Abstract
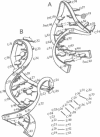
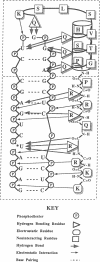
Transcription in the human immunodeficiency virus type 1 (HIV-1) retrovirus is regulated by binding the viral Tat protein (trans-acting transcriptional activator) to the trans-activation response (TAR) RNA sequence. Here, vacuum UV circular dichroism (VUV-CD) is used to study the structure of TAR and its complex with two peptide fragments that are important for Tat binding to TAR. The VUV-CD spectrum of TAR is typical of A-form RNA and is minimally perturbed when bound to either the short or the long Tat peptide. The CD spectra of the complexes indicate an extended structure in the arginine-rich region of Tat from amino acid residue 47 through residue 58 and a short alpha-helix within the adjacent 59-72 region. Models of TAR and its peptide complexes are constructed to integrate these spectroscopic results with current biochemical data. The model suggests that (i) the arginine-rich 49-58 region is primarily responsible for electrostatic interactions with the phosphates of the RNA, (ii) the arginine side chains can additionally interact with substituent groups of the nucleotide bases to confer base recognition in the complex, (iii) the recognition of uracil-23 in TAR is facilitated by the peptide backbone, and (iv) the glutamine-rich face of an alpha-helix within the 59-72 region pairs to bases UGG at nucleotide positions 31-33 in the TAR loop and thus provides an additional motif in the Tat trans-activating protein to recognize TAR RNA.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arya S. K., Guo C., Josephs S. F., Wong-Staal F. Trans-activator gene of human T-lymphotropic virus type III (HTLV-III). Science. 1985 Jul 5;229(4708):69–73. doi: 10.1126/science.2990040. [DOI] [PubMed] [Google Scholar]
- Berkhout B., Jeang K. T. Detailed mutational analysis of TAR RNA: critical spacing between the bulge and loop recognition domains. Nucleic Acids Res. 1991 Nov 25;19(22):6169–6176. doi: 10.1093/nar/19.22.6169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braddock M., Chambers A., Wilson W., Esnouf M. P., Adams S. E., Kingsman A. J., Kingsman S. M. HIV-1 TAT "activates" presynthesized RNA in the nucleus. Cell. 1989 Jul 28;58(2):269–279. doi: 10.1016/0092-8674(89)90841-6. [DOI] [PubMed] [Google Scholar]
- Calnan B. J., Biancalana S., Hudson D., Frankel A. D. Analysis of arginine-rich peptides from the HIV Tat protein reveals unusual features of RNA-protein recognition. Genes Dev. 1991 Feb;5(2):201–210. doi: 10.1101/gad.5.2.201. [DOI] [PubMed] [Google Scholar]
- Calnan B. J., Tidor B., Biancalana S., Hudson D., Frankel A. D. Arginine-mediated RNA recognition: the arginine fork. Science. 1991 May 24;252(5009):1167–1171. doi: 10.1126/science.252.5009.1167. [DOI] [PubMed] [Google Scholar]
- Cordingley M. G., LaFemina R. L., Callahan P. L., Condra J. H., Sardana V. V., Graham D. J., Nguyen T. M., LeGrow K., Gotlib L., Schlabach A. J. Sequence-specific interaction of Tat protein and Tat peptides with the transactivation-responsive sequence element of human immunodeficiency virus type 1 in vitro. Proc Natl Acad Sci U S A. 1990 Nov;87(22):8985–8989. doi: 10.1073/pnas.87.22.8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cullen B. R. The HIV-1 Tat protein: an RNA sequence-specific processivity factor? Cell. 1990 Nov 16;63(4):655–657. doi: 10.1016/0092-8674(90)90129-3. [DOI] [PubMed] [Google Scholar]
- Cullen B. R. Trans-activation of human immunodeficiency virus occurs via a bimodal mechanism. Cell. 1986 Sep 26;46(7):973–982. doi: 10.1016/0092-8674(86)90696-3. [DOI] [PubMed] [Google Scholar]
- Dingwall C., Ernberg I., Gait M. J., Green S. M., Heaphy S., Karn J., Lowe A. D., Singh M., Skinner M. A., Valerio R. Human immunodeficiency virus 1 tat protein binds trans-activation-responsive region (TAR) RNA in vitro. Proc Natl Acad Sci U S A. 1989 Sep;86(18):6925–6929. doi: 10.1073/pnas.86.18.6925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng S., Holland E. C. HIV-1 tat trans-activation requires the loop sequence within tar. Nature. 1988 Jul 14;334(6178):165–167. doi: 10.1038/334165a0. [DOI] [PubMed] [Google Scholar]
- Garcia J. A., Harrich D., Soultanakis E., Wu F., Mitsuyasu R., Gaynor R. B. Human immunodeficiency virus type 1 LTR TATA and TAR region sequences required for transcriptional regulation. EMBO J. 1989 Mar;8(3):765–778. doi: 10.1002/j.1460-2075.1989.tb03437.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gatignol A., Buckler-White A., Berkhout B., Jeang K. T. Characterization of a human TAR RNA-binding protein that activates the HIV-1 LTR. Science. 1991 Mar 29;251(5001):1597–1600. doi: 10.1126/science.2011739. [DOI] [PubMed] [Google Scholar]
- Gaynor R., Soultanakis E., Kuwabara M., Garcia J., Sigman D. S. Specific binding of a HeLa cell nuclear protein to RNA sequences in the human immunodeficiency virus transactivating region. Proc Natl Acad Sci U S A. 1989 Jul;86(13):4858–4862. doi: 10.1073/pnas.86.13.4858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman M., Verdini A. S., Toniolo C., Phillips W. D., Bovey F. A. Sensitive criteria for the critical size for helix formation in oligopeptides. Proc Natl Acad Sci U S A. 1969 Oct;64(2):444–450. doi: 10.1073/pnas.64.2.444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harper J. W., Logsdon N. J. Refolded HIV-1 tat protein protects both bulge and loop nucleotides in TAR RNA from ribonucleolytic cleavage. Biochemistry. 1991 Aug 13;30(32):8060–8066. doi: 10.1021/bi00246a026. [DOI] [PubMed] [Google Scholar]
- Hauber J., Malim M. H., Cullen B. R. Mutational analysis of the conserved basic domain of human immunodeficiency virus tat protein. J Virol. 1989 Mar;63(3):1181–1187. doi: 10.1128/jvi.63.3.1181-1187.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson W. C., Jr Circular dichroism and its empirical application to biopolymers. Methods Biochem Anal. 1985;31:61–163. doi: 10.1002/9780470110522.ch2. [DOI] [PubMed] [Google Scholar]
- Kamine J., Loewenstein P., Green M. Mapping of HIV-1 Tat protein sequences required for binding to Tar RNA. Virology. 1991 Jun;182(2):570–577. doi: 10.1016/0042-6822(91)90598-6. [DOI] [PubMed] [Google Scholar]
- Kuppuswamy M., Subramanian T., Srinivasan A., Chinnadurai G. Multiple functional domains of Tat, the trans-activator of HIV-1, defined by mutational analysis. Nucleic Acids Res. 1989 May 11;17(9):3551–3561. doi: 10.1093/nar/17.9.3551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loret E. P., Vives E., Ho P. S., Rochat H., Van Rietschoten J., Johnson W. C., Jr Activating region of HIV-1 Tat protein: vacuum UV circular dichroism and energy minimization. Biochemistry. 1991 Jun 18;30(24):6013–6023. doi: 10.1021/bi00238a027. [DOI] [PubMed] [Google Scholar]
- Manavalan P., Johnson W. C., Jr Variable selection method improves the prediction of protein secondary structure from circular dichroism spectra. Anal Biochem. 1987 Nov 15;167(1):76–85. doi: 10.1016/0003-2697(87)90135-7. [DOI] [PubMed] [Google Scholar]
- Marciniak R. A., Calnan B. J., Frankel A. D., Sharp P. A. HIV-1 Tat protein trans-activates transcription in vitro. Cell. 1990 Nov 16;63(4):791–802. doi: 10.1016/0092-8674(90)90145-5. [DOI] [PubMed] [Google Scholar]
- Marciniak R. A., Garcia-Blanco M. A., Sharp P. A. Identification and characterization of a HeLa nuclear protein that specifically binds to the trans-activation-response (TAR) element of human immunodeficiency virus. Proc Natl Acad Sci U S A. 1990 May;87(9):3624–3628. doi: 10.1073/pnas.87.9.3624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marciniak R. A., Sharp P. A. HIV-1 Tat protein promotes formation of more-processive elongation complexes. EMBO J. 1991 Dec;10(13):4189–4196. doi: 10.1002/j.1460-2075.1991.tb04997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muesing M. A., Smith D. H., Capon D. J. Regulation of mRNA accumulation by a human immunodeficiency virus trans-activator protein. Cell. 1987 Feb 27;48(4):691–701. doi: 10.1016/0092-8674(87)90247-9. [DOI] [PubMed] [Google Scholar]
- Nelbock P., Dillon P. J., Perkins A., Rosen C. A. A cDNA for a protein that interacts with the human immunodeficiency virus Tat transactivator. Science. 1990 Jun 29;248(4963):1650–1653. doi: 10.1126/science.2194290. [DOI] [PubMed] [Google Scholar]
- Pabo C. O., Sauer R. T. Protein-DNA recognition. Annu Rev Biochem. 1984;53:293–321. doi: 10.1146/annurev.bi.53.070184.001453. [DOI] [PubMed] [Google Scholar]
- Rounseville M. P., Kumar A. Binding of a host cell nuclear protein to the stem region of human immunodeficiency virus type 1 trans-activation-responsive RNA. J Virol. 1992 Mar;66(3):1688–1694. doi: 10.1128/jvi.66.3.1688-1694.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy S., Delling U., Chen C. H., Rosen C. A., Sonenberg N. A bulge structure in HIV-1 TAR RNA is required for Tat binding and Tat-mediated trans-activation. Genes Dev. 1990 Aug;4(8):1365–1373. doi: 10.1101/gad.4.8.1365. [DOI] [PubMed] [Google Scholar]
- Roy S., Parkin N. T., Rosen C., Itovitch J., Sonenberg N. Structural requirements for trans activation of human immunodeficiency virus type 1 long terminal repeat-directed gene expression by tat: importance of base pairing, loop sequence, and bulges in the tat-responsive sequence. J Virol. 1990 Mar;64(3):1402–1406. doi: 10.1128/jvi.64.3.1402-1406.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheline C. T., Milocco L. H., Jones K. A. Two distinct nuclear transcription factors recognize loop and bulge residues of the HIV-1 TAR RNA hairpin. Genes Dev. 1991 Dec;5(12B):2508–2520. doi: 10.1101/gad.5.12b.2508. [DOI] [PubMed] [Google Scholar]
- Sumner-Smith M., Roy S., Barnett R., Reid L. S., Kuperman R., Delling U., Sonenberg N. Critical chemical features in trans-acting-responsive RNA are required for interaction with human immunodeficiency virus type 1 Tat protein. J Virol. 1991 Oct;65(10):5196–5202. doi: 10.1128/jvi.65.10.5196-5202.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weeks K. M., Ampe C., Schultz S. C., Steitz T. A., Crothers D. M. Fragments of the HIV-1 Tat protein specifically bind TAR RNA. Science. 1990 Sep 14;249(4974):1281–1285. doi: 10.1126/science.2205002. [DOI] [PubMed] [Google Scholar]
- Weeks K. M., Crothers D. M. RNA recognition by Tat-derived peptides: interaction in the major groove? Cell. 1991 Aug 9;66(3):577–588. doi: 10.1016/0092-8674(81)90020-9. [DOI] [PubMed] [Google Scholar]