Abstract
Methylation at histone 3, lysine 36 (H3K36) is a conserved epigenetic mark regulating gene transcription, alternative splicing and DNA repair. Genes encoding H3K36 methyltransferases (KMTases) are commonly overexpressed, mutated or involved in chromosomal translocations in cancer. Molecular biology studies have demonstrated that H3K36 KMTases regulate oncogenic transcriptional programs. Structural studies of the catalytic SET domain of H3K36 KMTases have revealed intriguing opportunities for design of small molecule inhibitors. Nevertheless, potent inhibitors for most H3K36 KMTases have not yet been developed, underlining the challenges associated with this target class. As we now have strong evidence linking H3K36 KMTases to cancer, drug development efforts are predicted to yield novel compounds in the near future.
Keywords: : cancer, chromosomal translocation, epigenetics, gene expression, genome integrity, histone methyltransferase, oncogenes, protein–protein interactions, structure-based drug design
Cellular development and differentiation are controlled by post-translational modifications on DNA and histone proteins, forming an epigenetic (above the genes) regulatory network. Disruption of epigenetic pathways is a nearly universal feature of cancer, causing aberrations in gene expression and genome integrity (reviewed in [1]). A key group of epigenetic enzymes disrupted in cancer is the lysine methyltransferases (KMTases), which install methyl groups on histone proteins. In cancer cells, methylation performed by KMTases drives proliferation and halts differentiation by modifying gene transcription and other DNA-templated processes [2–4]. Over the past decade, KMTases have emerged as important drug targets for both industrial and academic research groups. Some progress has been made, most notably by selective inhibitors for the EZH2 and DOT1L KMTases that have reached clinical trials for the treatment of non-Hodgkin lymphoma and acute leukemia, respectively [5,6]. Nevertheless, selective and cell permeable inhibitors for many KMTases remain unavailable.
Methylation at H3K36 represents a particularly important chromatin mark implicated in diverse forms of cancer. KMTases with specificity toward H3K36 are overexpressed in cancer cells and have been characterized as regulators of cell growth, differentiation, stemness and DNA repair pathways [7–9]. However, very few selective and cell-active small molecule inhibitors of H3K36-specfic KMTases have been reported to date. In this article, we provide an overview of cellular pathways involving H3K36 methylation and discuss the diverse functions carried out by the eight different human H3K36-specific KMTases. Then we analyze structural characteristics of the catalytic SET domain of H3K36 KMTases and evaluate prospects for their inhibition by small molecules. We conclude with a discussion of the challenges and opportunities for targeting these proteins.
H3K36 methylation regulates diverse processes implicated in cancer
H3K36 methylation participates in a wide variety of nuclear pathways. Many of these processes, including transcriptional regulation, alternative splicing and DNA repair, are dysregulated in cancer.
Transcriptional regulation
On a genome scale, H3K36 methylation is enriched within the transcribed region of active genes rather than regulatory sites [10,11], with the highest levels at the 3′ end of genes [12]. H3K36 methylation plays a key role within gene bodies by preventing cryptic intragenic transcription, which may generate toxic proteins or waste cell resources [13]. In this way, H3K36 methylation is associated with active genes but actually has a repressive effect on chromatin structure within these genes. In yeast, the sole H3K36 methyltransferase is Set2, which is recruited to actively transcribed genes by binding to the hyperphosphorylated C-terminal domain (CTD) of RNA polymerase II [14]. Methylation of H3K36 by Set2 recruits the Rpd3S histone deacetylase (Figure 1A), which produces chromatin compaction and prevents aberrant transcription from cryptic intragenic promoters in the wake of polymerase II [15]. H3K36 methylation further suppresses cryptic transcription by preventing the incorporation of new, acetylated histones over open reading frames. This role depends on methylated H3K36 recruiting chromatin remodelers Isw1b and Chd1 and preventing H3 binding to the histone chaperone Asf1 [16,17].
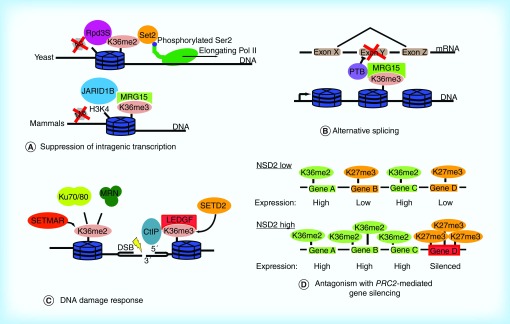
Figure 1. . The functions of H3K36 methylation in biology.
(A) H3K36 methylation plays a conserved role in suppressing intragenic transcription. Methylated H3K36 recruits histone deacetylases (yeast) or histone demethylases (humans) to maintain a suppressive transcriptional environment within gene bodies. (B) Splicing regulators including pyrimidine tract binding protein are recruited to loci by H3K36 methylation. (C) Methyl marks installed by the SETMAR and SETD2 KMTases are important for recruiting mediators of the DNA damage response including Ku70/Ku80, MRE11-RAD50-NBS1 (MRN) and CtIP. (D) Methylated H3K36 antagonizes polycomb-repressive complex 2 mediated H3K27 methylation and gene silencing. In NSD2-low cells, active genes are marked by H3K36 methylation, whereas lower expressed genes are marked by H3K27 methylation. In NSD2-high cells (as in t(4;14) multiple myeloma), overexpression of NSD2 increases genome-wide levels of H3K36 methylation, forcing accumulation of H3K27 methylation at a subset of silenced loci.
Adapted with permission from [18].
H3K36 methylation similarly prevents cryptic intragenic transcription in mammals. The human H3K36 trimethyltransferase and Set2 homolog SETD2 is required for preventing intragenic transcription initiation in 11% of genes [19]. This function does not depend on histone deacetylases as in yeast, but on recruitment of histone demethylases and chaperones. In embryonic stem cells, the H3K36me3 reader protein MRG15 recruits the JARID1B histone demethylase to intragenic regions, where it removes H3K4me3 to prevent cryptic intragenic transcription and facilitate transcriptional elongation of functional full-length mRNAs [20] (Figure 1A). Interestingly, human LSD2, a second H3K4 demethylase that maintains a repressive chromatin environment in gene bodies to facilitate optimal transcriptional elongation, forms a complex with the H3K36 KMTase NSD3 [21]. Additionally, H3K36me3-modified chromatin recruits the FAcilitates Chromatin Transcription histone chaperone complex, which helps restore chromatin structure following transcriptional elongation [19].
Alternative splicing
Splicing pathways are deregulated in cancer, resulting in expression of gene isoforms that contribute to oncogenic processes [22]. H3K36me3-modified nucleosomes are enriched within exons [23], and accumulating evidence indicates that H3K36 methylation signals to the splicing machinery. As an example, the human FGF receptor 2 (FGFR2) gene undergoes tissue-specific alternative splicing that is controlled by pyrimidine tract binding protein (PTB). Knockdown or overexpression of the H3K36 trimethyltransferase SETD2 causes inclusion or exclusion, respectively, of the PTB-regulated exon in FGFR2. Further experiments showed that PTB is specifically recruited to exons labeled with H3K36me3 by interacting with the MRG15 H3K36 methyl-reader protein [24] (Figure 1B). In addition to the H3K36me3/MRG15/PTB alternative splicing pathway, multiple PWWP domain-containing proteins that recognize H3K36me3 have been implicated in alternative splicing [25,26]. For example, the PWWP domain-containing protein ZMYND11 localizes primarily to gene bodies due to specific recognition of K36me3 on histone variant H3.3. ZMYND11 promotes intron retention at hundreds of sites by interacting with and antagonizing the function of the splicing regulator EFTUD2 [26]. On the other hand, alternative splicing has been shown to regulate H3K36 methylation. Deletion of splicing sites or pharmacological inhibition of splicing leads to redistribution of H3K36 trimethylation, reduces overall levels of H3K36 trimethylation and blocks SETD2 recruitment to chromatin [27,28]. These findings suggest regulatory cross-talk between H3K36 methylation and alternative splicing pathways.
DNA repair
Genome instability is a key characteristic of cancer, and H3K36 methylation has a conserved role in DNA repair. In yeast, H3K36 methylation mediated by Set2 promotes the nonhomologous end joining (NHEJ) pathway of DNA repair during the G1 phase of the cell cycle, whereas H3K36 acetylation mediated by Gcn5 promotes homologous recombination (HR) repair during the S and G2 phases [29,30]. The mammalian Set2 homolog SETD2 also regulates DNA repair, but in the opposite direction from Set2, as SETD2 promotes HR rather than NHEJ. Histones trimethylated at H3K36 by SET2D provide a constitutive-binding site for the LEDGF PWWP domain. Once a DNA double-strand break occurs, LEDGF recruits CtIP, which performs 5′ end resection to produce a single-stranded 3′ overhang (Figure 1C). Recruitment of major HR repair factors downstream of CtIP, such as RPA and RAD51, also depends on SETD2-mediated H3K36 trimethylation [31]. In cells with depleted SETD2, double-strand break repair proceeds via the error-prone microhomology-mediated end-joining pathway, leading to deletions between sequences of microhomology [31]. SETD2 is also responsible for regulating DNA mismatch repair through its interaction with MutSα [32]. These studies provide mechanistic basis for the genome instability observed in SETD2-deficient cancers (see the ‘SETD2: an important tumor suppressor’ section).
Another H3K36 KMTase involved in DNA repair is SETMAR, a protein that evolved from gene fusion of a SET domain with a transposase domain. Upon radiation damage, H3K36me2 levels are increased in a SETMAR-dependent manner. Depleting SETMAR or expression of H3 with mutated K36 decreases Ku70 and NBS1 recruitment, thereby decreasing efficiency of the NHEJ repair pathway [33] (Figure 1C). The SET domain of SETMAR is also required for the DNA damage response at stalled replication forks and efficient restarting of stalled replication forks [34]. Interestingly, SETMAR does not methylate recombinant nucleosomes in vitro [35], suggesting that SETMAR may indirectly produce H3K36me2 after DNA damage.
The reciprocal relationship between H3K36 & H3K27 methylation
How different epigenetic marks interact with each other to produce signaling outputs has important implications for epigenetic inhibitor development. H3K36 methylation interacts in an antagonistic fashion with H3K27 trimethylation, a repressive mark mediated by polycomb repressive complex 2 (PRC2). For example, in Drosophila the H3K36-specific KMTase Ash1 activates Hox genes during development by functioning as an antirepressor and antagonizing repressive H3K27 methylation installed by PRC2 [36]. In human HeLa cells, H3K36 methylation and H3K27 methylation are rarely found together on the same histone peptide. In fact, H3K36 premethylation inhibits PRC2 H3K27-KMTase activity in in vitro assays [37]. Conversely, H3K36 KMTases are inhibited by ubiquitinated H2A, a mark made by polycomb repressive complex 1 [38]. As development proceeds, however, the PRC2 complex must invade active, H3K36-methylated chromatin to silence certain genes. In this case, PRC2 targeting and spreading is mediated by Polycomb-like proteins with Tudor domains that specifically recognize H3K36me3 [39]. Disruption of the balance between the H3K36 and H3K27 methylation pattern is observed in multiple cancers (see the ‘NSD2 (MMSET/WHSC1): an oncogenic driver in MM’ section) (Figure 1D). In particular, cancers with chromosomal fusions involving H3K36 KMTases have disruptions in H3K27 methylation that drive oncogenesis, in addition to aberrant H3K36 methylation [8,18].
H3K36 KMTases play important & varying roles in carcinogenesis
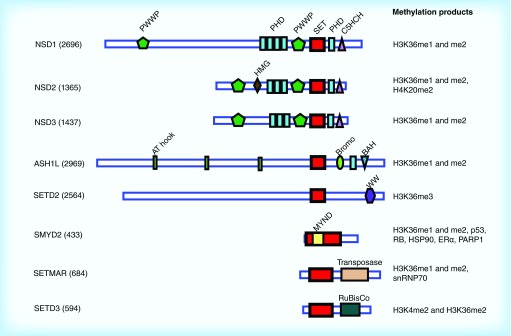
Given the importance of H3K36 methylation in diverse cellular processes, it is not surprising that H3K36 KMTases have been implicated both as oncogenes and tumor suppressors in cancer. The human genome encodes at least eight H3K36 KMTases (Figure 2), each of which contains a SET domain responsible for transferring a methyl group from the S-adenosyl methionine (SAM) cofactor to the substrate lysine. Although these proteins have all been reported to methylate H3K36, they vary in the number of methyl groups transferred as well as whether they methylate additional substrates. NSD1–3, ASH1L and SETD2 have closely related catalytic SET domains and show H3K36 methylation specificity in vitro and in vivo, while SMYD2, SETMAR and SETD3 have more distantly related SET domains with less well-characterized activities toward H3K36 (Figure 2). Of these KMTases, the NSD proteins are perhaps the best characterized oncoproteins, as they play important roles in multiple cancer types and are found in fusion proteins due to chromosomal translocations in acute myeloid leukemia (AML) and multiple myeloma (MM) [3,8]. The ASH1L KMTase is also overexpressed in cancer and regulates stem cell potential [7]. SMYD2 has oncogenic activity in multiple cancers [40–44], but whether this function depends on KMTase activity toward H3K36 or another of its many substrates is uncertain.
Figure 2. . Domain organization of human H3K36 methyltransferases.
SET domain is shown in red.
NSD1: activator of HOX genes in leukemia
NSD1 is a mono- and di- H3K36 KMTase with functions in development and cancer. Mutations in NSD1 cause Sotos syndrome, a condition of childhood overgrowth and intellectual disability, with a 2.4% increased risk of childhood malignancy [45,46]. A chromosomal translocation resulting in the NUP98-NSD1 fusion protein is found in 16% of cytogenetically normal pediatric AML and in a smaller portion of adult AML [47]. More than 90% of NUP98-NSD1-positive leukemias are also positive for internal tandem duplication mutation of the FLT3 tyrosine kinase, and the two genetic lesions exhibit potent cooperativity resulting in a 3-year-survival rate of 31% [48]. NUP98-NSD1 induces AML in vivo, sustains self-renewal of myeloid stem cells in vitro and enforces expression of the HOXA7, HOXA9, HOXA10 and MEIS1 proto-oncogenes [8]. Expression of the HOXA and MEIS1 oncogenes appears to be responsible for the transforming activity of NUP98-NSD1, as inhibition of the DOT1L–AF10 complex in NUP98-NSD1 leukemia decreases HOXA gene expression and triggers differentiation and apoptosis [49].
NSD1 has also been reported to methylate nonhistone proteins, including the p65 subunit of NF-κB at Lys218 and Lys221. In response to cytokines such as IL-1β and TNF-α, NSD1-mediated methylation enhances NF-κB's transcriptional activation and DNA-binding activities [50], which are active in most cancer cells and regulate genes that control proliferation, resistance to apoptosis, angiogenesis, invasion and metastasis [51]. Conversely, these activating marks on NF-κB are removed by the FBXL11 demethylase, and increasing methylation at Lys218 and Lys221 by depleting FBXL11 enhances cell proliferation and colony formation of colon cancer cells [50]. In addition, mutation of Lys218 and Lys221 on NF-κB showed that lysine methylation is required for activating the majority of NF-κB target genes in mouse embryonic fibroblasts, including cancer-relevant genes such as IGF1 and engulfment and cell motility 1 (ELMO1) [50].
In other contexts, however, NSD1 has been reported to function as a tumor suppressor. In bladder cancer cells, for example, inhibition of NF-κB by small molecule causes NSD1-dependent activation of pro-apoptotic BIM and the cell cycle regulator p21 [52]. Although it is not known if NSD1 KMTase activity is involved here, NF-κB inhibition does increase global levels of H3K36 trimethylation in addition to NSD1 expression [52]. Moreover, NSD1 frequently undergoes inactivating mutations causing premature termination of the protein in head and neck squamous cell carcinoma [53], and NSD1 is epigenetically silenced in neuroblastoma and glioma [54]. Finally, missense mutations in NSD1 are found in AML outside of the context of the NUP98-NSD1 fusion [55]. Although the functional consequences of these mutations are not known, these observations suggest variable roles for NSD1 in different cancer types.
NSD2 (MMSET/WHSC1): an oncogenic driver in MM
NSD2 (also known as MMSET and WHSC1) is an important developmental regulator and oncogene. Germline deletion of NSD2 causes Wolf–Hirschhorn syndrome, a developmental disorder characterized by craniofacial defects, growth retardation and microcephaly [56]. NSD2 has in vivo mono- and di-methyltransferase activity toward H3K36 [3]. Interestingly, NSD2 has also been reported to dimethylate H4K20, which was proposed to signal the recruitment of the DNA damage response regulator 53BP1 to sites of DNA damage [9,57], but other groups have not found evidence supporting this model [58,59].
NSD2 is a potentially attractive target for drug development due to its well characterized role in MM. In 15% of MM, the t(4;14)(p16.3;q32.3) translocation places the NSD2 gene under control of the immunoglobulin heavy-chain promoter/enhancer, leading to overexpression of NSD2, which is believed to be the key transforming factor [60]. In human myeloma cells harboring the t(4;14) translocation, overexpressed NSD2 leads to aberrant patterns of H3K36 methylation, causing a shift away from normal plasma cell gene expression programs and increased expression of cancer-associated genes [3]. Expression of NSD2 is sufficient to rescue tumor growth of myeloma cells in which the t(4:14) translocation has been inactivated [3], and knockdown of NSD2 in a t(4;14) mouse xenograft model causes dramatic reduction in tumor growth [18]. Importantly, the KMTase activity of the NSD2 SET domain is required for the epigenetic changes and oncogenic effects caused by NSD2 overexpression [3,18].
Beyond MM, expression of NSD2 is sufficient to transform mouse embryonic fibroblast cells lacking the p19ARF tumor suppressor, suggestive of it having a role as a general oncoprotein [3]. Indeed, NSD2 is overexpressed in neuroblastoma, lung, colon and bladder cancer [61,62], and is required for proliferation of neuroblastoma cells and brain-derived neural stem cells [62]. An E1099K mutation in the SET domain of NSD2 was identified in a range of human cancer cell lines and tumor samples, including acute lymphoblastic leukemia, chronic lymphocytic leukemia, lung cancer and stomach cancer [63,64]. The mutation appears to result in a hyperactive enzyme, as H3K36me2 levels are increased in cell lines harboring E1099K NSD2 [63,64]. Thus, the t(4;14) translocation in MM and the E1099K mutation in other cancers both result in increased H3K36me2 levels that drive oncogenesis.
NSD2's oncogenic role may partially reflect an imbalance between H3K36 and H3K27 methylation pathways. For example, in t(4;14) MM cells, a global loss of H3K27me3 is observed concurrently with a global increase in H3K36me2 [18], as one might predict due to the antagonistic relationship between these two marks. However, the excess H3K36me2 forces local accumulation of H3K27me3 at a subset of genes, including tumor suppressors. MM cells with t(4;14) translocation are sensitive to EZH2 inhibitors, indicating that the accumulation of EZH2 and H3K27me3 at tumor suppressor loci enhances myeloma cell proliferation [18,65]. Interestingly, in other cancers besides MM, EZH2 and NSD2 are coregulated in an EZH2/NSD2 oncogenic axis [66]. EZH2 represses expression of a set of miRNAs that repress NSD2, leading to EZH2/NSD2 coregulation. In prostate cancer for example, the EZH2/NSD2 axis confers increased cell proliferation, migration, invasion, stem cell-like properties and tumor growth in a mouse xenograft model [66]. Further studies in prostate cancer have shown that NSD2 acts as a co-activator with NF-κB and androgen receptor [67,68], and regulates the epithelial to mesenchymal transition by dimethylating H3K36 at the TWIST1 locus and activating TWIST1 expression [69].
NSD3: frequently overexpressed in diverse tumor types
NSD3, also known as WHSC1L1, is overexpressed in cancer [70] and catalyzes mono- and di-methylation at H3K36 in vitro [71]. In cells NSD3 functions as a transcriptional activator, but NSD3's KMTase activity may contribute only partially to its gene-activating role. For example, NSD3 regulates neural crest specification and migration and is required for activation of neural crest transcription factors Sox10, Snail2, Sox9 and FoxD3, but the only locus at which NSD3 is responsible for H3K36 dimethylation is Sox10 [72]. Similar to NSD1, NUP98-NSD3 fusion proteins have been reported in AML and myelodysplastic syndrome [73,74]. NSD3 undergoes copy number amplification in 21% of lung squamous cell carcinomas and 15% of breast invasive carcinomas, with a high correlation between copy number and mRNA expression, suggesting that NSD3 may function as an oncogenic driver [70]. Indeed, RNAi knockdown of NSD3 in non-small-cell lung cancer, colorectal cancer, bladder cancer and breast cancer cell lines with NSD3 overexpression causes reduced cell proliferation due to increased apoptosis or cell cycle arrest in the various cancer cell lines [70,75,76]. The breast epithelial cell line MCF10A is normally highly dependent on growth factors and forms small acinar-like structures in 3D Matrigel culture, but these cells can be transformed by expression of NSD3, which confers growth factor-independent proliferation, colony-forming ability in soft agar and expanded and disorganized growth in 3D culture [76]. In the transformed cells, NSD3 overexpression causes activation of IRX3, a homeobox gene family member, and downregulation of TGFBI, a putative tumor suppressor [76]. These findings, combined with frequent overexpression of NSD3 in breast cancer, make NSD3 an enticing therapeutic target.
ASH1L: HOX gene activator with emerging role in cancer
Multiple studies have found that ASH1L is an important transcriptional regulator during development, and the protein's role in cancer is beginning to be characterized. ASH1L is homologous to Drosophila Ash1, a trithorax group protein that activates genes involved in development and differentiation [77]. In mammals, ASH1L deficiency causes a major reduction in long-term hematopoietic stem cells (HSCs) in bone marrow, but surprisingly has very modest effects on peripheral blood counts due to increased proliferation of progenitors downstream of HSCs [7]. ASH1L-deficient HSCs are also unable to reconstitute bone marrow output when transplanted into lethally irradiated mice [7]. These findings indicate that ASH1L maintains quiescence and self-renewal potential of long-term HSCs, providing a foundation for a recent study suggesting that ASH1L may also regulate the stemness properties of leukemic stem cells [78].
ASH1L has been linked to liver fibrosis, facioscapulohumeral muscular dystrophy and cancer through its role in gene activation [79,80]. ASH1L activates HOXA genes and MEIS1 in mouse HSCs [7] and activates HOXB and HOXC genes in human erythroleukemic K562 cells [81]. These findings are highly relevant because HOX genes are oncogenic drivers in many different blood and solid tumors [82]. For example, overexpression of HOXA9 is highly associated with a poor prognosis in AML [83], and HOXA9 and its collaborator MEIS1 are required for survival of MLL-rearranged leukemia cells [84,85]. Indeed, a recent study showed that ASH1L is required for HOXA9 and MEIS1 expression in MLL-rearranged leukemia, and knockdown of ASH1L induces differentiation and prolongs survival in a mouse model of the disease [78]. The authors propose that H3K36 dimethylation catalyzed by ASH1L is bound by the H3K36me2 reader protein LEDGF, which recruits MLL to maintain target gene expression [78]. Additional evidence that ASH1L KMTase activity is important for its gene activating function comes from a mouse model with a targeted in-frame deletion of the ASH1L SET domain [86]. Differentiating mouse embryonic stem cells lacking the ASH1L SET domain show a loss of expression of 152 genes, including members of the Hox and Wnt families [86]. Together, these studies suggest that targeted inhibition of ASH1L KMTase activity could be a potential therapeutic approach in cancers driven by high HOX expression.
In addition to its role in leukemia, ASH1L is overexpressed in a variety of solid tumors, including thyroid and breast cancer [87,88]. In thyroid cancer, ASH1L is overexpressed in tumor-specific truncated forms, although what parts of the protein are truncated or what processing events produce them has not been explored. Expression of the miRNA miR-142–3p suppresses ASH1L protein expression by binding to the ASH1L 3′UTR, an effect correlated with inhibition of colony formation and slowing of thyroid cancer cell growth [87]. Interestingly, in mouse embryonic stem cells ASH1L is regulated by a different miRNA, miR-290, which downregulates ASH1L expression and prevents aberrant overexpression of Hoxa, Hoxb, Hoxc and Hoxd genes [89].
SETD2: an important tumor suppressor
SETD2 stands out among the human H3K36 KMTases as the sole mediator of H3K36 trimethylation [90] and as a tumor suppressor in most contexts. In human development, germline mutations in SETD2 cause an overgrowth condition with features similar to Sotos syndrome [91]. Underexpression and mutation of SETD2 are associated with poor prognosis in breast and renal cancer, GI stromal tumors and acute leukemia [92–96]. SETD2 mutations are more common in leukemias with MLL rearrangements (22%) than leukemias without such rearrangements (5%), and SETD2 loss cooperates with MLL fusions such as MLL-AF9 and MLL-NRIP to create a more aggressive leukemia with increased self-renewal of leukemia stem cells [97].
Numerous studies have examined the tumor suppressor function of SETD2 in kidney tumors, in which SETD2 is mutated in 3–12% of clear cell renal cell carcinoma (ccRCC) [94,98] and a similar proportion of papillary renal cell carcinoma [99]. Multiple different loss-of-function mutations in SETD2 occur within spatially distinct regions of a single renal tumor, suggesting strong selective pressure for SETD2 inactivation [100]. Furthermore, miRNA-mediated repression of SETD2 expression occurs broadly in ccRCC tumor samples and cell lines, causing reductions in SETD2 function where overt mutations may not be present [101]. The molecular mechanisms underlying SETD2's suppression of renal cancer reflect the physiological functions of H3K36 methylation in transcript processing and genome integrity, as outlined above. Briefly, loss of function of SETD2 in ccRCC results in defects in DNA repair, nucleosome dynamics, RNA processing and DNA methylation [102–105].
Other H3K36 methyltransferases: SMYD2, SETMAR & SETD3
As a member of the SMYD family of proteins, SMYD2 contains a bipartite SET domain split by a zinc-finger MYND domain. SMYD2 promotes cancer cell proliferation in head and neck squamous-cell carcinoma and esophageal squamous cell carcinoma [41,42], and overexpression of SMYD2 is a poor prognostic factor in pediatric acute lymphoblastic leukemia, gastric cancer and bladder cancer [40,43,44]. SMYD2 methylates H3K4 and H3K36 in vitro [106], and SMYD2-mediated H3K36me2 was reported to repress transcription of pro-inflammatory cytokines IL-6 and TNF-α in macrophages [107]. However, inhibition or knockdown of SMYD2 does not change global H3K36 or H3K4 mono-, di- or tri-methylation, and most of SMYD2 is found in the cytoplasm, suggesting SMYD2's activity on chromatin may be minimal [108]. Indeed, SMYD2 methylates many nonhistone substrates including p53, RB, HSP90, estrogen receptor α and PARP1 [109–113], conveying wide-ranging effects on transcriptional regulation, protein homeostasis, apoptosis and the DNA damage response.
The SETMAR gene evolved from fusion of a SET domain to a transposase domain [114]. As described in the ‘DNA repair’ section above, the SETMAR protein has been linked to cancer via its role in DNA repair, which requires SETMAR's KMTase activity [33]. In colon cancer SETMAR was further found to activate genes involved in repair, synthesis and methylation of DNA, as well as stemness markers [115], but the contribution of KMTase activity to this function was not determined. Interestingly, purified SETMAR protein is not active on nucleosomes in vitro, raising the possibility that SETMAR may function by methylating nonhistone substrates in vivo [35]. In addition, SETMAR undergoes automethylation near the active site of the transposase domain [116], but how automethylation regulates its DNA repair function is unknown.
SETD3 is a poorly characterized putative tumor suppressor that methylates H3K4 and H3K36 in vitro and in SETD3-transfected cells [117]. SETD3 expression is lower in renal cell tumors than normal renal tissues, and low expression of SETD3 is associated with shorter survival in renal cell carcinoma patients [118]. SETD3 has been implicated in DNA replication and repair due to its interaction with proliferating cell nuclear antigen, a conserved factor involved in DNA synthesis [119].
H3K36-specific SET domains: structural considerations for inhibitor development
Structural details of target proteins are extremely valuable in the development of potent and specific inhibitors. For the H3K36-specific KMTases, structural studies of the catalytic SET domain have revealed valuable information to jumpstart inhibitor development. An important feature shared among the NSD, ASH1L and SETD2 SET domains is an autoinhibitory loop blocking access of histone substrate to the active site [120–122]. This loop must undergo a conformational change to accommodate substrate binding. In contrast, structural studies of the more distantly related SMYD2 SET domain showed that substrate peptides bind this SET domain with minimal conformational changes [123,124].
Structural studies of NSD, ASH1L & SETD2 SET domains demonstrate feasibility of inhibitor design
The SET domains of NSD1–3, ASH1L and SETD2 consist of four subdomains: associated with SET, core SET, SET-insertion (SET-I) and post-SET (Figure 3A). Associated with SET is a less-structured region at the N-terminus that does not participate directly in catalysis, but the latter three subdomains contribute to the active site. The β-sheet-rich core SET region is split by SET-I, a sequence-variable subdomain that has been proposed to play a role in substrate specificity [125]. Post-SET is a cysteine-rich subdomain that contains the autoinhibitory loop (Figure 3A). The methyl donor SAM binds in a pocket at the junction of core SET, SET-I and post-SET, which make extensive contacts to SAM including hydrogen bonds and van der Waals contacts. Other important interactions involve tyrosine and carbonyl oxygens that form CH···O hydrogen bonds with the SAM methyl group [126].
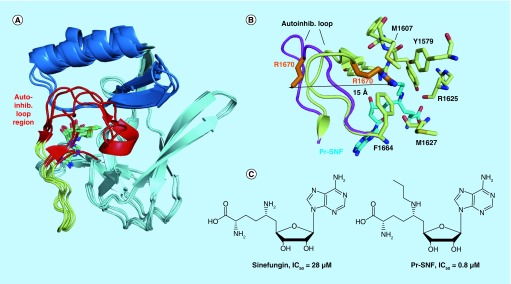
Figure 3. . Structures of H3K36-specific S-adenosyl methionine domains and SAM-competitive inhibitors.
(A) Overlay of the core SET (pale cyan), SET-I (sky blue) and post-SET (pale yellow) subdomains of NSD1 (PDB code 3OOI), NSD3 (4YZ8), ASH1L (4YNM) and SETD2 (4H12). The structurally variable autoinhibitory loop region is colored in red. (B) Binding of the N-propyl sinefungin (Pr-SNF) inhibitor to SETD2 causes opening of the autoinhibitory loop. Autoinhibitory loop conformation is shown with S-adenosyl homocysteine bound (pale yellow, 4H12) and with Pr-SNF bound (magenta, 4FMU). Steric clash between the propyl moiety of Pr-SNF (cyan) and the Arg1670 sidechain (orange) causes Arg1670 to flip out a distance of 15 Å. Residues stabilizing Arg1670 in the putative substrate lysine-binding channel in the S-adenosyl homocysteine bound form of SETD2 are shown in pale yellow sticks. (C) Chemical structures of SETD2 inhibitors sinefungin and Pr-SNF. In vitro IC50 values for SETD2 are listed.
The autoinhibitory loop must undergo a significant conformational change to accommodate nucleosome substrate, and interestingly this loop undergoes dynamics even without nucleosome present. Molecular dynamics simulations, crystallographic and NMR studies have shown that the NSD1 and ASH1L autoinhibitory loops experience conformational heterogeneity in the absence of nucleosome substrate [127,128]. In none of the observed conformations does the autoinhibitory loop completely open to allow nucleosome binding, suggesting that interaction with nucleosome is required to induce loop opening. Nevertheless, conformational dynamics of the loop may be important for facilitating the larger conformational change that must occur upon nucleosome binding [127,128]. Interestingly, the autoinhibitory loop in the crystal structure of the related NSD3 SET domain is disordered [129], further suggesting that autoinhibitory loop dynamics are characteristic of the NSD and related SET domains. In the case of SETD2, both auto-inhibited and open conformations of the SET domain were observed by crystallography [122] (Figure 3B). When bound to S-adenosyl homocysteine (SAH), SETD2 adopts an autoinhibited conformation similar to NSD1 and ASH1L, with the sidechain of Arg1670 in the autoinhibitory loop extending into the putative substrate lysine-binding channel (Figure 3B). In contrast, N-propyl sinefungin (Pr-SNF) was identified as a structural probe that replaces SAH and stabilizes an open conformation of SETD2 (Figure 3B & C). Pr-SNF partially occupies the lysine-binding channel and causes a dramatic conformational change in the autoinhibitory loop, as this loop moves away from core SET and Arg1670 flips 15 Å away from the lysine-binding channel [122].
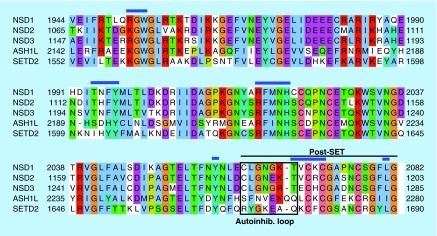
The autoinhibitory loop region of the NSD and related KMTases provides an intriguing opportunity for H3K36 KMTase inhibitor development. The conformational heterogeneity observed in this region may create transient pockets not directly observed in crystal structures into which small molecules could bind. These ligands could function as allosteric inhibitors by preferentially interacting with and stabilizing the autoinhibited conformation of the SET domain. Importantly, the autoinhibitory loop is poorly conserved in primary sequence and in structure among NSD1, ASH1L and SETD2 (Figure 3A & Figure 4), suggesting that targeting this region could result in specific inhibitors for the different KMTases. In NSD1, the autoinhibitory loop is stabilized by a β-turn within the loop as well as hydrophobic contacts with SET-I [121]. In contrast, no β-turn exists in the ASH1L autoinhibitory loop, and there are longer range contacts to SET-I [120,127]. In the SETD2 autoinhibitory loop, there is also no β-turn to stabilize the loop, but the interaction of Arg1670 at the beginning of the loop with the substrate lysine-binding channel pulls the loop into contact with SET-I [122] (Figure 3B). Thus, the autoinhibitory loop region could be exploited for designing specific inhibitors of H3K36 KMTases.
Figure 4. . Sequence alignment of core SET and post-SET regions of the related NSD, ASH1L and SETD2 KMTases.
Blue lines indicate conserved contacts to S-adenosyl methionine cofactor. Black box indicates the autoinhibitory loop, black line indicates post-SET subdomain.
SMYD2 structures: U-shaped substrate binding
Crystal structures of full-length SMYD2 show an overall structure of two lobes or domains separated by a deep groove (Figure 5A). The N-terminal lobe contains the core SET region surrounded by an MYND zinc finger, the SET-I subdomain and the cysteine-rich post-SET subdomain. The CTD is made up of seven antiparallel α-helices in a structure reminiscent of the tetratricopeptide repeat, a motif involved in protein–protein interactions. The CTD regulates KMTase activity of the SET domain, possibly via interactions the CTD forms with the substrate protein [123]. The CTD also controls activity and substrate specificity via an interaction with the SMYD2 cofactor Hsp90 [106,130]. The SAM cofactor binds in the crevasse between the N- and C-terminal lobes and makes direct contacts with core SET, SET-I and post-SET (Figure 5A). Regulatory cross-talk between the CTD and SET domain active site was suggested by the observation that binding of the SAM analog sinefungin induces a long range conformational change in the first two α-helices of the CTD, resulting in a more open conformation of SMYD2 [130,131].
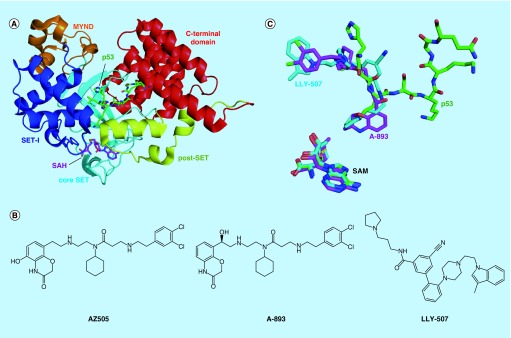
Figure 5. . Peptide substrate-competitive inhibitors of the SMYD2 KMTase.
(A) Crystal structure of SMYD2 bound to p53 peptide (PDB code 3TG5). (B) Chemical structures of SMYD2 inhibitors. (C) Overlay of binding modes of SMYD2 inhibitors LLY-507 (4WUY) and A-893 (4YND) and p53 peptide. The weaker compound AZ505 binds to the same site but has been omitted for clarity. S-adenosyl methionine position included for reference.
SAM: S-adenosyl methionine.
Ternary complexes of SMYD2 have been solved with SAM and p53 peptide, SAH and methylated p53 peptide and SAM and ER-α peptide (Figure 5A). The p53 and ER-α peptides bind in a U-shaped conformation at the hinge region between the N- and C-terminal lobes, with the target lysine at the base of the ‘U’ [123,124,132]. The target lysine projects deep into a hydrophobic cavity, with the lysine ϵ-amino group oriented by the hydroxyl groups of two conserved tyrosine residues and several main chain carbonyl groups. Although the CTD does not directly interact with SAM cofactor, residues from the CTD are involved in binding to substrate peptides. Remarkably, given the narrow groove between the N- and C-terminal lobes and the previously described flexibility in the CTD, only minor structural changes in SMYD2 occur upon binding to substrate peptides [123,124].
Inhibitors of H3K36 methyltransferases
Inhibitors for two members of the H3K36 methyltransferases, SETD2 and SMYD2, have been reported. Zheng et al. developed Pr-SNF and N-benzyl sinefungin, analogs of the broad-spectrum KMTase inhibitor sinefungin, as specific inhibitors of SETD2. Pr-SNF and N-benzyl sinefungin inhibit SETD2 KMTase activity in vitro with IC50 values of 0.8 and 0.5 μM, respectively (Figure 3C). These values represent activities more than tenfold better than unmodified sinefungin, and with at least twofold selectivity over a panel of related KMTases. A cocrystal structure of SETD2 in complex with Pr-SNF revealed that the N-propyl moiety partially occupies the lysine-binding channel and causes a dramatic conformational change in the autoinhibitory loop [122]. No activity for Pr-SNF has been reported in cells, and sinefungin analogs in general have poor cell membrane permeability. Nevertheless, combined occupancy of the SAM and substrate lysine-binding pockets by small molecules is an intriguing strategy that warrants further study in other KMTases.
The first specific SMYD2 inhibitor, AZ505, was discovered using an AlphaLISA-based high-throughput screen of 1.23 million compounds from the AstraZeneca library [124] (Figure 5B). The compound binds with Kd of 0.5 μM to the peptide-binding groove of SMYD2 and is greater than 100-fold selective over related KMTases including SMYD3. The AZ505-binding site partially overlaps with that of p53 peptide (Figure 5C). Interestingly, the benzooxazinone moiety of AZ505 occupies the substrate lysine-binding channel, indicating that this channel may accommodate much larger and more rigid groups than a lysine sidechain. The cyclohexyl group of AZ505 is positioned in a hydrophobic pocket occupied by Leu369 of the p53 peptide. However, the dichlorophenethyl group of AZ505 extends into a second hydrophobic pocket that is not occupied by the p53 substrate peptide.
AZ505 was optimized to improve potency while retaining cellular permeability. Nguyen et al. developed the SMYD2 inhibitor LLY-507 by making substantial changes to AZ505, using a pyrrolidine group rather than benzooxazinone to occupy the substrate lysine-binding channel (Figure 5B & C). LLY-507 inhibits SMYD2 methyltransferase activity in an in vitro assay with IC50 <15 nM but maintains an excellent selectivity profile over other KMTase and non-KMTase enzymes. LLY-507 inhibits p53 methylation in HEK293 and U2OS cells overexpressing SMYD2 and/or p53, and the compound inhibits proliferation of esophageal squamous cell carcinoma, hepatocellular carcinoma and breast cancer cell lines [108].
A second SMYD2 inhibitor, A-893, was also inspired by the AstraZeneca high-throughput screening hit but makes fewer changes to the AZ505 scaffold (Figure 5B & C). A-893 inhibits SMYD2 KMTase activity in vitro with an IC50 of 2.8 nM and blocks p53 methylation in the A549 lung carcinoma cell line. This large increase in activity over AZ505 was achieved by moving a hydroxyl group from the benzooxazinone to the diethylamine linker. The increase in potency is likely mediated by a hydrogen bond that the relocated hydroxyl group forms with the backbone carbonyl of Tyr240 [133].
Challenges & opportunities in developing specific inhibitors of H3K36 KMTases
Targeting the catalytic SET domain
The H3K36 KMTases are highly relevant to biology and medicine, and development of potent and specific inhibitors for this class of proteins is urgently needed. However, very few specific inhibitors of H3K36 KMTases have been reported, highlighting difficulties associated with this target class. First, developing in vitro methylation assays for H3K36 KMTases can be challenging because the SET domain may require chromatin-interacting domains and/or cofactor proteins for optimal activity on nucleosome substrate. For example, the isolated ASH1L SET domain has weak KMTase activity on nucleosomes, but constructs including the PHD, Bromo and BAH domains have increased activity [127]. An additional obstacle is that inhibitors of KMTases appear to be rare in current high-throughput screening libraries, as three different pharmaceutical companies published SAM-competitive inhibitors of the EZH2 KMTase with similar pyridone-containing scaffolds [4]. KMTase-privileged screening libraries, which could be based on known KMTase inhibitors such as sinefungin and chaetocin [122,134], may help increase the number of screening hits.
In contrast to screening through large compound libraries, a more focused approach to developing KMTase inhibitors could proceed by modifying the SAM cofactor. Structural chemical analysis of ten different human KMTase SET domains suggested that the SAM-binding pocket represents a druggable region because a low proportion of the SAM molecular surface is accessible to solvent when bound to the SET domain. Moreover, although the conformation of bound SAM cofactor is very similar across different SET domains due to a precise hydrogen bond network, SAM-binding pockets contain enough structural diversity surrounding this network to allow development of specific inhibitors for different KMTases [135]. Indeed, chemical modification of SAM or closely related compounds such as sinefungin has led to potent and specific inhibitors for several KMTases [122,136,137]. An additional consideration for designing SAM-derivative inhibitors is that the SAM-binding pocket is immediately adjacent to the narrow substrate lysine-binding channel. Therefore, weak or nonspecific SAM derivatives may be improved to target a specific KMTase by adding hydrophobic groups that interact with the lysine-binding channel. Proof of principle for this hypothesis was demonstrated by sinefungin analogs that specifically inhibit SETD2 [122]. The potency of SAM-derivative inhibitors could be further enhanced by including moieties that mimic the transition state for the methyl transfer reaction, as enzyme theory predicts that KMTases should bind the transition state more tightly than the individual SAM and histone substrates [138]. Kinetic isotope effects were recently used to generate a model of the NSD2 transition state, providing an opportunity for development of transition state mimics [139]. Thus far, poor membrane permeability of the available SAM-derivative inhibitors has proven a challenge in applying these compounds to cellular systems [140].
Alternative druggable regions of H3K36 KMTase proteins
Most H3K36-specific KMTases are large epigenetic regulators that contain multiple protein–protein interacting (PPI) domains in addition to the catalytic SET domain. In a growing number of cases, PPI domains of KMTase proteins have been found essential for recruiting the SET domain to its methylation targets. PPI domains may also have SET domain-independent oncogenic roles. Structural studies have revealed that PPI domains could be highly druggable targets [141,142]. Therefore, to overcome challenges associated with inhibitor development for H3K36 SET domains, an alternative approach is to target the PPI domains.
For example, PPI domains of NSD2 are essential for the protein's oncogenic activity. The N-terminal PWWP domain of NSD2 preferentially binds H3K36me2 and is important for recruiting NSD2 to its target genes [143]. Point mutations in the PWWP domain that prevent recognition of methyllysine inhibit NSD2's ability to increase H3K36 methylation and cancer cell proliferation. The fact that the NSD2 PWWP domain recognizes the same mark produced by the NSD2 SET domain suggests a role for the PWWP domain in spreading H3K36 dimethylation to neighboring genomic regions and to daughter cells during DNA replication [143]. NSD2 also contains four PHD domains, a family of epigenetic reader modules that bind methylated lysines on H3 [144]. The NSD2 PHD domains are required for epigenetic changes and oncogenic activity mediated by NSD2 due to their role in recruiting NSD2 to chromatin [18,145]. Structural studies have shown that PWWP and PHD domains interact with histone via an aromatic cage that surrounds methyllysine, as well as a hydrogen bonding network with nearby histone residues (Figure 6A & B) [141,142]. Although no well-validated inhibitors of PWWP or PHD domains have yet been reported, the histone-binding surface of these domains provides opportunities for development of small molecules that compete with histone [144,146].
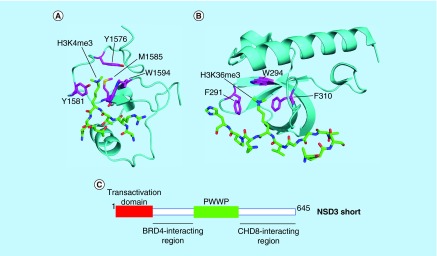
Figure 6. . Targeting protein–protein interacting domains of H3K36 KMTases as an alternative to the SET domain.
Examples of PHD and PWWP domains: (A) Third PHD domain of MLL1 (cyan) bound to H3K4me3 peptide (green) (PDB code 3LQJ) and (B) PWWP domain of ZMYND11 (cyan) bound to H3.3K36me3 peptide (green) (4N4I). Residues on the protein–protein interacting domain that form the aromatic cage are colored magenta. (C) Protein–protein interacting regions in the N-terminus of NSD3 that are required for leukemogenesis.
Adapted with permission from [147].
In some cases, KMTase proteins may engage in PPIs that drive oncogenesis independently of the SET domain. For example, recent studies have shown that NSD3 engages in a rich repertoire of PPIs with other chromatin readers and remodelers. In a subset of NUT midline carcinoma (NMC) cases, an NSD3-NUT fusion protein, which lacks the NSD3 SET domain, blocks differentiation [148]. The NSD3 N-terminus in the fusion protein interacts with the bromodomain-containing protein BRD4 to drive oncogenesis, and NSD3-NUT NMC cells are sensitive to the bromodomain inhibitor JQ1. In addition, wild-type NSD3 is required for proliferation of NMC cells harboring a BRD4-NUT fusion protein, demonstrating that the NSD3/BRD4 interaction is key to the pathogenesis of NMC in general [148]. The interaction between NSD3 and BRD4 was further characterized in AML, in which a short isoform of NSD3 lacking the SET domain drives leukemogenesis by serving as a bridge between BRD4 and the chromatin remodeler CHD8. An N-terminal transactivation domain and PWWP domain are also necessary for NSD3 to activate transcription and maintain leukemia (Figure 6C) [147]. Thus, in some contexts PPIs rather than KMTase activity are key to NSD3's oncogenic role.
These studies reveal that H3K36 KMTases harbor a variety of potentially druggable domains and regions in addition to the SET domain. Precise targeting of individual domains with small molecule inhibitors will ideally leave other protein functions unaffected and may reveal highly specific cancer dependencies. On the other hand, in some cases the entire KMTase protein may need to be eliminated to achieve anticancer activity. To overcome this challenge, one could use small molecule ligands capable of targeting proteins for proteolytic degradation [149,150]. For example, Winter et al. have recently found that coupling a ligand of a target protein to thalidomide results in engagement of the cellular ubiquitin ligase machinery and degradation of the target protein [149]. This method could be applied to KMTases, such that thalidomide conjugates of KMTase ligands could be used to degrade an entire KMTase protein.
Conclusion & future perspective
The H3K36-specific KMTases are prominent drug targets due to numerous studies linking them to oncogenesis. However, large knowledge gaps remain surrounding the mechanisms by which H3K36-specific KMTases drive cancer growth, whether it is by controlling transcription, splicing, DNA repair or other cellular processes. Small molecule inhibitors for H3K36-specific KMTases will be invaluable chemical tools to better understand the role of these proteins in cancer. Developing potent and specific inhibitors for this challenging target class will require creative and nontraditional approaches. Derivatives of SAM and sinefungin have shown promise as specific inhibitors of KMTases and this compound class could be further expanded to cover H3K36-specific KMTases. In addition, the autoinhibitory loop is a unique feature of H3K36-specific SET domains that could be exploited for inhibitor development. Furthermore, some PPI domains of KMTase proteins are required for oncogenic functions and provide excellent opportunities for targeting by small molecules. Major progress on the structure and function of H3K36 KMTases in recent years leads us to predict that the KMTase inhibitor field lies on the precipice of many innovative and exciting discoveries.
Executive summary.
Functions of H3K36 methylation
H3K36 methylation plays important roles in preventing cryptic intragenic transcription, alternative splicing pathways and regulation of DNA repair.
H3K36 methylation antagonizes H3K27 methylation, such that overexpressed NSD2 and high H3K36me2 levels cause aberrant H3K27 methylation patterns that drive oncogenesis.
H3K36 KMTases are implicated in cancer
NSD1 is fused to NUP98 in AML, leading to activation of HOXA and MEIS1 oncogenes.
NSD2 is the key oncogenic driver in t(4;14) MM and has hyperactive KMTase activity or is overexpressed in other solid tumors.
NSD3 is overexpressed in breast cancer and can transform breast epithelial cells in culture.
ASH1L maintains quiescence of hematopoietic stem cells and activates oncogenic HOX genes.
SETD2 is the sole human H3K36 trimethyltransferase and functions as a tumor suppressor.
SMYD2, SETMAR and SETD3 are more distantly related KMTases with uncertain specificity toward H3K36. SYMD2 regulates cancer-associated pathways by methylating nonhistone substrates.
Structural features of H3K36-specific SET domains
NSD, ASH1L and SETD2 SET domains contain an autoinhibitory loop that may be exploited in the design of novel allosteric inhibitors.
Ternary structures of SMYD2 with methyl donor and substrate peptides reveal a U-shaped substrate peptide-binding mode.
Inhibitors for H3K36 KMTases
Specific inhibitors have been reported for SETD2 and SMYD2 SET domains.
Additional inhibitors could be developed by making derivatives of SAM or sinefungin that extend into the substrate lysine-binding channel or mimic the catalytic transition state.
Protein–protein interacting domains of H3K36 KMTase proteins may represent alternative druggable regions.
Acknowledgements
The authors thank J Pollock and M Morgan for critical reading of the manuscript.
Footnotes
Financial & competing interests disclosure
This research has been supported by the Leukemia and Lymphoma Society TRP grants 6111-14 to T Cierpicki and 6485-16 to J Grembecka, NIH 1R01CA160467 to J Grembecka and 1R01CA181185 to T Cierpicki. J Grembecka and T Cierpicki are Leukemia and Lymphoma Society Scholars (grants 1215-14 and 1340-17). DS Rogawski acknowledges training grant support from the University of Michigan Chemistry–Biology Interface (CBI) training program (NIH Grant 5T32GM008597) and from the University of Michigan Medical Scientist Training Program (NIH Grant 5T32GM007863). J Grembecka and T Cierpicki receive research support from Kura Oncology. They are also receiving compensation as members of the scientific advisory board of Kura Oncology, and they have an equity ownership in the company. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
References
Papers of special note have been highlighted as: • of interest; •• of considerable interest
- 1.Dawson MA, Kouzarides T. Cancer epigenetics: from mechanism to therapy. Cell. 2012;150(1):12–27. doi: 10.1016/j.cell.2012.06.013. [DOI] [PubMed] [Google Scholar]
- 2.Daigle SR, Olhava EJ, Therkelsen CA, et al. Selective killing of mixed lineage leukemia cells by a potent small-molecule DOT1L inhibitor. Cancer Cell. 2011;20(1):53–65. doi: 10.1016/j.ccr.2011.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuo AJ, Cheung P, Chen K, et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol. Cell. 2011;44(4):609–620. doi: 10.1016/j.molcel.2011.08.042. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Demonstrates that NSD2 KMTase activity is required for oncogenesis in t(4;14) multiple myeloma (MM).
- 4.McCabe MT, Creasy CL. EZH2 as a potential target in cancer therapy. Epigenomics. 2014;6(3):341–351. doi: 10.2217/epi.14.23. [DOI] [PubMed] [Google Scholar]
- 5.Waters NJ, Smith SA, Olhava EJ, et al. Metabolism and disposition of the DOT1L inhibitor, pinometostat (EPZ-5676), in rat, dog and human. Cancer Chemother. Pharmacol. 2016;77(1):43–62. doi: 10.1007/s00280-015-2929-y. [DOI] [PubMed] [Google Scholar]
- 6.Kuntz KW, Campbell JE, Keilhack H, et al. The importance of being me: magic methyls, methyltransferase inhibitors, and the discovery of tazemetostat. J. Med. Chem. 2016;59(4) doi: 10.1021/acs.jmedchem.5b01501. acs.jmedchem.5b01501. [DOI] [PubMed] [Google Scholar]
- 7.Jones M, Chase J, Brinkmeier M, et al. Ash1l controls quiescence and self-renewal potential in hematopoietic stem cells. J. Clin. Invest. 2015;125(5):2007–2020. doi: 10.1172/JCI78124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang GG, Cai L, Pasillas MP, Kamps MP. NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat. Cell Biol. 2007;9(7):804–812. doi: 10.1038/ncb1608. [DOI] [PubMed] [Google Scholar]; • Shows that NSD1 KMTase activity is required for HOX gene activation and oncogenesis in leukemia with NUP98-NSD1 translocation.
- 9.Pei H, Zhang L, Luo K, et al. MMSET regulates histone H4K20 methylation and 53BP1 accumulation at DNA damage sites. Nature. 2011;470(7332):124–128. doi: 10.1038/nature09658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sein H, Värv S, Kristjuhan A. Distribution and maintenance of histone H3 lysine 36 trimethylation in transcribed locus. PLoS ONE. 2015;10(3):e0120200. doi: 10.1371/journal.pone.0120200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rao B, Shibata Y, Strahl BD, Lieb JD. Dimethylation of histone H3 at lysine 36 demarcates regulatory and nonregulatory chromatin genome-wide. Mol. Cell Biol. 2005;25(21):9447–9459. doi: 10.1128/MCB.25.21.9447-9459.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ernst J, Kellis M. Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 2010;28(8):817–825. doi: 10.1038/nbt.1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sen P, Dang W, Donahue G, et al. H3K36 methylation promotes longevity by enhancing transcriptional fidelity. Genes Dev. 2015;29(13):1362–1376. doi: 10.1101/gad.263707.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kizer KO, Phatnani HP, Shibata Y, Hall H, Greenleaf AL, Strahl BD. A novel domain in Set2 mediates RNA polymerase II interaction and couples histone H3 K36 methylation with transcript elongation. Mol. Cell Biol. 2005;25(8):3305–3316. doi: 10.1128/MCB.25.8.3305-3316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Carrozza MJ, Li B, Florens L, et al. Histone H3 methylation by Set2 directs deacetylation of coding regions by Rpd3S to suppress spurious intragenic transcription. Cell. 2005;123(4):581–592. doi: 10.1016/j.cell.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 16.Venkatesh S, Smolle M, Li H, et al. Set2 methylation of histone H3 lysine 36 suppresses histone exchange on transcribed genes. Nature. 2012;489(7416):452–455. doi: 10.1038/nature11326. [DOI] [PubMed] [Google Scholar]
- 17.Smolle M, Venkatesh S, Gogol MM, et al. Chromatin remodelers Isw1 and Chd1 maintain chromatin structure during transcription by preventing histone exchange. Nat. Struct. Mol. Biol. 2012;19(9):884–892. doi: 10.1038/nsmb.2312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Popovic R, Martinez-Garcia E, Giannopoulou EG, et al. Histone methyltransferase MMSET/NSD2 alters EZH2 binding and reprograms the myeloma epigenome through global and focal changes in H3K36 and H3K27 methylation. PLoS Genet. 2014;10(9):e1004566. doi: 10.1371/journal.pgen.1004566. [DOI] [PMC free article] [PubMed] [Google Scholar]; •• Reveals important changes in H3K27 methylation that occur together with a global increase in H3K36 methylation in t(4;14) MM.
- 19.Carvalho S, Raposo AC, Martins FB, et al. Histone methyltransferase SETD2 coordinates FACT recruitment with nucleosome dynamics during transcription. Nucleic Acids Res. 2013;41(5):2881–2893. doi: 10.1093/nar/gks1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xie L, Pelz C, Wang W, et al. KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 2011;30(8):1473–1484. doi: 10.1038/emboj.2011.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang R, Barbera AJ, Xu Y, et al. Human LSD2/KDM1b/AOF1 regulates gene transcription by modulating intragenic H3K4me2 methylation. Mol. Cell. 2010;39(2):222–233. doi: 10.1016/j.molcel.2010.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Oltean S, Bates DO. Hallmarks of alternative splicing in cancer. Oncogene. 2013;33(46):5311–5318. doi: 10.1038/onc.2013.533. [DOI] [PubMed] [Google Scholar]
- 23.Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat. Struct. Mol. Biol. 2009;16(9):990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- 24.Luco RF, Pan Q, Tominaga K, Blencowe BJ, Pereira-Smith OM, Misteli T. Regulation of alternative splicing by histone modifications. Science. 2010;327(5968):996–1000. doi: 10.1126/science.1184208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pradeepa MM, Sutherland HG, Ule J, Grimes GR, Bickmore WA. Psip1/Ledgf p52 binds methylated histone H3K36 and splicing factors and contributes to the regulation of alternative splicing. PLoS Genet. 2012;8(5):e1002717. doi: 10.1371/journal.pgen.1002717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo R, Zheng L, Park JW, et al. BS69/ZMYND11 reads and connects histone H3.3 lysine 36 trimethylation-decorated chromatin to regulated pre-mRNA processing. Mol. Cell. 2014;56(2):298–310. doi: 10.1016/j.molcel.2014.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kim S, Kim H, Fong N, Erickson B, Bentley DL. Pre-mRNA splicing is a determinant of histone H3K36 methylation. Proc. Natl Acad. Sci. USA. 2011;108(33):13564–13569. doi: 10.1073/pnas.1109475108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.de Almeida SF, Grosso AR, Koch F, et al. Splicing enhances recruitment of methyltransferase HYPB/Setd2 and methylation of histone H3 Lys36. Nat. Struct. Mol. Biol. 2011;18(9):977–983. doi: 10.1038/nsmb.2123. [DOI] [PubMed] [Google Scholar]
- 29.Pai C-C, Deegan RS, Subramanian L, et al. A histone H3K36 chromatin switch coordinates DNA double-strand break repair pathway choice. Nat. Commun. 2014;5:4091. doi: 10.1038/ncomms5091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jha DK, Strahl BD. An RNA polymerase II-coupled function for histone H3K36 methylation in checkpoint activation and DSB repair. Nat. Commun. 2014;5:3965. doi: 10.1038/ncomms4965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pfister SX, Ahrabi S, Zalmas L-P, et al. SETD2-dependent histone H3K36 trimethylation is required for homologous recombination repair and genome stability. Cell Rep. 2014;7(6):2006–2018. doi: 10.1016/j.celrep.2014.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Mechanistic study of SETD2's role in DNA repair.
- 32.Li F, Mao G, Tong D, et al. The histone mark H3K36me3 regulates human DNA mismatch repair through its interaction with MutSα. Cell. 2013;153(3):590–600. doi: 10.1016/j.cell.2013.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fnu S, Williamson EA, De Haro LP, et al. Methylation of histone H3 lysine 36 enhances DNA repair by nonhomologous end-joining. Proc. Natl Acad. Sci. USA. 2010;108(2):540–545. doi: 10.1073/pnas.1013571108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kim H-S, Kim S-K, Hromas R, Lee S-H. The SET domain is essential for metnase functions in replication restart and the 5′ end of SS-overhang cleavage. PLoS ONE. 2015;10(10):e0139418. doi: 10.1371/journal.pone.0139418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carlson SM, Moore KE, Sankaran SM, Reynoird N, Elias JE, Gozani O. A proteomic strategy identifies lysine methylation of splicing factor snRNP70 by the SETMAR enzyme. J. Biol. Chem. 2015;290(19):12040–12047. doi: 10.1074/jbc.M115.641530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dorighi KM, Tamkun JW. The trithorax group proteins Kismet and ASH1 promote H3K36 dimethylation to counteract Polycomb group repression in Drosophila . Development. 2013;140(20):4182–4192. doi: 10.1242/dev.095786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B. H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J. Biol. Chem. 2011;286(10):7983–7989. doi: 10.1074/jbc.M110.194027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yuan G, Ma B, Yuan W, et al. Histone H2A ubiquitination inhibits the enzymatic activity of H3 lysine 36 methyltransferases. J. Biol. Chem. 2013;288(43):30832–30842. doi: 10.1074/jbc.M113.475996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cai L, Rothbart SB, Lu R, et al. An H3K36 methylation-engaging Tudor motif of polycomb-like proteins mediates PRC2 complex targeting. Mol. Cell. 2013;49(3):571–582. doi: 10.1016/j.molcel.2012.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Komatsu S, Ichikawa D, Hirajima S, et al. Overexpression of SMYD2 contributes to malignant outcome in gastric cancer. Br. J. Cancer. 2015;112(2):357–364. doi: 10.1038/bjc.2014.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Komatsu S, Imoto I, Tsuda H, et al. Overexpression of SMYD2 relates to tumor cell proliferation and malignant outcome of esophageal squamous cell carcinoma. Carcinogenesis. 2009;30(7):1139–1146. doi: 10.1093/carcin/bgp116. [DOI] [PubMed] [Google Scholar]
- 42.Ohtomo-Oda R, Komatsu S, Mori T, et al. SMYD2 overexpression is associated with tumor cell proliferation and a worse outcome in human papillomavirus-unrelated nonmultiple head and neck carcinomas. Hum. Pathol. 2016;49:145–155. doi: 10.1016/j.humpath.2015.08.025. [DOI] [PubMed] [Google Scholar]
- 43.Sakamoto LHT, de Andrade RV, Felipe MSS, Motoyama AB, Pittella Silva F. SMYD2 is highly expressed in pediatric acute lymphoblastic leukemia and constitutes a bad prognostic factor. Leuk. Res. 2014;38(4):496–502. doi: 10.1016/j.leukres.2014.01.013. [DOI] [PubMed] [Google Scholar]
- 44.Cho H-S, Hayami S, Toyokawa G, et al. RB1 methylation by SMYD2 enhances cell cycle progression through an increase of RB1 phosphorylation. Neoplasia. 2012;14(6):476–486. doi: 10.1593/neo.12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kurotaki N, Imaizumi K, Harada N, et al. Haploinsufficiency of NSD1 causes Sotos syndrome. Nat. Genet. 2002;30(4):365–366. doi: 10.1038/ng863. [DOI] [PubMed] [Google Scholar]
- 46.Hersh JH, Cole TRP, Bloom AS, Bertolone SJ, Hughes HE. Risk of malignancy in Sotos syndrome. J. Pediatr. 1992;120(4):572–574. doi: 10.1016/s0022-3476(10)80004-6. [DOI] [PubMed] [Google Scholar]
- 47.Hollink IHIM, van den Heuvel-Eibrink MM, Arentsen-Peters STCJM, et al. NUP98/NSD1 characterizes a novel poor prognostic group in acute myeloid leukemia with a distinct HOX gene expression pattern. Blood. 2011;118(13):3645–3656. doi: 10.1182/blood-2011-04-346643. [DOI] [PubMed] [Google Scholar]
- 48.Ostronoff F, Othus M, Gerbing RB, et al. NUP98/NSD1 and FLT3/ITD coexpression is more prevalent in younger AML patients and leads to induction failure: a COG and SWOG report. Blood. 2014;124(15):2400–2407. doi: 10.1182/blood-2014-04-570929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deshpande AJ, Deshpande A, Sinha AU, et al. AF10 regulates progressive H3K79 methylation and HOX gene expression in diverse AML subtypes. Cancer Cell. 2014;26(6):896–908. doi: 10.1016/j.ccell.2014.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lu T, Jackson MW, Wang B, et al. Regulation of NF-kappaB by NSD1/FBXL11-dependent reversible lysine methylation of p65. Proc. Natl Acad. Sci. USA. 2010;107(1):46–51. doi: 10.1073/pnas.0912493107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Prasad S, Ravindran J, Aggarwal BB. NF-kappaB and cancer: how intimate is this relationship. Mol. Cell Biochem. 2010;336(1–2):25–37. doi: 10.1007/s11010-009-0267-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nakshatri H, Appaiah HN, Anjanappa M, et al. NF-κB-dependent and -independent epigenetic modulation using the novel anti-cancer agent DMAPT. Cell Death Dis. 2015;6:e1608. doi: 10.1038/cddis.2014.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Lawrence MS, Sougnez C, Lichtenstein L, et al. Comprehensive genomic characterization of head and neck squamous cell carcinomas. Nature. 2015;517(7536):576–582. doi: 10.1038/nature14129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berdasco M, Ropero S, Setien F, et al. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc. Natl Acad. Sci. USA. 2009;106(51):21830–21835. doi: 10.1073/pnas.0906831106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yan X-J, Xu J, Gu Z-H, et al. Exome sequencing identifies somatic mutations of DNA methyltransferase gene DNMT3A in acute monocytic leukemia. Nat. Genet. 2011;43(4):309–315. doi: 10.1038/ng.788. [DOI] [PubMed] [Google Scholar]
- 56.Bergemann AD, Cole F, Hirschhorn K. The etiology of Wolf–Hirschhorn syndrome. Trends Genet. 2005;21(3):188–195. doi: 10.1016/j.tig.2005.01.008. [DOI] [PubMed] [Google Scholar]
- 57.Hajdu I, Ciccia A, Lewis SM, Elledge SJ. Wolf–Hirschhorn syndrome candidate 1 is involved in the cellular response to DNA damage. Proc. Natl Acad. Sci. USA. 2011;108(32):13130–13134. doi: 10.1073/pnas.1110081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hsiao K-Y, Mizzen CA. Histone H4 deacetylation facilitates 53BP1 DNA damage signaling and double-strand break repair. J. Mol. Cell Biol. 2013;5(3):157–165. doi: 10.1093/jmcb/mjs066. [DOI] [PubMed] [Google Scholar]
- 59.Hartlerode AJ, Guan Y, Rajendran A, et al. Impact of histone H4 lysine 20 methylation on 53BP1 responses to chromosomal double strand breaks. PLoS ONE. 2012;7(11):e49211. doi: 10.1371/journal.pone.0049211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mirabella F, Wu P, Wardell CP, et al. MMSET is the key molecular target in t(4;14) myeloma. Blood Cancer J. 2013;3:e114. doi: 10.1038/bcj.2013.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Hudlebusch HR, Santoni-Rugiu E, Simon R, et al. The histone methyltransferase and putative oncoprotein MMSET is overexpressed in a large variety of human tumors. Clin. Cancer Res. 2011;17(9):2919–2933. doi: 10.1158/1078-0432.CCR-10-1302. [DOI] [PubMed] [Google Scholar]
- 62.Hudlebusch HR, Skotte J, Santoni-Rugiu E, et al. MMSET is highly expressed and associated with aggressiveness in neuroblastoma. Cancer Res. 2011;71(12):4226–4235. doi: 10.1158/0008-5472.CAN-10-3810. [DOI] [PubMed] [Google Scholar]
- 63.Oyer JA, Huang X, Zheng Y, et al. Point mutation E1099K in MMSET/NSD2 enhances its methyltranferase activity and leads to altered global chromatin methylation in lymphoid malignancies. Leukemia. 2014;28(1):198–201. doi: 10.1038/leu.2013.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jaffe JD, Wang Y, Chan HM, et al. Global chromatin profiling reveals NSD2 mutations in pediatric acute lymphoblastic leukemia. Nat. Genet. 2013;45(11):1386–1391. doi: 10.1038/ng.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Min D-J, Ezponda T, Kim MK, et al. MMSET stimulates myeloma cell growth through microRNA-mediated modulation of c-MYC. Leukemia. 2012;27(3):686–694. doi: 10.1038/leu.2012.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Asangani IA, Ateeq B, Cao Q, et al. Characterization of the EZH2-MMSET histone methyltransferase regulatory axis in cancer. Mol. Cell. 2013;49(1):80–93. doi: 10.1016/j.molcel.2012.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Kang H-B, Choi Y, Lee JM, et al. The histone methyltransferase, NSD2, enhances androgen receptor-mediated transcription. FEBS Lett. 2009;583(12):1880–1886. doi: 10.1016/j.febslet.2009.05.038. [DOI] [PubMed] [Google Scholar]
- 68.Yang P, Guo L, Duan ZJ, et al. Histone methyltransferase NSD2/MMSET mediates constitutive NF-κB signaling for cancer cell proliferation, survival, and tumor growth via a feed-forward loop. Mol. Cell Biol. 2012;32(15):3121–3131. doi: 10.1128/MCB.00204-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ezponda T, Popovic R, Shah MY, et al. The histone methyltransferase MMSET/WHSC1 activates TWIST1 to promote an epithelial-mesenchymal transition and invasive properties of prostate cancer. Oncogene. 2013;32(23):2882–2890. doi: 10.1038/onc.2012.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Chen Y, McGee J, Chen X, et al. Identification of druggable cancer driver genes amplified across TCGA datasets. PLoS ONE. 2014;9(5):e98293. doi: 10.1371/journal.pone.0098293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Li Y, Trojer P, Xu C-F, et al. The target of the NSD family of histone lysine methyltransferases depends on the nature of the substrate. J. Biol. Chem. 2009;284(49):34283–34295. doi: 10.1074/jbc.M109.034462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Jacques-Fricke BT, Gammill LS. Neural crest specification and migration independently require NSD3-related lysine methyltransferase activity. Mol. Biol. Cell. 2014;25(25):4174–4186. doi: 10.1091/mbc.E13-12-0744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Rosati R, La Starza R, Veronese A, et al. NUP98 is fused to the NSD3 gene in acute myeloid leukemia associated with t(8;11)(p11.2;p15) Blood. 2002;99(10):3857–3860. doi: 10.1182/blood.v99.10.3857. [DOI] [PubMed] [Google Scholar]
- 74.Taketani T, Taki T, Nakamura H, Taniwaki M, Masuda J, Hayashi Y. NUP98-NSD3 fusion gene in radiation-associated myelodysplastic syndrome with t(8;11)(p11;p15) and expression pattern of NSD family genes. Cancer Genet. Cytogenet. 2009;190(2):108–112. doi: 10.1016/j.cancergencyto.2008.12.008. [DOI] [PubMed] [Google Scholar]
- 75.Kang D, Cho H-S, Toyokawa G, et al. The histone methyltransferase Wolf–Hirschhorn syndrome candidate 1-like 1 (WHSC1L1) is involved in human carcinogenesis. Genes Chromosomes Cancer. 2013;52(2):126–139. doi: 10.1002/gcc.22012. [DOI] [PubMed] [Google Scholar]
- 76.Yang Z-Q, Liu G, Bollig-Fischer A, Giroux CN, Ethier SP. Transforming properties of 8p11–12 amplified genes in human breast cancer. Cancer Res. 2010;70(21):8487–8497. doi: 10.1158/0008-5472.CAN-10-1013. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Identifies NSD3 as a potent transforming oncogene in breast cancer.
- 77.Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G. Genome regulation by polycomb and trithorax proteins. Cell. 2007;128(4):735–745. doi: 10.1016/j.cell.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 78.Zhu L, Li Q, Wong SHK, et al. ASH1L links histone H3 lysine 36 di-methylation to MLL leukemia. Cancer Discov. 2016;7(6):770–783. doi: 10.1158/2159-8290.CD-16-0058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Cabianca DS, Casa V, Bodega B, et al. A long ncRNA links copy number variation to a polycomb/trithorax epigenetic switch in FSHD muscular dystrophy. Cell. 2012;149(4):819–831. doi: 10.1016/j.cell.2012.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Perugorria MJ, Wilson CL, Zeybel M, et al. Histone methyltransferase ASH1 orchestrates fibrogenic gene transcription during myofibroblast transdifferentiation. Hepatology. 2012;56(3):1129–1139. doi: 10.1002/hep.25754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Tanaka Y, Kawahashi K, Katagiri ZI, Nakayama Y, Mahajan M, Kioussis D. Dual function of histone H3 lysine 36 methyltransferase ASH1 in regulation of hox gene expression. PLoS ONE. 2011;6(11):e28171. doi: 10.1371/journal.pone.0028171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shah N, Sukumar S. The Hox genes and their roles in oncogenesis. Nat. Rev. Cancer. 2010;10(5):361–371. doi: 10.1038/nrc2826. [DOI] [PubMed] [Google Scholar]
- 83.Andreeff M, Ruvolo V, Gadgil S, et al. HOX expression patterns identify a common signature for favorable AML. Leukemia. 2008;22(11):2041–2047. doi: 10.1038/leu.2008.198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Faber J, Krivtsov AV, Stubbs MC, et al. HOXA9 is required for survival in human MLL-rearranged acute leukemias. Blood. 2009;113(11):2375–2385. doi: 10.1182/blood-2007-09-113597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Kroon E, Krosl J, Thorsteinsdottir U, Baban S, Buchberg AM, Sauvageau G. Hoxa9 transforms primary bone marrow cells through specific collaboration with Meis1a but not Pbx1b. EMBO J. 1998;17(13):3714–3725. doi: 10.1093/emboj/17.13.3714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Miyazaki H, Higashimoto K, Yada Y, et al. Ash1l methylates Lys36 of histone H3 independently of transcriptional elongation to counteract polycomb silencing. PLoS Genet. 2013;9(11):e1003897. doi: 10.1371/journal.pgen.1003897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Colamaio M, Puca F, Ragozzino E, et al. miR-142–3p downregulation contributes to thyroid follicular tumorigenesis by targeting ASH1L and MLL1. J. Clin. Endocrinol. Metab. 2014;100(1):E59–E69. doi: 10.1210/jc.2014-2280. [DOI] [PubMed] [Google Scholar]
- 88.Liu L, Kimball S, Liu H, Holowatyj A, Yang Z-Q. Genetic alterations of histone lysine methyltransferases and their significance in breast cancer [Internet] Oncotarget. 6(4):2466–2482. doi: 10.18632/oncotarget.2967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Kanellopoulou C, Gilpatrick T, Kilaru G, et al. Reprogramming of polycomb-mediated gene silencing in embryonic stem cells by the miR-290 family and the methyltransferase Ash1l. Stem Cell Rep. 2015;5(6):971–978. doi: 10.1016/j.stemcr.2015.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Edmunds JW, Mahadevan LC, Clayton AL. Dynamic histone H3 methylation during gene induction: HYPB/Setd2 mediates all H3K36 trimethylation. EMBO J. 2008;27(2):406–420. doi: 10.1038/sj.emboj.7601967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Luscan A, Laurendeau I, Malan V, et al. Mutations in SETD2 cause a novel overgrowth condition. J. Med. Genet. 2014;51(8):512–517. doi: 10.1136/jmedgenet-2014-102402. [DOI] [PubMed] [Google Scholar]
- 92.Al Sarakbi W, Sasi W, Jiang WG, Roberts T, Newbold RF, Mokbel K. The mRNA expression of SETD2 in human breast cancer: correlation with clinico-pathological parameters. BMC Cancer. 2009;9:290. doi: 10.1186/1471-2407-9-290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Hakimi AA, Ostrovnaya I, Reva B, et al. Adverse outcomes in clear cell renal cell carcinoma with mutations of 3p21 epigenetic regulators BAP1 and SETD2: a report by MSKCC and the KIRC TCGA research network. Clin. Cancer Res. 2013;19(12):3259–3267. doi: 10.1158/1078-0432.CCR-12-3886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Dalgliesh GL, Furge K, Greenman C, et al. Systematic sequencing of renal carcinoma reveals inactivation of histone modifying genes. Nature. 2010;463(7279):360–363. doi: 10.1038/nature08672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mar BG, Bullinger LB, McLean KM, et al. Mutations in epigenetic regulators including SETD2 are gained during relapse in paediatric acute lymphoblastic leukaemia. Nat. Commun. 2014;5:3469. doi: 10.1038/ncomms4469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Huang KK, McPherson JR, Tay ST, et al. SETD2 histone modifier loss in aggressive GI stromal tumours. Gut. 2015 doi: 10.1136/gutjnl-2015-309482. Epub ahead of print. [DOI] [PubMed] [Google Scholar]
- 97.Zhu X, He F, Zeng H, et al. Identification of functional cooperative mutations of SETD2 in human acute leukemia. Nat. Genet. 2014;46(3):287–293. doi: 10.1038/ng.2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kandoth C, McLellan MD, Vandin F, et al. Mutational landscape and significance across 12 major cancer types. Nature. 2013;502(7471):333–339. doi: 10.1038/nature12634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Kovac M, Navas C, Horswell S, et al. Recurrent chromosomal gains and heterogeneous driver mutations characterise papillary renal cancer evolution. Nat. Commun. 2015;6:6336. doi: 10.1038/ncomms7336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Gerlinger M, Rowan AJ, Horswell S, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N. Engl. J. Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Xiang W, He J, Huang C, et al. miR-106b-5p targets tumor suppressor gene SETD2 to inactive its function in clear cell renal cell carcinoma [Internet] Oncotarget. 2015;6(6):4066–4079. doi: 10.18632/oncotarget.2926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Kanu N, Grönroos E, Martinez P, et al. SETD2 loss-of-function promotes renal cancer branched evolution through replication stress and impaired DNA repair. Oncogene. 2015;34(46):5699–5708. doi: 10.1038/onc.2015.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Grosso AR, Leite AP, Carvalho S, et al. Pervasive transcription read-through promotes aberrant expression of oncogenes and RNA chimeras in renal carcinoma. Elife. 2015;4:pii: e09214. doi: 10.7554/eLife.09214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Simon JM, Hacker KE, Singh D, et al. Variation in chromatin accessibility in human kidney cancer links H3K36 methyltransferase loss with widespread RNA processing defects. Genome Res. 2014;24(2):241–250. doi: 10.1101/gr.158253.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Tiedemann RL, Hlady RA, Hanavan PD, et al. Dynamic reprogramming of DNA methylation in SETD2-deregulated renal cell carcinoma. Oncotarget. 2015;7(2):1927–1946. doi: 10.18632/oncotarget.6481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Abu-Farha M, Lambert J-P, Al-Madhoun AS, Elisma F, Skerjanc IS, Figeys D. The tale of two domains: proteomics and genomics analysis of SMYD2, a new histone methyltransferase. Mol. Cell Proteomics. 2008;7(3):560–572. doi: 10.1074/mcp.M700271-MCP200. [DOI] [PubMed] [Google Scholar]
- 107.Xu G, Liu G, Xiong S, Liu H, Chen X, Zheng B. The histone methyltransferase Smyd2 is a negative regulator of macrophage activation by suppressing interleukin 6 (IL-6) and tumor necrosis factor α (TNF-α) production. J. Biol. Chem. 2015;290(9):5414–5423. doi: 10.1074/jbc.M114.610345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Nguyen H, Allali-Hassani A, Antonysamy S, et al. LLY-507, a cell-active, potent, and selective inhibitor of protein-lysine methyltransferase SMYD2. J. Biol. Chem. 2015;290(22):13641–13653. doi: 10.1074/jbc.M114.626861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Huang J, Perez-Burgos L, Placek BJ, et al. Repression of p53 activity by Smyd2-mediated methylation. Nature. 2006;444(7119):629–632. doi: 10.1038/nature05287. [DOI] [PubMed] [Google Scholar]
- 110.Saddic LA, West LE, Aslanian A, et al. Methylation of the retinoblastoma tumor suppressor by SMYD2. J. Biol. Chem. 2010;285(48):37733–37740. doi: 10.1074/jbc.M110.137612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Abu-Farha M, Lanouette S, Elisma F, et al. Proteomic analyses of the SMYD family interactomes identify HSP90 as a novel target for SMYD2. J. Mol. Cell Biol. 2011;3(5):301–308. doi: 10.1093/jmcb/mjr025. [DOI] [PubMed] [Google Scholar]
- 112.Zhang X, Tanaka K, Yan J, et al. Regulation of estrogen receptor α by histone methyltransferase SMYD2-mediated protein methylation. Proc. Natl Acad. Sci. USA. 2013;110(43):17284–17289. doi: 10.1073/pnas.1307959110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Piao L, Kang D, Suzuki T, et al. The histone methyltransferase SMYD2 methylates PARP1 and promotes poly(ADP-ribosyl)ation activity in cancer cells. Neoplasia. 2014;16(3):257–264. doi: 10.1016/j.neo.2014.03.002. 264.e2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Cordaux R, Udit S, Batzer MA, Feschotte C. Birth of a chimeric primate gene by capture of the transposase gene from a mobile element. Proc. Natl Acad. Sci. USA. 2006;103(21):8101–8106. doi: 10.1073/pnas.0601161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Apostolou P, Toloudi M, Kourtidou E, et al. Potential role for the Metnase transposase fusion gene in colon cancer through the regulation of key genes. PLoS ONE. 2014;9(10):e109741. doi: 10.1371/journal.pone.0109741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Williamson EA, Rasila KK, Corwin LK, et al. The SET and transposase domain protein Metnase enhances chromosome decatenation: regulation by automethylation. Nucleic Acids Res. 2008;36(18):5822–5831. doi: 10.1093/nar/gkn560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Eom GH, Kim K-B, Kim JH, et al. Histone methyltransferase SETD3 regulates muscle differentiation. J. Biol. Chem. 2011;286(40):34733–34742. doi: 10.1074/jbc.M110.203307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Pires-Luís AS, Vieira-Coimbra M, Vieira FQ, et al. Expression of histone methyltransferases as novel biomarkers for renal cell tumor diagnosis and prognostication. Epigenetics. 2015;10(11):1033–1043. doi: 10.1080/15592294.2015.1103578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Cooper SE, Hodimont E, Green CM. A fluorescent bimolecular complementation screen reveals MAF1, RNF7 and SETD3 as PCNA-associated proteins in human cells. Cell Cycle. 2015;14(15):2509–2519. doi: 10.1080/15384101.2015.1053667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.An S, Yeo KJ, Jeon YH, Song JJ. Crystal structure of the human histone methyltransferase ASH1L catalytic domain and its implications for the regulatory mechanism. J. Biol. Chem. 2011;286(10):8369–8374. doi: 10.1074/jbc.M110.203380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Qiao Q, Li Y, Chen Z, Wang M, Reinberg D, Xu R-M. The structure of NSD1 reveals an autoregulatory mechanism underlying histone H3K36 methylation. J. Biol. Chem. 2011;286(10):8361–8368. doi: 10.1074/jbc.M110.204115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Zheng W, Ibáñez G, Wu H, et al. Sinefungin derivatives as inhibitors and structure probes of protein lysine methyltransferase SETD2. J. Am. Chem. Soc. 2012;134(43):18004–18014. doi: 10.1021/ja307060p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Wang L, Li L, Zhang H, et al. Structure of human SMYD2 protein reveals the basis of p53 tumor suppressor methylation. J. Biol. Chem. 2011;286(44):38725–38737. doi: 10.1074/jbc.M111.262410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Ferguson AD, Larsen NA, Howard T, et al. Structural basis of substrate methylation and inhibition of SMYD2. Structure. 2011;19(9):1262–1273. doi: 10.1016/j.str.2011.06.011. [DOI] [PubMed] [Google Scholar]
- 125.Xiao B, Jing C, Kelly G, et al. Specificity and mechanism of the histone methyltransferase Pr-Set7. Genes Dev. 2005;19(12):1444–1454. doi: 10.1101/gad.1315905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Horowitz S, Yesselman JD, Al-Hashimi HM, Trievel RC. Direct evidence for methyl group coordination by carbon-oxygen hydrogen bonds in the lysine methyltransferase SET7/9. J. Biol. Chem. 2011;286(21):18658–18663. doi: 10.1074/jbc.M111.232876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Rogawski DS, Ndoj J, Cho HJ, Maillard I, Grembecka J, Cierpicki T. Two loops undergoing concerted dynamics regulate the activity of the ASH1L histone methyltransferase. Biochemistry. 2015;54(35):5401–5413. doi: 10.1021/acs.biochem.5b00697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Graham SE, Tweedy SE, Carlson HA. Dynamic behavior of the post-SET loop region of NSD1: implications for histone binding and drug development. Protein Sci. 2016;25(5):1021–1029. doi: 10.1002/pro.2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Tempel W, Yu W, Dong A. PDB ID: 4YZ8. methyltransferase domain of human Wolf–Hirschhorn Syndrome Candidate 1-Like protein 1 (WHSC1L1) www.rcsb.org/pdb/explore.do?structureId=4YZ8
- 130.Jiang Y, Sirinupong N, Brunzelle J, Yang Z. Crystal structures of histone and p53 methyltransferase SmyD2 reveal a conformational flexibility of the autoinhibitory C-terminal domain. PLoS ONE. 2011;6(6):e21640. doi: 10.1371/journal.pone.0021640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xu S, Zhong C, Zhang T, Ding J. Structure of human lysine methyltransferase Smyd2 reveals insights into the substrate divergence in Smyd proteins. J. Mol. Cell Biol. 2011;3(5):293–300. doi: 10.1093/jmcb/mjr015. [DOI] [PubMed] [Google Scholar]
- 132.Jiang Y, Trescott L, Holcomb J, et al. Structural insights into estrogen receptor α methylation by histone methyltransferase SMYD2, a cellular event implicated in estrogen signaling regulation. J. Mol. Biol. 2014;426(20):3413–3425. doi: 10.1016/j.jmb.2014.02.019. [DOI] [PubMed] [Google Scholar]
- 133.Sweis RF, Wang Z, Algire M, et al. Discovery of A-893, a new cell-active benzoxazinone inhibitor of lysine methyltransferase SMYD2. ACS Med. Chem. Lett. 2015;6(6):695–700. doi: 10.1021/acsmedchemlett.5b00124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Greiner D, Bonaldi T, Eskeland R, Roemer E, Imhof A. Identification of a specific inhibitor of the histone methyltransferase SU(VAR)3–9. Nat. Chem. Biol. 2005;1(3):143–145. doi: 10.1038/nchembio721. [DOI] [PubMed] [Google Scholar]
- 135.Campagna-Slater V, Mok MW, Nguyen KT, Feher M, Najmanovich R, Schapira M. Structural chemistry of the histone methyltransferases cofactor binding site. J. Chem. Inf. Model. 2011;51(3):612–623. doi: 10.1021/ci100479z. [DOI] [PubMed] [Google Scholar]
- 136.Devkota K, Lohse B, Liu Q, et al. Analogues of the natural product sinefungin as inhibitors of EHMT1 and EHMT2. ACS Med. Chem. Lett. 2014;5(4):293–297. doi: 10.1021/ml4002503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Yao Y, Chen P, Diao J, et al. Selective inhibitors of histone methyltransferase DOT1L: design, synthesis, and crystallographic studies. J. Am. Chem. Soc. 2011;133(42):16746–16749. doi: 10.1021/ja206312b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schramm VL. Transition states, analogues, and drug development. ACS Chem. Biol. 2013;8(1):71–81. doi: 10.1021/cb300631k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Poulin MB, Schneck JL, Matico RE, et al. Transition state for the NSD2-catalyzed methylation of histone H3 lysine 36. Proc. Natl Acad. Sci. USA. 2016;113(5):1197–1201. doi: 10.1073/pnas.1521036113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Zhang J, Zheng YG. SAM/SAH analogs as versatile tools for SAM-dependent methyltransferases. ACS Chem. Biol. 2015;11(3):583–597. doi: 10.1021/acschembio.5b00812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Wang Z, Song J, Milne TA, et al. Pro isomerization in MLL1 PHD3-bromo cassette connects H3K4me readout to CyP33 and HDAC-mediated repression. Cell. 2010;141(7):1183–1194. doi: 10.1016/j.cell.2010.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Wen H, Li Y, Xi Y, et al. ZMYND11 links histone H3.3K36me3 to transcription elongation and tumour suppression. Nature. 2014;508(7495):263–268. doi: 10.1038/nature13045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sankaran SM, Wilkinson AW, Gozani O. A PWWP domain of histone-lysine N-methyltransferase NSD2 binds to dimethylated Lys36 of histone H3 and regulates NSD2 function at chromatin. J. Biol. Chem. 2016;291(16):8465–8474. doi: 10.1074/jbc.M116.720748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Sanchez R, Zhou M-M. The PHD finger: a versatile epigenome reader. Trends Biochem. Sci. 2011;36(7):364–372. doi: 10.1016/j.tibs.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Huang Z, Wu H, Chuai S, et al. NSD2 is recruited through its PHD domain to oncogenic gene loci to drive multiple myeloma. Cancer Res. 2013;73(20):6277–6288. doi: 10.1158/0008-5472.CAN-13-1000. [DOI] [PubMed] [Google Scholar]
- 146.Eidahl JO, Crowe BL, North JA, et al. Structural basis for high-affinity binding of LEDGF PWWP to mononucleosomes. Nucleic Acids Res. 2013;41(6):3924–3936. doi: 10.1093/nar/gkt074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Shen C, Ipsaro JJ, Shi J, et al. NSD3-Short is an adaptor protein that couples BRD4 to the CHD8 chromatin remodeler. Mol. Cell. 2015;60(6):847–859. doi: 10.1016/j.molcel.2015.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]; • Characterizes role of protein–protein interacting domains of NSD3 in leukemogenesis.
- 148.French CA, Rahman S, Walsh EM, et al. NSD3-NUT fusion oncoprotein in NUT midline carcinoma: implications for a novel oncogenic mechanism. Cancer Discov. 2014;4(8):928–941. doi: 10.1158/2159-8290.CD-14-0014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Winter GE, Buckley DL, Paulk J, et al. Drug development. Phthalimide conjugation as a strategy for in vivo target protein degradation. Science. 2015;348(6241):1376–1381. doi: 10.1126/science.aab1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Bondeson DP, Mares A, Smith IED, et al. Catalytic in vivo protein knockdown by small-molecule PROTACs. Nat. Chem. Biol. 2015;11(8):611–617. doi: 10.1038/nchembio.1858. [DOI] [PMC free article] [PubMed] [Google Scholar]