Abstract
The neuroprotective property of quercetin is well reported against hypoxia and ischemia in past studies. This property of quercetin lies in its antioxidant property with blood-brain barrier permeability and anti-inflammatory capabilities. µ-Calpain, a calcium ion activated intracellular cysteine protease causes serious cellular insult, leading to cell death in various pathological conditions including hypoxia and ischemic stroke. Hence, it may be considered as a potential drug target for the treatment of hypoxia induced neuronal injury. As the inhibitory property of µ-calpain is yet to be explored in details, hence, in the present study, we investigated the interaction of quercetin with µ-calpain through a molecular dynamics simulation study as a tool through clarifying the molecular mechanism of such inhibition and determining the probable sites and modes of quercetin interaction with the µ-calpain catalytic domain. In addition, we also investigated the structure-activity relationship of quercetin with μ-calpain. Affinity binding of quercetin with µ-calpain had a value of –28.73 kJ/mol and a Ki value of 35.87 µM that may be a probable reason to lead to altered functioning of µ-calpain. Hence, quercetin was found to be an inhibitor of µ-calpain which might have a possible therapeutic role in hypoxic injury.
Keywords: nerve regeneration, quercetin, μ-calpain, molecular docking, molecular dynamics simulation, hypoxia, neuroprotection, neural regeneration
Introduction
Calpain belongs to the group of intracellular cysteine proteases that need calcium ions for their activation (Nakagawa and Yuan, 2000). Even though their physiological role is still incompletely understood, several lines of evidence supports their implication in a variety of calcium-regulated cellular processes such as signal transduction, cell proliferation, cell cycle progression, differentiation, apoptosis, membrane fusion, and platelet activation (Sorimachi and Ishiura, 1997; Goll et al., 2003; Suzuki et al., 2003; Glantz et al., 2007). The disturbance in the regulation of calpain activity also contributes to various pathological conditions such as neuronal degeneration and Alzheimer's disease (Huang and Wang, 2001; Suzuki et al., 2003). Calpain 1 (microcalpain or μ-calpain) and calpain 2 (milli calpain or m-calpain) are among the best characterized calpains that show widest tissue distribution among the members of this family. These calcium-dependent cysteine proteases play a significant role in a huge set of intracellular processes (Carafoli and Molinari, 1998; Wang, 2000; Huang and Wang, 2001) particularly in the selective proteolysis of factors involved in the cell cycle (Raynaud et al., 2004), during apoptosis in association with caspases (Yamashima, 2004) or in the cleavage of membrane-cytoskeleton complexes during cell motility phases (Lebart and Benyamin, 2006). There is evidence that the increased production of free radicals and modifications in detoxifying enzymes also lead to apoptosis in cultured rat forebrain neurons exposed to transient hypoxia and can be prevented by antioxidants (Ishige et al., 2001; Lievre et al., 2001).
In recent years, neutraceutical (polyphenolic phytochemicals i.e., flavonoids) such as quercetin has gained much attention for its use as a potent drug targeting hypoxia and ischemic insult. The molecular structure of quercetin has been shown in Figure 1. Flavonoids have been reported to exhibit antioxidant activity by three mechanisms: (1) increasing intracellular glutathione (GSH) levels, (2) scavenging reactive oxygen species (ROS), (3) blocking Ca2+ influx (Ishige et al., 2001). The anti-oxidant properties of the flavonoids have shown neuroprotective effects in the brain. In spite of its in vitro anti-oxidant property and cytoprotective activity in cell cultures, it has also been reported that quercetin showed beneficial effects in ischemia mediated cerebral injury (Dajas et al., 2001). The tremendous potential of quercetin to inhibit cancer is also well documented (Seufi et al., 2009). Therefore, in the present study, the effect of quercetin on µ-calpain was investigated by molecular docking and molecular dynamics simulation as tools.
Figure 1.
Structure of quercetin a polyphenol used as ligand for docking with μ-calpain.
Computational Methods
Molecular docking
The molecular docking was performed using Autodock 4.0 software (http://autodock.scripps.edu). The crystal structure of µ-calpain (pdb id. 2G8J) was obtained from Research Collaboratory for Structural Bioinformatics (RCSB) protein data bank (Cuerrie et al., 2006). The structures and smile notation of ligands quercetin was generated by ACDLAB free software (www.acdlabs.com) and pdb file was built by online smile translator (https://cactus.nci.nih.gov/translate). All hetero atoms, including water molecules, except calcium were removed from pdb file of calpain.
Addition of hydrogen atoms to the calpain molecule and merging of all non polar hydrogen atoms were performed using Autodock program. Search parameters were based on adaptive local search using Lamarkian genetic algorithm. Short range vanderwaal and electrostatic interactions, hydrogen bonding, entropy losses were considered for energy based Autodock scoring function (Morris et al., 1998; Sudhamalla et al., 2010). The lamarkian GA parameters used in the study were: number of runs, 30; population size, 200; maximum no. of evals, 2,500,000; number of generations, 27,000; rate of gene mutation, 0.02 and crossover rate, 0.8. Blind docking was carried out using grid size 126, 126, 126 along the X, Y and Z axes with 0.0469 nm spacing. The grid center was set to –0.9193, 0.1203 and –1.29 nm, respectively.
Molecular dynamics simulation in water
Molecular dynamics (MD) simulation of the complex was carried out with the GROMACS 4.5.4 package (http://www.gromacs.org/) with GROMOS96 43A1 force field (Van Gunsteren et al., 1996; Lindah et al., 2001). Lowest binding energy obtained from Autodock i.e., docking conformation with most negative denomination was taken as initial conformation to carry out the molecular dynamics simulation study. The Gromacs program was used to generate topology parameters of proteins while the Dundee PRODRG server (http://davapc1.bioch.dundee.ac.uk) was used to build the topology parameters of quercetin. The complex was immersed in an octahedron box of extended simple point charge (SPC) water molecules (Grand and Owen, 1991; Schuler et al., 2001). The solvated system included µ-calpain, quercetin and water was neutralized by adding 8 Na+ ions. Energy minimization was performed using the steepest descent method of 10,000 steps followed by the conjugate gradient method for 10,000 steps to release conflicting contacts. As molecular dynamics simulation studies consist of equilibration and production phases therefore in the first stage of equilibration, the solute (protein, counter ions, and quercetin) was fixed and the position-restrained dynamics simulation of the system was intervened. Lastly, the full system was subjected to MD production run at 26.85°C temperature and 1 bar pressure with isotropic molecule based scaling for 10,000 ps. PYMOL (www.pymol.org) was used to generate all the structural images.
Results
Molecular docking
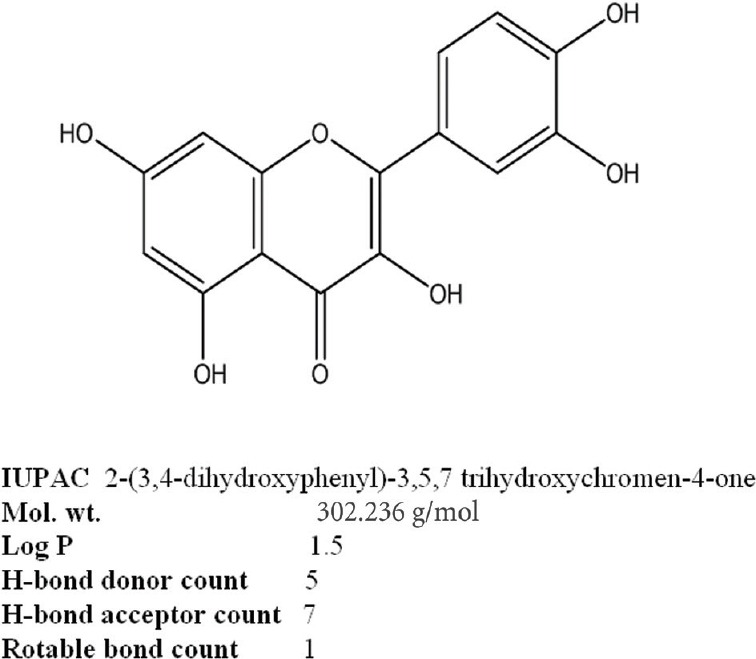
Quercetin binds with the active site having higher negative binding energy of –28.74408 kJ/mol with a Ki value of 35.87 µM. Binding energy was calculated as the summation of four different energies in terms of intermolecular energy, total internal energy, torsional free energy and unbound system energy. The intermolecular energy includes van der Waals, hydrogen bond, desolvation energy, and electrostatic energy. Quercetin was found to be engaged in H-bonding with active site residues Glu 72, Ser 206, Gln 109 and Trp 298 as shown in Figure 2.
Figure 2.
Interaction of quercetin with active site residues of μ-calpain.
(A) Position of quercetin in binding pocket (having residues Glu 72, Ser 206, Gln 109 and Trp 298) of μ-calpain at dII domain for its inhibition. (B) Interaction of quercetin with catalytic triad of calpain (green color shows the interacting residue of calpain with quercetin (rainbow color). (C) Closer view of catalytic site (having residues Glu 72, Ser 206, Gln 109 and Trp 298) occupied by quercetin in μ-calpain. (D) Closer view of quercetin interacting with the side chain of active site residue (yellow dashes indicate hydrogen bonding with active site residues).
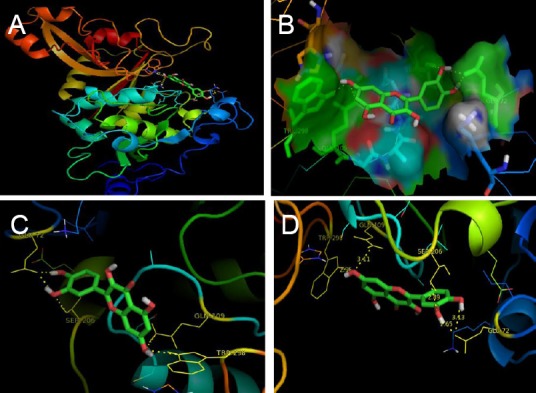
Comparative docking studies resulting from docking with known potent synthetic inhibitor (alpha-mercaptoacrylic acid derivative, i.e., PD151746 and PD150606) as shown in Figure 3 and Table 1, we revealed that the binding energies of PD151746 and PD150606 with µ-calpain were found to be significantly lower than that of quercetin which clearly indicates the strong inhibitory property of natural polyphenol quercetin. Wang et al. (1996) previously reported that alpha-mercaptoacrylic acid derivative such as PD151746 and PD150606 were selective nonpeptide cell-permeable calpain inhibitors and had a neuroprotective role. In comparative docking, PD151746 was engaged in H-bonding with active site residue Arg 326 of µ-calpain with –17.82384 kJ/mol binding energy and a Ki value of 759.23 µM whereas PD150606 was engaged in H-bonding with active site residue Lys 79 of µ-calpain with –16.98704 kJ/mol binding energy and a Ki value of 1.05 mM (Table 1). These findings suggest that the affinity of quercetin with µ-calpain active site as an inhibitor is higher than that of other synthetic inhibitors which enhances the applicability of quercetin as a potent calpain inhibitor against ischemic injury. On the basis of this comparative study and our previous in vitro study in which we explored the possible neuroprotective role of quercetin under different hypoxic conditions (Pandey et al., 2013), we further carried out a molecular dynamics simulation study involving µ-calpain-quercetin complex.
Figure 3.
Interaction of PD150606 with active site residues of μ-calpain.
(A) Position of PD150606 bound to active site residue Lys 79 of μ-calpain. (B) Interaction of PD150606 with catalytic domain of calpain (pink color shows the interacting residue of calpain with PD150606 (rainbow color). (C) Closer view of catalytic site occupied by PD151746 by forming H-bond with residue ARG 326 (green color shows the interacting residue of calpain with PD151746 (rainbow color). (D) Closer view of PD151746 interacting with the side chain of active site residue (yellow dashes indicate hydrogen bonding with active site residue).
Table 1.
Comparative study with known potent synthetic inhibitors of μ-calpain

Molecular dynamics simulation
µ-Calpain-quercetin complex with –26.73576 kJ/mol binding energy was obtained by molecular docking using Autodock and further processed for a molecular dynamics simulation study. A molecular dynamics simulation study represents a suitable method for examining the transformations in gestures of residues/atoms that are exposed to structural and chemical changes. In the present study, we considered the time-dependent activities of molecular dynamics trajectories for µ-calpain and ligand including root mean square deviation (RMSD) for whole backbone atoms and ligand along with the average residue fluctuations of the residues (RMSF). The RMSD of backbone atoms was calculated with respect to their initial conformation and µ-calpain-quercetin complex was changed with time to judge the effect of the binding of the ligand with their conformational stability of µ-calpain-quercetin complex throughout the simulation.
The RMSD trajectories were captured at every 0.5 ps until the simulation time reached 10,000 ps. The RMSD trajectories were found to vary from 0.18 nm to 0.39 nm in the unbound state of protein whereas 0.21 nm to 0.29 nm in the bound state of protein i.e., µ-calpain-quercetin complex. In case of unbound state of µ-calpain a sequential increase profile was observed at ~3,770 ps with slight up and down activities. After down movement for very small interval, sequential increase was observed again at 4,300 ps and reached to 0.38 nm at 10,000 ps. The RMSD analysis of µ-calpain-quercetin complex indicates the stability of complex with minor elevation in activity, i.e., 0.35 nm on 5,400 ps and 0.29 nm at 10,000 ps while RMSD analysis of quercetin showed notable stability throughout the simulation as shown in Figure 4.
Figure 4.
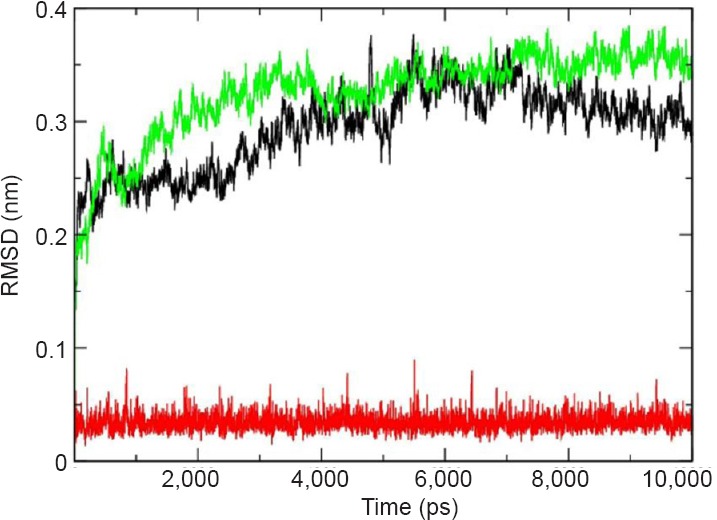
Plots of root mean square deviation (RMSD) of backbone of μ-calpain unbound (green), μ-calpain complex (black) and quercetin (red).
RMSDs were calculated using the initial structures as templates. The trajectories were captured at every 0.5 ps until the simulation time reached 10,000 ps. The RMSD of complex is less than the unbound calpain.
The thermodynamic stability of the complex was determined on the basis of the fluctuation in potential energy. The trajectories of potential energy were found to be higher as compared to unbound protein with persistent values of change up and down for the μ-calpain-quercetin complex during the entire simulation as shown in Figure 5. The number of H-bonds formed during MD simulation between quercetin and μ-calpain was also calculated and variable profile was observed which fluctuated between 0 and 4 with an average value of 1.12 (Figure 6).
Figure 5.
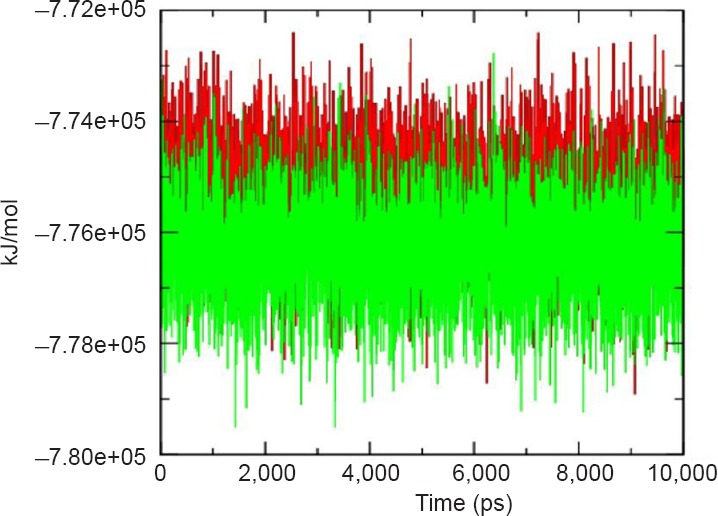
Potential energy profile of μ-calpain (green) and μ-calpain-quercetin complex (red) during 10,000 ps molecular dynamics simulation.
Figure 6.
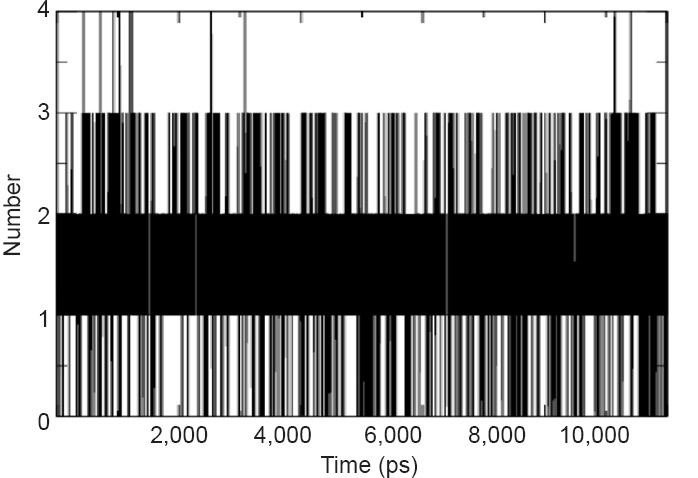
Number of H-bonds formed between quercetin and μ-calpain during 10,000 ps molecular dynamics simulation.
Significant number of H-bonds is 3 for 10,000 ps.
To define the effect of quercetin on the folding of μ-calpain, radius of gyration (Rg) of μ-calpain and quercetin was investigated. Rg values of complexes were observed with approximate similarity with endless ups and downs during simulation run represented in Figure 7. RMSF of backbone residues from its time averaged position was also analyzed to distinguish the flexible zones of the proteins. We observed that μ-calpain-quercetin complex flourished distinctly higher fluctuations in relation to μ-calpain itself as shown in Figure 8.
Figure 7.
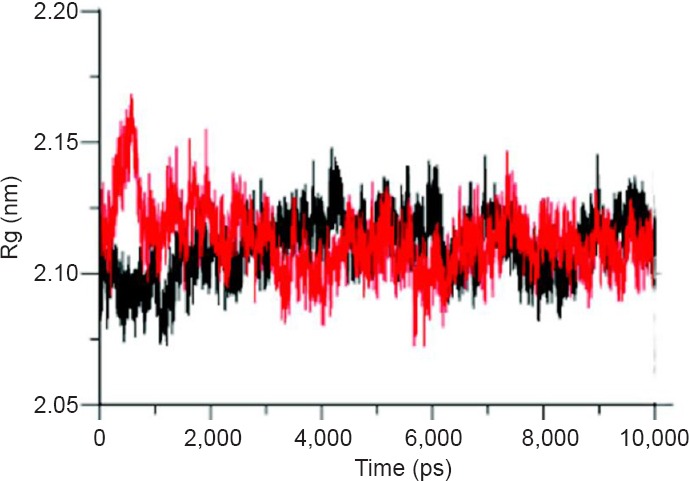
Radius of gyration (Rg) of μ-calpain unbound (black) and μ-calpain-quercetin complex (red).
Figure 8.
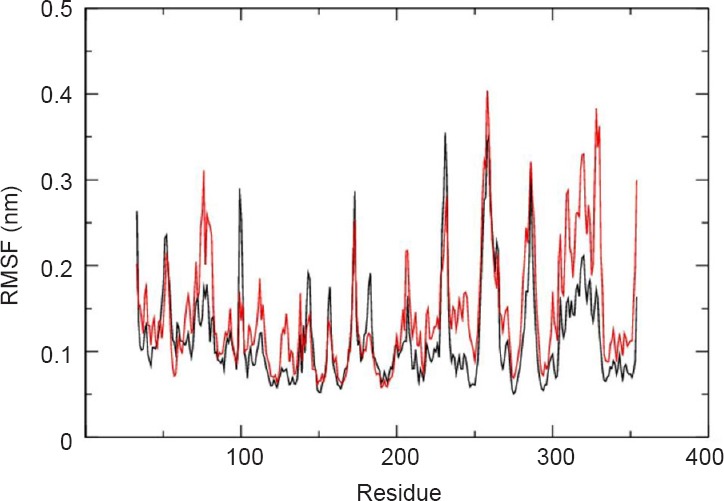
Root mean square fluctuations (RMSF) of μ-calpain backbone (black) and μ-calpain-quercetin backbone (red).
By reducing the dimensionality of the data obtained from a molecular dynamics simulation study, one can identify the configurational space that contains a few degrees of freedom in which an harmonic motion can occur. Principal components analysis (PCA) helps take the trajectory of a molecular dynamics simulation and extracts the dominant modes in the motion of the molecule (Van Aalten et al., 1995; Amadei et al., 1996; Yamaguchi et al., 1998). In this methodology, the most important motions of the protein were extracted from the trajectory by principal component analysis of the Cartesian coordinate covariance matrix, yielding eigenvectors and corresponding eigenvalues. The eigenvectors with the largest associated eigenvalues describes the indispensable subspace where most of the protein dynamics take place. The instant decay in the plot of sorted eigenvalues indicates that a small number of eigenvectors suffice to describe the motions of the μ-calpain i.e., the protein backbone motion can be described by a few essential modes. Principal component analysis was applied to the backbone atoms in quercetin bound and unbound μ-calpain. Eigenvalues of the covariance matrix resulting from the simulations of the μ-calpain-quercetin complex and μ-calpain are shown in Figure 9.
Figure 9.
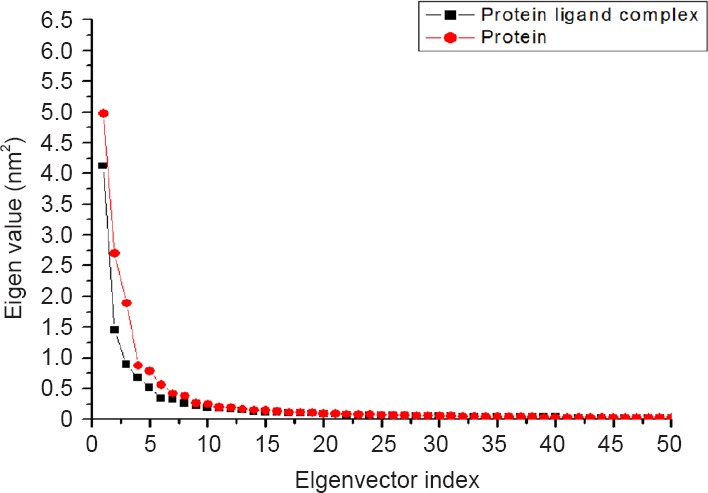
Eigen values of the covariance matrix resulting from the simulations of the μ-calpain-quercetin complex (black) and μ-calpain (red).
The correlation matrix describes the linear correlation between any pairs of backbone atoms as they move around their regular location during dynamics. At a qualitative level, a positive correlation between two atoms reflects a concerted motion along the same direction, whereas a negative correlation indicates an opposite direction motion (Van Aalten et al., 1995). Analysis of correlation matrices of backbone revealed that quercetin binding reduces opposing correlated motion in μ-calpain as compared to unbound form (Figure 10). Cross-correlations give us an idea to identify the most correlated, rationally correlated and negatively correlated regions between fluctuated residues. In the backbone of μ-calpain docked with quercetin (Figure 10), a highly negatively correlated motion was noticed as compared to the backbone of μ-calpain alone (Figure 10) which is due to the folding and conformational changes of hydrophobic groove of domain II. A large number of negatively charged residues of this loop bound to the calcium ion in distant proximity have been reported (Strobl et al., 2000). Our findings suggest that quercetin strongly binds to the hydrophobic dII domains and changes the structure with the induction of conformational changes.
Figure 10.
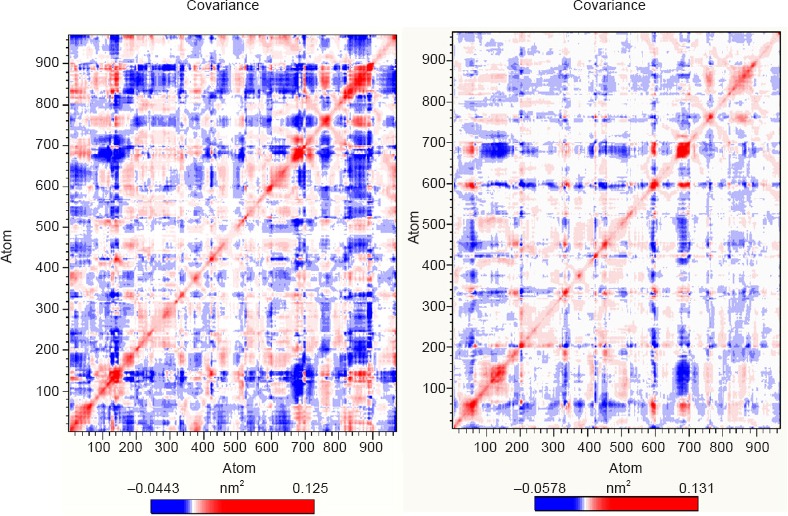
Covariance matrix of μ-calpain and μ-calpain-quercetin complex.
The color scale runs from blue to red (–0.443 to 0.125) for μ-calpain whereas scale runs from blue to red (–0.578 to 0.131) for quercetin docked μ-calpain.
Discussion
The myriad of calcium-dependent enzymes like neuronal nitric oxide synthase (nNOS), calpain, phospholipase, xanthine oxidase, ligases and DNases are activated due to elevation in the level of intraneuronal Ca2+. These enzymes are involved in the synthesis of free radicals and catabolism of proteins, phospholipids and nucleic acids (Mehta et al., 2007). Further, the inhibitory outcome of quercetin on calcium mediated rise in NO levels (Pandey et al., 2012) also supports the neuroprotective effect following hypoxic insult. An elevation in intraneuronal Ca2+ serves as a signal for the initiation of Ca2+ calmodulin and protein Kinase C regulated neuronal nitric oxide synthase (nNOS). Activation of nNOS produces nitric oxide (NO) which is a physiological mediator of vasodilation by elevating the level of 3′,5′-cyclic guanosine monophosphate (cGMP) in vascular smooth muscles (Cornwell et al., 1994).
Glutamate neurotoxicity might occur either due to the over activation of glutamate receptor or without activation of glutamate receptor that results in the Ca2+ influx through the voltage gated Ca2+ channels (Das et al., 2005). These calcium influxes might help to activate calpain, a Ca2+ dependent cysteine protease and degrade the key cytoskeleton protein such as spectrin leading to apoptosis (Jatzke et al., 2002). Cao et al. (2007) reported that calcium dysregulation is nearly a ubiquitous facet of calpain activation and neuronal cell death. Under the hypoxic condition, elevated level of ROS overwhelms the antioxidant capacity of neurons. This in turn causes membrane lipid peroxidation and decreased mitochondrial membrane potential which leads to alterations in Ca2+ homeostasis, DNA damage and apoptosis. Lactic acidosis follows as a result of anaerobic metabolism due to which Ca2+ gets dissociated from calmodulin, further activating calpain and leading to neuronal damage and apoptosis. Concurrently, in the other pathways, changed osmolarity and ATP depletion due to hypoxia lead to necrosis. Earlier studies also suggest that activation of acid sensing ion channels can lead to an activation of calpain during hypoxic insult (Aki et al., 2002).
Calpain activation has a prominent role in neuronal cell death either by the elevation of glutamate level or hypoxia induced oxidative stress mediated calcium influx. In the brain, calpain I (μ-calpain) is mainly neuronal, existing at high levels in the dendrites and cell bodies, whereas relatively higher levels of calpain II (m-calpain) have been observed in axons and glia (Onizuka et al., 1995; Ozben et al., 2005). Although calpain is also found outside cells yet its function is not known (Adachi et al., 1990). Calpain II is generally activated in response to cellular signals, whereas calpain I is often active constitutively. Crystal structure analysis revealed that calpain has one larger and several smaller subunits. The larger subunit of calpain I (catalytic subunit) consists of 4 domains (d1–d4). Domain d2 formed active site which was mainly composed of Cys 115, His 272, Asn 296 and Trp residue at position 298 and these residues form a catalytic triad of calpain cysteine proteases (Khorchid and Ikura, 2002; Goll et al., 2003). We found that quercetin occupied the active site of calpain by the hydrogen binding with active side residue Glu 72, Ser 206, Gln 109, Trp 298 during our docking study. The binding energy and Ki value of quercetin-calpain complex advocate their tendency of stable binding. It has been already well documented from our previous study that quercetin has antioxidant and Ca2+ adaptable capacity against hypoxic injury (Wang et al., 1996; Khorchid and Ikura, 2002; Cuerrier et al., 2006; Khatri and Man, 2013; Pandey et al., 2013; Machado et al., 2015; Sundaramoorthy et al., 2015; Lalkovicová and Danielisová, 2016; Tafani et al., 2016; Zeitouni et al., 2016). Quercetin having antioxidant and anti-inflammatory properties along with ASIC1a reducing capacity proves to be a future molecule of choice for ischemic stroke therapy (Cuerrier et al., 2006; Khatri and Man, 2013; Pandey et al., 2013; Machado et al., 2015; Sundaramoorthy et al., 2015; Lalkovicová and Danielisová, 2016; Tafani et al., 2016; Zeitouni et al., 2016). Present in silico studies suggest that quercetin might be a competitive inhibitor of calpain. The comparative binding energies of quercetin with two other known calpain inhibitors (PD151746 and PD150606) as shown in Table 1 reveal that quercetin might act as a potential calpain inhibitor. The schematic representation of possible mode of neuroprotection of quercetin in hypoxic injury is shown in Figure 11 (Cuerrier et al., 2006; Khatri and Man, 2013; Machado et al., 2015; Sundaramoorthy et al., 2015; Lalkovicová and Danielisová, 2016; Tafani et al., 2016; Zeitouni et al., 2016).
Figure 11.
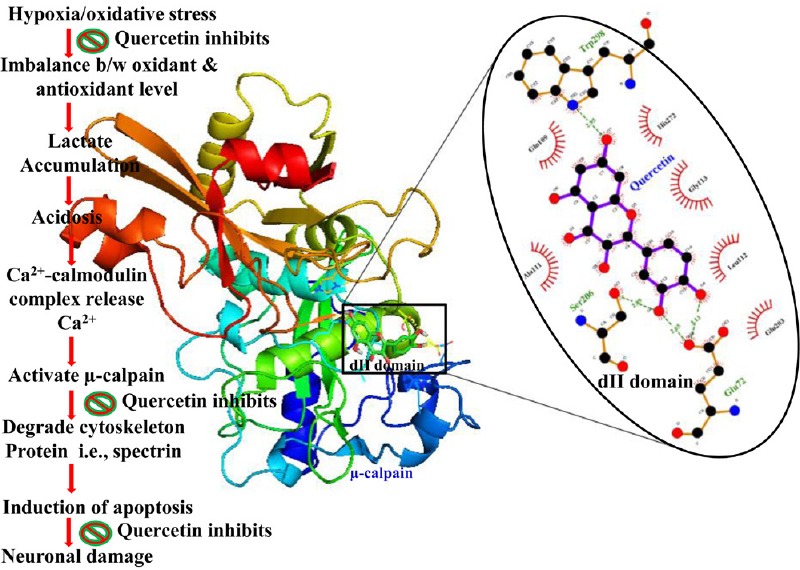
Systematic representation of biochemical changes.
Under hypoxic condition, increased production of reactive oxygen species overwhelms the antioxidant capacity. Lactic acidosis then occurs as a consequence of anaerobic metabolism due to which dissociated Ca2+ from the calmodulin activates calpain and leads to neuronal damage and apoptosis.
Taken together, the mechanism of stable quercetin-calpain complex was proposed in the present study. This stable complex was obtained by molecular dynamic stimulation resulting in 4 H-bonds between μ-calpain and quercetin at residues Glu 72, Ser 206, Gln 109 and Trp 298 in binding pocket with the binding energy of –28.74408 kJ/mol. Such results showed strong binding and inhibition of μ-calpain which is a potent target in our concern for hypoxia, thus providing full proof evidence to support our efforts. On the basis of these observations, it can be hypothesized that quercetin exhibits neuroprotective property by decreasing calpain activity against hypoxia mediated cellular insult.
Footnotes
Conflicts of interest: None declared.
Plagiarism check: This paper was screened twice using Cross-Check to verify originality before publication.
Peer review: This paper was double-blinded and stringently reviewed by international expert reviewers.
Copyedited by Li CH, Song LP, Zhao M
References
- Adachi E, Mukaiyama T, Sasai K, Hayashi T, Kawashima S, Kasai Y, Hayashi M, Hashimoto PH. Immunohistochemical evidence of the extracellular localization of calcium-activated neutral protease (CANP) in rabbit skeletal muscle lung and aorta. Arch Histol Cytol. 1990;53:413–422. doi: 10.1679/aohc.53.413. [DOI] [PubMed] [Google Scholar]
- Aki T, Yoshida K, Fujimiya T. Phosphoinositide 3-Kinase Accelerates Calpain Dependent Proteolysis of Fodrin during Hypoxic Cell Death. J Biochem. 2002;132:921–926. doi: 10.1093/oxfordjournals.jbchem.a003305. [DOI] [PubMed] [Google Scholar]
- Amadei A, Linssen AB, Groot BL, van Aalten DM, Berendsen HJ. An efficient method for sampling the essential subspace of proteins. J Biomol Struct Dyn. 1996;13:615–625. doi: 10.1080/07391102.1996.10508874. [DOI] [PubMed] [Google Scholar]
- Cao G, Xing J, Xiao X, Liou AK, Gao Y, Yin XM, Clark RS, Graham SH, Chen J. Critical role of calpain I in mitochondrial release of apoptosis-inducing factor in ischemic neuronal injury. J Neurosci. 2007;27:9278–9293. doi: 10.1523/JNEUROSCI.2826-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carafoli E, Molinari M. Calpain: a protease in search of a function? Biochem Biophys Res Commun. 1998;247:193–203. doi: 10.1006/bbrc.1998.8378. [DOI] [PubMed] [Google Scholar]
- Cornwell TL, Arnold E, Boerthand NJ, Lincoln TM. Inhibition of smooth muscle cell growth by nitric oxide and activation of cAMP-dependent protein kinase by cGMP. Am J Physiol Cell Physiol. 1994;267:C1405–1413. doi: 10.1152/ajpcell.1994.267.5.C1405. [DOI] [PubMed] [Google Scholar]
- Cuerrier D, Moldoveanu T, Inoue J, Davies PL, Campbell RL. Calpain inhibition by alpha-ketomide and cyclic hemiacetal inhibitors revealed by X-ray crystallograpgy. Biochemistry. 2006;45:7446–7452. doi: 10.1021/bi060425j. [DOI] [PubMed] [Google Scholar]
- Dajas F, Costa G, Abin-Carriguiry JA, Echeverry C, Martinez- Borges A, Dajas-Bailder F. Antioxidants and cholinergic neuroprotective mechanisms in experimentative Parkinsonism. Funct Neurol. 2001;17:37–44. [PubMed] [Google Scholar]
- Das A, Sribnick EA, Wingrave JM, Del Re AM, Woodward JJ, Appel SH, Banik NL, Ray SK. Calpain activation in apoptosis of ventral spinal cord 4.1 (VSC4.1) motoneurons exposed to glutamate: calpain inhibition provides functional neuroprotection. J Neurosci Res. 2005;81:551–562. doi: 10.1002/jnr.20581. [DOI] [PubMed] [Google Scholar]
- Glantz SB, Cianci CD, Iyer R, Pradhan D, Wang KK, Morrow JS. Sequential degradation of αII and βII spectrin by calpain in glutamate or maitotoxin-stimulated cells. Biochemistry. 2007;16:502–513. doi: 10.1021/bi061504y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goll DE, Thompson VF, Li H, Wei W, Cong J. The calpain system. Physiol Rev. 2003;83:731–801. doi: 10.1152/physrev.00029.2002. [DOI] [PubMed] [Google Scholar]
- Grand RJ, Owen D. The biochemistry of ras p21. Biochem J. 1991;279:609–631. doi: 10.1042/bj2790609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y, Wang KK. The calpain family in human disease. Trends Mol Med. 2001;7:355–362. doi: 10.1016/s1471-4914(01)02049-4. [DOI] [PubMed] [Google Scholar]
- Ishige K, Schubertm D, Sagaram Y. Flavonoids protect neuronal cells from oxidative stress by three distinct mechanisms. Free Radic Biol Med. 2001;30:443–446. doi: 10.1016/s0891-5849(00)00498-6. [DOI] [PubMed] [Google Scholar]
- Jatzke C, Watanabe J, Wollmuth LP. Voltage and concentration dependence of Ca2+ permeability in recombinant glutamate receptor subtypes. J Physiol. 2002;538:25–39. doi: 10.1113/jphysiol.2001.012897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatri N, Man H. Synaptic activity and bioenergy homeostasis: implications in brain trauma and neurodegenerative diseases. Front Neurol. 2013;4:1–11. doi: 10.3389/fneur.2013.00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khorchid A, Ikura M. How calpain is activated by calcium. Nat Struct Biol. 2002;9:239–241. doi: 10.1038/nsb0402-239. [DOI] [PubMed] [Google Scholar]
- Lalkovicová M, Danielisová V. Neuroprotection and antioxidants. Neural Regen Res. 2016;11:865–874. doi: 10.4103/1673-5374.184447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lebart MC, Benyamin Y. Calpain involvement in the remodeling of cytoskeletal anchorage complexes. FEBS J. 2006;273:3415–3426. doi: 10.1111/j.1742-4658.2006.05350.x. [DOI] [PubMed] [Google Scholar]
- Lievre V, Bechuwe P, Bianchi A, Bossenmeyer P, Koziel V, Franck VP, Nicolos P, Dauca MB, Daval JC. Intracellular generation of free radicals and modifications of detoxifying enzymes in cultured neurons from the developing rat forebrain in response to transient hypoxia. Neuroscience. 2001;105:287–297. doi: 10.1016/s0306-4522(01)00189-0. [DOI] [PubMed] [Google Scholar]
- Lindah E, Hess B, Van der Spoel D. GROMACS 3.0: a package for molecular simulation and trajectory analysis. J Mol Model. 2001;7:306–317. [Google Scholar]
- Machado VM, Morte MI, Carreira BP, Azevedo MM, Takano J, Iwata N, Saido TC, Asmussen H, Horwitz AR, Carvalho CM, Araújo IM. Involvement of calpains in adult neurogenesis: implications for stroke. Front Cell Neurosci. 2015;9:22. doi: 10.3389/fncel.2015.00022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta SL, Manhas N, Raghubir R. Molecular targets in cerebral ischemia for developing novel therapeutics. Brain Res Rev. 2007;54:34–66. doi: 10.1016/j.brainresrev.2006.11.003. [DOI] [PubMed] [Google Scholar]
- Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK, Olson AJ. Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem. 1998;19:1639–1662. [Google Scholar]
- Nakagawa T, Yuan J. Cross-talk between two cysteine protease families. Activation of caspase-12 by calpain in apoptosis. J Cell Biol. 2000;150:887–894. doi: 10.1083/jcb.150.4.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onizuka K, Kunimatsu M, Ozaki Y, Muramatsu K, Sasaki M, Nishino H. Distribution of mu-calpain proenzyme in the brain and other neural tissues in the rat. Brain Res. 1995;697:179–186. doi: 10.1016/0006-8993(95)00838-h. [DOI] [PubMed] [Google Scholar]
- Ozben T, Balkan E, Balkan S, Serteser M, Gumuslu S. Effects of MK-801 on nitrite and cGMP levels during focal cerebral ischemia in rats. Nitric Oxide. 2005;13:210–215. doi: 10.1016/j.niox.2005.07.005. [DOI] [PubMed] [Google Scholar]
- Pandey AK, Patnaik R, Muresanu DF, Sharma A, Sharma HS. Quercetin in hypoxia-induced oxidative stress: novel target for neuroprotection. Int Rev Neurobiol. 2012;102:107–146. doi: 10.1016/B978-0-12-386986-9.00005-3. [DOI] [PubMed] [Google Scholar]
- Pandey AK, Bhattacharya P, Shukla SC, Paul S, Patnaik R. Neuroprotective effects of quercetin in chemical hypoxia: in-silico evaluation of the hypothesis exploring PKC inhibition mediated pharmacotherapy. Med Chem Res. 2013;22:4836–4841. [Google Scholar]
- Raynaud F, Carnac G, Marcilhac A, Benyamin Y. m-Calpain implication in cell cycle during muscle precursor cell activation. Exp Cell Res. 2004;298:48–57. doi: 10.1016/j.yexcr.2004.03.053. [DOI] [PubMed] [Google Scholar]
- Schuler LD, Daura X, Van Gusteren WF. An improved GROMOS96 force field for aliphatic hydrocarbons in the condensed phase. J Comput Chem. 2001;22:1205–1218. [Google Scholar]
- Seufi AE, Ibrahim SS, Elmaghraby TK, Hafez EE. Preventive effect of the flavonoid quercetin, on hepatic cancer in rats via oxidant/antioxidant activity: molecular and histological evidences. J Exp Clin Cancer Res. 2009;28:80. doi: 10.1186/1756-9966-28-80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sorimachi H, Ishiura S. Suzuki K Structure and physiological function of calpains. Biochem J. 1997;328:721–732. doi: 10.1042/bj3280721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sudhamalla B, Gokara M, Ahalawat N, Amooru DG, Subramanyam R. Molecular dynamics simulation and binding studies of β- sitosterol with human serum albumin and its biological relevance. Phys Chem B. 2010;114:9054–9062. doi: 10.1021/jp102730p. [DOI] [PubMed] [Google Scholar]
- Sundaramoorthy P, Sim JJ, Jang YS, Mishra SK, Jeong KY, Mander P, Chul OB, Shim WS, Oh SH, Nam KY, Kim HM. Modulation of intracellular calcium levels by calcium lactate affects colon cancer cell motility through calcium-dependent calpain. PLoS One. 2015;10:e0116984. doi: 10.1371/journal.pone.0116984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki K, Hata S, Kawabata Y, Sorimachi H. Structure activation and biology of calpain. Diabetes. 2003;53:S12–18. doi: 10.2337/diabetes.53.2007.s12. [DOI] [PubMed] [Google Scholar]
- Tafani M, Sansone L, Limana F, Arcangeli T, De Santis E, Polese M, Fini M, Russo MA. The interplay of reactive oxygen species hypoxia, inflammation, and sirtuins in cancer initiation and progression. Oxid Med Cell Longev 2016. 2016 doi: 10.1155/2016/3907147. 3907147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Aalten DM, Findlay JB, Amadei A, Berendsen HJ. Essential dynamics of the cellular retinol-binding protein–evidence for ligand-induced conformational changes. Protein Eng. 1995;8:1129–1135. doi: 10.1093/protein/8.11.1129. [DOI] [PubMed] [Google Scholar]
- Van Gunsteren WF, Billeter SR, Eising AA, Hunenberger PH, Kruger P, Mark AE, Scott WR, Tironi IG. Biomolecular simulation: The GROMOS96 manual and user guide. Verlag der Fachvereine, Zürich. 1996 [Google Scholar]
- Wang KK. Calpain and caspase: can you tell the difference? Trends Neurosci. 2000;23:20–26. doi: 10.1016/s0166-2236(99)01536-2. [DOI] [PubMed] [Google Scholar]
- Wang KK, Nath R, Posner A, Raser KJ, Buroker-kilgore M, Hajimohammadreza I, Probert AW, Marcoux FW, Ye Q, Takano E, Hatanaka M, Maki M, Canerii H, Collinsii JL, Fergusii A, Leeii KS, Lunney EA, Haystt SJ, Yuen P. An alpha-mercaptoacrylic acid derivative is a selective nonpeptide cell-permeable calpain inhibitor and is neuroprotective (calpain/protease inhibitor/calcium-binding protein/protease/excitotoxicity) Neurobiology. 1996;93:6687–6692. doi: 10.1073/pnas.93.13.6687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashima T. Ca2+-dependent proteases in ischemic neuronal death; a conserved “calpain-cathepsin cascade” from nematodes to primates. Cell Calcium. 2004;36:285–293. doi: 10.1016/j.ceca.2004.03.001. [DOI] [PubMed] [Google Scholar]
- Yamaguchi H, van Aalten DM, Pinak M, Furukawa A, Osman R. Essential dynamics of DNA containing a cis. syn cyclobutane thymine dimer lesion. Nucleic Acids Res. 1998;26:1939–1946. doi: 10.1093/nar/26.8.1939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitouni NE, Dersch P, Naim HY, von Köckritz-Blickwede M. Hypoxia decreases invasin-mediated yersinia enterocolitica internalization into Caco-2 cells. PLoS One. 2016;11:e0146103. doi: 10.1371/journal.pone.0146103. [DOI] [PMC free article] [PubMed] [Google Scholar]


