Abstract
Objectives
To evaluate whether radiation exposure from cardiac computed tomographic angiography is associated with DNA damage and whether damage leads to programmed cell death and activation of genes involved in apoptosis and DNA repair.
Background
Exposure to radiation from medical imaging has become a public health concern, but whether it causes significant cell damage remains unclear.
Methods
We conducted a prospective cohort study in 67 patients undergoing cardiac computed tomographic angiography (CTA) between January 2012 and December 2013 in two US medical centers. Median blood radiation exposure was estimated using phantom dosimetry. Biomarkers of DNA damage and apoptosis were measured by flow cytometry, whole genome sequencing, and single cell polymerase chain reaction.
Results
The median DLP was 1535.3 mGy·cm (969.7 – 2674.0 mGy·cm). The median radiation dose to the blood was 29.8 milliSieverts (18.8 – 48.8 mSv). Median DNA damage increased 3.39% (1.29 – 8.04%, P<0.0001) post-radiation. Median apoptosis increased 3.1-fold (1.4 – 5.1-fold, P<0.0001) post-radiation. Whole genome sequencing revealed changes in the expression of 39 transcription factors involved in the regulation of apoptosis, cell cycle, and DNA repair. Genes involved in mediating apoptosis and DNA repair were significantly changed post-radiation, including DDB2 [1.9-fold (1.5 – 3.0-fold), P<0.001], XRCC4 [3.0-fold (1.1 – 5.4-fold), P=0.005], and BAX [1.6-fold (0.9 – 2.6-fold), P<0.001]. Exposure to radiation was associated with DNA damage [OR: 1.8 (1.2 – 2.6), P=0.003]. DNA damage was associated with apoptosis [OR: 1.9 (1.2 – 5.1), P<0.0001] and gene activation [OR: 2.8 (1.2 – 6.2), P=0.002].
Conclusions
Patients exposed to radiation from cardiac CTA had evidence of DNA damage, which was associated with programmed cell death and activation of genes involved in apoptosis and DNA repair.
Keywords: [30] CT/MRI, [142] Gene expression, [143] Gene regulation, [150] Imaging
Introduction
The application of cardiac computed tomographic angiography (CTA) has risen dramatically over the last decade (1–3). Cardiac CTA is now commonly used to manage patients with suspected coronary artery disease (4), aortic stenosis in preparation for transcatheter aortic valve replacement (5), atrial fibrillation prior to ablation (6), and aortic dissection post surgical repair (7). Radiation exposure from these procedures can be significant because of the need for gating to compensate for cardiac motion. A single cardiac CTA can expose patients to a radiation dose equivalent to having at ≥150 chest x-rays (8). Not surprisingly, the widespread use of this procedure has raised concern among physicians and patients about the potential deleterious effects of radiation exposure from cardiac CTA (9).
It is well known that exposure of cells to therapeutic doses of radiation triggers a complex network of signal transduction pathways that induce changes in gene expression and protein structure (10), resulting in apoptosis (e.g., programmed cell death), cell cycle arrest or progression, and DNA repair to minimize the risk of mutagenesis (11). Whether radiation doses from medical imaging tests (less than 100 millisieverts) causes similar damage and activates these biological pathways is less certain. Although previous studies have demonstrated that proteins involved in the DNA damage response pathway are phosphorylated after exposure to radiation from medical imaging (12–14), these clinical studies have been limited by the use of semi-qualitative measures, specifically counting the formation of gamma H2AX foci in a small subset of cells (12–14). Furthermore, prior studies have not measured the effects of radiation exposure from medical imaging on other key signaling proteins in the DNA damage response pathways, which are also altered after exposure to therapeutic doses of radiation (15), nor have they determined whether radiation from medical imaging is associated with programmed cell death. Finally, no human studies to date have measured the effects of radiation exposure from medical imaging on changes in gene expression in vivo (16), which have been shown to be significantly up-regulated after radiation therapy (17–19).
The purpose of our prospective study is to determine whether radiation exposure from cardiac CTA is associated with DNA damage, and whether the extent of damage is associated with the activation of pathways responsible for repairing or eliminating cells to minimize mutation risk. The results of this study will help clinicians better understand the risks associated with radiation exposure from medical imaging so they can better inform their patients. This study will also help determine whether additional strategies are needed to protect patients against radiation exposure from CTA.
METHODS
Please refer to the Supplemental Methods (Online Supplement) for a more detailed description of the methods.
Patients and diagnostic imaging studies
Adult patients aged ≥18 years old who underwent a clinically indicated cardiac CTA were recruited from Stanford Hospital (Stanford, CA) and the Veterans Affairs Palo Alto Health Care System (Palo Alto, CA). The study complies with the institutional review boards of Stanford University and the Veterans Affairs Health Care System Palo Alto. All subjects gave informed consent.
Estimation of radiation dose
Radiation dose to the body and blood was estimated using phantom dosimetery, the ImPACT Computed Tomography Patient Dosimetry Calculator spreadsheet (ImPACT, London, England) (13, 20). Only doses calculated from the ImPACT Computed Tomography Patient Dosimetry Calculator were used in the analysis.
Sample collection for in vivo studies
Whole blood was collected at baseline and multiple time points after cardiac CTA, as detailed in the Online Supplement.
Proteomic biomarker assays
Analyses of protein biomarkers of DNA damage and apoptosis by flow cytometry and immunohistochemistry were performed using standard protocols. DNA damage biomarkers included phosphorylated H2AX, ATM, and p53. Ten thousand cells were evaluated. Biomarkers of apoptosis including annexin and BAX were measured by flow cytometry and immunohistochemistry, respectively. One hundred thousand cells and 100 cells were evaluated for the expression of apoptotic markers by flow cytometry and immunohistochemistry, respectively.
Genomic biomarker assays
Whole genome profiling using RNA-sequencing (n=3) and single cell polymerase chain reaction (n=51) of selected genes (Supplemental Table 1) were performed using standard protocols.
Statistical analysis
Continuous variables with normal distribution and those that were not normally distributed were expressed as mean ± standard deviation and median (first quartile – third quartile), respectively. Observations from dichotomous variables were summarized as proportions. Differences in continuous variables that were normally distributed, continuous variables that were not normally distributed, and proportions were compared using Student’s t-, Wilcoxon-sign ranked and Fisher’s exact tests, respectively. Spearman correlations (rho, 95% confidence intervals, P value) were used to assess associations between continuous variables. Analysis of whole genome sequencing data is detailed in the Supplemental Methods. Multivariate logistic regression models were used to evaluate the association between mean blood radiation dose, DNA damage, programmed cell death, and gene activation. In addition to mean blood radiation dose, the following 7 covariates were evaluated for the presence of a significant association or correlation to the 3 outcomes identified above: 1) age, 2) sex, 3) body mass index (BMI), 4) race, 5) history of smoking, 6) history of cancer, and 7) mean iodine content. Only those covariates with significant associations with the three outcomes were included in the model. Statistical analysis was performed using Intercooled Stata, version 12.1 (Stata Corp, College Station, Texas). Tests had α level for significance set at P <0.05 for single comparisons. Bonferonni method was used to adjust the P-values for multiple testing. Unadjusted P-values are shown and an asterisk is noted when comparisons were significant.
RESULTS
Clinical and scan parameters
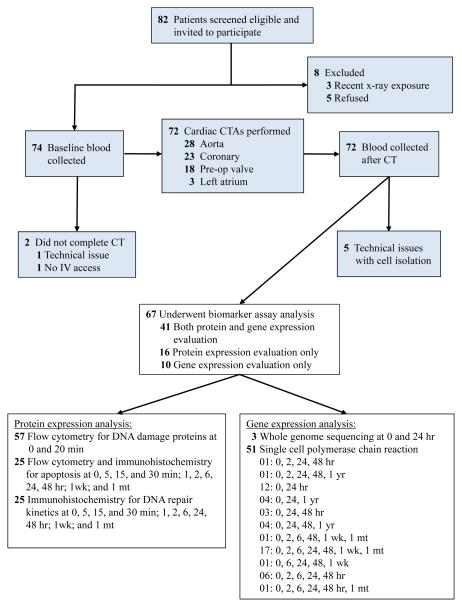
Of the 82 patients who were eligible to participate in the study, 67 patients underwent biomarker analysis before and after exposure to cardiac CTA (Figure 1), using standard scanning protocols detailed in Supplemental Table 2 and in the Online Supplement. Clinical and scan parameters are detailed in Table 1.
Figure 1.
Consort diagram. A consort flow diagram summarizing the participant flow through the study.
Table 1.
Clinical and scan parameters for patients with and without DNA damage
| Total (n=57) | No DNA Damage (n=21) | DNA Damage (n=36) | p-value | |
|---|---|---|---|---|
| Clinical Parameters | ||||
| Age at enrollment, years | 67 (56–79) | 73 (63–77) | 66.5 (54–88) | 0.67 |
| Sex | 0.33 | |||
| Male (%) | 78.9 (45/57) | 71.4 (15/21) | 83.3 (30/36) | |
| Female (%) | 21.1 (12/57) | 28.6 (6/21) | 16.7 (6/36) | |
| BMI (kg/m2) | 26.5 (24.4 – 29.4) | 25.7 (24.2 – 30.1) | 27.1 (24.4 – 30.2) | 0.66 |
| Race | ||||
| White (%) | 80.7 (46/57) | 71.4 (15/21) | 86.1 (31/36) | 0.18 |
| Non-White (%) | 19.3 (11/57) | 28.6 (6/21) | 13.9 (5/36) | |
| Current smoking (%) | 21.1 (12/57) | 14.2 (3/21) | 25.0 (9/36) | 0.27 |
| History of cancer (%) | 22.8 (13/57) | 23.8 (5/21) | 22.2 (8/36) | 0.57 |
| Scan parameters* | ||||
| Mean DLP (mGy·cm) | 1511.0 (969.7 – 2589.1) | 1105.9 (568.3 – 1431) | 2137.0 (1293.0 – 2740.9) | <0.0001 |
| Mean total effective dose (mSv) | 36.9 (26.1 – 61.3) | 30.2 (17.0 – 45.1) | 49.2 (32.6 – 71.3) | 0.004* |
| Mean blood radiation dose (mSv) | 29.8 (18.8 – 48.8) | 23.2 (8.8 – 31.5) | 39.6 (23.6 – 53.8) | <0.0001* |
| Mean iodine content (g) | 38.6 (33.3 – 46.9) | 35.0 (25.9 – 37.8) | 41.4 (33.3 – 48) | 0.01 |
Scan parameters reflect only the cohort of patients that underwent biomarker testing for DNA damage (n=57) and does not reflect the entire cohort (n=67).
Extent of DNA damage and its association with clinical and scan parameters
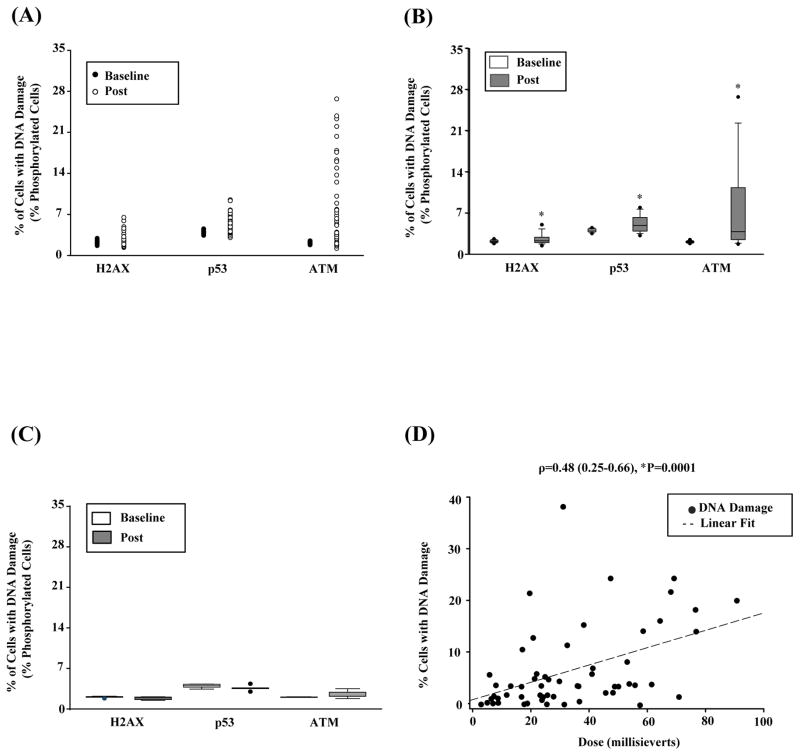
Seventy percent of patients (36/57) had ≥2% increase in phosphorylation of at least one DNA damage marker post-radiation exposure (Figure 2A–B), indicating that at least 200/10,000 cells per patient had evidence of DNA damage after cardiac CTA. A 2% cutoff was chosen based on findings in patients undergoing echocardiography (n=9), an imaging study that produces no radiation (data not shown). Patients who underwent echocardiography had <1% change in phosphorylation; thus, this cut-off is well above the level detected in our negative control group. The median change in phosphorylation of any DNA damage marker was 3.39% (1.29 – 8.04%, *P<0.0001). Although H2AX is more commonly used to estimate the extent of DNA damage (12–14, 21), the median change in phosphorylation was higher for ATM (1.7% [0.7 – 8.0%]; *P<0.0001) than both H2AX (0.2% [0 – 0.7%], *P<0.0001) and p53 (0.5% [−0.2 – 2.2], *P<0.0001), suggesting that phosphorylated ATM may be a more sensitive biomarker for DNA damage. Median change in phosphorylation of any DNA damage marker was higher in patients exposed to ≥20 mSv of radiation compared to those exposed to <20 mSv (3.6% [1.6 – 12.6%] vs. 1.3% [0.1 – 3.7%]; *P=0.03). Importantly, patients receiving radiation doses ≤7.5 mSv had no significant changes in phosphorylation, suggesting an absence of detectable DNA damage at very low doses (P>0.05, Figure 2C). Table 1 presents descriptive and bivariate analysis of patients with or without DNA damage with clinical and scan parameters. Of the clinical and demographic parameters evaluated, only radiation dose was significantly associated with the presence or absence of DNA damage (39.6 mSv [23.6 mSv – 53.8 mSv] vs. 23.2 mSv [8.8mSv – 31.5 mSv]; *P<0.0001). The extent of DNA damage was also correlated with the amount of radiation exposure (r=0.48, *P=0.0001; Figure 2D, Supplemental Figure 1). Although mean iodine dose was higher in patients who had evidence of DNA damage, this parameter was not significant after adjustment for multiple comparisons. In order to assess whether DNA damage was primarily due to radiation and not contrast effects, we performed in vitro whole blood irradiation experiments in the absence of contrast. Similar to our in vivo findings, biomarkers of DNA damage were consistently increased at radiation doses above 25 mSv (Supplemental Figure 5).
Figure 2.
DNA damage detected after radiation exposure. A) Scatter plot graph of levels of phosphorylated H2AX, ATM, p53 at baseline and after exposure to radiation from cardiac CTA (left, n=57). (B) Quantitative assessment of protein biomarkers of DNA damage (i.e., levels of phosphorylated H2AX, ATM, and p53) in all patients undergoing cardiac CTA (right, n=57). The horizontal line within the box marks the median, and the length of the box represents the interquartile range, which is the difference between the 75th and 25th percentiles. Whiskers (error bars) above and below the box indicate the 90th and 10th percentiles. In addition, outliers are indicated by closed circle. *Statistically significant at P<0.01. (C) Quantitative assessment of protein biomarkers of DNA damage in patients receiving very low doses of radiation (e.g., ≤7.5 mSv). (D) Scatterplot graph of the correlation analysis between DNA damage estimate and radiation dose (n=57).
Extent of programmed cell death and its association with DNA damage and radiation dose
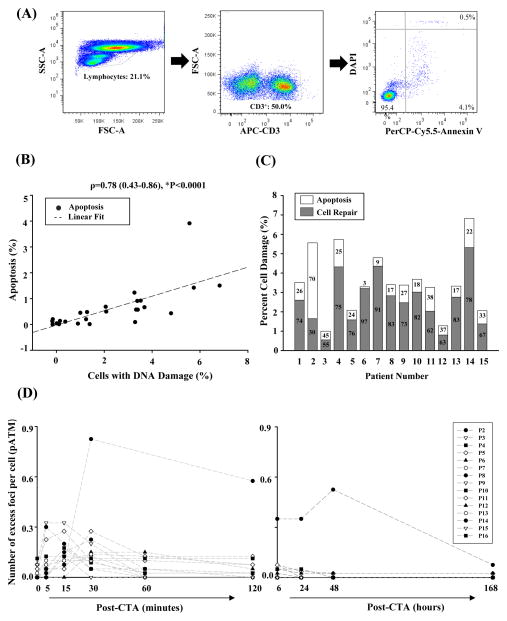
We next measured levels of apoptotic cell death in a subset of patients before and after undergoing cardiac CTA (n=25). Sixty percent (15/25) of patients had at least 2-fold increase in apoptosis. The median increase in apoptosis was 3.1-fold [(1.4 – 5.1-fold], P<0.0001) post- radiation. In absolute terms, however, the median number of cells undergoing programmed cell death was estimated at 0.7% [0.5 – 1.28%], which is equivalent to death of 700/100,000 lymphocytes evaluated (Figure 3A). Median fold apoptosis was highest in patients exposed to ≥20 mSv compared to those exposed to <20 mSv [2.3-fold (1.8 – 3.0-fold) vs. 0.7-fold (0.7 – 1.0-fold), *P=0.03) (Supplemental Figure 2A–C). The degree of apoptosis was more strongly correlated with the extent of DNA damage (r=0.78, *P<0.0001; Figure 3B) than the amount of radiation exposure (r=0.42, P=0.03; Supplemental Figure 2D). The majority of damaged cells, however, were repaired (Figure 3C). Although the rate of response of repair and apoptotic pathways to DNA damage (i.e., disappearance of excess foci counts per nucleus) varied across individuals, most patients did not have detectable DNA damage 2 hours after exposure to radiation from cardiac CTA (Figure 3D; Supplemental Table 3), which is consistent with a previous study (14).
Figure 3.
Apoptosis detected after radiation exposure. (A) Sample flow cytometry dot plot from a patient after cardiac CTA. In order to identify apoptotic cells, lymphocytes were first isolated using forward and side scatter profiles (left). CD3+ T cells were then identified from within this subset (middle). Lastly, apoptotic T cells were detected using Annexin V, an early marker of apoptosis, and DAPI, an intracellular marker for dead cells, which are either necrotic or at the later stages of apoptosis (right). The estimate of apoptotic cell death includes the sum of cells that express Annexin only or Annexin-DAPI. This patient had 1.8% of apoptotic cells that express Annexin only and 0.2% of dead cells that express both Annexin and DAPI. (B) Scatterplot graph of the correlation analysis between the estimates of apoptosis and DNA damage (n=25). (C) Scatter plot graph of the disappearance of the phosphorylated ATM (pATM) over time as damaged cells are repaired or eliminated (n=25). (D) Bar graph displaying the relative percentages of cells that were repaired or had undergone programmed cell death in individual patients with evidence of cell death by flow cytometry (n=15). The remainder of the patients had no cell death and complete repair (n=10) (not shown in graph).
Whole genome profiling to evaluate changes in biological pathways after radiation exposure from cardiac CTA
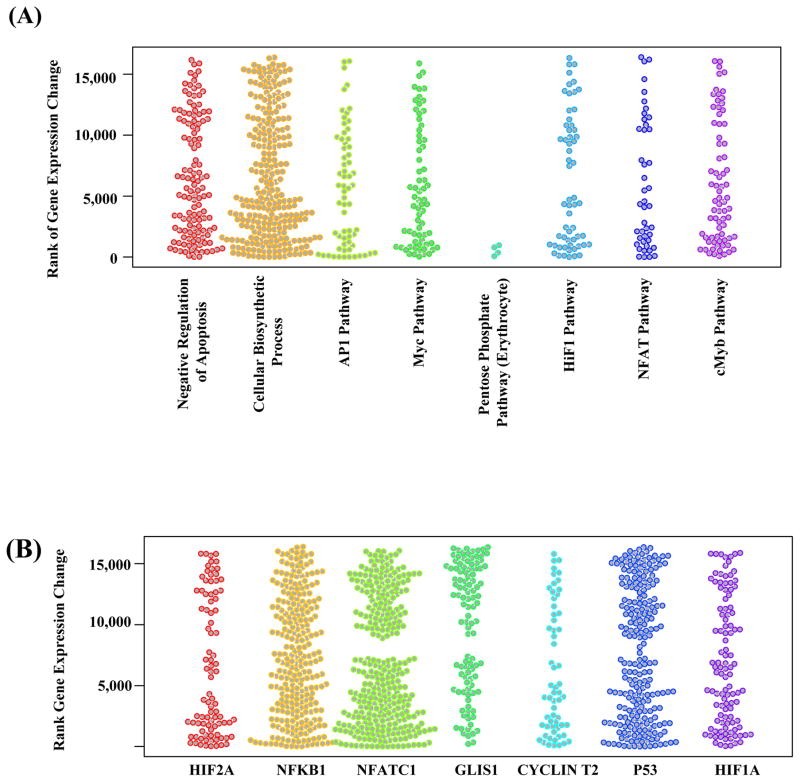
Using the transcriptome data of T lymphocytes cells from 3 patients (Supplemental Table 4), we performed gene functional enrichment analyses to identify biological processes, signaling/metabolic pathways, and transcription factors that were significantly associated with radiation exposure. In total, 33 signaling/metabolic pathways, 39 transcription factors, and 17 biological processes were significantly changed after multiple test correction (q-value cutoff 0.1, p-values obtained by Fisher’s exact test (Figure 4, Supplemental Figure 3, Supplemental Tables 5–8). The active transcription factors formed a densely connected regulatory network (Supplemental Figure 3B), with many transcriptional regulations among them, suggesting that the activity change of some of the significant transcription factors may be driven by other upstream active transcription factors.
Figure 4.
Stripcharts showing the associations of selected gene sets with the transcriptional response of blood cells to cardiac CTA exposure. (A) Functional gene sets for biological processes, signaling and metabolic pathways. (B) Regulation gene sets for transcription factor target genes. The transcriptome data is obtained through RNA-sequencing of 3 selected individuals before and 24 hours after cardiac CTA. The color groups the genes (shown as dots) in different biological processes/pathways (A) or regulated by different transcription factors (B). The y-axis corresponds to the rank of the gene’s expression changes among 16,486 protein-coding genes. Low ranks correspond to decreased expression of genes after cardiac CTA exposure, high ranks correspond to increased expression. When a set of genes are not associated with cardiac CTA exposure, we expect the ranks of the genes’ expression changes to be evenly distributed, hence an uneven distribution of the dots along the y-axis indicates association of a gene set with cardiac CTA exposure.
Changes in expression of individual genes associated with DNA repair and apoptosis
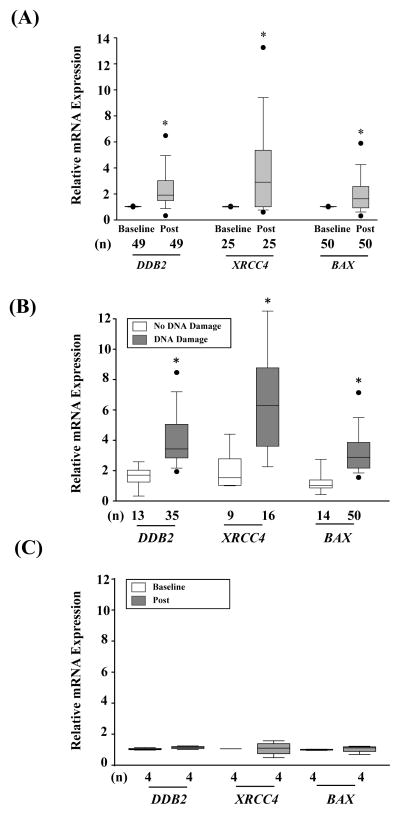
To validate the overall differences in gene expression after radiation found in the RNA-sequencing analysis, we next measured the expression levels of a select number of genes involved in the regulation of apoptosis, cell cycle, and cell repair that were found to be up-regulated in the whole genome transcription analysis (Supplemental Table 1, Supplemental Table 9–11). Radiation exposure from cardiac CTA elicited a statistically significant change in the expression levels of DDB2, XRCC4, and BAX over time (*P≤0.01) as shown in Supplemental Figure 4 and Supplemental Table 10. These genes are known to play important roles in response to DNA damage. DDB2, for example, facilites DNA binding for nuclear excision repair and regulates cell fate by promoting cell cycle progression and programmed cell death (22), XRCC4, on the other hand, has been found to complex with DNA ligase IV to complete the final steps of nonhomolous repair of DNA double stranded breaks (23). Finally, BAX mediates p53-dependent apoptosis by inserting into the mitochondrial membrane and releasing pro-apoptotic factors (24). The maximum relative fold change in gene activation of these genes were significantly increased after cardiac CTA: DDB2: 1.9-fold [1.5 – 3.0-fold], *P<0.001; XRCC4: 3.0-fold [1.1 – 5.4-fold],*P=0.005; BAX: 1.6-fold [0.9 – 2.6-fold],*P<0.001; Figure 5). Although DNA repair was complete within two hours post radiaiton exposure, peak changes in gene expression occurred 24 hours post-radiation expousre. This finding is consistent with previous studies that report that transcription is arrested by DNA damage but subsequently recovers after repair is complete to prevent the production of aberrant transcripts and interference between the transcription and repair machinery (25–27). Although gene changes did not vary significantly by dose (Supplemental Figure 4), patients with evidence of DNA damage had significantly higher activation of these genes compared to those with no DNA damage (DDB2: 2.55-fold [1.74 – 4.42-fold] vs. 1.26-fold [1.16 – 1.77-fold], *P=0.003; XRCC4: 4.9-fold [2.8 – 6.6-fold] vs. 1.0-fold [0.9– 1.9-fold], *P=0.005; BAX: 2.1-fold [1.1 – 2.9-fold] vs. 1.0-fold [0.9 – 1.3-fold], *P=0.001, Figure 5B). In support of these findings, gene activation could not be detected in patients receiving a radiation dose of ≤7.5 mSv who also had no evidence of DNA damage as detailed above (P>0.05, Figure 5C). These findings suggest that more extensive DNA damage is associated with greater transcriptional changes in repair and apoptotic genes. To assess whether changes were primarly due to radiation and not contrast effects, experiments were repeated in vitro in the absence of contrast. Similar to our findings in vivo, increased expression of several genetic biomarkers associated with cell repeair and death were found after radiation exposure (Supplemental Figure 6). Importantly, patients who underwent echocardiography (n=11) showed no significant change in gene expression post imaging (data not shown).
Figure 5.
Individual genes involved in apoptosis and repair altered after radiation exposure. (A) Maximum median change in gene expression of DDB2, XRCC4, and BAX post-radiation compared to baseline in all patients. (B) Bar graph comparing the relative change in gene expression in patients with or without DNA damage. DNA damage is measured as the maximum change of any of the three DNA damage biomarkers 30 minutes post-radiation. Apoptosis is the maximum apoptotic cell death detected at any time point within 1-week post-radiation. (C) Maximum median change in gene expression of DDB2, XRCC4, and BAX post-radiation compared to baseline in the subset of patients who received a radiation dose of ≤7.5 mSv. * Statistically significant at P<0.01.
Multivariate regression analysis
Only dose and mean iodine content were associated with DNA damage, programmed cell death, and gene activation. Findings from the bivariate analysis suggest that mean iodine content may be a potential confounder although the difference between the mean iodine content in patients who did and did not have DNA damage was not significant after adjustment for multiple comparisons (Table 1). Mean iodine content was found to correlate with dose (r=0.67, *P<0.0001). To determine the effects of mean iodine content on the relationship between dose and DNA damage, we performed the following set of regressions: 1) DNA damage on dose alone, 2) DNA damage on mean iodine content alone, and 3) DNA damage on dose, iodine, and their interaction. Although both were significant (P<0.01) when analyzed separately, only DNA damage remained significant when both were analyzed together. Given the confounding effects of mean iodine content, subsequent analyses excluded this covariate. Results of separate multivariate regressions are summarized in Table 2. Higher radiation dose was associated with greater odds of having DNA damage (OR: 1.8 [1.2 – 2.6], P=0.003*). Patients with a greater extent of DNA damage had greater odds of gene activation (OR: 2.8 [1.2 – 6.2], P=0.002*) and apoptosis (OR: 1.9 [1.2 – 5.1], *P<0.0001). Patients with greater extent of DNA damage and at least a 2-fold activation of at least one gene had greater odds of apoptosis (OR: 1.6 [1.2 – 2.7], *P<0.0001). Radiation dose was not a statistically significant predictor of gene activation or apoptosis. Taken together, it appears that an estimate of radiation dose does not capture the entire spectrum of biological changes associated with radiation exposure from cardiac CTA.
Table 2.
Multivariate regression model of the association between DNA damage, gene activation, and apoptosis with dose
| Outcome | Multivariate Logistic Regression | Exact Logistic Regression | |
|---|---|---|---|
| DNA Damage (n=57) | Gene Activation (n=51) | Apoptosis (n=25) | |
| OR (95% CI) | 1.8 (1.2–2.6)* for dose | 1.0 (0.99–1.1) for dose 2.8 (1.2 – 6.2)* for DNA damage1 |
1.1 (1.0 – 1.2) for dose 1.9 (1.2 – 5.1)* for DNA damage1 1.6 (1.2 – 2.7)* for gene activation |
| R2 | 0.2 | 0.16 | -- |
| Model Score | 13.1# | 8.2# | 16.4 |
| P value | 0.003 | 0.002 | <0.0001 |
χ2 for Likelihood Ratio Test;
P<0.01.
Model that replaces dose with DNA damage given DNA damage captures the effects of dose.
DISCUSSION
In this study, we demonstrated that patients undergoing cardiac CTA at doses ≥7.5 mSv had evidence of DNA damage, which was associated with activation of genes involved in regulating cell repair and programmed cell death. Although most damaged cells are repaired, a small percentage of cells die. These findings raise the possibility that radiation exposure from cardiac CTA may cause DNA damage that can lead to mutations if damaged cells are not repaired or eliminated properly. Cumulative cell death after repeated exposures may also be problematic.
Although previous studies have reported cross-sectional associations between radiation exposure from medical imaging and DNA damage, these studies were conducted in either the pediatric population (12) or in small groups of adult patients undergoing CTA using a single biomarker of damage (13, 14, 28, 29). The relationship between dosage and DNA damage has also not been fully explored in adult patients undergoing cardiac CTA (30). In this study, we performed a comprehensive evaluation of cellular effects of radiation exposure from diagnostic imaging including measurements of multiple parameters of DNA damage and apoptosis as well as transcription changes in over 50 patients at serial time points. In a multivariate analysis, we found that radiation dose was associated with DNA damage although the relationship was only roughly linear (31, 32). Dose was not predictive of gene activation or programmed cell death. These results are not surprising, given that we currently rely on complicated computer simulations and mathematical models to calculate the “absorbed dose” and “biological equivalent dose” (33), and then use these values to extrapolate biological risk (i.e., cellular injury and cancer risk) (34). Our findings suggest that measurement of the degree of DNA damage is more predictive of both gene activation and apoptosis than estimated radiation dose and may be a better marker of biological risk.
Importantly, in this study, we purposely recruited patients who had a wide range of doses to determine the effects of dose on cellular damage. Patients with radiation exposure greater than ≈20 mSv either underwent a coronary tomographic angiography performed in a traditional scanner equipped with older technology (e.g., Discovery 750HD, GE, Milwaukee, WI) and retrospective gating or underwent an evaluation of their entire aorta (e.g., patients with a history of dissection or pre-TAVR) using a state-of-the-art dual source scanner (e.g., Sensation Dual Source, Siemens Medical Solutions, Forchheim, Germany) and prospective gating. In patients undergoing coronary angiography using a dual source scanner where radiation doses were ≤7.5 mSV, no damage was observed, further supporting a dose-response relationship. Overall, these data are consistent with our hypothesis that higher radiation dose leads to more damage.
In addition to evaluating the relationship between dose and damage, we provide a comprehensive analysis of how cells respond to damage, which has not been evaluated in prior studies. Our study found that exposure to radiation from CTA, like exposure to therapeutic doses of radiation (17, 18, 35, 36), activates biological pathways and genes and increases the activity of transcription factors involved in the regulation of cell repair, cell cycle progression, and apoptosis, which are critical in preventing the development of mutations, a finding that has not yet been previously reported. Although cells with DNA damage are mostly repaired, a small number of cells die after radiation exposure from CTA. These findings are consistent with known biological responses to DNA damage (11). Cells with insignificant damage can be repaired, whereas those with extensive damage undergo programmed cell death to minimize the risk of mutation. We found a complete resolution of DNA damage occurred within two hours of exposure in the majority of patients, which is consistent with prior studies (14). A few patients, however, had cells with residual DNA damage and continued activation of cellular response pathways detectable up to 1 month post-exposure, which has been reported after exposure to radiation doses as low as 1 mGy in vitro, supporting a possible lack of efficient activation of repair mechanisms at dosing levels used in medical imaging (21). If residual cells are not eventually repaired or eliminated, they can potentially retain mutations. The number of cells with residual DNA damage after radiation exposure from CTA, however, is small (<1%). The number of lymphocytes that died after imaging is also small (<1%). Nevertheless, cumulative cell death from repeated exposure can be potentially harmful, especially in elderly patients who may have a limited pool of naïve T cells that respond to novel pathogens (37). Further study is needed to evaluate whether the subpopulation of naïve T cells is affected by radiation exposure from medical imaging.
A potential limitation of the study is that we did not directly measure DNA damage. However, a direct measure of small changes in DNA damage is not possible with current techniques (38). This study also did not measure the risk of cancer from radiation exposure from cardiac CTA because only cells that evade cellular repair and programmed cell death survive and produce cancer long-term. Because identifying these cells is not feasible, interpretation of these findings should be limited to the cellular effects of radiation from CTA in the short-term. Finally, this study was not designed to assess cancer risk. Nevertheless, it provides valuable insight into the biological response to radiation from medical imaging.
In summary, patients undergoing cardiac CTA have evidence of DNA damage in T lymphocytes, which is associated with death of a small fraction of cells and activation of biological pathway, transcription factors and genes involved in cell repair and apoptosis. Although cardiac CTA is a valuable clinical tool in the management of patients with cardiovascular disease, awareness among physicians and patients that DNA damage and apoptosis can occur even after diagnostic imaging may encourage greater adherence to dose reduction strategies and perhaps further research to develop novel agents to protect patients from the potential adverse effects of radiation exposure from cardiac CTA.
Supplementary Material
Acknowledgments
Funding Sources: NIH, American Heart Association
SOURCES OF FUNDING
This work was supported by the American Heart Association [10SDG4280129], National Institutes of Health [HL093172 and HL09551], and Stanford Cardiovascular Institute. We would like to thank the Stanford Functional Genomics Facility and the Neuroscience Microscopy Service (Andrew Olson) for their assistance with the RNA-sequencing experiment and the analysis of the immunohistochemistry data, respectively. We thank Jarrett Rosenberg for his assistance with the statistical analysis. We thank Blake Wu and Ian Chen for their assistance with editing of this manuscript, respectively.
Abbreviations and Acronyms
- BAX
BCL2-associated X protein
- BSA
bovine serum albumin
- CTA
computed tomographic angiography
- CTDI
computed tomographic dose index
- DAPI
4′,6-diamidino-2-phenylindole
- DDB2
damage-specific DNA binding protein 2
- DLP
dose length product
- DPBS
Dulbecco’s phosphate buffered saline
- FACS
fluorescence-activated cell sorting
- GO
gene ontology
- H2AX
H2A histone family, member X
- mGy
milliGray
- mSv
milliSieverts
- pATM
phosphorylated ataxia telangiectasia mutated
- PBMC
peripheral blood mononuclear cell
- PBS
phosphate buffered saline
- PCR
polymerase chain reaction
- p53
tumor protein p53
- RT-STA
reverse transcription-specific target amplification
- 7-AAD
7-Amino-Actinomycin D
- TFTG
transcription factor target genes
- XRCC4
X-ray repair complementing defective repair in Chinese hamster cells 4
Footnotes
Relationship to Industry: None
CONFLICTS OF INTERESTS
None
References
- 1.Brenner DJ, Hall EJ. Computed tomography--an increasing source of radiation exposure. N Engl J Med. 2007;357(22):2277–2284. doi: 10.1056/NEJMra072149. [DOI] [PubMed] [Google Scholar]
- 2.Fazel R, Krumholz HM, Wang Y, et al. Exposure to low-dose ionizing radiation from medical imaging procedures. N Engl J Med. 2009;361(9):849–857. doi: 10.1056/NEJMoa0901249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Einstein AJ, Moser KW, Thompson RC, Cerqueira MD, Henzlova MJ. Radiation dose to patients from cardiac diagnostic imaging. Circulation. 2007;116(11):1290–1305. doi: 10.1161/CIRCULATIONAHA.107.688101. [DOI] [PubMed] [Google Scholar]
- 4.Mark DB, Berman DS, Budoff MJ, et al. ACCF/ACR/AHA/NASCI/SAIP/SCAI/SCCT 2010 expert consensus document on coronary computed tomographic angiography. Catheter Cardiovasc Interv. 2010;76(2):E1–42. doi: 10.1002/ccd.22495. [DOI] [PubMed] [Google Scholar]
- 5.Bloomfield GS, Gillam LD, Hahn RT, et al. A practical guide to multimodality imaging of transcatheter aortic valve replacement. JACC Cardiovasc Imaging. 2012;5(4):441–455. doi: 10.1016/j.jcmg.2011.12.013. [DOI] [PubMed] [Google Scholar]
- 6.Martinez MW, Kirsch J, Williamson EE, et al. Utility of nongated multidetector computed tomography for detection of left atrial thrombus in patients undergoing catheter ablation of atrial fibrillation. JACC Cardiovasc Imaging. 2009;2(1):69–76. doi: 10.1016/j.jcmg.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 7.Hiratzka LF, Bakris GL, Beckman JA, et al. 2010 ACCF/AHA/AATS/ACR/ASA/ SCA/SCAI/SIR/STS/SVM guidelines for the diagnosis and management of patients with thoracic aortic disease. J Am Coll Cardiol. 2010;55(14):e27–e129. doi: 10.1016/j.jacc.2010.02.015. [DOI] [PubMed] [Google Scholar]
- 8.Hausleiter J, Meyer T, Hermann F, et al. Estimated radiation dose associated with cardiac ct angiography. JAMA. 2009;301(5):500–507. doi: 10.1001/jama.2009.54. [DOI] [PubMed] [Google Scholar]
- 9.Douglas P, Iskandrian AE, Krumholz HM, et al. Achieving quality in cardiovascular imaging. J Am Coll Cardiol. 2006;48(10):2141–2151. doi: 10.1016/j.jacc.2006.06.076. [DOI] [PubMed] [Google Scholar]
- 10.Marchetti F, Coleman MA, Jones IM, Wyrobek AJ. Candidate protein biodosimeters of human exposure to ionizing radiation. Int J Radiat Biol. 2006;82(9):605–639. doi: 10.1080/09553000600930103. [DOI] [PubMed] [Google Scholar]
- 11.Jackson SP, Bartek J. The DNA-damage response in human biology and disease. Nature. 2009;461(7267):1071–1078. doi: 10.1038/nature08467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beels L, Bacher K, De Wolf D, Werbrouck J, Thierens H. Gamma-h2ax foci as a biomarker for patient x-ray exposure in pediatric cardiac catheterization: Are we underestimating radiation risks? Circulation. 2009;120(19):1903–1909. doi: 10.1161/CIRCULATIONAHA.109.880385. [DOI] [PubMed] [Google Scholar]
- 13.Rothkamm K, Balroop S, Shekhdar J, Fernie P, Goh V. Leukocyte DNA damage after multi-detector row CT: A quantitative biomarker of low-level radiation exposure. Radiology. 2007;242(1):244–251. doi: 10.1148/radiol.2421060171. [DOI] [PubMed] [Google Scholar]
- 14.Lobrich M, Rief N, Kuhne M, et al. In vivo formation and repair of DNA double-strand breaks after computed tomography examinations. PNAS. 2005;102(25):8984–8989. doi: 10.1073/pnas.0501895102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kodama M, Otsubo C, Hirota T, Yokota J, Enari M, Taya Y. Requirement of atm for rapid p53 phosphorylation at ser46 without ser/thr-gln sequences. Molecular and Cellular biology. 2010;30(7):1620–1633. doi: 10.1128/MCB.00810-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nosel I, Vaurijoux A, Barquinero JF, Gruel G. Characterization of gene expression profiles at low and very low doses of ionizing radiation. DNA Repair. 2013;12(7):508–517. doi: 10.1016/j.dnarep.2013.04.021. [DOI] [PubMed] [Google Scholar]
- 17.Amundson SA, Lee RA, Koch-Paiz CA, et al. Differential responses of stress genes to low dose-rate gamma irradiation. MCR. 2003;1(6):445–452. [PubMed] [Google Scholar]
- 18.Amundson SA, Fornace AJ., Jr Microarray approaches for analysis of cell cycle regulatory genes. Methods in Molecular Biology. 2004;241:125–141. doi: 10.1385/1-59259-646-0:125. [DOI] [PubMed] [Google Scholar]
- 19.Paul S, Amundson SA. Development of gene expression signatures for practical radiation biodosimetry. International Journal of Radiation Oncology, Biology, Physics. 2008;71(4):1236–1244. doi: 10.1016/j.ijrobp.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Einstein AJ, Elliston CD, Arai AE, et al. Radiation dose from single-heartbeat coronary ct angiography performed with a 320-detector row volume scanner. Radiology. 2010;254(3):698–706. doi: 10.1148/radiol.09090779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rothkamm K, Lobrich M. Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. PNAS. 2003;100(9):5057–5062. doi: 10.1073/pnas.0830918100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bakkenist CJ, Kastan MB. DNA damage activates atm through intermolecular autophosphorylation and dimer dissociation. Nature. 2003;421(6922):499–506. doi: 10.1038/nature01368. [DOI] [PubMed] [Google Scholar]
- 23.Drouet J, Delteil C, Lefrancois J, Concannon P, Salles B, Calsou P. DNA-dependent protein kinase and xrcc4-DNA ligase IV mobilization in the cell in response to DNA double strand breaks. JBC. 2005;280(8):7060–7069. doi: 10.1074/jbc.M410746200. [DOI] [PubMed] [Google Scholar]
- 24.Chipuk JE, Green DR. Dissecting p53-dependent apoptosis. Cell Death Differ. 2006;13(6):994–1002. doi: 10.1038/sj.cdd.4401908. [DOI] [PubMed] [Google Scholar]
- 25.Svejstrup JQ. The interface between transcription and mechanisms maintaining genome integrity. Trends Biochem Sci. 2010;35(6):333–338. doi: 10.1016/j.tibs.2010.02.001. [DOI] [PubMed] [Google Scholar]
- 26.Adam S, Polo SE, Almouzni G. Transcription recovery after DNA damage requires chromatin priming by the h3.3 histone chaperone hira. Cell. 2013;155(1):94–106. doi: 10.1016/j.cell.2013.08.029. [DOI] [PubMed] [Google Scholar]
- 27.Heine GF, Horwitz AA, Parvin JD. Multiple mechanisms contribute to inhibit transcription in response to DNA damage. J Biol Chem. 2008;283(15):9555–9561. doi: 10.1074/jbc.M707700200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuefner MA, Grudzenski S, Hamann J, et al. Effect of ct scan protocols on x-ray-induced DNA double-strand breaks in blood lymphocytes of patients undergoing coronary ct angiography. Eur Radiol. 2010;20(12):2917–2924. doi: 10.1007/s00330-010-1873-9. [DOI] [PubMed] [Google Scholar]
- 29.Brand M, Sommer M, Achenbach S, et al. X-ray induced DNA double-strand breaks in coronary ct angiography: Comparison of sequential, low-pitch helical and high-pitch helical data acquisition. Eur J Radiol. 2012;81(3):e357–362. doi: 10.1016/j.ejrad.2011.11.027. [DOI] [PubMed] [Google Scholar]
- 30.Beels L, Bacher K, Smeets P, Verstraete K, Vral A, Thierens H. Dose-length product of scanners correlates with DNA damage in patients undergoing contrast CT. Eur J Radiol. 2012;81(7):1495–1499. doi: 10.1016/j.ejrad.2011.04.063. [DOI] [PubMed] [Google Scholar]
- 31.Tubiana M, Feinendegen LE, Yang C, Kaminski JM. The linear no-threshold relationship is inconsistent with radiation biologic and experimental data. Radiology. 2009;251(1):13–22. doi: 10.1148/radiol.2511080671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Little MP, Wakeford R, Tawn EJ, Bouffler SD, Berrington de Gonzalez A. Risks associated with low doses and low dose rates of ionizing radiation: Why linearity may be (almost) the best we can do. Radiology. 2009;251(1):6–12. doi: 10.1148/radiol.2511081686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McCollough CH, Christner JA, Kofler JM. How effective is effective dose as a predictor of radiation risk? AJR. 2010;194(4):890–896. doi: 10.2214/AJR.09.4179. [DOI] [PubMed] [Google Scholar]
- 34.Einstein AJ, Henzlova MJ, Rajagopalan S. Estimating risk of cancer associated with radiation exposure from 64-slice computed tomography coronary angiography. JAMA. 2007;298(3):317–323. doi: 10.1001/jama.298.3.317. [DOI] [PubMed] [Google Scholar]
- 35.Smirnov DA, Brady L, Halasa K, Morley M, Solomon S, Cheung VG. Genetic variation in radiation-induced cell death. Genome Res. 2012;22(2):332–339. doi: 10.1101/gr.122044.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smirnov DA, Morley M, Shin E, Spielman RS, Cheung VG. Genetic analysis of radiation-induced changes in human gene expression. Nature. 2009;459(7246):587–591. doi: 10.1038/nature07940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fagnoni FF, Vescovini R, Passeri G, et al. Shortage of circulating naive cd8(+) t cells provides new insights on immunodeficiency in aging. Blood. 2000;95(9):2860–2868. [PubMed] [Google Scholar]
- 38.Vral A, Fenech M, Thierens H. The micronucleus assay as a biological dosimeter of in vivo ionising radiation exposure. Mutagenesis. 2011;26(1):11–17. doi: 10.1093/mutage/geq078. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.