Abstract
Study Objectives:
Low or excessive sleep duration has been associated with multiple outcomes, but the biology behind these associations remains elusive. Specifically, genetic studies in children are scarce. In this study, we aimed to: (1) estimate the proportion of genetic variance of sleep duration in children attributed to common single nucleotide polymorphisms (SNPs), (2) identify novel SNPs associated with sleep duration in children, and (3) investigate the genetic overlap of sleep duration in children and related metabolic and psychiatric traits.
Methods:
We performed a population-based molecular genetic study, using data form the EArly Genetics and Life course Epidemiology (EAGLE) Consortium. 10,554 children of European ancestry were included in the discovery, and 1,250 children in the replication phase.
Results:
We found evidence of significant but modest SNP heritability of sleep duration in children (SNP h2 0.14, 95% CI [0.05, 0.23]) using the LD score regression method. A novel region at chromosome 11q13.4 (top SNP: rs74506765, P = 2.27e-08) was associated with sleep duration in children, but this was not replicated in independent studies. Nominally significant genetic overlap was only found (rG = 0.23, P = 0.05) between sleep duration in children and type 2 diabetes in adults, supporting the hypothesis of a common pathogenic mechanism.
Conclusions:
The significant SNP heritability of sleep duration in children and the suggestive genetic overlap with type 2 diabetes support the search for genetic mechanisms linking sleep duration in children to multiple outcomes in health and disease.
Citation:
Marinelli M, Pappa I, Bustamante M, Bonilla C, Suarez A, Tiesler CM, Vilor-Tejedor N, Zafarmand MH, Alvarez-Pedrerol M, Andersson S, Bakermans-Kranenburg MJ, Estivill X, Evans DM, Flexeder C, Forns J, Gonzalez JR, Guxens M, Huss A, van IJzendoorn MH, Jaddoe VW, Julvez J, Lahti J, López-Vicente M, Lopez-Espinosa MJ, Manz J, Mileva-Seitz VR, Perola M, Pesonen AK, Rivadeneira F, Salo PP, Shahand S, Schulz H, Standl M, Thiering E, Timpson NJ, Torrent M, Uitterlinden AG, Smith GD, Estarlich M, Heinrich J, Räikkönen K, Vrijkotte TG, Tiemeier H, Sunyer J. Heritability and genome-wide association analyses of sleep duration in children: the EAGLE consortium. SLEEP 2016;39(10):1859–1869.
Keywords: SNP heritability, genome-wide association study (GWAS), meta-analysis, childhood sleep duration, pathway analysis
Significance.
Sleep duration has been an alluring research topic, yet its genetic underpinnings in children remain mainly uncovered. The present study used novel population-based molecular genetic methods, in the largest sample of children available to date, to provide an unbiased estimate of SNP (single nucleotide polymorphism) heritability of sleep duration in children (measured by parent-rated questionnaires). Moreover, new chromosomal regions associated with sleep duration in children were identified and may serve as subject to further research. Finally, this study extended the epidemiological findings of association between sleep duration and impaired glucose metabolism, providing evidence of a common genetic infrastructure, beginning early in life.
INTRODUCTION
Sleep duration, defined as the total amount of sleep obtained across a 24-hour period, is a general indicator of sleep need and overall well-being.1 The effects of insufficient or excessive sleep duration have been extensively investigated in child and adult samples, and strong associations have been found with a variety of medical2–7 and psychosocial outcomes.8–14 What is still unknown is whether common genetic variants explain part of the individual variation of sleep duration in children, and whether these genetic variants provide a mechanism delineating the central role of sleep duration in health and disease.
Sleep duration is considered a complex phenomenon that can be influenced by several extrinsic factors, such as the use of medication and other substances,15,16 or a variety of environmental and medical conditions,17 with substantial individual variability across all ages.18 This individual variability in sleep duration could be due to an underlying genetic component. Twin studies in adults provide evidence of low to moderate heritability of sleep duration (ranging from 0.00–0.55),19–22 and a genome-wide linkage study further supported this finding (0.17, 95% CI [0.01–0.33]).23 The few twin studies in children indicate moderate heritability of sleep duration (ranging from 0.26–0.58).24–26 However, recent approaches have been developed (i.e., Genome-wide Complex Trait Analysis (GCTA)27 and LD score regression28), which can estimate heritability based on single nucleotide polymorphisms (SNP heritability, SNP h2) and that can be used to verify these preliminary findings in population-based samples.
A number of studies have tried to identify the specific genetic variants associated with sleep duration, mainly in adults. Candidate gene studies provide evidence of association between sleep duration and genetic variants within DEC2,29 CLOCK,30 ARNTL,31 and SORCS132 genes, among others. Additionally, a small genome-wide linkage study (N = 749) indicated a chromosomal region (chromosome 3, 71.3Mb) related to sleep duration,23 while a Finnish genome-wide association study (GWAS) of N = 8,554 individuals identified genetic variation near the KLF6 (i.e., rs2031573) and the PCDH7-CENTD1 (i.e., rs1037079) genes associated with sleep duration, via differential gene expression.33
Another GWAS including seven European samples (total N = 4,251) identified an intronic genetic variant within ABCC9 (i.e., rs1104205) associated with sleep duration,34 but the result could not be replicated in an independent sample of young adults (N = 952).35 Recently, a large GWAS meta-analysis of 47,180 individuals of European ancestry reported a genome-wide significant association at rs1823125 with sleep duration (upstream PAX8, a transcription factor important for development and maintenance of thyroid function), which was replicated in an independent sample of 4,747 African-American individuals.36 However, little attention has been given to genome-wide approaches of sleep duration in children. Since young children's sleep patterns are less influenced by external factors than adults (e.g., use of alarm clock, usage of caffeine/ sleeping pills, working shifts),16,37 genetic studies in children could provide us useful insights into the mechanisms of normal variation of sleep duration.
In this study, we analysed data from five population-based cohorts assessing parent-reported sleep duration in N discovery = 10,554 children of European ancestry. First, a GWAS meta-analysis was performed to identify novel SNPs associated with sleep duration in children, and a replication of our top findings was attempted in two independent samples (Nreplication = 1,250). Second, we performed gene-set enrichment analyses to identify possible functional pathways associated with sleep duration in children. Third, we estimated SNP heritability of sleep duration in children using approaches implemented in the GCTA27 and LD score regression methods.28 Finally, we formally compared the results of the GWAS meta-analysis in children with the results of previously published genetic studies of sleep duration in adults, and with other related metabolic, and psychiatric traits. To our knowledge, this is the largest study to date investigating the genetic component of sleep duration in children using multiple, partly novel methods.
METHODS
Subjects
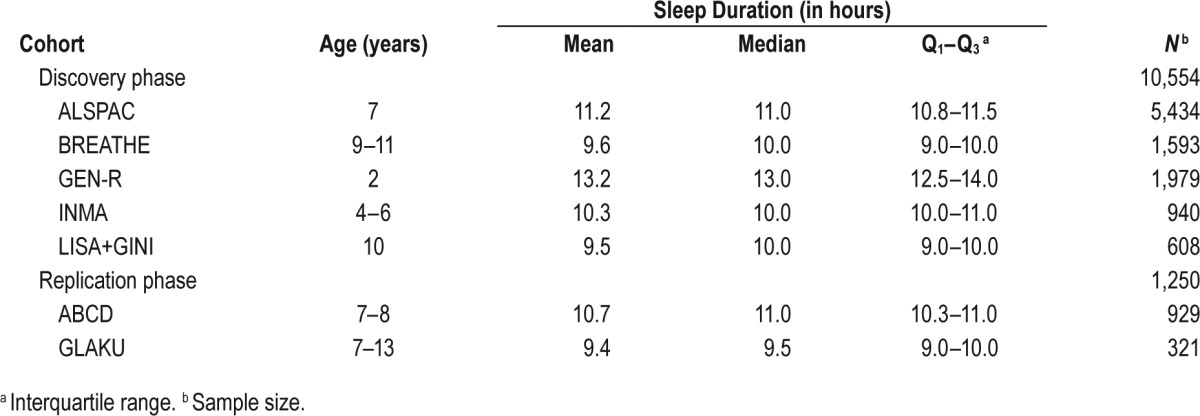
This study was performed within the framework of the EArly Genetics and Lifecourse Epidemiology (EAGLE) Consortium.38 The study was divided in two phases: discovery and replication. In the discovery phase, we combined data from five population-based cohorts (i.e., the Avon Longitudinal Study of Parents and Children [ALSPAC], the Brain development and Air pollution ultrafine particles in school children [BREATHE], the Generation R Study [GEN-R], the Infancia y Medio Ambiente project [INMA], and the influence of Lifestyle factors on the development of the Immune System and Allergies in East and West Germany PLUS the influence of traffic emissions and genetics study and the German Infant study on the influence of Nutrition Intervention PLUS environmental and genetic influences on allergy development [LISA+GINI]). In the replication phase, data from two population-based cohorts were used (i.e., Amsterdam Born Children and their Development [ABCD] and Glycyrrhizin in Licorice [GLAKU]). Information from the participating cohorts is summarized in Table 1, and a more detailed description can be found in the supplemental material. Cohorts could participate if they fulfilled the following inclusion criteria: European descent, and age of the children between 2 to 14 years at the time of assessment. This age range was selected to avoid developmental periods that influence sleep duration in children younger than 2 years,38 and in adolescents (> 14 years).39 Each study was conducted with appropriate institutional ethics approval, and written informed consent was obtained from all participants.
Table 1.
Characteristics of all cohorts participating in the GWAS meta-analysis (discovery and replication phase) of sleep duration in children.
Phenotype Definition
In all cohorts (except in GLAKU), child sleep duration was assessed by a single, parent-rated, open question, “How many hours does your child sleep per day including naps?” In GLAKU, parents were asked about the usual bed and rise times during school days, from which the total sleep duration could be estimated. A detailed description of all questions used to evaluate sleep duration in children can be found in supplemental material (under Individual study cohort descriptions).
Genotyping, Quality Control, and Imputation
DNA was extracted from whole blood or buccal cells. Genotype information within each cohort was collected using high-density SNP arrays on Illumina platforms for ALSPAC, ABCD, BREATHE, GEN-R, GLAKU, and INMA cohorts, while for LISA+GINI the Affymetrix platform was used. In all cohorts participating in this study, basic quality checks were performed (supplemental material, Table S1). Samples were also checked for excess heterozygosity, sex accuracy, relatedness, and missing data. Following these quality control steps, phased genotype data were imputed to 1000 GENOME release March 2012 reference panel (http://mathgen.stats.ox.ac.uk/impute/ALL_1000G_phase1integrated_v3_impute.tgz), resulting in up to ∼30 million SNPs for GWAS analysis.
GWAS analyses, Quality Control (QC), and Meta-Analysis
GWAS analyses in unrelated individuals were performed within each of the five cohorts participating in the discovery phase. Association analyses were performed using two linear regression models. In the first model (basic model), sleep duration (in hours) was regressed on age, sex, and principal components of the genetic data to account for population stratification, and allele dosage (obtained from the imputed data, for more details see supplemental material, Table S1). The second model (body mass index [BMI] adjusted model) was additionally adjusted for BMI, based on previous reports on genetic overlap of shorter sleep duration and increased BMI in adults.40
Prior to the GWAS meta-analysis, rigorous quality control (QC) was performed by two independent researchers, using the EasyQC software package41 or a custom code in R.42 SNPs with minor allele frequency (MAF) ≥ 0.01 and accurate imputation (MACH r2 ≥ 0.4 or IMPUTE2 INFO ≥ 0.3) were considered for further analysis. Due to the limited sample size of some of the cohorts, additional filtering based on expected minor allele counts (EMAC) was done. This parameter is related to both the sample size (N) and the quality of imputation (2*N*MAF*quality of imputation). SNPs that did not reach an EMAC ≥ 100 were excluded. Genomic inflation as estimated by the lambda (λ) parameter was calculated in each study. After the QC filters were implemented, between 5 to 8 million SNPs were kept for further analysis in each cohort. Before the meta-analysis, marker names and alleles were harmonized among cohorts and the effect allele frequencies were compared.
After quality control, a fixed-effect inverse variance weighted meta-analysis of sleep duration in the five discovery cohorts was performed. This method is implemented in METAL (Meta-Analysis Helper).43 First we meta-analysed the data from our first model and then we proceeded to the second, BMI adjusted model. Only SNPs evaluated in at least 3 cohorts were considered. A threshold of P ≤ 5.00e-08 was used to define genome-wide levels of significance. A threshold of P ≤ 1.00e-05 was used to indicate suggestive associations of a SNP with the outcome. Quantile-quantile (Q-Q) plots, estimations of lambda (λ) and Manhattan plots were performed in R. A regional association plot of the top SNP was performed with Locus Zoom.44
Genetic Annotation and Enrichment Analysis
Genetic variants annotation (nearest gene, position in the gene, expression quantitative trait loci (eQTLs),and regulatory features) was performed with the HaploRegv3 program.45 In addition, eQTLs were investigated using data from two available public databases: the Genotype-Tissue Expression (GTEx) project46 and through the web tool of the UK Brain Expression Consortium (BRAINEAC, http://www.braineac.org/).
For the enrichment analysis, we selected the loci that passed the suggestive level of genome-wide significance (P < 1.00e-05), resulting in 20 lead SNPs. The suggestive cutoff was selected because these SNPs may represent true positives that did not reach genome-wide significance due to power issues.33 Molecular and functional enrichment analyses were performed using several tools to allow comparison. First, the identification of tissues/cell types where genes from associated loci are highly expressed was evaluated with DEPICT.47 DEPICT was also used to identify enriched gene-sets. Second, the PANTHER pathway-classification tool was used to sort the annotated genes derived from our GWAS meta-analysis efforts into functional pathways.48 For the overrepresentation of these genes in specific pathways, the PANTHER Gene Ontology (GO)-Slim Biological Process annotation dataset was used. Both approaches use the False Discovery Rate (FDR) to correct for multiple-testing.
SNP Heritability of Sleep Duration and Genetic Correlation Estimations
To estimate SNP heritability (SNP h2) of sleep duration in children, we used two approaches: LD score regression based on GWAS meta-analysis summary statistics, and Genome-Wide Complex Trait Analysis (GCTA) on directly genotyped data, in single cohorts. The LD score regression method has been explained previously in detail,49 but the basic principle is to estimate SNP heritability from the correlation between the marginal effect size of a SNP and a measure of its linkage disequilibrium (LD) with other SNPs. For LD score regression, the summary statistics of our GWAS meta-analysis in children (basic and BMI adjusted models) were used. Since genomic control (GC) can bias the heritability estimates downwards,50 we applied no GC for LD score regression in the meta-analysis.
In addition, GCTA was performed in the largest cohort participating in the discovery phase of this study (i.e., ALSPAC), to quantify the variance tagged by common SNPs in each cohort. The GCTA method has been explained in detail elsewhere.27 In summary, a genetic relatedness matrix (GRM) of unrelated individuals was estimated in the ALSAPC cohort. Related participants (pairwise genetic relatedness > 0.025) were excluded from further analysis. Finally, a restricted maximum likelihood method (REML) was used to partition the phenotypic similarity between unrelated individuals into a genetic and residual component. GCTA estimate was adjusted for age, sex and the principal components of the genotype data.
To estimate SNP h2 of sleep duration in adults, the LD score regression method was applied on the summary statistics of the largest GWAS meta-analysis of sleep duration in adults (Nadults = 47,180).36
Finally, LD score regression50 was performed to estimate the genetic correlation between sleep duration in children and related traits in adults and children. For these analyses, we first used the summary statistics from the largest GWAS meta-analysis on sleep duration in adults (Nadults = 47,180)36 and compared these with the summary statistics of the GWAS meta-analysis in children (this study, Nchildren = 10,554). Second, we computed the genetic correlation between sleep duration in children and common metabolic traits (i.e., obesity in children51 [available by the Early Growth Genetics Consortium, EGG; 5,530 cases and 8,318 controls], type 2 diabetes52 [available by the DIAGRAM consortium; 12,171 cases and 56,862 controls], and 2-hour glucose levels53 [available by the Meta-analyses of Glucose and Insulin-related traits Consortium, MAGIC; 46,186 non-diabetic subjects]). Third, we computed the genetic correlation between sleep duration in children and common psychiatric traits (i.e., bipolar disorder54 [7,481 cases and 9,250 controls], major depression55 [9,240 cases and 9,519 controls], and schizophrenia56 [31,335 cases and 38,765 controls] in adults, and attention deficit/hyperactivity disorder [ADHD] in children/family trios57 [896 cases and 2,455 controls] using available GWAS summary statistics from the Psychiatric Genomics Consortium [PGC]).
Replication of SNPs Previously Reported in the Literature
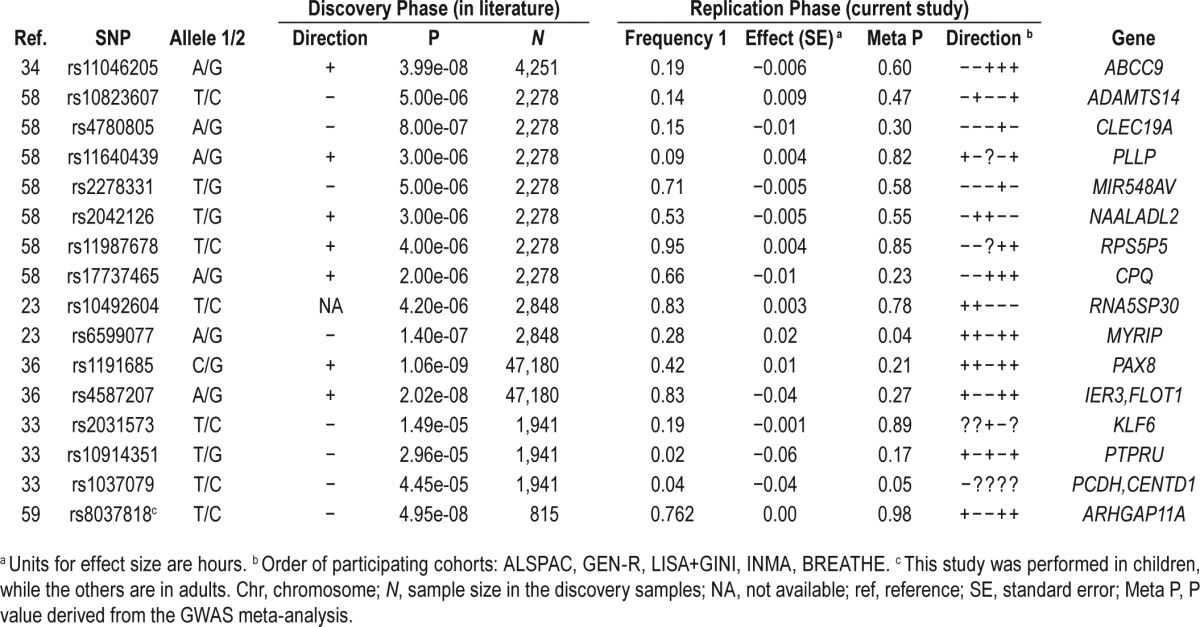
To compare the results of the current GWAS meta-analysis in children with previously reported associations of genetic variants with sleep duration, an extensive literature search was performed. We identified all the GWAS of sleep duration reported in humans23,33,34,36,58,59 and selected the lead SNP in all reported loci, regardless of the genome-wide significance. We ended up with 16 SNPs and tested whether they could be replicated in the summary statistics of our GWAS meta-analysis (basic model).
Finally, we selected SNPs associated with sleep duration in adults (P value < 1.00e-3 and at least 10,000) from the largest GWAS meta-analysis on sleep duration in adults,36 and compared their association with results from this study in children.
RESULTS
GWAS Meta-Analysis
In the discovery phase, five cohorts contributed data yielding a total of N = 10,554 children from 2 to 11 years (Table 1). As expected, the mean sleep duration in young children (GEN-R, 2 years old) was higher than in older children (BREATHE, 9–11 years old). The distribution of sleep duration in children was approximately normal in all participating cohorts. No additional transformation was applied. Study-level and meta-level QC was implemented in all studies (supplemental material, Table S1, Figure S1, and Figure S2), and the summary statistics from the five cohorts were meta-analysed. For the basic model (adjusted for age, sex, and principal components), the meta-analysis revealed a genome-wide significant locus at chromo-some 11q13.4 (top SNP: rs74506765, P = 2.27e-08). There was no evidence of heterogeneity between studies (heterogeneity P = 0.30), and the direction of the effect was positive for all cohorts (i.e., the minor allele C was associated with increased sleep duration in children). The top SNP (rs74506765) is located in an intronic region of the ARAP1 gene. The regional association plot of this locus is presented in Figure 1, and shows a linkage disequilibrium block covering ARAP1 gene. All suggestive SNPs (P < 1.00e-05) are summarized in Table 2. The Manhattan and Q-Q plots of the GWAS meta-analysis (basic model), are presented in Figure 2 and Figure 3, respectively.
Figure 1.
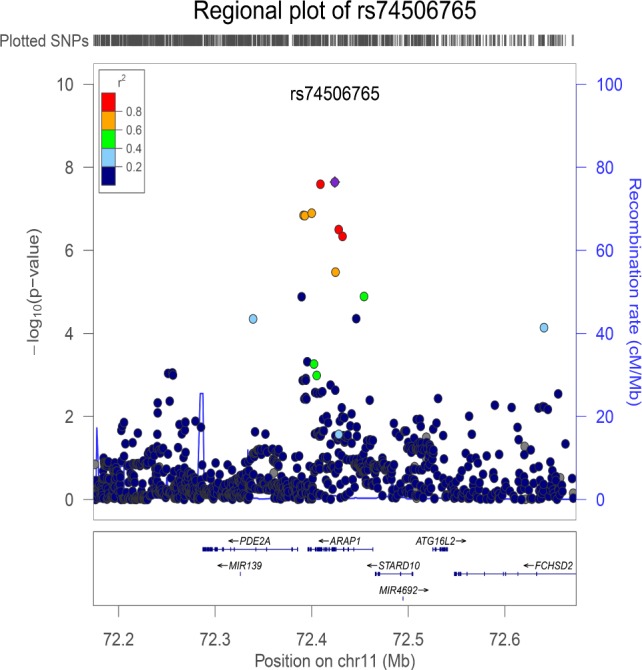
Regional association plot of the chromosome 11 locus (top SNP: rs74506765) with sleep duration in children (N = 10,554). The top SNP is indicated with a diamond and the flanking SNPs in circles, colored according to their linkage disequilibrium (LD). The plot was constructed by 1000 Genomes, CEU population (Northern and Western European ancestry).
Table 2.
Top signals that reached suggestive genome wide significance (P < 1.00e-05), sorted by ascending P, in the discovery phase (N = 10,554), for the basic model.
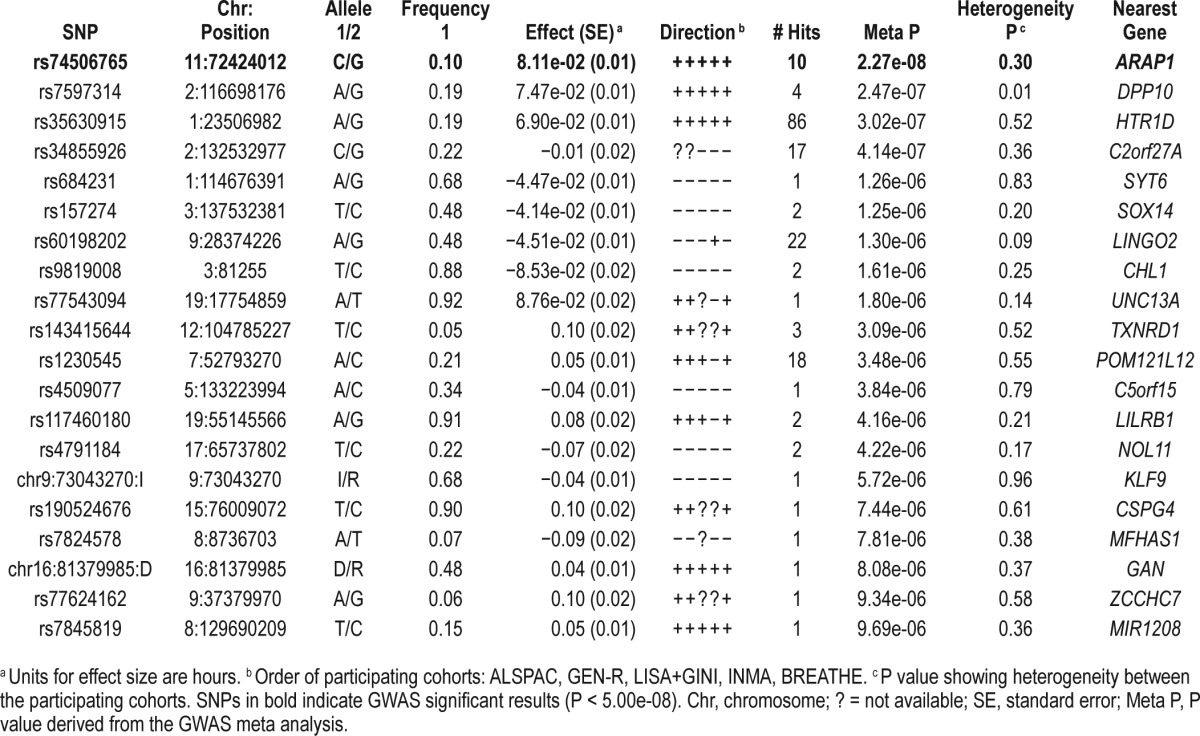
Figure 2.
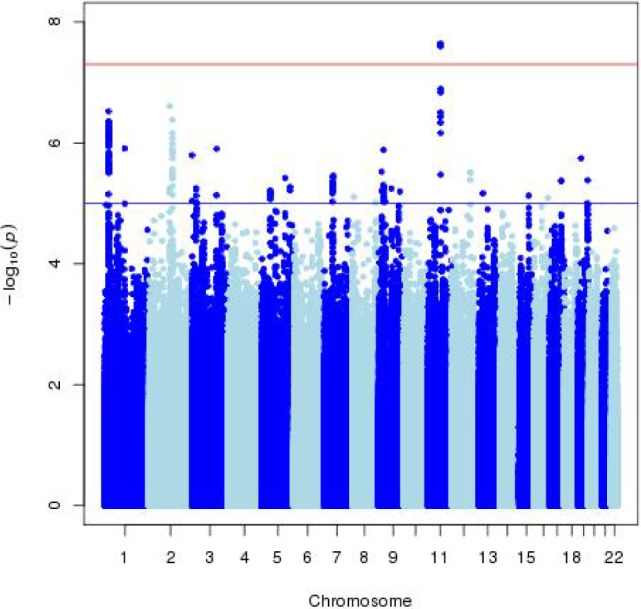
Manhattan plot of the GWAS meta-analysis of sleep duration in children, in the discovery phase, for the basic model (N = 10,554). The x-axis represents the autosomal chromosomes and the y-axis represents the –log10(p). The red line indicates genome-wide significance (P = 5.00e-08) and the blue line indicates suggestive genome-wide significance (P = 1.00e-05)
Figure 3.
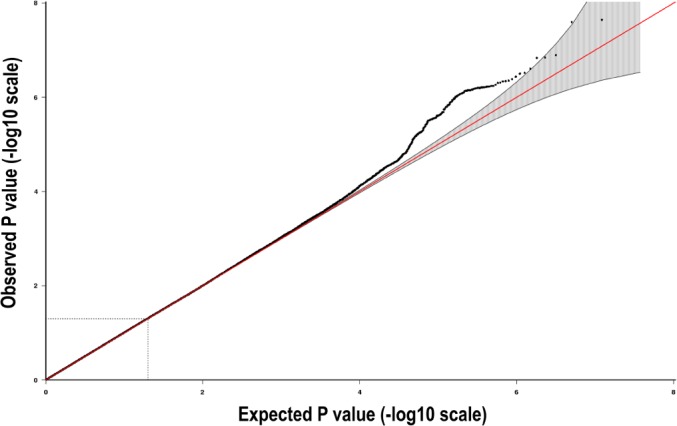
Quantile-quantile (QQ) plot showing the probability values from GWAS meta-analysis of sleep duration, in the discovery phase, for the basic model (N = 10,554). The red line indicates the distribution under the null hypothesis and the shaded area indicates the 95% confidence band
The GWAS meta-analysis of the BMI adjusted model (i.e., additionally adjusted for body mass index), in N = 10,502 children yielded similar results to the basic model (top SNP: rs7121351, chromosome 11q13.4, P = 2.29e-08). The suggestive SNPs, as well as the Manhattan and Q-Q plot of the GWAS meta-analysis on sleep duration for the BMI adjusted model, are presented in the supplemental material (Table S2, Figure S3, and Figure S4, respectively).
Replication Analyses
Replication efforts were attempted for the significant top signal (rs74506765) derived from our GWAS meta-analysis in two independent, population-based cohorts (i.e., ABCD and GLAKU). The characteristics of these cohorts are summarized in Table 1. The association analyses did not replicate our initial finding, indicating a non-significant association for the ABCD sample (beta = 0.01, SE = 0.08, P = 0.95, N = 929), and a nominally significant, but opposite direction association for the GLAKU sample (beta = −0.27, SE = 0.11, P = 0.015, N = 321).The results of the replication analysis are presented in more detail in supplemental material (Table S3a). The combined P value of our meta-analysis (discovery and replication sample, N = 11,804) was 2.70e-06 (not genome-wide significant). Figure 4 shows the forest plot of the association ofrs74506765 with sleep duration across all participating cohorts (i.e., in both discovery and replication phase).
Figure 4.
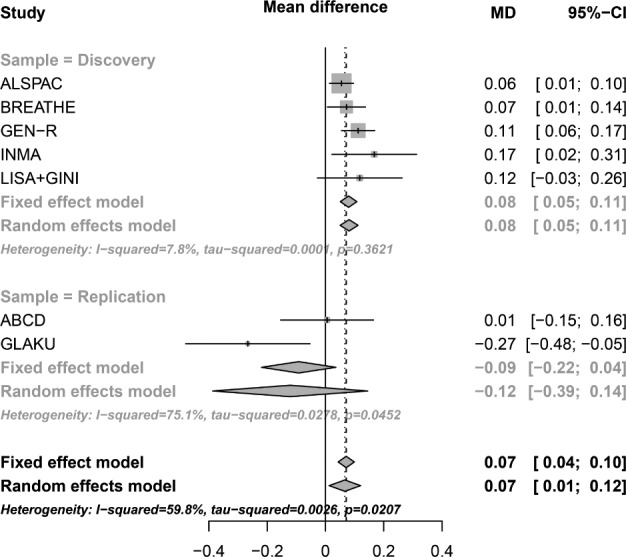
Forest plot for the top hit of the GWAS meta-analysis on sleep duration in children. In the vertical panel, the studies participating in the discovery or replication phase are presented. In the horizontal lines, the boxes represent precision and the lines the confidence intervals. The diamond shapes represent the pooled effect estimates per allele, for both the fixed- and random-effect models. The horizontal axis shows the scale of the effect estimates. MD, mean difference; CI, confidence interval.
We also investigated whether our genome-wide significant locus would be replicated in adult samples, making use the available summary statistics of the largest GWAS meta-analysis of sleep duration in adults (Nadults = 47,180).36 Since the top SNP (rs74506765) was not found in the GWAS meta-analysis of sleep duration in adults, we searched for proxies (r2 threshold ≥ 0.6) within 1 Mb, using the SNAP (SNP annotation and Proxy Search) tool.60 We found no association between the proxy SNPs and sleep duration in adults, although there was a trend of association for rs7121351 (beta = 0.02, SE = 0.01, P = 0.07, N = 38,398, r2 = 0.93). These results are presented in more detail in supplemental material (Table S3b).
Annotation and Enrichment Analysis
The lead SNP from the genome-wide significant locus at chromosome 11q13.4 (top SNP: rs74506765) was not in linkage disequilibrium with any non-synonymous variant or genome-wide eQTLs, according to HaploReg v3, GTEX and BRAINEAC.DEPICT analyses did not show any relevant tissue/cell type enriched among the suggestive locus (supplemental material, Table S4). Several gene-sets were nominally enriched, but none of them passed multiple-testing. The top 10 pathways are presented in supplemental material (Table S5). Similarly, PANTHER pathway analysis did not indicate any significantly enriched pathways associated with sleep duration in children (data not shown).
SNP Heritability of Sleep Duration and Genetic Correlation Estimations
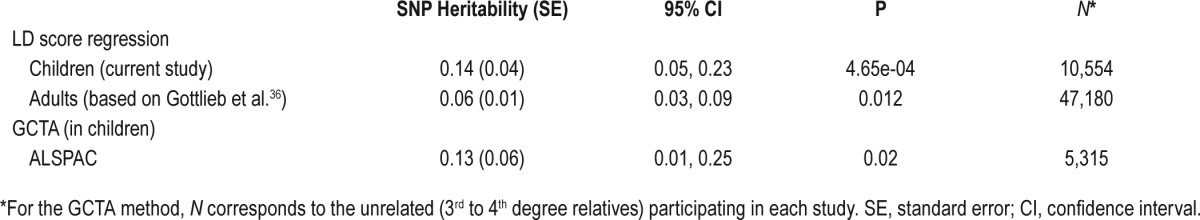
First, we used LD score regression on the summary statistics of our GWAS meta-analysis in N = 10,554 children (basic model) to estimate SNP heritability (SNP h2). Our results indicate a significant but modest SNP h2 (95% CI [0.05–0.23], P < 0.05) of sleep duration in children. The estimated SNP h2 using the GCTA method in the largest cohort participating in this study supported the evidence of low to modest SNP h2 (95% CI [0.01–0.25], Nunrelated = 5,315). The estimates of heritability, using both methods, are described in more detail in Table 3. Finally, we estimated SNP h2 of sleep duration in adults by using LD score regression in Nadults = 47,180, and found also modest heritability (95% CI [0.03–0.09], P < 0.05).
Table 3.
Estimates of SNP heritability of sleep duration in children and adults, based on LD score regression and GCTA methods.
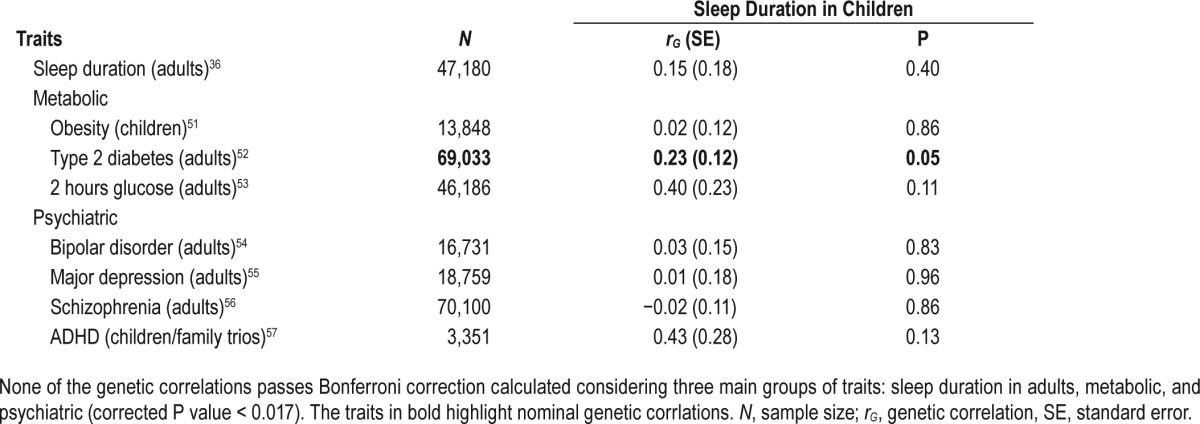
To estimate the genetic correlation between sleep duration in children and other relative traits, cross-trait LD score regression was used. We found no significant genetic correlation between sleep duration in children and sleep duration in adults, nor between sleep duration in children and common psychiatric traits (i.e., bipolar disorder, major depression, and schizophrenia). The genetic correlation of sleep duration in children and ADHD (rG = 0.43, SE = 0.28, P = 0.13) although nonsignificant, was larger than the genetic correlation with other psychiatric traits, indicating potentially a substantial genetic overlap which we were unable to estimate precisely due to low statistical power. No significant genetic correlation was found between sleep duration in children and obesity in children. A nominally significant genetic correlation was however found between sleep duration in children and type 2 diabetes (T2D) in adults (rG = 0.23, SE = 0.12, P = 0.05), and a trend was found between sleep duration in children and 2-h glucose levels in adults (rG = 0.40, SE = 0.26, P = 0.11). The genetic correlation between sleep duration in children and T2D in adults remained unchanged (rG = 0.22, SE = 0.12, P = 0.05) after exclusion of our top loci (chromosome 11q13.4) and after correcting for BMI. The estimates of the genetic correlations based on our basic model are presented in more detail in Table 4. Similar genetic correlations of sleep duration in children and related traits were also obtained by the summary statistics of our BMI adjusted model. These results are presented in supplemental material (Table S6).
Table 4.
Genetic correlations among sleep duration in children (derived by the GWAS summary statistics of the current study, under the basic model, (N = 10,554) and common metabolic and psychiatric traits, using LD score regression and GWAS summary statistics data, available in the literature.
Replication of SNPs Previously Reported in the Literature
Our literature search identified 16 SNPs previously reported in the literature which are associated (with suggestive or genome-wide significance) with individual differences in sleep duration mainly in adults. A comparison of these SNPs with the results of our GWAS meta-analysis on sleep duration in children (basic model) is presented in Table 5. None of the SNPs tested was associated with sleep duration in children.
Table 5.
Replication of SNPs previously associated with individual differences in sleep duration, with summary statistics reported in the current study (GWAS meta-analysis of sleep duration in children, basic model, N = 10,554).
Furthermore, we selected top SNPs (P < 1.00e-03 and sample size > 10,000) from Gottlieb et al., the largest meta-GWAS study of sleep duration performed in adults.36 Among the 3,436 shared SNPs among both studies, only 49 of them were nominally associated and in the same direction in children. Figure S5 shows the Q-Q plot of the association in children of the 3,436 top SNPs reported in adults.
DISCUSSION
In this study, we presented the first GWAS meta-analysis and SNP heritability estimates of sleep duration in N = 10,554 children of European ancestry. We identified a genome wide significant locus at chromosome 11q13.4 covering several SNPs, without significant heterogeneity among the participating cohorts. The minor allele of the top SNP within this locus (rs74506765, beta = 8.11e-02, SE = 0.01, P = 2.27e-08) was associated with longer sleep duration in children. This polymorphism is located in an intronic region of ARAP1 gene, a phosphatidylinositol 3,4,5-triphosphate-dependent GTPase-activating gene that has been previously associated type 2 diabetes (T2D), and other related metabolic traits,61–65 and with rheumatoid arthritis in Japanese samples.66 However, the linkage disequilibrium between SNPs reported in the literature and the one detected in this study is limited (r2 < 0.1). The rs74506765 variant was not replicated as a top hit in two independent samples of children, indicating either a false positive finding in the discovery phase, or lack of statistical power in the replication phase given the small size of the replication studies (13% estimated power using post hoc power analysis).
To further investigate the genetic architecture of sleep duration in children, we also performed gene-enrichment and pathway analyses, using the results of our GWAS meta-analysis. Several epidemiological studies have reported observational correlations between sleep duration and adverse metabolic traits in children and/or adults (e.g., T2D, obesity).67 In line with these studies, we identified nominally significant enrichment of the “attenuation of insulin receptor signalling cascade” gene-set, which however did not survive after multiple testing correction.
So far, twin studies have been the only source of information regarding the heritability of sleep duration in children, indicating moderate levels of heritability (ranging from 0.26–0.58).24–26 In this study, we extended the previous findings by estimating SNP heritability (SNP h2) based both on GWAS meta-analysis results using the LD score regression method,28 and on directly genotyped SNPs derived from the largest individual cohort participating in this study using the GCTA method.27 SNP h2 based on our GWAS meta-analysis summary statistics indicated low to moderate heritability of sleep duration in children (95% CI [0.05, 0.23]). SNP h2 based on individual cohort data supported the finding of low to moderate heritability (95% CI [0.01- 0.25], Nunrelated = 5,315 for the largest ALSPAC sample). This SNP h2 estimation indicates that large sample sizes are needed to more accurately estimate the heritability of sleep duration in children.
The estimations of SNP h2 presented in this study are lower than the estimations previously described in twin studies.24–26 Since the previous twin studies have been performed in very young children (6 to 48 months age), a possible explanation is that over age, the influences of external factors (e.g., school schedule, parental behaviors) on sleep duration increases, while the relative contribution of genetic variants decreases. Indeed, SNP h2 estimations based on the summary statistics of the largest GWAS meta-analysis on sleep duration in adults36 indicated modest heritability (95% CI [0.03, 0.09]). An alternative or additional explanation could be that the heritability estimates from twin studies are inflated because of the use of a single informant (i.e., parent) or because of the violation of the assumption of equal environments in monozygotic and dizygotic twins.68 Finally, SNP h2 is an underestimation of the total heritability of any single trait, because it does not take into account rare polymorphic and structural variation, and epistatic effects.69
Previous epidemiological studies have identified associations between variation of sleep duration and metabolic traits, such as obesity in children,2 and T2D6 and glucose metabolism70 in adults. Furthermore, differences in sleep duration have been associated with psychiatric traits, such as ADHD in children,13 and bipolar disorder,10 major depression,71 and schizophrenia72 in adults. These associations could signify the simple coexistence/comorbidity of metabolic and psychiatric traits with variation of sleep duration, or they could signify a common underlying pathogenic mechanism. In this study, we estimated the genetic correlations between sleep duration in children and the above-mentioned metabolic and psychiatric traits. We found one nominally significant genetic overlap between T2D and sleep duration in children (rG = 0.23, P = 0.05), implying that the same genetic variants influence longer sleep duration in children and increase the possibility of developing T2D later in life. Similar genetic overlap was also found between sleep duration in adults and T2D (rG = 0.24, P = 0.05), indicating stability of the genetic factors influencing both sleep duration and impaired glucose metabolism throughout life (data from personal communication, manuscript in preparation).
We estimated the genetic correlation between sleep duration in children and in adults, but we failed to find statistically significant overlap (rG = 0.15, P = 0.40). This finding may indicate that different genetic variants influence sleep duration in children and in adults. Indeed, SNPs previously associated with sleep duration in adults were not replicated in our GWAS meta-analysis on sleep duration in children. In addition, no enrichment of significant associations was observed in children among the top SNPs described by Gottlieb et al. in adults (P < 1.00e-03). However, we note that the nonsignificant genetic correlation between sleep duration in children and other metabolic and psychiatric traits could be due to low power, and larger GWAS studies are needed to more accurately estimate the genetic overlap of related traits. In fact, the only nominally significant genetic correlation observed for sleep duration was with T2D, the phenotype with the largest GWAS dataset.
The current study is not without limitations. First, sleep duration was assessed by parent-reported open question(s), and the responses were mostly whole number values. Although parent- and self-reported questionnaires have widely been used to assess sleep duration in children and adults,23,33,36 it is possible that these measurements lack the necessary accuracy to estimate the small effects of single genetic variants (e.g., each copy of the minor allele of rs1823125 is associated with an increase in sleep duration of 3.1 minutes36). More objective and accurate qualitative measurements, such as polysomnography, actigraphy, sleep electroencephalogram (EEG) and/ or multiple sleep latency tests could be used to estimate sleep duration.73,74 Indeed, substantial heritability has been reported for sleep EEG patterns.75 Second, both the discovery and the replication phase of our GWAS meta-analysis is likely under-powered. Although it is evident that large samples are needed to identify replicable genetic associations in the general population,76 studies on child populations are limited. For this study, we used all known available child cohorts with both genetic data and data on sleep duration. More and larger studies, with good quality data on sleep duration, could extend the findings of this first study in the future.
In summary, this is the first GWAS meta-analysis of sleep duration in children. We identified a novel region (i.e., chromosome 11q13.4) associated with sleep duration in children, which was not replicated. More and larger studies should follow to verify our initial finding. Second, we rejected the null hypothesis of no SNP heritability of parent-or self-rated questionnaires on sleep duration, by showing evidence of modest but significant heritability of sleep duration in both children and adults, which may decrease by age. Finally, there was recurring evidence of an association between sleep duration in children and impaired glucose metabolism, as identified by both gene-enrichment analysis of our GWAS meta-analysis results and by the estimation of significant genetic correlation between sleep duration in children and type 2 diabetes in adults. These results give input to new genetic studies of sleep duration, especially in child populations, to unravel the genetic mechanisms associating sleep duration with health and disease.
DISCLOSURE STATEMENT
This was not an industry supported study. The authors have indicated no financial conflicts of interest.
ACKNOWLEDGMENTS
The authors thank all parents and children participating in each cohort study. Detailed acknowledgements regarding each participating cohort are included in the supplemental material.
REFERENCES
- 1.Allada R, Siegel JM. Unearthing the phylogenetic roots of sleep. Curr Biol. 2008;18:R670–R9. doi: 10.1016/j.cub.2008.06.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cappuccio FP, Taggart FM, Kandala NB, et al. Meta-analysis of short sleep duration and obesity in children and adults. Sleep. 2008;31:619–26. doi: 10.1093/sleep/31.5.619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gangwisch JE, Heymsfield SB, Boden-Albala B, et al. Short sleep duration as a risk factor for hypertension - Analyses of the first national health and nutrition examination survey. Hypertension. 2006;47:833–9. doi: 10.1161/01.HYP.0000217362.34748.e0. [DOI] [PubMed] [Google Scholar]
- 4.Ayas NT, White DP, Manson JE, et al. A prospective study of sleep duration and coronary heart disease in women. Arch Intern Med. 2003;163:205–9. doi: 10.1001/archinte.163.2.205. [DOI] [PubMed] [Google Scholar]
- 5.Leng Y, Cappuccio FP, Wainwright NWJ, et al. Sleep duration and risk of fatal and nonfatal stroke: a prospective study and meta-analysis. Neurology. 2015;84:1072–9. doi: 10.1212/WNL.0000000000001371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shan ZL, Ma HF, Xie ML, et al. Sleep duration and risk of type 2 diabetes: a meta-analysis of prospective studies. Diabetes Care. 2015;38:529–37. doi: 10.2337/dc14-2073. [DOI] [PubMed] [Google Scholar]
- 7.Cappuccio FP, D'Elia L, Strazzullo P, Miller MA. Sleep duration and all-cause mortality: a systematic review and meta-analysis of prospective studies. Sleep. 2010;33:585–92. doi: 10.1093/sleep/33.5.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Astill RG, Van der Heijden KB, Van IJzendoorn MH, Van Someren EJ. Sleep, Cognition, and behavioral problems in school-age children: a century of research meta-analyzed. Psychol Bull. 2012;138:1109–38. doi: 10.1037/a0028204. [DOI] [PubMed] [Google Scholar]
- 9.Watson NF, Harden KP, Buchwald D, et al. Sleep duration and depressive symptoms: a gene-environment interaction. Sleep. 2014;37:351–8. doi: 10.5665/sleep.3412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perlman CA, Johnson SL, Mellman TA. The prospective impact of sleep duration on depression and mania. Bipolar Disord. 2006;8:271–4. doi: 10.1111/j.1399-5618.2006.00330.x. [DOI] [PubMed] [Google Scholar]
- 11.Barnes JC, Meldrum RC. The impact of sleep duration on adolescent development: a genetically informed analysis of identical twin pairs. J Youth Adolesc. 2015;44:489–506. doi: 10.1007/s10964-014-0137-4. [DOI] [PubMed] [Google Scholar]
- 12.Liu XC. Sleep and adolescent suicidal behavior. Sleep. 2004;27:1351–8. doi: 10.1093/sleep/27.7.1351. [DOI] [PubMed] [Google Scholar]
- 13.Hvolby A. Associations of sleep disturbance with ADHD: implications for treatment. Atten Defic Hyperact Disord. 2015;7:1–18. doi: 10.1007/s12402-014-0151-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anderson B, Storfer-Isser A, Taylor HG, Rosen CL, Redline S. Associations of executive function with sleepiness and sleep duration in adolescents. Pediatrics. 2009;123:E701–7. doi: 10.1542/peds.2008-1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cohrs S, Rodenbeck A, Riemann D, et al. Impaired sleep quality and sleep duration in smokers-results from the German Multicenter Study on Nicotine Dependence. Addict Biol. 2014;19:486–96. doi: 10.1111/j.1369-1600.2012.00487.x. [DOI] [PubMed] [Google Scholar]
- 16.Calamaro CJ, Mason TBA, Ratcliffe SJ. Adolescents living the 24/7 lifestyle: effects of caffeine and technology on sleep duration and daytime functioning. Pediatrics. 2009;123:E1005–10. doi: 10.1542/peds.2008-3641. [DOI] [PubMed] [Google Scholar]
- 17.Krueger PM, Friedman EM. Sleep duration in the United States: a cross-sectional population-based study. Am J Epidemiol. 2009;169:1052–63. doi: 10.1093/aje/kwp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Blair PS, Humphreys JS, Gringras P, et al. Childhood sleep duration and associated demographic characteristics in an English cohort. Sleep. 2012;35:353–60. doi: 10.5665/sleep.1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.de Castro JM. The influence of heredity on self-reported sleep patterns in free-living humans. Physiol Behav. 2002;76:479–86. doi: 10.1016/s0031-9384(02)00699-6. [DOI] [PubMed] [Google Scholar]
- 20.Heath AC, Kendler KS, Eaves LJ, Martin NG. Evidence for genetic influences on sleep disturbance and sleep pattern in twins. Sleep. 1990;13:318–35. doi: 10.1093/sleep/13.4.318. [DOI] [PubMed] [Google Scholar]
- 21.Watson NF, Buchwald D, Vitiello MV, Noonan C, Goldberg J. A twin study of sleep duration and body mass index. J Clin Sleep Med. 2010;6:11–7. [PMC free article] [PubMed] [Google Scholar]
- 22.Partinen M, Kaprio J, Koskenvuo M, Putkonen P, Langinvainio H. Genetic and environmental determination of human sleep. Sleep. 1983;6:179–85. doi: 10.1093/sleep/6.3.179. [DOI] [PubMed] [Google Scholar]
- 23.Gottlieb DJ, O'Connor GT, Wilk JB. Genome-wide association of sleep and circadian phenotypes. BMC Med Genet. 2007;8(Suppl 1):S9. doi: 10.1186/1471-2350-8-S1-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brescianini S, Volzone A, Fagnani C, et al. Genetic and environmental factors shape infant sleep patterns: a study of 18-month-old twins. Pediatrics. 2011;127:E1296–302. doi: 10.1542/peds.2010-0858. [DOI] [PubMed] [Google Scholar]
- 25.Touchette E, Dionne G, Forget-Dubois N, et al. Genetic and environmental influences on daytime and nighttime sleep duration in early childhood. Pediatrics. 2013;131:E1874–80. doi: 10.1542/peds.2012-2284. [DOI] [PubMed] [Google Scholar]
- 26.Fisher A, van Jaarsveld CH, Llewellyn CH, Wardle J. Genetic and environmental influences on infant sleep. Pediatrics. 2012;129:1091–6. doi: 10.1542/peds.2011-1571. [DOI] [PubMed] [Google Scholar]
- 27.Yang JA, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88:76–82. doi: 10.1016/j.ajhg.2010.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bulik-Sullivan BK, Loh PR, Finucane HK, et al. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat Genet. 2015;47:291–5. doi: 10.1038/ng.3211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.He Y, Jones CR, Fujiki N, et al. The transcriptional repressor DEC2 Regulates sleep length in mammals. Science. 2009;325:866–70. doi: 10.1126/science.1174443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Allebrandt KV, Teder-Laving M, Akyol M, et al. CLOCK gene variants associate with sleep duration in two independent populations. Biol Psychiat. 2010;67:1040–7. doi: 10.1016/j.biopsych.2009.12.026. [DOI] [PubMed] [Google Scholar]
- 31.Kripke DF, Kline LE, Nievergelt CM, et al. Genetic variants associated with sleep disorders. Sleep Med. 2015;16:217–24. doi: 10.1016/j.sleep.2014.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Scheinfeldt LB, Gharani N, Kasper RS, et al. Using the Coriell Personalized Medicine Collaborative Data to conduct a genome-wide association study of sleep duration. Am J Med Genet B Neuropsychiatr Genet. 2015;168:697–705. doi: 10.1002/ajmg.b.32362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ollila HM, Kettunen J, Pietilainen O, et al. Genome-wide association study of sleep duration in the Finnish population. J Sleep Res. 2014;23:609–18. doi: 10.1111/jsr.12175. [DOI] [PubMed] [Google Scholar]
- 34.Allebrandt KV, Amin N, Muller-Myhsok B, et al. A K-ATP channel gene effect on sleep duration: from genome-wide association studies to function in Drosophila. Mol Psychiatr. 2013;18:122–32. doi: 10.1038/mp.2011.142. [DOI] [PubMed] [Google Scholar]
- 35.Parsons MJ, Lester KJ, Barclay NL, Nolan PM, Eley TC, Gregory AM. Replication of genome-wide association studies (GWAS) loci for sleep in the British G1219 cohort. Am J Med Genet B. 2013;162B:431–8. doi: 10.1002/ajmg.b.32106. [DOI] [PubMed] [Google Scholar]
- 36.Gottlieb DJ, Hek K, Chen TH, et al. Novel loci associated with usual sleep duration: the CHARGE consortium genome-wide association study. Mol Psychiatry. 2015;20:1232–9. doi: 10.1038/mp.2014.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Akerstedt T. Shift work and disturbed sleep/wakefulness. Occup Med-Oxford. 2003;53:89–94. doi: 10.1093/occmed/kqg046. [DOI] [PubMed] [Google Scholar]
- 38.Iglowstein I, Jenni OG, Molinari L, Largo RH. Sleep duration from infancy to adolescence: reference values and generational trends. Pediatrics. 2003;111:302–7. doi: 10.1542/peds.111.2.302. [DOI] [PubMed] [Google Scholar]
- 39.Maslowsky J, Ozer EJ. Developmental trends in sleep duration in adolescence and young adulthood: evidence from a national United States sample. J Adolesc Health. 2014;54:691–7. doi: 10.1016/j.jadohealth.2013.10.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Watson NF, Harden KP, Buchwald D, et al. Sleep duration and body mass index in twins: a gene-environment interaction. Sleep. 2012;35:597–603. doi: 10.5665/sleep.1810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Winkler TW, Day FR, Croteau-Chonka DC, et al. Quality control and conduct of genome-wide association meta-analyses. Nat Protoc. 2014;9:1192–212. doi: 10.1038/nprot.2014.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.R Core Team. R: a language and environment for statistical computing. 2013. [Accessed 2015]. Available from: http://www.R-project.org/
- 43.Willer CJ, Li Y, Abecasis GR. METAL: fast and efficient meta-analysis of genomewide association scans. Bioinformatics. 2010;26:2190–1. doi: 10.1093/bioinformatics/btq340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010;26:2336–7. doi: 10.1093/bioinformatics/btq419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ward LD, Kellis M. HaploReg: a resource for exploring chromatin states, conservation, and regulatory motif alterations within sets of genetically linked variants. Nucleic Acids Res. 2012;40:D930–4. doi: 10.1093/nar/gkr917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ardlie KG, DeLuca DS, Segre AV, et al. The genotype-tissue expression (GTEx) pilot analysis: multitissue gene regulation in humans. Science. 2015;348:648–60. doi: 10.1126/science.1262110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pers TH, Karjalainen JM, Chan Y, et al. Biological interpretation of genome-wide association studies using predicted gene functions. Nat Commun. 2015;6:5890. doi: 10.1038/ncomms6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Mi HY, Muruganujan A, Thomas PD. PANTHER in 2013: modeling the evolution of gene function, and other gene attributes, in the context of phylogenetic trees. Nucleic Acids Res. 2013;41:D377–86. doi: 10.1093/nar/gks1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Finucane HK, Bulik-Sullivan B, Gusev A, et al. Partitioning heritability by functional annotation using genome-wide association summary statistics. Nat Genet. 2015;47:1228–35. doi: 10.1038/ng.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bulik-Sullivan B, Finucane HK, Anttila V, et al. An atlas of genetic correlations across human diseases and traits. Nat Genet. 2015;47:1236–41. doi: 10.1038/ng.3406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bradfield JP, Taal HR, Timpson NJ, et al. A genome-wide association meta-analysis identifies new childhood obesity loci. Nat Genet. 2012;44:526–31. doi: 10.1038/ng.2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Morris AP, Voight BF, Teslovich TM, et al. Large-scale association analysis provides insights into the genetic architecture and pathophysiology of type 2 diabetes. Nat Genet. 2012;44:981–90. doi: 10.1038/ng.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Saxena R, Hivert MF, Langenberg C, et al. Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet. 2010;42:142–U75. doi: 10.1038/ng.521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sklar P, Ripke S, Scott LJ, et al. Large-scale genome-wide association analysis of bipolar disorder identifies a new susceptibility locus near ODZ4. Nat Genet. 2011;43:977–U162. doi: 10.1038/ng.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sullivan PF, Daly MJ, Ripke S, et al. A mega-analysis of genome-wide association studies for major depressive disorder. Mol Psychiatr. 2013;18:497–511. doi: 10.1038/mp.2012.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ripke S, Neale BM, Corvin A, et al. Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014;511:421–7. doi: 10.1038/nature13595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mick E, Todorov A, Smalley S, et al. Family-based genome-wide association scan of attention-deficit/hyperactivity disorder. J Am Acad Child Psy. 2010;49:898–905. doi: 10.1016/j.jaac.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Byrne EM, Gehrman PR, Medland SE, et al. A genome-wide association study of sleep habits and insomnia. Am J Med Genet B. 2013;162B:439–51. doi: 10.1002/ajmg.b.32168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Comuzzie AG, Cole SA, Laston SL, et al. Novel genetic loci identified for the pathophysiology of childhood obesity in the Hispanic population. PLoS One. 2012;7:e51954. doi: 10.1371/journal.pone.0051954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O'Donnell CJ, de Bakker PIW. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics. 2008;24:2938–9. doi: 10.1093/bioinformatics/btn564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mahajan A, Go MJ, Zhang WH, et al. Genome-wide trans-ancestry meta-analysis provides insight into the genetic architecture of type 2 diabetes susceptibility. Nat Genet. 2014;46:234–44. doi: 10.1038/ng.2897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Manning AK, Hivert MF, Scott RA, et al. A genome-wide approach accounting for body mass index identifies genetic variants influencing fasting glycemic traits and insulin resistance. Nat Genet. 2012;44:659–U81. doi: 10.1038/ng.2274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Strawbridge RJ, Dupuis J, Prokopenko I, et al. Genome-Wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes. 2011;60:2624–34. doi: 10.2337/db11-0415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42:579–89. doi: 10.1038/ng.609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kulzer JR, Stitzel ML, Morken MA, et al. A common functional regulatory variant at a type 2 diabetes locus upregulates ARAP1 expression in the pancreatic beta cell. Am J Hum Genet. 2014;94:186–97. doi: 10.1016/j.ajhg.2013.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Okada Y, Terao C, Ikari K, et al. Meta-analysis identifies nine new loci associated with rheumatoid arthritis in the Japanese population. Nat Genet. 2012;44:511–6. doi: 10.1038/ng.2231. [DOI] [PubMed] [Google Scholar]
- 67.Schmid SM, Hallschmid M, Schultes B. The metabolic burden of sleep loss. Lancet Diabetes Endo. 2015;3:52–62. doi: 10.1016/S2213-8587(14)70012-9. [DOI] [PubMed] [Google Scholar]
- 68.Felson J. What can we learn from twin studies? A comprehensive evaluation of the equal environments assumption. Soc Sci Res. 2014;43:184–99. doi: 10.1016/j.ssresearch.2013.10.004. [DOI] [PubMed] [Google Scholar]
- 69.Visscher PM, Brown MA, McCarthy MI, Yang J. Five years of GWAS discovery. Am J Hum Genet. 2012;90:7–24. doi: 10.1016/j.ajhg.2011.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Gottlieb DJ, Punjabi NM, Newman AB, et al. Association of sleep time with diabetes mellitus and impaired glucose tolerance. Arch Intern Med. 2005;165:863–8. doi: 10.1001/archinte.165.8.863. [DOI] [PubMed] [Google Scholar]
- 71.Furihata R, Uchiyama M, Suzuki M, et al. Association of short sleep duration and short time in bed with depression: a Japanese general population survey. Sleep Biol Rhythms. 2015;13:136–45. [Google Scholar]
- 72.Monti JM, Monti D. Sleep disturbance in schizophrenia. Int Rev Psychiatry. 2005;17:247–53. doi: 10.1080/09540260500104516. [DOI] [PubMed] [Google Scholar]
- 73.Lauderdale DS, Knutson KL, Yan LL, Liu K, Rathouz PJ. Sleep duration: how well do self-reports reflect objective measures? The CARDIA Sleep Study. Epidemiology. 2008;19:838–45. doi: 10.1097/EDE.0b013e318187a7b0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Van Den Berg JF, Van Rooij FJA, Vos H, et al. Disagreement between subjective and actigraphic measures of sleep duration in a population-based study of elderly persons. J Sleep Res. 2008;17:295–302. doi: 10.1111/j.1365-2869.2008.00638.x. [DOI] [PubMed] [Google Scholar]
- 75.Ambrosius U, Lietzenmaier S, Wehrle R, et al. Heritability of sleep electroencephalogram. Biol Psychiat. 2008;64:344–8. doi: 10.1016/j.biopsych.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 76.Chabris CF, Hebert BM, Benjamin DJ, et al. Most reported genetic associations with general intelligence are probably false positives. Psychol Sci. 2012;23:1314–23. doi: 10.1177/0956797611435528. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


