Abstract
Study Objectives:
We sought to examine how much of the heritability of self-report sleep duration is tagged by common genetic variation in populations of European ancestry and to test if the common variants contributing to sleep duration are also associated with other diseases and traits.
Methods:
We utilized linkage disequilibrium (LD)-score regression to estimate the heritability tagged by common single nucleotide polymorphisms (SNPs) in the CHARGE consortium genome-wide association study (GWAS) of self-report sleep duration. We also used bivariate LD-score regression to investigate the genetic correlation of sleep duration with other publicly available GWAS datasets.
Results:
We show that 6% (SE = 1%) of the variance in self-report sleep duration in the CHARGE study is tagged by common SNPs in European populations. Furthermore, we find evidence of a positive genetic correlation (rG) between sleep duration and type 2 diabetes (rG = 0.26, P = 0.02), and between sleep duration and schizophrenia (rG = 0.19, P = 0.01).
Conclusions:
Our results show that increased sample sizes will identify more common variants for self-report sleep duration; however, the heritability tagged is small when compared to other traits and diseases. These results also suggest that those who carry variants that increase risk to type 2 diabetes and schizophrenia are more likely to report longer sleep duration.
Citation:
Byrne EM, Gehrman PR, Trzaskowski M, Tiemeier H, Pack AI. Genetic correlation analysis suggests association between increased self-reported sleep duration in adults and schizophrenia and type 2 diabetes. SLEEP 2016;39(10):1853–1857.
Keywords: genetics, sleep duration, GWAS, type 2 diabetes
Significance.
Sleep duration is an important epidemiological variable and both short and long sleep have been associated with a range of negative outcomes. In this paper, we demonstrate that genetic variants that are common in populations of European ancestry explain 6% of the total variance in sleep duration and more genetic variants that highlight biological pathways underlying sleep duration will be found with larger studies. Moreover, we find that genetic variants that increase sleep duration also show evidence of increasing risk to schizophrenia and type 2 diabetes. This suggests that association between short sleep and diabetes may be due to nongenetic factors. Future studies should follow up known genetic variants for schizophrenia and diabetes to test for a role in sleep regulation
INTRODUCTION
Both short and long sleep duration have been associated with a number of negative health outcomes including obesity,1–5 cardiovascular disease,6–8 hypertension,2,9 depression,10 and diabetes mellitus.11,12 However, the mechanisms behind these associations remain elusive. It is unclear whether environmental changes such as increased stress lead to a change in sleep patterns and thus increased risk to disease, or whether there are shared biological mechanisms between sleep regulation and a range of diseases.
A nontrivial proportion of the variance in sleep duration is due to genetic variation. A recent genome-wide association study (GWAS) from the CHARGE consortium that included more than 47,000 participants from 18 population-based cohorts identified 2 genome-wide significant signals on chromo-some 2 and chromosome 6, respectively.13 The signal from chromosome 2, found between the PAX8 and CBWD2 genes, replicated in a cohort of 4,400 African Americans. Nominal associations for the associated variants were found with metabolic traits such as glycated hemoglobin and also with attention deficit hyperactivity disorder, thus highlighting potential shared etiology between sleep duration and metabolic and psychiatric disorders.
Despite such a large sample size, it is perhaps surprising that only two genome-wide significant loci were found in the study. This raises questions about the genetic architecture of sleep duration. Is the reason for not finding more common variants associated due to the sample size being too small, or is the majority of the heritability explained by rare variants not well tagged by GWAS chips used in the CHARGE analysis? The aim of this study was to investigate how much of the overall heritability of sleep duration that is explained by common single nucleotide polymorphisms (SNPs) and not yet found by GWAS, and to investigate whether variants that predispose to short or long sleep also predispose or protect against diseases and disease-related traits.
METHODS
We utilized a recently developed method called linkage disequilibrium (LD)-score regression for assessing the heritability explained by common SNPs. This method can distinguish between population stratification and polygenicity in GWAS studies and requires only summary statistics rather than actual genotypes.14
LD-score regression has also been extended to permit estimation of the genetic correlation between traits using GWAS summary statistics.15 We obtained the summary statistics from a number of large GWAS studies across a range of traits of potential relevance to sleep duration. In each instance, the GWAS summary statistics were freely available for download for academic research purposes. We could therefore test whether there is evidence for shared genetic risk factors between sleep duration and a number of disease-related outcomes. It has been noted that genetic correlations are likely to only be meaningful where the heritability z-score of the individual traits estimated from univariate LD-score regression are greater than 4.15 We therefore only include studies where the z-score was greater than 4. For each trait, we calculated the genetic correlation (rG) with the CHARGE sleep duration results.
We obtained summary statistics from the CHARGE GWAS study of self-report sleep duration. The study is described in detail in the published primary GWAS study. Briefly, the discovery sample included 18 community-based cohorts that included only participants of European ancestry. Phenotype data were obtained from standardized questionnaires and self-completion questionnaires. The measure of sleep duration varied across studies. Where possible, weekday sleep duration was used as the measure for analysis. We use the genome-wide summary statistics from approximately 45,000 individuals.
We applied the same quality control steps as described by Bulik-Sullivan et al.15 to select SNPs for inclusion from each study. Only SNPs with minor allele frequency (MAF) ≥ 5% were used. As imputation quality scores were not available for the CHARGE study, HapMap 3 SNPs were used because of their ability to be well imputed in most studies. All of the studies included only individuals of European ancestry.
RESULTS
Using LD-score regression with SNPs included in HapMap 3 (950,000 SNPs), we estimate that 6.2% of the variance in sleep duration is explained by common variants (SE = 1.4%). This corresponds to a heritability z-score of 4.38.
Genetic Correlations with Sleep Duration
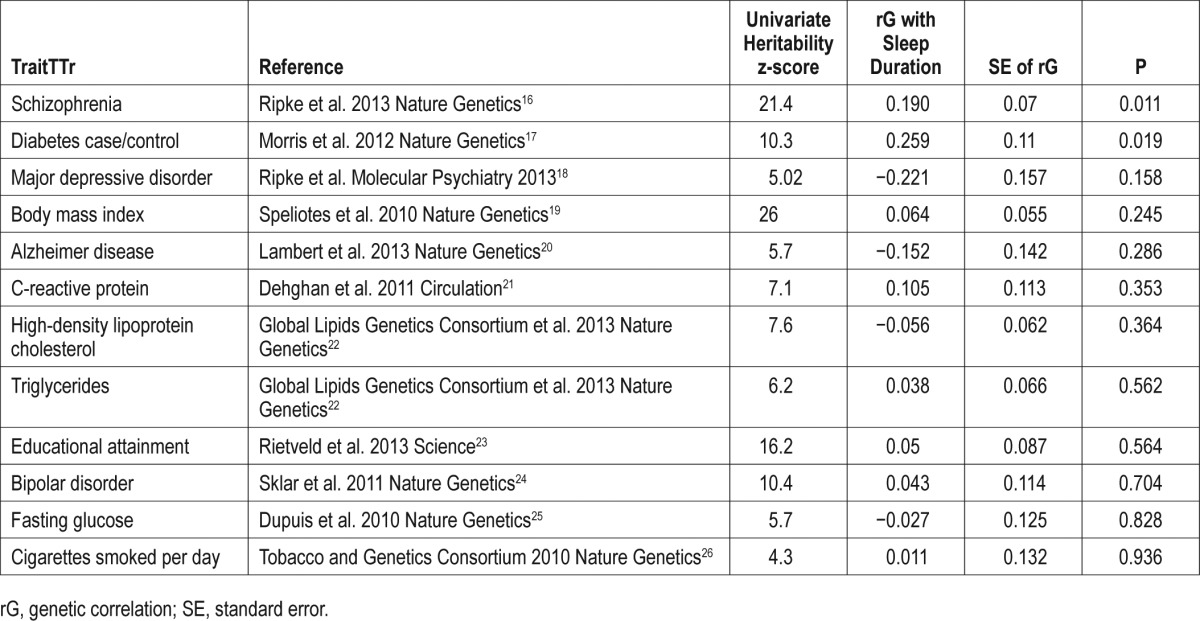
The results of the genetic correlation analysis16–26 are presented in Table 1. The results have been sorted to present those with the highest statistical significance first.
Table 1.
Results from LD-score regression bivariate analyses by trait.
DISCUSSION
We estimate that 6% of the variance of sleep duration in the CHARGE meta-analysis is explained by common SNPs. This indicates that studies that include even larger sample sizes than the CHARGE study will identify more common variants associated with sleep duration and reveal more about the underlying biological regulation of sleep.
When compared to other traits and diseases, however, 6% is on the smaller end of estimates of variance explained by variants on GWAS chips. For example, the variance explained by common SNPs for Crohn disease is 23%,27 for intelligence it is 40%,28 for body mass index it is 21%,29 and for bipolar disorder it is 35%.27
There are a number of reasons why the estimate is comparably small. In the CHARGE study, genomic control was applied to each cohort individually prior to the meta-analysis. As described by Bulik-Sullivan et al.14, this may have led to some polygenic signal being removed. Therefore, the estimate given here may have been lower than the true value. It is also possible that the heritability across all of the cohorts in CHARGE is comparably small or that the genetic architecture of sleep duration is skewed toward rare variants not tagged by GWAS chips. For nearly every trait studied so far, common variants explain nearly one-half of the overall heritability. Currently, there is no reason to believe that sleep duration has an architecture different from other traits. It is more likely that the overall heritability of sleep duration across the entire meta-analysis is low because of the heterogeneity of the individual studies. As described in the GWAS, there were differences in how sleep duration was assessed and some studies only had information on weekend sleep duration.
Twin studies have estimated the narrow-sense heritability of sleep duration to be between 0.09 and 0.44.30,31 However, twin studies rely on correlations between individuals of the same age to estimate the genetic contribution to sleep duration. It is known that sleep duration declines with age and environmental influences change throughout life. It has not been demonstrated that the heritability is the same across different age groups and it is possible that some of the genetic variants that influence sleep duration in young adulthood are not the same as those that influence it in older adults. There is substantial variation in the mean age among cohorts included in the CHARGE study. For instance, the mean age of the Young Finns Study is 37.7 and the Queensland Institute of Medical Research sample is 34.5, whereas the mean age in the Cardiovascular Health Study is 77.9 and in the Rotterdam Study I, it is 76.1. The genome-wide significant variant was found to have a higher effect size in the older cohorts compared to the younger ones. It is reasonable to conclude that combining data from multiple cohorts from different countries, with different age profiles and different questionnaires for assessing sleep duration, leads to an increase in the proportion of environmental variance and thus there is less genetic signal to detect.
In order to test this hypothesis, LD-score analysis would need to be applied in a very large cohort with a less variable age profile and where the same method of assessing sleep duration is used. Finding a higher estimate than the 6% found in this study would give credence to the idea that different variants might be affecting sleep duration in different age groups, and that the within-sample heritability of sleep duration is higher than that across all samples. This is an important direction of future research for understanding the genetics of self-report sleep duration.
Furthermore self-report sleep duration is a somewhat unreliable measure of sleep. Previous studies have found the correlation of self-report sleep with objectively measured sleep to be 0.45.32 Objectively measured sleep duration may be a more heritable phenotype and have a higher contribution to the heritability from common SNPs.
Genetic Correlations with Sleep Duration
None of the genetic correlations were significant at a Bonferroni-corrected significance threshold of P < 0.004. However, some of the tests are correlated and so Bonferroni correction is likely overly conservative in this instance. The most statistically significant genetic correlations with sleep duration are with schizophrenia (P = 0.011) and type 2 diabetes (P = 0.019). Intriguingly, in both cases, the estimate of the genetic correlation is positive, implying that genes that increase sleep duration also increase risk of diabetes and schizophrenia. It should be noted that the statistical significance of the estimate is influenced by the sample size of the GWAS for each trait, in addition to how much variance is explained by common SNPs for that trait. For example, the GWAS of schizophrenia included approximately 14,000 cases and 18,000 controls and the proportion of variance explained by the SNPs is 30%. By contrast, common variants only tag approximately 3% of the variation in cigarettes smoked per day.
Despite these differences that affect power, it is informative to look at the estimates of rG. In addition to the positive correlations with diabetes and schizophrenia, there are also large negative correlation estimates with major depressive disorder (MDD) (−0.221) and Alzheimer disease (−0.152). The negative correlation suggests that genetic variants that increase sleep duration decrease risk of MDD or Alzheimer disease. Changes in sleep form part of an MDD diagnosis. This can include both insomnia and hypersomnia; however, insomnia is far more commonly reported in MDD than hypersomnia.
It has previously been observed that there are shared genetic risk factors between psychiatric disorders.33 MDD shares a substantial amount of genetic risk with schizophrenia, bipolar disorder, etc. It might therefore be expected that the genetic correlations with sleep duration would be similar across disorders. However, this is not the case. Genetic variants for schizophrenia and MDD appear to have opposite correlations with sleep duration, whereas there is little evidence of a correlation between bipolar disorder and sleep duration. A possible explanation for this is that the genetic risk that is unique to MDD and not shared with other psychiatric disorders influences sleep duration.
There have been a number of studies of the relationship between sleep quality and schizophrenia, and they have consistently shown that self-reported sleep quality is poorer in cases than in controls. Less focus has been placed on sleep duration specifically. One study that used actigraphy to measure sleep in cases and controls found that although schizophrenia cases took longer to fall asleep than controls, their total sleep duration was significantly longer.34 Another GWAS study of sleep duration identified an associated variant in the DRD2 gene that increased sleep duration and also increased risk to schizophrenia.35 A polygenic risk score constructed from 12 SNPs genome-wide significant for schizophrenia was also associated with increased sleep duration, supporting the findings of the genetic correlation analysis.
The relationship between type 2 diabetes risk and sleep duration has been investigated in a number of studies. A recent meta-analysis of 10 studies confirmed that there is a U-shaped distribution of risk with sleep duration.36 Both short and long sleepers are at increased risk of the development of type 2 diabetes compared to those who sleep 7 to 8 h per night. The results presented here suggest that the genetic variants that predispose to type 2 diabetes also predispose to longer sleep duration. Therefore, there may be environmental factors that lead to short sleep duration and risk of the development of diabetes. One previous study evaluated whether type 2 diabetes risk variants are associated with sleep duration and did not find an association.37 However, the study was based on 27 genome-wide significant SNPs and was not a genome-wide analysis, which may explain the difference in findings to the current study.
The genetic correlation with type 2 diabetes contrasts with the finding that one of the genome-wide significant variants from the CHARGE GWAS that is associated with increased sleep duration is also associated with a better metabolic profile. The genetic correlation analysis uses a set of SNPs that are representative of common SNPs across the genome. Therefore, the genetic correlation suggests an overall pattern of increased risk of type 2 diabetes for alleles that lengthen self-report sleep; some individual alleles may have opposite effects.
The negative correlation with Alzheimer disease supports findings from a number of studies showing decreases in sleep duration and efficiency associated with an increasing load of β-amyloid.37,38 The results presented here suggest that shared genetic risk factors play a role in this relationship.
A number of other disorders and traits that have previously been found to be associated with decreased sleep duration such as body mass index, cigarette smoking, triglycerides, and fasting glucose do not show evidence of genetic correlation with sleep duration. Because of statistical power, it is not possible to rule out shared genetic factors, but our results suggest that any association is likely to be due to environmental factors.
The results of this study have implications for future research. Genetic variants associated with diabetes and schizophrenia should be investigated further for their potential role in sleep regulation. Although no variants have yet been found for MDD, it may be useful to incorporate measures of sleep into future studies of depression in order to help identify genetic associations. Future studies with larger, more homogeneous populations may help untangle the genetic architecture of sleep duration further. Large studies with objective measures of sleep duration are also needed.
DISCLOSURE STATEMENT
This was not an industry supported study. Dr. Byrne is supported by an Early Career Fellowship from the National Health and Medical Research Council of Australia. Dr. Gehrman has received research support from Merck and has consulted for General Sleep Corp. The other authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
The authors thank all of the consortia for making the results publicly available.
REFERENCES
- 1.Buxton OM, Marcelli E. Short and long sleep are positively associated with obesity, diabetes, hypertension, and cardiovascular disease among adults in the United States. Soc Sci Med. 2010;71:1027–36. doi: 10.1016/j.socscimed.2010.05.041. [DOI] [PubMed] [Google Scholar]
- 2.Altman NG, Izci-Balserak B, Schopfer E, et al. Sleep duration versus sleep insufficiency as predictors of cardiometabolic health outcomes. Sleep Med. 2012;13:1261–70. doi: 10.1016/j.sleep.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Klingenberg L, Sjodin A, Holmback U, Astrup A, Chaput JP. Short sleep duration and its association with energy metabolism. Obes Rev. 2012;13:565–77. doi: 10.1111/j.1467-789X.2012.00991.x. [DOI] [PubMed] [Google Scholar]
- 4.Kong AP, Wing YK, Choi KC, et al. Associations of sleep duration with obesity and serum lipid profile in children and adolescents. Sleep Med. 2011;12:659–65. doi: 10.1016/j.sleep.2010.12.015. [DOI] [PubMed] [Google Scholar]
- 5.Knutson KL. Does inadequate sleep play a role in vulnerability to obesity? Am J Hum Biol. 2012;24:361–71. doi: 10.1002/ajhb.22219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cappuccio FP, Cooper D, D'Elia L, Strazzullo P, Miller MA. Sleep duration predicts cardiovascular outcomes: a systematic review and meta-analysis of prospective studies. Eur Heart J. 2011;32:1484–92. doi: 10.1093/eurheartj/ehr007. [DOI] [PubMed] [Google Scholar]
- 7.Cappuccio FP, Miller MA. Are short bad sleep nights a hindrance to a healthy heart? Sleep. 2011;34:1457–8. doi: 10.5665/sleep.1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cappuccio FP, Stranges S, Kandala NB, et al. Gender-specific associations of short sleep duration with prevalent and incident hypertension: the Whitehall II Study. Hypertension. 2007;50:693–700. doi: 10.1161/HYPERTENSIONAHA.107.095471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim SJ, Lee SK, Kim SH, et al. Genetic association of short sleep duration with hypertension incidence--a 6-year follow-up in the Korean genome and epidemiology study. Circ J. 2012;76:907–13. doi: 10.1253/circj.cj-11-0713. [DOI] [PubMed] [Google Scholar]
- 10.Watson NF, Harden KP, Buchwald D, et al. Sleep duration and depressive symptoms: a gene-environment interaction. Sleep. 2014;37:351–8. doi: 10.5665/sleep.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morselli LL, Guyon A, Spiegel K. Sleep and metabolic function. Pflugers Arch. 2012;463:139–60. doi: 10.1007/s00424-011-1053-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zizi F, Pandey A, Murrray-Bachmann R, et al. Race/ethnicity, sleep duration, and diabetes mellitus: analysis of the National Health Interview Survey. Am J Med. 2012;125:162–7. doi: 10.1016/j.amjmed.2011.08.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gottlieb DJ, Hek K, Chen TH, et al. Novel loci associated with usual sleep duration: the CHARGE Consortium Genome-Wide Association Study. Mol Psychiatry. 2015;20:1232–9. doi: 10.1038/mp.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bulik-Sullivan BK, Loh PR, Finucane HK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bulik-Sullivan B, Finucane HK, Anttila V, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ripke S, O'Dushlaine C, Chambert K, et al. Genome-wide association analysis identifies 13 new risk loci for schizophrenia. Nat Genet. 2013;45:1150–9. doi: 10.1038/ng.2742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ripke S, Wray NR, Lewis CM, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatry. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Speliotes EK, Willer CJ, Berndt SI, et al. Association analyses of 249,796 individuals reveal 18 new loci associated with body mass index. Nat Genet. 2010;42:937–48. doi: 10.1038/ng.686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambert JC, Ibrahim-Verbaas CA, Harold D, et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer's disease. Nat Genet. 2013;45:1452–8. doi: 10.1038/ng.2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dehghan A, Dupuis J, Barbalic M, et al. Meta-analysis of genome-wide association studies in > 80 000 subjects identifies multiple loci for C-reactive protein levels. Circulation. 2011;123:731–8. doi: 10.1161/CIRCULATIONAHA.110.948570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Global Lipids Genetics Consortium. Willer CJ, Schmidt EM, et al. Discovery and refinement of loci associated with lipid levels. Nat Genet. 2013;45:1274–83. doi: 10.1038/ng.2797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rietveld CA, Medland SE, Derringer J, et al. GWAS of 126,559 individuals identifies genetic variants associated with educational attainment. Science. 2013;340:1467–71. doi: 10.1126/science.1235488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sklar P, Ripke S, Scott LJ, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–83. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42:105–16. doi: 10.1038/ng.520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tobacco and Genetics Consortium. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–7. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee SH, Wray NR, Goddard ME, Visscher PM. Estimating missing heritability for disease from genome-wide association studies. Am J Hum Genet. 2011;88:294–305. doi: 10.1016/j.ajhg.2011.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davies G, Tenesa A, Payton A, et al. Genome-wide association studies establish that human intelligence is highly heritable and polygenic. Mol Psychiatry. 2011;16:996–1005. doi: 10.1038/mp.2011.85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Locke AE, Kahali B, Berndt SI, et al. Genetic studies of body mass index yield new insights for obesity biology. Nature. 2015;518:197–206. doi: 10.1038/nature14177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heath AC, Kendler KS, Eaves LJ, Martin NG. Evidence for genetic influences on sleep disturbance and sleep pattern in twins. Sleep. 1990;13:318–35. doi: 10.1093/sleep/13.4.318. [DOI] [PubMed] [Google Scholar]
- 31.Partinen M, Kaprio J, Koskenvuo M, Putkonen P, Langinvainio H. Genetic and environmental determination of human sleep. Sleep. 1983;6:179–85. doi: 10.1093/sleep/6.3.179. [DOI] [PubMed] [Google Scholar]
- 32.Lauderdale DS, Knutson KL, Yan LL, Liu K, Rathouz PJ. Self-reported and measured sleep duration: how similar are they? Epidemiology. 2008;19:838–45. doi: 10.1097/EDE.0b013e318187a7b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cross-Disorder Group of the Psychiatric Genomics C. Lee SH, Ripke S, et al. Genetic relationship between five psychiatric disorders estimated from genome-wide SNPs. Nat Genet. 2013;45:984–94. doi: 10.1038/ng.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wulff K, Dijk DJ, Middleton B, Foster RG, Joyce EM. Sleep and circadian rhythm disruption in schizophrenia. Br J Psychiatry. 2012;200:308–16. doi: 10.1192/bjp.bp.111.096321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cade BE, Gottlieb DJ, Lauderdale DS, et al. Common variants in DRD2 are associated with sleep duration: the CARe consortium. Hum Mol Genet. 2016;25:167–79. doi: 10.1093/hmg/ddv434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shan Z, Ma H, Xie M, et al. Sleep duration and risk of type 2 diabetes: a meta-analysis of prospective studies. Diabetes Care. 2015;38:529–37. doi: 10.2337/dc14-2073. [DOI] [PubMed] [Google Scholar]
- 37.Mander BA, Marks SM, Vogel JW, et al. β-amyloid disrupts human NREM slow waves and related hippocampus-dependent memory consolidation. Nat Neurosci. 2015;18:1051–7. doi: 10.1038/nn.4035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spira AP, Gamaldo AA, An Y, et al. Self-reported sleep and beta-amyloid deposition in community-dwelling older adults. JAMA Neurol. 2013;70:1537–43. doi: 10.1001/jamaneurol.2013.4258. [DOI] [PMC free article] [PubMed] [Google Scholar]


