Abstract
Background
The purpose of our research was to determine the prognostic impact and clinicopathological feature of c-MYC and β-catenin overexpression in colorectal cancer (CRC) patients.
Methods
Using immunohistochemistry (IHC), we measured the c-MYC and β-catenin expression in 367 consecutive CRC patients retrospectively (cohort 1). Also, c-MYC expression was measured by mRNA in situ hybridization. Moreover, to analyze regional heterogeneity, three sites of CRC including the primary, distant and lymph node metastasis were evaluated in 176 advanced CRC patients (cohort 2).
Results
In cohort 1, c-MYC protein and mRNA overexpression and ß-catenin nuclear expression were found in 201 (54.8 %), 241 (65.7 %) and 221 (60.2 %) of 367 patients, respectively, each of which was associated with improved prognosis (P = 0.011, P = 0.012 and P = 0.033, respectively). Moreover, co-expression of c-MYC and ß-catenin was significantly correlated with longer survival by univariate (P = 0.012) and multivariate (P = 0.048) studies. Overexpression of c-MYC protein was associated with mRNA overexpression (ρ, 0.479; P < 0.001) and nuclear ß-catenin expression (ρ, 0.282; P < 0.001). Expression of c-MYC and ß-catenin was heterogeneous depending on location in advanced CRC patients (cohort 2). Nevertheless, both c-MYC and ß-catenin expression in primary cancer were significantly correlated with improved survival in univariate (P = 0.001) and multivariate (P = 0.002) analyses. c-MYC and ß-catenin expression of lymph node or distant metastatic tumor was not significantly correlated with patients’ prognosis (P > 0.05).
Conclusions
Co-expression of c-MYC and ß-catenin was independently correlated with favorable prognosis in CRC patient. We concluded that the expression of c-MYC and ß-catenin might be useful predicting indicator of CRC patient’s prognosis.
Electronic supplementary material
The online version of this article (doi:10.1186/s12885-016-2770-7) contains supplementary material, which is available to authorized users.
Keywords: Colorectal cancer, c-MYC, ß-catenin, Immunohistochemistry, mRNA in situ hybridization, Prognosis
Background
The c-MYC protein encode by c-MYC gene, acts as transcription factor for variable cellular function including proliferation, differentiation, metabolism, survival, and apoptosis [1, 2]. The c-MYC gene can promote tumorigenesis in various malignant tumors [3, 4] and mediate the critical role in the colorectal cancer (CRC) progression [5, 6]. Deregulation of c-MYC is a consequence of mutations in APC, a central hub in early colorectal carcinogenesis [7].
c-MYC gene amplification, translocation, and alteration of regulatory molecules are major causes of c-MYC protein overexpression [8, 9]. Previously, other group indicated that c-MYC amplification and overexpression was showed in approximately 10 and 70 % in CRC, respectively [10]. These studies have deduced that overexpression of c-MYC is controlled by mechanisms other than gene amplification [10]. In recent years, it has been evident that the mechanism of c-MYC overexpression is not restricted to genetic alterations, such as amplification or translocation, but can also occur as a consequence of abnormalities in regulatory molecules [11]; in CRC, ß-catenin is one such regulatory molecule. It is now well established that APC gene mutation, a key driver of adenoma-carcinoma transition, often leads to altered ß-catenin regulation via the well-studied Wnt signaling pathway [12–14]. Regulation of this pathway occurred while changing in nuclear ß-catenin protein levels. A destruction complex maintains a low cytoplasmic concentration of ß-catenin when the Wnt signaling pathway is inactivated. On the contrary, the destruction complex degrades and ß-catenin increases in the cytoplasm, leading to its migration to the nucleus, where it work like a transcriptional factor for c-MYC and cyclin D1 [15, 16]. Recent studies reported CRCs with marked WNT and c-MYC signaling activation as a distinct molecular subtype by gene expression-based CRC classifications, which was associated with relatively better prognosis [17, 18]. It suggests that CRCs with activated c-MYC via Wnt signaling pathway have distinct clinicopathologic characteristics, but it has not been confirmed.
Nevertheless, there were a few researches that reported clinicopathological impact of c-MYC and ß-catenin status in CRC. Their prognostic value for CRC patients remains debatable. A recent study reported that c-MYC protein overexpression obtained by immunohistochemistry (IHC) was significantly correlated with better survival of CRC patients [19]. In contrast, other researchers conducted a meta-analysis showing that the accumulation of nuclear ß-catenin could be a biomarker for advanced stage and worse survival of CRC [20]. However, the correlation between immunohistochemical nuclear ß-catenin expression and patient prognosis is quite controversial. Consequently, it is necessary to further evaluate c-MYC and ß-catenin expression to reach a conclusion about their prognostic value.
Recently, the systemic chemotherapy in CRC has made a remarkable development, and targeted therapy has been used to increase survival in advanced CRC patients [21]. However, targeted therapy has no effect in some CRC patients, despite presenting positivity for target-therapy specific molecular examination [22]. Several researchers have demonstrated that CRC shows a regional heterogeneity in KRAS, EGFR, and BRAF mutation, thus tumor heterogeneity may explain this discrepancy between molecular alteration and responses of targeted therapy [23–25]. Therefore, molecular alterations between the metastatic and primary lesions need to be discovered to enhance the treatment effect of metastatic CRCs.
The aim of our research was to evaluate the clinical implication of c-MYC and ß-catenin in CRC and evaluate their heterogeneity in primary and distant metastatic tumors. We also analyzed the association between c-MYC and ß-catenin status.
Methods
Collection of samples
A total of 543 CRC cases of this study had been collected in our previous study [26]. To investigate the clinicopathological significance of c-MYC and ß-catenin expression, we collected 367 consecutive CRC patients who underwent surgery between 2005 and 2006 at Seoul National University Bundang Hospital (cohort 1). Additionally, to evaluate the locational heterogeneity of c-MYC and ß-catenin expression, we collected synchronous or metachronous metastatic 176 CRC patients with who had received surgery between 2003 and 2004, as well as between 2007 and 2009 excluding any patient already enrolled in cohort 1 (cohort 2). Pathologists K.S.L and H.S.L reviewed all the cases. Cancer stage was determined from the American Joint Committee on Cancer (AJCC) 7th edition. Clinical and pathologic information was acquired from hospital medical records including patient’ outcome and survival.
Tissue array method
Tissue microarray (TMA) was constructed with representative lesions of the donor formalin-fixed paraffin-embedded (FFPE) CRC tissues as previously described [27].
Immunohistochemistry
c-MYC IHC analysis was performed using an antibody against c-MYC (clone Y69, catalog ab32072, Abcam, Burlingame, CA, USA). ß-catenin IHC used a commercially available antibody against ß-catenin (clone CAT-5H10, Invitrogen, Camarillo, CA, USA). The staining process was performed using an automated immunostainer (BenchMark XT, Ventana Medical Systems), according to the manufacturer’s recommendations. Normal colonic mucosa cells were considered as internal negative controls. Normal mucosa was negative for c-MYC nuclear immunostaining. ß-catenin was negative in inflammatory cells, but expressed in colonic epithelium in three patterns: membrane, cytoplasm, and nucleus. We only found ß-catenin nuclear expression in malignant cells. For statistical analysis, c-MYC and ß-catenin immunostaining were regarded as positive when they were expressed in more than 10 % of neoplastic nucleus in any intensity (Fig. 1) [19, 28]. Negative controls were obtained omitting the primary antibody for each immunostaining.
Fig. 1.
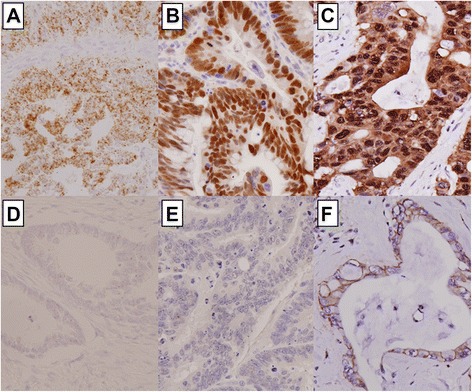
Representative figures of c-MYC status detected by in situ hybridization (a and d) and immunohistochemistry (IHC; b and e), and of ß-catenin expression by IHC (c and f), in colorectal cancer patients. a Score 4 mRNA (40×); b c-MYC overexpression (40×); c Nuclear ß-catenin expression (40×); d Score 0 mRNA (40×); e No c-MYC expression (40×); f Membranous ß-catenin expression (40×)
mRNA in situ hybridization
For the detection of c-MYC mRNA transcripts, the RNAscope 2.0 HD detection kit (Advanced Cell Diagnostics, Hayward, CA, USA) was used according to the manufacturer’s protocols. The experimental data was interpreted according to the manual in the RNAscope FFPE assay kit: no staining or less than 1 dot/cell at 40× objective view (score of 0); staining in 1–3 dots/cell visible at 20–40× objective view (score of 1); staining in 4–10 dots/cell with no or very few dot clusters visible at 20–40× objective view (score of 2); staining in >10 dots/cell with less than 10 % of positive cells having dot clusters visible at 20× objective view (score of 3); staining in >10 dots/cell with more than 10 % of positive cells having dot clusters visible at 20× objective view (score of 4). A score of 2–4 indicates c-MYC mRNA overexpression (Fig. 1). UBC (ubiquitin C) and dapB (a bacterial gene) were used for positive and negative controls. Tissues were regarded as appropriate when the UBC mRNA signals were visible without difficulty at 10× magnification and the dapB signal was not visible.
Microsatellite instability
Microsatellite instability (MSI) examination using fragmentation assay of ABI-3130xl with five microsatellite markers (BAT-26, BAT-25, D5S346, D17S250, and D2S123) were analyzed according to the instruction demonstrated previously [29]. MSI examination was evaluated in available 519 cases.
Statistical analyses
All statistical analysis was performed with the SPSS version 21 (IBM, Armonk, NY, USA) software. The Chi-square test or Fisher’s exact test was used for evaluating the correlation between clinicopathological characteristics and c-MYC and ß-catenin expression. The Pearson correlation coefficient was used for analyzing comparison of detection methods. The Kaplan-Meier method with the log-rank test and multivariate regression were performed to assess survival difference. The survival results were determined with hazard ratio (HR) and its 95 % confidence interval (CI). P < 0.05 was considered statistically significant.
Results
Clinicopathological impacts of c-MYC and ß-catenin expression in consecutive CRC patients
In 367 patients (cohort 1), a c-MYC mRNA in situ hybridization score of 0 was observed in 34 (9.3 %), a score of 1 in 92 (25.1 %), a score of 2 in 123 (33.5 %), a score of 3 in 93 (25.3 %), and a score of 4 in 25 (6.8 %). Consequentially, overexpression of c-MYC mRNA (a score of 2–4) was observed in 241 patients (65.7 %). c-MYC protein overexpression was observed in 201 (54.8 %), and ß-catenin nuclear overexpression was observed in 221 (60.2 %) patients.
Table 1 demonstrates the correlations between c-MYC and ß-catenin overexpression and clinicopathological parameters. c-MYC protein overexpression was associated with non-aggressive characteristics, including early pT stage, low-grade differentiation, absence of perineural invasion, and smaller tumor size (P < 0.001, P = 0.007, P = 0.025 and P < 0.001, respectively). In addition, c-MYC protein overexpression was associated with a tumor location in the recto-sigmoid colon. Increased levels of the c-MYC mRNA transcript were associated with microsatellite stable CRC (P = 0.019), located in the sigmoid colon and rectum, and with less aggressive features, similarly to c-MYC protein overexpression. Likewise, ß-catenin nuclear expression was frequently detected in tumors of the recto-sigmoid colon, of low-grade differentiation (P = 0.006), of small size (P = 0.007) and microsatellite stable CRC (P < 0.001).
Table 1.
The association between clinicopathological parameters and expression of c-MYC and ß-catenin in 367 CRC patients (cohort1)
| Total | c-Myc IHC | P-Value | c-Myc RNA ISH (score) | P-Value | ß-catenin IHC (%) | P-Value | ||||
|---|---|---|---|---|---|---|---|---|---|---|
| Negative | Positive | Negative | Positive | Negative | Positive | |||||
| Age | ||||||||||
| mean | 64.2 | 64.6 | 63.9 | 0.537 | 63.4 | 64.7 | 0.334 | 64.1 | 63.7 | 0.257 |
| Sex | ||||||||||
| male | 205 | 89 (43.4 %) | 116 (56.6 %) | 0.431 | 72 (35.1 %) | 133 (64.9 %) | 0.720 | 82 (40.0 %) | 123 (60.0 %) | 0.924 |
| female | 162 | 77 (47.5 %) | 85 (52.5 %) | 54 (33.3 %) | 108 (66.7 %) | 64 (39.5 %) | 98 (60.5 %) | |||
| Location | ||||||||||
| cecum | 13 | 12 (92.3 %) | 1 (7.7 %) | 0.002 | 12 (92.3 %) | 1 (7.7 %) | 0.039 | 11 (84.6 %) | 2 (15.4 %) | 0.103 |
| ascending colon | 55 | 39 (70.9 %) | 16 (29.1 %) | 37 (67.3 %) | 18 (32.7 %) | 36 (65.5 %) | 19 (34.5 %) | |||
| hepatic flexure | 22 | 14 (63.6 %) | 8 (36.4 %) | 17 (77.3 %) | 5 (22.7 %) | 14 (63.6 %) | 8 (36.4 %) | |||
| transverse colon | 16 | 10 (62.5 %) | 6 (37.5 %) | 11 (68.8 %) | 5 (31.3 %) | 11 (68.8 %) | 5 (31.3 %) | |||
| splenic flexure | 6 | 5 (83.3 %) | 1 (16.7 %) | 4 (66.7 %) | 2 (33.3 %) | 4 (66.7 %) | 2 (33.3 %) | |||
| descending colon | 18 | 15 (83.3 %) | 3 (16.7 %) | 15 (83.3 %) | 3 (16.7 %) | 13 (72.2 %) | 5 (27.8 %) | |||
| sigmoid colon | 114 | 53 (46.5 %) | 61 (53.5 %) | 64 (56.1 %) | 50 (43.9 %) | 62 (54.4 %) | 52 (45.6 %) | |||
| rectum | 123 | 71 (57.7 %) | 52 (42.3 %) | 89 (72.4 %) | 34 (27.6 %) | 61 (49.6 %) | 62 (50.4 %) | |||
| pT stage | ||||||||||
| 0–2 | 58 | 14 (24.1 %) | 44 (75.9 %) | <0.001 | 14 (24.1 %) | 44 (75.9 %) | 0.075 | 17 (29.3 %) | 41 (70.7 %) | 0.076 |
| 3–4 | 309 | 152 (49.2 %) | 157 (50.8 %) | 112 (36.2 %) | 197 (63.8 %) | 129 (41.7 %) | 180 (58.3 %) | |||
| Differentiation | ||||||||||
| LG | 331 | 142 (42.9 %) | 189 (57.1 %) | 0.007 | 101 (30.5 %) | 230 (69.5 %) | <0.001 | 124 (37.5 %) | 207 (62.5 %) | 0.006 |
| HG | 36 | 24 (66.7 %) | 12 (33.3 %) | 25 (69.4 %) | 11 (30.6 %) | 22 (61.1 %) | 14 (38.9 %) | |||
| LN metastasis | ||||||||||
| absent | 168 | 67 (39.9 %) | 101 (60.1 %) | 0.058 | 58 (34.5 %) | 110 (65.5 %) | 0.943 | 63 (37.5 %) | 105 (62.5 %) | 0.412 |
| present | 199 | 99 (49.7 %) | 100 (50.3 %) | 68 (34.2 %) | 131 (65.8 %) | 83 (41.7 %) | 116 (58.3 %) | |||
| Lymphatic invasion | ||||||||||
| absent | 158 | 63 (39.9 %) | 95 (60.1 %) | 0.073 | 51 (32.3 %) | 107 (67.7 %) | 0.471 | 61 (38.6 %) | 97 (61.4 %) | 0.689 |
| present | 209 | 103 (49.3 %) | 106 (50.7 %) | 75 (35.9 %) | 134 (64.1 %) | 85 (40.7 %) | 124 (59.3 %) | |||
| Perineural invasion | ||||||||||
| absent | 154 | 49 (31.8 %) | 105 (68.2 %) | 0.025 | 78 (30.7 %) | 176 (69.3 %) | 0.028 | 99 (39.0 %) | 155 (61.0 %) | 0.636 |
| present | 113 | 61 (54.0 %) | 52 (46.0 %) | 48 (42.5 %) | 65 (57.5 %) | 47 (41.6 %) | 66 (58.4 %) | |||
| Venous invasion | ||||||||||
| absent | 296 | 133 (44.9 %) | 163 (55.1 %) | 0.814 | 101 (34.1 %) | 195 (65.9 %) | 0.862 | 121 (40.9 %) | 175 (59.1 %) | 0.381 |
| present | 71 | 33 (46.5 %) | 38 (53.5 %) | 25 (35.2 %) | 46 (64.8 %) | 25 (35.2 %) | 46 (64.8 %) | |||
| Tumor border | ||||||||||
| expanding | 60 | 25 (41.7 %) | 35 (58.3 %) | 0.544 | 27 (45.0 %) | 33 (55.0 %) | 0.057 | 24 (40.0 %) | 36 (60.0 %) | 0.970 |
| infiltrative | 307 | 141 (45.9) | 166 (54.1 %) | 99 (32.2 %) | 208 (67.8 %) | 122 (39.7 %) | 185 (60.3 %) | |||
| Size (cm) | ||||||||||
| mean | 5.3 | 5.8 | 4.8 | <0.001 | 6.0 | 4.9 | <0.001 | 5.6 | 5.0 | 0.007 |
| Distant metastasis | ||||||||||
| absent | 299 | 131 (43.8 %) | 168 (56.2 %) | 0.252 | 97 (32.4 %) | 202 (67.6 %) | 0.110 | 115 (38.5 %) | 184 (61.5 %) | 0.278 |
| present | 68 | 35 (51.5 %) | 33 (48.5 %) | 29 (42.6 %) | 39 (57.4 %) | 31 (45.6 %) | 37 (54.4 %) | |||
| pTNM stage | ||||||||||
| I, II | 162 | 64 (39.5 %) | 98 (60.5 %) | 0.050 | 55 (34.0 %) | 107 (66.0 %) | 0.891 | 59 (37.1 %) | 100 (62.9 %) | 0.399 |
| III, IV | 205 | 102 (49.8 %) | 103 (50.2 %) | 71 (34.6 %) | 134 (65.4 %) | 85 (41.5 %) | 120 (58.5 %) | |||
| MSI status | ||||||||||
| MSS/MSI-L | 323 | 141 (43.7 %) | 182 (56.3 %) | 0.490 | 105 (32.5 %) | 218 (67.5 %) | 0.019 | 177 (54.8 %) | 146 (45.2 %) | <0.001 |
| MSI-H | 32 | 16 (50.0 %) | 16 (50.0 %) | 17 (53.1 %) | 15 (46.9 %) | 28 (87.5 %) | 4 (12.5 %) | |||
| Chemotherapy status | ||||||||||
| none | 97 | 41 (42.3 %) | 56 (57.7 %) | <0.001 | 67 (69.1 %) | 30 (30.9 %) | 0.748 | 49 (50.5 %) | 48 (49.5 %) | 0.175 |
| pre- | 0 | 0 (0.0 %) | 0 (0.0 %) | 0 (0.0 %) | 0 (0.0 %) | 0 (0.0 %) | 0 (0.0 %) | |||
| post- | 269 | 177 (65.8 %) | 92 (34.2 %) | 181 (67.3 %) | 88 (32.7 %) | 162 (60.2 %) | 107 (39.8 %) | |||
| Pre- and post- | 1 | 1 (100.0 %) | 0 (0.0 %) | 1 (100.0 %) | 0 (0.0 %) | 1 (100.0 %) | 0 (0.0 %) | |||
P-values are calculated by using χ2-test or Fisher’s exact test
Abbreviations: CRC colorectal cancer, T tumor, LG low grade, HG high grade, LN lymph node, MSS microsatellite stable, MSI-L microsatellite instability-low, MSI-H microsatellite instability-high, IHC immunohistochemistry, ISH in-situ hybridization
Correlation between c-MYC and ß-catenin expression in consecutive CRC patients
In cohort 1, c-MYC protein overexpression was correlated with mRNA overexpression (ρ, 0.479; P < 0.001), which was classified as moderate correlation [30]. ß-catenin nuclear expression was weakly associated with c-MYC protein overexpression and mRNA overexpression (ρ, 0.282; P < 0.001 and 0.211; P < 0.001, respectively).
Locational heterogeneity of c-MYC and ß-catenin status
For analysis the locational heterogeneity of c-MYC and ß-catenin expression, we investigated cancer from three lesion, including the primary, distant and lymph node metastasis (cohort 2). All 176 cases had distant metastatic lesions. Among them, 142 cases had lymph node metastases, even though we dissected more than 20 lymph nodes in all CRC patients respectively. The clinicopathological features of the cohort 2 are indicated in Table 2 as previously reported [31]. Not every cohort 2 patients are stage IV due to metachronous metastasis which develops consequently after treatment of the first primary tumor. The distant metastatic sites were described below: liver in 82 cases (46.6 %), lung in 37 cases (21.0 %), peritoneal seeding in 38 cases (21.6 %), distant lymph nodes in 3 cases (1.7 %), and ovary in 16 cases (9.0 %).
Table 2.
The clinicopathologic parameters of 176 advanced CRC patients with synchronous or metachronous metastases (cohort2)
| Total (n = 176) | Metastasis | ||||
|---|---|---|---|---|---|
| Synchronous (n = 118) | Metachronous (n = 58) | P-Value | |||
| Sex | male | 96 | 56 (47.5 %) | 40 (69.0 %) | 0.006 |
| female | 80 | 62 (52.5 %) | 18 (31.0 %) | ||
| Metastatic site | liver | 82 | 61 (52.1 %) | 21 (36.2 %) | <0.001 |
| lung | 37 | 10 (8.5 %) | 27 (46.6 %) | ||
| peritoneal seeding | 38 | 32 (26.5 %) | 6 (10.3 %) | ||
| distant lymph node | 3 | 2 (1.7 %) | 1 (1.7 %) | ||
| ovary | 16 | 13 (11.1 %) | 3 (5.2 %) | ||
| Location of primary tumor | appendix | 1 | 1 (0.9 %) | 0 (0.0 %) | 0.293 |
| cecum | 7 | 7 (6.0 %) | 0 (0.0 %) | ||
| ascending colon | 17 | 14 (12.0 %) | 3 (5.2 %) | ||
| hepatic flexure | 13 | 9 (7.7 %) | 4 (6.9 %) | ||
| transverse colon | 9 | 7 (6.0 %) | 2 (3.4 %) | ||
| splenic flexure | 7 | 4 (3.4 %) | 3 (5.2 %) | ||
| descending colon | 8 | 6 (5.1 %) | 2 (3.4 %) | ||
| sigmoid colon | 50 | 28 (23.9 %) | 22 (37.9 %) | ||
| rectum | 64 | 42 (35.0 %) | 22 (37.9 %) | ||
| T stage | T1 | 1 | 1 (0.9 %) | 0 (0.0 %) | <0.001 |
| T2 | 3 | 1 (0.9 %) | 2 (3.4 %) | ||
| T3 | 107 | 60 (50.4 %) | 47 (81.0 %) | ||
| T4 | 65 | 56 (47.9 %) | 9 (15.5 %) | ||
| N stage | N0 | 34 | 13 (11.1 %) | 21 (36.2 %) | <0.001 |
| N1 | 62 | 37 (31.6 %) | 25 (43.1 %) | ||
| N2 | 80 | 68 (57.3 %) | 12 (20.7 %) | ||
| Stage | I | 2 | 0 (0.0 %) | 2 (3.4 %) | <0.001 |
| II | 18 | 0 (0.0 %) | 18 (31.0 %) | ||
| III | 45 | 8 (6.8 %) | 37 (63.8 %) | ||
| IV | 111 | 110 (93.2 %) | 1 (1.7 %) | ||
| Differentiation | low grade | 158 | 104 (88.0 %) | 54 (93.1 %) | 0.299 |
| high grade | 18 | 14 (12.0 %) | 4 (6.9 %) | ||
| Lymphatic invasion | absent | 62 | 38 (32.5 %) | 24 (41.4 %) | 0.247 |
| present | 114 | 80 (67.5 %) | 34 (58.6 %) | ||
| Venous invasion | absent | 123 | 77 (65.0 %) | 46 (79.3 %) | 0.052 |
| present | 53 | 41 (35.0 %) | 12 (20.7 %) | ||
| Perineural invasion | absent | 90 | 56 (47.9 %) | 34 (58.6 %) | 0.180 |
| present | 86 | 62 (52.1 %) | 24 (41.4 %) | ||
| Tumor border | expanding | 13 | 6 (5.1 %) | 7 (12.1 %) | 0.099 |
| infiltrative | 163 | 112 (94.9 %) | 51 (87.9 %) | ||
P-values are calculated by using χ2-test or Fisher’s exact test
Abbreviations: T tumor; N lymph node
In the primary tumors of cohort 2, c-MYC protein overexpression, mRNA overexpression and nuclear ß-catenin expression was detected in 57.6 % (102 out of 176), 77.4 % (137 out of 176) and 61.0 % (108 out of 176) of tumors, respectively. In distant metastatic tumors, c-MYC protein overexpression, mRNA overexpression, and nuclear ß-catenin expression was detected in 37.3 % (66 out of 176), 74.6 % (132 out of 176) and 47.5 % (84 out of 176) of tumors, respectively. In 142 lymph node metastases, we performed c-MYC and ß-catenin analysis in 111 cases which paraffin blocks were available. c-MYC protein overexpression, mRNA overexpression, and nuclear ß-catenin expression was detected in 66.7 % (74 out of 111), 77.5 % (86 out of 111) and 58.6 % (65 out of 111) of tumors, respectively.
The locational heterogeneity of c-MYC and ß-catenin status is demonstrated in Table 3. Discordance of c-MYC protein overexpression between the primary and distant metastatic cancer was detected in 45.5 % (80 out of 176) of cases, and discordance between the primary and lymph node metastatic cancer was observed in 31.5 % (35 out of 111) of cases. Discordance of c-MYC mRNA overexpression between the primary and distant metastatic cancer was detected in 25.6 % (45 out of 176) of cases, and discordance between the primary and lymph node metastatic cancer was observed in 30.6 % (34 out of 111) of cases. Discordance of nuclear ß-catenin expression between the primary and distant metastatic cancer was detected in 29.0 % (51 out of 176) of cases, while discordance between the primary and lymph node metastatic cancer was observed in 26.1 % (29 out of 111) of cases. Consequently, locational heterogeneity of c-MYC and ß-catenin expression was frequently seen in advanced CRC.
Table 3.
Heterogeneity of c-MYC and ß-catenin with respect to tumor location in advanced CRC (cohort 2)
| cMYC IHC | Distant metastasis | LN metastasis | |||
| Negative | Positive | Negative | Positive | ||
| Primary | Negative | 52 (29.5 %) | 22 (12.5 %) | 23 (20.7 %) | 21 (18.9 %) |
| Positive | 58 (33.0 %) | 44 (25.0 %) | 14 (12.6 %) | 53 (47.7 %) | |
| cMYC mRNA ISH | Distant metastasis | LN metastasis | |||
| Negative | Positive | Negative | Positive | ||
| Primary | Negative | 19 (10.8 %) | 20 (11.4 %) | 8 (7.2 %) | 17 (15.3 %) |
| Positive | 25 (14.2 %) | 112 (63.6 %) | 17 (15.3 %) | 69 (62.2 %) | |
| ß-catenin IHC | Distant metastasis | LN metastasis | |||
| Negative | Positive | Negative | Positive | ||
| Primary | Negative | 55 (31.3 %) | 14 (8.0 %) | 30 (27.0 %) | 13 (11.7 %) |
| Positive | 37 (21.0 %) | 70 (39.8 %) | 16 (14.4 %) | 52 (46.8 %) | |
Abbreviations: IHC immunohistochemistry, ISH in-situ hybridization
Prognostic impact of c-MYC and ß-catenin expression in CRC
All CRC patients of our study were successfully included survival analysis (Fig. 2 and Additional file 1: Table S1). In the consecutive cohort (cohort 1), the median follow-up was 55 months (1–85 months) as previous reported [26]. c-MYC protein overexpression, mRNA overexpression and nuclear ß-catenin expression were significantly correlated with an improved survival in Kaplan-Meier analysis (P = 0.011, P = 0.012 and P = 0.033, respectively). When prognostic analysis was performed using the combined status of c-MYC and ß-catenin expression, positivity for both proteins (c-MYC/ß-catenin: +/+) was observed 84/367 (22.9 %) cases and was significantly correlated with an improved survival (P = 0.012). We additionally investigated the c-MYC protein overexpression by density of staining - scoring each tumor as low (0–1) to high (2–3) in cohort 1. The percentage of positive neoplastic cells was correlated with density of staining of neoplastic cells (ρ, 0.789; P < 0.001), which was categorized as strong correlation [30]. However, the staining density of c-MYC protein was not significantly correlated with patients’ prognosis (P = 0.070, Additional file 2: Figure S1).
Fig. 2.
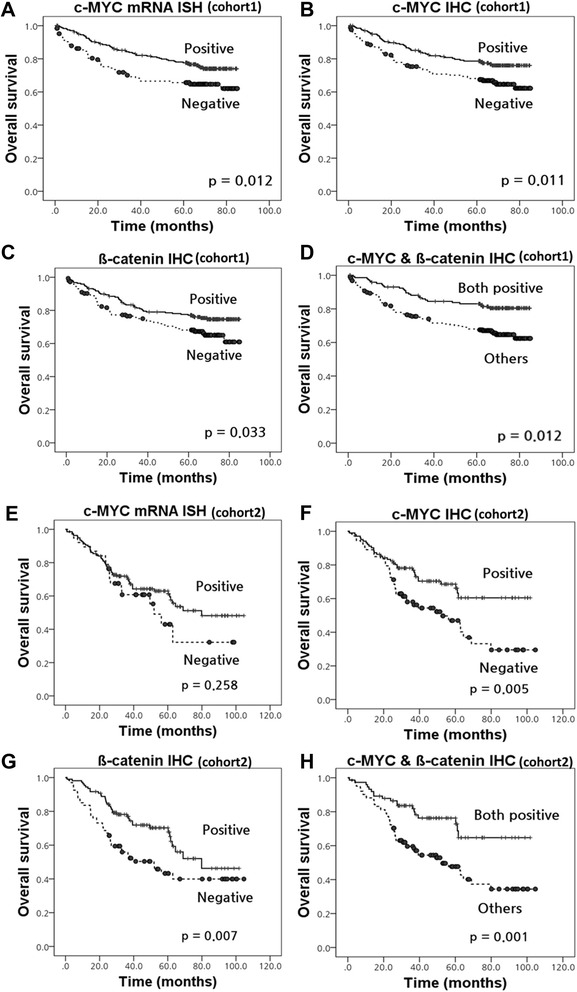
Kaplan–Meier survival curves illustrating the prognostic effects of c-MYC status in colorectal cancer. a-d Cohort 1; a c-MYC mRNA overexpression; b c-MYC protein overexpression; c Nuclear ß-catenin expression; d Co-expression of c-MYC and ß-catenin; e-h Primary tumor of cohort 2; e c-MYC mRNA overexpression; f c-MYC protein overexpression; g Nuclear ß-catenin expression; h Co-expression of c-MYC and ß-catenin
In the cohort with metastases (cohort 2), the median follow-up was 43 months (1–105 months), as previous reported [31]. In the primary cancer, Kaplan-Meier analysis showed that c-MYC protein overexpression and nuclear ß-catenin expression were significantly correlated with improved prognosis (P = 0.005 and P = 0.007, respectively), but mRNA overexpression was not (P = 0.258). Co-expression of c-MYC and ß-catenin (c-MYC/ß-catenin: +/+) also predicted better prognosis (P = 0.001). The c-MYC and ß-catenin expression status in distant or lymph node metastatic cancer did not significantly associated with patients’ survival (P > 0.05, data not shown). The presence of locational heterogeneity of c-MYC and ß-catenin expression was not associated with survival (P > 0.05; data not shown). In addition, we evaluated the Kaplan–Meier survival for MSI status, stage, chemotherapy status, site of primary cancer and site of distant metastasis (Additional file 2: Figure S1).
The multivariate Cox’s proportional hazards regression model of c-MYC and ß-catenin expression was described in Table 4, and indicated that co-expression of c-MYC and ß-catenin was an independent prognostic factor for better survival in both cohort 1 and primary tumor of cohort 2 (P = 0.048 and P = 0.002, respectively). However, individual analysis of c-MYC protein overexpression, mRNA overexpression, and nuclear ß-catenin expression did not independently predict better prognosis.
Table 4.
Multivariate Cox proportional hazard models for the predictors of overall survival
| Factors | Univariate survival analysis | Multivariate survival analysis | ||
|---|---|---|---|---|
| HR (95 % CI) | P value | HR (95 % CI) | P value | |
| Cohort 1 | ||||
| co-expression of c-MYC and ß-catenin | 0.482 (0.310–0.749) | 0.012 | 0.629 (0.397–0.996) | 0.048 |
| Age | 1.026 (1.008–1.045) | 0.005 | 1.022 (1.004–1.040) | 0.015 |
| Size | 1.244 (1.059–1.244) | 0.001 | 1.049 (0.946–1.163) | NS (0.362) |
| Histologic grade (high vs. low) | 3.143 (1.904–5.188) | <0.001 | 2.842 (1.608–5.023) | <0.001 |
| Stage (3/4 vs. 1/2) | 6.151 (3.494–10.829) | <0.001 | 2.942 (1.538–5.628) | 0.001 |
| Lymphatic invasion | 3.661 (2.242–5.980) | <0.001 | 1.338 (0.763–2.349) | NS (0.310) |
| Perineural invasion | 3.942 (2.648–5.870) | <0.001 | 2.337 (1.487–3.673) | <0.001 |
| Venous invasion | 3.985 (2.671–5.946) | <0.001 | 2.163 (1.390–3.366) | 0.001 |
| c-MYC mRNA expression | 0.599 (0.403–0.891) | 0.011 | 0.703 (0.464–1.066) | NS (0.097) |
| Age | 1.026 (1.008–1.045) | 0.005 | 1.024 (1.006–1.042) | 0.008 |
| Size | 1.244 (1.059–1.244) | 0.001 | 1.056 (0.953–1.169) | NS (0.297) |
| Histologic grade (high vs. low) | 3.143 (1.904–5.188) | <0.001 | 2.785 (1.562–4.968) | 0.001 |
| Stage (3/4 vs. 1/2) | 6.151 (3.494–10.829) | <0.001 | 3.017 (1.574–5.782) | 0.001 |
| Lymphatic invasion | 3.661 (2.242–5.980) | <0.001 | 1.327 (0.756–2.330) | NS (0.324) |
| Perineural invasion | 3.942 (2.648–5.870) | <0.001 | 2.438 (1.554–3.827) | <0.001 |
| Venous invasion | 3.985 (2.671–5.946) | <0.001 | 2.102 (1.356–3.259) | 0.001 |
| c-MYC protein expression | 0.593 (0.400–0.880) | 0.010 | 0.751 (0.498–1.132) | NS (0.171) |
| Age | 1.026 (1.008–1.045) | 0.005 | 1.023 (1.005–1.041) | 0.012 |
| Size | 1.244 (1.059–1.244) | 0.001 | 1.054 (0.951–1.169) | NS (0.317) |
| Histologic grade (high vs. low) | 3.143 (1.904–5.188) | <0.001 | 2.967 (1.681–5.236) | <0.001 |
| Stage (3/4 vs. 1/2) | 6.151 (3.494–10.829) | <0.001 | 2.959 (1.549–5.653) | 0.001 |
| Lymphatic invasion | 3.661 (2.242–5.980) | <0.001 | 1.344 (0.766–2.356) | NS (0.303) |
| Perineural invasion | 3.942 (2.648–5.870) | <0.001 | 2.388 (1.522–3.746) | <0.001 |
| Venous invasion | 3.985 (2.671–5.946) | <0.001 | 2.105 (1.356–3.269) | 0.001 |
| ß-catenin protein expression | 0.666 (0.449–0.986) | 0.042 | 0.740 (0.497–1.101) | NS (0.138) |
| Age | 1.026 (1.008–1.045) | 0.005 | 1.023 (1.005–1.041) | 0.012 |
| Size | 1.244 (1.059–1.244) | 0.001 | 1.068 (0.966–1.180) | NS (0.197) |
| Histologic grade (high vs. low) | 3.143 (1.904–5.188) | <0.001 | 2.897 (1.645–5.102) | <0.001 |
| Stage (3/4 vs. 1/2) | 6.151 (3.494–10.829) | <0.001 | 3.034 (1.587–5.801) | 0.001 |
| Lymphatic invasion | 3.661 (2.242–5.980) | <0.001 | 1.356 (0.771–2.383) | NS (0.290) |
| Perineural invasion | 3.942 (2.648–5.870) | <0.001 | 2.367 (1.508–3.717) | <0.001 |
| Venous invasion | 3.985 (2.671–5.946) | <0.001 | 2.071 (1.331–3.224) | 0.001 |
| Primary tumor of cohort 2 | ||||
| co-expression of c-MYC and ß-catenin | 0.430 (0.254–0.726) | 0.001 | 0.440 (0.259–0.747) | 0.002 |
| Age | 1.023 (1.001–1.045) | 0.037 | 1.025 (1.004–1.047) | 0.020 |
| Stage (3/4 vs. 1/2) | 7.894 (1.922–32.418) | 0.004 | 5.731 (1.380–23.800) | 0.016 |
| Lymphatic invasion | 2.480 (1.402–4.386) | 0.002 | 1.712 (0.940–3.117) | NS (0.079) |
| Perineural invasion | 2.119 (1.313–3.421) | 0.002 | 1.724 (1.049–2.831) | 0.032 |
P-values are calculated by using χ2-test or Fisher’s exact test
Abbreviations: HR hazard ratio
Discussion
There were several studies on c-MYC status in various malignancies. Some malignant tumors with c-MYC overexpression including gastric carcinoma, esophageal squamous cell carcinoma, and soft tissue leiomyosarcoma are associated with poor survival [32–34]. Likewise, several cancers with c-MYC gene amplification tend to be correlated with poor survival [35–37]. Interestingly, c-MYC mRNA overexpression in CRC was reported to be correlated with improved survival [5], but this was opposite result to previous other study [38]. Nevertheless, recent research indicated that immunohistochemical c-MYC overexpression was significantly associated with better prognosis of CRC patients in univariate model, but not in multivariate model [19]. In addition, many studies have shown that ß-catenin is crucial part of the Wnt signaling pathway in CRC development [39]. Recently, a meta-analysis study showed that nuclear overexpression of ß-catenin appeared to be associated with progressive disease for CRC patients [20]. However, prognostic value of nuclear overexpression of ß-catenin in CRC patients remains controversial [40, 41].
In our study, overexpression of c-MYC protein in the consecutive cohort was significantly correlated with an improved prognosis in univariate model, but not in multivariate model. The prognostic significance of nuclear ß-catenin expression is similar to that of c-MYC protein overexpression. We performed a combined analysis of c-MYC and ß-catenin expression because these proteins are closely related. Astonishingly, co-expression of c-MYC and ß-catenin correlated with an improved prognosis by univariate and multivariate analysis. Although the advanced cohort was mainly consisted of stage IV CRC patients (111 cases; 63.1 %), co-expression of c-MYC and ß-catenin was independently predicted favorable prognosis. Furthermore, overexpression of c-MYC and ß-catenin—except c-MYC mRNA—in the advanced cohort was significantly correlated with better prognosis using a univariate model. Consequently, co-expression of c-MYC and ß-catenin determined by IHC might be of use in the assessment of CRC patients.
We also demonstrated that ß-catenin nuclear expression significantly associated with its target molecule c-MYC in CRC patients (ρ, 0.282; P < 0.001). Overexpression of c-MYC can be caused by complex regulatory pathways and multiple communications with other factors, rather than just c-MYC gene alterations [42]. An example of such a mechanism is signaling via ß-catenin, a c-MYC regulator whose nuclear accumulation is correlated with c-MYC overexpression [7, 43]. ß-catenin increases in the cytoplasm and undergoes translocation to the nucleus, where it plays as a transcription factor for target genes such as c-MYC [16]. These processes explain that nuclear expression of ß-catenin is partly responsible for c-MYC overexpression. As a result, co-expression of c-MYC and ß-catenin can be considered as c-MYC overexpression via ß-catenin in CRC. The APC gene mutation is the initial step of CRC oncogenesis [44] and often lead to deregulation of ß-catenin [45]. Thus, ß-catenin-dependent c-MYC overexpression can be suggested in early colorectal carcinogenesis. In addition, recent studies suggested that high-level nuclear β-catenin in CRC was significantly correlated with high Ki67 expression [46], and indicated that tumor proliferative activity was inversely related to CRC aggressiveness and metastases [47, 48]. For this reason, c-MYC overexpression via ß-catenin might have an influence on improved prognosis of CRC patients. Moreover, E Melo et al. reported that CpG island methylation interrupts several Wnt target genes, including ASCL2 and LGR5 during CRC progression and promoter methylation of Wnt target genes is a powerful predictive factor for CRC relapse [16]. Therefore, silencing of ß-catenin/Wnt pathway by methylation generates CRC progression and worse prognosis. It is noteworthy that our result adds clinical evidence to support these previous studies [16–18, 46–48]. The lack of ß-catenin expression in CRC patients with presenting c-MYC overexpression shows a rather worse survival, presumably because, in these patients, the c-MYC is controlled by other regulatory factors. Future studies will be required to dissect the different mechanisms of ß-catenin-mediated c-MYC overexpression.
In the advanced cohort, regional heterogeneity of c-MYC and ß-catenin expression was frequently observed in advanced CRC. Nonetheless, c-MYC and ß-catenin expression was associated with better prognosis in the primary, not distant and lymph node metastatic cancer. Consequently, when we evaluate prognosis with c-MYC and ß-catenin, tissue from primary CRC should be used. In daily practice for pathologists, metastatic cancer tissue can only occasionally be obtained. Some researchers suggest that regional heterogeneity should be considered as a potential limitation to the evaluation of prognostic and therapeutic value in tissue from metastatic lesions [49, 50]. Therefore, the importance of regional heterogeneity must be assessed in biomarker research.
Conclusions
Our study comprehensively evaluated the c-MYC and ß-catenin status of CRC patients. Overexpression of c-MYC protein, mRNA, and ß-catenin nuclear expression were observed in 54.8, 65.7, and 60.2 % of consecutive CRC patients, respectively. c-MYC protein overexpression was significantly correlated with mRNA overexpression and with ß-catenin nuclear expression. Interestingly, co-expression of c-MYC and ß-catenin—in other words, c-MYC overexpression via ß-catenin—was an independent improved prognostic factor in both the consecutive and advanced cohort. These findings indicate that c-MYC and ß-catenin IHC can be used as prognostic marker of CRC patients. However, further investigations on the detailed mechanism of connection between c-MYC and ß-catenin and its impact on patients’ outcome in CRC are needed.
Acknowledgments
Not applicable.
Funding
This study was funded through a grant of the Korea Health Technology R&D Project through the Korea Health Industry Development Institute (KHIDI), funded by the Ministry of Health & Welfare, Republic of Korea (grant number : HI14C1813).
Availability of data and materials
Datasets of c-MYC and ß-catenin status is available in supplementary files (Additional files 3 and 4).
Authors’ contributions
KSL scored immunohistochemistry and mRNA in situ hybridization, performed data interpretation, statistical analysis and drafted the manuscript; YK participated in the data interpretation; KHN participated in the mRNA in situ hybridization; DWK and SBK operated on colorectal cancer; GC and WHK participated in the design of the study and reviewed the final manuscript; HSL reviewed cases, statistical analysis, conceived the study and study design, reviewed and approved the final manuscript. All authors have read and approved the manuscript.
Competing interests
The authors declare that they have no competing interests.
Consent for publication
Not applicable.
Ethics approval and consent to participate
The Institutional Review Board (IRB) of Seoul National University Bundang Hospital approved the use of medical record data and tissue samples for this study (IRB No: B-1210/174-301). The need for consent of participates who received surgery before February 2013 has been waived by an IRB. All specimens and medical record were anonymized.
Abbreviations
- CRC
Colorectal cancer
- IHC
Immunohistochemistry
- FFPE
Formalin-fixed and paraffin-embedded
- MSI
Microsatellite instability
- PCR
Polymerase chain reaction
- HR
Hazard ratio
- CI
Confidence interval
Additional files
The number at risk in each category, at each interval for all Kaplan-Meier plots in Fig. 2. (DOCX 21 kb)
Kaplan–Meier survival curves illustrating the prognostic effects of clinicopathological parameter. (A-E) Cohort 1; (A) MSI status; (B) stage I, II versus III, IV; (C) chemotherapy status; (D) site of primary cancer; (E) c-MYC protein overexpression by staning density (F) Primary of tumor of cohort 2; site of distant metastasis. (TIF 416 kb)
Raw data of c-MYC and ß-catenin status obtained from cohort1. (XLSX 51 kb)
Raw data of c-MYC and ß-catenin status obtained from cohort2. (XLSX 21 kb)
Contributor Information
Kyu Sang Lee, Email: tigerkyu@gmail.com.
Yoonjin Kwak, Email: dbswls131@naver.com.
Kyung Han Nam, Email: kyunghannam@gmail.com.
Duck-Woo Kim, Email: KDW@snubh.org.
Sung-Bum Kang, Email: kangsb@snubh.org.
Gheeyoung Choe, Email: gychoe@snu.ac.kr.
Woo Ho Kim, Email: woohokim@snu.ac.kr.
Hye Seung Lee, Email: hye2@snu.ac.kr.
References
- 1.Nesbit CE, Tersak JM, Prochownik EV. MYC oncogenes and human neoplastic disease. Oncogene. 1999;18(19):3004–3016. doi: 10.1038/sj.onc.1202746. [DOI] [PubMed] [Google Scholar]
- 2.Beroukhim R, Mermel CH, Porter D, Wei G, Raychaudhuri S, Donovan J, Barretina J, Boehm JS, Dobson J, Urashima M, et al. The landscape of somatic copy-number alteration across human cancers. Nature. 2010;463(7283):899–905. doi: 10.1038/nature08822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Secombe J, Pierce SB, Eisenman RN. Myc: a weapon of mass destruction. Cell. 2004;117(2):153–156. doi: 10.1016/S0092-8674(04)00336-8. [DOI] [PubMed] [Google Scholar]
- 4.Singh AM, Dalton S. The cell cycle and Myc intersect with mechanisms that regulate pluripotency and reprogramming. Cell Stem Cell. 2009;5(2):141–149. doi: 10.1016/j.stem.2009.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith DR, Goh HS. Overexpression of the c-myc proto-oncogene in colorectal carcinoma is associated with a reduced mortality that is abrogated by point mutation of the p53 tumor suppressor gene. Clin Cancer Res. 1996;2(6):1049–1053. [PubMed] [Google Scholar]
- 6.Erisman MD, Rothberg PG, Diehl RE, Morse CC, Spandorfer JM, Astrin SM. Deregulation of c-myc gene expression in human colon carcinoma is not accompanied by amplification or rearrangement of the gene. Mol Cell Biol. 1985;5(8):1969–1976. doi: 10.1128/MCB.5.8.1969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281(5382):1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 8.Junttila MR, Westermarck J. Mechanisms of MYC stabilization in human malignancies. Cell Cycle. 2008;7(5):592–596. doi: 10.4161/cc.7.5.5492. [DOI] [PubMed] [Google Scholar]
- 9.Luscher B, Vervoorts J. Regulation of gene transcription by the oncoprotein MYC. Gene. 2012;494(2):145–160. doi: 10.1016/j.gene.2011.12.027. [DOI] [PubMed] [Google Scholar]
- 10.Rochlitz CF, Herrmann R, de Kant E. Overexpression and amplification of c-myc during progression of human colorectal cancer. Oncology. 1996;53(6):448–454. doi: 10.1159/000227619. [DOI] [PubMed] [Google Scholar]
- 11.Oster SK, Ho CS, Soucie EL, Penn LZ. The myc oncogene: MarvelouslY Complex. Adv Cancer Res. 2002;84:81–154. doi: 10.1016/S0065-230X(02)84004-0. [DOI] [PubMed] [Google Scholar]
- 12.Powell SM, Zilz N, Beazer-Barclay Y, Bryan TM, Hamilton SR, Thibodeau SN, Vogelstein B, Kinzler KW. APC mutations occur early during colorectal tumorigenesis. Nature. 1992;359(6392):235–237. doi: 10.1038/359235a0. [DOI] [PubMed] [Google Scholar]
- 13.Chan SK, Griffith OL, Tai IT, Jones SJ. Meta-analysis of colorectal cancer gene expression profiling studies identifies consistently reported candidate biomarkers. Cancer Epidemiol Biomark Prev. 2008;17(3):543–552. doi: 10.1158/1055-9965.EPI-07-2615. [DOI] [PubMed] [Google Scholar]
- 14.Morin PJ, Sparks AB, Korinek V, Barker N, Clevers H, Vogelstein B, Kinzler KW. Activation of beta-catenin-Tcf signaling in colon cancer by mutations in beta-catenin or APC. Science. 1997;275(5307):1787–1790. doi: 10.1126/science.275.5307.1787. [DOI] [PubMed] [Google Scholar]
- 15.Morin PJ. beta-catenin signaling and cancer. BioEssays. 1999;21(12):1021–1030. doi: 10.1002/(SICI)1521-1878(199912)22:1<1021::AID-BIES6>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- 16.de Sousa EMF, Colak S, Buikhuisen J, Koster J, Cameron K, de Jong JH, Tuynman JB, Prasetyanti PR, Fessler E, van den Bergh SP, et al. Methylation of cancer-stem-cell-associated Wnt target genes predicts poor prognosis in colorectal cancer patients. Cell Stem Cell. 2011;9(5):476–485. doi: 10.1016/j.stem.2011.10.008. [DOI] [PubMed] [Google Scholar]
- 17.Guinney J, Dienstmann R, Wang X, de Reynies A, Schlicker A, Soneson C, Marisa L, Roepman P, Nyamundanda G, Angelino P, et al. The consensus molecular subtypes of colorectal cancer. Nat Med. 2015;21(11):1350–1356. doi: 10.1038/nm.3967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.De Sousa EMF, Wang X, Jansen M, Fessler E, Trinh A, de Rooij LP, de Jong JH, de Boer OJ, van Leersum R, Bijlsma MF, et al. Poor-prognosis colon cancer is defined by a molecularly distinct subtype and develops from serrated precursor lesions. Nat Med. 2013;19(5):614–618. doi: 10.1038/nm.3174. [DOI] [PubMed] [Google Scholar]
- 19.Toon CW, Chou A, Clarkson A, DeSilva K, Houang M, Chan JC, Sioson LL, Jankova L, Gill AJ. Immunohistochemistry for myc predicts survival in colorectal cancer. PLoS One. 2014;9(2):e87456. doi: 10.1371/journal.pone.0087456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z, He X, Jia M, Liu Y, Qu D, Wu D, Wu P, Ni C, Zhang Z, Ye J, et al. beta-catenin overexpression in the nucleus predicts progress disease and unfavourable survival in colorectal cancer: a meta-analysis. PLoS One. 2013;8(5):e63854. doi: 10.1371/journal.pone.0063854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Knijn N, Tol J, Punt CJ. Current issues in the targeted therapy of advanced colorectal cancer. Discov Med. 2010;9(47):328–336. [PubMed] [Google Scholar]
- 22.Van Cutsem E. Optimizing administration of epidermal growth factor receptor-targeted agents in the treatment of colorectal cancer. Clin Colorectal Cancer. 2007;6(Suppl 2):S60–S65. doi: 10.3816/CCC.2007.s.004. [DOI] [PubMed] [Google Scholar]
- 23.Albanese I, Scibetta AG, Migliavacca M, Russo A, Bazan V, Tomasino RM, Colomba P, Tagliavia M, La Farina M. Heterogeneity within and between primary colorectal carcinomas and matched metastases as revealed by analysis of Ki-ras and p53 mutations. Biochem Biophys Res Commun. 2004;325(3):784–791. doi: 10.1016/j.bbrc.2004.10.111. [DOI] [PubMed] [Google Scholar]
- 24.Park JH, Han SW, Oh DY, Im SA, Jeong SY, Park KJ, Kim TY, Bang YJ, Park JG. Analysis of KRAS, BRAF, PTEN, IGF1R, EGFR intron 1 CA status in both primary tumors and paired metastases in determining benefit from cetuximab therapy in colon cancer. Cancer Chemother Pharmacol. 2011;68(4):1045–1055. doi: 10.1007/s00280-011-1586-z. [DOI] [PubMed] [Google Scholar]
- 25.Baldus SE, Schaefer KL, Engers R, Hartleb D, Stoecklein NH, Gabbert HE. Prevalence and heterogeneity of KRAS, BRAF, and PIK3CA mutations in primary colorectal adenocarcinomas and their corresponding metastases. Clin Cancer Res. 2010;16(3):790–799. doi: 10.1158/1078-0432.CCR-09-2446. [DOI] [PubMed] [Google Scholar]
- 26.Lee KS, Kwak Y, Nam KH, Kim DW, Kang SB, Choe G, Kim WH, Lee HS. c-MYC Copy-Number Gain Is an Independent Prognostic Factor in Patients with Colorectal Cancer. PLoS One. 2015;10(10):e0139727. doi: 10.1371/journal.pone.0139727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee HS, Cho SB, Lee HE, Kim MA, Kim JH, Park do J, Kim JH, Yang HK, Lee BL, Kim WH. Protein expression profiling and molecular classification of gastric cancer by the tissue array method. Clin Cancer Res. 2007;13(14):4154–4163. doi: 10.1158/1078-0432.CCR-07-0173. [DOI] [PubMed] [Google Scholar]
- 28.Iwamoto M, Ahnen DJ, Franklin WA, Maltzman TH. Expression of beta-catenin and full-length APC protein in normal and neoplastic colonic tissues. Carcinogenesis. 2000;21(11):1935–1940. doi: 10.1093/carcin/21.11.1935. [DOI] [PubMed] [Google Scholar]
- 29.Boland CR, Thibodeau SN, Hamilton SR, Sidransky D, Eshleman JR, Burt RW, Meltzer SJ, Rodriguez-Bigas MA, Fodde R, Ranzani GN, et al. A National Cancer Institute Workshop on Microsatellite Instability for cancer detection and familial predisposition: development of international criteria for the determination of microsatellite instability in colorectal cancer. Cancer Res. 1998;58(22):5248–5257. [PubMed] [Google Scholar]
- 30.Dancey C. Statistics without maths for psychology: using SPSS for windows. London: Prentice Hall; 2004. [Google Scholar]
- 31.Kwak Y, Lee HE, Kim WH, Kim DW, Kang SB, Lee HS. The clinical implication of cancer-associated microvasculature and fibroblast in advanced colorectal cancer patients with synchronous or metachronous metastases. PLoS One. 2014;9(3):e91811. doi: 10.1371/journal.pone.0091811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tsiatis AC, Herceg ME, Keedy VL, Halpern JL, Holt GE, Schwartz HS, Cates JM. Prognostic significance of c-Myc expression in soft tissue leiomyosarcoma. Mod Pathol. 2009;22(11):1432–1438. doi: 10.1038/modpathol.2009.113. [DOI] [PubMed] [Google Scholar]
- 33.Wang W, Xue L, Wang P. Prognostic value of beta-catenin, c-myc, and cyclin D1 expressions in patients with esophageal squamous cell carcinoma. Med Oncol. 2011;28(1):163–169. doi: 10.1007/s12032-010-9436-0. [DOI] [PubMed] [Google Scholar]
- 34.Ninomiya I, Yonemura Y, Matsumoto H, Sugiyama K, Kamata T, Miwa K, Miyazaki I, Shiku H. Expression of c-myc gene product in gastric carcinoma. Oncology. 1991;48(2):149–153. doi: 10.1159/000226915. [DOI] [PubMed] [Google Scholar]
- 35.Choi JS, Seo J, Jung EJ, Kim EJ, Lee GK, Kim WH. c-MYC amplification in mucinous gastric carcinoma: a possible genetic alteration leading to deeply invasive tumors. Anticancer Res. 2012;32(11):5031–5037. [PubMed] [Google Scholar]
- 36.Dimova I, Raitcheva S, Dimitrov R, Doganov N, Toncheva D. Correlations between c-myc gene copy-number and clinicopathological parameters of ovarian tumours. Eur J Cancer. 2006;42(5):674–679. doi: 10.1016/j.ejca.2005.11.022. [DOI] [PubMed] [Google Scholar]
- 37.Seo AN, Yang JM, Kim H, Jheon S, Kim K, Lee CT, Jin Y, Yun S, Chung JH, Paik JH. Clinicopathologic and prognostic significance of c-MYC copy number gain in lung adenocarcinomas. Br J Cancer. 2014;110(11):2688–2699. doi: 10.1038/bjc.2014.218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Erisman MD, Litwin S, Keidan RD, Comis RL, Astrin SM. Noncorrelation of the expression of the c-myc oncogene in colorectal carcinoma with recurrence of disease or patient survival. Cancer Res. 1988;48(5):1350–1355. [PubMed] [Google Scholar]
- 39.Gough NR. Focus issue: Wnt and beta-catenin signaling in development and disease. Sci Signal. 2012;5(206):eg2. doi: 10.1126/scisignal.2002806. [DOI] [PubMed] [Google Scholar]
- 40.Wong SC, Lo ES, Lee KC, Chan JK, Hsiao WL. Prognostic and diagnostic significance of beta-catenin nuclear immunostaining in colorectal cancer. Clin Cancer Res. 2004;10(4):1401–1408. doi: 10.1158/1078-0432.CCR-0157-03. [DOI] [PubMed] [Google Scholar]
- 41.Chung GG, Provost E, Kielhorn EP, Charette LA, Smith BL, Rimm DL. Tissue microarray analysis of beta-catenin in colorectal cancer shows nuclear phospho-beta-catenin is associated with a better prognosis. Clin Cancer Res. 2001;7(12):4013–4020. [PubMed] [Google Scholar]
- 42.Lin CY, Loven J, Rahl PB, Paranal RM, Burge CB, Bradner JE, Lee TI, Young RA. Transcriptional amplification in tumor cells with elevated c-Myc. Cell. 2012;151(1):56–67. doi: 10.1016/j.cell.2012.08.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tetsu O, McCormick F. Beta-catenin regulates expression of cyclin D1 in colon carcinoma cells. Nature. 1999;398(6726):422–426. doi: 10.1038/18884. [DOI] [PubMed] [Google Scholar]
- 44.Kinzler KW, Vogelstein B. Lessons from hereditary colorectal cancer. Cell. 1996;87(2):159–170. doi: 10.1016/S0092-8674(00)81333-1. [DOI] [PubMed] [Google Scholar]
- 45.Willert K, Nusse R. Beta-catenin: a key mediator of Wnt signaling. Curr Opin Genet Dev. 1998;8(1):95–102. doi: 10.1016/S0959-437X(98)80068-3. [DOI] [PubMed] [Google Scholar]
- 46.Melling N, Kowitz CM, Simon R, Bokemeyer C, Terracciano L, Sauter G, Izbicki JR, Marx AH. High Ki67 expression is an independent good prognostic marker in colorectal cancer. J Clin Pathol. 2016;69(3):209–214. doi: 10.1136/jclinpath-2015-202985. [DOI] [PubMed] [Google Scholar]
- 47.Anjomshoaa A, Nasri S, Humar B, McCall JL, Chatterjee A, Yoon HS, McNoe L, Black MA, Reeve AE. Slow proliferation as a biological feature of colorectal cancer metastasis. Br J Cancer. 2009;101(5):822–828. doi: 10.1038/sj.bjc.6605229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Anjomshoaa A, Lin YH, Black MA, McCall JL, Humar B, Song S, Fukuzawa R, Yoon HS, Holzmann B, Friederichs J, et al. Reduced expression of a gene proliferation signature is associated with enhanced malignancy in colon cancer. Br J Cancer. 2008;99(6):966–973. doi: 10.1038/sj.bjc.6604560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Couvelard A, Deschamps L, Ravaud P, Baron G, Sauvanet A, Hentic O, Colnot N, Paradis V, Belghiti J, Bedossa P, et al. Heterogeneity of tumor prognostic markers: a reproducibility study applied to liver metastases of pancreatic endocrine tumors. Mod Pathol. 2009;22(2):273–281. doi: 10.1038/modpathol.2008.177. [DOI] [PubMed] [Google Scholar]
- 50.Koelzer VH, Herrmann P, Zlobec I, Karamitopoulou E, Lugli A, Stein U. Heterogeneity analysis of Metastasis Associated in Colon Cancer 1 (MACC1) for survival prognosis of colorectal cancer patients: a retrospective cohort study. BMC Cancer. 2015;15:160. doi: 10.1186/s12885-015-1150-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Datasets of c-MYC and ß-catenin status is available in supplementary files (Additional files 3 and 4).


