Abstract
Background
The red palm weevil, Rhynchophorus ferrugineus, is a lethal pest of the palms. The identification of odorant binding protein (OBP) genes will be helpful for clarifing the mechanism of odorant detection of this pest. By sequencing the full length cDNA library of its antenne, 11 OBP genes (RferOBP1-11) were identified.
Findings
The result showed RferOBP1-7 and RferOBP8-11 belonged to the minus-C and classic family, respectively qPCR revealed that RferOBP1-10 highly transcribed in the antennae, of which RferOBP1, RferOBP4, RferOBP8 and RferOBP10 were obviously male-biased expression. RferOBP7 and RferOBP11 exhibited highly expression in female head and male thorax. RferOBP2, RferOBP5 and RferOBP6 were highly expressed in the female thorax, leg and abdomen respectively.
Conclusions
The results paved the way towards a future understanding of the olfaction in this species.
Keywords: Expression pattern, Odorant binding protein, Olfactory, Rhynchophorus ferrugineus
Background
The red palm weevil, Rhynchophorus ferrugineus, belongs to Coleoptera: Dryophthoridae family and is native to tropical Asian regions which have become a most threatening pest of palm trees worldwide (Yan et al. 2015). R. ferrugineus is a concealed tissue borer that resides inside the trunk of the palms with a highly aggregated population distribution pattern and tree injuries are often severe when it is discovered (Abraham et al. 1998). The widespread nature of this pest is due to their ability of adoption to a broader host range, to a wider variety of climates, and practice of shipping the palm trees between different territories (Murphy and Briscoe 1999). In the newly invaded areas, this pest generally causes serious damage to the trees and significant economic losses are associated by minimizing the production of palms, and increases cost management in eradication of the pest (El-Sabea et al. 2009). Due to its concealed nature and exceptional colonization capability, integrated pest management strategies play an important role in controlling the red palm weevil (Vacas et al. 2014). With respect to this, large quantities of insecticides are still applied to prevent and control R. ferrugineus infestations (Llácer et al. 2013). However, trapping systems which include aggregation pheromone, date fruits, bait traps have been effectively and environmentally used to detect and control this pest (Abuagla and Al-Deeb 2012). New management strategies from the aspect of chemical ecology should be developed by understanding the R. ferrugineus olfactory system, which will be efficient in reducing the weevil populations and the usage of pesticides. However, most of the olfactory genes have been deciphered by transcriptome sequencing are fragmental and their expression patterns are not well known (Yan et al. 2015; Antony et al. 2016). Thus, the information regarding the molecular mechanisms underlying the olfaction in this species is scarce.
Odorant-binding proteins (OBPs) are hydrophilic soluble proteins which are composed of approximately 130 amino acids and typically contain six conserved cysteine residues (Leal et al. 1999) which are secreted by the accessory cells around the olfactory neurons and are accumulated in the sensillar lymph (Klein 1987). OBPs are essential for insect olfaction which acts by transporting hydrophobic compounds through aqueous sensillar lymph to the receptors embedded on dendritic membranes of olfactory receptor neurons from the external environment and is thought to be the first step in the recognition of chemical signals (Krieger and Breer 1999; Leal and Leal 2015). Hence, it is necessary to understand the mechanism of olfaction of R. ferrugineus OBPs which has an important role. Hence, in our study we reported the identification of 11 OBP genes from the antennae of this pest and analyzed their tissue expression patterns.
Methods
Insects and sample collection
Rhynchophorus ferrugineus was collected from cocoons but was originally collected in naturally infested palms in the suburbs of Hainan province. The lab colony was maintained as described in the study by Yan et al. (2014). The antennae, heads (without antenna), thoraxes, abdomens, and legs of male and female were collected separately and frozen immediately in liquid nitrogen.
cDNA library construction and sequencing
Total RNA was extracted from approximately 100 antennae of both sexes after 5 days of emergence by Trizol regent. The quantity and quality of the total RNA was validated using spectrophotometer. 5 μg of total RNA was subjected to construct full length cDNA library using Creator SMART cDNA Library Construction Kit (Clontech, Mountain View, CA, USA) according to the manufacturer’s instructions. During the library construction, 400–1200 bp fragments were selected. About 1000 clones were randomly sequenced from 5′-end of the gene. After BlastX annotation, the genes were assigned to the OBP family and were completely sequenced to obtain their full length cDNA. The signal peptide of the protein sequences were predicted using SignalP 4.0 server version (Petersen et al. 2011). Sequence alignments were performed using the program ClustalX (Thompson et al. 1997). Phylogenetic trees were constructed by MEGA version 5 based on the neighbor-joining algorithm method at bootstrap 1000 (Tamura et al. 2011).
qPCR analysis
Total RNA from different tissues were extracted as mentioned above. After DNase treatment, the cDNA was synthesized from 1 μg total RNA using a GoScript Reverse Transcription System (Promega, USA). qPCR was performed using gene-specific primers (Table 1) by SYBR Premix EX Taq™ (TaKaRa, Dalian, Liaoning, China) in three biological and technical replicates with different samples. PCR reactions were performed in a 3-step amplification process at 94 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. Expression levels of the genes were calculated relative to the control gene 18S RNA expression using the method (Livak and Schmittgen 2001). The total expression of one gene in all the tissues was set as 100 % and the percentage of one gene in each tissue was used to measure the expression level.
Table 1.
qPCR primers
| Primer name | Sequence (5′ → 3′) | |
|---|---|---|
| RferOBP1 | Reverse | TCCTCGCCCAACATTAC |
| Forward | TTTGACCGCCTCCTTTA | |
| RferOBP2 | Reverse | TAGTCCAAGCGGATCTCACA |
| Forward | CGTAGCACCAGTTTCCTC | |
| RferOBP3 | Reverse | TTTTCAGCGACTCACCA |
| Forward | GACATTTATCTAGCATAGCG | |
| RferOBP4 | Reverse | TGGAGAACTCACCGACTC |
| Forward | CGAACAACATAATCCCTTT | |
| RferOBP5 | Reverse | CTGAACAACGCCAGAGG |
| Forward | TCATTCCCAAACATACCA | |
| RferOBP6 | Reverse | CTGAACAACGCCAGAGG |
| Forward | TCATTCCCAAACATACCA | |
| RferOBP7 | Reverse | TGGTGTCGGCCATCTCA |
| Forward | CTTCGCCCTCGTCGTTT | |
| RferOBP8 | Reverse | TGATGGTATGTGGGACTT |
| Forward | ATGGTGGAGCCTGAGTT | |
| RferOBP9 | Reverse | AGGCGACTGGGAGGTAG |
| Forward | TGTGCGTCTGCGGATTT | |
| RferOBP10 | Reverse | GGTACTCCTCGCTGTTT |
| Forward | ATCCATAGATCCCGTTT | |
| RferOBP11 | Reverse | AACAGGAGCAACAGAAGAT |
| Forward | ATTACTGGCGGTAGGGT | |
Results and discussion
Characteristics of OBP sequences
In total, 11 full length OBP genes from 36 clones named as RferOBP1-11 were identified from the antennal cDNA library sequencing, which have been deposited in the NCBI under the accession numbers KR780571 to KR780581 (Table 2). Compared to the 49 OBPs identified in the genome of Tribolium castaneum, more OBP genes might be expected to be identified in R. ferrugineus by massive sequencing strategies and by using more different tissues that may also express OBPs (Tribolium Genome Sequencing Consortium 2008; Zhu et al. 2013). The cDNA length ranged from 497 bp to 579 bp. The open reading frame (ORF) ranged from 396 to 441 bp, which encoded approximately 130 amino acid residues, with a predicted molecular weight of about 15 kDa. Except for RferOBP1, 8 and 9, the predicted pH of others were acidic. The amino acid sequences identity of RferOBP5 and RferOBP6 was 99 %, while their DNA sequence homology was only 62 % (Fig. 1). RferOBP5 and RferOBP6 might be the result of gene duplication events. Similar to the characteristics of OBP with a rather low level of sequence similarity (Forêt and Maleszka 2006; Zhu et al. 2012), RferOBP1-4 and RferOBP7-11 shared 8–65 % amino acid residual identities with each other. In addition, they contained the characteristic sequence features for these proteins, such as conserved cysteine residues and signal peptide. All identified OBPs were used to construct a phylogenetic tree clustering with OBPs from T. castaneum (Fig. 2), which was the only species with known genomic sequence from Coleoptera and were distributed into all three groups. The phylogenetic tree had relatively large minus-C and classic OBP, and one plus-C OBP branches. Minus-C OBPs lack the second and fifth cysteine residues, and plus-C OBPs contained three additional conserved cysteines and one conserved proline (Xu et al. 2009). Four (RferOBP8-11) of the candidate sequences represented classic OBPs, each containing the six conserved cysteine residues. RferOBP1-7 showed minus-C OBPs, and there was no plus-C OBP. Of the 12 OBPs, RferOBP2-6 were clustered with each other together, rather than those from other insect species suggesting that they may be the products of recent gene duplications (Li et al. 2015a, b). These OBPs from different clades may have different functions, which are interesting to be determined.
Table 2.
Characteristics of Rhynchophorus ferrugineus OBP genes
| Gene name | Nucleotide length (bp) | ORF length (bp) | Complete protein (aa) | Signal peptide (aa) | Molecular weight (kDa) | Isoelectric point | Accession number |
|---|---|---|---|---|---|---|---|
| RferOBP1 | 556 | 405 | 135 | 19 | 15.27 | 8.43 | KR780571 |
| RferOBP2 | 569 | 396 | 132 | 18 | 15.18 | 4.42 | KR780572 |
| RferOBP3 | 568 | 402 | 134 | 20 | 15.03 | 5.25 | KR780573 |
| RferOBP4 | 497 | 396 | 132 | 18 | 14.86 | 5.02 | KR780574 |
| RferOBP5 | 509 | 399 | 133 | 17 | 15.21 | 4.43 | KR780575 |
| RferOBP6 | 533 | 399 | 133 | 17 | 15.16 | 4.43 | KR780576 |
| RferOBP7 | 580 | 441 | 147 | 19 | 16.68 | 6.59 | KR780577 |
| RferOBP8 | 556 | 432 | 144 | 18 | 16.16 | 7.01 | KR780578 |
| RferOBP9 | 579 | 432 | 144 | 21 | 15.56 | 7.78 | KR780579 |
| RferOBP10 | 570 | 429 | 143 | 19 | 15.82 | 4.6 | KR780580 |
| RferOBP11 | 571 | 402 | 134 | 18 | 14.96 | 4.29 | KR780581 |
Fig. 1.
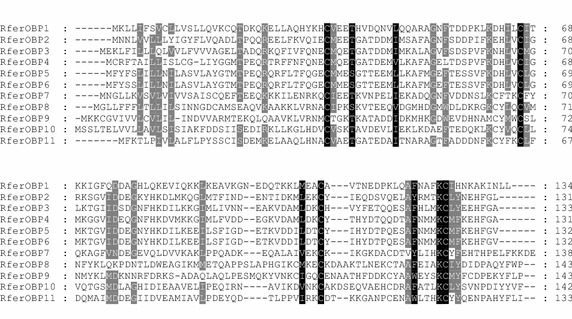
Alignment of the amino acid sequences of Rhynchophorus ferrugineus OBPs
Fig. 2.
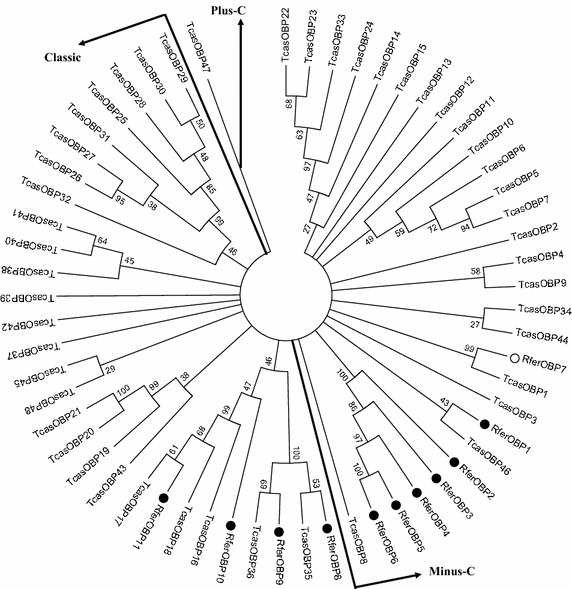
Neighbor-joining tree of OBP proteins from Rhynchophorus ferrugineus and Tribolium castaneum. The phylogenetic tree was presented with 25 % cut-off bootstrap value
Tissue expression pattern
To uncover the tissue expression pattern, we used qPCR to assess the transcriptional abundance of the identified 11 OBP genes which showed varying degrees of expression in the antennae, head (without antenna), thorax, abdomen, and leg (Table 3). Except for RferOBP11 with abundant expression in female head and male thorax, RferOBP1-10 showed a relatively higher expression in the antennae suggesting that the OBP genes identified in current study may play an important role in the olfaction (Leal 2013). Of them, RferOBP1, RferOBP4, RferOBP8 and RferOBP10 were obviously male biased which may play the same role as pheromone binding proteins and could play a role in odorant perception of certain plant volatiles (Zhang et al. 2015). RferOBP3 was highly expressed in the legs with near identical expression with the antennae. With respect to the other genes, RferOBP2, RferOBP5 and RferOBP6 were highly expressed in the female thorax, leg and abdomen, respectively. As head, thorax and leg have taste sensilla and other olfactory sensilla, OBP expressed in these tissues may function in the perception of non-volatile host chemicals, gustatory reorganization and other olfactory function (Mitaka et al. 2011). These results were similar to the previous studies which showed that OBPs of some insect are expressed primarily or exclusively in non-antennae tissues or in larvae, which may have physiological functions independent of olfaction (Li et al. 2015a, b). This also reflects that the identification of these OBPs solely depend on the structural similarities and not function. Nonetheless, why OBPs are expressed in abdomen remains a mystery. RferOBP7 exhibited similar expression patterns with that of RferOBP11 and was expressed at a very high level in the female head and male thorax. The expression patterns make an important contribution to our understanding of OBPs in R. ferrugineus and might facilitate for their future functional characterization.
Table 3.
Tissue expression pattern of Rhynchophorus ferrugineus OBP genes
| Gene | Female | Male | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Anntena | Head | Thorax | Abdomen | Leg | Anntena | Head | Thorax | Abdomen | Leg | |
| RferOBP1 | 33.21 ± 1.01 | 2.66 ± 0.22 | 0.45 ± 0.03 | 0.24 ± 0.04 | 0.16 ± 0.01 | 61.54 ± 1.23 | 0.07 ± 0.01 | 1.12 ± 0.02 | 0.39 ± 0.04 | 0.17 ± 0.01 |
| RferOBP2 | 18.06 ± 1.54 | 9.88 ± 0.26 | 23.67 ± 0.01 | 0.93 ± 0.00 | 17.17 ± 0.01 | 11.04 ± 1.68 | 0.71 ± 0.03 | 4.24 ± 0.12 | 1.01 ± 0.01 | 13.28 ± 0.02 |
| RferOBP3 | 29.79 ± 0.07 | 5.83 ± 0.44 | 0.20 ± 1.11 | 1.05 ± 0.16 | 4.96 ± 0.79 | 29.53 ± 0.75 | 1.74 ± 0.04 | 0.02 ± 0.22 | 0.33 ± 0.12 | 26.55 ± 0.40 |
| RferOBP4 | 24.91 ± 1.15 | 5.11 ± 0.82 | 4.07 ± 0.01 | 0.50 ± 0.13 | 10.45 ± 0.22 | 39.00 ± 2.14 | 1.11 ± 0.09 | 0.90 ± 0.00 | 0.92 ± 0.03 | 13.04 ± 0.58 |
| RferOBP5 | 15.05 ± 0.54 | 1.07 ± 0.21 | 5.65 ± 0.29 | 15.94 ± 0.05 | 35.88 ± 0.47 | 12.86 ± 1.28 | 0.40 ± 0.17 | 1.06 ± 0.02 | 0.47 ± 0.13 | 11.62 ± 1.28 |
| RferOBP6 | 15.30 ± 1.94 | 1.47 ± 0.08 | 1.03 ± 0.29 | 27.38 ± 1.87 | 15.27 ± 2.67 | 22.20 ± 1.10 | 0.04 ± 0.04 | 2.99 ± 0.04 | 2.48 ± 0.04 | 11.84 ± 1.09 |
| RferOBP7 | 12.30 ± 1.33 | 28.06 ± 0.08 | 2.38 ± 0.07 | 0.91 ± 4.53 | 1.78 ± 0.32 | 9.05 ± 0.98 | 1.31 ± 0.01 | 28.85 ± 0.35 | 8.87 ± 0.16 | 6.50 ± 1.79 |
| RferOBP8 | 30.21 ± 0.71 | 2.18 ± 1.18 | 0.39 ± 0.16 | 0.16 ± 0.12 | 0.09 ± 0.16 | 65.51 ± 1.23 | 0.05 ± 0.18 | 0.83 ± 2.40 | 0.45 ± 0.50 | 0.13 ± 0.93 |
| RferOBP9 | 30.92 ± 1.37 | 17.68 ± 0.13 | 0.78 ± 0.04 | 3.20 ± 0.02 | 4.73 ± 0.01 | 23.60 ± 1.35 | 0.83 ± 0.01 | 5.44 ± 0.04 | 8.83 ± 0.04 | 4.00 ± 0.00 |
| RferOBP10 | 32.84 ± 0.24 | 10.23 ± 1.17 | 0.03 ± 0.04 | 0.58 ± 0.20 | 3.64 ± 0.66 | 43.84 ± 1.11 | 1.51 ± 0.18 | 0.22 ± 0.86 | 4.08 ± 0.85 | 3.03 ± 0.33 |
| RferOBP11 | 1.17 ± 0.09 | 48.84 ± 2.22 | 3.60 ± 0.03 | 2.92 ± 0.18 | 0.52 ± 0.02 | 3.48 ± 0.11 | 1.99 ± 0.18 | 36.33 ± 2.14 | 0.30 ± 0.03 | 0.85 ± 0.13 |
Conclusion
The work presented here brings an identification of 11 OBP genes from red palm weevil by sequencing the antennal full length cDNA library. In particular, the expression profile studies provided a clear map of these genes, which may further facilitate other functional studies on these genes. These data enables a basis to reveal the molecular mechanisms of olfactory functions in the red palm weevil.
Authors’ contributions
Conceived and designed the experiments: WY, LL and WQQ. Performed the experiments: YQL and NH. Analyzed the data: WY and LL. Contributed reagents/materials/analysis tools: WY, XZM and MI. Helped and advised preparations of the manuscript: YQL, XZM and NH. Wrote the paper: WY, LL and WQQ. All authors read and approved the final manuscript.
Acknowledgements
This work was supported by the Key Program for Science and Technology and the Major Projects of Hainan, China (ZDZX2013008), and the Scientific Research Project of CATAS (1630012015023).
Competing interests
The authors declare that they have no competing interests.
Contributor Information
Wei Yan, Email: andy206@126.com.
Li Liu, Email: liulicatas@126.com.
Weiquan Qin, Email: qwq268@126.com.
Youqing Luo, Email: youqingluo@126.com.
Xuezhong Ma, Email: maxz@cafunds.net.
Nabil Haider, Email: NMHAIDER@dm.gov.ae.
Muhanad Inayeh, Email: agriclub@outlook.com.
References
- Abraham VA, Al Shuaibi MA, Faleiro JR, Abozuhairah RA, Vidyasagar PSPV. An integrated management approach for red palm weevil, Rhynchophorus ferrugineus Oliv.—a key pest of date palm in the Middle East. Sultan Qaboos Univ J Sci Res Agric Sci. 1998;3:77–83. [Google Scholar]
- Abuagla AM, Al-Deeb MA. Effect of bait quantity and trap color on the trapping efficacy of the pheromone trap for the red palm weevil, Rhynchophorus ferrugineus. J Insect Sci. 2012;12:120. doi: 10.1673/031.012.12002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Antony B, Soffan A, Jakše J, Abdelazim MM, Aldosari SA, Aldawood AS, Pain A. Identification of the genes involved in odorant reception and detection in the palm weevil Rhynchophorus ferrugineus, an important quarantine pest, by antennal transcriptome analysis. BMC Genomics. 2016;17:69. doi: 10.1186/s12864-016-2362-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Sabea AMR, Faleiro JR, Abo-El-Saad MM. The threat of red palm weevil Rhynchophorus ferrugineus to date plantations of the Gulf region in the Middle-East: an economic perspective. Outlooks Pest Manag. 2009;20:131–134. doi: 10.1564/20jun11. [DOI] [Google Scholar]
- Forêt S, Maleszka R. Function and evolution of a gene family encoding odorant binding-like proteins in a social insect, the honey bee (Apis mellifera) Genome Res. 2006;16(11):1404–1413. doi: 10.1101/gr.5075706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein U. Sensillum-lymph proteins from antennal olfactory hairs of the moth Antheraea polyphemus (Saturniidae) Insect Biochem. 1987;17:1193–1204. doi: 10.1016/0020-1790(87)90093-X. [DOI] [Google Scholar]
- Krieger J, Breer H. Olfactory reception in invertebrates. Science. 1999;286:720–723. doi: 10.1126/science.286.5440.720. [DOI] [PubMed] [Google Scholar]
- Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- Leal GM, Leal WS. Binding of a fluorescence reporter and a ligand to an odorant-binding protein of the yellow fever mosquito, Aedes aegypti. F1000Research. 2015;3:305. doi: 10.12688/f1000research.5879.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leal WS, Nikonova L, Peng G. Disulfide structure of the pheromone binding protein from the silkworm moth, Bombyx mori. FEBS Lett. 1999;464:85–90. doi: 10.1016/S0014-5793(99)01683-X. [DOI] [PubMed] [Google Scholar]
- Li X, Ju Q, Jie W, Li F, Jiang X, Hu J, Qu M. Chemosensory gene families in adult antennae of Anomala corpulenta Motschulsky (Coleoptera: Scarabaeidae: Rutelinae) PLoS One. 2015;10(4):e0121504. doi: 10.1371/journal.pone.0121504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XM, Zhu XY, Wang ZQ, Wang Y, He P, Chen G, Sun L, Deng DG, Zhang YN. Candidate chemosensory genes identified in Colaphellus bowringi by antennal transcriptome analysis. BMC Genomics. 2015;16:1028. doi: 10.1186/s12864-015-2236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Llácer E, Santiago-Álvarez C, Jacas JA. Could sterile males be used to vector a microbiological control agent? The case of Rhynchophorus ferrugineus and Beauveria bassiana. Bull Entomol Res. 2013;103(2):241–250. doi: 10.1017/S0007485312000582. [DOI] [PubMed] [Google Scholar]
- Mitaka H, Matsuo T, Miura N, Ishikawa Y. Identification of odorant-binding protein genes from antennal expressed sequence tags of the onion fly, Delia antiqua. Mol Biol Rep. 2011;38(3):1787–1792. doi: 10.1007/s11033-010-0293-x. [DOI] [PubMed] [Google Scholar]
- Murphy ST, Briscoe BR. The red palm weevil as an alien invasive: biology and the prospects for biological control as a component of IPM. Biocontrol. 1999;20:35–45. [Google Scholar]
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods. 2011;8(10):785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28:2731–2739. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG. The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tribolium Genome Sequencing Consortium The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452(7190):949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- Vacas S, Abad-Payá M, Primo J, Navarro-Llopis V. Identification of pheromone synergists for Rhynchophorus ferrugineus trapping systems from Phoenix canariensis palm volatiles. J Agric Food Chem. 2014;62:6053–6064. doi: 10.1021/jf502663y. [DOI] [PubMed] [Google Scholar]
- Xu YL, He P, Zhang L, Fang SQ, Dong SL, Zhang YJ, Li F. Large-scale identification of odorant-binding proteins and chemosensory proteins from expressed sequence tags in insects. BMC Genom. 2009;10:632. doi: 10.1186/1471-2164-10-632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan W, Li L, Li CX, Huang SC, Liu L, Qin WQ, Peng ZQ, Luo YQ. An artificial diet for the red palm weevil, Rhynchophorus ferrugineus Oliver. Chin J Appl Entomol. 2014;51(5):1387–1392. [Google Scholar]
- Yan W, Liu L, Qin WQ, Li CX, Peng ZQ. Transcriptomic identification of chemoreceptor genes in the red palm weevil Rhynchophorus ferrugineus. Genet Mol Res. 2015;14(3):7469–7480. doi: 10.4238/2015.July.3.23. [DOI] [PubMed] [Google Scholar]
- Zhang J, Walker WB, Wang G. Pheromone reception in moths: from molecules to behaviors. Prog Mol Biol Transl Sci. 2015;130:109–128. doi: 10.1016/bs.pmbts.2014.11.005. [DOI] [PubMed] [Google Scholar]
- Zhu JY, Zhao N, Yang B (2012) Global transcriptional analysis of olfactory genes in the head of pine shoot beetle, Tomicus yunnanensis. Comp Funct Genom. Article ID 491748 [DOI] [PMC free article] [PubMed]
- Zhu JY, Zhang LF, Ze SZ, Wang DW, Yang B. Identification and tissue distribution of odorant binding protein genes in the beet armyworm, Spodoptera exigua. J Insect Physiol. 2013;59(7):722–728. doi: 10.1016/j.jinsphys.2013.02.011. [DOI] [PubMed] [Google Scholar]


