Abstract
The sodium-pumping NADH:ubiquinone oxidoreductase (Na+-NQR) is a bacterial respiratory enzyme that obtains energy from the redox reaction between NADH and ubiquinone and uses this energy to create an electrochemical Na+ gradient across the cell membrane. A number of acidic residues in transmembrane helices have been shown to be important for Na+ translocation. One of these, Asp-397 in the NqrB subunit, is a key residue for Na+ uptake and binding. In this study, we show that when this residue is replaced with asparagine, the enzyme acquires a new sensitivity to K+; in the mutant, K+ both activates the redox reaction and uncouples it from the ion translocation reaction. In the wild-type enzyme, Na+ (or Li+) accelerates turnover while K+ alone does not activate. In the NqrB-D397N mutant, K+ accelerates the same internal electron transfer step (2Fe-2S → FMNC) that is accelerated by Na+. This is the same step that is inhibited in mutants in which Na+ uptake is blocked. NqrB-D397N is able to translocate Na+ and Li+, but when K+ is introduced, no ion translocation is observed, regardless of whether Na+ or Li+ is present. Thus, this mutant, when it turns over in the presence of K+, is the first, and currently the only, example of an uncoupled Na+-NQR. The fact the redox reaction and ion pumping become decoupled from each other only in the presence of K+ provides a switch that promises to be a useful experimental tool.
Graphical Abstract
The sodium-pumping NADH:ubiquinone oxidoreductase (Na+-NQR) is a redox-driven sodium pump found in a wide range of different bacteria, including many marine and pathogenic species.1 Na+-NQR selectively pumps Na+ across the cell membrane, generating a sodium motive force that provides energy for a number of key physiological functions, including rotation of the flagella, uptake of nutrients, ion homeostasis, and extrusion of virulence factors and antibiotics. Some of these functions are essential for pathogens such as Vibrio cholerae to flourish in diverse environments.2–6 The energy for this electrogenic ion translocation process is provided by the transfer of electrons from NADH to ubiquinone, as the first step in respiratory electron transport.7,8 Thus, the primary function of Na+-NQR is to be an energy transducer; it couples together two fundamentally different reactions in such a way that the chemical energy of the redox reaction is converted into the energy of the Na+ gradient.9–11 Mechanistically, this coupling of two reactions means that the enzyme must ensure the energy-releasing “driving” reaction cannot proceed efficiently unless the energy requiring “driven” reaction also occurs.
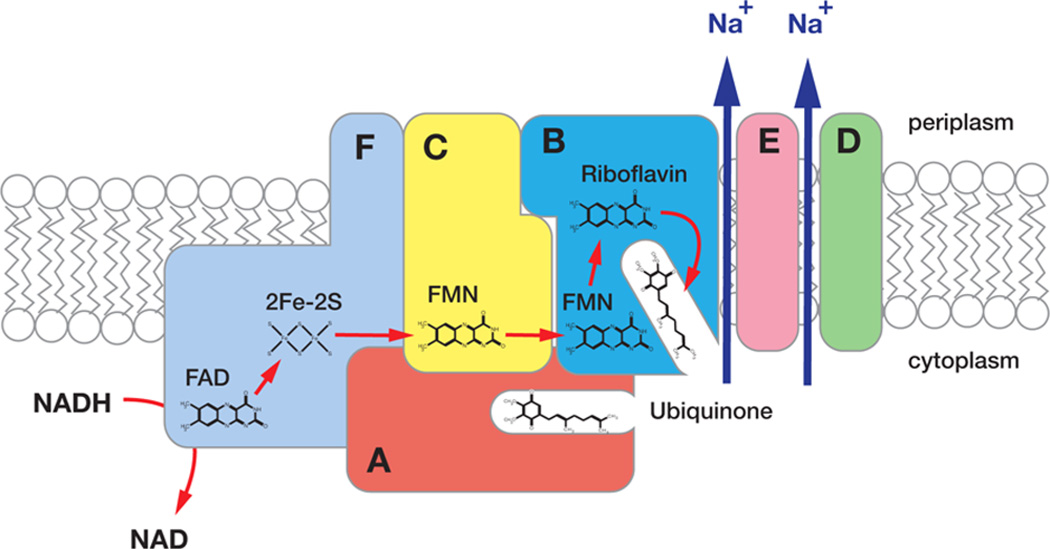
Na+-NQR is composed of six subunits (NqrA–F), all of which, except for NqrA, have transmembrane helices.12–14 Electrons are transported through the enzyme from NADH to ubiquinone along a linear pathway consisting of five different cofactors, in the following order (Figure 1): a noncovalently bound FAD and a 2Fe-2S center, both located in the NqrF subunit,15–17 two covalently attached FMN’s in NqrC and NqrB, known as FMNC and FMNB, respectively,18–20 and a noncovalently bound riboflavin, likely located in NqrB.21–23 The flow of electrons along this pathway is thought to be controlled by a series of conformational changes that expose ion binding sites to opposite sides of the membrane, for capture and release of sodium.11,24,25 Stopped-flow kinetic measurements have shown that the first Na+-dependent step in the pathway is the one in which electrons move from the 2Fe-2S center to FMNC. This redox process is also inhibited by mutations that interfere with Na+ binding on the cytosolic side of the membrane (see below).11 On the basis of these results it has been concluded that Na+ uptake occurs during this step. However, this reaction step is not electrogenic, and other measurements, with mutants that are missing specific redox cofactors, have shown that the first step in the reaction that generates membrane potential (ΔΨ) is the one in which electrons move from FMNB to riboflavin. This step is also inhibited by mutations that remove candidate Na+ binding residues on the periplasmic side of the membrane, and it has been concluded that this is the step in which the electrogenic transport of the Na+ ion through the membrane dielectric and likely its release to the periplasm take place.11,26 Thus, the processes of Na+ uptake and its transport across the membrane take place in distinct reaction steps, linked to electron transfer processes that do not share any cofactor. This has been interpreted as strong evidence against a single-site directly coupled mechanism for Na+ pumping in Na+-NQR.11 It should be noted that the redox midpoint potential of FMNC is dependent on bulk Na+ concentration, but that the magnitude of this effect, on its own, is not sufficient to account for the energetic requirements of Na+ translocation.26
Figure 1.
Scheme of the Na+-NQR complex showing the subunit composition and redox cofactors. NADH is oxidized by FAD in NqrF. Electrons move trough the enzyme to the final electron ubiquinone in the following order: FAD → 2Fe-2S center → FMNC → FMNB → riboflavin. The 2Fe-2S center → FMNC step controls Na+ uptake, and the FMNB → riboflavin step controls Na+ release. The arrows indicate the translocation of Na+ through NqrB, -D, and -E.
For efficient turnover, Na+-NQR requires its redox substrates, NADH and ubiquinone, but also its ion-pumping substrate, Na+ (or under laboratory conditions Li+).27,28 In the absence of Na+, the enzyme runs slowly, but turnover is activated up to 8-fold by Na+. In contrast, K+ alone does not activate turnover, although it has an allosteric activating effect when Na+ is present. Although other monovalent cations are not translocated by the enzyme, some such as potassium and rubidium influence the reaction allosterically. K+ behaves as a nonessential activator in the presence of sodium, lowering the Kmapp by as much as 50% and slightly increasing the kcat. Rb+ binds at the same site as K+ but has an inhibitory effect.28
To function as a Na+ pump, Na+-NQR must have structures that allow it to bind Na+ on the cytosolic side of the cell membrane and carry this highly charged ion through the hydrophobic center of the membrane, so that it can be released on the periplasmic side. Because Na+ carries a positive charge, the structures involved in Na+ binding and transport are likely to include negatively charged amino acid residues, in particular, the acidic groups Asp and Glu. Our group’s earlier studies identified 17 conserved acidic residues in the transmembrane helices of Na+-NQR, of which seven are essential for enzyme activity.27 Three of these residues are located near the periplasmic side of the membrane and appear to have a role in Na+ transport and release, but not Na+ uptake; replacing any of them with residues that do not have a negative charge leads to a decreased kcat, but no change in Kmapp for Na+. The four remaining residues are located near the cytoplasmic side of the membrane, where they are involved in Na+ uptake; replacing any of the four residues results in changes in the Kmapp for Na+.27
Further examination of the four cytoplasmic residues showed that Asp-397 in the NqrB subunit (NqrB-D397) is a key residue for the uptake of Na+ by the enzyme.27,29 Replacement of this residue with an uncharged (Ala) or positively charged (Lys) residue slowed enzyme turnover and weakened Na+ affinity (Kmapp changes from 2.5 mM for the wild-type enzyme to >100 mM), confirming its importance in Na+ binding. The turnover rate in these mutants is so slow that Na+ translocation, if present, could not be detected. Transient kinetic measurements showed that this decrease in turnover rate is due to slowing of the electron transfer from the 2Fe-2S center to FMNC,11,29 the step whose rate is controlled by Na+ in the wild-type enzyme.11
Replacement of Asp-397 with glutamate, a larger residue, but one with the same charge, produced a large decrease in kcat, to ~¼ of that of the wild-type enzyme, but interestingly had no major effect on Na+ affinity.29 This suggests that, in this mutant (NqrB-D397E), at least one of the two proposed sodium binding sites remains largely unchanged. Qualitatively similar changes in activity were observed when this aspartate was replaced with serine or asparagine residues that, while not charged, are sufficiently polar to contribute to ion binding. Possibly the most informative substitution was to cysteine. NqrB-D397C has activity slightly higher than those of the other mutants; however, the affinity of one of the two apparent binding sites in the wild-type enzyme has been drastically weakened, and the positive cooperativity changed to negative cooperativity.29 Activity with Li+, instead of Na+, is generally less severely affected by these mutations, consistent with lithium’s smaller atomic radius. These results were interpreted in terms of a model in which Na+-NQR has two Na+ uptake sites, where NqrB-D397 functions as a bidentate ligand with one “tooth” in each of the two binding sites. Replacement with cysteine, which would be monodentate, severely disrupts one of the two binding sites, leaving the other site largely intact.29
In this work, the NqrB-D397N mutant is examined in detail. In contrast to that in the wild type, redox turnover in the mutant is accelerated by K+, even in the absence of Na+. The fact that Na+ is not needed for activation by K+ shows that it is a distinct phenomenon from the allosteric effect of K+ in the wild-type enzyme described previously.28 Although increasing the K+ concentration results in an increase in the rate of redox turnover, K+ is not translocated. Furthermore, in the absence of K+, the mutant is able to pump Na+, but Na+ transport is abolished when K+ is introduced. Thus, in NqrB-D397N, in the presence of K+, the redox reaction is uncoupled from the Na+ pump; this is the first report of such an uncoupling mutation in Na+-NQR.
MATERIALS AND METHODS
Growth of Bacterial Strains
Vibrio cholerae O395N1 cells, with the nqr operon deleted (Δnqr), containing the mutant NqrB-D397N in the nqr operon, were cloned into the pBAD expression plasmid, which includes a six-histidine tag at the end of NqrF. Cells were grown in Luria-Bertani medium, in 30 L fermenters with constant agitation and aeration with 100 µg/mL ampicillin and 50 µg/mL streptomycin. Arabinose was used to induce plasmid expression as previously reported.12
Purification of NqrB-D397N
Cells were harvested and washed by centrifugation and lysed in a microfluidizer.12 Membranes were recovered by ultracentrifugation as previously reported. Membranes were solubilized with 0.3% (w/v) n-dodecyl β-d-maltoside, and the enzyme was purified using Ni-NTA and DEAE chromatography, as reported previously.16 Preparations were stored in liquid nitrogen after purification.
Steady State Kinetics
The NADH dehydrogenase and ubiquinone-1 reductase (CoQ-red) activities of Na+-NQR were measured spectrophotometrically at 340 and 287 nm, respectively, as described previously.11 All steady state kinetic reactions were conducted in TEG buffer, containing 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, 5% (v/v) glycerol, and 0.05% (w/v) n-dodecyl β-d-maltoside with different concentrations of NaCl, KCl, and LiCl.
Fast Kinetics Measurements
The kinetics of reduction of the NqrB-D397N mutant by NADH (final concentration of 300 µM) were followed using a stopped-flow spectrophotometer, as reported previously.11 The experiments were conducted using the purified mutant enzyme (30–40 µM) in TEG buffer with different concentrations of K+.
Reconstitution of the Enzyme into Proteoliposomes and Measurement of Membrane Potential (ΔΨ)
The purified NqrB-D397N enzyme was mixed with n-octyl glucoside (Anatrace) and Escherichia coli phospholipids (Avanti Lipids) in a 10:1 (w/w) lipid:protein ratio in a buffer containing 300 mM sorbitol, 50 mM Tris-HCl (pH 7.5), and 1 mM EDTA. To promote proteoliposome assembly, detergent was slowly removed via the addition of SM Bio-Beads (Bio-Rad), following the protocol described by Verkhovskaya et al.30 The voltage-sensitive dye oxonol VI was used to follow changes in membrane potential (ΔΨ) and thus track cation (Na+, Li+, or K+) transport.11 Oxonol VI absorption was followed spectrophotometrically at 625–587 nm in a reaction buffer containing 5 µM oxonol VI, 200 µM NADH, 100 µM CoQ-1, 300 mM sorbitol, 50 mM Tris-HCl (pH 7.5), and 1 mM EDTA, along with the cation(s) of interest.
RESULTS
Stimulation of the Quinone Reductase Activity of NqrB-D397N by K+
When wild-type Na+-NQR turns over at steady state in the presence of Na+ (or Li+), addition of K+ can accelerate ubiquinone reductase activity, to as much as 1.5 times the original level. This activation is not observed in the absence of Na+ (or Li+). Kinetic analysis indicates that K+ is acting as an allosteric activator, binding to the enzyme in a location distinct from the two Na+ uptake sites.28 In the NqrB-D397N mutant, however, K+ is able to activate the enzyme even in the absence of Na+ (Table 1).
Table 1.
Ubiquinone Reductase (CoQ-1 red) Activity of NqrB-D397 Mutants in the Absence of Cations and in the Presence of Saturating Concentrationsa of Na+, Li+, and K+b
| CoQ red (s−1) | ||||
|---|---|---|---|---|
| enzyme | no cation | NaCl | LiCl | KCl |
| wild type | 66.3 | 528.5 | 205.5 | 62.4 |
| NqrB-D397C | 61.9 ± 8.7 | 162.2 ± 12.3 | 110.6 ± 5.9 | 62.1 ± 10.3 |
| NqrB-D397E | 66.0 ± 4.8 | 129.6 ± 8.8 | 96.4 ± 10.3 | 67.4 ± 10.1 |
| NqrB-D397N | 59.7 ± 2.5 | 122.9 ± 4.3 | 95.5 ± 6.2 | 97.0 ± 7.4c |
| NqrB-D397S | 57.7 ± 4.1 | 101.3 ± 8.3 | 80.7 ± 5.0 | 60.3 ± 2.8 |
In 100 mM NaCl, LiCl, or KCl.
All experiments were repeated at least seven times with three independent preparations. The average and standard deviation are presented.
p < 0.05.
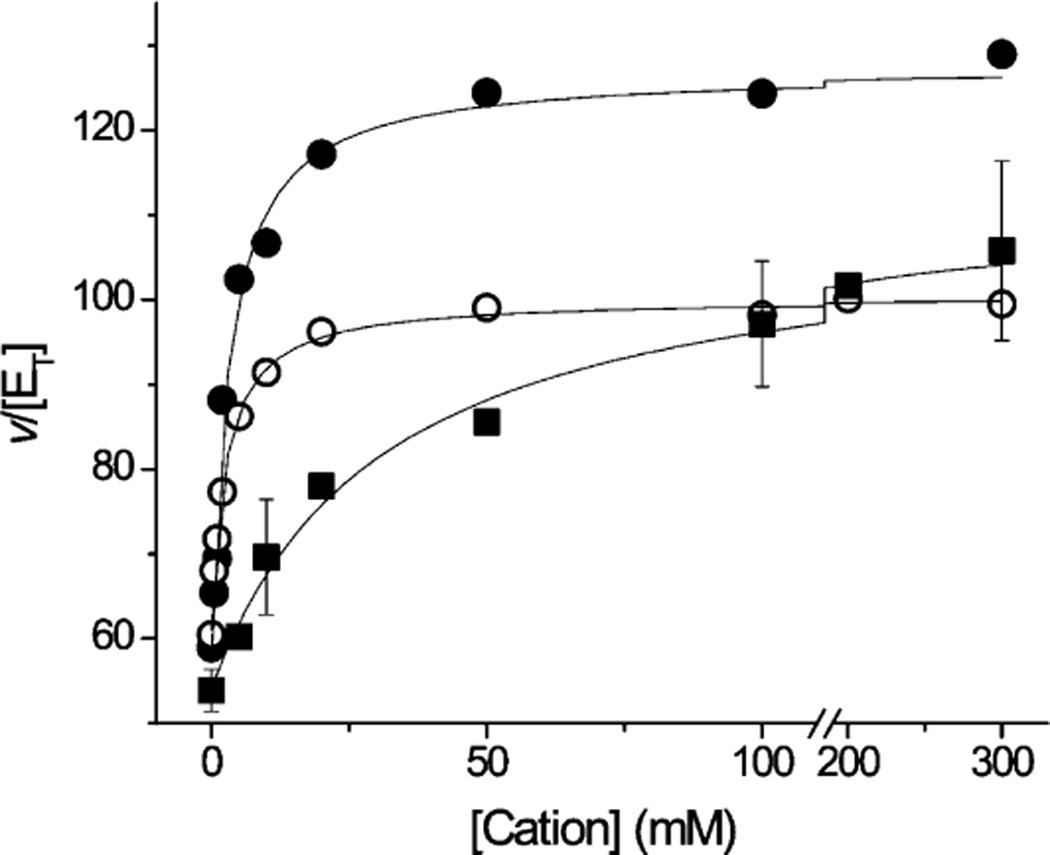
To investigate this further, the steady state activity of the mutant enzyme was studied as a function of sodium, lithium, and potassium concentrations (Figure 2). With each of the three different ions, Michaelis–Menten behavior (hyperbolic saturation kinetics) is observed, with similar kcat values for the three ions (Table 2). Although the turnover rate of the mutant enzyme is much lower than that of the wild type, the effect of K+ on the rate is very similar to those of Na+ and Li+. These results, together with the fact that NqrB-D397 is known to be part of at least one of the enzyme’s two Na+ uptake sites, suggest that, in the mutant, K+ is exerting its effects by binding to one of the sites where Na+ and Li+ bind. The apparent binding affinities (Kmapp) for Na+ and Li+ are unaffected by the mutation, but the Kmapp for K+ is significantly higher than for either Na+ or Li+ (Table 2). This is what would be expected if the mutation had altered one of the two Na+ uptake sites, allowing K+ to interact, albeit weakly, while Na+ and Li+ could bind tightly to the other site. Interestingly, the turnover rate of the NqrB-D397N mutant is even higher when both K+ and Na+ are present than with either ion alone.
Figure 2.
Dependence of the quinone reductase activity of NqrB-D397N on cation concentration for Na+ (●), Li+ (○), and K+ (■). The Co-Q1 red activity was in TEG buffer, containing 50 mM Tris-HCl (pH 8.0), 1 mM EDTA, 5% (v/v) glycerol, 0.05% (w/v) n-dodecyl β-d-maltoside, and NADH with different concentrations of NaCl, KCl, and LiCl. Saturating amounts of K2-NADH and Co-Q1 were used (150 and 50 µM, respectively) Standard deviations are listed in Table 1.
Table 2.
Kinetic Constants of the Quinone Reductase Activity for NqrB-D397N and Wild-Type Na+-NQR Obtained in the Presence of Na+, Li+, and K+a
| kcat (s−1) | Km (mM) | kcat/Km (mM/s) | ||
|---|---|---|---|---|
| NaCl | wild type | 500 ± 42 | 2.5 ± 0.9 | 200 ± 73.9 |
| NqrB-D397N | 127 ± 15 | 3.1 ± 1.1 | 41 ± 15.3 | |
| LiCl | wild type | 180 ± 25 | 3.5 ± 1.1 | 51.4 ± 17.67 |
| NqrB-D397N | 100 ± 15 | 2.4 ± 1.2 | 41.6 ± 21.75 | |
| KC1 | wild type | NDb | NDb | NDb |
| NqrB-D397N | 109 ± 8.4 | 53.7 ± 16.8 | 2.03 ± 0.65 |
The kinetic constants were determined from the saturation curves using the Michaelis–Menten equation. Each titration curve is the result of at least seven independent repetitions. The standard deviations are presented. In each repetition, every point was measured in triplicate. Conditions of the measurements are described in Materials and Methods.
Not detected.
Fast Kinetics of Reduction of NqrB-D397N in the Presence of K+
The electron transfer reactions of the wild-type enzyme have been studied extensively, and it has been established that Na+ uptake occurs in the 2Fe-2S → FMNC redox step.11 This is the first Na+-dependent process and the main rate-limiting step in the reduction reaction, when the enzyme reacts with NADH. The steady state kinetics show that in the NqrB-D397N mutant, K+ activates the redox reactions much like Na+, and it was thus important to determine whether K+ accelerates the same redox step as Na+. Because the 2Fe-2S → FMNC step is the rate-limiting step in the pathway, its rate is most easily followed by observing the arrival of electrons at riboflavin, the final redox carrier. Riboflavin undergoes a one-electron reduction from a neutral flavosemiquinone to its fully reduced form, which can be followed at 575 nm, where there are only minimal contributions from other cofactors.23 The overall progress of the reaction can also be followed at 460 nm, where the initial two-electron reduction of FAD as well as the reduction of the two FMN’s can be observed.
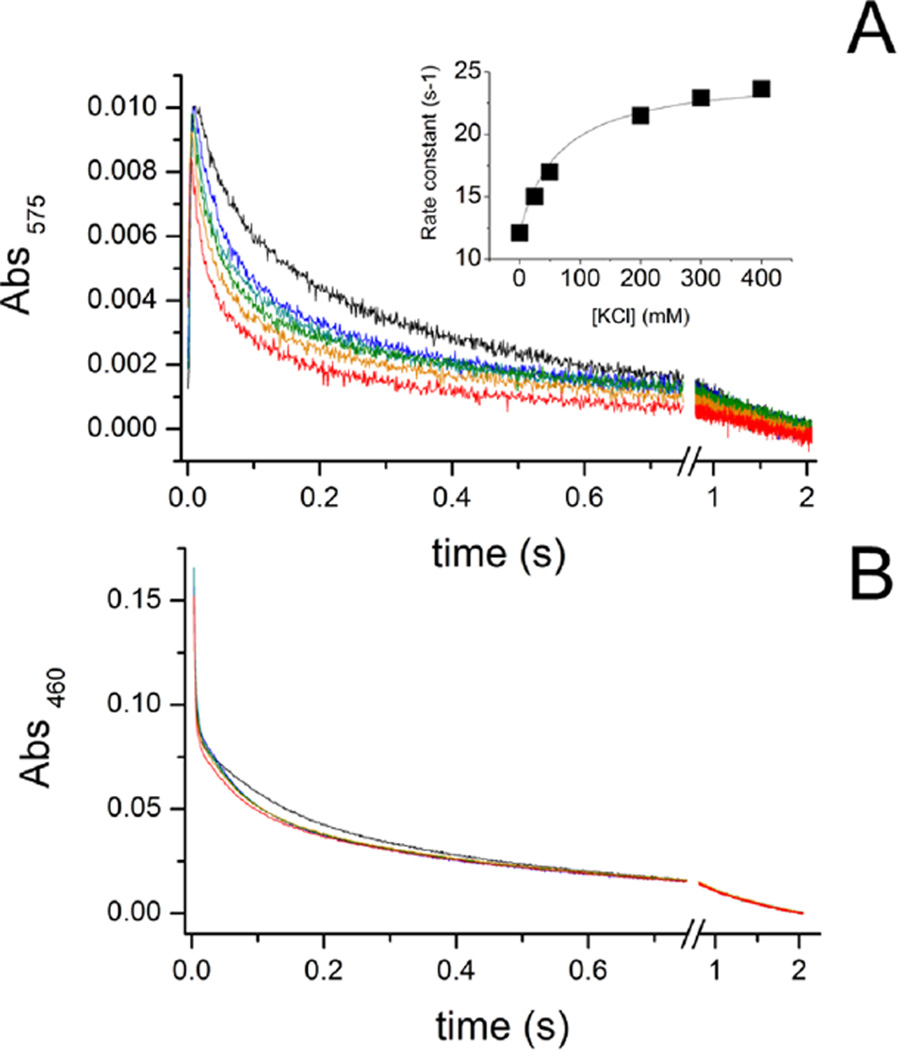
Figure 3 shows the time course of this reaction at 575 nm (A) and 460 nm (B) in the presence of different concentrations of KCl, from 0 to 400 mM. As the K+ concentration is increased, the rate of reduction of riboflavin approximately doubles (Table 3). This dependence shows saturation behavior, with a Km of 50 mM (inset). The rate increase and the Kmapp are in good agreement with values determined in the steady state measurements of ubiquinone reductase activity (Tables 1 and 2), suggesting that the acceleration by K+ of this step in the redox reaction is responsible for the effect of K+ on the overall redox reaction. The time course at 460 nm in the mutant (Figure 3B) is very similar to that observed in the wild-type enzyme and shows the reduction of FAD as a fast phase, with a rate constant of >300 s−1 (Table 3). As in the wild-type enzyme, this phase is insensitive to changes in cation concentration. At this wavelength, it is also possible to observe a much slower component with a rate that is also stimulated by K+, which corresponds to the formation of one or two FMN anionic flavosemiquinone radicals, as electrons fill in the intervening redox carriers after reduction of riboflavin.11,24
Figure 3.
Time course of reduction of the NqrB-D397N mutant by NADH (300 µM) in the presence of different concentrations of KCl at 575 nm (A) and 460 nm (B): no K+ (black), 25 mM KCl (blue), 50 mM KCl (cyan), 200 mM KCl (green), 300 mM KCl (orange), and 400 mM KCl (red). The inset shows the rate constant of the main K+-dependent phase at 575 nm as a function of K+ concentration. All concentrations after mixing.
Table 3.
Rate Constants of the Reduction of NqrB-D397N by NADH in the Presence of Different Concentrations of KCla
| rate (s−1) | |||
|---|---|---|---|
| [KCl] (mM) | FAD → FADH2 | RibH• → RibH2 | FMNc•− → FMNcH2 |
| 0 | 321 ± 2.3 | 12.1 ± 1.6 | 1.85 ± 0.34 |
| 25 | 325 ± 3.5 | 15 ± 3.2 | 2.13 ± 0.48 |
| 50 | 312 ± 10.4 | 17 ± 0.55 | 2.6 ± 0.56 |
| 200 | 336 ± 7.7 | 21.5 ± 4.6 | 3.2 ± 0.78 |
| 300 | 331 ± 5.6 | 23 ± 8.2 | 3.7 ± 0.36 |
| 400 | 334 ± 2.3 | 23.7 ± 5.3 | 3.9 ± 1.2 |
Each experiment corresponds to at least 12 kinetic traces. Three independent experiments were averaged (corresponding to at least 36 individual traces).
Ion Translocation by NqrB-D397N
Having determined that, in the NqrB-D397N mutant, K+ is able to take the place of Na+ in activating the redox reaction, we next asked whether if K+ is translocated the enzyme can translocate K+ across the membrane. To this end, the enzyme was reconstituted into phospholipid vesicles, and the formation of membrane potential was measured by following the change in the absorbance of oxonol VI (625 nm minus 587 nm).27 Enzyme turnover was initiated by the addition of NADH and ubiquinone-1. Measurements were taken in the presence of varying amounts of Na+, K+, and Li+.
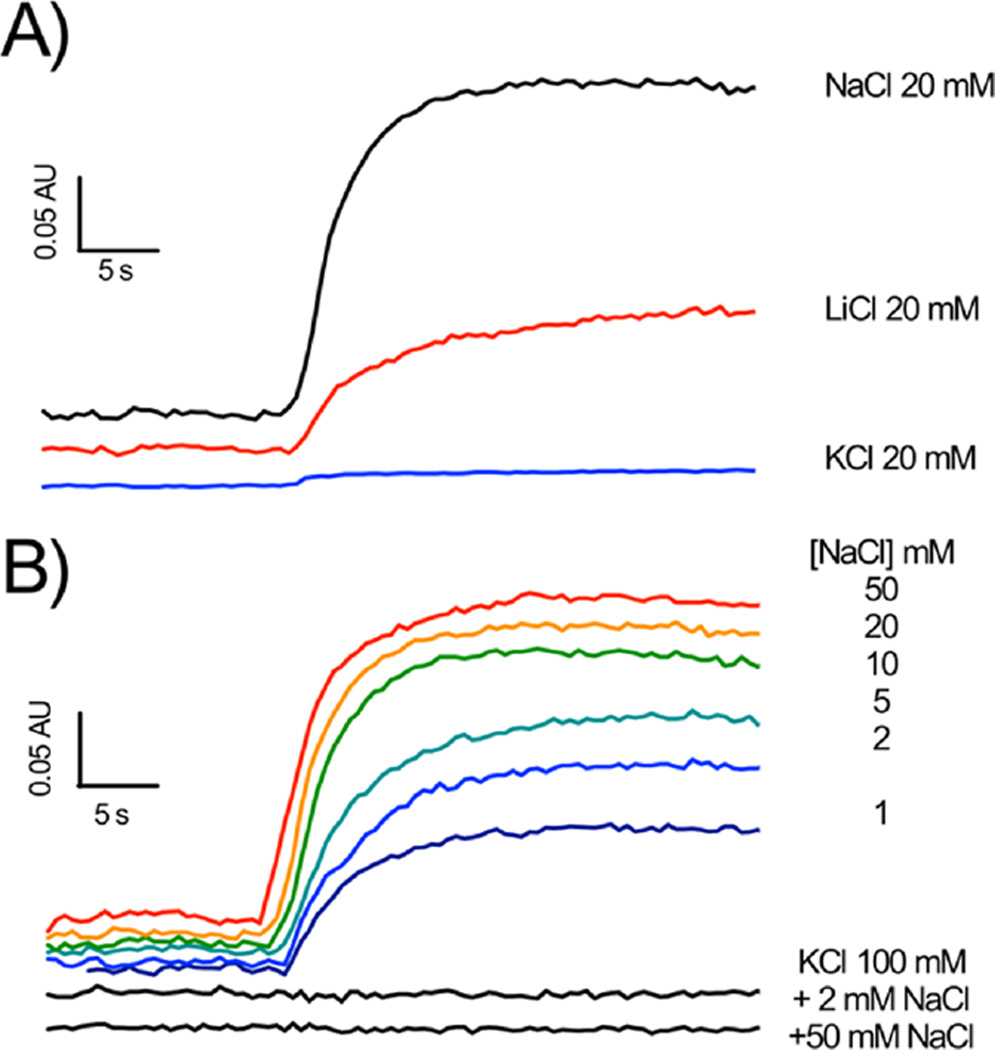
As shown in Figure 4A, turnover of NqrB-D397N, in the presence of Na+, leads to a clear increase in absorbance at 625 nm minus 587 nm, showing the generation of an electrical potential, thus confirming that the mutant is able to translocate Na+ across the membrane. It is worth noting that the redox reaction of Na+-NQR is not itself electrogenic, and a ΔΨ clearly indicates Na+ translocation. The mutant is also able to generate a potential with Li+, although as in the case of the wild-type enzyme, the rate and amplitude of the observed increase are both smaller. Thus, this aspect of ion selectivity is retained in the mutant. In the presence of K+, no change in absorbance could be detected (Figure 4A), indicating that there is no translocation of K+ by the enzyme. Thus, in the presence of K+, NqrB-D397N is effectively uncoupled: K+ binds to the enzyme, accelerating the redox reaction, but this does not result in ion pumping. This is the first instance of a mutant with an uncoupling phenotype in Na+-NQR. This is different from the case for the wild-type enzyme in which K+ alone has no effect on the redox reaction, and thus, there is no expectation that it would be translocated.
Figure 4.
Generation of membrane potential (ΔΨ) by NqrB-D397N reconstituted into proteolipsomes. The essay buffer contained 5 µM oxonol VI, 200 µM NADH, 100 µM CoQ-1, 300 mM sorbitol, 50 mM Tris-HCl (pH 7.5), and 1 mM EDTA, along with the cation(s) of interest (A) using 20 mM NaCl (black), 20 mM LiCl (red), and 20 mM KCl (blue) and (B) (top) NaCl titration (1 to 50 mM) and (bottom) effect of KCl (100 mM) on the membrane potential in the presence of 2 mM NaCl and 50 mM NaCl.
Further experiments were performed to understand the uncoupling effect of K+. Membrane potential was measured in the presence and absence of a fixed concentration of K+ (100 mM) with varying concentrations of Na+. In the absence of potassium, increasing the concentration of sodium accelerated the formation of membrane potential (Figure 4B, top). The concentration dependence showed saturation behavior with a Kmapp of 1–2 mM, close to that observed for Na+ activation of redox turnover in the isolated enzyme.28 When the same titration was conducted in the presence of 100 mM KCl, no membrane potential was formed at any concentration of Na+ (2 and 50 mM NaCl are shown in the bottom portion of Figure 4B). Thus, K+ causes the NqrB-D397N mutant to be uncoupled, not only with respect to translocation of K+ itself but also with respect to pumping of Na+.
DISCUSSION
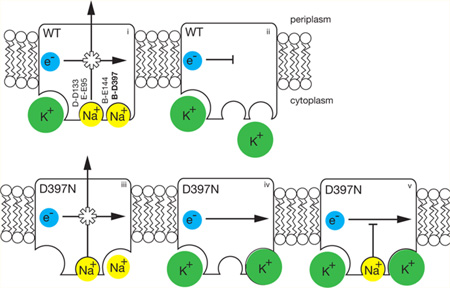
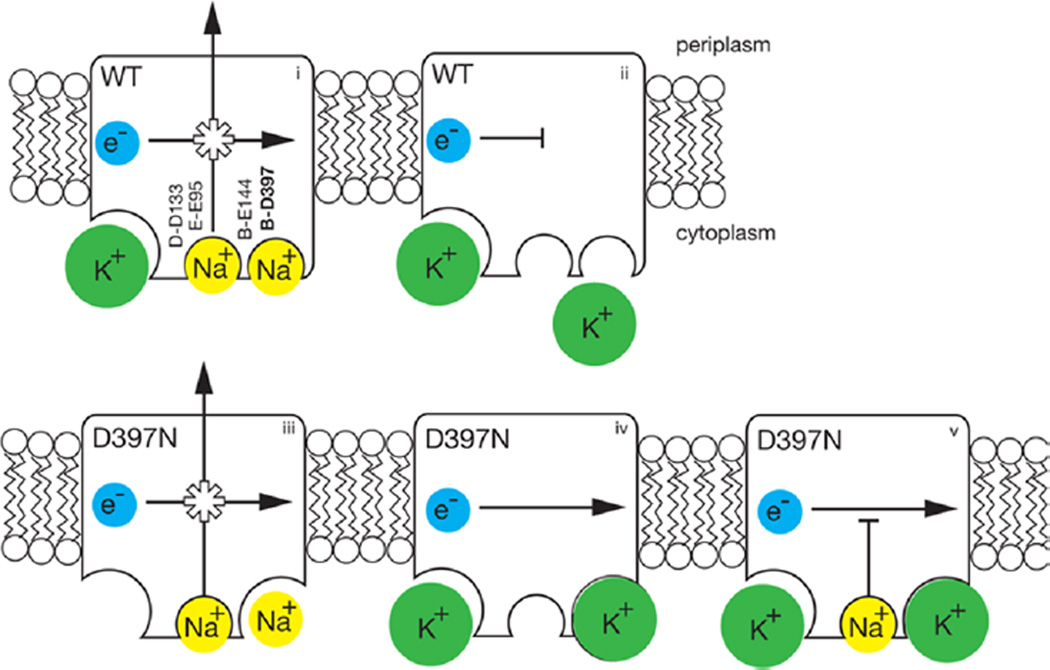
The function of Na+-NQR is to harness energy released by electron transfer from NADH to quinone, and to use this energy to translocate Na+ across the cell membrane, creating an electrochemical gradient. In the presence of potassium ions, the NqrB-D397N mutant of Na+-NQR becomes uncoupled; the redox reaction is able to proceed without ion pumping. This is the first and, thus far, only example in Na+-NQR in which the linkage between the redox and ion pumping functions of the enzyme appears to be broken. In other enzyme systems, uncoupled mutants have proven to be important experimental tools with which to study the central mechanistic question of coupling, and how energy is transduced from one form to another in biology. The NqrB-D397N mutant has the added feature that the linkage between the redox and ion pumping processes is apparently severed only when K+ is added to the system. This can provide a potentially useful experimental switch.
Substituting the conserved aspartic acid, NqrB-397, with asparagine has a number of effects on the enzyme. The redox reaction is still activated by Na+ and Li+, but to a lesser degree than in the wild-type enzyme. As in the wild type, the level of activation by Li+ is lower than that by Na+. Also, the apparent Kmapp values for both Na+ and Li+ activation are essentially the same as in the wild type, suggesting that the mutation does not cause any large changes at the relevant binding site.
One striking feature of NqrB-D397N is that the redox reaction can also be activated by K+ alone. In the wild type, addition of K+ does not affect the turnover rate unless Na+ or Li+ is present, in which case there is a weak activation caused by K+ binding at an allosteric site. In the NqrB-D397N mutant, K+ can activate the enzyme in the absence of Na+ or Li+. Once K+ is introduced, regardless of whether Na+ or Li+ is present, the enzyme becomes uncoupled, and redox activity proceeds without ion translocation. In the mutant, the Kmapp for activation by K+ is much higher than for either Na+ or Li+, indicating significantly weaker binding. Activation of the mutant by K+ and Na+ is synergistic; the maximal turnover rate with K+ and Na+ together is higher than with either ion alone (data not shown), suggesting that allosteric activation by K+ is also operating.
It appears that the primary effect of K+ in activating the NqrB-D397N mutant is exerted at a mutated Na+ uptake site, and not at the allosteric K+ binding site. Investigation of other substitutions of NqrB-D397 has shown that this residue is part of at least one Na+ uptake site and that it is unlikely to be part of the allosteric K+ site. For example, in the NqrB-D397C mutant, the Kmapp for Na+ is increased, indicating that Na+ binding at the uptake site is significantly weakened.29 Like the wild-type enzyme, the NqrB-D397C mutant is not activated by K+ alone, and redox and Na+ translocation activities continue to be coupled, even in the presence of K+. However, in this mutant, the Na+-dependent activity can still be activated by K+, with a Kmapp that is approximately the same as in the wild-type enzyme, indicating that the allosteric K+ binding site is not affected by a mutation at NqrB-D397 that causes a significant perturbation at a Na+ uptake site.
Stopped-flow kinetic measurements of the reaction of NqrB-D397N with NADH showed that activation by K+ occurs at the same step in the internal redox process as activation by Na+. This suggests that in the mutant K+ activates the enzyme by interacting in a manner similar to that of Na+.
Finally, in the presence of K+, NqrB-D397N does not generate a membrane potential, indicating that no ion translocation is taking place, in spite of the fact the redox reaction proceeds and, in fact, is activated by the cation. In the presence of Na+ or Li+, without K+, the mutant is able to generate a membrane potential, albeit with a rate and an amplitude somewhat smaller than those of the wild type, but when K+ is introduced in addition to Na+ or Li+, ion pumping does not occur, although the redox reaction is not inhibited. Figure 5 illustrates these findings, comparing the coupled wild-type enzyme to the NqrB-D397N mutant. It is worth noting that on the basis of the kcat values with Na+ and K+ and the amplitudes of the oxonol responses (Figure 5), we can estimate that cation transport in the case of K+ is no more than 0.06 times cation transport in the presence of Na+; this means that in the presence of K+ the redox turnover in the mutant is at least 94% uncoupled. In experiments with K+ diffusion potentials in vesicles made by the same protocol (data not shown), we obtained a straight line to 0 mV; there is no detectable threshold above the noise of the measurement.
Figure 5.
Scheme illustrating properties of the NqrB-D397N mutant compared to those of the wild-type enzyme. In red are shown the acidic residues involved in Na+ uptake. In the wild-type enzyme (top row, left to right), (i) electron flow is coupled to Na+ pumping, K+ binds to a different, allosteric site (bottom left corner) and accelerates the reaction (ii) in the absence of Na+, and electron flow is inhibited, confirming that the redox and pumping processes are coupled. In the NqrB-D397N mutant (bottom row, left to right), (iii) in the absence of K+, electron flow is still coupled to Na+ pumping; (iv) in the presence of K+ (in the absence of Na+), electron flow proceeds, but without ion pumping, meaning that coupling has been disrupted; and (v) K+ causes the mutant to run uncoupled, even in the presence of Na+. Of the acidic residues involved in cation uptake, shown in bold, only NqrB-D397 is associated with the modified binding site.
The reasons for the behavior of the mutant are not yet clear. An earlier study indicated that Na+-NQR has two uptake/binding sites for Na+ and other translocated ions (i.e., Li+), and that NqrB-D397 may provide ligands in one of these sites, or possibly both, because aspartate is potentially a bidentate ligand.31 Because the Kmapp values for activation of the redox reaction by Na+ and Li+ are unchanged from that of the wild type, it is likely that at least one of these binding sites is largely unchanged by the mutation. On the other hand, K+ appears to activate the redox reaction in much the same way as Na+, suggesting that one Na+ binding site may have been altered by the mutation so that it binds K+, though somewhat weakly. It thus appears that, in the mutant, translocation of K+ does not take place because a second ion selectivity filter, beyond the initial ion binding site, prevents K+ from passing through the ion pump. However, why this would result in an uncoupled enzyme, rather than an inactive one, remains to be elucidated.
Acknowledgments
Funding
This work was supported by National Science Foundation Grant MCB1052234.
Footnotes
The authors declare no competing financial interest.
REFERENCES
- 1.Hase CC, Fedorova ND, Galperin MY, Dibrov PA. Sodium ion cycle in bacterial pathogens: Evidence from cross-genome comparisons. Microbiol. Mol. Biol. Rev. 2001;65:353–370. doi: 10.1128/MMBR.65.3.353-370.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dibrov PA, Kostryko VA, Lazarova RL, Skulachev VP, Smirnova IA. The sodium cycle. I. Na+-dependent motility and modes of membrane energization in the marine alkalotolerant Vibrio alginolyticus. Biochim. Biophys. Acta. 1986;850:449–457. doi: 10.1016/0005-2728(86)90113-1. [DOI] [PubMed] [Google Scholar]
- 3.Dibrov PA, Lazarova RL, Skulachev VP, Verkhovskaya ML. The sodium cycle. II. Na+-coupled oxidative phosphorylation in Vibrio alginolyticus cells. Biochim. Biophys. Acta. 1986;850:458–465. doi: 10.1016/0005-2728(86)90114-3. [DOI] [PubMed] [Google Scholar]
- 4.Terashima H, Kojima S, Homma M. Flagellar motility in bacteria structure and function of flagellar motor. Int. Rev. Cell Mol. Biol. 2008;270:39–85. doi: 10.1016/S1937-6448(08)01402-0. [DOI] [PubMed] [Google Scholar]
- 5.Dashper SG, Brownfield L, Slakeski N, Zilm PS, Rogers AH, Reynolds EC. Sodium ion-driven serine/threonine transport in Porphyromonas gingivalis. J. Bacteriol. 2001;183:4142–4148. doi: 10.1128/JB.183.14.4142-4148.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huda MN, Morita Y, Kuroda T, Mizushima T, Tsuchiya T. Na+-driven multidrug efflux pump VcmA from Vibrio cholerae non-O1, a non-halophilic bacterium. FEMS Microbiol. Lett. 2001;203:235–239. doi: 10.1111/j.1574-6968.2001.tb10847.x. [DOI] [PubMed] [Google Scholar]
- 7.Tokuda H, Asano M, Shimamura Y, Unemoto T, Sugiyama S, Imae Y. Roles of the respiratory Na+ pump in bioenergetics of Vibrio alginolyticus. J. Biochem. 1988;103:650–655. doi: 10.1093/oxfordjournals.jbchem.a122323. [DOI] [PubMed] [Google Scholar]
- 8.Tokuda H, Unemoto T. A respiration-dependent primary sodium extrusion system functioning at alkaline pH in the marine bacterium Vibrio alginolyticus. Biochem. Biophys. Res. Commun. 1981;102:265–271. doi: 10.1016/0006-291x(81)91516-3. [DOI] [PubMed] [Google Scholar]
- 9.Bertsova YV, Bogachev AV. The origin of the sodium-dependent NADH oxidation by the respiratory chain of Klebsiella pneumoniae. FEBS Lett. 2004;563:207–212. doi: 10.1016/S0014-5793(04)00312-6. [DOI] [PubMed] [Google Scholar]
- 10.Bogachev AV, Bertsova YV, Aitio O, Permi P, Verkhovsky MI. Redox-dependent sodium binding by the Na+-translocating NADH:quinone oxidoreductase from Vibrio harveyi. Biochemistry. 2007;46:10186–10191. doi: 10.1021/bi700440w. [DOI] [PubMed] [Google Scholar]
- 11.Juarez O, Morgan JE, Nilges MJ, Barquera B. The energy transducing redox steps of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 2010;107:12505–12510. doi: 10.1073/pnas.1002866107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barquera B, Hellwig P, Zhou W, Morgan JE, Hase CC, Gosink KK, Nilges MJ, Bruesehoff PJ, Roth A, Lancaster CR, Gennis RB. Purification and characterization of the recombinant Na+-translocating NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry. 2002;41:3781–3789. doi: 10.1021/bi011873o. [DOI] [PubMed] [Google Scholar]
- 13.Duffy EB, Barquera B. Membrane topology mapping of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae by PhoA-green fluorescent protein fusion analysis. J. Bacteriol. 2006;188:8343–8351. doi: 10.1128/JB.01383-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bogachev AV, Verkhovsky MI. Na+-translocating NADH:quinone oxidoreductase: Progress achieved and prospecst of investigations. Biochemistry (Moscow) 2005;70:143–149. doi: 10.1007/s10541-005-0093-4. [DOI] [PubMed] [Google Scholar]
- 15.Rich PR, Meunier B, Ward FB. Predicted structure and possible ionmotive mechanism of the sodium-linked NADH-ubiquinone oxidoreductase of Vibrio alginolyticus. FEBS Lett. 1995;375:5–10. doi: 10.1016/0014-5793(95)01164-a. [DOI] [PubMed] [Google Scholar]
- 16.Barquera B, Nilges MJ, Morgan JE, Ramirez-Silva L, Zhou W, Gennis RB. Mutagenesis study of the 2Fe-2S center and the FAD binding site of the Na+-translocating NADH:ubiquinone oxidoreductase from Vibrio cholerae. Biochemistry. 2004;43:12322–12330. doi: 10.1021/bi048689y. [DOI] [PubMed] [Google Scholar]
- 17.Bogachev AV, Belevich NP, Bertsova YV, Verkhovsky MI. Primary steps of the Na+-translocating NADH:ubiquinone oxidoreductase catalytic cycle resolved by the ultrafast freeze-quench approach. J. Biol. Chem. 2009;284:5533–5538. doi: 10.1074/jbc.M808984200. [DOI] [PubMed] [Google Scholar]
- 18.Hayashi M, Nakayama Y, Yasui M, Maeda M, Furuishi K, Unemoto T. FMN is covalently attached to a threonine residue in the NqrB and NqrC subunits of Na+-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 2001;488:5–8. doi: 10.1016/s0014-5793(00)02404-2. [DOI] [PubMed] [Google Scholar]
- 19.Nakayama Y, Yasui M, Sugahara K, Hayashi M, Unemoto T. Covalently bound flavin in the NqrB and NqrC subunits of Na+-translocating NADH-quinone reductase from Vibrio alginolyticus. FEBS Lett. 2000;474:165–168. doi: 10.1016/s0014-5793(00)01595-7. [DOI] [PubMed] [Google Scholar]
- 20.Barquera B, Hase CC, Gennis RB. Expression and mutagenesis of the NqrC subunit of the NQR respiratory Na+ pump from Vibrio cholerae with covalently attached FMN. FEBS Lett. 2001;492:45–49. doi: 10.1016/s0014-5793(01)02224-4. [DOI] [PubMed] [Google Scholar]
- 21.Barquera B, Zhou W, Morgan JE, Gennis RB. Riboflavin is a component of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. Proc. Natl. Acad. Sci. U.S.A. 2002;99:10322–10324. doi: 10.1073/pnas.162361299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Casutt MS, Huber T, Brunisholz R, Tao M, Fritz G, Steuber J. Localization and function of the membrane-bound riboflavin in the Na+-translocating NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae. J. Biol. Chem. 2010;285:27088–27099. doi: 10.1074/jbc.M109.071126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juarez O, Nilges MJ, Gillespie P, Cotton J, Barquera B. Riboflavin is an active redox cofactor in the Na+-pumping NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae. J. Biol. Chem. 2008;283:33162–33167. doi: 10.1074/jbc.M806913200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Juárez O, Morgan JE, Barquera B. The electron transfer pathway of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. J. Biol. Chem. 2009;284:8963–8972. doi: 10.1074/jbc.M809395200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Neehaul Y, Juarez O, Barquera B, Hellwig P. Infrared spectroscopic evidence of a redox-dependent conformational change involving ion binding residue NqrB-D397 in the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry. 2013;52:3085–3093. doi: 10.1021/bi4000386. [DOI] [PubMed] [Google Scholar]
- 26.Neehaul Y, Juarez O, Barquera B, Hellwig P. Thermodynamic contribution to the regulation of electron transfer in the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. Biochemistry. 2012;51:4072–4077. doi: 10.1021/bi300343u. [DOI] [PubMed] [Google Scholar]
- 27.Juarez O, Athearn K, Gillespie P, Barquera B. Acid residues in the transmembrane helices of the Na+-pumping NADH:quinone oxidoreductase (Na+-NQR) from Vibrio cholerae involved in sodium translocation. Biochemistry. 2009;48:9516–9524. doi: 10.1021/bi900845y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Juárez O, Shea ME, Makhatadze GI, Barquera B. The role and specificity of the catalytic and regulatory cation-binding sites of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae. J. Biol. Chem. 2011;286:26383–26390. doi: 10.1074/jbc.M111.257873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shea ME, Juarez O, Cho J, Barquera B. Aspartic acid 397 in subunit B of the Na+-pumping NADH:quinone oxidoreductase from Vibrio cholerae forms part of a sodium-binding site, is involved in cation selectivity, and affects cation-binding site cooperativity. J. Biol. Chem. 2013;43:31241–31249. doi: 10.1074/jbc.M113.510776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Verkhovskaya ML, Garcìa-Horsman A, Puustinen A, Rigaud JL, Morgan JE, Verkhovsky MI, Wikström M. Glutamic acid 286 in subunit I of cytochrome bo3 is involved in proton translocation. Proc. Natl. Acad. Sci. U.S.A. 1997;94:10128–10131. doi: 10.1073/pnas.94.19.10128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Nayal M, Di Cera E. Valence screening of water in protein crystals reveals potential Na+ binding sites. J. Mol. Biol. 1996;256:228–234. doi: 10.1006/jmbi.1996.0081. [DOI] [PubMed] [Google Scholar]