Abstract
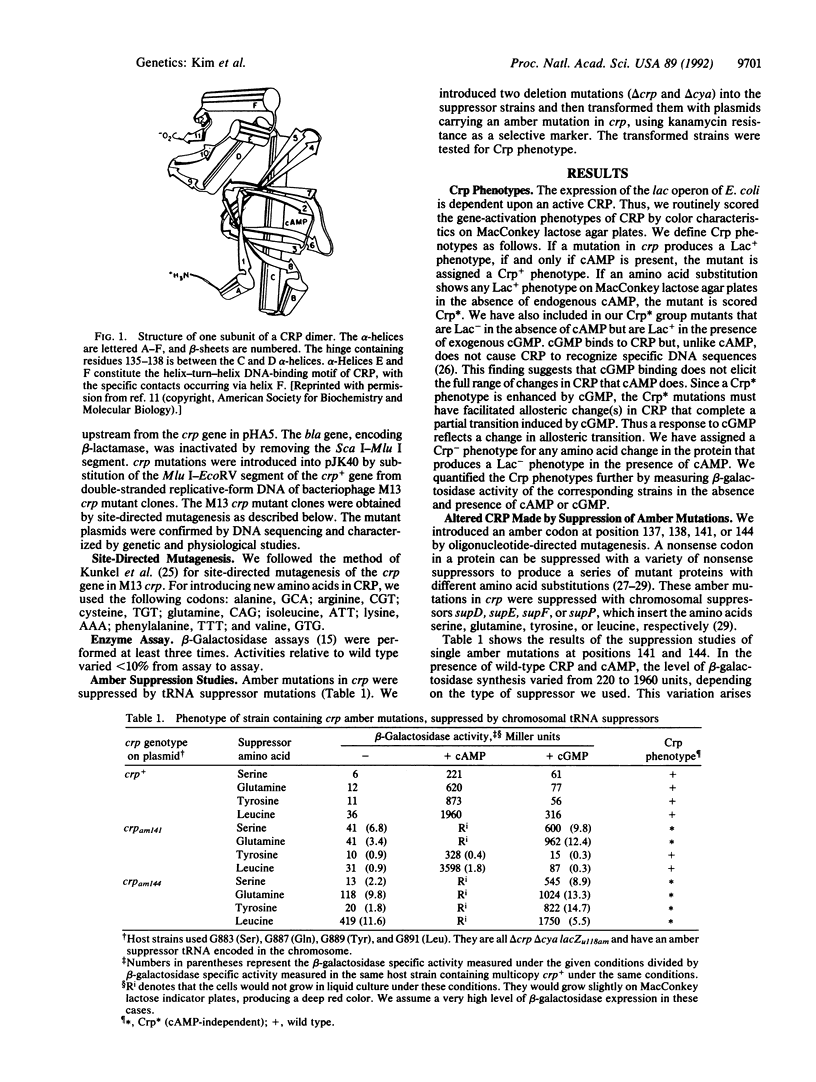
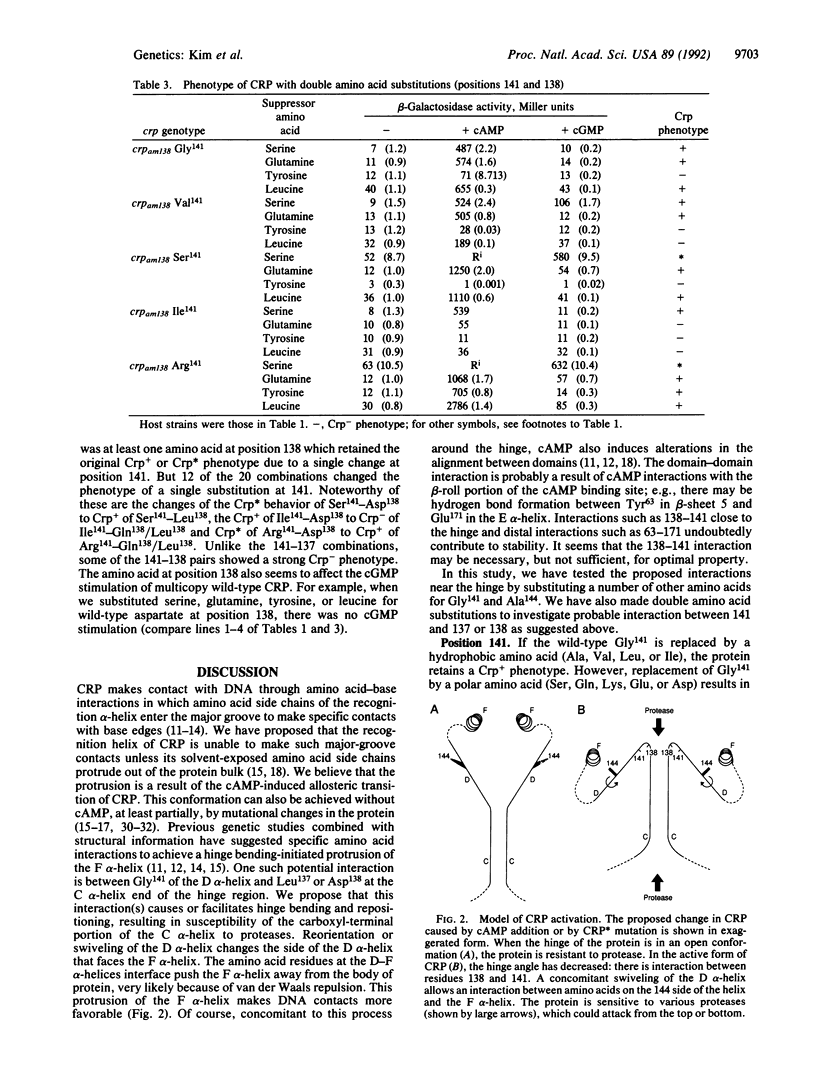
The cAMP receptor protein (CRP) of Escherichia coli is a dimer of a two-domain subunit. It requires binding of cAMP for a conformational change in order to function as a site-specific DNA-binding protein that regulates gene activity. The hinge region connecting the cAMP-binding domain to the DNA-binding domain is involved in the cAMP-induced allosteric change. We studied the structural changes in CRP that are required for gene regulation by making a large number of single and double amino acid substitutions at four different positions in or near the hinge. To achieve cAMP-independent transcription by CRP, amino acid residues 138 (located within the hinge region) and 141 (located in the D alpha-helix adjacent to the hinge) must be polar. This need for polar residues at positions 138 and 141 suggests an interaction that causes the C and D alpha-helices to come together. As a consequence, the F alpha-helix is released from the D alpha-helix and can interact with DNA. At position 144 in the D alpha-helix and within interacting distances of the F alpha-helix, replacement of alanine by an amino acid with a larger side chain, regardless of its nature, allows cAMP independence. This result indicates that pushing against the F alpha-helix may be a way of making the helix available for DNA binding. We believe that the cAMP-induced allosteric change involves similar hinge reorientation to adjust the C and D alpha-helices, allowing outward movement of the F alpha-helix.
Full text
PDF


Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Aiba H., Fujimoto S., Ozaki N. Molecular cloning and nucleotide sequencing of the gene for E. coli cAMP receptor protein. Nucleic Acids Res. 1982 Feb 25;10(4):1345–1361. doi: 10.1093/nar/10.4.1345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aiba H., Nakamura T., Mitani H., Mori H. Mutations that alter the allosteric nature of cAMP receptor protein of Escherichia coli. EMBO J. 1985 Dec 1;4(12):3329–3332. doi: 10.1002/j.1460-2075.1985.tb04084.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson W. B., Perlman R. L., Pastan I. Effect of adenosine 3',5'-monophosphate analogues on the activity of the cyclic adenosine 3',5'-monophosphate receptor in Escherichia coli. J Biol Chem. 1972 May 10;247(9):2717–2722. [PubMed] [Google Scholar]
- Bolivar F. Construction and characterization of new cloning vehicles. III. Derivatives of plasmid pBR322 carrying unique Eco RI sites for selection of Eco RI generated recombinant DNA molecules. Gene. 1978 Oct;4(2):121–136. doi: 10.1016/0378-1119(78)90025-2. [DOI] [PubMed] [Google Scholar]
- DeGrazia H., Harman J. G., Tan G. S., Wartell R. M. Investigation of the cAMP receptor protein secondary structure by Raman spectroscopy. Biochemistry. 1990 Apr 10;29(14):3557–3562. doi: 10.1021/bi00466a019. [DOI] [PubMed] [Google Scholar]
- Ebright R. H., Cossart P., Gicquel-Sanzey B., Beckwith J. Mutations that alter the DNA sequence specificity of the catabolite gene activator protein of E. coli. Nature. 1984 Sep 20;311(5983):232–235. doi: 10.1038/311232a0. [DOI] [PubMed] [Google Scholar]
- Eilen E., Krakow J. S. Cyclic AMP-mediated intersubunit disulfide crosslinking of the cyclic AMP receptor protein of Escherichia coli. J Mol Biol. 1977 Jul;114(1):47–60. doi: 10.1016/0022-2836(77)90282-0. [DOI] [PubMed] [Google Scholar]
- Garges S., Adhya S. Cyclic AMP-induced conformational change of cyclic AMP receptor protein (CRP): intragenic suppressors of cyclic AMP-independent CRP mutations. J Bacteriol. 1988 Apr;170(4):1417–1422. doi: 10.1128/jb.170.4.1417-1422.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garges S., Adhya S. Sites of allosteric shift in the structure of the cyclic AMP receptor protein. Cell. 1985 Jul;41(3):745–751. doi: 10.1016/s0092-8674(85)80055-6. [DOI] [PubMed] [Google Scholar]
- Gorini L. Informational suppression. Annu Rev Genet. 1970;4:107–134. doi: 10.1146/annurev.ge.04.120170.000543. [DOI] [PubMed] [Google Scholar]
- Harman J. G., McKenney K., Peterkofsky A. Structure-function analysis of three cAMP-independent forms of the cAMP receptor protein. J Biol Chem. 1986 Dec 15;261(35):16332–16339. [PubMed] [Google Scholar]
- Hyde C. C., Ahmed S. A., Padlan E. A., Miles E. W., Davies D. R. Three-dimensional structure of the tryptophan synthase alpha 2 beta 2 multienzyme complex from Salmonella typhimurium. J Biol Chem. 1988 Nov 25;263(33):17857–17871. [PubMed] [Google Scholar]
- Kleina L. G., Masson J. M., Normanly J., Abelson J., Miller J. H. Construction of Escherichia coli amber suppressor tRNA genes. II. Synthesis of additional tRNA genes and improvement of suppressor efficiency. J Mol Biol. 1990 Jun 20;213(4):705–717. doi: 10.1016/S0022-2836(05)80257-8. [DOI] [PubMed] [Google Scholar]
- Krakow J. S., Pastan I. Cyclic adenosine monophosphate receptor: loss of cAMP-dependent DNA binding activity after proteolysis in the presence of cyclic adenosine monophosphate. Proc Natl Acad Sci U S A. 1973 Sep;70(9):2529–2533. doi: 10.1073/pnas.70.9.2529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S. A., Murthy N. S., Krakow J. S. Ligand-induced change in the radius of gyration of cAMP receptor protein from Escherichia coli. FEBS Lett. 1980 Jan 1;109(1):121–124. doi: 10.1016/0014-5793(80)81324-x. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Luisi B. F., Sigler P. B. The stereochemistry and biochemistry of the trp repressor-operator complex. Biochim Biophys Acta. 1990 Apr 6;1048(2-3):113–126. doi: 10.1016/0167-4781(90)90047-6. [DOI] [PubMed] [Google Scholar]
- Matthews B. W. Genetic and structural analysis of the protein stability problem. Biochemistry. 1987 Nov 3;26(22):6885–6888. doi: 10.1021/bi00396a001. [DOI] [PubMed] [Google Scholar]
- McKay D. B., Weber I. T., Steitz T. A. Structure of catabolite gene activator protein at 2.9-A resolution. Incorporation of amino acid sequence and interactions with cyclic AMP. J Biol Chem. 1982 Aug 25;257(16):9518–9524. [PubMed] [Google Scholar]
- Murgola E. J., Yanofsky C. Selection for new amino acids at position 211 of the tryptophan synthetase alpha chain of Escherichia coli. J Mol Biol. 1974 Jul 15;86(4):775–784. doi: 10.1016/0022-2836(74)90353-2. [DOI] [PubMed] [Google Scholar]
- Nagata S., Hyde C. C., Miles E. W. The alpha subunit of tryptophan synthase. Evidence that aspartic acid 60 is a catalytic residue and that the double alteration of residues 175 and 211 in a second-site revertant restores the proper geometry of the substrate binding site. J Biol Chem. 1989 Apr 15;264(11):6288–6296. [PubMed] [Google Scholar]
- Normanly J., Masson J. M., Kleina L. G., Abelson J., Miller J. H. Construction of two Escherichia coli amber suppressor genes: tRNAPheCUA and tRNACysCUA. Proc Natl Acad Sci U S A. 1986 Sep;83(17):6548–6552. doi: 10.1073/pnas.83.17.6548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ren Y. L., Garges S., Adhya S., Krakow J. S. Cooperative DNA binding of heterologous proteins: evidence for contact between the cyclic AMP receptor protein and RNA polymerase. Proc Natl Acad Sci U S A. 1988 Jun;85(12):4138–4142. doi: 10.1073/pnas.85.12.4138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz S. C., Shields G. C., Steitz T. A. Crystal structure of a CAP-DNA complex: the DNA is bent by 90 degrees. Science. 1991 Aug 30;253(5023):1001–1007. doi: 10.1126/science.1653449. [DOI] [PubMed] [Google Scholar]
- Tan G. S., Kelly P., Kim J., Wartell R. M. Comparison of cAMP receptor protein (CRP) and a cAMP-independent form of CRP by Raman spectroscopy and DNA binding. Biochemistry. 1991 May 21;30(20):5076–5080. doi: 10.1021/bi00234a034. [DOI] [PubMed] [Google Scholar]
- Tokeson J. P., Garges S., Adhya S. Further inducibility of a constitutive system: ultrainduction of the gal operon. J Bacteriol. 1991 Apr;173(7):2319–2327. doi: 10.1128/jb.173.7.2319-2327.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weber I. T., Steitz T. A. Structure of a complex of catabolite gene activator protein and cyclic AMP refined at 2.5 A resolution. J Mol Biol. 1987 Nov 20;198(2):311–326. doi: 10.1016/0022-2836(87)90315-9. [DOI] [PubMed] [Google Scholar]
- Wolfenden R. V., Cullis P. M., Southgate C. C. Water, protein folding, and the genetic code. Science. 1979 Nov 2;206(4418):575–577. doi: 10.1126/science.493962. [DOI] [PubMed] [Google Scholar]
- Wu C. W., Wu F. Y. Conformational transitions of cyclic adenosine monophosphate receptor protein of Escherichia coli. A temperature-jump study. Biochemistry. 1974 Jun 4;13(12):2573–2578. doi: 10.1021/bi00709a016. [DOI] [PubMed] [Google Scholar]
- Wu F. Y., Nath K., Wu C. W. Conformational transitions of cyclic adenosine monophosphate receptor protein of Escherichia coli. A fluorescent probe study. Biochemistry. 1974 Jun 4;13(12):2567–2572. doi: 10.1021/bi00709a015. [DOI] [PubMed] [Google Scholar]
- Zimmerman J. M., Eliezer N., Simha R. The characterization of amino acid sequences in proteins by statistical methods. J Theor Biol. 1968 Nov;21(2):170–201. doi: 10.1016/0022-5193(68)90069-6. [DOI] [PubMed] [Google Scholar]
- de Crombrugghe B., Busby S., Buc H. Cyclic AMP receptor protein: role in transcription activation. Science. 1984 May 25;224(4651):831–838. doi: 10.1126/science.6372090. [DOI] [PubMed] [Google Scholar]


