Abstract
Purpose
We assessed the independent predictive values of the serum markers free prostate specific antigen, proenzyme prostate specific antigen, neuroendocrine marker and Dickkopf-1 compared to serum prostate specific antigen and other standard risk factors for early prostate cancer detection.
Materials and Methods
From the prospectively collected SABOR cohort 250 prostate cancer cases, and 250 mean age matched and proportion of African-American race/ethnicity matched controls were selected who had a prior available prostate specific antigen and digital rectal examination. Serum samples were obtained, and free prostate specific antigen, [−2]proenzyme prostate specific antigen, Dickkopf-1 and neuroendocrine marker were measured. AUC, sensitivities and specificities were calculated, and multivariable logistic regression was used to assess the independent predictive value compared to prostate specific antigen, digital rectal examination, family history, prior biopsy history, race/ethnicity and age.
Results
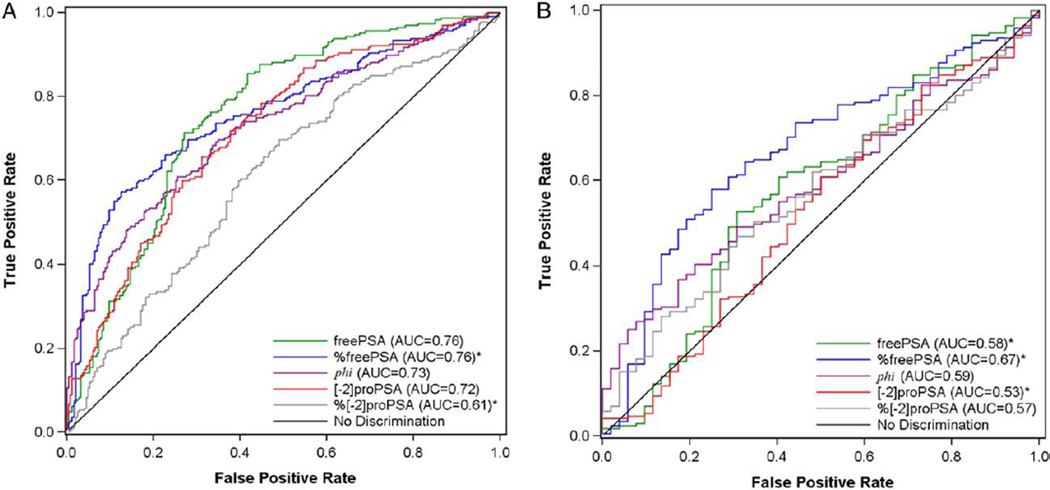
The AUCs (95% CI) were 0.76 (0.71, 0.8) for free prostate specific antigen, 0.72 (0.67, 0.76) for [−2]proenzyme prostate specific antigen, 0.76 (0.72, 0.8) for %free prostate specific antigen, 0.61 (0.56, 0.66) for %[−2]proenzyme prostate specific antigen, 0.73 (0.68, 0.77) for prostate health index, 0.53 (0.48, 0.58) for Dickkopf-1 and 0.53 (0.48, 0.59) for neuroendocrine marker. In the 2 to 10 ng/ml prostate specific antigen range the AUCs (95% CI) were 0.58 (0.49, 0.67) for free prostate specific antigen, 0.53 (0.44, 0.62) for [−2]proenzyme prostate specific antigen, 0.67 (0.59, 0.75) for %free prostate specific antigen, 0.57 (0.49, 0.65) for %[−2]proenzyme prostate specific antigen and 0.59 (0.51, 0.67) for phi. Only %free prostate specific antigen retained independent predictive value compared to the traditional risk factors.
Conclusions
Free prostate specific antigen retained independent diagnostic usefulness for prostate cancers detected through prostate specific antigen and digital rectal examination screening. Prostate specific antigen isoforms are highly correlated with prostate specific antigen. Future research is needed to identify new markers associated with prostate cancer through different mechanisms.
Keywords: prostatic neoplasms, prostate-specific antigen, DKK1 protein, human, biological markers
In the United States 1 in 6 men will be diagnosed with prostate cancer during their lifetime and 1 in 35 will die of the disease. Despite its high prevalence, the uncertain course of PCa combined with a high risk of over detection and the lack of sensitive screening tests has resulted in confusion over the application of PCa screening. Recently the American Cancer Society revised its guidelines, no longer recommending mass screening, but rather recommending individualized cancer risk discussions with patients, resulting in informed decisions regarding PCa testing.1 The Society reiterated the importance of finding new biomarkers for PCa, particularly for men at risk for high grade cancer. In this study we evaluated the operating characteristics of a panel of recently identified serum markers, including freePSA, proPSA, NEM and DKK1 for screen detected PCa, as identified in a cohort of men undergoing routine PSA and DRE screening. The correlation of these markers to PSA, and their independent diagnostic value to PSA and DRE were evaluated.
Materials and Methods
Subjects
SABOR is a National Cancer Institute, Early Detection Research Network sponsored Clinical Validation Center comprised of more than 3,700 San Antonio area male residents without a prior PCa diagnosis. Participation in the study involves annual screening with serum PSA and DRE, as well as referral to biopsy for identification of PCa for high risk participants with PSA exceeding 2.5 ng/ml, abnormal DRE or a family history of PCa. The 12-core ultrasound guided biopsy was performed on the majority of the participants. From the SABOR cohort a nested case-control population was selected, comprising 250 PCa cases with serum PSA measured at or within 2.5 years before diagnosis, and 250 mean age matched and proportion African-American matched controls with at least 5 years of followup with no PCa detection. Serum samples for controls were taken at the first visit. Through this sampling plan controls were not necessarily biopsy confirmed negative (19.4% of controls and 47.8% of high risk controls were biopsy confirmed) and cases included screen detected PCa.
Specimens and Laboratory Analysis
Informed consent approved by the institutional review board at the University of Texas Health Science Center at San Antonio was obtained for each SABOR participant. Participants had blood drawn in an 8 ml red top Vacutainer® tube before DRE at each visit. Whole blood was allowed to clot for 30 minutes at room temperature before serum was separated by centrifugation for 15 minutes at 10C, distributed into approximately 1 ml aliquots and immediately frozen at −80C. Samples were thawed and distributed into smaller aliquots. A set of serum samples was provided to Beckman Coulter, Inc. (Chaska, Minnesota) to measure or compute several isoforms and transformations of the isoforms, including freePSA, [−2]proPSA, %freePSA (freePSA/PSA×100), %[−2]proPSA ([−2]proPSA/[10×freePSA]) and phi ([−2]proPSA/freePSA×√PSA).
The commercially available freePSA assay and the research use only [−2]proPSA assay use dual monoclonal antibodies in sandwich assay formats with chemiluminescent detection. A set of serum samples was used to measure DKK1 using the enzyme-linked immunosorbent assay (ELISA) method as previously described by Tian et al.2 A set of serum samples was used to measure NEM by a displacement ELISA using monoclonal antibody NEM3.3 A total of 20 blinded duplicate samples (10 cases, 10 controls) were randomly mixed among the samples for quality control analysis of the individual markers.
Sample Size and Power
A total of 224 cases was necessary to achieve 80% power to detect a difference between a sensitivity of 40% for PSA reported from prior studies without verification bias4 and an estimated improved sensitivity of 58% for a new marker, using the midpoint method based on McNemar’s test with a Bonferroni correction for a type I error of 0.05.5 The number of subjects identified for study was inflated to 250 cases and 250 controls. Of the 500 subjects 26 (23 cases, 3 controls) were excluded from analysis due to various laboratory test flags and missing data. The remaining 474 subjects (227 cases and 247 controls) provided an adequate number of cancer cases (more than 224) to ensure sufficient power to detect an increase of 18% or more in the sensitivity of a new marker compared to PSA at a cut point which achieves 80% specificity.
Analysis
Spearman’s correlation coefficient (corr) was calculated to examine the reproducibility of the biomarkers in 20 blinded duplicate samples. Descriptive statistics were used to summarize patient and biomarker characteristics, as well as all clinical risk factors for PCa.
AUC was calculated as the Wilcoxon statistic, and tests of differences among AUCs were performed via the nonparametric U-statistic method with or without Bonferroni correction for multiple comparisons.6 For those markers whose average values were higher in cancer cases, sensitivity was calculated as the proportion of cancer cases with a biomarker value equal to or greater than the cut point, and specificity as the proportion of controls with a biomarker value less than the cut point. For those markers whose average values were lower in cancer cases, ie under-expressed in PCa, sensitivity was calculated as the proportion of cancer cases with a biomarker value equal to or less than the cut point and specificity as the proportion of controls with a biomarker value greater than the cut point. Fisher exact 95% CIs for sensitivity and specificity were calculated, and sensitivities and specificities were compared across different markers using McNemar’s test with or without Bonferroni correction.
Backward selected multiple logistic regression was used to identify risk factors that significantly contributed to predicting PCa while controlling for all other markers and standard PCa risk factors. Spearman’s correlation coefficients were calculated among the PSA isoforms for cases and controls separately, and successively used to remove isoforms highly correlated with each other to reduce multicollinearity in the model building process. The Kruskal-Wallis rank test or Mann-Whitney U test was used to examine the relationship between new biomarkers and Gleason score when appropriate. In addition, secondary analyses were performed for all cases and controls with PSA in the subrange of 2 to 10 ng/ml. All statistical tests were performed at the significance level of 0.05 (2-sided) and all statistical analyses were conducted using SAS® (version 9.2).
RESULTS
FreePSA and [−2]proPSA were reproducibly measured on 20 duplicated samples while reliabilities for DKK1 and NEM were lower but acceptable (freePSA, corr >0.99; [−2]proPSA, corr 0.96; DKK1, corr 0.63 and NEM, corr 0.61). Participant characteristics are summarized in table 1. There were significant differences between the cases and the controls with respect to PSA, race/ethnicity, DRE and family history of PCa, but no differences between the 2 groups in prior negative biopsy history or age. The proportion of African-American race/ethnicity in cases and controls (approximately 14%) was the same. Differences between cases and controls for PSA, DRE and family history reflected that cancers were screen detected (by referral to biopsy for high risk participants). PSA ranged from 0.5 to 49.8 ng/ml for subjects with an abnormal DRE and from 0.3 to 54.8 ng/ml for subjects with a family history of PCa.
Table 1.
Patient characteristics
| Cases | Controls | All Subjects | p Value | |||||
|---|---|---|---|---|---|---|---|---|
| Mean pt age (SD) | 64.1 | (8.4) | 64.2 | (8.6) | 64.1 | (8.5) | 0.64* | |
| No. pt age (%): | 0.90† | |||||||
| 40–49 | 12 | (5.3) | 13 | (5.3) | 25 | (5.3) | ||
| 50–59 | 61 | (26.9) | 65 | (26.3) | 126 | (26.6) | ||
| 60–69 | 102 | (44.9) | 105 | (42.5) | 207 | (43.7) | ||
| 70 | 52 | (22.9) | 64 | (25.9) | 116 | (24.5) | ||
| No. race (%): | 0.02† | |||||||
| White | 129 | (56.8) | 167 | (67.6) | 296 | (62.4) | ||
| Black | 32 | (14.1) | 35 | (14.2) | 67 | (14.1) | ||
| Hispanic | 65 | (28.6) | 43 | (17.4) | 108 | (22.8) | ||
| Other | 1 | (0.4) | 2 | (0.8) | 3 | (0.6) | ||
| No. prior neg biopsy (%): | 0.30† | |||||||
| Never | 177 | (78) | 203 | (82.2) | 380 | (80.2) | ||
| At least 1 | 50 | (22) | 44 | (17.8) | 94 | (19.8) | ||
| No. DRE (%): | <0.001† | |||||||
| Normal | 160 | (70.5) | 240 | (97.2) | 400 | (84.4) | ||
| Abnormal | 67 | (29.5) | 7 | (2.8) | 74 | (15.6) | ||
| No. family history (%): | <0.001† | |||||||
| No | 171 | (75.3) | 219 | (88.7) | 390 | (82.3) | ||
| Yes | 56 | (24.7) | 28 | (11.3) | 84 | (17.7) | ||
| PSA (ng/ml): | <0.001* | |||||||
| Mean (SD) | 5.1 | (8.8) | 1.5 | (1.3) | 3.2 | (6.4) | ||
| Range | 0.3–93.8 | 0.1–8.4 | 0.1–93.8 | |||||
| No. Gleason score (%): | ||||||||
| 5 | 6 | (2.6) | ||||||
| 6 | 142 | (62.6) | ||||||
| 7 | 52 | (22.9) | ||||||
| 8 | 9 | (4) | ||||||
| 9 | 8 | (3.5) | ||||||
| Missing | 10 | (4.4) | ||||||
Mann-Whitney U test.
Fisher’s exact test.
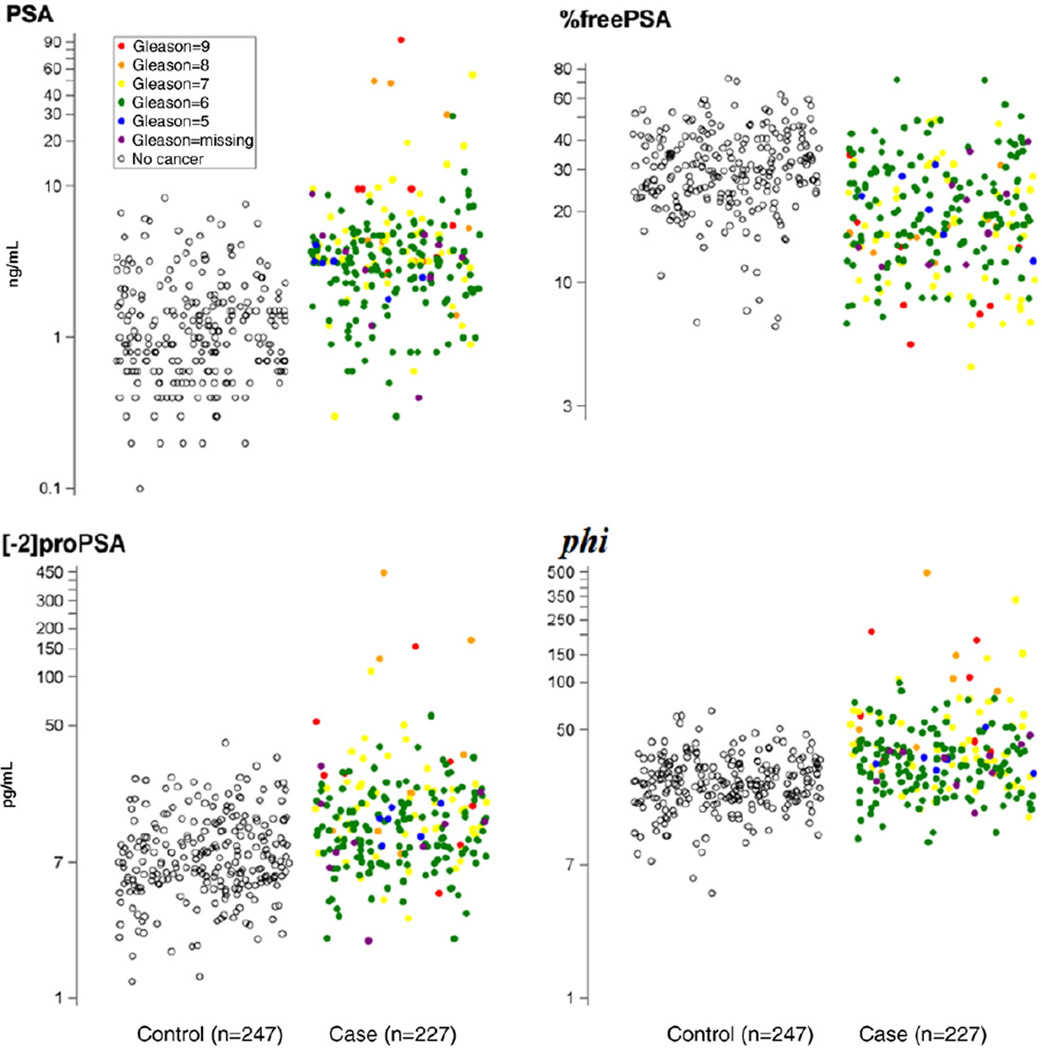
In the 474 subjects with complete measurements of the 7 new biomarkers, freePSA, [−2]proPSA and phi were significantly higher in cases (p <0.001), %freePSA and %[−2]proPSA were significantly lower (p<0.001), and DKK1 and NEM did not differ (table 2, fig. 1). The AUC for PSA, 0.84 (95% CI 0.81–0.88), was higher than typically reported because many cancer cases were referred to biopsy due to high PSA (sampling bias). Of the new markers freePSA and %freePSA obtained the highest AUC (0.76), which was significantly higher than proPSA without Bonferroni adjustment (AUC 0.72, p = 0.01, table 2, fig. 2, A). The new transform phi, which combines PSA, freePSA and proPSA, did not significantly differ from freePSA or proPSA (p = 0.27, p = 0.64). The percentage form of proPSA performed ostensibly worse than the raw form (AUC 0.61, p = 0.004). The AUCs and 95% CIs of DKK1 and NEM indicated that they performed no better than flipping a coin, which would obtain an AUC of 0.50 for predicting cancer on biopsy. The ROC curves and corresponding sensitivities at matched specificities of 70% to 80% revealed superior accuracy of %freePSA to freePSA (table 3). For example at 80% specificity, %freePSA achieves a sensitivity of 62%, which is 16% sensitivity points higher than freePSA with a corresponding sensitivity of 46% (p = 0.002). The ROC curve of %freePSA consistently fell above freePSA for all false-positive rates less than 20% (fig. 2).
Table 2.
Biomarker characteristics
| Controls | Cases | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean (SD) | Median (Q1, Q3) | Mean (SD) | Median (Q1, Q3) | AUC (95% CI) | |||||
| All Participants: | |||||||||
| freePSA (ng/ml) | 0.5 | (0.4) | 0.3 | (0.2, 0.5) | 0.9 | (1.1) | 0.6 | (0.5, 1) | 0.76 (0.71, 0.8) |
| [−2]proPSA (pg/ml) | 8.9 | (5.7) | 7.2 | (5.3, 11) | 18.1 | (34.5) | 11.8 | (8.3, 18.3) | 0.72 (0.67, 0.76) |
| phi | 24.5 | (9.7) | 22.8 | (18.2, 29.7) | 42.9 | (45) | 32.9 | (23.8, 47) | 0.73 (0.68, 0.77) |
| %freePSA* | 31.9 | (11.6) | 30.4 | (24, 39) | 21.6 | (11.6) | 18.5 | (13.5, 27.2) | 0.76 (0.72, 0.8) |
| %[−2]proPSA* | 2.2 | (0.8) | 2.1 | (1.6, 2.6) | 2 | (0.9) | 1.8 | (1.4, 2.3) | 0.61 (0.56, 0.66) |
| DKK1 (ng/ml)* | 1.24 (0.66) | 1.15 (0.76, 1.61) | 1.2 | (0.73) | 1.08 (0.75, 1.43) | 0.53 (0.48, 0.58) | |||
| NEM (ng/ml) | 0.93 (3.15) | 0.13 (0.05, 0.44) | 1.14 (4.23) | 0.17 (0.06, 0.61) | 0.53 (0.48, 0.59) | ||||
| PSA 2–10 ng/ml: | |||||||||
| freePSA (ng/ml)* | 1 | (0.6) | 0.9 | (0.6, 1.3) | 0.8 | (0.5) | 0.7 | (0.5, 1.1) | 0.58 (0.49, 0.67) |
| [−2]proPSA (pg/ml)* | 15 | (7.3) | 14.4 | (9.2, 20.3) | 14.7 | (8.1) | 12.9 | (9, 18.7) | 0.53 (0.44, 0.62) |
| phi | 32.4 | (10) | 32.1 | (24.9, 37.7) | 39.7 | (21) | 35.5 | (27, 47) | 0.59 (0.51, 0.67) |
| %freePSA* | 23.9 | (8.1) | 24.1 | (18.7, 28.2) | 19.4 | (9) | 17.7 | (12.4, 24.5) | 0.67 (0.59, 0.75) |
| %[−2]proPSA | 1.7 | (0.6) | 1.5 | (1.3, 2) | 1.9 | (0.8) | 1.8 | (1.4, 2.2) | 0.57 (0.49, 0.65) |
Marker is under-expressed in cancer cases.
Figure 1.
PSA isoform values for controls and prostate cancer cases
Figure 2.
ROC curves for all markers in all 474 subjects (A) and in 223 subjects with PSA in 2 to 10 ng/ml range (B). Asterisk indicates markers were lower in cancer group vs noncancer group. Therefore, sensitivity was defined as proportion of cancer cases with biomarker value equal to or less than cut point and specificity as proportion of controls with biomarker value exceeding cut point.
Table 3.
Sensitivities of biomarkers at various specificity levels
| 70% Specificity | 80% Specificity | 90% Specificity | 95% Specificity | |||||
|---|---|---|---|---|---|---|---|---|
| Cutoff | Sensitivity (95% CI) | Cutoff | Sensitivity (95% CI) | Cutoff | Sensitivity (95% CI) | Cutoff | Sensitivity (95% CI) | |
| All Subjects: | ||||||||
| freePSA (ng/ml) | 0.49 | 0.72 (0.69, 0.78) | 0.67 | 0.46 (0.43, 0.53) | 0.90 | 0.31 (0.28, 0.38) | 1.31 | 0.14 (0.12, 0.2) |
| [−2]proPSA (pg/ml) | 9.93 | 0.61 (0.58, 0.68) | 12.58 | 0.47 (0.44, 0.54) | 17.22 | 0.29 (0.26, 0.35) | 20.79 | 0.16 (0.14, 0.22) |
| phi | 28.05 | 0.62 (0.59, 0.69) | 31.58 | 0.53 (0.5, 0.6) | 36.45 | 0.42 (0.39, 0.49) | 42.27 | 0.29 (0.26, 0.36) |
| %freePSA* | 25.23 | 0.7 (0.67, 0.76) | 22.75 | 0.62 (0.59, 0.69) | 18.87 | 0.53 (0.5, 0.6) | 15.58 | 0.34 (0.31, 0.41) |
| %[−2]proPSA* | 1.72 | 0.43 (0.4, 0.5) | 1.53 | 0.33 (0.3, 0.4) | 1.31 | 0.19 (0.17, 0.25) | 1.10 | 0.11 (0.09, 0.16) |
| DKK1 (ng/ml)* | 0.84 | 0.31 (0.28, 0.38) | 0.68 | 0.21 (0.19, 0.27) | 0.53 | 0.14 (0.12, 0.2) | 0.40 | 0.05 (0.04, 0.09) |
| NEM (ng/ml) | 0.28 | 0.37 (0.34, 0.44) | 0.64 | 0.25 (0.22, 0.31) | 1.93 | 0.07 (0.06, 0.12) | 4.36 | 0.04 (0.03, 0.08) |
| PSA 2–10 ng/ml: | ||||||||
| freePSA (ng/ml)* | 0.68 | 0.49 (0.42, 0.64) | 0.52 | 0.24 (0.19, 0.39) | 0.39 | 0.07 (0.04, 0.18) | 0.28 | 0.02 (0.01, 0.11) |
| [−2]proPSA (pg/ml)* | 10.00 | 0.33 (0.27, 0.48) | 8.05 | 0.19 (0.14, 0.33) | 5.83 | 0.05 (0.03, 0.15) | 5.46 | 0.04 (0.02, 0.14) |
| phi | 36.88 | 0.46 (0.39, 0.61) | 39.09 | 0.38 (0.32, 0.53) | 43.47 | 0.27 (0.22, 0.43) | 51.06 | 0.22 (0.16, 0.36) |
| %freePSA* | 19.46 | 0.61 (0.54, 0.75) | 17.70 | 0.51 (0.44, 0.66) | 14.08 | 0.29 (0.23, 0.44) | 7.37 | 0.04 (0.02, 0.13) |
| %[−2]proPSA | 1.85 | 0.44 (0.38, 0.6) | 2.12 | 0.3 (0.24, 0.46) | 2.45 | 0.19 (0.14, 0.33) | 2.59 | 0.15 (0.11, 0.29) |
Marker is under-expressed in cancer cases.
In the PSA subrange of 2 to 10 ng/ml %freePSA ranged from 6.56 to 42.07 in controls and 5.50 to 50.59 in cases. %FreePSA was significantly lower in cases while phi was significantly higher (table 2). However, there were no significant differences in freePSA, [−2]proPSA or %[−2]proPSA between cases and controls. The operating characteristics of all markers decreased in this range, and with the decrease in power (171 cases, 52 controls) most 95% CIs of the AUCs included 50% (no diagnostic discrimination) or bordered next to it (fig. 2, B).
The PSA isoforms were highly correlated with each other and with PSA (freePSA vs [−2]proPSA, corr 0.84; freePSA vs PSA, corr 0.81; [−2]proPSA vs phi, corr 0.74 and [−2]proPSA vs PSA, corr 0.7). This implied that all would most likely not contribute independent diagnostic information in terms of composite risk modeling. Several factors contributed significant independent predictive value to PCa diagnosis on biopsy including log %freePSA (OR 0.53, 95% CI 0.29–0.98, p = 0.04), family history (OR 3.56, 95% CI 1.85–6.86, p <0.001), DRE (OR 22.64, 95% CI 8.53–62.12, p <0.001) and log PSA (OR 6.4, 95% CI 4.07–10.06, p <0.001). The predictive values of PSA, DRE and family history were exaggerated due to sampling bias. In a multivariable model excluding these factors several factors became significant including log phi (OR 20.57, 95% CI 9.08–46.63, p <0.001), log %freePSA (OR 0.5, 95% CI 0.27–0.91, p = 0.02) and log %[−2]proPSA (OR 0.05, 95% CI 0.02–0.11, p <0.001).
Of the 217 cases (excluding 10 missing Gleason score) 69 (32%) had high grade cancer (Gleason score 7 or greater). Median PSA, freePSA, [−2]proPSA, phi and %[−2]proPSA were significantly higher in the high grade cancer group vs the low grade cancer group while median %freePSA was significantly lower. In addition, trend tests indicated that %freePSA significantly decreased by Gleason score and the rest of the markers significantly increased by Gleason score, except for DKK1 and NEM (table 4, fig. 1).
Table 4.
Trend analysis by Gleason score
| Gleason 5 | Gleason 6 | Gleason 7 | Gleason 8 | Gleason 9 | p Value (difference)* |
p Value (trend)† |
||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| No. Cases | 6 | 142 | 52 | 9 | 8 | |||||||
| freePSA (ng/ml): | ||||||||||||
| Mean (SD) | 0.7 | (0.2) | 0.7 | (0.5) | 0.9 | (0.7) | 3.1 | (3.8) | 1.7 | (2.2) | 0.131 | <0.001 |
| Median (Q1, Q3) | 0.7 | (0.6, 0.9) | 0.6 | (0.5, 0.9) | 0.8 | (0.5, 1.1) | 0.8 | (0.6, 6.1) | 0.9 | (0.5, 1.9) | ||
| [−2]proPSA (pg/ml): | ||||||||||||
| Mean (SD) | 12.9 | (2.9) | 12.8 | (7.8) | 18.2 | (16.2) | 93.7 | (145.5) | 39.6 | (49.1) | 0.002 | <0.001 |
| Median (Q1, Q3) | 13.2 | (10.1, 15.5) | 11.1 | (7.5, 16.7) | 14.9 | (10.1, 20.3) | 19 | (11, 130.3) | 24.8 | (12.4, 41.4) | ||
| phi: | ||||||||||||
| Mean (SD) | 33.9 | (9.4) | 33.5 | (16.4) | 52.2 | (49) | 113.8 | (149.1) | 89.8 | (71.4) | <0.001 | <0.001 |
| Median (Q1, Q3) | 31.5 | (27.9, 33.6) | 29.5 | (22.2, 40) | 37.5 | (27.9, 61.6) | 50.3 | (39.1, 106.4) | 52.3 | (38.8, 146.8) | ||
| %freePSA: | ||||||||||||
| Mean (SD) | 22 | (7.3) | 23.3 | (12.1) | 18.8 | (11) | 17.5 | (5.6) | 13.7 | (9.5) | 0.006 | <0.001 |
| Median (Q1, Q3) | 21.8 | (16, 28.2) | 20 | (14.8, 29.1) | 16 | (9.8, 24.6) | 16.2 | (15.5, 17.4) | 11.1 | (7.7, 16.1) | ||
| %[−2]proPSA: | ||||||||||||
| Mean (SD) | 1.9 | (0.7) | 1.8 | (0.7) | 2.1 | (0.9) | 2.8 | (1.9) | 2.6 | (1.3) | 0.113 | 0.002 |
| Median (Q1, Q3) | 1.6 | (1.5, 2) | 1.8 | (1.4, 2.1) | 1.9 | (1.5, 2.5) | 2.1 | (2, 2.7) | 2.2 | (1.6, 3.4) | ||
| DKK1 (ng/ml): | ||||||||||||
| Mean (SD) | 1.07 | (0.49) | 1.18 | (0.6) | 1.17 | (0.98) | 1.24 | (0.45) | 1.62 | (1.24) | 0.57 | 0.544 |
| Median (Q1, Q3) | 1.05 (0.95, 1.13) | 1.16 (0.76, 1.44) | 0.96 (0.67, 1.35) | 1.11 | (0.89, 1.58) | 1.09 | (0.83, 2.18) | |||||
| NEM (ng/ml): | ||||||||||||
| Mean (SD) | 0.21 | (0.23) | 0.95 | (4.08) | 1.33 | (3.3) | 5.31 | (10.97) | 0.22 | (0.2) | 0.486 | 0.372 |
| Median (Q1, Q3) | 0.08 (0.07, 0.37) | 0.18 (0.06, 0.64) | 0.15 (0.05, 0.61) | 0.57 | (0.12, 1.15) | 0.13 | (0.07, 0.39) | |||||
Test of difference among Gleason score categories using the Kruskal-Wallis test.
Test of linear trend of log transformed markers across Gleason score categories.
DISCUSSION
Roles and functions of PSA isoforms have been studied for more than 10 years.7–13 PSA exists unbound (free PSA) or bound to other proteins. The higher the percentage of freePSA as a part of total PSA, the less likely it is that cancer is present and the more likely the PSA is increased due to benign prostate hyperplasia.14 The proenzyme forms of PSA that are collectively called proPSA have been shown to be associated with PCa, and appear to have potential as tumor markers. Recent studies involving [−2]proPSA, in particular, have shown promise to improve the role of PSA in detecting PCa.15–18
The clinical usefulness of proPSA to detect PCa in men with PSA less than 10 ng/ml has been assessed in several studies.8,18–21 These studies used ratios involving freePSA and [−2]proPSA, and revealed the greatest improvement in PCa detection among men with serum PSA between 2 and 10 ng/ml. Results of studies that assess potential associations between proPSA and aggressive PCa (as defined by high Gleason scores) have been mixed.9,16,18,22 The predictive value of [−2]proPSA, freePSA, %[−2]proPSA, %freePSA and phi has recently been evaluated by researchers using serum samples from 2 European studies within the PSA range of 2 to 10 ng/ml and age 50 years or older.17 Their analysis showed that for PCa prediction, phi performed significantly better than PSA and %freePSA, and the addition of [−2]proPSA to a logistic regression model consisting of PSA and freePSA significantly increased PCa predictive value and specificity. However, overall no large differences among the variables regarding the specific detection of high grade PCa were observed.
In this case-control study we evaluated the diagnostic performance of a panel of biomarkers throughout the entire PSA range and in the restricted 2 to 10 ng/ml range, and found in both analyses that %freePSA provided independent diagnostic information. An important limitation is that the performance of PSA (AUC 84%) is biased in this study due to the biopsy referral dependence on PSA values (greater than 2.5 ng/ml) in the SABOR cohort from which cancer cases and controls were selected. This may have impacted the relative performance of other markers, especially those which include PSA in the derivation. Reported AUCs of PSA in populations with no verification bias range between 65% and 70%.23–25 In addition, controls in this study were not necessarily biopsy confirmed. This is not necessarily a disadvantage because biopsy confirmed controls would not represent average healthy men undergoing screening, but rather a clinical population. Another limitation is that for some cases, serum collections were not performed immediately before the date of diagnosis, although 91.2% of cases had the blood drawn within 1 year before diagnosis. The AUCs of other markers decreased in the PSA 2 to 10 ng/ml range vs the entire PSA range, although our study was not powered to conduct the secondary subgroup analyses in this PSA range (table 2).
DKK1, a secreted inhibitor of the Wnt/β-catenin signaling pathway, has been described as a novel biomarker for several epithelial carcinomas26 and has been shown to be increased in early PCa cells with a subsequent decrease during progression.27,28 NEM, a novel gene transcript, has been selectively localized in the malignant epithelium of the prostate. Preliminary studies on NEM noted a 3-fold increase in serum levels of NEM in patients with PCa compared to those with benign prostatic hyperplasia, other genitourinary cancers or controls.3 However, in this case-control study serum DKK1 and NEM did not prove to be clinically relevant diagnostic markers for the detection of PCa, and added no independent predictive value to the standard PCa risk factors.
CONCLUSIONS
This study provides several conclusions regarding biomarkers for PCa detection. Care must be taken as new biomarkers are evaluated in the case-control setting. Initial results may not be confirmed or, alternatively, may be confirmed but the biomarkers may not provide independent value to that already established by the standard risk factors and the leading PCa biomarker, PSA, already in place for screening. In addition, many markers, in particular PSA isoforms, are highly correlated with PSA, meaning that when PSA is in the gray zone of 2 to 10 ng/ml they are also more likely to be in their own gray zone and, thus, will also have decreased operating characteristics in terms of AUC. PSA has led to substantial over detection of insignificant PCa and overtreatment,29,30 and by including screen detected PCa as the end point, this study has shown that a panel of new markers will continue to over detect PCa. Finally, this study demonstrated that serum NEM and DKK1 had no potential to discriminate between PCa and no PCa. Newer markers more biologically linked with PCa or, even better, with high grade or significant PCa via different mechanisms will be required to substantially improve PCa risk evaluation.
Acknowledgments
Study received institutional review board approval.
Supported by National Institutes of Health Grants CA086402 and CA054174.
Abbreviations and Acronyms
- DKK1
Dickkopf-1
- DRE
digital rectal examination
- freePSA
free prostate specific antigen
- NEM
neuroendocrine marker
- PCa
prostate cancer
- phi
prostate health index
- proPSA
proenzyme prostate specific antigen
- PSA
prostate specific antigen
- SABOR
San Antonio center for Biomarkers Of Risk for prostate cancer
- [−2]proPSA
proenzyme form of PSA
Footnotes
Nothing to disclose.
Financial interest and/or other relationship with Mission Pharmacal, National Cancer Institute, Southwest Oncology Group and Ferring Pharmaceuticals.
REFERENCES
- 1.Wolf AM, Wender RC, Etzioni RB, et al. American Cancer Society guideline for the early detection of prostate cancer: update 2010. CA Cancer J Clin. 2010;60:70. doi: 10.3322/caac.20066. [DOI] [PubMed] [Google Scholar]
- 2.Tian E, Zhan F, Walker R, et al. The role of the Wnt-signaling antagonist DKK1 in the development of osteolytic lesions in multiple myeloma. N Engl J Med. 2003;349:2483. doi: 10.1056/NEJMoa030847. [DOI] [PubMed] [Google Scholar]
- 3.Shah GV, Srivastava AK, Iczkowski K. Neuroendocrine marker: a novel, reliable early stage marker for prostate cancer; Presented at the 100th annual meeting of the American Association for Cancer Research; April 18–22, 2009; Denver, Colorado. abstract 4826. [Google Scholar]
- 4.Thompson IM, Ankerst DP, Chi C, et al. The operating characteristics of prostate-specific antigen in a population with initial PSA of 3.0 ng/ml or lower. JAMA. 2005;294:66. doi: 10.1001/jama.294.1.66. [DOI] [PubMed] [Google Scholar]
- 5.Lachenbruch PA. On the sample size for studies based upon McNemar’s test. Stat Med. 1992;11:1521. doi: 10.1002/sim.4780111110. [DOI] [PubMed] [Google Scholar]
- 6.DeLong ER, DeLong DM, Clarke-Pearson DL. Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics. 1988;44:837. [PubMed] [Google Scholar]
- 7.Mikolajczyk SD, Marker KM, Millar LS, et al. A truncated precursor form of prostate-specific antigen is a more specific serum marker of prostate cancer. Cancer Res. 2001;61:6958. [PubMed] [Google Scholar]
- 8.Mikolajczyk SD, Marks LS, Partin AW, et al. Free prostate-specific antigen in serum is becoming more complex. Urology. 2002;50:797. doi: 10.1016/s0090-4295(01)01605-3. [DOI] [PubMed] [Google Scholar]
- 9.Mikolajczyk SD, Rittenhouse HG. Pro PSA: a more cancer specific form of prostate specific antigen for the early detection of prostate cancer. Keio J Med. 2003;52:86. doi: 10.2302/kjm.52.86. [DOI] [PubMed] [Google Scholar]
- 10.Mikolajczyk SD, Catalona WJ, Evans CL, et al. Proenzyme forms of prostate-specific antigen in serum improve the detection of prostate cancer. Clin Chem. 2004;50:1017. doi: 10.1373/clinchem.2003.026823. [DOI] [PubMed] [Google Scholar]
- 11.Mikolajczyk SD, Song Y, Wong JR, et al. Are multiple markers the future of prostate cancer diagnostics? Clin Biochem. 2004;37:519. doi: 10.1016/j.clinbiochem.2004.05.016. [DOI] [PubMed] [Google Scholar]
- 12.Balk SP, Ko YJ, Bubley GJ. Biology of prostate- specific antigen. J Clin Oncol. 2003;21:383. doi: 10.1200/JCO.2003.02.083. [DOI] [PubMed] [Google Scholar]
- 13.Haese A, Graefen M, Huland H, et al. Prostate-specific antigen and related isoforms in the diagnosis and management of prostate cancer. Curr Urol Rep. 2004;5:231. doi: 10.1007/s11934-004-0042-6. [DOI] [PubMed] [Google Scholar]
- 14.Finne P, Auvinen A, Aro J, et al. Estimation of prostate cancer risk on the basis of total and free prostate-specific antigen, prostate volume and digital rectal examination. Eur Urol. 2002;41:619. doi: 10.1016/s0302-2838(02)00179-3. [DOI] [PubMed] [Google Scholar]
- 15.Sokoll LJ, Want Y, Feng Z, et al. [−2]Proenzyme prostate specific antigen for prostate cancer detection: a National Cancer Institute Early Detection Research Network validation study. J Urol. 2008;180:539. doi: 10.1016/j.juro.2008.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stephan C, Kahrs AM, Cammann H, et al. A [−2]proPSA-based artificial neural network significantly improves differentiation between prostate cancer and benign prostatic diseases. Prostate. 2009;69:198. doi: 10.1002/pros.20872. [DOI] [PubMed] [Google Scholar]
- 17.Jansen FH, van Schaik RH, Kurstjens J, et al. Prostate-specific antigen (PSA) isoform p2PSA in combination with total PSA and free PSA improves diagnostic accuracy in prostate cancer detection. Eur Urol. 2010;57:921. doi: 10.1016/j.eururo.2010.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Sokoll LJ, Sanda MG, Feng Z, et al. A prospective, multicenter, National Cancer Institute Early Detection Research Network study of [−2]proPSA: improving prostate cancer detection and correlating with cancer aggressiveness. Cancer Epidemiol Biomarkers Prev. 2010;19:1193. doi: 10.1158/1055-9965.EPI-10-0007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naya Y, Fritsche HA, Bhadkamkar VA, et al. Evaluation of precursor prostate-specific antigen isoform ratios in the detection of prostate cancer. Urol Oncol. 2005;23:16. doi: 10.1016/j.urolonc.2004.07.017. [DOI] [PubMed] [Google Scholar]
- 20.Sokoll LJ, Chan DW, Mikolajczyk SD, et al. Proenzyme PSA for the early detection of prostate cancer in the 2.5– 4.0 ng/ml total PSA range: primary analysis. Urology. 2003;1:274. doi: 10.1016/s0090-4295(02)02398-1. [DOI] [PubMed] [Google Scholar]
- 21.Catalona WJ, Bartsch G, Rittenhouse HG, et al. Serum pro prostate specific antigen improves cancer detection compared to free and complexed prostate specific antigen in men with prostate specific antigen 2 to 4 ng/ml. J Urol. 2003;170:2181. doi: 10.1097/01.ju.0000095460.12999.43. [DOI] [PubMed] [Google Scholar]
- 22.Catalona WJ, Bartsch G, Rittenhouse HG, et al. Serum pro-prostate specific antigen preferentially detects aggressive prostate cancers in men with 2 to 4 ng/ml prostate specific antigen. J Urol. 2004;171:2239. doi: 10.1097/01.ju.0000127737.94221.3e. [DOI] [PubMed] [Google Scholar]
- 23.Thompson IM, Ankerst DP, Chi C, et al. Assessing prostate cancer risk: results from the Prostate Cancer Prevention Trial. J Natl Cancer Inst. 2006;98:529. doi: 10.1093/jnci/djj131. [DOI] [PubMed] [Google Scholar]
- 24.Parekh DJ, Ankerst DP, Higgins BA, et al. External validation of the Prostate Cancer Prevention Trial risk calculator in a screened population. Urology. 2006;68:1152. doi: 10.1016/j.urology.2006.10.022. [DOI] [PubMed] [Google Scholar]
- 25.Eyre SJ, Ankerst DP, Wei JT, et al. Validation in a multiple urology practice cohort of the Prostate Cancer Prevention Trial calculator for predicting prostate cancer detection. J Urol. 2009;182:2653. doi: 10.1016/j.juro.2009.08.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sato N, Yamabuki T, Takano A, et al. Wnt inhibitor Dickkopf-1 as a target for passive cancer immunotherapy. Cancer Res. 2010;70:5326. doi: 10.1158/0008-5472.CAN-09-3879. [DOI] [PubMed] [Google Scholar]
- 27.Yamabuki T, Takano A, Hayama S, et al. Dikkopf-1 as a novel serologic and prognostic biomarker for lung and esophageal carcinomas. Cancer Res. 2007;67:2517. doi: 10.1158/0008-5472.CAN-06-3369. [DOI] [PubMed] [Google Scholar]
- 28.Hall CL, Daignault SD, Shah RB, et al. Dickkopf-1 expression increases early in prostate cancer development and decreases during progression from primary tumor to metastasis. Prostate. 2008;68:1396. doi: 10.1002/pros.20805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Thompson IM, Pauler DK, Goodman PJ, et al. Prevalence of prostate cancer among men with a prostate-specific antigen level < or = 4.0 ng per milliliter. N Engl J Med. 2004;350:2239. doi: 10.1056/NEJMoa031918. [DOI] [PubMed] [Google Scholar]
- 30.Schroder FH, van den Bergh RC, Wolters T, et al. Eleven-year outcome of patients with prostate cancers diagnosed during screening after initial negative sextant biopsies. Eur Urol. 2010;57:256. doi: 10.1016/j.eururo.2009.10.031. [DOI] [PubMed] [Google Scholar]