Abstract
Bacterial lipoproteins (Lpp) represent a major class of membrane proteins. They are distinguished by a lipid moiety at the N-terminus by which they are anchored either in the outer leaflet of the cytoplasmic membrane or, in Gram-negative bacteria, also in the inner leaflet of the outer membrane. In Gram-positive bacteria Lpp significantly contribute to nutrient transport, Toll-like receptor 2 activation and pathogenicity. Here we examine the Lpp of Staphylococcus aureus USA300, as a prototype for a multiple antibiotic resistant and community-acquired pathogen that is rapidly spreading worldwide. The compiled Lpp were grouped according to the postulated function and dissemination of homologs in the genus Staphylococcus and beyond. Based on this evaluation we also point out Lpp as promising vaccine candidates.
Keywords: Staphylococcus, S. aureus USA300, lipoprotein, lipoprotein functions, lipoprotein dissemination, ion transporters, pathogenicity
Introduction
Bacterial lipoproteins (Lpp) are a distinctive class of membrane-anchored proteins. They contain a N-terminal lipid modification, the N-acyl-S-diacyl-glyceryl-cysteine (Hantke and Braun, 1973). There are three cytoplasmic membrane localized enzymes involved in the biogenesis of Lpp: the phosphatidylglycerol-prolipoprotein diacylglyceryl transferase (Lgt) (Sankaran and Wu, 1994), the specific signal peptidase II (Lsp) that recognizes the diacylglyceryl modification and cleaves between the amino acid at position −1 and the lipid-modified cysteine residue (Hussain et al., 1982), and finally the N-acyltransferase (Lnt) to form N-acyl diacylglyceryl cysteine (Gan et al., 1995). This maturation of Lpp is uniform in Gram-negative and—positive bacteria. In Gram-positive bacteria Lpp are anchored in the outer leaflet of the cytoplasmic membrane and may extend into the cell wall and beyond. Although their anchoring at the cell envelope is completely different from the covalently cell wall anchored proteins mediated by the sortase (Mazmanian et al., 2001), there is an overlap which have both in common, namely, the binding to external components, such as nutrients and host proteins. Particularly this function is affected when the maturation of pre-Lpp by the Lgt and Lsp does not take place.
The crucial role of maturation of pre-Lpp by the Lgt and Lsp for virulence and TLR2 signaling has been reviewed recently (Nguyen and Götz, 2016). For example the Δlgt mutants of various S. aureus strains were severely affected in immune stimulation and pathogenicity (Stoll et al., 2005; Schmaler et al., 2009); and the Δlgt mutants were also affected in iron acquisition under infectious conditions (Schmaler et al., 2009, 2010). In many other Gram-positive bacteria like Mycobacterium tuberculosis (Sander et al., 2004), Streptococcus pneumoniae (Petit et al., 2001; Khandavilli et al., 2008), Streptococcus agalactiae (Henneke et al., 2008; Bray et al., 2009), Streptococcus pyogenes, and Streptococcus equi (Sutcliffe and Harrington, 2002; Hamilton et al., 2006; Weston et al., 2009; Sutcliffe et al., 2012) or Listeria monocytogenes (Baumgärtner et al., 2007; Machata et al., 2008) deletion of the lgt or the lsp gene likewise impaired growth and pathogenicity.
By screening the S. aureus N315 genome with the new Lpp search program ParSeq (Schmollinger et al., 2004), more than 70 putative Lpp were identified, but only 55 contained a signal peptide with the right length (Stoll et al., 2005). 35 of the Lpp could be annotated as transporters for iron, zinc, amino acid, oligopeptide, glycine betaine, sugar, and teichoic acid, other had enzymatic functions such heme/copper-type cytochrome/quinol oxidase, protein-disulfide isomerase, peptidyl-prolyl cis/trans isomerase (PrsA), or pyruvate-format-lyase-activating enzyme. In this report it has also been shown that SitC was one of the most abundant Lpp and that in a lgt mutant only 20–25% retained in the membrane, while the majority was released into the supernatant (Stoll et al., 2005). In the very detailed review by Sibbald et al. 43 core and a similar amount of variant Lpp were analyzed and they also mention that the translocation pathway is mostly Sec- but in some cases also Tat-mediated (Sibbald et al., 2006).
In the meantime the knowledge as to the function of Lpp in S. aureus has increased, justifying a reevaluation of the data. We carefully analyzed the Lpp in strain USA300 as an epidemic prototype and traced the distribution of each Lpp homolog in the S. aureus species, the Staphylococcus genus and beyond. This allows us to unravel housekeeping and virulence associated Lpp. It turned out that particularly pathogenic strains have a number of additional Lpp serving as transporters for nutrients and contributing to virulence and fitness.
Materials and methods
Bioinformatic study
We collected all the predicted Lpp generated by different programs, for example Hidden Markov Model (Bagos et al., 2008), LipoP (Juncker et al., 2003), G+LPP (Sutcliffe and Harrington, 2002), G+LPPv2 (Rahman et al., 2008), LIPPREP (Taylor et al., 2006), Dolop (Babu et al., 2006), and Von Heijne (von Heijne, 1989). These lipoproteins were screened carefully by combination of the different values including hydrophobic plot (http://gcat.davidson.edu/DGPB/kd/kyte-doolittle.htm), lipobox sequence (http://www.mrc-lmb.cam.ac.uk/genomes/dolop/lipobox.shtml) and the cleaving site by PREP-LIPO (http://bioinformatics.biol.uoa.gr/PRED-LIPO/) or LipoP (http://www.cbs.dtu.dk/services/LipoP/). All Lpp sequences were blasted with other staphylococcal species and other genera by following programs NCBI protein blast (http://blast.ncbi.nlm.nih.gov/Blast.cgi?PAGE=Proteins) or KEGG (http://www.genome.jp/kegg/ko.html).
Results and discussion
Evaluation of lipoproteins (Lpp) in Staphylococcus aureus USA300
Here, we re-evaluated the Lpp of a pathogenic S. aureus strain and categorized the Lpp according to the function and dissemination. Methicillin-resistant S. aureus USA300 was chosen as a model strain as it is a major source of community-acquired infections almost worldwide (Diep et al., 2006). The common Lpp prediction programs yielded different numbers of proposed Lpp in the 2560 genes of USA300 (http://biophysics.biol.uoa.gr/PRED-LIPO-results/): DOLOP (52), G+LPP (54), von heijne (66), PS51257 (68), PS00013 (65) and LipoP (68) and PRED-LIPO (63). Because of this heterogeneity each proposed Lpp was examined for the presence of an unambiguous Lpp signal peptide (LSP), which must fulfill three criteria: length between 16 and 40 amino acids, presence of a hydrophobic domain, followed by the lipobox. Based on these criteria we propose 67 Lpp in USA300, which is 2.57% of all genes (Table 1). Three of the identified Lpp in USA300 (no 5, 65, and 64 of Table 1) were incorrectly annotated by using the wrong start codon; these Lpp were also recently detected in the supernatant of S. aureus Newman by proteomic analysis (Vu et al., 2016). The size of the Lpp ranged from 6 to 89 kDa, however, the average size was between 30 and 50 kDa.
Table 1.
Lpp of S. aureus USA300.
| No | Locus tag | Function/Annotation | PFAM | SP (aa) | Lipobox | Mass (KDa) | Disseminationa | References |
|---|---|---|---|---|---|---|---|---|
| Fe transport | ||||||||
| 01 | SAUSA300_1978 | Ferric hydroxamate receptor/FhuD1 | Peripla_BP_2 | 17 | LTA C | 34 | 15* | Sebulsky and Heinrichs, 2001; Sebulsky et al., 2004 |
| 02 | SAUSA300_2235 | Fe ABC transporter/FhuD2 | Peripla_BP_2, ABC2_membrane_3 | 17 | LAA C | 34 | 16 | Sebulsky and Heinrichs, 2001; Sebulsky et al., 2004; Mariotti et al., 2013 |
| 03 | SAUSA300_0721 | Transferrin receptor/SstD | Peripla_BP_2 | 18 | LAA C | 38 | 14 | Morrissey et al., 2000 |
| 04 | SAUSA300_0117 | Fe ABC transporter/SirA | Peripla_BP_2 | 20 | LAG C | 37 | 10 | Heinrichs et al., 1999 |
| 05 | SAUSA300_1032 | Fe ABC transporter/IsdE | Peripla_BP_2 | 19 | LTS C | 32 | 10 | Mazmanian et al., 2002, 2003; Grigg et al., 2007 |
| 06 | SAUSA300_0344 | FepA, Fe-binding protein, part of fepABC and tat-AC cluster | Peptidase_M75 | 17 | IAA C | 32 | 10 | Biswas et al., 2009 |
| 07 | SAUSA300_2136 | Fe ABC transporter | Peripla_BP_2 | 21 | VAA C | 36 | 14 | |
| 08 | SAUSA300_0219 | Iron Binding Protein | SBP_bac_1, 6, 8, 11 | 17 | LSA C | 36 | 4* | |
| Other cation transport | ||||||||
| 09 | SAUSA300_0618 | Manganese-binding protein MntC (SitC) | ZnuA, Nit_Regul_Hom | 17 | VAA C | 34 | 19 | Cockayne et al., 1998; Müller et al., 2010; Diep et al., 2014 |
| 10 | SAUSA300_2351 | Zinc-binding, adcA-like | ZnuA, ZinT | 20 | LAA C | 57 | 10 | |
| 11 | SAUSA300_2411 | Cobalt and nickel transporter Cnt (Opp1A) | SBP_bac_5 | 20 | LTG C | 59 | 10 | Remy et al., 2013 |
| 12 | SAUSA300_0231 | Nickel ABC transporter | SBP_bac_5 | 18 | LSG C | 55 | 10* | |
| 13 | SAUSA300_0203 | Nickel-Peptide/transporter substrate-binding protein | SBP_bac_5 | 18 | LSG C | 66 | + | |
| 14 | SAUSA300_2230 | Molybdenum ABC transporter (ModA) | SBP_bac_11, 1, PBP_like_2 | 19 | LAG C | 29 | 15 | Neubauer et al., 1999 |
| Anion transport | ||||||||
| 15 | SAUSA300_1283 | Phosphate ABC transporter | PBP_like_2, PBP_like | 20 | LGA C | 36 | 15 | |
| 16 | SAUSA300_0145 | Phosphonate ABC transporter | Phosphonate-bd, SBP_bac_3 | 20 | AAA C | 35 | 10 | |
| 17 | SAUSA300_0175 | Nitrate ABC transporter substrate-binding protein | NMT1_2, | 17 | ITG C | 36 | 4 | |
| AA and Peptide transport | ||||||||
| 18 | SAUSA300_2391 | Glycine betaine /carnitine/ choline ABC transporter (OpuCc) | OpuAC | 20 | LSG C | 37 | 19 | |
| 19 | SAUSA300_2359 | Amino acid ABC transporter | SBP_bac_3 | 17 | LAA C | 13 | 12 | |
| 20 | SAUSA300_0073 | Peptide ABC transporter | SBP_bac_5 | 19 | LAG C | 57 | 11 | |
| 21 | SAUSA300_0891 | Oligopeptide ABC transporter (Opp3A) | SBP_bac_5 | 20 | LSG C | 61 | 11 | Hiron et al., 2007 |
| 22 | SAUSA300_0892 | Oligopeptide ABC transporter (Opp4A) | SBP_bac_5 | 20 | LSA C | 63 | 5 | Hiron et al., 2007 |
| 23 | SAUSA300_0437 | NLPA/ D-Methionine binding (GmpC) | Lipoprotein_9 (NLPA) | 17 | LAA C | 31 | 9 | Williams et al., 2004 |
| 24 | SAUSA300_0798 | D-Methionine ABC transporter | OpuAC, Lipoprotein_9 | 19 | LAA C | 30 | 15 | |
| Sugar transport | ||||||||
| 25 | SAUSA300_0209 | Maltose ABC transporter | SBP_bac_1, 8 | 20 | VTA C | 47 | 6 | |
| Miscellaneous functions | ||||||||
| Biosynthesis | ||||||||
| 26 | SAUSA300_1884 | CamS sex pheromone biosynthesis | CamS | 17 | LAA C | 44 | 14 | |
| Respiration | ||||||||
| 27 | SAUSA300_0963 | Quinol oxidase, subunit II (QoxA) | COX2 | 19 | LSG C | 41 | 21 | |
| 28 | SAUSA300_0693 | Electron transfer domain/SaeP | CfAFP, DM13 | 20 | LGA C | 16 | 22 | Makgotlho et al., 2013 |
| Chaperone-Foldases | ||||||||
| 29 | SAUSA300_1790 | Foldase protein PrsA | Rotamase, | 20 | LGA C | 36 | 15 | Heikkinen et al., 2009; Jousselin et al., 2012 |
| 30 | SAUSA300_2354 | Thioredoxin/Protein disulfide-isomerase | Thioredoxin_2, 4, 5 | 18 | LTA C | 22 | 15* | |
| Protein translocation | ||||||||
| 31 | SAUSA300_2046 | YidC (OxaA)–essential protein | OATP, 60KD_IMP | 19 | LAG C | 32 | 25 | |
| Phage and plasmid encoded Lpp | ||||||||
| 32 | SAUSA300_1436 | PhiSLT ORF144-like | DUF1510, Zip, Presenilin | 17 | LTA C | 16 | 2* | |
| 33 | pUSA300_HOUMR0011 | Membrane bound penicillinase BlaZ | 16 | LSA C | 31 | 11 | Nielsen and Lampen, 1982b | |
| Lpl cluster | ||||||||
| 34 | SAUSA300_0410 | Lpl-1 νSaα specific | DUF576 | 32 | IAG C | 30 | + | Nguyen et al., 2015b |
| 35 | SAUSA300_0411 | Lpl-2 νSaα specific | DUF576 | 22 | IIG C | 30 | + | Nguyen et al., 2015b |
| 36 | SAUSA300_0413 | Lpl-3 νSaα specific | DUF576 | 23 | IIG C | 30 | + | Nguyen et al., 2015b |
| 37 | SAUSA300_0414 | Lpl-4 νSaα specific | DUF576 | 22 | VTS C | 28 | + | Nguyen et al., 2015b |
| 38 | SAUSA300_0415 | Lpl-5 νSaα specific | DUF576 | 22 | IMG C | 29 | + | Nguyen et al., 2015b |
| 39 | SAUSA300_0416 | Lpl-6 νSaα specific | DUF576 | 20 | MAG C | 29 | + | Nguyen et al., 2015b |
| 40 | SAUSA300_0417 | Lpl-7 νSaα specific | DUF576 | 23 | IVG C | 30 | + | Nguyen et al., 2015b |
| 41 | SAUSA300_0418 | Lpl-8 νSaα specific | DUF576 | 22 | VTS C | 29 | + | Nguyen et al., 2015b |
| 42 | SAUSA300_0419 | Lpl-9 νSaα specific | DUF576 | 22 | IGG C | 30 | + | Nguyen et al., 2015b |
| 43 | SAUSA300_2429 | Tandem lpp | DUF576 | 22 | IGG C | 16 | 3* | |
| 44 | SAUSA300_2430 | Tandem lpp | DUF576 | 23 | IGA C | 29 | + | |
| 45 | SAUSA300_0100 | Tandem lpp/Conserved staphylococcal antigen 1A (Csa1A) | DUF576 | 24 | TAG C | 28 | + | Schluepen et al., 2013 |
| 46 | SAUSA300_0101 | Tandem lpp | DUF576 | 24 | TAG C | 28 | + | |
| 47 | SAUSA300_0102 | Tandem lpp | DUF576 | 24 | TAG C | 28 | + | |
| 48 | SAUSA300_0103 | Tandem lpp | DUF576 | 23 | TAG C | 28 | + | |
| Unknown function | ||||||||
| 49 | SAUSA300_0079 | Unknown function | DUF1541 | 18 | LSA C | 20 | 17 | |
| 50 | SAUSA300_0372 | Unknown function | PepSY | 18 | LTA C | 21 | 17* | |
| 51 | SAUSA300_0377 | Unknown function | DUF1748 | 19 | LTG C | 23 | 14 | |
| 52 | SAUSA300_1492 | Unknown function | 16 | LAG C | 13 | 15* | ||
| 53 | SAUSA300_0992 | Cell-wall binding lipoprotein | YkyA, EzrA | 19 | LAG C | 23 | 13* | |
| 54 | SAUSA300_2403 | Unknown function | DUF1307 | 20 | LSA C | 17 | 12* | |
| 55 | SAUSA300_0724 | Unknown function | IncA, TarH | 19 | ISA C | 32 | 12 | |
| 56 | SAUSA300_2315 | Unknown function | PA26, IncA, CLN3 | 17 | LAA C | 23 | 11* | |
| 57 | SAUSA300_2614 | Unknown function | DUF_1980 | 20 | LYS C | 42 | 6 | |
| 58 | SAUSA300_0663 | Unknown function | PA26, IncA | 17 | LTG C | 15 | 5 | |
| 59 | SAUSA300_1106 | Unknown function | FAM176 | 18 | VAG C | 35 | 5* | |
| 60 | SAUSA300_0303 | Unknown function | DUF4467 | 17 | LAG C | 14 | 5* | |
| 61 | SAUSA300_1478 | Unknown function | DUF4467 | 17 | LSA C | 13 | 3* | |
| 62 | SAUSA300_1376 | Unknown function | DUF1672 | 17 | LSG C | 34 | 2* | |
| 63 | SAUSA300_1379 | Unknown function | DUF1672 | 17 | LSG C | 34 | 2* | |
| 64 | SAUSA300_1440 | Unknown function | DUF1672 | 17 | LGG C | 34 | 2* | |
| 65 | SAUSA300_1742 | Unknown function | 18 | LTA C | 23 | 2* | ||
| 66 | SAUSA300_1741 | Unknown function | ETRAMP, Myco_19_kDa | 18 | LTA C | 6 | + | |
| 67 | SAUSA300_0769 | Unknown function | DUF5067 | 17 | LGA C | 27 | + |
The number indicates the number of staphylococcal species in which the corresponding homologues gene/protein (more than 40% identity over the whole protein length) is present.
+S. aureus specific gene.
Staphylococcal specific gene.
The corresponding gene has been compared in 34 staphylococcal species and other genera in which genome sequence has been available. Abbreviations: 60KD_IMP, 60Kd inner membrane protein; ABC2_membrane_3, ABC-2 family transporter protein; CamS, CamS sex pheromone cAM373 precursor; CfAFP, Choristoneura fumiferana antifreeze protein (CfAFP); CLN3, CLN3 protein; DM13, Electron transfer DM13; COX2, Cytochrome C oxidase subunit II; DUF, Domain Unknown Function; ETRAMP, Malarial early transcribed membrane protein (ETRAMP); EzrA, Septation ring formation regulator, IncA, IncA protein; Myco_19_kDa, Mycobacterium 19 kDa lipoprotein antigen; NMT1_2, NMT1-like family; Nit_Regul_Hom, Uncharacterized protein, homolog of nitrogen regulatory protein PII; OATP, Organic Anion Transporter Polypeptide (OATP) family; OpuAC, Substrate binding domain of ABC-type glycine betaine transport system; PA26, PA26 p53-induced protein (sestrin); PBP_like_2, PBP superfamily domain; PepSY, Peptidase propeptide and YPEB domain; Peripla_BP_2, Periplasmic binding protein; Pfam, Protein Families; Phosphonate-bd, ABC transporter, phosphonate, periplasmic substrate-binding protein; Rotamase, PPIC-type PPIASE domain; SBP_bac, Bacterial extracellular solute-binding protein; SP, Signal Peptide; TarH, Tar ligand binding domain homolog; YkyA, Putative cell-wall binding lipoprotein; ZnuA, Zinc-uptake complex component A periplasmic; ZinT, (YodA) periplasmic lipocalin-like zinc-recruitment; Zip, ZIP Zinc transporter.
Functional categorization of Lpp
We grouped the Lpp according to their function. Most of the functions were deduced from studies of homologous counterparts in other bacteria, but in 12 cases the ascribed functions were confirmed by molecular/biochemical studies in S. aureus.
Iron transporters
The first group in Table 1 represent Lpp that are involved in iron acquisition which is extremely important in order for pathogenic bacteria to obtain some of the limited free iron ions during infection. For this reason intricate iron transport and iron regulatory systems have evolved in pathogenic bacteria to guarantee sufficient iron supply (Braun, 2001). It is therefore not surprising that 8 Lpp are involved iron acquisition or utilization of host-derived heme iron (isd operon) as an iron source. Under iron limitation, iron(III)-hydroxamate siderophores are excreted as iron chelators by the FhuCBG system (Sebulsky et al., 2000) and the iron-loaded chelators are bound by two Lpp (FhuD1 and FhuD2) acting as receptors and representing the first step in iron acquisition. Although both receptors are homologous (41% identity) they exhibit different activities. FhuD2 is conserved in other species and binds a broad spectrum of Fe chelators, such as ferric hydroxamate, and various siderophores, such as ferrichrome, ferrioxamineB, aerobactin, and coporgen. FhuD1, on the other hand, is only found in staphylococcal species and binds only ferrichrome and ferrioxamine B, and exhibits a lower affinity for hydroxamate siderophore binding than FhuD2 (Sebulsky and Heinrichs, 2001; Sebulsky et al., 2004). The Lpp SirA belongs to the iron regulated SirABC operon (Heinrichs et al., 1999). Mutants of either sirA or sirB are unable to take up iron complexes, such as ferric hydroxamates, ferric enterobactin or ferric citrate but they retain the ability to produce staphylobactin (Dale et al., 2004). During infection and iron limitation S. aureus is also able to use heme as an iron source via the complex isdCDEFsrtBisdG operon. IsdA and IsdB are involved in heme uptake, which is derived from the host hemoglobin. IsdC is a cell wall localized transporter, while IsdD, IsdE, and IsdF are the membrane translocation factors, and IsdG, is the cytoplasmic heme-iron binding protein (Mazmanian et al., 2003). While IsdA, IsdB, and IsdC are covalently cell-wall bound by sortase anchoring, IsdE is the heme-binding Lpp (Mazmanian et al., 2002; Grigg et al., 2007). Another iron-limited expressed siderophore transport system is composed by 4 proteins SstA,B,C, and D where SstD is the Lpp (Morrissey et al., 2000). The twin-arginine translocation (Tat) pathway, is present in only some staphylococcal species, and is composed of TatA and TatC (Biswas et al., 2009). The tatAC operon is associated with the fepABC operon. FepA is a Lpp and mediates iron binding, FepB with its typical TAT signal peptide is the iron-dependent peroxidase (FepB), and FepC is supposed to be the high affinity iron permease (Biswas et al., 2009). The advantage of the fep-tat cluster could lie in iron uptake and in the external detoxification of reactive oxygens. Indeed, the iron uptake under iron limitation was significantly decreased in the fep-tat mutants, which were less virulent in a mouse kidney abscess model (Biswas et al., 2009). The last two Lpp in this group (7 and 8) were annotated as iron binding Lpp, however, their function is unknown. The USA300_2136 (7) is encoded in the operon with two other genes, both were annotated as ion ABC transporter permease proteins containing the same motifs as FecCD and ABC3. The USA300_0219 (8) contains a SBP_bac motif (Table 1).
Other cation and anion transporters
Beside iron transporters there are other cation transporters for Co, Cu, Mn, Mo, Ni, Zn ions, which become important when these trace elements are limited. In some cases, they are crucial in infection. One of the most abundant Lpp in S. aureus is the 33 kDa MntC (SitC) (Stoll et al., 2005). Originally it was referred to as SitC because its protein sequence shares 77% identity to SitC of S. epidermidis, where it has been described as being involved in iron transport (Cockayne et al., 1998). However, instead of being involved in iron transport, SitC has recently been found to have a role in manganese (Mn) transport and is part of the MntABC system (Horsburgh et al., 2002; Diep et al., 2014). To avoid confusion we use here the term MntC (SitC). Three Lpp were annotated as Ni transporters. The Opp1 transport system is involved in cobalt and nickel uptake and has been renamed as Cnt (Remy et al., 2013). There is a Cnt related protein (25% identity) encoded by SAUSA300_0203 that was annotated as peptide/nickel transport system. The Lpp, ModA, is part of the molybdate transporter complex (ModABC) (Neubauer et al., 1999).
Based on annotation the following Lpp are involved in phosphate (USA300_1283), phosphonate (USA300_0145) and nitrate transport (USA300_0175). They are encoded with other genes in the same operon involved in the transport apparatus.
Amino acid and peptide transporters
There are 7 Lpp involved in amino acid and peptide transport. Opp3 is a main system that provides oligopeptides as a nutritional source (Hiron et al., 2007), and GmpC binds the dipeptide glycyl-methionine and is part of an ABC transporter system (Williams et al., 2004).
Sugar transport
There is only one Lpp involved in sugar transport, the maltose ABC transporter.
Lpp with miscellaneous functions
The group of miscellaneous Lpp comprise Lpp with diverse functions. They are involved in sex pheromone biosynthesis, in terminal electron transfer to oxygen (QoxA) or in global regulation (SaeP). QoxA is part of the terminal cytochrome aa3 quinol oxidase encoded by qoxABCD (Götz and Mayer, 2013; Hammer et al., 2013). The protein complex SaePQ activates the phosphatase activity of sensor kinase SaeS in the SaeRS two-component system of S. aureus (Jeong et al., 2012). The Sae system controls the expression of numerous virulence factors, such as the extracellular adherence protein (Eap), which facilitates host cell invasion (Makgotlho et al., 2013). The chaperon, PrsA, belongs to the parvulin PPIase family (pepdidyl-prolyl cis/trans isomerase) that assists posttranslocational folding at the outer surface of the cytoplasmic membrane (Heikkinen et al., 2009). In Bacillus, PrsA is an essential protein. In S. aureus it is not essential, but a prsA deletion mutant is impaired in post-transcriptional maturation of PBP2A and shows therefore decreased methicillin resistance (Jousselin et al., 2012, 2015). There is another chaperone annotated as a thioredoxin/protein disulfide-isomerase related to DsbA that catalyzes disulfide formation and isomerization and that acts simultaneously as a chaperone by preventing protein aggregation (Kouwen et al., 2007). YidC (short form of OxaI-like protein) acts as a membrane integrase for Sec-dependent substrates, such as ATP synthase subunit a (Foa) or cytochrome bo3 oxidase CyoA subunit, but can also act as a chaperone and as an assembly site for membrane protein folding (Wang and Dalbey, 2011). YidC is highly conserved in bacteria and appears to be essential; at least we were unable to delete the gene in S. aureus. There are also pro-phage and plasmid encoded Lpp. The function of the prophage encoded PhiSLT is unknown. However, the plasmid encoded Lpp in USA300 encodes for a beta-lactamase (BlaZ), which was one of the first Lpp discovered in S. aureus (Nielsen and Lampen, 1982a). This gene is plasmid encoded in many other S. aureus strains such as JH1, JH9, N315, but it may also be chromosomally located as in MRSA252 or Bmb9393. The advantage of the membrane-anchored penicillinase is that it is retained at the cell surface and is not diluted in the environment like the secreted ones.
Paralogous tandem lpp gene cluster
USA300 belongs to the hypervirulent clonal complex CC8 (Robinson et al., 2005; Cockfield et al., 2007), Most of these virulent strains carry a conserved genomic island termed νSaα that encodes a number of homologous lpp arranged in tandem, referred to as “lipoprotein-like” (lpl) (Babu et al., 2006). Most likely, the lpl cluster represents an example of paralogous genes in S. aureus, which are homologs genes in this species and that have diverged after a duplication event. USA300 carries nine such lpl genes. The exact activity of the Lpl proteins is unknown. However, recently it has been shown that this lpl cluster triggers host cell invasion, increases pathogenicity, and it has been speculated that the lpl cluster contributes to epidemic of the CC8 and CC5 strains (Nguyen et al., 2015a). Moreover, there are two other clusters of paralogous lpp genes that also contain the same conserved motifs as DUF567 of unknown function, and which are taxonomically restricted to staphylococci. With one of the paralogous Lpp Csa1A the structure has been determined that revealed a new structure family (Schluepen et al., 2013).
Lpp with unknown function
Among the 67 Lpp there are 19 (28%) with unknown function. A graphical representation of the grouped Lpp is shown in Figure 1.
Figure 1.
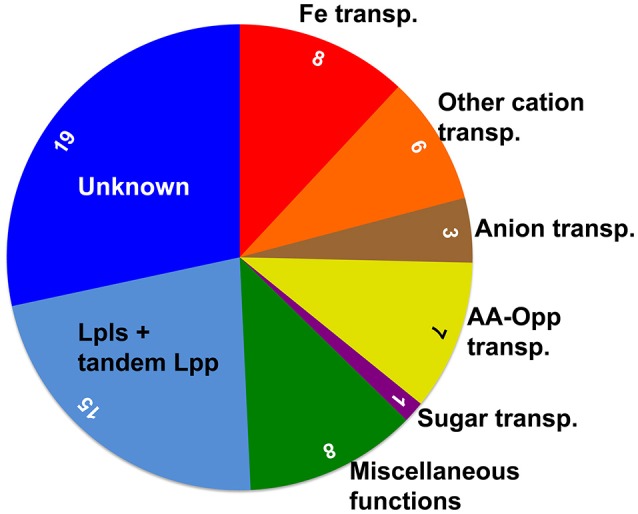
Schematic representation of the functional distribution of the 67 Lpp in S. aureus USA300. The area of the sectors, with the inserted number of Lpp, indicates the proportional distribution. Alone 25 (36%) of the Lpp are involved in ion and nutrient transport, and the ’miscellaneous’ group contains important enzymes and chaperones. USA300 carries relatively high number of 15 (22%) tandem Lpp, to which also the 9 Lpl proteins belong that are encoded by the νSaα genomic island and which play a role in pathogenicity; however, their exact function is still unknown. Finally, the function of 28% of the Lpp is completely unknown.
Dissemination of Lpp in the genus Staphylococcus and beyond
We questioned how widely distributed the various Lpp are. Do they only occur in single strains, the entire species of S. aureus, the whole genus, or even beyond? Therefore, the amino acid sequence of each Lpp was analyzed by BLAST to identify corresponding homologs. As a cut off we have chosen ≥40% identity over the entire protein sequence. The comparison was restricted to those species and their representatives where a genome was available. With regards to their abundance we categorized the Lpp of USA300 into four groups: Group A comprises Lpp that were widely distributed in the entire Staphylococcus genus and in many other genera. Group B comprises Staphylococcus genus specific Lpp. Group C represent Lpp that are mainly found only in S. aureus, and Group D represents only a few Lpp that are essentially USA300 specific. How these 4 groups differ from each other is illustrated in Figure 2, which shows one example of each Lpp group.
Figure 2.
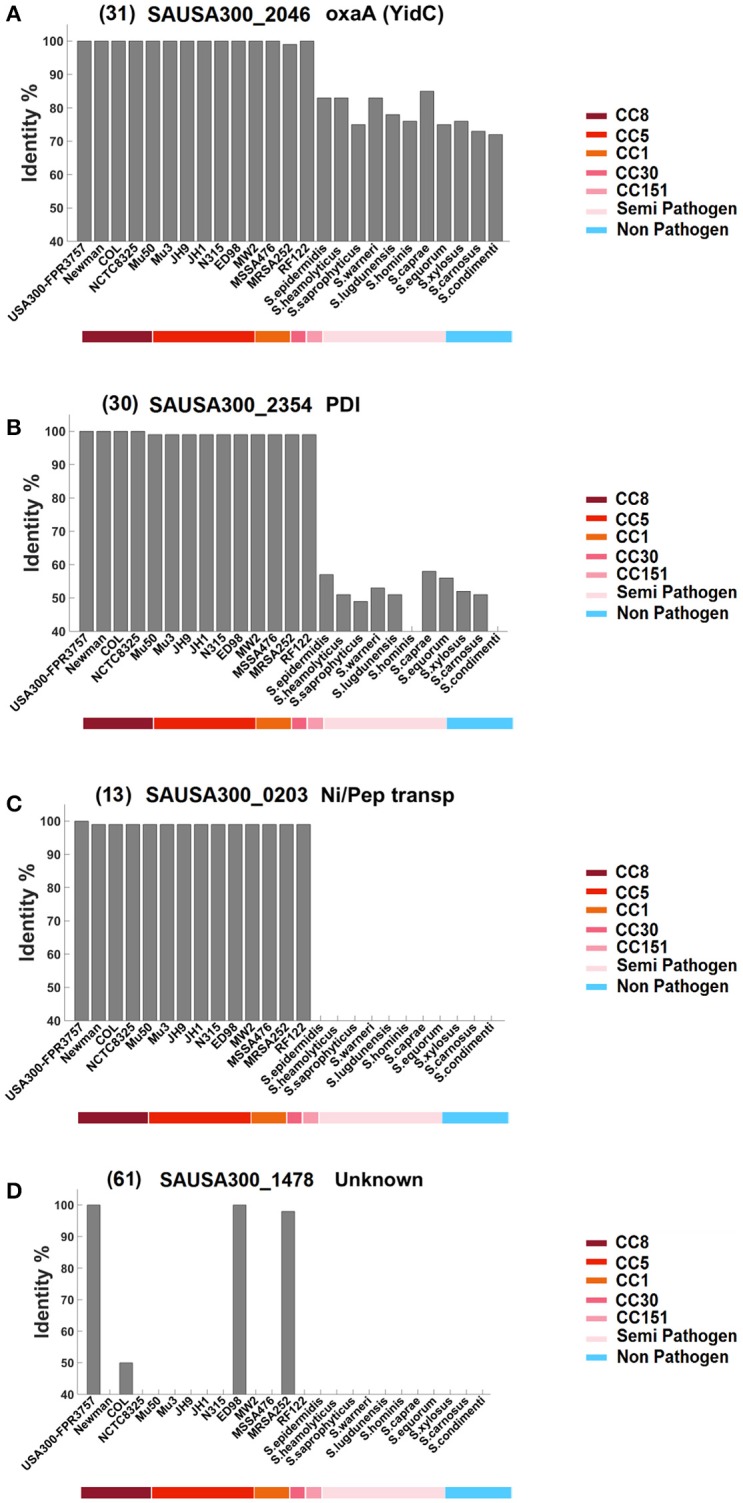
Classification of the Lpp into four groups based on their similarity and dissemination in bacteria. Each Lpp was blasted against the indicated S. aureus strains and other staphylococcal species representatives. The cut off was ≥40% identity over the entire protein sequence. The colored bar below the listed strains indicates clonal complexes (CC-types) of S. aureus (first half), as well as other staphylococcal species representatives, grouped in semi-pathogenic and non-pathogenic (second half). Group (A) represents Lpp that are highly conserved in the Staphylococcus genus but also in many other genera; an example for this group is YidC, an essential protein in many bacteria. Group (B) represents Lpp that are mainly found in the genus Staphylococcus; the example for this group is PDI, a proposed thioredoxin disulfide-isomerase. Group (C) represents Lpp mainly found in the S. aureus species; an example is the proposed nickel-peptide transporter. Group (D) represents strain-specific Lpp essentially occurring in the strain USA300; the example is an unknown Lpp. The number in front of the gene ID refers to the corresponding numbering in Table 1.
The first half of the strains represent S. aureus strains; the underlined colored bar indicates their association to the corresponding clonal complexes (CC-types). The second halve of the strains represent a selection of other staphylococcal species; the underlined colored bar indicates whether they are regarded as semi-pathogenic or non-pathogenic. In the A and B group the USA300 Lpp are highly conserved in the S. aureus species (>95 identity) thus comprising a homogeneous block. Members of the A group are distinguished by a broad dissemination and are highly conserved in other staphylococcal species (>70% identity) and in many other genera (≥40% identity). The group B Lpp are less conserved in other staphylococcal species and are only occasionally found in other genera. Group C lists Lpp that essentially occur in the species S. aureus apart from a few examples that show a slightly broader dissemination. And, finally, group D lists Lpp that are essentially strain specific for USA300 and that are not found in other S. aureus strains apart from few exceptions. A complete listing of all 67 Lpp is shown in Supplementary Figure 1.
Group A Lpp comprise highly conserved Lpp with a broad dissemination (Table 2A). Most likely they play an important role in metabolism or are involved in basic cellular processes. The majority of these Lpp are annotated as transporters. The corresponding homologous proteins are found in many, but not all, other staphylococcal species, and they are also found in many other genera. Examples of the most conserved Lpp that were even found in completely unrelated genera such Gram-negatives or high-GC Gram-positives are: Fe transporter-SirA (04), IsdE (05), FepA (06), MntC (SitC) (09), Phosphate ABC transporter (15), Glycine betaine transporter (18), Amino acid ABC transporter (19), D-Methionine transporter (24), Quinol oxidase (QoxA) (27), Electron transfer domain/SaeP (28), YidC (31), BlaZ (33), or the Lpp with unknown function (49). The latter Lpp is unusual in so far as it is only found in USA300 but not in other S. aureus strains; however, it is present in many other staphylococcal species and even in other genera. The most disseminated Lpp is the D-Methionine ABC transporter (24). In E. coli the MetNIQ transporter, belonging to ABC type permease superfamily, is involved in the uptake of both D- and L-methionine. MetN is the putative ATPase, MetI is a membrane spanning protein and MetQ (the Lpp) is the major binding protein for both L- and D-methionine as well as their analogs, such as N-formyl methionine (Merlin et al., 2002). In complex medium the transporter is not essential for growth, which also applies for most other Lpp. However, under certain nutrient limitations and environmental conditions they are crucial for growth and survival. So far we know only one Lpp that is essential even in complex medium, and that is YidC (31). YidC is evolutionarily conserved and is involved in membrane biogenesis in bacteria, mitochondria, and chloroplasts. It comprises several activities, such as acting as a protein insertase, as chaperone, and as an assembly factor for transmembrane proteins (Wang and Dalbey, 2011).
Table 2A.
USA300 Lpp with broad dissemination.
| No1 | Function/annotation | Staphylococcal species and other genera2 |
|---|---|---|
| 02 | Fe ABC transporter/FhuD2 | S.aureus, S.capitis, S.carnosus, S.equorum, S.gallinarum, S.haemolyticus, S.hyicus, S.lugdunensis, S.pasteuri, S.pettenkoferi, S.pseudintermedius, S.saprophyticus, S.schleiferi, S.simiae, S.warneri, |
| S.xylosus, | ||
| Bacillus, Exiguobacterium, Macrococcus, Salinicoccus | ||
| 04 | Fe ABC transporter/SirA | S.aureus, S.agnetis, S.arlettae, S.delphini, S.equorum, S.hyicus, S.intermedius, S.lugdunensis, S.pseudintermedius, S.schleiferi |
| Advenella, Bacillus, Chromohalobacter, Clostridium, Gynuella, Hahella, Halobacteroides, Halomonas, Haloplasma, Jeotgalibacillus, Lysinibacillus, Marinomonas, Paenibacillus, Pectobacterium, Planococcus, Sporosarcina | ||
| 05 | Fe ABC transporter/IsdE | S.aureus, S.auricularis, S.capitis, S.caprae, S.condimenti, S.lentus, S.lugdunensis, S.pasteuri, S.sciuri, S.simulans |
| Bacillus, Carnobacterium, Coprococcus, Eubacterium, Lactobacillus, Listeria, Lysinibacillus, Paenibacillus, Solibacillus, Streptococcus, Terribacillus | ||
| 06 | FepA, Fe-binding protein, part of fep ABC and tat-AC cluster | S.aureus, S.capitis, S.carnosus, S.condimenti, S.epidermidis, S.haemolyticus, S.lugdunensis, S.pasteuri, S.simiae, S.warneri |
| Actinobacillus, Actinoplanes, Bacillus, Bibersteinia, Brevibacillus, Clavibacter, Corynebacterium, Dermacoccus, Exiguobacterium, Gallibacterium, Hyphomicrobium, Kineococcus, Kitasatospora, Kyrpidia, Leptotrichia, Listeria, Lysinibacillus, Mannheimia, Moraxella, Mycobacterium, Neisseria, Paenibacillus, Planococcus, Pseudomonas, Streptococcus, Streptomyces, Yersinia | ||
| 09 | Manganese-binding protein MntC (SitC) | S.aureus, S.capitis, S.caprae, S.carnosus, S.condimenti, S.epidermidis, S.equorum, S.haemolyticus, S.hominis, S.hyicus, S.lugdunensis, S.pasteuri, S.pseudintermedius, S.saprophyticus, S.schleiferi, S.simiae, S.simulans, S.warneri, S.xylosus |
| Aerococcus, Amphibacillus, Bacillus, Carnobacterium, Clostridium, Enterococcus, Exiguobacterium, Finegoldia, Lactobacillus, Macrococcus, Oceanobacillus, Paenibacillus, Streptomyces, Terribacillus, Tetragenococcus | ||
| 10 | Zinc-binding, adcA-like | S.aureus, S.capitis, S.haemolyticus, S.hyicus, S.pasteuri, S.pseudintermedius, S.saprophyticus, S.schleiferi, S.warneri, S.xylosus |
| Bacillus, Enterococcus, Halobacillus, Streptococcus | ||
| 11 | Cobalt and nickel transporter Cnt (Opp1A) | S.aureus, S.epidermidis, S. hyicus, S.lugdunensis, S.pasteuri, S.pseudintermedius, S.saprophyticus, S.schleiferi, S.warneri, S.xylosus, |
| Aggregatibacter, Bacillus, Brevibacillus, Eubacterium, Mannheimia, Methanosarcina, Paenibacillus, Proteus | ||
| 14 | Molybdenum ABC transporter (ModA) | S.aureus, S.capitis, S.caprae, S.carnosus, S.epidermidis, S.haemolyticus, S.hyicus, S.lugdunensis, S.pasteuri, S.pseudintermedius, S.saprophyticus, S.schleiferi, S.simiae, S.warneri, S.xylosus |
| Bacillus, Clostridium, Macrococcus, Paenibacillus, Salinicoccus, Syntrophobotulus | ||
| 15 | Phosphate ABC transporter | S.aureus, S.capitis, S.caprae, S.carnosus, S.epidermidis, S.haemolyticus, S.hyicus, S.lugdunensis, S.pasteuri, S.pseudintermedius, S.saprophyticus, S.schleiferi, S.simiae, S.warneri, S.xylosus |
| Alkaliphilus, Anoxybacillus, Bacillus, Clostridium, Enterobacter, Escherichia, Eubacterium, Exiguobacterium, Geobacillus, Halobacillus, Jeotgalibacillus, Lysinibacillus, Macrococcus, Maribacter, Paenibacillus, Planococcus, Pleurocapsa, Salinicoccus, Sebaldella, Streptomyces, Thermobacillus, Thermosediminibacter | ||
| 16 | Phosphonate ABC transporter | S.aureus, S.capitis, S.caprae, S.epidermidis, S.haemolyticus, S.hominis, S.lugdunensis, S.pasteuri, S.saprophyticus, S.warneri |
| Bacillus, Carnobacterium, Enterobacter, Enterococcus, Lactobacillus, Melissococcus, Paenibacillus, Terribacillus, Weissella | ||
| 17 | Nitrate ABC transporter | S.aureus, S.pseudintermedius, S.saprophyticus, S.schleiferi |
| Bacillus, Clostridium, Geobacillus, Methanosarcina, Ruminiclostridium, Solibacillus | ||
| 18 | Glycine betaine /carnitine/choline ABC transporter (OpuCc) | S.aureus, S.capitis, S.carnosus, S.cohnii, S.condimenti, S.epidermidis, S.equorum, S.gallinarum, S.haemolyticus, S.hyicus, S.lugdunensis, S.pasteuri, S.saprophyticus, S.schleiferi, S.simiae, S.simulans, S.succinus, S.warneri, S.xylosus |
| Bacillus, Carnobacterium, Enterococcus, Lactobacillus, Listeria, Macrococcus, Pediococcus, Solibacillus, Streptococcus, Tetragenococcus, Virgibacillus | ||
| 19 | Amino acid ABC transporter | S.aureus, S.capitis, S.caprae, S.carnosus, S.epidermidis, haemolyticus, S.lugdunensis, S.pasteuri, S.saprophyticus, S.simiae, S.warneri, S. S.xylosus |
| Aggregatibacter, Anoxybacillus, Arthrobacter, Bacillus, Brevibacillus, Brucella, Campylobacter, Clostridium, Ensifer, Gallibacterium, Geobacillus, Haemophilus, Hafnia, Halobacillus, Jeotgalibacillus, Macrococcus, Mesorhizobium, Neisseria, Neorhizobium, Nocardia, Ochrobactrum, Paenibacillus, Photorhabdus, Raoultella, Rhizobium, Sinorhizobium, Streptococcus, Xenorhabdus | ||
| 21 | Oligopeptide ABC transporter (Opp3A) | S.aureus, S.capitis, S.carnosus, S.epidermidis, S.haemolyticus, S.lugdunensis, S.pasteuri, S.pseudintermedius, S.saprophyticus, S.schleiferi, S.xylosus |
| Anoxybacillus, Bacillus, Carnobacterium, Geobacillus, Macrococcus | ||
| 23 | NLPA/D-methionine binding, (GmpC) | S.aureus, S.epidermidis, S.simiae, S.capitis, S.lugdunensis, S.xylosus, S.carnosus, S.schleiferi, S.pseudintermedius |
| Aerococcus, Amphibacillus, Bacillus, Carnobacterium, Clostridium, Enterococcus, Exiguobacterium, Finegoldia, Lactobacillus, Listeria, Macrococcus, Megasphaera, Oceanobacillus, Paenibacillus, Ralstonia, Streptococcus, Streptomyces, Terribacillus, Tetragenococcus | ||
| 24 | D-Methionine ABC transporter | S.aureus, S.capitis, S.caprae, S.carnosus, S.cohnii, S.epidermidis, S.haemolyticus, S.hyicus, S.lugdunensis, S.pasteuri, S.pseudintermedius, S.saprophyticus, S.schleiferi, S.warneri, S.xylosus |
| Acetohalobium, Acidaminococcus, Actinoplanes, Advenella, Amphibacillus, Anoxybacillus, Arcobacter, Azospirillum, Bacillus, Bifidobacterium, Caldicellulosiruptor, Campylobacter, Carnobacterium, Cedecea, Citrobacter, Clostridium, Cronobacter, Desulfitobacterium, Desulfosporosinus, Enterobacter, Enterobacteriaceae, Enterococcus, Erwinia, Exiguobacterium, Finegoldia, Geobacillus, Halobacillus, Jeotgalibacillus, Ketogulonicigenium, Klebsiella, Kluyvera, Kosakonia, Lactobacillus, Leptotrichia, Lipotes, Listeria, Macrococcus, Megamonas, Megasphaera, Melissococcus, Paenibacillus, Pectobacterium, Pediococcus, Pelosinus, Planococcus, Pseudomonas, Salinicoccus, Sebaldella, Selenomonas, Serratia, Sodalis, Solibacillus, Streptomyces, Sulfurospirillum, Terribacillus, Thermoanaerobacterium, Veillonella, Virgibacillus, Xanthomonas | ||
| 25 | Maltose ABC transporter | S.aureus, S.delphini, S.intermedius, S.lentus, S.pseudintermedius, S.sciuri |
| Bacillus, Geobacillus, Salinicoccus | ||
| 26 | CamS sex pheromone biosynthesis | S.aureus, S.capitis, S.carnosus, S.epidermidis, S.haemolyticus, S.hyicus, S.lugdunensis, S.pasteuri, S.pseudintermedius, S.saprophyticus, S.schleiferi, S.simiae, S.warneri, S.xylosus |
| Escherichia, Macrococcus, Salinicoccus | ||
| 27 | Quinol oxidase, subunit II (QoxA) | S.aureus, S.agnetis, S.auricularis, S.capitis, S.caprae, S.carnosus, S.chromogenes, S.cohnii, S.epidermidis, S.haemolyticus, S.hominis, S.hyicus, S.lugdunensis, S.pasteuri, S.pettenkoferi, S.pseudintermedius, S.saprophyticus, S.schleiferi, S.simiae, S.warneri, S.xylosus, |
| Bacillus, Geobacillus, Halobacillus, Listeria, Lysinibacillus, Paenibacillus, Salinicoccus, Streptococcus, Virgibacillus | ||
| 28 | Electron transfer domain/SaeP | S.aureus, S.auricularis, S.capitis, S.caprae, S.carnosus, S.condimenti, S.delphini, S.epidermidis, S.haemolyticus, S.intermedius, S.lentus, S.lugdunensis, S.microti, S.pettenkoferi, S.pseudintermedius, S.saprophyticus, S.schleiferi, S.sciuri, S.simiae, S.simulans, S.vitulinus, S.xylosus |
| Bacillus, Brochothrix, Carnobacterium, Enterococcus, Fictibacillus, Listeria, Mycobacterium, Paenibacillus | ||
| 31 | YidC (OxaA) - essential protein | S.aureus, S.arlettae, S.capitis, S.caprae, S.carnosus, S.cohnii, S.condimenti, S.delphini, S.epidermidis, S.equorum, S.gallinarum, S.haemolyticus, S.hominis, S.hyicus, S.lugdunensis, S.massiliensis, S.pasteuri, S.pseudintermedius, S.saprophyticus, S.schleiferi, S.simiae, S.simulans, S.succinus, S.warneri, S.xylosus |
| Bacillus, Halobacillus, Lysinibacillus, Macrococcus, Planococcus, Salinicoccus, Solibacillus, Xylanimonas | ||
| 33 | Membrane bound penicillinases BlaZ | S.aureus, S.agnetis, S.capitis, S.epidermidis, S.equorum, S.haemolyticus, S.lugdunensis, S.pseudintermedius, S.saprophyticus, S.warneri, S.xylosus, |
| Acidaminococcus, Bacillus, Carnobacterium, Enterococcus, Escherichia, Kribbella, Macrococcus, Paenibacillus, Salinicoccus, Streptococcus, Streptomyces | ||
| 49 | Unknown function | USA300, S.arlettae, S.capitis, S.carnosus, S.cohnii, S.epidermidis, S.equorum, S.haemolyticus, S.hominis, S.lugdunensis, S.pasteuri, S.pettenkoferi, S.pseudintermedius, S.saprophyticus, S.sciuri, S.warneri, S.xylosus |
| Bacillus, Carnobacterium, Corynebacterium, Dermacoccus, Enterococcus, Exiguobacterium, Geobacillus, Jeotgalibacillus, Kocuria, Kytococcus, Listeria, Lysinibacillus, Macrococcus, Oceanobacillus, Paenibacillus, Planococcus, Salinicoccus, Solibacillus, Streptococcus, Terribacillus, Virgibacillus (see Table 2D) | ||
| 57 | Unknown function | S.aureus, S.agnetis, S.capitis, S.haemolyticus, S.hyicus, S.simiae, |
| Bacillus, Clostridium, Enterococcus, Herbinix, Lactobacillus, Leuconostoc, Streptococcus |
Numbering and proposed function are the same as listed in Table 1.
The other genera are in bold letters.
Group B represents Lpp that are mainly found in the genus Staphylococcus. Only exceptionally they were also found in the related genera Salinicoccus and Bacillus, or in the unrelated genus Xylanimonas, a high-GC Gram-positive cocci belonging to the Actinomycetales (Table 2B). Only 8 of the 16 Lpp in this list have an annotated function such as the ferric hydroxamate receptor (FhuD1) (01), nickel- (12), oligopeptide (Opp4A) (22) transporters, as well as thioredoxin disulfide-isomerase (30). The functions of the other 8 Lpp are unknown.
Table 2B.
USA300 Lpp mainly occurring in the genus Staphylococcus.
| No1 | Function/annotation | Staphylococcal species and other genera2 |
|---|---|---|
| 01 | Ferric hydroxamate receptor/FhuD1 | S.aureus, S.capitis, S.carnosus, S.equorum, S.gallinarum, S.haemolyticus, S.hyicus, S.lugdunensis, S.pasteuri, S.saprophyticus, S.schleiferi, S.sciuri, S.succinus, S.warneri, S.xylosus |
| 03 | Transferrin receptor/SstD | S.aureus, S.carnosus, S.condimenti, S.epidermidis, S.haemolyticus, S.hyicus, S.lugdunensis, S.pasteuri, S.saprophyticus, S.schleiferi, S.simiae, S.simulans, S.warneri, S.xylosus, |
| Bacillus, Macrococcus | ||
| 07 | Fe ABC transporter | S.aureus, S.capitis, S.carnosus, S.epidermidis, S.haemolyticus, S.hyicus, S.lugdunensis, S.pasteuri, S.pseudintermedius, S.saprophyticus, S.schleiferi, S.simiae, S.warneri, S.xylosus Bacillus, Paenibacillus |
| 12 | Nickel ABC transporter | S.aureus, S.capitis, S.carnosus, S.epidermidis, S.pasteuri, S.pseudintermedius, S.saprophyticus, S.schleiferi, S.warneri, S.xylosus |
| 20 | Peptide ABC transporter | USA300, S.caprae, S.chromogenes, S.delphini, S.epidermidis, S.haemolyticus, S.hominis, S.lugdunensis, S.pasteuri, S.schleiferi, S.warneri |
| Brevibacillus, Streptococcus (see Table 2D) | ||
| 22 | OligopeptideABC transporter (Opp4A) | S.aureus, S.carnosus, S.hyicus, S.pseudintermedius, S.schleiferi Bacillus |
| 29 | Foldase protein PrsA | S.aureus, S.capitis, S.caprae, S.carnosus, S.epidermidis, S.haemolyticus, S.hyicus, S.lugdunensis, S.pasteuri, S.pseudintermedius, S.saprophyticus, S.schleiferi, S.simiae, S.warneri, S.xylosus |
| Macrococcus, Salinicoccus | ||
| 30 | Thioredoxin/Protein disulfide-isomerase | S.aureus, S.capitis, S.caprae, S.carnosus, S.epidermidis, S.equorum, S.gallinarum, S.haemolyticus, S.hyicus, S.lugdunensis, S.pasteuri, S.saprophyticus, S.simiae, S.warneri, S.xylosus |
| 50 | Unknown function | S.aureus, S.carnosus, S.condimenti, S.delphini, S.equorum, S.gallinarum, S.lugdunensis, S.pasteuri, S.pettenkoferi, S.pseudintermedius, S.schleiferi, S.sciuri, S.simiae, S.simulans, S.succinus, S.warneri, S.xylosus |
| 51 | Unknown function | S.aureus, S.auricularis, S.capitis, S.caprae, S.carnosus, S.epidermidis, S.haemolyticus, S.hominis, S.lugdunensis, S.pasteuri, S.saprophyticus, S.simiae, S.warneri, S.xylosus |
| Xylanimonas | ||
| 52 | Unknown function | S.aureus, S.arlettae, S.capitis, S.caprae, S.carnosus, S.cohnii, S.epidermidis, S.haemolyticus, S.lugdunensis, S.pasteuri, S.saprophyticus, S.simiae, S.succinus, S.warneri, S.xylosus |
| 53 | Cell-wall binding lpp | S.aureus, S.capitis, S.caprae, S.epidermidis, S.haemolyticus, S.hominis, S.lugdunensis, S.pasteuri, S.saprophyticus, S.simiae, S.succinus, S.warneri, S.xylosus |
| 54 | Unknown function | S.aureus, S.capitis, S.caprae, S.carnosus, S.epidermidis, S.haemolyticus, S.hominis, S.lugdunensis, S.pasteuri, S.simiae, S.simulans, S.warneri |
| 55 | Unknown function | S.aureus, S.capitis, S.caprae, S.carnosus, S.epidermidis, S.haemolyticus, S.lugdunensis, S.pasteuri, S.saprophyticus, S.schleiferi, S.warneri, S.xylosus |
| Escherichia, Streptococcus | ||
| 56 | Unknown function | S.aureus, S.capitis, S.caprae, S.epidermidis, S.hyicus, S.intermedius, S.lugdunensis, S.pasteuri, S.pseudintermedius, S.simiae, S.warneri |
| 58 | Unknown function | S.aureus, S.epidermidis, S.haemolyticus, S.pasteuri, S.warneri, |
| Fusarium, Nasonia |
Numbering and proposed function are the same as listed in Table 1.
The other genera are in bold letters.
Group C represent Lpp that are mainly found in the S. aureus species (Table 2C). Only two Lpp are ascribed a function, the iron binding protein (08) and the nickel-peptide transporter (13). Besides S. aureus, this latter Lpp is also found in various Bacillus genera, suggesting that the corresponding gene was exchanged by horizontal transfer and was maintained in S. aureus probably because of its beneficial effect in infection. Interestingly, all 9 Lpl proteins encoded on the νSaα island, as well as two other tandem Lpp clusters, were only found in S. aureus. These genes likely arose by gene duplication, an important mechanism for acquiring new genes and creating genetic novelty in organisms (Magadum et al., 2013). It is striking why only S. aureus accumulated and preserved these Lpl and tandem Lpp. Their contribution in virulence has been shown for the Lpl proteins; they modulate the innate immunity, and enhance host cell invasion and pathogenicity (Nguyen et al., 2015a). Finding out their precise function warrants further investigation.
Table 2C.
USA300 Lpp essentially occurring in the S. aureus species.
| No1 | Function/annotation | S. aureus and few other species/genera2 |
|---|---|---|
| 08 | Iron Binding Protein | S.aureus, S.epidermidis, S.pseudintermedius, S.schleiferi |
| 13 | Nickel-Peptide/transporter | S. aureus only - but also in: |
| Amphibacillus, Bacillus, Lysinibacillus, Solibacillus | ||
| 34 | Lpl-1 νSaα specific | S.aureus |
| 35 | Lpl-2 νSaα specific | S.aureus |
| 36 | Lpl-3 νSaα specific | S.aureus |
| 37 | Lpl-4 νSaα specific | S.aureus |
| 38 | Lpl-5 νSaα specific | S.aureus |
| 39 | Lpl-6 νSaα specific | S.aureus |
| 40 | Lpl-7 νSaα specific | S.aureus |
| 41 | Lpl-8 νSaα specific | S.aureus |
| 42 | Lpl-9 νSaα specific | S.aureus |
| 43 | Tandem lpp | S.aureus, S.heamolyticus, S.lugdunensis |
| 44 | Tandem lpp | S.aureus |
| 45 | Tandem lpp (Csa1A) | S.aureus |
| 46 | Tandem lpp | S.aureus |
| 47 | Tandem lpp | S.aureus |
| 48 | Tandem lpp | S.aureus |
| 59 | Unknown function | S.aureus, S.epidermidis, S.lugdunensis S.pasteuri, S.warneri |
| 60 | Unknown function | S.aureus, S.caprae, S.epidermidis, S.lugdunensis, S.warneri |
| 62 | Unknown function | S.aureus, S.epidermidis |
| 63 | Unknown function | S.aureus, S.epidermidis |
| 64 | Unknown function | S.aureus, S.epidermidis |
| 65 | Unknown function | S.aureus, S.epidermidis |
| 66 | Unknown function | S.aureus |
| Bacillus | ||
| 67 | Unknown function | S.aureus |
Numbering and proposed function are the same as listed in Table 1.
The other genera are in bold letters.
Group D represents Lpp that are essentially USA300 specific and are not found in other S. aureus strains, but some are found in few other staphylococcal species or even in other genera as the Lpp (49) (Table 2D). The prophage encoded PhiSLT (32) is only found in USA300 and a limited number of other S. aureus strains as well as in S. xylosus, suggesting a limited distribution of this prophage in the genus.
Table 2D.
USA300 Lpp essentially occurring in strain USA300.
| 20 | Peptide ABC transporter | Only USA300 not in other S. aureus strains, but other species and genera (see Table 2B) |
| 32 | PhiSLT ORF144-like | Only USA300, ED98, MW2, MSSA476, MRSA252 and S.xylosus |
| 49 | Unknown function | Only USA300 not in other S. aureus strains, but other species and genera (see Table 2A) |
| 61 | Unknown function | Only USA300, COL, ED98, MRSA252 |
Correlation between the number of Lpp and pathogenicity
We compared the total number of Lpp (based on PREP-LIPO method) in various S. aureus strains belonging to different clonal complex groups (CC 8, 5, 1, 30, 151) and in representatives of other staphylococcal species (Table 3). As can be seen, the highly epidemic categorized strains (S. aureus USA300, Newman, Mu50, Mu3, JH1, JH9, N315) contain more than 60 putative Lpp, whereas the moderate epidemic strains (S. aureus NCTC8325, S. epiderminis RP62A, ATCC_12228, S. haemolyticus JCSC1435, S. saprophyticus ATCC 15305) contain only around 50 putative Lpp. Although speculative, this suggests that epidemic/pathogenic strains are distinguished by a higher number of Lpp.
Table 3.
The number of Lpp in different staphylococcal strains and species.
| Species | Strains | Number of Lpp | |
|---|---|---|---|
| S.aureus | CC8 | USA300 | 67 |
| Newman | 64 | ||
| COL | 61 | ||
| NCTC8325 | 50 | ||
| CC5 | Mu50 | 66 | |
| Mu3 | 66 | ||
| JH1 | 65 | ||
| JH9 | 65 | ||
| N315 | 64 | ||
| CC1 | MSSA476 | 62 | |
| MW2 | 60 | ||
| CC30 | MRSA252 | 57 | |
| CC151 | RF122 | 56 | |
| S. epidermidis | RP62A | 53 | |
| ATCC_12228 | 48 | ||
| S. haemolyticus | JCSC1435 | 51 | |
| S. saprophyticus | ATCC 15305 | 42 | |
| S. carnosus | TM300 | 58 | |
Lpp as TLR2 agonists
It is long known that bacterial Lpp are recognized by Toll-like receptor 2 (TLR2) of the innate immune system and are sensed at very low concentrations (Zähringer et al., 2008). Dependent on the degree of acylation Lpp are recognized by different TLR2 heterodimers: Diacylated Lpp are recognized by TLR2 and TLR6 (Bulut et al., 2001; Takeuchi et al., 2001), while triacylated Lpp by TLR2 and TLR1 heterodimers (Takeda et al., 2002; Takeuchi et al., 2002). In Gram-negative bacteria Lpp biogenesis consists of three successive reactions catalyzed by prolipoprotein diacylglyceryl transferase (Lgt), signal peptidase II (Lsp), and apolipoprotein N-acyltransferase (Lnt) (Sankaran and Wu, 1994; Buddelmeijer, 2015). A Lnt homolog has so far not been found in staphylococci. However, Kurokawa and colleagues showed that MntC (SitC) is triacylated in exponential growth phase but becomes diacylated (lacking the alpha-aminoacylation) in post-exponential phase (Kurokawa et al., 2009, 2012). The occurrence of triacylated MntC (SitC) indicates that S. aureus has a apolipoprotein N-acyltransferase, which adds a fatty acid to the amino group of S-(diacylglyceryl) cysteine residue. MntC (SitC) was also the first native Lpp for which its TLR2 activation and its co-localization with TLR2 have been demonstrated (Kurokawa et al., 2009; Müller et al., 2010). The structure of the lipid moiety of Lpp has an enormous influence on the immune response. For example it has been shown that di- but not tri-acylated Lpp suppress immune responses and might play a role in immune tolerance (Skabytska et al., 2014).
Lpp as vaccine candidates
In principle all Lpp listed in Table 1 are potential vaccine candidates, however, some are more or less suitable. All Lpp listed in Table 2C are promising vaccine candidates because they occur essentially in the S. aureus species. Promising vaccine candidates would be the 9 Lpl proteins (34–42 in Table 1) because they share a highly conserved (92% similarity) core region of 44 amino acids (Nguyen et al., 2015a). A vaccine against this core domain would simultaneously cross-react with several antigens. There is also a set of 4 tandem Lpp (45–48 in Table 1) that show a very high similarity (>80%) over the entire protein length; most likely these tandem Lpp arose by gene duplication. However, both these tandem Lpp and Lpl share also a conserved 38 amino acid long domain Figure 3. Therefore, one can expect that antibodies against this conserved domain would cross-react with 13 Lpp. The advantage of these tandem Lpp is also that they are highly present only in S. aureus. A vaccine would therefore not target staphylococcal skin residents such as S. epidermidis and other species. Another parameter for a promising Lpp-antigen is its size. The size should be large enough that the protein part can penetrate the cell wall to be accessible for antibodies. The size of the Lpp varies enormously, ranging from 6 to 89 kDa (Table 1). All Lpp with a mass >40 kDa are also good candidates like the zinc-binding (10), cobalt and nickel transporter Cnt (11), nickel ABC transporter (12), nickel-Peptide/ transporter (13), peptide ABC transporter (20), oligopeptide ABC transporter (Opp3A) (21), CamS sex pheromone biosynthesis (26), quinol oxidase, subunit II (QoxA) (27).
Figure 3.
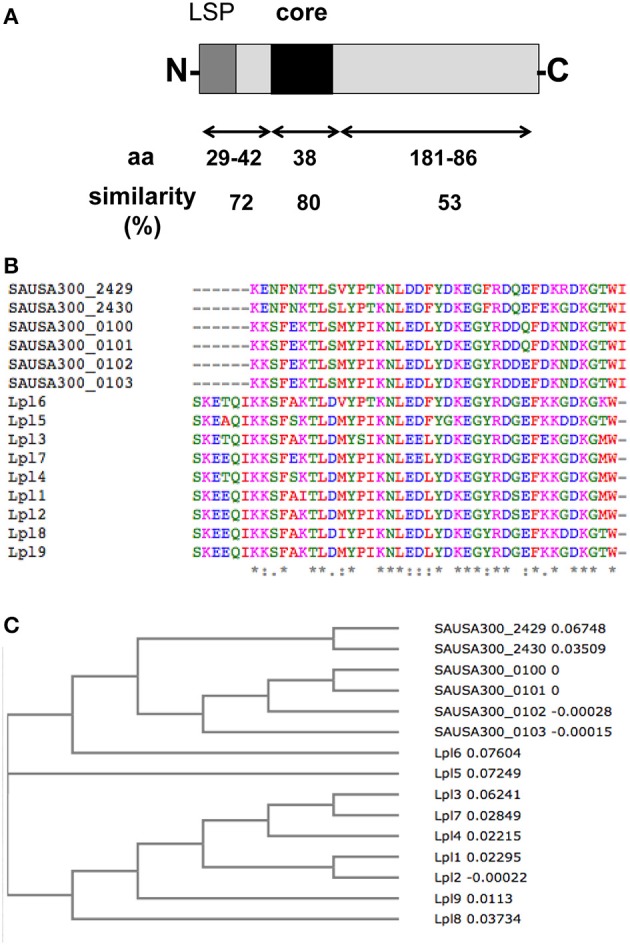
Conserved sequence motif in Lpl and the tandem Lpp. (A) Alignment of the 9 Lpl and the 4 tandem Lpp revealed a core sequence over 38 amino acids with 80% similarity. (B) Sequence alignment of the core region of 15 tandem Lpp by using Clustal Omega program. (C) Phylogenetic tree of the core region of 15 tandem Lpp by using Clustal Omega program.
A third parameter for a promising vaccine candidate is the abundance of an antigen. The most abundant Lpp is MntC (SitC), which is essential for MRSA virulence during murine systemic infection (Kehl-Fie et al., 2013). It was therefore concluded that MntABC might be a potential vaccine candidate (Diep et al., 2014). Another potential vaccine candidate is the FhuD2 (02) involved in ferric-hydroxamate uptake. FhuD2 binds ferrichrome with nanomolar affinity and the structure of FhuD2-ferrichrome has been determined (Mariotti et al., 2013; Podkowa et al., 2014). Immunization with FhuD2 alone or together with hydroxamate siderophores was protective in a murine staphylococcal infection model (Mariotti et al., 2013). However, a break-through was reported recently with a combination of five antigens, including FhuD2 (02), and Csa1A (45), formulated with a novel adjuvant containing a TLR7-dependent agonist adsorbed to alum. This vaccine provided close to 100% protection against four different staphylococcal strains (Bagnoli et al., 2015). As can seen, certain Lpp have been already turned out experimentally as promising vaccine candidates.
Conclusion
The re-evaluation of the chromosomal encoded Lpp in S. aureus USA300 was necessary, as none of the Lpp prediction tools of the public domain yielded the complete inventory of Lpp. The 67 identified Lpp constituted a solid basis for a systematic analysis. A large proportion of the Lpp is involved in the uptake of essential ions and nutrients. Frequently they function as receptors for the target molecule and are part of ABC transporter complexes. Generally one can say that 39% of the Lpp are involved in ion and nutrient transport, indicating, that this is one of their major physiological task. However, two Lpp in the miscellaneous group should be mentioned, QoxA and YidC, which play important functions in respiration and folding of membrane proteins.
Regarding the dissemination of Lpp within the S. aureus species and in other species we can group them into four categories: (i) those that are highly conserved and broadly disseminated, (ii) that are mainly found in the genus Staphylococcus, (iii) that are mainly found in the species S. aureus, and (iv) a few Lpp are rather strain specific. Particularly the group of S. aureus specific Lpp are promising vaccine candidates for future work. Our data show that the relatively high number of lpp genes (>60) in the S. aureus species is mainly due to the tandem lpp. There was only one S. aureus strain, NCTC8325, that shows only 50 lpp. As this strain is used for almost 50 years as a lab strain, it is conceivable that under non-selective conditions a reversal of the lpp amplification occurred. Indeed, this strain contains only three tandem lpl genes in the νSaα island, which might represent an example of adaptive evolution. There are many open questions regarding the Lpp in S. aureus and also the Lpp of the staphylococcal skin microbiota that need to be studied in the future.
Author contributions
FG designed the work, SS and MN analyzed and interpreted data, SS and MN prepared the draft of the Manuscript (MS), figures and tables; and FG wrote the MS.
Funding
This work was supported by grants from the Deutsche Forschungsgemeinschaft (DFG; GO 371/9-1, SFB766 and TRR34) and Open Access Publishing Fund of Tuebingen University.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Ralf Rosenstein for his useful suggestion and discussion.
Glossary
Abbreviations
- Lpp
Lipoproteins
- lgt
diacylglyceryl transferase enzyme encoding gene
- Lpl
lipoprotein-like
- S
Staphylococcus
- TLR
Toll-like receptor
- CC
clonal complex
- AA
Amino acid Numbers behind Lpp in parenthesis refer to the numbers in Table 1.
Supplementary material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01404
References
- Babu M. M., Priya M. L., Selvan A. T., Madera M., Gough J., Aravind L., et al. (2006). A database of bacterial lipoproteins (DOLOP) with functional assignments to predicted lipoproteins. J. Bacteriol. 188, 2761–2773. 10.1128/Jb.188.8.2761-2773.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagnoli F., Fontana M. R., Soldaini E., Mishra R. P., Fiaschi L., Cartocci E., et al. (2015). Vaccine composition formulated with a novel TLR7-dependent adjuvant induces high and broad protection against Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 112, 3680–3685. 10.1073/pnas.1424924112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bagos P. G., Tsirigos K. D., Liakopoulos T. D., Hamodrakas S. J. (2008). Prediction of lipoprotein signal peptides in Gram-positive bacteria with a Hidden Markov Model. J. Proteome Res. 7, 5082–5093. 10.1021/pr800162c [DOI] [PubMed] [Google Scholar]
- Baumgärtner M., Kärst U., Gerstel B., Loessner M., Wehland J., Jänsch L. (2007). Inactivation of Lgt allows systematic characterization of lipoproteins from Listeria monocytogenes. J. Bacteriol. 189, 313–324. 10.1128/jb.00976-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas L., Biswas R., Nerz C., Ohlsen K., Schlag M., Schäfer T., et al. (2009). Role of the twin-arginine translocation pathway in Staphylococcus. J. Bacteriol. 191, 5921–5929. 10.1128/jb.00642-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braun V. (2001). Iron uptake mechanisms and their regulation in pathogenic bacteria. Int. J. Med. Microbiol. 291, 67–79. 10.1078/1438-4221-00103 [DOI] [PubMed] [Google Scholar]
- Bray B. A., Sutcliffe I. C., Harrington D. J. (2009). Impact of lgt mutation on lipoprotein biosynthesis and in vitro phenotypes of Streptococcus agalactiae. Microbiology 155(Pt 5), 1451–1458. 10.1099/mic.0.025213-0 [DOI] [PubMed] [Google Scholar]
- Buddelmeijer N. (2015). The molecular mechanism of bacterial lipoprotein modification–how, when and why? FEMS Microbiol. Rev. 39, 246–261. 10.1093/femsre/fuu006 [DOI] [PubMed] [Google Scholar]
- Bulut Y., Faure E., Thomas L., Equils O., Arditi M. (2001). Cooperation of Toll-like receptor 2 and 6 for cellular activation by soluble tuberculosis factor and Borrelia burgdorferi outer surface protein A lipoprotein: role of Toll-interacting protein and IL-1 receptor signaling molecules in Toll-like receptor 2 signaling. J. Immunol. 167, 987–994. 10.4049/jimmunol.167.2.987 [DOI] [PubMed] [Google Scholar]
- Cockayne A., Hill P. J., Powell N. B., Bishop K., Sims C., Williams P. (1998). Molecular cloning of a 32-kilodalton lipoprotein component of a novel iron-regulated Staphylococcus epidermidis ABC transporter. Infect. Immun. 66, 3767–3774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cockfield J. D., Pathak S., Edgeworth J. D., Lindsay J. A. (2007). Rapid determination of hospital-acquired meticillin-resistant Staphylococcus aureus lineages. J. Med. Microbiol. 56(Pt 5), 614–619. 10.1099/jmm.0.47074-0 [DOI] [PubMed] [Google Scholar]
- Dale S. E., Sebulsky M. T., Heinrichs D. E. (2004). Involvement of SirABC in iron-siderophore import in Staphylococcus aureus. J. Bacteriol. 186, 8356–8362. 10.1128/jb.186.24.8356-8362.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diep B. A., Gill S. R., Chang R. F., Phan T. H., Chen J. H., Davidson M. G., et al. (2006). Complete genome sequence of USA300, an epidemic clone of community-acquired meticillin-resistant Staphylococcus aureus. Lancet 367, 731–739. 10.1016/S0140-673668231-7 [DOI] [PubMed] [Google Scholar]
- Diep B. A., Phung Q., Date S., Arnott D., Bakalarski C., Xu M., et al. (2014). Identifying potential therapeutic targets of methicillin-resistant Staphylococcus aureus through in vivo proteomic analysis. J. Infect. Dis. 209, 1533–1541. 10.1093/infdis/jit662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gan K., Sankaran K., Williams M. G., Aldea M., Rudd K. E., Kushner S. R., et al. (1995). The umpA gene of Escherichia coli encodes phosphatidylglycerol:prolipoprotein diacylglyceryl transferase (lgt) and regulates thymidylate synthase levels through translational coupling. J. Bacteriol. 177, 1879–1882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Götz F., Mayer S. (2013). Both terminal oxidases contribute to fitness and virulence during organ-specific Staphylococcus aureus colonization. MBio 4, e00976–e00913. 10.1128/mBio.00976-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grigg J. C., Vermeiren C. L., Heinrichs D. E., Murphy M. E. (2007). Heme coordination by Staphylococcus aureus IsdE. J. Biol. Chem. 282, 28815–28822. 10.1074/jbc.M704602200 [DOI] [PubMed] [Google Scholar]
- Hamilton A., Robinson C., Sutcliffe I. C., Slater J., Maskell D. J., Davis-Poynter N., et al. (2006). Mutation of the maturase lipoprotein attenuates the virulence of Streptococcus equi to a greater extent than does loss of general lipoprotein lipidation. Infect. Immun. 74, 6907–6919. 10.1128/IAI.01116-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer N. D., Reniere M. L., Cassat J. E., Zhang Y., Hirsch A. O., Indriati Hood M., et al. (2013). Two heme-dependent terminal oxidases power Staphylococcus aureus organ-specific colonization of the vertebrate host. MBio 4:e00241–13. 10.1128/mBio.00241-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hantke K., Braun V. (1973). Covalent binding of lipid to protein. Diglyceride and amide-linked fatty acid at the N-terminal end of the murein-lipoprotein of the Escherichia coli outer membrane. Eur. J. Biochem. 34, 284–296. [DOI] [PubMed] [Google Scholar]
- Heikkinen O., Seppala R., Tossavainen H., Heikkinen S., Koskela H., Permi P., et al. (2009). Solution structure of the parvulin-type PPIase domain of Staphylococcus aureus PrsA–implications for the catalytic mechanism of parvulins. BMC Struct. Biol. 9:17. 10.1186/1472-6807-9-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinrichs J. H., Gatlin L. E., Kunsch C., Choi G. H., Hanson M. S. (1999). Identification and characterization of SirA, an iron-regulated protein from Staphylococcus aureus. J. Bacteriol. 181, 1436–1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henneke P., Dramsi S., Mancuso G., Chraibi K., Pellegrini E., Theilacker C., et al. (2008). Lipoproteins are critical TLR2 activating toxins in group B streptococcal sepsis. J. Immunol. 180, 6149–6158. 10.4049/jimmunol.180.9.6149 [DOI] [PubMed] [Google Scholar]
- Hiron A., Borezée-Durant E., Piard J. C., Juillard V. (2007). Only one of four oligopeptide transport systems mediates nitrogen nutrition in Staphylococcus aureus. J. Bacteriol. 189, 5119–5129. 10.1128/jb.00274-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh M. J., Wharton S. J., Cox A. G., Ingham E., Peacock S., Foster S. J. (2002). MntR modulates expression of the PerR regulon and superoxide resistance in Staphylococcus aureus through control of manganese uptake. Mol. Microbiol. 44, 1269–1286. 10.1046/j.1365-2958.2002.02944.x [DOI] [PubMed] [Google Scholar]
- Hussain M., Ichihara S., Mizushima S. (1982). Mechanism of signal peptide cleavage in the biosynthesis of the major lipoprotein of the Escherichia coli outer membrane. J. Biol. Chem. 257, 5177–5182. [PubMed] [Google Scholar]
- Jeong D. W., Cho H., Jones M. B., Shatzkes K., Sun F., Ji Q., et al. (2012). The auxiliary protein complex SaePQ activates the phosphatase activity of sensor kinase SaeS in the SaeRS two-component system of Staphylococcus aureus. Mol. Microbiol. 86, 331–348. 10.1111/j.1365-2958.2012.08198.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousselin A., Manzano C., Biette A., Reed P., Pinho M., Rosato A., et al. (2015). The Staphylococcus aureus chaperone PrsA is a new auxiliary factor of oxacillin resistance affecting Penicillin-binding protein 2A. Antimicrob. Agents Chemother. 60, 1656–1666. 10.1128/AAC.02333-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousselin A., Renzoni A., Andrey D. O., Monod A., Lew D. P., Kelley W. L. (2012). The posttranslocational chaperone lipoprotein PrsA is involved in both glycopeptide and oxacillin resistance in Staphylococcus aureus. Antimicrob. Agents Chemother. 56, 3629–3640. 10.1128/AAC.06264-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juncker A. S., Willenbrock H., Von Heijne G., Brunak S., Nielsen H., Krogh A. (2003). Prediction of lipoprotein signal peptides in Gram-negative bacteria. Protein Sci. 12, 1652–1662. 10.1110/ps.0303703 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kehl-Fie T. E., Zhang Y., Moore J. L., Farrand A. J., Hood M. I., Rathi S., et al. (2013). MntABC and MntH contribute to systemic Staphylococcus aureus infection by competing with calprotectin for nutrient manganese. Infect. Immun. 81, 3395–3405. 10.1128/IAI.00420-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khandavilli S., Homer K. A., Yuste J., Basavanna S., Mitchell T., Brown J. S. (2008). Maturation of Streptococcus pneumoniae lipoproteins by a type II signal peptidase is required for ABC transporter function and full virulence. Mol. Microbiol. 67, 541–557. 10.1111/j.1365-2958.2007.06065.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kouwen T. R., van der Goot A., Dorenbos R., Winter T., Antelmann H., Plaisier M. C., et al. (2007). Thiol-disulphide oxidoreductase modules in the low-GC Gram-positive bacteria. Mol. Microbiol. 64, 984–999. 10.1111/j.1365-2958.2007.05707.x [DOI] [PubMed] [Google Scholar]
- Kurokawa K., Kim M. S., Ichikawa R., Ryu K. H., Dohmae N., Nakayama H., et al. (2012). Environment-mediated accumulation of diacyl lipoproteins over their triacyl counterparts in Staphylococcus aureus. J. Bacteriol. 194, 3299–3306. 10.1128/JB.00314-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurokawa K., Lee H., Roh K. B., Asanuma M., Kim Y. S., Nakyama H., et al. (2009). The triacylated ATP binding cluster transporter substrate-binding lipoprotein of Staphylococcus aureus functions as a native ligand for the toll-like receptor 2. J. Biol. Chem. 284, 8406–8411. 10.1074/jbc.M809618200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machata S., Tchatalbachev S., Mohamed W., Jänsch L., Hain T., Chakraborty T. (2008). Lipoproteins of Listeria monocytogenes are critical for virulence and TLR2-mediated immune activation. J. Immunol. 181, 2028–2035. 10.4049/jimmunol.181.3.2028 [DOI] [PubMed] [Google Scholar]
- Magadum S., Banerjee U., Murugan P., Gangapur D., Ravikesavan R. (2013). Gene duplication as a major force in evolution. J. Genet. 92, 155–161. 10.1007/s12041-013-0212-8 [DOI] [PubMed] [Google Scholar]
- Makgotlho P. E., Marincola G., Schäfer D., Liu Q., Bae T., Geiger T., et al. (2013). SDS interferes with SaeS signaling of Staphylococcus aureus independently of SaePQ. PLoS ONE 8:e71644. 10.1371/journal.pone.0071644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mariotti P., Malito E., Biancucci M., Lo Surdo P., Mishra R. P., Nardi-Dei V., et al. (2013). Structural and functional characterization of the Staphylococcus aureus virulence factor and vaccine candidate FhuD2. Biochem. J. 449, 683–693. 10.1042/BJ20121426 [DOI] [PubMed] [Google Scholar]
- Mazmanian S. K., Skaar E. P., Gaspar A. H., Humayun M., Gornicki P., Jelenska J., et al. (2003). Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299, 906–909. 10.1126/science.1081147 [DOI] [PubMed] [Google Scholar]
- Mazmanian S. K., Ton-That H., Schneewind O. (2001). Sortase-catalysed anchoring of surface proteins to the cell wall of Staphylococcus aureus. Mol. Microbiol. 40, 1049–1057. 10.1046/j.1365-2958.2001.02411.x [DOI] [PubMed] [Google Scholar]
- Mazmanian S. K., Ton-That H., Su K., Schneewind O. (2002). An iron-regulated sortase anchors a class of surface protein during Staphylococcus aureus pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 99, 2293–2298. 10.1073/pnas.032523999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merlin C., Gardiner G., Durand S., Masters M. (2002). The Escherichia coli metD locus encodes an ABC transporter which includes Abc (MetN), YaeE (MetI), and YaeC (MetQ). J. Bacteriol. 184, 5513–5517. 10.1128/JB.184.19.5513-5517.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrissey J. A., Cockayne A., Hill P. J., Williams P. (2000). Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus. Infect. Immun. 68, 6281–6288. 10.1128/IAI.68.11.6281-6288.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller P., Müller-Anstett M., Wagener J., Gao Q., Kaesler S., Schaller M., et al. (2010). The Staphylococcus aureus lipoprotein SitC colocalizes with Toll-like receptor 2 (TLR2) in murine keratinocytes and elicits intracellular TLR2 accumulation. Infect. Immun. 78, 4243–4250. 10.1128/IAI.00538-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubauer H., Pantel I., Lindgren P. E., Götz F. (1999). Characterization of the molybdate transport system ModABC of Staphylococcus carnosus. Arch. Microbiol. 172, 109–115. [DOI] [PubMed] [Google Scholar]
- Nguyen M. T., Götz F. (2016). Lipoproteins of Gram-Positive bacteria: key players in the immune response and virulence. Microbiol. Mol. Biol. Rev. 80, 891–903. 10.1128/MMBR.00028-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M. T., Hanzelmann D., Hartner T., Peschel A., Götz F. (2015a). Skin-specific unsaturated fatty acids boost the Staphylococcus aureus innate immune response. Infect. Immun. 84, 205–215. 10.1128/IAI.00822-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen M. T., Kraft B., Yu W., Demicrioglu D. D., Hertlein T., Burian M., et al. (2015b). The νSaα specific Lipoprotein Like Cluster (lpl) of S. aureus USA300 contributes to immune stimulation and invasion in human cells. PLoS Pathog. 11:e1004984. 10.1371/journal.ppat.1004984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J. B., Lampen J. O. (1982a). Glyceride-cysteine lipoproteins and secretion by Gram-positive bacteria. J. Bacteriol. 152, 315–322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J. B., Lampen J. O. (1982b). Membrane-bound penicillinases in Gram-positive bacteria. J. Biol. Chem. 257, 4490–4495. [PubMed] [Google Scholar]
- Petit C. M., Brown J. R., Ingraham K., Bryant A. P., Holmes D. J. (2001). Lipid modification of prelipoproteins is dispensable for growth in vitro but essential for virulence in Streptococcus pneumoniae. FEMS Microbiol. Lett. 200, 229–233. 10.1111/j.1574-6968.2001.tb10720.x [DOI] [PubMed] [Google Scholar]
- Podkowa K. J., Briere L. A., Heinrichs D. E., Shilton B. H. (2014). Crystal and solution structure analysis of FhuD2 from Staphylococcus aureus in multiple unliganded conformations and bound to ferrioxamine-B. Biochemistry 53, 2017–2031. 10.1021/bi401349d [DOI] [PubMed] [Google Scholar]
- Rahman O., Cummings S. P., Harrington D. J., Sutcliffe I. C. (2008). Methods for the bioinformatic identification of bacterial lipoproteins encoded in the genomes of Gram-positive bacteria. World J. Microbiol. Biotechnol. 24, 2377–2382. 10.1007/s11274-008-9795-2 [DOI] [Google Scholar]
- Remy L., Carrière M., Derré-Bobillot A., Martini C., Sanguinetti M., Borezee-Durant E. (2013). The Staphylococcus aureus Opp1 ABC transporter imports nickel and cobalt in zinc-depleted conditions and contributes to virulence. Mol. Microbiol. 87, 730–743. 10.1111/mmi.12126 [DOI] [PubMed] [Google Scholar]
- Robinson D. A., Kearns A. M., Holmes A., Morrison D., Grundmann H., Edwards G., et al. (2005). Re-emergence of early pandemic Staphylococcus aureus as a community-acquired meticillin-resistant clone. Lancet 365, 1256–1258. 10.1016/S0140-673674814-5 [DOI] [PubMed] [Google Scholar]
- Sander P., Rezwan M., Walker B., Rampini S. K., Kroppenstedt R. M., Ehlers S., et al. (2004). Lipoprotein processing is required for virulence of Mycobacterium tuberculosis. Mol. Microbiol. 52, 1543–1552. 10.1111/j.1365-2958.2004.04041.x [DOI] [PubMed] [Google Scholar]
- Sankaran K., Wu H. C. (1994). Lipid modification of bacterial prolipoprotein. Transfer of diacylglyceryl moiety from phosphatidylglycerol. J. Biol. Chem. 269, 19701–19706. [PubMed] [Google Scholar]
- Schluepen C., Malito E., Marongiu A., Schirle M., McWhinnie E., Lo Surdo P., et al. (2013). Mining the bacterial unknown proteome: identification and characterization of a novel family of highly conserved protective antigens in Staphylococcus aureus. Biochem. J. 455, 273–284. 10.1042/BJ20130540 [DOI] [PubMed] [Google Scholar]
- Schmaler M., Jann N. J., Ferracin F., Landolt L. Z., Biswas L., Götz F., et al. (2009). Lipoproteins in Staphylococcus aureus mediate inflammation by TLR2 and iron-dependent growth in vivo. J. Immunol. 182, 7110–7118. 10.4049/jimmunol.0804292 [DOI] [PubMed] [Google Scholar]
- Schmaler M., Jann N. J., Götz F., Landmann R. (2010). Staphylococcal lipoproteins and their role in bacterial survival in mice. Int. J. Med. Microbiol. 300, 155–160. 10.1016/j.ijmm.2009.08.018 [DOI] [PubMed] [Google Scholar]
- Schmollinger M., Fischer I., Nerz C., Pinkenburg S., Götz F., Kaufmann M., et al. (2004). ParSeq: searching motifs with structural and biochemical properties. Bioinformatics 20, 1459–1461. 10.1093/bioinformatics/bth083 [DOI] [PubMed] [Google Scholar]
- Sebulsky M. T., Heinrichs D. E. (2001). Identification and characterization of fhuD1 and fhuD2, two genes involved in iron-hydroxamate uptake in Staphylococcus aureus. J. Bacteriol. 183, 4994–5000. 10.1128/JB.183.17.4994-5000.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebulsky M. T., Hohnstein D., Hunter M. D., Heinrichs D. E. (2000). Identification and characterization of a membrane permease involved in iron-hydroxamate transport in Staphylococcus aureus. J. Bacteriol. 182, 4394–4400. 10.1128/JB.182.16.4394-4400.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebulsky M. T., Speziali C. D., Shilton B. H., Edgell D. R., Heinrichs D. E. (2004). FhuD1, a ferric hydroxamate-binding lipoprotein in Staphylococcus aureus: a case of gene duplication and lateral transfer. J. Biol. Chem. 279, 53152–53159. 10.1074/jbc.M409793200 [DOI] [PubMed] [Google Scholar]
- Sibbald M. J., Ziebandt A. K., Engelmann S., Hecker M., de Jong A., Harmsen H. J., et al. (2006). Mapping the pathways to staphylococcal pathogenesis by comparative secretomics. Microbiol. Mol. Biol. Rev. 70, 755–788. 10.1128/MMBR.00008-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Skabytska Y., Wölbing F., Gunther C., Koberle M., Kaesler S., Chen K. M., et al. (2014). Cutaneous innate immune sensing of Toll-like receptor 2–6 ligands suppresses T cell immunity by inducing myeloid-derived suppressor cells. Immunity 41, 762–775. 10.1016/j.immuni.2014.10.009 [DOI] [PubMed] [Google Scholar]
- Stoll H., Dengjel J., Nerz C., Götz F. (2005). Staphylococcus aureus deficient in lipidation of prelipoproteins is attenuated in growth and immune activation. Infect. Immun. 73, 2411–2423. 10.1128/IAI.73.4.2411-2423.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe I. C., Harrington D. J. (2002). Pattern searches for the identification of putative lipoprotein genes in Gram-positive bacterial genomes. Microbiology 148(Pt 7), 2065–2077. 10.1099/00221287-148-7-2065 [DOI] [PubMed] [Google Scholar]
- Sutcliffe I. C., Harrington D. J., Hutchings M. I. (2012). A phylum level analysis reveals lipoprotein biosynthesis to be a fundamental property of bacteria. Protein Cell 3, 163–170. 10.1007/s13238-012-2023-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K., Takeuchi O., Akira S. (2002). Recognition of lipopeptides by Toll-like receptors. J. Endotoxin Res. 8, 459–463. 10.1179/096805102125001073 [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Kawai T., Mühlradt P. F., Morr M., Radolf J. D., Zychlinsky A., et al. (2001). Discrimination of bacterial lipoproteins by Toll-like receptor 6. Int. Immunol. 13, 933–940. 10.1093/intimm/13.7.933 [DOI] [PubMed] [Google Scholar]
- Takeuchi O., Sato S., Horiuchi T., Hoshino K., Takeda K., Dong Z., et al. (2002). Cutting edge: role of Toll-like receptor 1 in mediating immune response to microbial lipoproteins. J. Immunol. 169, 10–14. 10.4049/jimmunol.169.1.10 [DOI] [PubMed] [Google Scholar]
- Taylor P. D., Toseland C. P., Attwood T. K., Flower D. R. (2006). LIPPRED: A web server for accurate prediction of lipoprotein signal sequences and cleavage sites. Bioinformation 1, 176–179. 10.6026/97320630001176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- von Heijne G. (1989). The structure of signal peptides from bacterial lipoproteins. Protein Eng. 2, 531–534. [DOI] [PubMed] [Google Scholar]
- Vu C. H., Kolata J., Stentzel S., Beyer A., Salazar M. G., Steil L., et al. (2016). Adaptive immune response to lipoproteins of Staphylococcus aureus in healthy subjects. Proteomics. [Epub ahead of print]. 10.1002/pmic.201600151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang P., Dalbey R. E. (2011). Inserting membrane proteins: the YidC/Oxa1/Alb3 machinery in bacteria, mitochondria, and chloroplasts. Biochim. Biophys. Acta 1808, 866–875. 10.1016/j.bbamem.2010.08.014 [DOI] [PubMed] [Google Scholar]
- Weston B. F., Brenot A., Caparon M. G. (2009). The metal homeostasis protein, Lsp, of Streptococcus pyogenes is necessary for acquisition of zinc and virulence. Infect. Immun. 77, 2840–2848. 10.1128/IAI.01299-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams W. A., Zhang R. G., Zhou M., Joachimiak G., Gornicki P., Missiakas D., et al. (2004). The membrane-associated lipoprotein-9 GmpC from Staphylococcus aureus binds the dipeptide GlyMet via side chain interactions. Biochemistry 43, 16193–16202. 10.1021/bi048877o [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zähringer U., Lindner B., Inamura S., Heine H., Alexander C. (2008). TLR2 — promiscuous or specific? A critical re-evaluation of a receptor expressing apparent broad specificity. Immunobiology 213, 205–224. 10.1016/j.imbio.2008.02.005 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


