Abstract
The Greater Middle East (GME) has been a central hub of human migration and population admixture. The tradition of consanguinity, variably practiced in the Gulf region, North Africa, and Central Asia 1–3, has resulted in an elevated burden of recessive disease4. Here we generated a whole exome GME variome from 1,111 unrelated subjects. We detected substantial diversity from sub-geographies, continental and subregional admixture, several ancient founder populations with little evidence of bottlenecks. Measured consanguinity was an order-of-magnitude above that of other sampled populations, and included an increased burden of runs of homozygosity (ROH), but no evidence for reduced burden of deleterious variation due to classically theorized ‘genetic purging’. Applying this database to unsolved GME recessive conditions reduced the number of potential disease-causing variants by 4–7-fold. These results reveal the variegated GME genetic architecture and support future human genetic discoveries in Mendelian and population genetics.
Keywords: Mutational load, whole exome sequencing, introgression, admixture, inbreeding coefficient, homozygous, derived allele frequency, consanguineous, selective pressure, runs of homozygosity
The Greater Middle East (GME), loosely defined as a large swath of Arab and non-Arab Muslim countries from Morocco in the west to as far east as Pakistan 5, is home to approximately 10% of the world’s population. Despite its invaluable contribution to our understanding of the genetic causes of inherited conditions, especially recessive conditions, and its critical hub as a crossroad to early civilizations, genetic architecture and extent of rare genetic variation remains poorly defined 6–8.
To address this shortcoming, the GME Variome Consortium collected whole-exome data on 1,794 self-reported nationals from GME regions participating in on-going genetics studies. In order to minimize selection bias or overrepresentation of disease alleles, we selected primarily healthy individuals from families, and wherever possible, removed from datasets the allele that brought the family to medical attention. Samples were jointly processed, and filtered for quality and familial relation, leaving 1,111 high-quality unrelated individuals.
We grouped the 1,111 GME exomes into six different GME subregions: Northwest Africa (NWA, 85 samples), Northeast Africa (NEA, 423 samples), Turkish Peninsula (TP, 140 samples), Syrian Desert (SD, 81 samples), Arabian Peninsula (AP, 214 samples), and Persia and Pakistan (PP, 168 samples) (Fig. S1, Table S1), which represent historic groupings, then compared with exomic data of nine established continental populations from 1000 Genomes (1000G) 9. Unbiased identity-by-state clustering showed that samples largely grouped according to the location of ascertainment, validating grouping criteria (Fig. S2).
To evaluate GME genetic substructure, we ran the unsupervised algorithm ADMIXTURE 10, where K=6 clusters minimized cross-validation error (Fig. S3). We found some overlap with the primary admixture components from Africa, Europe and East Asia at the edges of geography, but also a large proportion not found in previous reference samples (Figs. 1a, S4). The admixture results also aligned with publications reporting common variation 11–13.
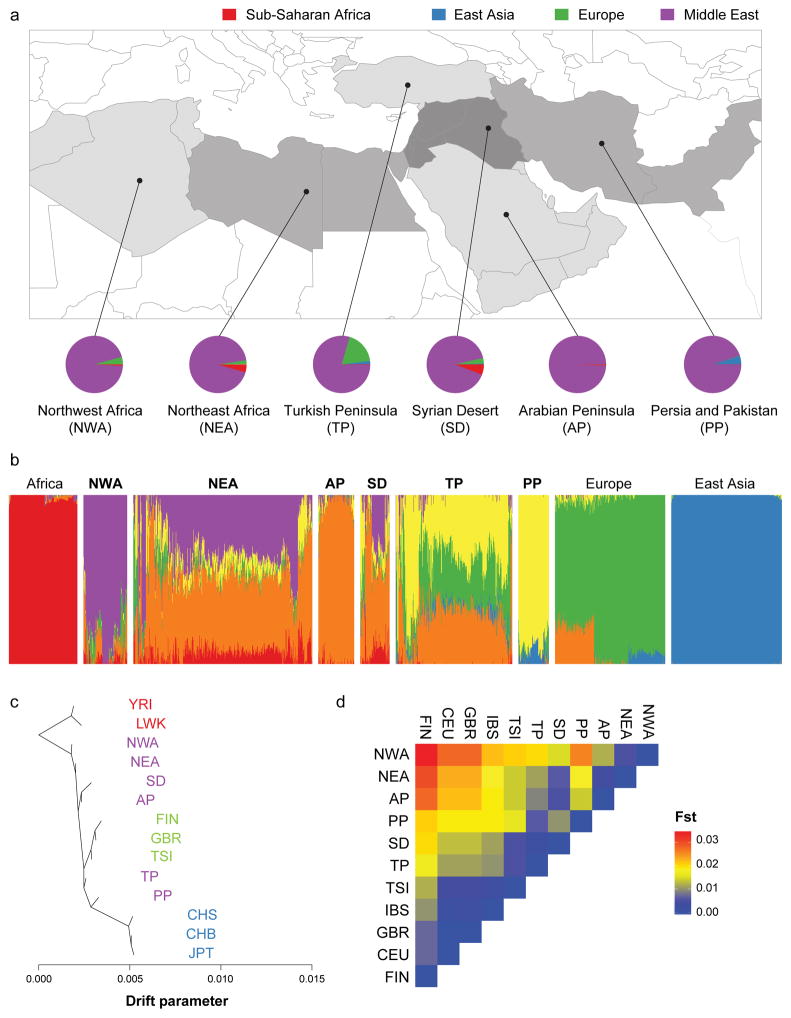
Figure 1. Greater Middle East Variome as a hub of human genetics.
a. Map of GME sub-regions. Lines define borders for admixture analysis from East Asia, Europe, Sub-Saharan Africa and the novel GME contribution (NWA: Northwest Africa, NEA: Northeast Africa, TP: Turkish Peninsula, SD: Syrian Desert, AP: Arabian Peninsula, PP: Persia and Pakistan). Pie charts: admixture proportions of 1000 Genomes Project (1000G) continental populations according to K=6 clusters.
b. Global ancestry proportions (K=6) for 1000G control populations with three distinct sources of contribution. 1000G population contributions: Africa (red), Europe (green) and East Asia (blue). GME populations from west to east: NWA (purple), AP (orange), and PP (yellow) derived from the GME.
c. TreeMix phylogeny of GME along with 1000G controls representing population divergence patterns. Length of the branch proportional to population drift. GME populations grouped around the African branch, but showed a substantial divergence. YRI: Yoruba in Ibadan, LWK: Luhya in Webuye Kenya, FIN: Finnish, GBR: Great Britain, TSI: Toscani, CHS: Southern Han Chinese, CHB: Han Chinese in Beijing, JPT: Japanese in Tokyo.
d. Wright’s Fixation Index (Fst) values for all pairs of GME and 1000G European populations, showing a smaller distance between GME and European populations compared with Sub-Saharan African populations. Greatest Fst value between any two GME populations was 0.026 (i.e. a quarter of the distance between FIN and JPT).
The least admixed samples were found in NWA, AP, and PP, suggesting these were founder populations, but showed inter-regional variation of GME-specific components suggesting local admixture (Fig. 1b), and potentially supporting historic events. The NWA component was found from west to east across North Africa, likely representing the presence of Berber genetic background 14. The AP component likely represented ancestral Arab populations and was observed in nearly all regions, possibly a result of the Arab conquests of the 7th century coincident with the expansion of the Arabic language now spoken over much of the region. Similarly, the Persian expansion into TP, SD, and parts of NEA in the 5th century was the most likely contributor of PP signal.
Additional sources of human heterogeneity derive from ancient introgression. We found similar patterns of Neanderthal introgression across all GME populations with the exception of NWA, which clustered closer to Sub-Saharan Africans (Fig. S5) 15–17. These data supports the reduced Neanderthal introgression observed in native African populations.
Patterns of human migration and drift were recapitulated using TreeMix among GME subregions, based upon 1000G control populations (Fig. 1c) 18. The inferred tree with no migration showed tight clusters of European and Asian populations, but much larger apparent divergence among GME regions. The ordering of GME subregions from the root corroborated much of the ‘out-of-Africa’ ordering of subsequent founder populations 13. Within the GME, the distance from the root emulated the west-to-east organization of GME samples, with PP showing the largest inferred drift parameter, supporting a west-to-east trajectory of human migrations.
Assessment of Wright’s fixation index (Fst) demonstrated that the GME grouped with European populations, agreeing with TreeMix results. This resulted in three distinct clusters with a low degree of differentiation (Figs. 1d, S6). PP and NWA represented the extremes of the identified subregions, and showed the highest degree of differentiation (Fst = 0.026) (>2x compared to the distance between Finnish (FIN) and Toscani (TSI) but smaller than intercontinental comparisons). Of the four measured 1000G European populations, GME Fst measurements were closest to TSI, especially SD and TP, consistent with higher levels of European admixture in these populations. Despite the contribution of admixture, these values suggested extended periods of isolation relative to 1000G populations within each subregion.
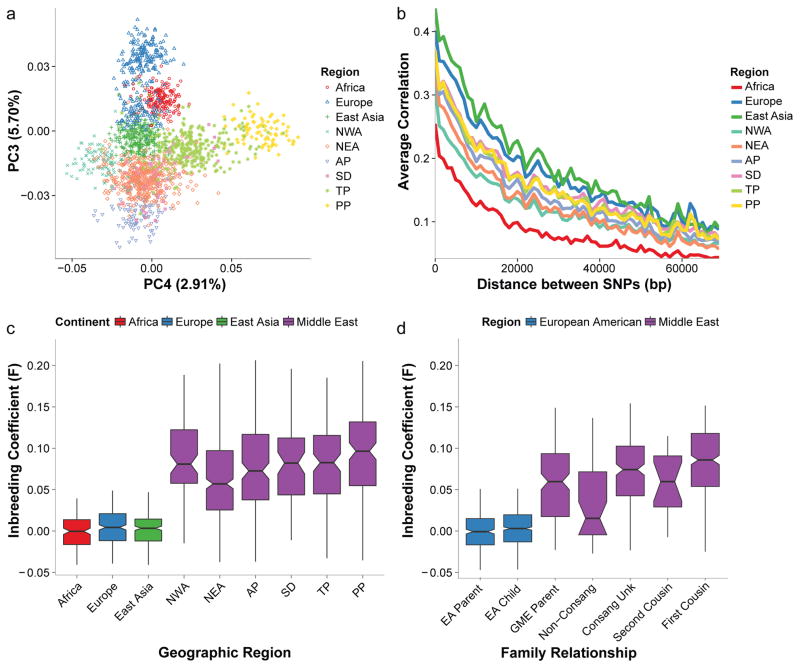
Inter-subregion relationships were tested using principal component analysis (PCA). As expected, the first two PCs separated along well-established geographic axes: PC1 separated Sub-Saharan Africans from all other populations, and PC2 separated Eurasian populations (Fig. S7). GME sub-regions fell between the 1000G African, East Asian, and European, supporting recent admixture. PP and TP were closer to East Asian, while NEA, NWA, and SD were closer to Sub-Saharan Africans. PC3 and PC4 separated samples along topographical north-south and east-west gradients, while exhibiting largely distinct but overlapping groups with a high-degree of inter-region diversity (Fig. 2a).
Figure 2. Wide diversity and high inbreeding coefficients in GME substructure.
a. Principal component analysis (PCA) for individuals from GME and 1000G populations. Individuals projected along PC3 and PC4 axes. Persia and Pakistan (PP), Northwest Africa (NWA) and Europe defined the limits from right, left, and top, as coinciding with geography. Arab Peninsula (AP) defined the bottom limit, and was closest to Northeast Africa (NEA) and Syrian Desert (SD).
b. GME populations had increased rates of linkage disequilibrium decay compared to 1000G European and East Asian populations. Mean variant correlations (r2) shown for each 1,000 basepair (bp) bin from 1,000–70,000 bp.
c. Inbreeding coefficient (F) distributions for GME and 1000G populations. GME populations (purple) showed elevated F values, consistent with increased rates of consanguineous marriages. Box plots show median (horizontal line), 25%ile (45° angle), 75%ile (90° angle), minimum and maximum observations (whiskers).
d. F distributions for family structures for GME and European American (EA) trios. Mean F values correlated with expected for consanguineous offspring. Unk=unknown.
To test if these populations were subject to bottlenecks, we calculated the mean linkage disequilibrium (LD) decay, as haplotypes should decay as a function of size more slowly with increased bottleneck (Fig. 2b). LD for each GME population decayed faster than European and East Asian but slower than African populations. LD decayed faster in NWA and NEA compared with other GME regions, in agreement with our TreeMix results. Diverse patterns of admixture across these regions suggested these trends were not predominantly due to intermixing, but instead argued for a historic common ancient bottleneck.
Between 20–50% of all GME marriages are consanguineous (compared with < 0.2% in the Americas and Western Europe) 1–3, with the majority being first cousin. This roughly 100X higher rate of consanguinity has correlated with roughly a doubling of the rate of recessive Mendelian disease 19,20. European, African and East Asian 1000G populations all had distributions of estimated inbreeding coefficients (F) ~0.005, whereas GME F values ranged from 0.059 to 0.098, but with high variance within each population (Fig. 2c). Thus, measured F was ~10–20X higher, reflecting the shared blocks common to all human populations. F values were dominated by immediate family structure rather than historic or population-wide data trends (Fig. S8) 21. Examining the larger set of 1,794 exomes that included many parent-child trios also showed an overwhelming influence of immediate family structure, in which offspring from first-cousin marriages displayed higher F values compared with non-consanguineous marriages (Fig. 2d).
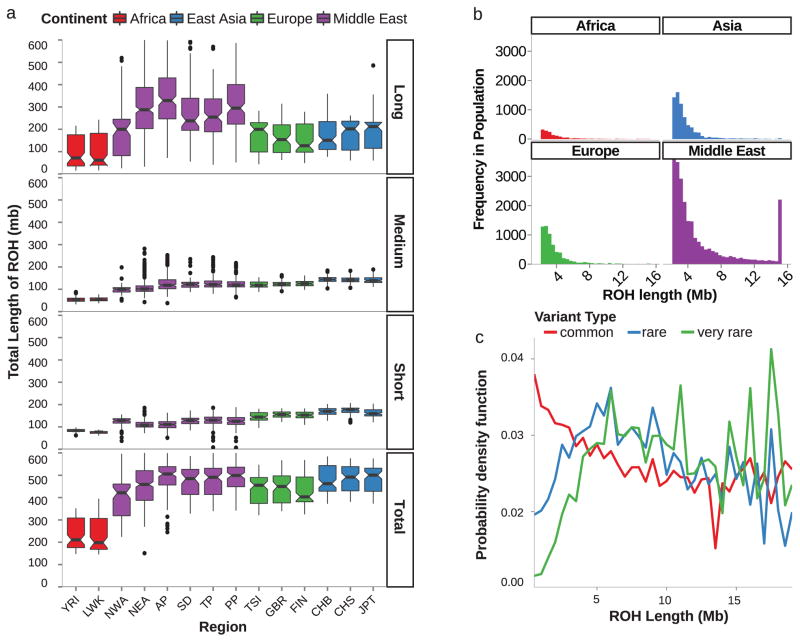
We expected that higher F values would correlate with an increased burden and length of ‘runs of homozygosity’ (ROH), defined as homozygous haplotypes as a function of length 22. 1000G sub-Saharan Africa displayed the smallest total ROH as expected 23, whereas the two other 1000G assessed populations were relatively similar to GME (Figs. 3a, S9), probably reflecting similar lengths of short (<0.515 Mb) and medium (0.516–1.606 Mb) ROH. Most striking was the increase in long ROH (>1.607 Mb), found nearly exclusively in GME samples, especially for those over 4 Mb (Fig. 3b). In the GME, there was an enrichment of rare and very rare variants (AF <.05, and AF <.01) in longer ROH, and of common variants (AF >= .05) in shorter ROH (Fig. 3c), suggesting that the longer ROH result from recent consanguinity 24
Figure 3. Distributions of short and long Runs of Homozygosity (ROH) correlates with patterns of bottlenecks and recent consanguinity.
a. Sample burdens of ROH grouped by length (Short: <0.155 Mb, Medium: 0.156–1.606 Mb, Long: >1.607 Mb). GME samples (purple) showed a unique contribution of long ROH compared with other populations (*), with less in short and medium bins compared to Europe and East Asia. Total ROH in GME sub-regions overlapped with European and East Asian likely due to greater bottlenecks in these populations.
b. Histograms of long ROH for GME, Africa, Europe, and East Asia. GME samples more frequently harbored runs >4 Mb compared to other populations. ROH >15 Mb are binned together (* peak unique to Middle East).
c. Longer GME ROH spans were enriched for rare variation, while shorter runs were enriched for more common variation. Proportion of variants binned by allele frequency for different sized ROH, binned by 0.5 Mb intervals. Probability density function calculated for each allele frequency class. Note that AFs for common alleles declined whereas AFs for rare and very rare alleles rose as ROH increased in size (Common: AF > .05, Rare: AF 0.05–0.01, Very Rare: AF < 0.01).
This increased ROH provided an opportunity to identify homozygous loss-of-function variants (LOF) in healthy humans. While these variants are only putatively LOF until experimentally verified, these exhibit the strongest signs of selective pressure and are the first checked as disease candidates25. Recently, among 2,636 sequenced and 101,584 chip-imputed Icelanders, 1,171 genes were predicted to be inactivated 26. From our 354 exomes on verified healthy adults, we found 301 genes with rare homozygous putative LOF variants (Table S2, S3), with only 50 genes overlapping with the Icelandic gene list. Similarly the ExAC dataset on 60,706 sequenced individuals identified 2068 genes inactivated, of which only 94 genes overlapped with our 301 genes. This suggests that the set of non-clinically relevant LOF variants is far from being exhausted. The GME represents an optimal population from which to identify homozygous variants due to the elevated consanguinity rates.
Darwin observed that rare self-fertilized orchid strains exhibited surprisingly higher fitness than founder strains, which he termed ‘hero strains’ 27. This led to the concept of ‘purging of recessive alleles’ by Haldane 28, referring to increased loss of deleterious alleles due to increased selective pressure in inbred populations. Purging was hypothesized to impact the GME genome due to the higher rates of birth defects incompatible with future reproduction 29, but has yet to be documented in humans. We compared the distribution of derived allele frequencies (DAF) in GME and 1000G populations 30. Variants were divided into 7 functional and PolyPhen-2 deleterious classes. We calculated mean DAFs using chimpanzee (PanTro2) as the common ancestor (Figs. S10, S11) 31. Neither autosomal nor X-linked variants showed significant differences (Fig. S12), arguing against a measurable effect on overall variant burden resulting from consanguinity.
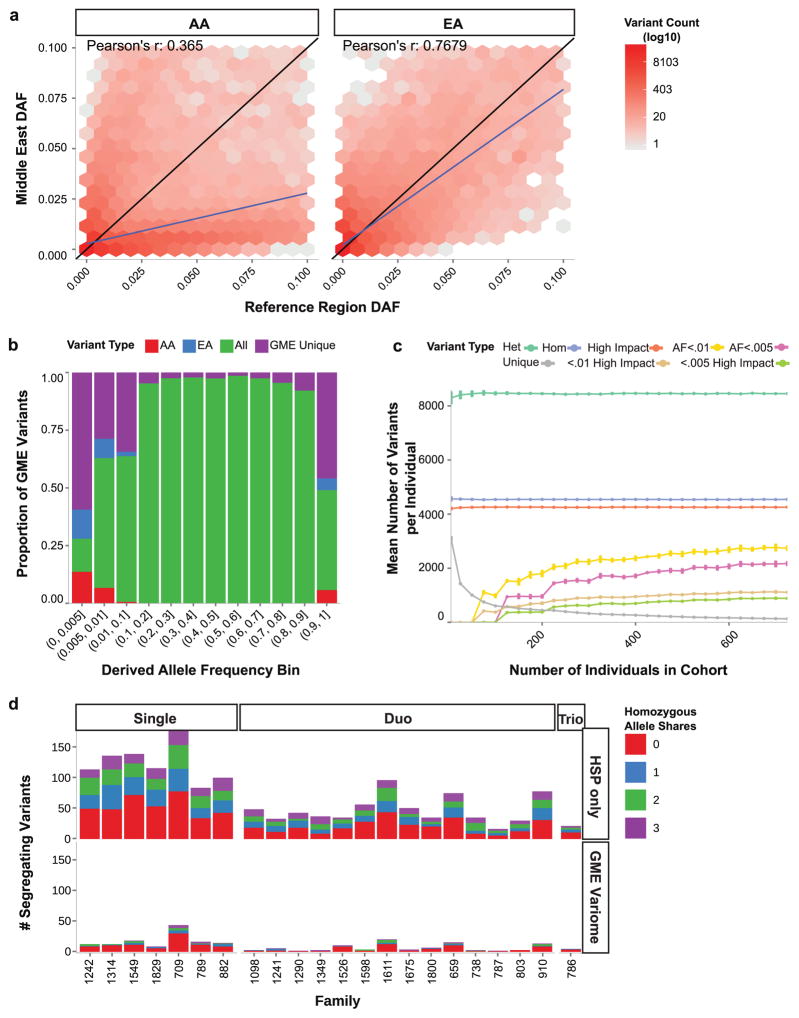
Numerous studies have relied on the increased power of GME-resident consanguineous families to identify causes of recessive disease, but the lack of an accessible variome has hindered progress. Efforts like the NHLBI GO Exome Sequencing Project (ESP) produced variomes for European American (EA) and African American (AA) populations, but poor correlation of DAFs between population pairs determined that neither were good estimators for GME DAFs (Pearson’s r 0.7979 GME vs. EA, 0.385 GME vs. AA, 0.1447 EA vs. AA, Figs. 4a, S13). Moreover, we found much of the GME variation to be poorly represented outside the GME (Fig. 4b), with the majority of variants in the rarest DAF bin found only in the GME.
Figure 4. GME Variome facilitates the discovery of Mendelian disease genes.
a–b. Comparison of rare derived allele frequencies (DAF) between GME and Exome Sequencing Project (ESP). AA: African American, EA: European-American. Hexagonal bins shaded by log number of variants within each bin. Pearson’s r suggests GME DAFs were not accurately estimated by AA or EA populations.
b. The majority of variants in the rarest DAF bins were unique to the GME. AA: found only in GME and AA. EA: found only in GME and EA. All: found in GME, EA and AA. GME Unique: found only in GME.
c. Change in per-individual burden of eight variant classes as a function of increasing the number of individuals incorporated into the GME Variome cohort. As sample size increased there was a drop in the number of unique variants, along with more accurate estimation of DAFs for rare variants. Bootstraps were sampled with replacement for 100 iterations to calculate standard errors. “High impact”: variants meeting predicted deleteriousness thresholds (see Methods).
d. Number of candidate variants for 20 families, meeting segregation and deleteriousness filtering criteria, using DAFs derived from Hereditary Spastic Paraplegia (HSP)-only families (top) or also incorporating the GME Variome (bottom). Single, Duo, Trio: families with one, two or three affected members. Colors: number of individuals sharing the variant. “0”: no other individuals carried the allele, etc. Analysis was performed using this threshold for the number of individuals sharing alleles (0,1,2,3). Note drop in number of segregating variants for any given family after the GME Variome was applied.
In order to assess how well the GME Variome captured extant exome variation, we sub-sampled the cohort for 100 iterations from 5 to 700 individuals, and for 8 variant classes (see methods, Fig. 4c). There was decay in the number of unique variants and accumulation of rare variants as sample size increased, due to a scaled ability to estimate prevalence. When sampled near 1,000 individuals, the change in mean of these values was negligible as new samples were added. Thus the GME Variome should allow accurate determination of population-level DAFs for all but the rarest alleles.
In order to investigate the potential of the Variome to expedite the discovery of new disease genes, we compared causal variant sets from GME families displaying recessive hereditary spastic paraplegia (HSP), where we recently established 17 new genetic forms of disease 32. For a disease like recessive HSP with a prevalence of 3–10 per 100,000 33 and where there are more than 40 genetic forms and hundreds of individual genetic mutations known, the expected allele frequency for any causative mutation should be <1:1000 (see methods). Select individuals from 20 representative families underwent whole exome sequencing. For each family, we calculated the number of alleles that passed standard filtering (i.e. LOF or otherwise potential ‘high impact’) 34 and were unique, both without and with the DAFs of the Variome (see website for Variome below). Using only exome data from the 20 families and public sources, there were on average 56, 20, and 11 unique variants passing filters from families with one, two or three sequenced affected members, respectively. In contrast, by accessing the Variome there were on average 13, 5, and 4 unique variants (Fig. 4d, Table S4), yielding a 4–7-fold reduction of the number of variants requiring further consideration. Loosening the allowable AFs to <1:500, <1:333 or <1:250 also showed substantial reduction in the number variants for consideration.
Here we have interrogated the fine-scale genomic structure across the GME, shaped by prehistoric as well as historic migrations, conquests, and cultural traditions. The degree of unique genetic variation represented in the GME was surprising given previous efforts to capture diversity, and speaks to the value of sampling of understudied populations. The data support records of migrations and conquests, but also suggest a previously unstudied GME contribution.
Despite millennia of elevated consanguinity in the GME, we detected no evidence for purging of recessive alleles. Instead, we detected large rare homozygous blocks, distinct from the small homozygous blocks found in other populations, supporting recent consanguineous matings, and allowing identification of genes harboring putatively high impact homozygous variants in healthy humans from this population. Applying the Variome to future sequencing projects for GME-originating subjects could aid in recessive gene identification across all classes of disease. GME Variome is a publicly accessible resource that will facilitate a broad range of genomic studies in the GME and globally.
Online Methods
1. Definition of the Greater Middle East
The term “greater Middle East” has been used to refer to a large swath of Arab and non-Arab Muslim countries, stretching from Morocco in the west to as far east as Pakistan in southeast Asia. However, no precise listing of designated countries has yet emerged. “U.S. Working Paper for G8 Sherpas,” Al-Hayat, February 13, 2004. Available online at [http://english.daralhayat.com/Spec/02-2004/Article-20040213-ac40bdaf-c0a8-01ed-004e-5e7ac897d678/story.html] and [https://www.fas.org/sgp/crs/mideast/RS22053.pdf]. Editable map of the Middle East was downloaded from [http://www.presentationmagazine.com].
2. Exome Resequencing
2.1 Study sample
The 2,497 individuals used in the analysis were selected from samples ascertained across three labs and recruited with the help of clinicians that constituted the GME Consortium. Although these individuals were not a random sample, they were ascertained within a wide variety of distinct phenotypes such that cohort-specific effects were not expected to bias patterns of variation. All study participants in each of the component studies provided written informed consent for the use of their DNA in studies aimed at identifying genetic risk variants for disease, and for broad data sharing. Institutional certification was obtained for each sample to allow deposition of genotype data in dbGaP and other purposes.
2.2 Exome resequencing, variant calling, and filtering
Blood DNA was extracted using Qiagen reagents, subjected to exome capture with the Agilent SureSelect Human All Exome 50 Megabase (Mb) kit, sequenced on an Illumina HiSeq2000 instrument, resulting in ~94% target coverage at > 30X depth 35–37. FASTQ files were reprocessed and jointly called to minimize batch effects and ensure consistent variant calling. using the GATK pipeline (version 3.1–1) adhering to best practices 38, eliminating duplicate reads. Paired-end reads were aligned to the human reference genome NCBI Build 37, using BWA (version 0.7.5) 39. Principal component analysis (PCA) was run on the resultant set of variants to identify potential batch effects between labs, sequencing centers, or collectively run groups of samples, then samples eliminated until no batch effects were observed.
We calculated four quality control (QC) metrics for each sample using PSEQ and identified statistical outliers. Metrics included: total number of variants, transition/transversion ratio, number of sequenced positions, and number of singletons. Due to possible reference distance bias, we considered samples grouped by geographic region independently. Samples were identified as outliers using a cutoff of >5 standard deviations from the mean threshold for each QC metric, removing 314 samples. The PCA based outlier analysis algorithm from the EIGENSOFT software library was also run, but failed to find any additional samples violating a standard deviation threshold of 5.0 40.
To ensure unbiased population structure statistics and allele frequency estimates, we removed close and cryptic relationships from the dataset. Kinship estimation was generated using KING, which calculated relatedness between all pairs of individuals and was robust to population structure 41. Using the 182,967 LD filtered SNPs, we ran KING following standard guidelines for a 3rd degree relationship (i.e. first cousins), using a kinship coefficient of 0.04419. When a cluster of related individuals was identified, we preferentially removed those to leave the largest number of samples. Of the remaining 2183 samples after outlier filtering, 667 samples were removed to reduce dataset relatedness, leaving a final cohort of 1516 non-related individuals. Remaining samples were rerun through the KING, which identified no additional kinships. Final continental sample counts after filtering: Sub-Saharan Africa: 19, America: 33, Europe: 378, Oceania: 1, and Middle East: 1111.
Coverage statistics were generated across all internal exome data sets using BEDTools, to calculate the average coverage across each exon 42. Exons were filtered from the analysis if greater than 5% of samples had less than 10x average coverage. Out of the initial 192,056 exons targeted by the Agilent SureSelect II capture kit, 170,032 exons were well covered in at least 95% of samples. Variants were filtered if identified outside of these genomic regions, leaving 32,967,859 bases under consideration (~1% of the human genome) within 17,800 genes.
Standard filters for variants that were called with posterior probability >99% (glfMultiples SNP quality > 20), were at least 5 bp away from an indel detected in the 1000G Pilot Project, were targeted in at least 95% individuals and had a total depth across samples between 6,823 to 6,823,000 (~1–1000 reads per sample on average) 9. Variant positions were filtered based on population statistics including a ‘missingness’ rate (referring to the percent of samples where information was missing) of less than 5%, and Hardy-Weinberg equilibrium (HWE) deviation p-value < 0.00005 43.
We generated a subset of variants in minimal linkage disequilibrium (LD) by pruning variants exhibiting pairwise linkage disequilibrium (r2). Variants were filtered to exclude SNPs with minor allele frequency (MAF) <5%, and all indels. Remaining SNPs were pruned adhering to a maximum threshold of 0.5 using PLINK’s ‘--indep-pairwise’ command 43. Of the initial 578,231 variants, 182,967 SNPs passed filters. This LD pruned dataset was used for population structure characterization including principal component analysis (PCA), Wright’s fixation index (Fst), admixture analysis, KING relationship testing, and estimation of inbreeding coefficient.
2.3 Geographic region assignment
Samples were recruited from 20 countries and territories across the GME and grouped into a set of six geographic regions: Northwest Africa (85 Samples), Northeast Africa (423 Samples), Arabian Peninsula (214 Samples), Syrian Desert (81 Samples), Turkish Peninsula (140 Samples), and Persia and Pakistan (168 Samples). Country boundaries were not used to group samples for two reasons: 1] Inconsistent sampling left several countries with too few samples to accurately represent the diversity of the population. Syria and Yemen, for example, were only represented by a few samples, due to ongoing conflicts. 2] Current country borders frequently fail to accurately separate ethnicities, due to a combination of recent migrations and recent political history. For example, south-eastern Arabian Peninsula Bedouin tribes do not distinguish between the relatively recently defined borders of Oman and the UAE.
Self-identified ethnicities were available for some samples, but incompleteness of this annotation, and the great diversity of populations affiliating as “Arab”, prompted use of geography for groupings. As much as possible we assigned location to the current residence, rather than ancestral residence or location where samples were drawn. While some reference GME ethnicities exist in public resources, such as the Human Genome Diversity Project (HGDP) 44, we found both the breadth of ascertained ethnicities and sample size insufficient to impute ethnicities where absent.
The original cohort was largely composed of samples from GME countries, but also included samples of African, European, and East Asian decent. To ensure consistency in our geographic designations we performed and linkage clustering, based on pairwise distances between samples using Plink’s ‘--distance-matrix’ command 43. We performed hierarchical clustering on all samples using Ward’s hierarchical clustering method (“ward.D2” option for the “hclust” algorithm in R)45.
3. Population Structure of GME
3.1 Data integration
Population structure was analyzed in the context of continental populations from the 1000 Genomes Phase I (1000G) dataset 9. As 1000G samples were generated from a combination of whole genome and exome sequencing, variants falling outside of RefSeq exonic regions +/− 30 base pairs (bp) were filtered using BedTools and merged with the GME cohort 46,47. Nine populations from 1000G data were used in comparative analyses: African populations YRI and LWK; East Asian populations CHB, CHS, and JPT; and European populations GBR, TSI, IBS, and FIN. Related 1000G samples were filtered by a KING analysis as previously described. A total of 1821 samples remained after filtering representing 15 geographic regions, 6 from the GME and 9 from 1000G.
3.2 Substructure analysis
To investigate the influence of admixture on the GME samples, we used the block relaxation algorithm implemented in ADMIXTURE to estimate individual ancestry proportions given K ancestral populations 48. Unsupervised ADMIXTURE was run using default settings (folds=5) on merged GME and 1000G samples and iterations of K values from 2 to 14. Minimum squared error values calculated from ADMIXTURE’s cross-validation procedure for evaluating fit of different values of K, found an optimum K = 6 for just GME samples, and 7 including 1000G control data.
3.3 PCA and Wright’s Fixation Index (Fst)
Principal component analysis (PCA) was used to investigate the affinities within human populations and the relationships between them. We performed PCA on GME and 1000G samples using the SmartPCA tool from the EIGENSOFT software library and the first four principal components compared graphically 40,49.
Wright’s fixation index (Fst) was used to explore the degree of differentiation between populations. Fst values and standard error for all pairs of populations were calculated using the estimator of Weir & Cockerham, also included in the EIGENSOFT software library. All plots were generated using ggplot2 50.
3.4 LD decay
Pairwise linkage disequilibrium among pairs of SNPs is an indicator of the past history of recombination and genetic drift. To calculate LD, we tallied pairwise r2 for SNP pairs for all GME and control populations using the Plink “r2” option 43. Correlations between all SNPs falling within each sliding-window of 70 kilobase (kb) were calculated with no lower limit on r2 values. Pairwise correlations were binned by genomic distance between SNPs (up to 70kb), and averages calculated for each bin. Control samples followed expected patterns of LD decay.
3.5 Estimation of inbreeding
The inbreeding coefficient of an individual (F) was used to represent the probability that two randomly chosen alleles at a homologous locus within an individual were identical by descent (IBD) with respect to a base reference population in which all alleles were independent. While the true inbreeding coefficient of an individual is often unknown, several estimation methods have been shown to give a reasonable estimate.
F estimates were calculated using the Plink “het” algorithm on LD pruned variants following authors guidelines 43. We compared results to the HMM algorithm Festim 51 and found the two estimates were very similar (Pearson’s r: 0.874) but frequently Festim failed to return results for samples with missing data. Negative F values were most likely the result of either biased variant sampling, a high-degree interracial marriage, or due to recent intermixing of previously disparate populations8.
3.6 Runs of homozygosity (ROH) estimation
To infer estimates of the autozygosity and relative recent population size, we estimated runs of homozygosity using the HMM algorithm H3M2 52. H3M2 was run directly on aligned BAM files, following authors recommendations for all parameters. Proportion of genome and exome falling within ROH was calculated for each sampling using BedTools. ROH length classes were based on published ranges 23, where the authors used machine learning to identify three ROH classes including: Short (<0.515 Mb), Medium (0.156–1.606 Mb), and Long (>1.607 Mb). We compared densities of ROH lengths from internal data and found a near identical distribution as the published values used to identify these classes.
4. Variant Annotation and Classification
4.1 Variant annotation
Functional annotation was performed for genetic purging and loss of function analyses. Variants were annotated using the ANNOVAR suite of scripts (version 2014Nov12) 53. ANNOVAR classified variants into eight coding region functional groups including: “frameshift_deletion”, “frameshift_insertion”, “nonframeshift_deletion”, “nonframeshift_insertion”, “nonsynonymous_SNV”, “stopgain”, “stoploss”, and “synonymous_SNV”. Non-coding variants are classified as “unknown”. Splicing defects were identified based on 2 base pair distance from the splice junction, either on the intronic or exonic side. A predicted deleteriousness classification was generated for each missense variant using PolyPhen-2 54. The functional designations for PolyPhen-2 include: B (Benign), P (Possibly Damaging), D (Probably Damaging). We compared these annotations to those generated by SNPEff 55, and while there were some differences, found distributions of calls from each sample to be consistent.
4.2 Ancestral allele identification
We used the Chimpanzee genome as the closest assembled out-group genome. Ancestral allele estimates were obtained by UCSC pairwise alignments between human reference hg19 and chimp references PanTro2 and PanTro4. Systematic lookups for all GME and 1000G variants were performed using UCSC Genome Browser tools and custom scripts to identify associated chimpanzee alleles. We compared PanTro2 and PanTro4 to assess the difference in correcting the apparent reference bias, but found both worked equally well.
Estimated ancestral alleles were used as the reference allele to calculate derived allele frequencies (DAF). DAFs were not calculated for variants where the ancestral allele was not present in the human germline.
4.3 Identity-by-state (IBS) distance to reference
To interrogate the potential biases that might result from reference selection we calculate the IBS distance between samples and multiple different references including hg19, and chimpanzee. The distance represents the proportion of positions that diverge from reference, and was calculated between all pairs of samples and references.
The IBS distance, d, represented the number of differing alleles between the two samples divided by the total number of alleles compared. More formally, d, between the two n-length vectors p, q (in our case where p is the reference sample and q is the sample being compared) in a vector space v, where v ={0,1,2} encoding the homozygote for the human reference allele, the heterozygote, and the homozygote for the alternate allele, respectively.
For any two samples, we calculate d as:
where (p,q) are vectors such that p = (p1, p2,…, pn)and q = (q1,q2,…,qn)
Each vector represented all genotype calls between the two samples, excluding filtered sites or missing positions.
The IBS distance was calculated for all GME and 1000G samples against the hg19 and chimpanzee reference genomes. All genotypes from the merged VCF file were coded based on a comparison to the hg19 reference. Variant positions were filtered to remove indels, due to the possibility of alignment errors, and non-biallelic sites. When comparing to hg19, vector p was represented by a vector of zeros.
4.4 Hereditary Spastic Paraplegia (HSP) candidate variant analysis
Samples from 20 consanguineous families displaying an autosomal recessive inheritance pattern of HSP were selected from a previously analyzed cohort 32, selected from a total cohort of 55 families because in these 20 there was a single genetic causes identified. Families were analyzed in adherence to published methods 32. Briefly, homozygous variants were filtered based on family structure to ensure variants segregated with the disease phenotype. We performed deleteriousness filtering using functional classes and GERP++ scores 56. All candidate variants were potentially LOF (frameshift, stop, or perturbing splicing) or a coding variant with a GERP score >4.
The maximum allele frequency for candidate variants were based on established rates of disease prevalence, estimated at 1:10,000 for clinical presentations classified of HSP 57. Approximately 50% of HSP is autosomal dominant, and of the remaining, about 50% is explained by mutations by SPG11 58, leaving only 1:40,000 with recessive HSP caused by other genes. At least 35 other genes are reported to cause recessive HSP. Thus, the contribution to HSP disease prevalence for any given gene is unlikely to be more than 1:1,000,000. While prevalence of HSP mutations is not expected to be uniform, we expect the maximum carrier frequency for any new causal variant to be no more than 1:1000, assuming full penetrance and a classic recessive inheritance, and is actuality is likely to be much rarer given allelic diversity.
With roughly 1000 individuals in our cohort, we calculated that variants with DAFs <1:1000 should not be observed commonly in our dataset, AFs <1:500 should be not be observed in more than 1 individual, AFs <1:333 in not more than 2 individuals and AFs <1:250 in not more than 3 individuals. Variants passing deleteriousness and allele frequency thresholds were treated as candidates to calculate the usefulness of the Variome to limit the number of deleterious variants considered as candidates.
5. Testing for the Influence of Genetic Purging
Consanguinity has been practiced in the GME for at least several centuries 59. Simulations of GME like populations have found sufficient time has past for purging to have been effective in reducing genetic load 29. Clinical studies aimed at comparing clinical rates of birth defect rates, premature births or miscarriages, between communities that practice consanguinity to those largely out-breeding populations have found all metrics have fallen within range for the rate of immediate form of consanguinity 21,60,61. More recent genetics studies investigating differential selective pressure across human populations focused on the role of population bottlenecks, neglecting the potential influence of consanguinity, and lacked representation from the GME 31,62. For these reasons, we sought to investigate the possibility that genetic purging has influenced variant burden in the GME.
In order to approach the question of variable selective pressure across human populations, we implemented a variation of the DAF comparison method 31. We assumed that any change in the efficacy of natural selection should be evident across populations in the mean DAF within each variant classes.
For all variants described across the GME and 1000G populations, we filtered for high quality calls, identified ancestral alleles (described in “Ancestral Allele” section), annotated variants for predicted function and PolyPhen-2 classes using ANNOVAR, down-sampled to achieve an equivalent numbers of chromosomes across populations, and calculated DAFs for all positions. Variants were grouped by class, and the DAF means were calculated for each population. Standard-errors were calculated by bootstrapping DAF means for 1000 iterations.
Recent studies using PolyPhen-2 demonstrated a deflation of deleteriousness scores for derived variants found in the hg19 reference, likely due to a training artifact 31,62. Before using PolyPhen-2 classes, this bias was corrected for all derived reference positions. Bias correction was implemented by grouping variants by DAF bins, and calculating the proportions of each PolyPhen-2 class per bin for ancestral reference positions. Using these proportions as expectations, and all derived reference positions were randomly reassigned a new PolyPhen-2 class based on a hypergeometric distribution within each DAF bin. DAF means across classes for all included 1000G and GME populations showed no deviation outside the standard-error for any two populations61.
6. Neanderthal and Denisovan Introgression Analysis
Neanderthal-derived variants are often subjected to strong negative selection, thereby making exome analysis inadequate for estimating age of introgression. Thus we calculated the proportion observed between extant populations 15,63.
To estimate introgression in exome samples, we identified aligned consensus calls for all human variant positions from the chimpanzee, Neanderthal and Denisovan reference genomes. Alignments of Neanderthal and Denisovan genomes to 1000G variant positions were downloaded from the Max Planck Institute for Evolutionary Anthropology FTP 15,64. Neanderthal and Denisovan alleles were identified from the hg19-ancestor alignment files. Chimpanzee alleles were identified as described in the “ancestral allele” section of these methods.
We projected GME and 1000G control populations on the principal components calculated using representative samples from Neanderthal, Denisova, and chimpanzee 16,65,66, and aligned the human samples to these ancestral populations. Principal components were computed using R’s “prcomp” function (see web resources), and projected vectors were calculated for all 1000G and GME samples. Distance from the re-adjusted origin to each species reflected the proportion of introgression observed in each sample. The limited number of SNPs that were examined in this analysis compared to similar genotype-based analysis likely inflated the sampling variance within populations, and limited the sensitivity of our analysis to smaller introgression proportions. Centroids for all populations were labeled with their abbreviated names. Similar to previous work, Europeans, East Asians, and GME populations overlapped, and demonstrated larger proportions of Neanderthal than African populations 65–67.
Supplementary Material
Acknowledgments
The authors thank Shamil Sunyaev and David Reich for help with PolyPhen-2 and DAF corrections, Michael Turchin for help with purging analysis, Joseph Pickrell for help with TreeMix, Vineet Bafna, Nicholas Schork, Stefano Bonissone for suggestions. Work was supported by grants from the National Institutes of Health (P01HD070494, R01NS048453), Qatari National Research Foundation (NPRP6-1463), Simons Foundation Autism Research Initiative (175303 and 275275) to JGG, the Yale Center for Mendelian Disorders (U54HG006504), the Broad Institute (U54HG003067), The Rockefeller University CTSA (5UL1RR024143-04), the Howard Hughes Medical Institute (to JGG and J-LC), Institut National de la Santé et de la Recherche Médicale, the St. Giles Foundation, and the Candidoser Association, R01AI088364, R37AI095983, P01AI061093, U01AI109697 (to J-LC), U01AI088685 to J-LC and LA, R21AI107508 (to E. Jouanguy), DHFMR Collaborative Research Grant and KACST 13-BIO1113-20 (to FSA).
Greater Middle Eastern Variome Consortium
Sohair Abdel Rahim, Sawsan Abdel-Hadi, Ghada Abdel-Salam, Ekram Abdel-Salam, Mohammed Abdou, Avinash Abhytankar, Parisa Adimi, Jamil Ahmad, Mustafa Akcakus, Guside Aksu, Sami Al Hajjar, Suliman Al Juamaah, Saleh Al Muhsen, Nouriya Al Sannaa, Salem Al Tameni, Jumana Al-Aama, Nasir Al-Allawi, Raidah Al-Baradie, Lihadh Al-Gazali, Amal Al-Hashem, Waleed Al-Herz, Deema Al-Jeaid, Asma Al-Tawari, Abdullah Alangari, Alexandre Alcais, Tariq S AlFawaz, Zobaida Alsum, Aomar Ammar-Khodja, Sepideh Amouian, Cigdem Arikan, Omid Aryani, Ayca Aslanger, Cigdem Aydogmus, Caner Aytekin, Matloob Azam, Boglarka Bansagi, Mohamed-Rhida Barbouche, Laila Bastaki, Tawfeg Ben-Omran, PS Bindu, Lizbeth Blancas, Stéphanie Boisson-Dupuis, Damien Bonnet, Omar Boudghene Stambouli, Aziz Bousfiha, Lobna Boussafara, Jeannette Boutros, Jacinta Bustamante, Huseyin Caksen, Yildiz Camcioglu, Emilie Catherinot, Fatma C Celik, Michael Ciancanelli, Funda E Cipe, Gary Clark, Aurélie Cobat, Sinan Comu, Angela Condie, Antonio Condino-Neto, Mukesh Desai, William Dobyns, Figen Dogu, Mohamed Domaia, Meltem Dorum, Odul Egritas, Safa El Azbaoui, Jamila El Baghdadi, Mona El Ruby, Ashraf El-Harouni, Reem A Elfeky, Gehad Elghazali, Eissa Faqeih, Elif Fenerci, Claire Fieschi, Cipe Funda, Iman Gamal, Umit Gelik, Fetah Genel, Alper Gezdirici, KM Girisha, Amy Goldstein, Padraic Grattan-Smith, Neerja Gupta, Jin Hahn, Nevin Hatipoglu, Raoul Hennekam, Massoud Houshmand, Philippe Ichai, Aydan Ikinciogullari, Samira Ismail, Chaim Jalas, Emmanuelle Jouanguy, Madhulika Kabra, Göknur Kalkan, Majdi Kara, Neslihan Karaca, Kadri Karaer, Ariana Kariminejad, Hulya Kayserili, Melike Keser-Emiroglu, Sara S Kilic, Najib Kissani, Cristina Kokron, Roshan Koul, Necil Kutukculer, Fanny Lanternier, Alireza Mahdaviani, Nizar Malhaoui, Lobna Mansour, Davood Mansouri, Lucia Margari, Enza Maria Valente, Naima Marzouki, Amira Masri, Amina Megahed, Hisham Megahed, Najla Mekki, Mehrnaz Mesdaghi, Mohd Mikati, Faezeh Mojahedi, John Mulley, Sheela Nampoothiri, Carmen Navarrete, Tarek Omar, Azza Oraby, Ayse Pandaluz, Nima Parvaneh, Turkan Patiroglu, Zeynep Peker Koc, Isabelle Pellier, Capucine Picard, Anne Puel, Annick Raas-Rothschild, Anna Rajab, Didier Raoult, Ismail Reisli, Nima Rezaei, Ayoub Sabri, Yasin Sahin, Laila Saleem, Fadia Salem, Najla Sameer AlSediq, Ozden Sanal, Terry Sanger, Hanan Shakankiry, Lei Shang, Nabil Shehata, Nuri Shembesh, Vared Shkalim, Ameen Softah, Sameera Sogaty, Neveen Soliman, Fatma Sonmez-Aunaci, Laszlo Sztriha, Lynda Taibi-Berrah, Samia Temtamy, Hasan Tonekaboni, Doris Trauner, Beyhan Tuysuz, Beyhan Tuysuz, Ali Varan, Guillaume Vogt, Christopher Walsh, Geoffrey Woods, Gozde Yesil, Alisan Yildiran, Basak Yildiz, Adnan Yuksel, Maha Zaki, Shen-Ying Zhang
Footnotes
Full list of Consortium contributors provided in Acknowledgements
URLs
ANNOVAR, http://annovar.openbioinformatics.org
Kinship-based INference for Gwas (KING), http://people.virginia.edu/~wc9c/KING/
Plink, http://pngu.mgh.harvard.edu/~purcell/plink/
PolyPhen-2, http://genetics.bwh.harvard.edu/pph2/
PSEQ, http://atgu.mgh.harvard.edu/plinkseq/pseq.shtml
SnpEff, http://snpeff.sourceforge.net/SnpEff_manual.html
UCSC Genome Browser, http://genome.ucsc.edu
1,000 Genomes Browser, http://browser.1000genomes.org
Consang.net, http://consang.net/index.php/Global_prevalence
Denisovan to Human alignment (FTP), http://www.eva.mpg.de/denisova
Neanderthal to Human alignment (FTP), http://cdna.eva.mpg.de/neandertal
GME Variome, http://gme.igm.ucsd.edu
Author contributions
E.M.S. performed analysis and generated all figures. A.H, Y.I, Y.H., M.A.A. consulted on analysis. E.G.S., A.B., B.B., A.A., F.S.A., J.-L.C., J.G.G. contributed subjects and jointly wrote and edited the manuscript. S.B.G. oversaw sequencing. A.G.C. consulted on population studies. GME Consortium identified subjects for study.
Competing financial interests
The authors declare no competing financial interests
References
- 1.Anwar WA, Khyatti M, Hemminki K. Consanguinity and genetic diseases in North Africa and immigrants to Europe. Eur J Public Health. 2014;24(Suppl 1):57–63. doi: 10.1093/eurpub/cku104. [DOI] [PubMed] [Google Scholar]
- 2.Al-Gazali L, Hamamy H, Al-Arrayad S. Genetic disorders in the Arab world. British Med J. 2006;333:831–4. doi: 10.1136/bmj.38982.704931.AE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hussain R, Bittles AH. The prevalence and demographic characteristics of consanguineous marriages in Pakistan. J Biosoc Sci. 1998;30:261–75. doi: 10.1017/s0021932098002612. [DOI] [PubMed] [Google Scholar]
- 4.Sheffield VC, Stone EM, Carmi R. Use of isolated inbred human populations for identification of disease genes. Trends Genet. 1998;14:391–6. doi: 10.1016/s0168-9525(98)01556-x. [DOI] [PubMed] [Google Scholar]
- 5.Sharp JM. The Broader Middle East and North Africa Initiative: An overview. CRS Report for Congress; 2005. [Google Scholar]
- 6.Hellenthal G, et al. A genetic atlas of human admixture history. Science. 2014;343:747–51. doi: 10.1126/science.1243518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ravindranath V, et al. Regional research priorities in brain and nervous system disorders. Nature. 2015;527:S198–206. doi: 10.1038/nature16036. [DOI] [PubMed] [Google Scholar]
- 8.Hunter-Zinck H, et al. Population genetic structure of the people of Qatar. Am J Hum Genet. 2010;87:17–25. doi: 10.1016/j.ajhg.2010.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Consortium GP, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Moreno-Estrada A, et al. Reconstructing the population genetic history of the Caribbean. PLoS Genet. 2013;9:e1003925. doi: 10.1371/journal.pgen.1003925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Botigue LR, et al. Gene flow from North Africa contributes to differential human genetic diversity in southern Europe. Proc Natl Acad Sci U S A. 2013;110:11791–6. doi: 10.1073/pnas.1306223110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li JZ, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–4. doi: 10.1126/science.1153717. [DOI] [PubMed] [Google Scholar]
- 13.Henn BM, et al. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet. 2012;8:e1002397. doi: 10.1371/journal.pgen.1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerard N, Berriche S, Aouizerate A, Dieterlen F, Lucotte G. North African Berber and Arab influences in the western Mediterranean revealed by Y-chromosome DNA haplotypes. Hum Biol. 2006;78:307–16. doi: 10.1353/hub.2006.0045. [DOI] [PubMed] [Google Scholar]
- 15.Green RE, et al. A draft sequence of the Neandertal genome. Science. 2010;328:710–22. doi: 10.1126/science.1188021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sankararaman S, et al. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 2014;507:354–7. doi: 10.1038/nature12961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Consortium STD, et al. Sequence variants in SLC16A11 are a common risk factor for type 2 diabetes in Mexico. Nature. 2014;506:97–101. doi: 10.1038/nature12828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pickrell JK, Pritchard JK. Inference of population splits and mixtures from genome-wide allele frequency data. PLoS Genet. 2012;8:e1002967. doi: 10.1371/journal.pgen.1002967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tadmouri GO, et al. Consanguinity and reproductive health among Arabs. Reprod Health. 2009;6:17. doi: 10.1186/1742-4755-6-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leutenegger AL, Sahbatou M, Gazal S, Cann H, Genin E. Consanguinity around the world: what do the genomic data of the HGDP-CEPH diversity panel tell us? Eur J Hum Genet. 2011;19:583–7. doi: 10.1038/ejhg.2010.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bittles AH, Black ML. Global patterns and tables of consanguinity. 2014 < http://consang.net>.
- 22.Pippucci T, Magi A, Gialluisi A, Romeo G. Detection of runs of homozygosity from whole exome sequencing data: state of the art and perspectives for clinical, population and epidemiological studies. Hum Hered. 2014;77:63–72. doi: 10.1159/000362412. [DOI] [PubMed] [Google Scholar]
- 23.Pemberton TJ, et al. Genomic patterns of homozygosity in worldwide human populations. Am J Hum Genet. 2012;91:275–92. doi: 10.1016/j.ajhg.2012.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Szpiech ZA, et al. Long runs of homozygosity are enriched for deleterious variation. Am J Hum Genet. 2013;93:90–102. doi: 10.1016/j.ajhg.2013.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.MacArthur DG, et al. A systematic survey of loss-of-function variants in human protein-coding genes. Science. 2012;335:823–8. doi: 10.1126/science.1215040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sulem P, et al. Identification of a large set of rare complete human knockouts. Nat Genet. 2015;47:448–52. doi: 10.1038/ng.3243. [DOI] [PubMed] [Google Scholar]
- 27.Jones S. The Darwin Archipelago. Yale University Press; New Haven: 2011. [Google Scholar]
- 28.Haldane JBS. The effect of variation of fitness. Am Nat. 1937;71:337–349. [Google Scholar]
- 29.Overall AD, Ahmad M, Nichols RA. The effect of reproductive compensation on recessive disorders within consanguineous human populations. Heredity. 2002;88:474–9. doi: 10.1038/sj.hdy.6800090. [DOI] [PubMed] [Google Scholar]
- 30.Neale BM, et al. Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature. 2012;485:242–5. doi: 10.1038/nature11011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Simons YB, Turchin MC, Pritchard JK, Sella G. The deleterious mutation load is insensitive to recent population history. Nat Genet. 2014;46:220–4. doi: 10.1038/ng.2896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Novarino G, et al. Exome sequencing links corticospinal motor neuron disease to common neurodegenerative disorders. Science. 2014;343:506–11. doi: 10.1126/science.1247363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Blackstone C, O’Kane CJ, Reid E. Hereditary spastic paraplegias: membrane traffic and the motor pathway. Nat Rev Neurosci. 2011;12:31–42. doi: 10.1038/nrn2946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.MacArthur DG, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014;508:469–76. doi: 10.1038/nature13127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dixon-Salazar TJ, et al. Exome sequencing can improve diagnosis and alter patient management. Sci Transl Med. 2012;4:138ra78. doi: 10.1126/scitranslmed.3003544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Okada S, et al. IMMUNODEFICIENCIES. Impairment of immunity to Candida and Mycobacterium in humans with bi-allelic RORC mutations. Science. 2015;349:606–13. doi: 10.1126/science.aaa4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alsalem AB, Halees AS, Anazi S, Alshamekh S, Alkuraya FS. Autozygome sequencing expands the horizon of human knockout research and provides novel insights into human phenotypic variation. PLoS Genet. 2013;9:e1004030. doi: 10.1371/journal.pgen.1004030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.DePristo MA, et al. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nat Genet. 2011;43:491–8. doi: 10.1038/ng.806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–95. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Patterson N, Price AL, Reich D. Population structure and eigenanalysis. PLoS Genet. 2006;2:e190. doi: 10.1371/journal.pgen.0020190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Manichaikul A, et al. Robust relationship inference in genome-wide association studies. Bioinformatics. 2010;26:2867–73. doi: 10.1093/bioinformatics/btq559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Quinlan AR, Hall IM. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–2. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Purcell S, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–75. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cann HM, et al. A human genome diversity cell line panel. Science. 2002;296:261–2. doi: 10.1126/science.296.5566.261b. [DOI] [PubMed] [Google Scholar]
- 45.Behar DM, et al. The genome-wide structure of the Jewish people. Nature. 2010;466:238–42. doi: 10.1038/nature09103. [DOI] [PubMed] [Google Scholar]
- 46.Danecek P, et al. The variant call format and VCFtools. Bioinformatics. 2011;27:2156–8. doi: 10.1093/bioinformatics/btr330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pruitt KD, et al. RefSeq: an update on mammalian reference sequences. Nucleic Acids Res. 2014;42:D756–63. doi: 10.1093/nar/gkt1114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res. 2009;19:1655–64. doi: 10.1101/gr.094052.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price AL, et al. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38:904–9. doi: 10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 50.Wickham H. ggplot2: Elegant graphics for data analysis. Springer Science & Business Media; 2009. [Google Scholar]
- 51.Polasek O, et al. Comparative assessment of methods for estimating individual genome-wide homozygosity-by-descent from human genomic data. BMC Genomics. 2010;11:139. doi: 10.1186/1471-2164-11-139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Magi A, et al. H3M2: detection of runs of homozygosity from whole-exome sequencing data. Bioinformatics. 2014;30:2852–9. doi: 10.1093/bioinformatics/btu401. [DOI] [PubMed] [Google Scholar]
- 53.Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Adzhubei IA, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7:248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cingolani P, et al. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Davydov EV, et al. Identifying a high fraction of the human genome to be under selective constraint using GERP++ PLoS Comput Biol. 2010;6:e1001025. doi: 10.1371/journal.pcbi.1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Erichsen AK, Koht J, Stray-Pedersen A, Abdelnoor M, Tallaksen CM. Prevalence of hereditary ataxia and spastic paraplegia in southeast Norway: a population-based study. Brain. 2009;132:1577–88. doi: 10.1093/brain/awp056. [DOI] [PubMed] [Google Scholar]
- 58.Stevanin G, et al. Mutations in SPG11 are frequent in autosomal recessive spastic paraplegia with thin corpus callosum, cognitive decline and lower motor neuron degeneration. Brain. 2008;131:772–84. doi: 10.1093/brain/awm293. [DOI] [PubMed] [Google Scholar]
- 59.Vardi-Saliternik R, Friedlander Y, Cohen T. Consanguinity in a population sample of Israeli Muslim Arabs, Christian Arabs and Druze. Ann Hum Biol. 2002;29:422–31. doi: 10.1080/03014460110100928. [DOI] [PubMed] [Google Scholar]
- 60.Shami SA, Qaisar R, Bittles AH. Consanguinity and adult morbidity in Pakistan. Lancet. 1991;338:954. doi: 10.1016/0140-6736(91)91828-i. [DOI] [PubMed] [Google Scholar]
- 61.Stoltenberg C, Magnus P, Lie RT, Daltveit AK, Irgens LM. Birth defects and parental consanguinity in Norway. Am J Epidemiol. 1997;145:439–48. doi: 10.1093/oxfordjournals.aje.a009126. [DOI] [PubMed] [Google Scholar]
- 62.Do R, et al. No evidence that selection has been less effective at removing deleterious mutations in Europeans than in Africans. Nat Genet. 2015;47:126–31. doi: 10.1038/ng.3186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Consortium STD, et al. Association of a low-frequency variant in HNF1A with type 2 diabetes in a Latino population. JAMA. 2014;311:2305–14. doi: 10.1001/jama.2014.6511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Meyer M, et al. A high-coverage genome sequence from an archaic Denisovan individual. Science. 2012;338:222–6. doi: 10.1126/science.1224344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Huerta-Sanchez E, et al. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 2014;512:194–7. doi: 10.1038/nature13408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Wang S, Lachance J, Tishkoff SA, Hey J, Xing J. Apparent variation in Neanderthal admixture among African populations is consistent with gene flow from Non-African populations. Genome Biol Evol. 2013;5:2075–81. doi: 10.1093/gbe/evt160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Lowery RK, et al. Neanderthal and Denisova genetic affinities with contemporary humans: introgression versus common ancestral polymorphisms. Gene. 2013;530:83–94. doi: 10.1016/j.gene.2013.06.005. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.