in females)] / Freq (47,XXX in females) | |
| Prevalence of SS in females with 47,XXX | |
| = [Freq (47,XXX in SS) *Freq (SS in female)] / Freq (47,XXX in females) | |
Population stratification was performed using SNP & Variation Suite software (Golden Helix) for SLE cases and controls. European ancestry and African ancestry were segregated within 3 s.d. of the mean of the first 2 principal components. Non-European or African ancestry subjects were dropped as well as the ones that their self-identified races did not match the principal component analysis results. Logistic regression modeling in R (Version 3.0.0) using glm function was performed to adjust odds ratio values for ancestry.
To confirm our results were not due to biases in the control groups, we also performed tests comparing 47,XXX enrichment in our case subjects to different control groups. The enrichment of 47,XXX in SLE versus non-affected SLE family controls was tested by two-tailed Fisher’s exact test using R (Version 3.2.2). The enrichment of 47,XXX in SS versus healthy non-auto-inflammatory controls was also tested by two-tailed Fisher’s exact test using R (Version 3.2.2). The enrichment of SLE and SS subjects versus a) controls without non-sarcoidosis subjects, b) controls without population controls and c) controls without non-sarcoidosis and population controls were also tested by two-tailed Fisher’s exact test using R (Version 3.2.2). Finally, the same analysis was used to test the enrichment of 47,XXX in SLE, SS, PBC, RA subjects versus unselected newborn infants controls as reported by Jacob et.al(34).
Results
Subjects
After quality control, we had 2826 SLE, 1033 SS, 1118 PBC, 1710 RA, 939 sarcoidosis and 7074 control subjects for our study. For controls, we had 2090 non-affected first degree family member of SLE patients, 1684 non-auto-inflammatory healthy control subjects, 2680 population control subjects and 620 non-sarcoidosis control subjects (Supplementary table 1a, 1b). There is no difference in prevalence of 47,XXX between SLE family control versus the rest controls by Fisher exact test (p=1, data not shown). A risk with the population controls is that some subjects might have the diseases of interest because this group represents a general population and was not specifically screened for the diseases of interest to this study. The non-sarcoidosis controls (n=620) may also contain subjects with diseases of interest (other than sarcoidosis) but the smaller number of subjects makes it less likely to contain SLE/SS/PBC/RA patients. Since the non-sarcoidosis control and population control each contains one of the two 47,XXX in the controls, we decided to retain them for modeling purpose. This decision potentially biases our results towards the null hypothesis. We therefore considered all three control cohorts in aggregate for our primary analysis.
47,XXX enrichment in systemic lupus erythematosus and primary Sjögren’s syndrome
Using single nucleotide polymorphism typing of the X chromosome, we found 47,XXX in seven of 2,826 women with SLE and in three of 1,033 women with SS, but only two in the 7,074 controls (p=0.003, OR=8.78, 95% CI: 1.67-86.79 and p=0.02, OR=10.29, 95% CI: 1.18-123.47; respectively; Table 1, Fig. 1). Five of the seven 47,XXX subjects with SLE were validated by FISH (Fig. 2). We validated one using frozen PBMCs and another four using lymphoblastoid cell lines (LCLs). Four subjects had 100% of 47,XXX cells, one had 98.5% of 47,XXX cells and 1.5% of 48,XXXX cells, which led us to consider this subject with the other 47,XXX subjects. We validated the two other 47,XXX subjects with SLE and the three with primary SS using q-PCR, which demonstrated a three X chromosome complement in all five DNA samples (Fig. 3). No DNA or cells were available from the two 47,XXX controls, but the complete fidelity between the genotyping screening and more traditional method supports the likelihood of accuracy in these samples.
Thus, we find that 1 in 404 (95% CI: 196-1004) women with SLE and 1 in 344 (95% CI: 115-1620) with SS have 47,XXX, ~2.5 and ~2.9 times higher, respectively, than the published population prevalence of 47,XXX which is ~1/1000(12, 13).
Controls in this study consisted of non-affected SLE family members, non-auto-inflammatory subjects, population controls, and non-sarcoidosis controls. The best controls for the SLE patients are the 2090 non-affected SLE family members. However, there are no 47,XXX participants found in this control cohort. There is a significant difference in 47,XXX enrichment when comparing SLE patients only to the family control cohort(Supplementary table 3). The non-affected SLE family members could possibly contain subjects with SS, so we tested 47,XXX enrichment in SS with only the 1684 healthy non-auto-inflammatory controls and the results remained significant (Supplementary table 4). The 2680 population controls and 620 non-sarcoidosis controls could potentially contain subjects with SLE or SS. We tested 47,XXX enrichment in SLE and SS with the removal of either or both of these control datasets and the results remained significant(Supplementary table 5a, 5b, 5c). We also compared 47,XXX in SLE and SS with a published unselected new born infants 47,XXX screening datasets (n=20,790) (34). Since a reported 47,XXX phenotype is increased anxiety and shyness combined with lower self-esteem(18), we feared 47,XXX participants potential may be less willing to participate in research studies, although this should equally affect case and control recruitment. Using the unselected new born infants’ dataset should have no recruitment bias but a potential bias in cases remains. We still have a significant enrichment of 47,XXX in SLE (p=0.036, OR=2.58, 95% CI: 0.92-6.36) and the same trend for 47,XXX enrichment in SS (p=0.093, OR=3.02, 95% CI: 0.57-10.22) (Supplementary table 6). We are therefore inclined to conclude that the comparison with the combination of all control cohorts is a fair representation of the enrichment in SLE and SS. The loss of significance in SS maybe due to small sample size.
Using the published population prevalence of 47,XXX in the population (~1/1000) and Bayes’ theorem(9, 12, 13), we predict that SLE exists in 1 of 404 women with 47,XXX and SS in 1of 69 women with 47,XXX. This calculation assumes that SLE is present in 1/1,000, and SS in 1/200 women in the USA. We estimate the prevalence of SLE and SS with 47,XXX is ~2.5 times higher than that with 46,XX and ~25 and ~41 times higher than 46,XY, respectively (Table 3).
Table 3.
Systemic lupus erythematosus (SLE) and primary Sjögren's syndrome (SS) relative risk with X chromosome numbers.
| X number | Karyotype | SLE Relative Risk | SS Relative Risk |
|---|---|---|---|
| 1 | 45X; 46XY | 1 | 1 |
| 2 | 46XX; 47XXY* | ~10 | ~14 |
| 3 | 47XXX | ~25 | ~41 |
47,XXY data with Sjögren's syndrome is not available here.
47,XXX risk is independent of ancestry
Individuals of African ancestry have a higher risk for SLE compared to those of European ancestry(35), and we observed a majority of the 47,XXX subjects self-identified as of African ancestry. While there are no studies on 47,XXX prevalence by ancestry, there is a study demonstrating 47,XXY differences by ancestry(34). Therefore, we asked whether the SLE risk associated with 47,XXX is dependent on ancestry. After population stratification, 375 samples were categorized as non-European/African ancestries and therefore excluded. An additional 144 samples were dropped because of a self-identified race mismatch with the principal component analysis, giving us a final of 1592 subjects of African ancestry (AA) and 1083 subjects of European ancestry (EA) with SLE, along with 3755 AA and 2951 EA control subjects. Among the seven 47,XXX women with SLE, five self-identified as being of African ancestry, and two of European ancestry. The two control 47,XXX women self-identified as of African ancestry. After principal component analysis for population stratification, one African ancestry SLE 47,XXX subject was removed as an admixed outlier (Table 2). We performed logistic regression analysis of SLE status with 47,XXX and ancestry as independent variables. The ancestry adjusted OR was 7.36 with p=0.01, similar to the unadjusted model (Supplementary Table 2). We also modeled using the interaction term between ancestry and 47,XXX to test for effect modification, but the interaction term was not significant. So we conclude that the odds of 47,XXX in SLE is independent of African ancestry in our data.
Table 2.
Numbers of 47,XXX by ancestry in SLE cases and controls.
| Disease | Race | 47,XXX | Sample Size | Total |
|---|---|---|---|---|
| Systemic lupus erythematosus | AA EA |
4 2 | 1592 1083 |
2675 |
| Controls | AA EA |
2 0 | 3755 2951 |
6706 |
| Total | AA EA |
6 2 |
5347 4034 |
9381 |
In addition, there was an increased incidence of 47,XXX among the SS patients. The SS patient cohort was almost entirely comprised of individuals of European ancestry (>95%); all three 47,XXX SS cases self-identified as being of European ancestry. This meant we were unable to adjust for ancestry effects in SS.
47, XXX in other autoimmune diseases
We found 47,XXX in 1 of 1,118 women with PBC, 1 of 1,710 with RA, and none of 939 with sarcoidosis (p>0.05, Table 1). The 47,XXX subject with PBC was validated to have three X chromosomes with q-PCR (Figure 3). No DNA or cells were available for the 47,XXX subject with RA. There was no statistically significant increase in prevalence of 47,XXX seen in PBC, RA or sarcoidosis compared to our controls. The comparison of 47,XXX in PBC, RA and sarcoidosis to the unselected newborn infants dataset also found no significant difference (Supplementary table 6).
Discussion
We showed that 47,XXX is more frequent in patients with SLE and SS. Indeed, this and previous work show that the risk of SLE and SS increases with each additional X chromosome (Table 3). The ~25-fold increase in predicted prevalence for SLE and ~41-fold increase for SS observed in 47,XXX women compared to 46,XY men reveals an effect on risk exceeding by many fold that of all other known genetic risk factors for SLE or SS. The importance of these findings is not for the few individuals with 47,XXX, but rather lies in the fact that rare events and phenotypes reveal insights into the mechanism for the general disease circumstances. Everyone has an X chromosome, and increased 47,XXX among women with either SLE or SS informs the potential mechanism underpinning the disparate risk of these diseases found for men and women with a normal sex chromosome complement. Because sexual development and sex hormones are normal in 47,XXX women, these data suggest that the number of X chromosomes is a key factor imparting the 10-fold risk difference between men and women.
Because 47,XXX is a rare event, in order to get a sufficiently large sample size, we included non-affected SLE family controls, non-auto-inflammatory subjects, population controls and non-sarcoidosis controls in our study. The 2090 non-affected SLE family controls may contain subjects with SS, we showed that when comparing 47,XXX enrichment in SS only with the healthy non-auto-inflammatory controls, the result is still significant (Supplementary table 4). The 2680 population control and 620 non-sarcoidosis controls may contain subjects with SLE or SS. We showed that even when removing these control groups, our 47,XXX enrichment in SLE and will still be significant if not more so (Supplementary table 5a,5b,5c). One possible phenotype associated with 47,XXX is anxiety, shyness and low self-esteem(18), a potential source of bias as 47,XXX subjects may be less willing to participate in research studies. This raises the question of whether 47,XXX would be less willing to participate in our study. We think this personality trait of 47,XXX women would lead to a non-differential error that would equally affect both case and control group recruitment. The majority (90%) of the 47,XXX are unrecognized(14), which means they only rarely exhibit a clear phenotype or that any phenotype is typically mild. We also compared our rate of 47,XXX in SLE, SS, RA, PBC, and sarcoidosis with a published unselected new born infants 47,XXX screening dataset(34). The newborn infant dataset has no bias in recruitment since consecutive births were studied (Supplementary table 6). Even in this case, we still have a significant enrichment of 47,XXX in SLE and a trend for 47,XXX enrichment in SS, which supports our hypothesis of X chromosome does effect in SLE and SS.
We did not find excess 47,XXX among women with the other autoimmune diseases studied here (PBC and RA) compared to our controls or the unselected newborn infant dataset (Table 1, Supplementary table 6). While it is possible that we had inadequate sample size for PBC and RA, the relationship of Turner’s syndrome (45,X) to autoimmunity also suggests heterogeneity in the mechanisms of the sex bias found in autoimmune diseases. Autoimmune thyroiditis, type 1 diabetes mellitus, and celiac disease are found in excess among Turner’s syndrome (45,X) patients(36). On the other hand, there is no evidence that prevalence of SLE or SS are increased in Turner’s syndrome. While rarer than the Turner’s-associated diseases, the paucity of Turner’s syndrome among SLE patients (37, 38) suggests that SLE risk in these patients is more similar to 46,XY males rather than to 46,XX females. Moreover, PBC, autoimmune thyroid disease (AITD) and systemic sclerosis have increased acquired X-monosomy but SLE does not(7, 8, 39), also suggesting existence of multiple mechanisms for sex bias in autoimmunity.
If SLE and 47,XXX both occur at 1 in ~1000 and were independent, then only 1 in a million women would have both SLE and 47,XXX. Our data suggest that both are coincident at 1 in ~40,000 women. The SLE families provide an interesting insight into how these risks may be modified. There are 7 women with SLE and 47,XXX in our sample of 2826 SLE cases, but no primary female relatives of SLE patients with 47,XXX among the 2090 subjects we examined. That this difference is significant (p=0.02) suggests that 47,XXX is a strong contributor to SLE when present within the family milieu of other genetics and environmental factors that establish risk for SLE (Supplementary table 3).
Sex hormones have long been considered a candidate mechanism of gender bias. We previously reported Klinefelter’s men (47,XXY) have a similar risk of SLE as women (46,XX) (9), while in the present study, 47,XXX women, who do not produce abnormal sex hormones(14, 40), have excess risk for SLE and SS compared to 46,XX women. Furthermore, there is no difference in the levels of sex hormones between SLE patients at disease onset and healthy controls(41). Other data suggest that sex hormone levels in men with SLE are abnormal, but similar to those found in men with other non-female-biased chronic illnesses(42). The female bias in SLE also exists before puberty with girls being five times more likely to develop SLE than boys(43). Similarly, the sex bias in SLE is unaffected by the lower estrogen levels after menopause, and is not statistically different from those with disease onset in the reproductive years(44). These observations suggest that X chromosome dose contributing to sex bias in SLE is independent of hormonal effects. X chromosome dose effect predisposes women to greater susceptibility to SLE and SS.
Perhaps the observation of a global reduction in methylation in SLE by Richardson and colleagues in activated T cells of SLE patients (45) offers possible explanations for a gene dose hypothesis. Each additional X chromosome, which is normally inactivated by methylation, could exert an effect if methylation is reversed or if additional X chromosomes affected methylation-related regulation of autosomal genes. Alternatively, a gene or gene(s) that escapes X inactivation (46), producing a gene-dose effect could be an explanation. Support for above mechanisms has not yet been developed.
To conclude, the available data strongly support a genetic cause of the powerful female bias in SLE and SS being located on the X chromosome with a mechanism that appears to be at least partially independent of circulating sex hormones. Our new data demonstrating increased risk of SLE and SS among 47,XXX women indicate that the female bias may be explained, at least in part, by an X chromosome gene dose effect. Identifying the specific properties of the X chromosome responsible for the increase in SLE and SS risk will be critical for advancing the understanding of these disorders.
Supplementary Material
Acknowledgments
We thank for AECG for SS data. We thank for Dr. Siminovitch’s group for kindly providing us PBC and part of the RA data. We thank for GENAR collaboration for providing us part of the RA data. We thank for Dr. Montgomery’s group for providing us the sarcoidosis data. We thank the Cincinnati control cohort group for allowing us to use the data. We thanks Dr. Bottinger for providing us the Mt. Sinai African American control dataset. Also, this research would not have been possible without the help and cooperation of the patients and others who participated in this study. We thank the cohort personnel, physicians, and genotyping core for their help with acquisition, referral, and processing of several thousand samples.
Funding:
This work was funded by the USA National Institutes of Health (AR062755, AI024717, AI031584 , AI062629 , AI063274, AI082714, AI083194, AI101934, AR042460, AR048204, AR048940, AR049084, AR052125, AR053483, AR053734, AR056360, AR058959, AR062277, AR043814, AR065626, DE015223, DE018209, RR020143, GM103510, GM104938, HG008667, TR001475 and HG006828), the Intramural Research Program of the National Institute of Dental and Craniofacial Research, the University of Oklahoma Health Sciences Center and its Clinical and Translational Science OCTSI Summer Scholar Program, the U.S. Department of Veterans Affairs (IMMA9 to RHS and JBH), the U.S. Department of Defense (PR094002), Alliance for Lupus Research (JBH), Mary Kirkland Scholar (JAJ and JBH), the Strategic Research Program at Helse Bergen, The Western Norway Regional Health Authority, The Broegelmann Foundation, EvASSESS PHRC (Programme Hospitalier de Recherche Clinique) 2006 from the French Ministry of Health, the Swedish Rheumatism Foundation, Arthritis Australia, an unrestricted grant from Research to Prevent Blindness to the Dean McGee Eye Institute and the Department of Ophthalmology, University of Oklahoma College of Medicine, the Medical Research Council, UK (G0800629), DFG KFO 250 WI 1031/6-1 (TW), a Senior Scientific Investigator Award to James Chodosh from Research to Prevent Blindness, Canadian Institutes for Health Research (MOP89955and MOP74621),nOntario Research Fund(RE05-075), Canada Research Chair(KAS), and the ISCIII (02558) through FEDER funds (MEAR).
Footnotes
Competing interests:
We declare that we have no conflict of interest.
Author contributions:
JB Harley and RH Scofield designed this study conception.
K Liu, BT Kurien, SL Zimmerman, KM Kaufman, DH Taft, JB Harley and RH Scofield participated in data analysis.
K Liu, BT Kurien, KM Kaufman, LC Kottyan, S. Lazaro, CA Weaver, JA Ice, AJ Adler, J Chodosh , L Radfar, A Rasmussen, DU Stone, KS Hefner, GD Houston, DM Lewis, S Li, KA Koelsch, A Igoe, M Dave, J Kumar, A D’souza, JS Maier-Moore, VM Harris, R Gopalakrishnan, R Jonsson, JA Lessard, X Lu, JE Gottenberg, JM Anaya, DS Cunninghame-Graham, AJW Huang, MT Brennan, P Hughes, GG. Illei, C Miceli-Richard, EC Keystone, VP Bykerk, G Hirschfield, G Xie, WF Ng, G Nordmark, P Eriksson, R Omdal, NL Rhodus, M Rischmueller, M Rohrer, BM Sega, TJ Vyse, M Wahren-Herlenius, T Witte, X Mariette, B Pons-Estel, ME Alarcon-Riquelme, JM Guthridge, JA James, CJ Lessard, JA Kelly, SD Thompson, PM Gaffney, CG Montgomery, KA Siminovitch, JC Edberg, RP Kimberly, GS Alarcón, CL Langefeld, GS Gilkeson, DL Kamen, BP Tsao, WJ McCune, JE Salmon, JT Merrill, MH Weisman, DJ Wallace, TO Utset, EP Bottinger, CI Amos, KL Sivils, JB Harley and RH Scofield participate in data acquisition.
K Liu, DH Taft, X Mariette, JB Harley and RH Scofield worked on manuscript preparation.
References
- 1.Eaton WW, Rose NR, Kalaydjian A, Pedersen MG, Mortensen PB. Epidemiology of autoimmune diseases in Denmark. Journal of autoimmunity. 2007;29(1):1–9. doi: 10.1016/j.jaut.2007.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Helmick CG, Felson DT, Lawrence RC, Gabriel S, Hirsch R, Kwoh CK, et al. Estimates of the prevalence of arthritis and other rheumatic conditions in the United States. Part I. Arthritis and rheumatism. 2008;58(1):15–25. doi: 10.1002/art.23177. [DOI] [PubMed] [Google Scholar]
- 3.Selmi C, Bowlus CL, Gershwin ME, Coppel RL. Primary biliary cirrhosis. Lancet. 2011;377(9777):1600–9. doi: 10.1016/S0140-6736(10)61965-4. [DOI] [PubMed] [Google Scholar]
- 4.Gabriel SE. The epidemiology of rheumatoid arthritis. Rheum Dis Clin North Am. 2001;27(2):269–81. doi: 10.1016/s0889-857x(05)70201-5. [DOI] [PubMed] [Google Scholar]
- 5.Massardo L, Pons-Estel BA, Wojdyla D, Cardiel MH, Galarza-Maldonado CM, Sacnun MP, et al. Early rheumatoid arthritis in Latin America: low socioeconomic status related to high disease activity at baseline. Arthritis care & research. 2012;64(8):1135–43. doi: 10.1002/acr.21680. [DOI] [PubMed] [Google Scholar]
- 6.Quintero OL, Amador-Patarroyo MJ, Montoya-Ortiz G, Rojas-Villarraga A, Anaya JM. Autoimmune disease and gender: plausible mechanisms for the female predominance of autoimmunity. Journal of autoimmunity. 2012;38(2-3):J109–19. doi: 10.1016/j.jaut.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 7.Invernizzi P, Miozzo M, Battezzati PM, Bianchi I, Grati FR, Simoni G, et al. Frequency of monosomy X in women with primary biliary cirrhosis. Lancet. 2004;363(9408):533–5. doi: 10.1016/S0140-6736(04)15541-4. [DOI] [PubMed] [Google Scholar]
- 8.Invernizzi P, Miozzo M, Selmi C, Persani L, Battezzati PM, Zuin M, et al. X chromosome monosomy: a common mechanism for autoimmune diseases. Journal of immunology (Baltimore, Md : 1950) 2005;175(1):575–8. doi: 10.4049/jimmunol.175.1.575. [DOI] [PubMed] [Google Scholar]
- 9.Scofield RH, Bruner GR, Namjou B, Kimberly RP, Ramsey-Goldman R, Petri M, et al. Klinefelter's syndrome (47,XXY) in male systemic lupus erythematosus patients: support for the notion of a gene-dose effect from the X chromosome. Arthritis and rheumatism. 2008;58(8):2511–7. doi: 10.1002/art.23701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Smith-Bouvier DL, Divekar AA, Sasidhar M, Du S, Tiwari-Woodruff SK, King JK, et al. A role for sex chromosome complement in the female bias in autoimmune disease. The Journal of experimental medicine. 2008;205(5):1099–108. doi: 10.1084/jem.20070850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sasidhar MV, Itoh N, Gold SM, Lawson GW, Voskuhl RR. The XX sex chromosome complement in mice is associated with increased spontaneous lupus compared with XY. Ann Rheum Dis. 2012;71(8):1418–22. doi: 10.1136/annrheumdis-2011-201246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nielsen J, Wohlert M. Chromosome abnormalities found among 34,910 newborn children: results from a 13-year incidence study in Arhus, Denmark. Hum Genet. 1991;87(1):81–3. doi: 10.1007/BF01213097. [DOI] [PubMed] [Google Scholar]
- 13.Jacobs PA, Browne C, Gregson N, Joyce C, White H. Estimates of the frequency of chromosome abnormalities detectable in unselected newborns using moderate levels of banding. J Med Genet. 1992;29(2):103–8. doi: 10.1136/jmg.29.2.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tartaglia NR, Howell S, Sutherland A, Wilson R, Wilson L. A review of trisomy X (47,XXX) Orphanet J Rare Dis. 2010;5:8. doi: 10.1186/1750-1172-5-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Devi A, Benn PA. X-chromosome abnormalities in women with premature ovarian failure. J Reprod Med. 1999;44(4):321–4. [PubMed] [Google Scholar]
- 16.Baronchelli S, Conconi D, Panzeri E, Bentivegna A, Redaelli S, Lissoni S, et al. Cytogenetics of premature ovarian failure: an investigation on 269 affected women. J Biomed Biotechnol. 2011;2011:370195. doi: 10.1155/2011/370195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jiao X, Qin C, Li J, Qin Y, Gao X, Zhang B, et al. Cytogenetic analysis of 531 Chinese women with premature ovarian failure. Hum Reprod. 2012;27(7):2201–7. doi: 10.1093/humrep/des104. [DOI] [PubMed] [Google Scholar]
- 18.Otter M, Schrander-Stumpel CT, Curfs LM. Triple X syndrome: a review of the literature. European journal of human genetics : EJHG. 2010;18(3):265–71. doi: 10.1038/ejhg.2009.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tan EM, Cohen AS, Fries JF, Masi AT, McShane DJ, Rothfield NF, et al. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis and rheumatism. 1982;25(11):1271–7. doi: 10.1002/art.1780251101. [DOI] [PubMed] [Google Scholar]
- 20.Shiboski SC, Shiboski CH, Criswell L, Baer A, Challacombe S, Lanfranchi H, et al. American College of Rheumatology classification criteria for Sjogren's syndrome: a data-driven, expert consensus approach in the Sjogren's International Collaborative Clinical Alliance cohort. Arthritis care & research. 2012;64(4):475–87. doi: 10.1002/acr.21591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lindor KD, Gershwin ME, Poupon R, Kaplan M, Bergasa NV, Heathcote EJ. Primary biliary cirrhosis. Hepatology. 2009;50(1):291–308. doi: 10.1002/hep.22906. [DOI] [PubMed] [Google Scholar]
- 22.Arnett FC, Edworthy SM, Bloch DA, McShane DJ, Fries JF, Cooper NS, et al. The American Rheumatism Association 1987 revised criteria for the classification of rheumatoid arthritis. Arthritis and rheumatism. 1988;31(3):315–24. doi: 10.1002/art.1780310302. [DOI] [PubMed] [Google Scholar]
- 23.Design of a case control etiologic study of sarcoidosis (ACCESS) ACCESS Research Group. Journal of clinical epidemiology. 1999;52(12):1173–86. doi: 10.1016/s0895-4356(99)00142-0. [DOI] [PubMed] [Google Scholar]
- 24.Rybicki BA, Hirst K, Iyengar SK, Barnard JG, Judson MA, Rose CS, et al. A sarcoidosis genetic linkage consortium: the sarcoidosis genetic analysis (SAGA) study. Sarcoidosis, vasculitis, and diffuse lung diseases : official journal of WASOG / World Association of Sarcoidosis and Other Granulomatous Disorders. 2005;22(2):115–22. [PubMed] [Google Scholar]
- 25.Iannuzzi MC, Maliarik MJ, Poisson LM, Rybicki BA. Sarcoidosis susceptibility and resistance HLA-DQB1 alleles in African Americans. American journal of respiratory and critical care medicine. 2003;167(9):1225–31. doi: 10.1164/rccm.200209-1097OC. [DOI] [PubMed] [Google Scholar]
- 26.Adrianto I, Lin CP, Hale JJ, Levin AM, Datta I, Parker R, et al. Genome-wide association study of African and European Americans implicates multiple shared and ethnic specific loci in sarcoidosis susceptibility. PloS one. 2012;7(8):e43907. doi: 10.1371/journal.pone.0043907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rasmussen A, Sevier S, Kelly JA, Glenn SB, Aberle T, Cooney CM, et al. The lupus family registry and repository. Rheumatology (Oxford, England) 2011;50(1):47–59. doi: 10.1093/rheumatology/keq302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hollenbach JA, Thompson SD, Bugawan TL, Ryan M, Sudman M, Marion M, et al. Juvenile idiopathic arthritis and HLA class I and class II interactions and age-at-onset effects. Arthritis and rheumatism. 2010;62(6):1781–91. doi: 10.1002/art.27424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hinks A, Cobb J, Marion MC, Prahalad S, Sudman M, Bowes J, et al. Dense genotyping of immune-related disease regions identifies 14 new susceptibility loci for juvenile idiopathic arthritis. Nature genetics. 2013;45(6):664–9. doi: 10.1038/ng.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gregersen PK, Amos CI, Lee AT, Lu Y, Remmers EF, Kastner DL, et al. REL, encoding a member of the NF-kappaB family of transcription factors, is a newly defined risk locus for rheumatoid arthritis. Nature genetics. 2009;41(7):820–3. doi: 10.1038/ng.395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lessard CJ, Li H, Adrianto I, Ice JA, Rasmussen A, Grundahl KM, et al. Variants at multiple loci implicated in both innate and adaptive immune responses are associated with Sjogren's syndrome. Nature genetics. 2013;45(11):1284–92. doi: 10.1038/ng.2792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dillon SP, Kurien BT, Li S, Bruner GR, Kaufman KM, Harley JB, et al. Sex chromosome aneuploidies among men with systemic lupus erythematosus. Journal of autoimmunity. 2012;38(2-3):J129–34. doi: 10.1016/j.jaut.2011.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vitali C, Bombardieri S, Jonsson R, Moutsopoulos HM, Alexander EL, Carsons SE, et al. Classification criteria for Sjogren's syndrome: a revised version of the European criteria proposed by the American-European Consensus Group. Ann Rheum Dis. 2002;61(6):554–8. doi: 10.1136/ard.61.6.554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jacobs PA. The incidence and etiology of sex chromosome abnormalities in man. Birth defects original article series. 1979;15(1):3–14. [PubMed] [Google Scholar]
- 35.Danchenko N, Satia JA, Anthony MS. Epidemiology of systemic lupus erythematosus: a comparison of worldwide disease burden. Lupus. 2006;15(5):308–18. doi: 10.1191/0961203306lu2305xx. [DOI] [PubMed] [Google Scholar]
- 36.Bonamico M, Pasquino AM, Mariani P, Danesi HM, Culasso F, Mazzanti L, et al. Prevalence and clinical picture of celiac disease in Turner syndrome. J Clin Endocrinol Metab. 2002;87(12):5495–8. doi: 10.1210/jc.2002-020855. [DOI] [PubMed] [Google Scholar]
- 37.Cooney CM, Bruner GR, Aberle T, Namjou-Khales B, Myers LK, Feo L, et al. 46,X,del(X)(q13) Turner's syndrome women with systemic lupus erythematosus in a pedigree multiplex for SLE. Genes and immunity. 2009;10(5):478–81. doi: 10.1038/gene.2009.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Takegami T, Nakao K, Nagayama Y, Fujita T, Hoshino T, Tsunematsu T, et al. [A case of SLE associated with Turner's syndrome of 45, XO/46, XXq+ mosaicism (author's transl)] Nihon Naika Gakkai Zasshi. 1980;69(7):861–6. doi: 10.2169/naika.69.861. [DOI] [PubMed] [Google Scholar]
- 39.Invernizzi P, Miozzo M, Oertelt-Prigione S, Meroni PL, Persani L, Selmi C, et al. X monosomy in female systemic lupus erythematosus. Ann N Y Acad Sci. 2007;1110:84–91. doi: 10.1196/annals.1423.010. [DOI] [PubMed] [Google Scholar]
- 40.Barr ML, Sergovich FR, Carr DH, Saver EL. The triplo-X female: an appraisal based on a study of 12 cases and a review of the literature. Can Med Assoc J. 1969;101(5):247–58. [PMC free article] [PubMed] [Google Scholar]
- 41.Chang DM, Chang CC, Kuo SY, Chu SJ, Chang ML. Hormonal profiles and immunological studies of male lupus in Taiwan. Clin Rheumatol. 1999;18(2):158–62. doi: 10.1007/s100670050075. [DOI] [PubMed] [Google Scholar]
- 42.Mackworth-Young CG, Parke AL, Morley KD, Fotherby K, Hughes GR. Sex hormones in male patients with systemic lupus erythematosus: a comparison with other disease groups. Eur J Rheumatol Inflamm. 1983;6(3):228–32. [PubMed] [Google Scholar]
- 43.Kamphuis S, Silverman ED. Prevalence and burden of pediatric-onset systemic lupus erythematosus. Nat Rev Rheumatol. 2010;6(9):538–46. doi: 10.1038/nrrheum.2010.121. [DOI] [PubMed] [Google Scholar]
- 44.Lalani S, Pope J, de Leon F, Peschken C. Clinical features and prognosis of late-onset systemic lupus erythematosus: results from the 1000 faces of lupus study. J Rheumatol. 2010;37(1):38–44. doi: 10.3899/jrheum.080957. [DOI] [PubMed] [Google Scholar]
- 45.Lu Q, Wu A, Tesmer L, Ray D, Yousif N, Richardson B. Demethylation of CD40LG on the inactive X in T cells from women with lupus. Journal of immunology (Baltimore, Md : 1950) 2007;179(9):6352–8. doi: 10.4049/jimmunol.179.9.6352. [DOI] [PubMed] [Google Scholar]
- 46.Carrel L, Willard HF. X-inactivation profile reveals extensive variability in X-linked gene expression in females. Nature. 2005;434(7031):400–4. doi: 10.1038/nature03479. [DOI] [PubMed] [Google Scholar]
- 47.Hirschfield GM, Liu X, Xu C, Lu Y, Xie G, Gu X, et al. Primary biliary cirrhosis associated with HLA, IL12A, and IL12RB2 variants. The New England journal of medicine. 2009;360(24):2544–55. doi: 10.1056/NEJMoa0810440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Juran BD, Hirschfield GM, Invernizzi P, Atkinson EJ, Li Y, Xie G, et al. Immunochip analyses identify a novel risk locus for primary biliary cirrhosis at 13q14, multiple independent associations at four established risk loci and epistasis between 1p31 and 7q32 risk variants. Human molecular genetics. 2012;21(23):5209–21. doi: 10.1093/hmg/dds359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lopez Herraez D, Martinez-Bueno M, Riba L, Garcia de la Torre I, Sacnun M, Goni M, et al. Rheumatoid arthritis in Latin Americans enriched for Amerindian ancestry is associated with loci in chromosomes 1, 12, and 13, and the HLA class II region. Arthritis and rheumatism. 2013;65(6):1457–67. doi: 10.1002/art.37923. [DOI] [PubMed] [Google Scholar]
- 50.Karlson EW, Sanchez-Guerrero J, Wright EA, Lew RA, Daltroy LH, Katz JN, et al. A connective tissue disease screening questionnaire for population studies. Annals of epidemiology. 1995;5(4):297–302. doi: 10.1016/1047-2797(94)00096-c. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.