Abstract
The nuclear factor erythroid‐derived two‐like 2‐antioxidant response element (Nrf2‐ARE) pathway and its downstream antioxidant enzyme heme oxygenase‐1 (HMOX1 or HO‐1) play essential roles in H2O2‐induced oxidative damage in human melanocytes. However, the link between Nrf2 promoter polymorphisms and susceptibility to oxidative stress‐related diseases such as vitiligo is unknown. This study evaluated the association of the Nrf2 and HO‐1 genes polymorphisms with vitiligo susceptibility. In this case–control study of 1136 Han Chinese vitiligo patients and 1200 controls, Nrf2 (rs35652124 and rs6721961) and HO‐1 (rs2071746) genes were genotyped by PCR‐restriction fragment length polymorphism analysis. Overall, a significantly decreased risk of vitiligo was found to be associated with Nrf2 rs35652124 CC and combined (CT+CC) genotypes [odds ratio (OR) 0.64, 95% confidence interval (CI) 0.50–0.83 and OR, 0.84, 95% CI 0.71–0.99, respectively], as well as among subgroups: female, onset age ≤20 and never smoker. We subsequently found that Nrf2 rs35652124 C allele had higher transcriptional activity in the luciferase reporter assay compared with Nrf2 rs35652124 T allele. Furthermore, we investigated serum HO‐1 activity was associated with the rs35652124 CT+CC genotype and lower in patients than in controls (P = 0.024). Logistic regression analysis showed a dose–response relationship between lower vitiligo risk and increased HO‐1 activity in rs35652124 CT+CC genotype carriers (P trend < 0.05). These findings indicate that the C allele of rs35652124 located in the promoter region of Nrf2 gene is associated with protective effect on vitiligo in a Han Chinese population.
Keywords: vitiligo, SNP, Nrf2
Introduction
Vitiligo, an acquired skin depigmentation/hypopigmentation disorder, affects 0.05–1% of the global population and approximately 0.09% of the Chinese population, independently of gender 1, 2. The age of onset is typically before 20 years. Although the exact cause is unknown, vitiligo is thought to result from complex pathogenetic mechanisms involving a combination of environmental and genetic risk factors. Some studies have suggested that oxidative stress triggers the degeneration of basal layers of epidermal melanocytes 3, 4. Reactive oxygen species (ROS) disrupt the homoeostasis of melanocytes and damage nucleic acids, lipids and proteins, leading to cell death 5; for instance, excessive accumulation of H2O2 is observed in the epidermis of vitiligo patients and plays an important role in the disease 6, 7. Meanwhile, accumulating lines of evidence have also revealed that the nuclear factor erythroid‐derived 2‐like 2 (Nrf2) is a pivotal transcription factor of the antioxidant response in oxidative stress‐related illnesses 8.
Nrf2 protein, a member of the Cap‐N‐Collar basic leucine zipper transcription factor family, dimerizes with musculoaponeurotic fibrosarcomaor members of the activator protein 1 family in the cytoplasm. In response to oxidative stress, Nrf2 is quickly translocates into the nucleus and binds to the antioxidant response element (ARE), which is located in the upstream promoter region of many antioxidative genes 9. Therefore, the activation of Nrf2 is an important clue for the inducible expression of cytoprotective genes. Some phase‐detoxifying enzymes encoded by these genes, such as heme oxygenase‐1 (HMOX1 or HO‐1), glutathione S‐transferase (GST), NADH quinone oxidoreductase‐1 and superoxide dismutase be confirmed to play a major role in oxidative stress 10, 11. As one of the crucial cytoprotective proteins which induced by activation of Nrf2, HO‐1 has been shown to protect from some oxidative related pathologies, including hypertension, atherosclerosis and kidney injury 12, 13, 14. In addition, our previous study found that the Nrf2‐ARE pathway and its downstream antioxidant enzyme HO‐1 can mitigate H2O2‐induced oxidative damage in human melanocytes 15. Taken together, these findings indicated that involvement of altered Nrf2 expression may closely correlated with the pathogenesis of vitiligo.
The human Nrf2 gene maps to chromosome 2q31 and consists of five exons and four introns (Fig. 1A). It was known that polymorphisms of the Nrf2 gene had effects on the affinity and expression of the Nrf2 proteins and activation of the Nrf2 16, 17. Among the previous studies on single nucleotide polymorphisms (SNPs) of the Nrf2 gene, the polymorphisms of Nrf2 promoter region paly a major role in affecting its transcriptional activity. Two polymorphisms in the promoter region – a T to C substitution at position −653 (Nrf2 rs35652124) and a G to T substitution at −617 (Nrf2 rs6721961) – are located in an ARE‐like element and the myeloid zinc finger gene 1 binding site respectively (Fig. 1B) 18. Human HO‐1 gene on chromosome 22q12 comprises five exons and four introns (Fig. 1D) 19. An A to T substitution at position −413 (HO‐1 rs2071746) of the promoter is associated with Parkinson's and Alzheimer's diseases and aspirin resistance (Fig. 1E) 20, 21, 22. Given that the activation of HO‐1 transcription by Nrf2 has a protective effect against a variety of pathologies, it is possible that Nrf2 and HO‐1 gene polymorphisms are associated with the development of vitiligo, although there have been no studies to date investigating this link. This study has been undertaken to fill this gap.
Figure 1.
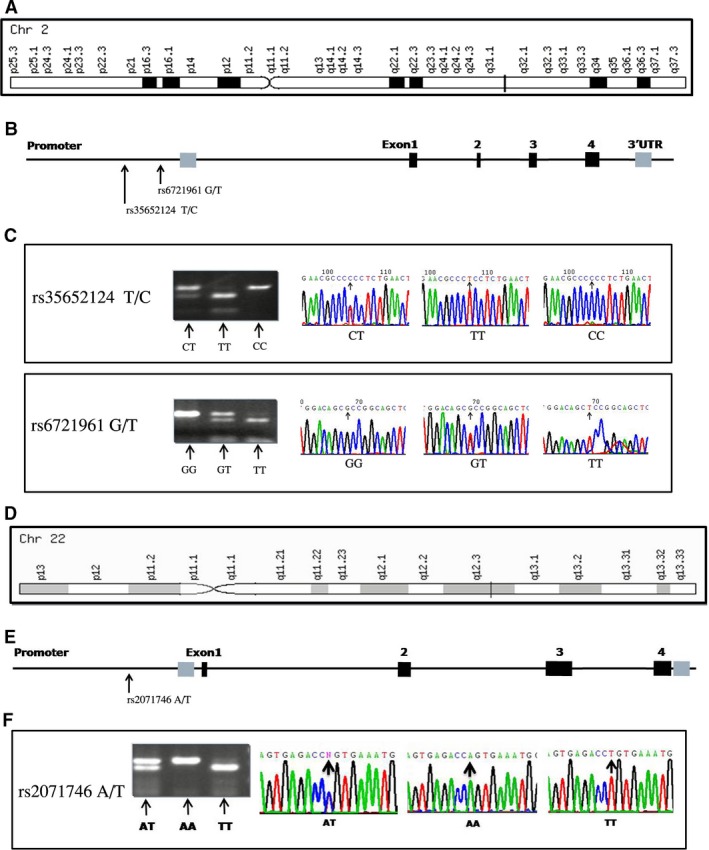
Location and detection of the Nrf2 gene and HO‐1 gene and SNPs. Nrf2, nuclear factor erythroid 2‐related factor 2; HO‐1, heme oxygenase‐1; SNPs, single nucleotide polymorphisms. (A) Location of the Nrf2 gene. (B) The Nrf2 gene structure and the location of the selected SNPs. (C) Genotypes of rs35652124 and rs6721961 in the Nrf2 gene and sequence analyses of the Nrf2 PCR products. (D) Location of the HO‐1 gene. (E) The HO‐1 gene structure and the location of the selected SNPs. (F) Genotypes of rs2071746 in the HO‐1 gene and sequence analyses of the HO‐1 PCR products.
As polymorphisms of the Nrf2 gene may influence Nrf2 function and expression of phase II metabolism genes, thereby affecting antioxidative targeting of melanocytes and increasing the risk of vitiligo. To test this hypothesis, we selected three SNPs from the Hapmap database with minor allele frequency >5% in Chinese population to evaluate the associations between the genetic variants in the Nrf2 and HO‐1 genes and vitiligo risk in Han Chinese subjects. Among these SNPs, we identified a protect‐associated Nrf2 rs35652124 T/C polymorphism in the promoter region by using PCR‐restriction fragment length polymorphism (PCR‐RFLP) method. And we further assessed the Nrf2 promoter activity related to −653T→C polymorphism by using transient transfections and luciferase assays. In addition, serum levels of HO‐1 and GST were measured in these subjects to evaluate potential associations between Nrf2 variants and clinical manifestations of vitiligo.
Materials and methods
Study subjects
The study protocol was approved by the Clinical Ethics Committee of Xijing Hospital and the experiments were conducted according to the principles outlined by the Declaration of Helsinki. From August 2007 to January 2011, vitiligo patients (n = 1136) and vitiligo‐free control subjects (n = 1200) were recruited from Xijing Hospital, the Fourth Military Medical University. Only Han Chinese subjects (more than 90% of the Chinese population) were included to avoid genotype frequency variations among ethnic groups. Patients that were diagnosed by dermatologists and had undergone treatment in the preceding 6 months were excluded. Active vitiligo was defined as the appearance of new lesions or the enlargement of existing lesions in the 3 months before presentation in our study project. The vitiligo‐free controls were excluded if they had received blood transfusions in the last 6 months, or if they had other autoimmune diseases or other depigmentation disorders, such as piebaldism and albinism. A questionnaire was provided to obtain demographic and other information, and patients and controls were matched for age (±5 years), sex and ethnicity (Table 1). The response rate among participants was nearly 90%. Prior to recruitment, each participant provided written, informed consent and donated 5 ml of blood collected in heparinized tubes for genomic DNA extraction.
Table 1.
Clinical characteristics of the selected variables in cases of vitiligo and controls
| Case, n (%); n = 1136 | Control, n (%); n = 1200 | P a | |
|---|---|---|---|
| Age (years, ≤20/>20) | 519 (45.7)/617 (54.3) | 545 (45.5)/655 '(54.6) | 0.896 |
| Gender (male/female) | 608 (53.5)/528 (46.5) | 644 (53.7)/556 (46.3) | 0.944 |
| Onset age (years, ≤20/>20) | 681 (59.9)/455 (40.1) | ||
| Type (segmental/nonsegmental) | 101 (8.9)/1035 (91.1) | ||
| Stage (stable/active) | 228 (20.1)/908 (79.9) | ||
| Family history (yes/no) | 174 (15.3)/962 (84.7) | ||
| Autoimmunity disease (yes/no) | 31 (2.7)/1105 (97.3) | ||
| Smoking status (never smokers/ex‐smokers/current smokers) | 651 (57.3)/94 (8.3)/391 (34.4) | 647 (53.9)/95 (7.9)/458 (38.2) | 0.169 |
| Drinking status (never/ever) | 1110 (97.7)/26 (2.3) | 1164 (97.0)/36 (3.0) | 0.285 |
P values based on two‐sided chi‐square test.
Genotyping
Genomic DNA was extracted from peripheral blood samples, using a DNA isolation kit (Tiangen, Beijing, China). DNA purity and concentration were evaluated by spectrophotometric measurements of absorbance at 260 and 280 nm. The PCR‐RFLP method was used to amplify Nrf2 (rs35652124, rs6721961) and HO‐1 (rs2071746) with the following forward and reverse primers: Nrf2 rs35652124 T/C, 5′‐CCT TGC CCT GCT TTT ATC TC‐3′ and 5′‐CTT CTC CGT TTG CCT TTG AC‐3′; Nrf2 rs6721961 G/T, 5′‐GAA AGG CGT TGG TGT AGG AG‐3′ and 5′‐GAA TGG AGA CAC GTG GGA GT‐3′; HO‐1 rs2071746 A/T, 5′‐GTT CCT GAT GTT GCC CAC CAA GC‐3′ and 5′‐CTG CAG GCT CTG GGT GTG ATT TTG‐3′, which generated products of 264, 278, and 151 bp, respectively, that were digested with BseRI (MBI Fermentas, Leon‐Rot, Germany), NgoMIV and HindIII (both from New England Biolabs, Ipswich, MA, USA) restriction enzymes respectively. The fragments that were generated by the digestion were 192 and 72 bp for Nrf2 rs35652124 T/C (Fig. 1C), 215 and 63 bp for Nrf2 rs6721961 (Fig. 1C), and 20 and 131 bp for HO‐1 rs2071746 (Fig. 1F). Results were confirmed by repeating the genotyping for >10% of randomly selected samples, yielding a concordance of 100%.
Cell culture
Human normal melanocyte PIG1 cells (generously provided by Dr Caroline Le Poole, Loyola University Chicago, Chicago, IL, USA) were cultured in Medium 254 (Cascade Biologics, Portland, OR, USA) supplemented with human melanocyte growth supplement (Cascade Biologics), 5% foetal bovine serum (Gibco‐Invitrogen, Carlsbad, CA, USA) under a humidified atmosphere containing 5% CO2 at 37°C. This cell line was immortalized by retroviral introduction of the human papillomavirus type 16 E6 and E7 genes. Human embryonic kidney 293T cells were cultured in DMEM (Gibco‐Invitrogen) with 10% foetal bovine serum (Gibco‐Invitrogen) under a humidified atmosphere containing 5% CO2 at 37°C. This cell line was transformed by adenovirus E1A gene.
Construction of reporter plasmids
Nrf2 promoter‐luciferase reporter plasmids containing either rs35652124 T or C sequences were prepared by amplifying the 731 bp promoter region (from −738 to −7) using the primers 5′‐CCGCTCGAG ACCACTCTCCGACCTAAAGG‐3′ (forward) and 5′‐CCGAAGCTTCGTCGGCGGCTCCTCCGGGCTC‐3′ (reverse) that included XhoI and HindIII (both from Cnservice Invitrogen, Shanghai, China) restriction sites. Amplified fragments were sequenced to confirm that there were no errors in matched nucleotides and that the plasmid contained the correct allele. The fragments were cloned into the pGL3‐basic vector (Promega, Madison, WI, USA) after both were cut with XhoI and HindIII enzymes. Vectors were sequenced to confirm the orientation and integrity of the inserts.
Transient transfections and luciferase assays
For the luciferase assay, 293T and PIG1 cells were seeded in 24‐well plates (1 × 105 cells per well) and transfect with 0.8 μg of the recombinant pGL3 reporter vector with either −653 T or −653 C allele using Lipofectamine 2000 Transfection Reagent (Invitrogen, Carlsbad, CA, USA). As an internal control, all plasmids were co‐transfect with 0.02 μg pRL‐SV40 (Promega). The pGL3‐Basic vector without an insert and pGL3‐Control vector (Promega) were used as a negative control and a positive control respectively. Cells were collected 48 h after transfection, and cell lysates were prepared according to the manufacturer's instructions. Luciferase activity was measured with a Dual‐Luciferase Reporter Assay System (Promega) and normalized to the activity of pRL‐SV40 with the Renilla luciferase gene. Independent triplicate experiments were performed for each construct.
Serum HO‐1 and GST enzymatic activity
Computer‐generated random numbers were used to select samples from patients with non‐segmental vitiligo and normal controls (n = 180 each) for serum HO‐1 and GST activity analysis. No statistically significant difference was observed between the 180 vitiligo patients and the total patient population in terms of epidemiologic features and genotype frequencies. Serum HO‐1 and GST activities were measured using kits (Xi Tang Biotechnology Co. Ltd, Shanghai, China and Cusabio Biotech Co. Ltd, Wuhan, China, respectively), according to the manufacturer's instructions. The absorbance of samples was measured at 450 nm.
Statistical analysis
Differences in the frequency distributions of demographic variables, each allele and genotypes of Nrf2 and HO‐1 polymorphisms, and serum HO‐1 and GST activities between patients and controls were evaluated with the chi‐square test. The statistical analysis of the relationship between Nrf2 rs35652124 T/C genotype and serum HO‐1 and GST activity in vitiligo patients was performed with GraphPad Prism 5 software (GraphPad, San Diego, CA, USA) with the unpaired t‐test. Hardy–Weinberg equilibrium of the genotype distribution of the controls was also tested by a goodness‐of‐fit chi‐square test. Associations between polymorphisms and vitiligo risk were determined by computing odds ratios (ORs) and 95% confidence intervals (CIs) from both uni‐ and multivariate logistic regression analyses, adjusting for the following subgroups: age, gender, stage, type, onset age, family history, related autoimmune disease status, and smoking and drinking status. For the luciferase reporter assay, differences in expression levels of the luciferase gene between constructs were evaluated by the Student's t‐test. Two‐tailed tests of statistical analyses were performed with SAS software version 9.1 (SAS Institute, Inc., Cary, NC, USA). Statistical significance was defined as P < 0.05.
Results
Characteristics of study subjects
This analysis included 1136 Han Chinese patients with vitiligo and 1200 controls (Table 1). Age and sex were well matched between patients and controls (P = 0.896 and P = 0.944, respectively). Patients were considered to have early‐onset vitiligo (n = 681, 59.9%) if the age of onset was prior to 20 years old. Of the patients with vitiligo, 101 (8.9%) had segmental vitiligo and 1035 (91.1%) had non‐segmental vitiligo; 908 (79.9%) had active vitiligo and 228 (20.1%) had stable vitiligo in the cohort. Patients with vitiligo were considered to have a family history (n = 174, 15.3%), if they had one or more first‐ to third‐degree relatives afflicted with the condition. In addition, 31 (2.7%) patients with vitiligo had an accompanying autoimmune disease such as hyperthyroidism, alopecia areata, diabetes, halo naevi or connective tissue disease. There were 651 (57.3%) never smokers, 94 (8.3%) ex‐smokers (had quit smoking for >1 year), and 391 (34.4%) current smokers among patients; and 647 (53.9%) never smokers, 95 (7.9%) ex‐smokers and 458 (38.2%) current smokers among controls. The frequency distributions of smoking status were similar between patients and controls (P = 0.169). The drinking status (defined as individuals who had consumed alcohol at least once in their lifetime) among patients and controls was 26 (2.3%) and 36 (3.0%) respectively (P = 0.285).
Association between Nrf2 and HO‐1 genotypes and vitiligo risk
Nrf2 and HO‐1 genotype distributions and their associations with vitiligo risk for patients and controls are presented in Table 2. Genotype frequencies among control subjects were in Hardy–Weinberg equilibrium (P = 0.649 for Nrf2 rs35652124 T/C; P = 0.406 for Nrf2 rs6721961 G/T and P = 0.363 for HO‐1 rs2071746 A/T). TT, CT and CC genotype frequencies for Nrf2 rs35652124 were 33.4%, 49.3% and 17.3%, respectively, in controls and 37.3%, 50.4%, and 12.3%, respectively, in patients. The Nrf2 rs35652124 variant C allele frequency was significantly lower among the vitiligo cases than among the controls (P = 0.002). When the TT genotype was used as the reference, a statistically significant decreased risk of vitiligo was associated with the CC (adjusted OR = 0.64; 95% CI = 0.50–0.83) and combined (CT+CC) genotype (adjusted OR = 0.84; 95% CI = 0.71–0.99), whereas the CT genotype was not associated with a decreased vitiligo risk (adjusted OR = 0.91; 95% CI = 0.76–1.09).
Table 2.
Genotypic frequency of the Nrf2 and HO‐1 polymorphisms between cases and controls and their associations with the risk of vitiligo
| Genotypes | Cases (n = 1136) | Controls (n = 1200)a | P b | Adjusted OR (95% CI)c | ||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| Nrf2 rs35652124 | 0.002 | |||||
| TT | 424 | 37.3 | 401 | 33.4 | 1.00 (reference) | |
| CT | 572 | 50.4 | 592 | 49.3 | 0.91 (0.76–1.09) | |
| CC | 140 | 12.3 | 207 | 17.3 | 0.64 (0.50–0.83) | |
| CT+CC | 712 | 62.7 | 799 | 66.6 | 0.048 | 0.84 (0.71–0.99) |
| C allele | 35.7 | 40.7 | 0.002 | |||
| Nrf2 rs6721961 | 0.447 | |||||
| GG | 508 | 44.7 | 561 | 46.8 | 1.00 (reference) | |
| GT | 508 | 44.7 | 528 | 44.0 | 1.06 (0.90–1.26) | |
| TT | 120 | 10.6 | 111 | 9.2 | 1.19 (0.90–1.59) | |
| GT+TT | 628 | 55.3 | 639 | 53.2 | 0.346 | 1.09 (0.92–1.28) |
| T allele | 49.1 | 45.4 | 0.221 | |||
| HO‐1 rs2071746 | 0.124 | |||||
| AA | 499 | 43.9 | 520 | 43.3 | 1.00 (reference) | |
| AT | 465 | 40.9 | 529 | 44.1 | 0.92 (0.77–1.09) | |
| TT | 172 | 15.0 | 151 | 12.6 | 1.19 (0.92–1.53) | |
| AT+TT | 637 | 56.1 | 680 | 56.7 | 0.773 | 0.98 (0.83–1.15) |
| T allele | 55.3 | 53.0 | 0.482 | |||
Nrf2, Nuclear factor erythroid 2‐related factor 2; HO‐1, Heme Oxygenase‐1; OR, odds ratio; CI, confidence interval.
The observed genotype frequencies among the controls were in agreement with the Hardy–Weinberg equilibrium (χ2 = 0.207, P = 0.649 for Nrf2 rs35652124; χ2 = 0.691, P = 0.406 for Nrf2 rs6721961; χ2 = 0.828, P = 0.363 for HO‐1 rs2071746).
Two‐sided chi‐square test for distributions of genotype and allele frequencies between the cases and controls.
Adjusted ORs were obtained from a multivariate logistic regression with adjustment for age and gender.
However, there was no evidence that the allele and genotype frequencies of Nrf2 rs6721961 G/T and HO‐1 rs2071746 A/T were associated with susceptibility to vitiligo (P = 0.447 for Nrf2 rs6721961; P = 0.124 for HO‐1 rs2071746).
Effects of selected Nrf2 variables in different subgroups and their association with vitiligo risk
To evaluate the effects of Nrf2 polymorphism on patient characteristics, stratification analyses of Nrf2 polymorphisms and vitiligo risk were performed (Table 3). The frequency of the Nrf2 rs35652124 CT+CC genotype was lower for the subgroups, including female, onset age ≤20 years and never smokers in patients but not in controls (P = 0.029, P = 0.001 and P = 0.013 respectively). When Nrf2 rs35652124 TT genotype was used as a reference, the CT+CC genotype frequency was lower among patient subgroups with the following characteristics: female (adjusted OR = 0.76, 95% CI = 0.59–0.97), onset age ≤20 years (adjusted OR = 0.65, 95% CI = 0.51–0.83) and never smokers (adjusted OR = 0.73, 95% CI = 0.57–0.94).
Table 3.
Stratification analysis of the Nrf2 rs35652124 genotypes and vitiligo risk by selected variables
| Variables | Nrf2 rs35652124 (case–control) | P a | Adjusted OR (95% CI)b | |||
|---|---|---|---|---|---|---|
| TT | CT+CC | |||||
| n | % | n | % | |||
| Sex | ||||||
| Male | 199/200 | 17.5/16.7 | 409/444 | 36.0/37.0 | 0.525 | 0.93 (0.73–1.15) |
| Female | 225/201 | 19.8/16.7 | 303/355 | 26.7/29.6 | 0.029 | 0.76 (0.59–0.97) |
| Onset age (years) | ||||||
| ≤20 | 228/184 | 20.1/15.3 | 291/361 | 25.6/30.1 | 0.001 | 0.65 (0.51–0.83) |
| >20 | 196/217 | 17.3/18.1 | 421/438 | 37.0/36.5 | 0.604 | 1.06 (0.84–1.35) |
| Type | ||||||
| Segmental | 38/401 | 3.3/33.4 | 63/799 | 5.6/66.6 | 0.390 | 0.83 (0.55–1.27) |
| Non‐segmental | 386/401 | 34.0/33.4 | 649/799 | 57.1/66.6 | 0.056 | 0.84 (0.71–1.00) |
| Stage | ||||||
| Stable | 90/401 | 7.9/33.4 | 138/799 | 12.2/66.6 | 0.078 | 0.77 (0.58–1.03) |
| Active | 334/401 | 29.4/33.4 | 574/799 | 50.5/66.6 | 0.108 | 0.86 (0.72–1.03) |
| Family history | ||||||
| Yes | 69/401 | 6.1/33.4 | 105/799 | 9.2/66.6 | 0.105 | 0.76 (0.55–1.06) |
| No | 355/401 | 31.3/33.4 | 607/799 | 53.4/66.6 | 0.091 | 0.86 (0.70–1.01) |
| Autoimmunity disease | ||||||
| Yes | 15/401 | 1.3/33.4 | 16/799 | 1.4/66.6 | 0.082 | 0.54 (0.26–1.09) |
| No | 409/401 | 36.0/33.4 | 696/799 | 61.3/66.6 | 0.071 | 0.85 (0.72–1.02) |
| Smoking status | ||||||
| Never‐smokers | 184/144 | 16.2/12.0 | 467/503 | 41.1/41.9 | 0.013 | 0.73 (0.57–0.94) |
| Ex‐smokers | 60/57 | 5.3/4.7 | 34/38 | 3.0/3.2 | 0.588 | 0.85 (0.47–'1.53) |
| Current‐smokers | 180/200 | 15.8/16.7 | 211/258 | 18.6/21.5 | 0.489 | 0.91 (0.69–1.19) |
| Drinking status | ||||||
| Never | 404/380 | 35.6/31.7 | 706/784 | 62.1/65.3 | 0.060 | 0.85 (0.71–1.01) |
| Ever | 20/21 | 1.8/1.7 | 6/15 | 0.5/1.3 | 0.127 | 0.42 (0.14–1.29) |
Nrf2, Nuclear factor erythroid 2‐related factor 2; OR, odds ratio; CI, confidence interval.
Two‐sided chi‐square test for distributions of genotype and allele frequencies between the cases and controls.
Adjusted ORs were obtained from a multivariate logistic regression with adjustment for age and gender.
Effects of Nrf2 rs35652124 polymorphism on transcriptional activity
To evaluate the promoter activity associated with Nrf2 rs35652124 T/C polymorphism, T or C promoter constructs were transiently transfect into human embryonic kidney 293T and human normal melanocyte PIG1 cells. A higher relative luciferase activity was observed for the C than for the T allele of rs35652124 in both cell lines (P = 0.017 for 293T cells, P = 0.035 for PIG1 cells) (Fig. 2). These results indicate that the T to C transition in the Nrf2 rs35652124 promoter region increases the transcriptional activity of the Nrf2 gene.
Figure 2.
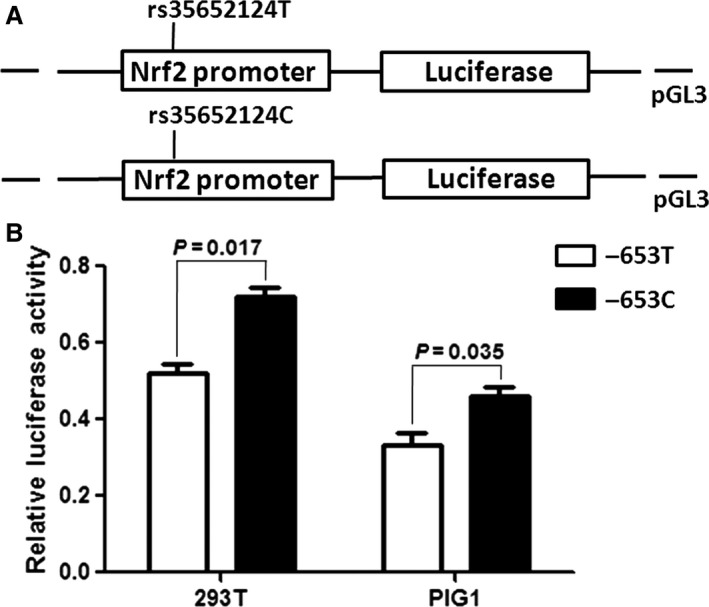
Effect of the rs35652124 T/C polymorphism in the Nrf2 promoter activity. Nrf2, nuclear factor erythroid 2‐related factor 2. (A) Schematic representation of reporter plasmids containing the rs35652124T or rs35652124C allele, which was inserted upstream of the luciferase reporter gene in the pGL3 basic plasmid. (B) Two constructs were transiently transfected into the 293 T and PIG1 cells, respectively. The luciferase activity of each construct was normalized against the internal control of Renilla luciferase. Columns, mean from two‐independent experiments; bars, SD. *P < 0.05 compared with the construct counterpart.
Serum HO‐1 and GST activities in vitiligo patients and controls
Serum HO‐1 activity was measured in 180 vitiligo patients and 180 normal controls, whose demographic, clinical characteristics and genotype frequencies corresponded to those of the study population (1136 cases and 1200 controls; data not shown). As shown in Fig. 3A, HO‐1 activity was lower in the vitiligo than in the control group (34.91 ± 12.12 ng/ml vs. 40.97 ± 14.91 ng/ml; P = 0.024). Similarly, serum GST activity was lower in 67 vitiligo patient samples than in age‐ and sex‐matched control samples (14.50 ± 8.31 ng/ml vs. 21.94 ± 11.76 ng/ml; P = 0.0001) (Fig. 3C).
Figure 3.
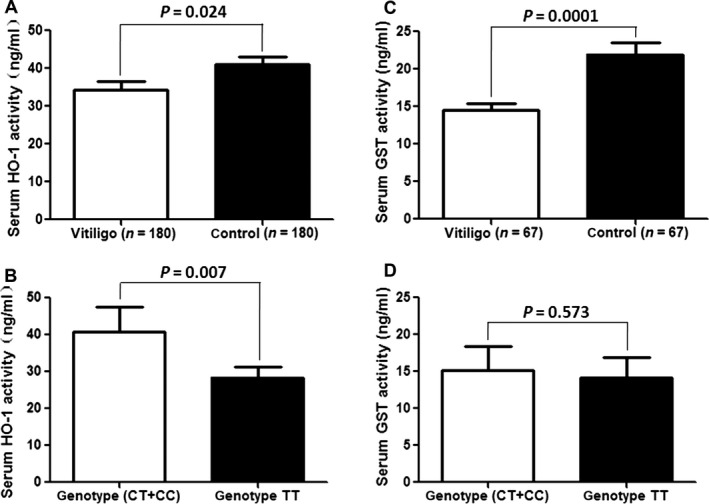
Serum HO‐1 activity and correlations to vitiligo genotype. Nrf2, nuclear factor erythroid 2‐related factor 2; HO‐1, Heme Oxygenase‐1. (A) The serum HO‐1 activity in vitiligo patients’ group is significantly lower than that in the normal control group (P < 0.05). (B) Compared with the Nrf2 rs35652124 combined TT genotype group, the protect combined genotype CT+CC group has the high serum HO‐1 activity (P < 0.05). (C) The serum GST activity in vitiligo patients’ group is significantly lower than that in the normal control group (P < 0.05). (D) There were no differences in serum GST activity among Nrf2 rs35652124 genotypes (P > 0.05).
Furthermore, an analysis of the relationship between Nrf2 rs35652124T/C genotype and serum HO‐1 activity in vitiligo patients showed that compared to TT, the combined genotype (CT+CC) genotype was associated with higher serum HO‐1 activity (P = 0.007) (Fig. 3B). However, there were no differences in serum GST activity among Nrf2 rs35652124 genotypes (P = 0.573) (Fig. 3D).
Logistic regression analysis of HO‐1 activity in vitiligo patients and controls
In a logistic regression analysis in which serum HO‐1 activity was dichotomized by median activity of the control group, a higher activity was associated with a 0.64‐fold reduction in vitiligo risk (P = 0.034, 95% CI = 0.42–0.97) (Table 4). Furthermore, when HO‐1 activity was divided into tertiles, according to the activity of the control group, a dose–response relationship between increased activity and decreased risk was apparent: suboptimal (upper tertile), intermediate (mid tertile) and efficient (lower tertile) activities had adjusted ORs of 1.00, 0.55 (P = 0.015, 95% CI = 0.33–0.90) and 0.43 (P = 0.001, 95% CI = 0.26–0.72) respectively.
Table 4.
Logistic regression analysis of HO‐1 activity in vitiligo patients and controls
| HO‐1 activity (ng/ml) | Case (n=180) | Control (n=180) | P a | Adjusted OR (95% CI)b | ||
|---|---|---|---|---|---|---|
| n | % | n | % | |||
| By median | ||||||
| <32.1 | 110 | 61.1 | 90 | 50.0 | 1.00 (reference) | |
| ≥32.1 | 70 | 38.9 | 90 | 50.0 | 0.034 | 0.64 (0.42–0.97) |
| By tertile | ||||||
| ≤26.3 | 91 | 57.2 | 59 | 32.8 | 1.00 (reference) | |
| 26.3–43.6 | 50 | 10.6 | 60 | 33.3 | 0.015 | 0.55 (0.33–0.90) |
| ≥43.6 | 39 | 32.2 | 61 | 33.9 | 0.001 | 0.43 (0.26–0.72) |
| Trend test | c P < 0.05 | |||||
Nrf2, Nuclear factor erythroid 2‐related factor 2; HO‐1, Heme Oxygenase‐1; OR, odds ratio; CI, confidence interval.
Two‐sided chi‐square test for distributions of genotype and allele frequencies between the cases and controls.
Adjusted ORs were obtained from a logistic regression with adjustment for age and gender.
Adjusted for age and sex.
Association between Nrf2 rs35652124 genotype and vitiligo risk based on HO‐1 activity
The association between vitiligo risk and Nrf2 rs35652124 genotypes was examined with respect to HO‐1 activities (Table 5). Nrf2 rs35652124 genotypes were divided into two categories: the 1‐2 (CT or TT) and 0 (CC) risk alleles. When carriers of the 1‐2 risk allele and with lower HO‐1 activity (≤32.1 pg/ml) were used as a reference, individuals with the 0 risk allele and higher HO‐1 activity (>32.1 pg/ml) had a decreased risk of vitiligo (adjusted OR = 0.43, 95% CI = 0.23–0.79). Consistent with these results, when the serum HO‐1 activity was divided into tertiles, individuals with the 0 risk allele and HO‐1 activity in the upper tertile (>43.6 pg/ml) had a larger decrease in vitiligo risk (adjusted OR = 0.41, 95% CI = 0.20–0.85) compared to those with the 1‐2 risk allele and lower tertile HO‐1 activity (≤26.3 pg/ml).
Table 5.
Risk of vitiligo associated with Nrf2 rs35652124 genotypes by HO‐1 activity
| HO‐1 activity (pg/ml) | Nrf2 rs35652124 (case–control) | |||||
|---|---|---|---|---|---|---|
| CT/TT (1‐2 risk genotype) | P a | OR (95% CI) | CC (0 risk genotypes) | P a | OR (95% CI) | |
| By median | ||||||
| <32.1 | 64/47 | 1.00 (reference) | 46/43 | 0.399 | 0.79 (0.45–1.38) | |
| ≥32.1 | 43/44 | 0.249 | 0.72 (0.41–1.26) | 27/46 | 0.006 | 0.43 (0.23–0.79) |
| By tertile | ||||||
| ≤26.3 | 42/32 | 1.00 (reference) | 49/29 | 0.446 | 1.29 (0.67–2.47) | |
| 26.3–43.6 | 30/33 | 0.286 | 0.69 (0.35–1.36) | 20/27 | 0.128 | 0.56 (0.27–1.18) |
| ≥43.6 | 20/26 | 0.157 | 0.59 (0.28–1.23) | 19/35 | 0.016 | 0.41 (0.20–0.85) |
Nrf2, Nuclear factor erythroid 2‐related factor 2; HO‐1, Heme Oxygenase‐1; OR, odds ratio; CI, confidence interval.
Two‐sided chi‐square test for distributions of genotype and allele frequencies between the cases and controls.
ORs were obtained from a multivariate logistic regression with adjustment for age and gender.
Discussion
Although the aetiology of vitiligo is not fully understood, the accumulation of H2O2 because of oxidative stress may be a trigger for melanocyte degeneration 23. Nrf2 is a redox‐sensitive transcription factor that has a protective role against oxidative stress, and Nrf2 variants may have a influential capacity for binding promoters, which undermines target gene expression 24.
This study demonstrated that whether certain genetic polymorphisms would contribute to the risk of developing vitiligo. In 2008, Guan et al. found that an Nrf2 promoter SNP at −650 position was associated with the development of vitiligo in China and the −650 A allele may be one of the risk factor 25. Their study provided genetic evidence for the relationship between Nrf2, an important antioxidant gene, and vitiligo. In addition to the −650 SNP loci, some other SNPs of human Nrf2 have been shown to be associated with the risk of various diseases, which prompted us to evaluate their association with vitiligo susceptibility in this study. As a result, we demonstrated that a statistically decreased risk of vitiligo was associated with the Nrf2 rs35652124 variant C allele, although no evident risk was associated with Nrf2 rs6721961 or HO‐1 rs2071746 variants, indicating that there was an intrinsic linkage between Nrf2 genetic variants and the risk of vitiligo. Moreover, a lower vitiligo risk was found in the female, early‐onset age and never smoker subgroups. Since smoking known to aggravate oxidative stress, our result supported Nrf2 gene polymorphism may play a key role for the systemic response to ROS in cigarette smoke exposure. Interestingly, the T to C substitution in Nrf2 rs35652124 increased the transcriptional activity of Nrf2 in vitro, and a higher serum HO‐1 activity was detected in vitiligo patients with the CT+CC rather than the TT genotypes. These data suggested that this variant of Nrf2 might affect Nrf2 function and the risk of developing vitiligo in a Han Chinese population.
The rs35652124 T/C polymorphism is located at position −653 of the Nrf2 gene. The transcription factor binding search analysis was performed with TRANSFAC® 7.0 public database (http://www.gene-regulation.com/cgi-bin/pub/programs/alibaba2/webbaba2.cgi) to ascertain the potential functional relevance of this SNP. Interestingly, this analysis was performed that the variant of Nrf2 rs35652124 T/C is situated in the core sequence of the putative binding site for the transcription factor, specificity protein 1 (Sp1); this T to C allele mutation might therefore contribute to the combination of Sp1 with the Nrf2 promoter region. Based on the in vitro reporter assay, the protective C allele of this polymorphism may enhance Nrf2 promoter activity through influencing the binding activity of transcriptional regulator Sp1 compared with T allele. Thus, the Nrf2 rs35652124 C allele may be expected to affect Nrf2 expression and the important regulatory roles in various cellular responses following oxidative damage, thereby conferring protection against vitiligo. This hypothesis is strongly supported by our results.
Oxidative stress activates transcription of a variety of genes encoding antioxidative enzymes through a cis‐acting sequence known as the ARE 26. The ARE is initially found in promoter regions of genes encoding phase II detoxification enzymes and antioxidant proteins 27. Nrf2 rs6721961 G/T polymorphism is located at position −617 of the promoter region, affecting the ARE binding site and possibly gene expression. Nrf2 functional polymorphisms have been linked to the risk of developing ALI after major trauma 28, childhood asthma caused by air pollution 29, and oxidative stress‐related defects in human spermatogenesis caused by heavy smoking 30. In addition, in one study of forearm vasodilatory responses, Caucasian subjects with the Nrf2 rs6721961 T allele had decreased blood flow and increased vascular resistance as compared to that in non‐carriers, while no such association was observed among African‐American subjects 31, suggesting an ethnic component to the distribution of the Nrf2 rs6721961 G/T polymorphism. In this study, there was no statistical correlation between rs6721961 and vitiligo risk. This may be due to sample size or variability across diseases and populations. Similarly, there was no evidence that HO‐1 rs2071746 A/T is associated with susceptibility to vitiligo.
It has been suggested that oxidative stress may be an important contributor to the pathogenesis of melanocyte death because of the increase in the production of oxygen free radicals and a deficiency in antioxidant defences mechanisms. Many studies have examined the status of antioxidants and antioxidant enzymes in vitiligo patients 32. Meanwhile, up‐regulation of HO‐1 may represent an attempt to minimize cellular injury 33. In our previous study, we investigated the serum HO‐1 activity in patients with vitiligo and controls, and a lower HO‐1 activity was observed in vitiligo patients than in controls. This finding suggested that a deficiency in HO‐1 might contribute to the development of vitiligo. Moreover, HO‐1 is a major antioxidant enzyme regulated by Nrf2 in vitiligo, which plays a role in protecting melanocytes against H2O2‐induced oxidative stress. Induction of these antioxidant enzymes is mediated largely by the transcription factor Nrf2 34. Among patients with vitiligo, we report for the first time that there was a dose–response relationship between HO‐1 activity and Nrf2 rs35652124 T/C genotype. Comprehensive analysis of HO‐1 activity and Nrf2 rs35652124 T/C genotypes suggested that, much as with antioxidants, the rs35652124 C variant genotype played an important role in protection from oxidative damage. Individuals with higher HO‐1 activity and the rs35652124 C variant genotype might have a minor risk for developing vitiligo compared with those of low HO‐1 activity and an rs35652124 T wild genotype. Consistent with this hypothesis, targeted disruption of Nrf2 changed antioxidant capacity in mice and thus increased susceptibility to pro‐oxidant and carcinogenic agents 35, 36.
Moreover, GSTs are a large family of enzymes participating in detoxification of endogenous. GST antioxidant activity in blood was proven to be linked to the risk of oxidative stress‐related type 2 diabetes mellitus and impaired glucose tolerance 37. These findings have led to the hypotheses that serum GST activity may be associated with vitiligo. We evaluated serum GST level, and a lower GST activity indeed existed in vitiligo patients than in controls. However, our results showed no association between serum GST and Nrf2 rs35652124 polymorphisms. This is not surprising, since many cytoprotective genes which could regulate the enzyme production and, a single abnormality, may not be adequate to affect the level of GST activity. The population‐based studies with a larger sample size are necessary to confirm these findings.
In summary, we report that the Nrf2 rs35652124 T/C polymorphism and serum HO‐1 activity affect susceptibility to vitiligo among Han Chinese population. Future studies will examine SNPs in other genes involved in oxidative stress to confirm the link between this process and melanocyte production in vitiligo. The present findings indicate that treatments strategies that regulate HO‐1 activity could potentially reverse melanocyte degeneration and depigmentation in vitiligo, ultimately leading to improved self‐esteem and better quality of life in patients. Furthermore, studies are necessary to evaluate whether other potential transcriptional mechanisms are associated with Nrf2 rs35652124 T/C polymorphisms.
Conflict of interest
The authors confirm that there are no conflicts of interest.
Acknowledgements
We thank Yajun Zhang, Qiong Shi, Xia Li and Xiuli Yi for helping recruit the subjects; Hua Wang for laboratory assistance. This study was supported by grants from the National Natural Science Foundation of China (No. 30972642, No.30972528 and No.81172749). As for the authors’ specific contributions to the work, Tianwen Gao, Chunying Li, and Gang Wang designed the research study; Ling Liu and Kai Li recruited the subjects; Pu Song, Xiaowen Wang and Zhe Jian performed the research; Pu Song, Kai Li and Weigang Zhang analysed the data and wrote the paper.
Contributor Information
Chunying Li, Email: lichying@fmmu.edu.cn.
Tianwen Gao, Email: gaotw75401@hotmail.com.
References
- 1. Ezzedine K, Lim HW, Suzuki T, et al Revised classification/nomenclature of vitiligo and related issues: the Vitiligo Global Issues Consensus Conference. Pigment Cell Melanoma Res. 2012; 25: E1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhang Z, Xu SX, Zhang FY, et al The analysis of genetics and associated autoimmune diseases in Chinese vitiligo patients. Arch Dermatol Res. 2009; 301: 167–73. [DOI] [PubMed] [Google Scholar]
- 3. Arican O, Kurutas EB. Oxidative stress in the blood of patients with active localized vitiligo. Acta Dermatovenerol Alp Pannonica Adriat. 2008; 17: 12–6. [PubMed] [Google Scholar]
- 4. Denat L, Kadekaro AL, Marrot L, et al Melanocytes as instigators and victims of oxidative stress. J Invest Dermatol. 2014; 134: 1512–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Sander CS, Hamm F, Elsner P, et al Oxidative stress in malignant melanoma and non‐melanoma skin cancer. Br J Dermatol. 2003; 148: 913–22. [DOI] [PubMed] [Google Scholar]
- 6. Schallreuter KU, Gibbons NC, Zothner C, et al Hydrogen peroxide‐mediated oxidative stress disrupts calcium binding on calmodulin: more evidence for oxidative stress in vitiligo. Biochem Biophys Res Commun. 2007; 360: 70–5. [DOI] [PubMed] [Google Scholar]
- 7. Shalbaf M, Gibbons NC, Wood JM, et al Presence of epidermal allantoin further supports oxidative stress in vitiligo. Exp Dermatol. 2008; 17: 761–70. [DOI] [PubMed] [Google Scholar]
- 8. Kong M, Mao J, Luo B, et al Role of transcriptional factor Nrf2 in the acute lung injury of mice. Int J Clin Exp Pathol. 2015; 8: 10929–34. [PMC free article] [PubMed] [Google Scholar]
- 9. Hayes JD, McMahon M, Chowdhry S, et al Cancer chemoprevention mechanisms mediated through the Keap1‐Nrf2 pathway. Antioxid Redox Signal. 2010; 13: 1713–48. [DOI] [PubMed] [Google Scholar]
- 10. Lee JS, Surh YJ. Nrf2 as a novel molecular target for chemoprevention. Cancer Lett. 2005; 224: 171–84. [DOI] [PubMed] [Google Scholar]
- 11. Andrews NC, Erdjument‐Bromage H, Davidson MB, et al Erythroid transcription factor NF‐E2 is a haematopoietic‐specific basic‐leucine zipper protein. Nature. 1993; 362: 722–8. [DOI] [PubMed] [Google Scholar]
- 12. Askenazi DJ, Halloran B, Patil N, et al Genetic polymorphisms of heme‐oxygenase 1 (HO‐1) may impact on acute kidney injury, bronchopulmonary dysplasia, and mortality in premature infants. Pediatr Res. 2015; 77: 793–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Luo Y, Sun G, Dong X, et al Isorhamnetin attenuates atherosclerosis by inhibiting macrophage apoptosis via PI3K/AKT activation and HO‐1 induction. PLoS ONE. 2015; 10: e120259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Varol F, Uzunoglu R, Erbas H, et al VEGFR‐1, Bcl‐2, and HO‐1 ratios in pregnant women with hypertension. Clin Appl Thromb Hemost. 2015; 21: 285–8. [DOI] [PubMed] [Google Scholar]
- 15. Jian Z, Li K, Liu L, et al Heme oxygenase‐1 protects human melanocytes from H2O2‐induced oxidative stress via the Nrf2‐ARE pathway. J Invest Dermatol. 2011; 131: 1420–7. [DOI] [PubMed] [Google Scholar]
- 16. Kim YC, Masutani H, Yamaguchi Y, et al Hemin‐induced activation of the thioredoxin gene by Nrf2. A differential regulation of the antioxidant responsive element by a switch of its binding factors. J Biol Chem. 2001; 276: 18399–406. [DOI] [PubMed] [Google Scholar]
- 17. Wang XR, Shi GX, Yang JW, et al Acupuncture ameliorates cognitive impairment and hippocampus neuronal loss in experimental vascular dementia through Nrf2‐mediated antioxidant response. Free Radic Biol Med. 2015; 89: 1077–84. [DOI] [PubMed] [Google Scholar]
- 18. Cho HY, Jedlicka AE, Reddy SP, et al Linkage analysis of susceptibility to hyperoxia. Nrf2 is a candidate gene. Am J Respir Cell Mol Biol. 2002; 26: 42–51. [DOI] [PubMed] [Google Scholar]
- 19. Schipper HM. Heme oxygenase‐1: transducer of pathological brain iron sequestration under oxidative stress. Ann N Y Acad Sci. 2004; 1012: 84–93. [DOI] [PubMed] [Google Scholar]
- 20. Infante J, Garcia‐Gorostiaga I, Sanchez‐Juan P, et al Synergistic effect of two oxidative stress‐related genes (heme oxygenase‐1 and GSK3beta) on the risk of Parkinson's disease. Eur J Neurol. 2010; 17: 760–2. [DOI] [PubMed] [Google Scholar]
- 21. Mateo I, Sanchez‐Juan P, Rodriguez‐Rodriguez E, et al Synergistic effect of heme oxygenase‐1 and tau genetic variants on Alzheimer's disease risk. Dement Geriatr Cogn Disord. 2008; 26: 339–42. [DOI] [PubMed] [Google Scholar]
- 22. Li XL, Cao J, Fan L, et al Genetic polymorphisms of HO‐1 and COX‐1 are associated with aspirin resistance defined by light transmittance aggregation in Chinese Han patients. Clin Appl Thromb Hemost. 2013; 19: 513–21. [DOI] [PubMed] [Google Scholar]
- 23. Passi S, Grandinetti M, Maggio F, et al Epidermal oxidative stress in vitiligo. Pigment Cell Res. 1998; 11: 81–5. [DOI] [PubMed] [Google Scholar]
- 24. Cho HY. Genomic structure and variation of nuclear factor (erythroid‐derived 2)‐like 2. Oxid Med Cell Longev. 2013; 2013: 286524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Guan CP, Zhou MN, Xu AE, et al The susceptibility to vitiligo is associated with NF‐E2‐related factor2 (Nrf2) gene polymorphisms: a study on Chinese Han population. Exp Dermatol. 2008; 17: 1059–62. [DOI] [PubMed] [Google Scholar]
- 26. Biswas M, Chan JY. Role of Nrf1 in antioxidant response element‐mediated gene expression and beyond. Toxicol Appl Pharmacol. 2010; 244: 16–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kwok SC, Samuel SP, Handal J. Atorvastatin activates heme oxygenase‐1 at the stress response elements. J Cell Mol Med. 2012; 16: 394–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. O'Mahony DS, Glavan BJ, Holden TD, et al Inflammation and immune‐related candidate gene associations with acute lung injury susceptibility and severity: a validation study. PLoS ONE. 2012; 7: e51104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ungvari I, Hadadi E, Virag V, et al Relationship between air pollution, NFE2L2 gene polymorphisms and childhood asthma in a Hungarian population. J Community Genet. 2012; 3: 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Yu B, Chen J, Liu D, et al Cigarette smoking is associated with human semen quality in synergy with functional NRF2 polymorphisms. Biol Reprod. 2013; 89: 5. [DOI] [PubMed] [Google Scholar]
- 31. Marczak ED, Marzec J, Zeldin DC, et al Polymorphisms in the transcription factor NRF2 and forearm vasodilator responses in humans. Pharmacogenet Genomics. 2012; 22: 620–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Liu L, Li C, Gao J, et al Promoter variant in the catalase gene is associated with vitiligo in Chinese people. J Invest Dermatol. 2010; 130: 2647–53. [DOI] [PubMed] [Google Scholar]
- 33. Muchova L, Vanova K, Suk J, et al Protective effect of heme oxygenase induction in ethinylestradiol‐induced cholestasis. J Cell Mol Med. 2015; 19: 924–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McMahon M, Itoh K, Yamamoto M, et al The Cap'n'Collar basic leucine zipper transcription factor Nrf2 (NF‐E2 p45‐related factor 2) controls both constitutive and inducible expression of intestinal detoxification and glutathione biosynthetic enzymes. Cancer Res. 2001; 61: 3299–307. [PubMed] [Google Scholar]
- 35. Ramos‐Gomez M, Kwak MK, Dolan PM, et al Sensitivity to carcinogenesis is increased and chemoprotective efficacy of enzyme inducers is lost in nrf2 transcription factor‐deficient mice. Proc Natl Acad Sci U S A. 2001; 98: 3410–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Kwak MK, Kensler TW. Targeting NRF2 signaling for cancer chemoprevention. Toxicol Appl Pharmacol. 2010; 244: 66–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Karaman A, Bayram F, Gundogan K, et al Prevalence of diabetes mellitus and glucose metabolism disorders in the first degree relatives of type 2 diabetic patients. Bratisl Lek Listy. 2012; 113: 361–7. [DOI] [PubMed] [Google Scholar]


