Abstract
Previous genome-wide association studies (GWAS) of prostate cancer risk focused on cases unselected for family history and have reported over 100 significant associations. The International Consortium for Prostate Cancer Genetics (ICPCG) has now performed a GWAS of 2511 (unrelated) familial prostate cancer cases and 1382 unaffected controls from 12 member sites. All samples were genotyped on the Illumina 5M+exome single nucleotide polymorphism (SNP) platform. The GWAS identified a significant evidence for association for SNPs in six regions previously associated with prostate cancer in population-based cohorts, including 3q26.2, 6q25.3, 8q24.21, 10q11.23, 11q13.3, and 17q12. Of note, SNP rs138042437 (p = 1.7e−8) at 8q24.21 achieved a large estimated effect size in this cohort (odds ratio = 13.3). 116 previously sampled affected relatives of 62 risk-allele carriers from the GWAS cohort were genotyped for this SNP, identifying 78 additional affected carriers in 62 pedigrees. A test for an excess number of affected carriers among relatives exhibited strong evidence for co-segregation of the variant with disease (p = 8.5e−11). The majority (92 %) of risk-allele carriers at rs138042437 had a consistent estimated haplotype spanning approximately 100 kb of 8q24.21 that contained the minor alleles of three rare SNPs (dosage minor allele frequencies <1.7 %), rs183373024 (PRNCR1), previously associated SNP rs188140481, and rs138042437 (CASC19). Strong evidence for co-segregation of a SNP on the haplotype further characterizes the haplotype as a prostate cancer pre-disposition locus.
Introduction
More than 100 SNPs have been identified through genome-wide association studies (GWAS) to date as being significantly associated with prostate cancer risk in case-control data sets. These studies mainly included prostate cancer cases that were unselected based on family history (Amundadottir et al. 2006; Duggan et al. 2007; Gudmundsson et al. 2007a, b, 2008, 2009, 2012; Haiman et al. 2007, 2011; Yeager et al. 2007; Eeles et al. 2008, 2013; Salinas et al.2008; Sun et al. 2008; Thomas et al. 2008; Kote-Jarai et al.2008, 2011; Schumacher et al. 2011; Al Olama et al. 2014, 2015). Three additional studies have identified another five SNPs associated with aggressive prostate cancer (Helfand et al. 2015; Berndt et al. 2015; Al Olama et al. 2013).
The ICPCG has now conducted a GWAS of familial prostate cancer in a large international case-control data set. Twelve member sites contributed one case from each studied high-risk prostate cancer pedigree along with matched unrelated population controls. A total of 2511 cases and 1382 geographically and frequency age-matched controls without a history of prostate cancer were genotyped on the Illumina 5M+exome SNP platform. A subset of 1394 of the prostate cancer cases were identified as having aggressive disease by clinically documented cancer features. Separate analyses for all prostate cancer cases and the aggressive prostate cancer cases were conducted.
A SNP in the 8q24.21 region, rs138042437, exhibiting strong association (odds ratio, OR = 13.3; p 1.7e−8) and in high LD [r2 = 1.0 (Gudmundsson et al. 2012] with a previously reported SNP, rs188140481, was an ideal candidate for further investigation. The rare allele of SNP rs130842437 was tested for co-segregation with disease in 116 affected relatives of 62 index cases from the GWAS study that were available from ICCPG member sites; 78 additional case carriers and 38 non-carriers were observed in 48 of 62 typed families. The marker achieved showed a strong association with disease (p = 8.5e−11) in a test for an excess number of affected carrier relatives (43.9 expected by chance), further characterizing the haplotype containing the SNP as a prostate cancer predisposition locus.
Subjects and methods
Sample cohort
The ICPCG consists of multiple international groups focused on the study of high-risk prostate cancer pedigrees and represents the largest available collect of familial prostate cancer cases and pedigrees. Twelve member sites of the ICPCG consortium provided samples for analysis. These sites included Cancer Council Victoria (Australia), Centre de Recherche sur les Pathologies Prostatiques (CeRePP), Fred Hutchinson Cancer Research Center—Seattle (FHCRC), University of Tampere (Finland), Johns Hopkins University (JHU), University of Ulm (Germany), Institute of Cancer Research, UK (ICR), Louisiana State University Health Sciences Center—New Orleans (LSU), The Mayo Clinic (Mayo), University of Michigan (Michigan), Northwestern University (Northwestern), and University of Utah (Utah). Selection of cases and controls was limited to Caucasians (race self-reported). Each site recruited study participants according to their own protocols; however, to define a set of familial cases, sites were asked to supply only one case from previously sampled pedigrees having at least three related cases with an average age at diagnosis ≤75 years. Sites selected the sampled case from each eligible pedigree that was clinically most aggressive or had the earliest age at diagnosis. Controls were selected to be male, unrelated to cases and each other, and with no prior diagnosis of cancer. Controls were not matched 1:1 to cases, but were selected, such that the overall distribution of race and birth-year was similar between selected cases and controls. Only 9 of 12 sites contributed controls. However, the principal components analysis (PCA) of genotype data identified that cases from the three sites without controls were found to be of similar genetic background to cases at other sites and were merged accordingly. The PCA outcomes that were used to merge sites are shown in Supplemental Figure 1. The specific merging of controls by site is indicated in Table 1, which also shows the composition of cases and controls contributed by each site. All sites collected samples under approval from institutional review boards at their respective institutions, and informed consent was obtained for all study participants.
Table 1.
Description of the study population by site
| GROUP NAME | Analysis grouping | Cases (QC) | Aggressive (QC) | Controls (QC) | Total (QC) |
|---|---|---|---|---|---|
| FINLAND | A | 112 (112) | 59 (59) | 94 (93) | 206 (205) |
| AUSTRALIA | B | 166 (163) | 92 (90) | 169 (165) | 335 (328) |
| LSU | C | 17 (17) | 17 (17) | 0 (0) | 17 (17) |
| MAYO | C | 174 (171) | 124 (122) | 135 (135) | 309 (306) |
| JHU | D | 169 (166) | 86 (85) | 0 (0) | 169 (166) |
| NORTHWESTERN | D | 217 (212) | 71 (70) | 179 (177) | 396 (389) |
| ICR | E | 565 (544) | 304 (296) | 339 (333) | 904 (877) |
| FHCRC | F | 127 (123) | 96 (93) | 122 (113) | 249 (236) |
| MICHIGAN | F | 394 (389) | 308 (303) | 0 (0) | 394 (389) |
| UTAH | G | 364 (352) | 63 (61) | 160 (142) | 524 (494) |
| CEREPP | H | 83 (83) | 69 (69) | 72 (72) | 155 (155) |
| GERMANY | I | 180 (179) | 129 (129) | 152 (152) | 332 (331) |
| Total | – | 2568 (2511) | 1418 (1394) | 1422 (1382) | 3990 (3893) |
Sites that did not contribute controls were analyzed with other sites of similar genetic background; groups analyzed together are denoted by capital letters in ‘Analysis grouping’. Numbers in the table indicate the number of cases or controls from each site, and the numbers in parentheses indicate those that passed quality control procedures
Phenotypes
Standardized criteria for determining prostate cancer status were imposed to address the potential for clinical heterogeneity across the 12 sites. All prostate cancer cases were confirmed by death certificate or medical record. Cases were designated as aggressive if they had: (1) regional or distant stage at diagnosis (stage T3, T4, N1, or M1, based on pathology if radical prostatectomy was done; otherwise, based on clinical stage), (2) tumor Gleason score 8–10 at diagnosis, or poorly differentiated or non-differentiated grade if Gleason score was unavailable, (3) diagnostic PSA >20 ng/ml, or (4) died from metastatic prostate cancer before age 65. These criteria were previously used to define aggressive disease by the ICPCG (Schaid et al.2006; Christensen et al. 2007). Cases not meeting these criteria were considered less-aggressive prostate cancer. According to this definition, there were 1394 aggressive cases and 1096 less-aggressive cases and 21 of ‘unknown’ aggressive status (not analyzed) among the set of cases passing quality control. Two phenotypes were analyzed for GWAS, all prostate cancer and aggressive cases versus less-aggressive cases, applying the ICPCG definition.
Genotype data
All participants were genotyped on the Illumina HumanOmni5Exome-4v1-1+exome SNP array at the Center for Inherited Disease Research (CIDR). The exome portion of the data set was analyzed and reported separately. Quality control was performed at the time of genotyping by CIDR in conjunction with the GENEVA Coordinating Center at the University of Washington (Laurie et al. 2010). The quality control procedure removed 43 participants because of previously undocumented relationships closer than third degree. PCA of approximately 150,000 SNPs identified a relatively homogeneous subset of the cases, and 53 outliers were removed. All individuals had call rates greater than 99 %. Quality control removed 57 cases and 39 controls, leaving 2511 cases and 1382 controls for analysis. SNP markers were filtered out according to HWE p value <0.0001, minor allele frequency (MAF < 0.01), >1 Mendelian error detected in a set of duplicated samples, and a missing call rate ≥2 %, which led to the removal of approximately 45 % of markers (of which a large proportion was monomorphic), leaving 2,567,713 SNPs to serve as the imputation basis. The genomic inflation parameter (λ) was 0.98 for the observed genotype data indicating that population stratification and cryptic relatedness between study subjects are well controlled in this data set (Price et al. 2010). The GENEVA Coordinating Center also imputed genotypes for SNPs that were observed in the 1000 Genomes Project that did not occur on the SNP array. Imputation used the IMPUTE2 software (Howie and Machini 2010; Delaneau et al. 2011) and estimated haplotypes using a reference panel of 1092 samples from International HapMap Consortium Phases 2 and 3 haplotypes. The target imputation set included approximately 27.7M SNPs, which were analyzed in GWAS. Approximately, 66 % of the resulting target SNPs had MAF <1 % (dosage) or IMPUTE INFO score ≤0.3, leaving approximately 9.4M SNPs for analysis.
GWAS was performed with the PLINK software v1.9 (Purcell et al. 2007) using the logistic regression analysis at each imputed marker and including study site assignment. The first four PCA eigenvectors estimated during quality control were included as covariates, because a test of association indicated a significant relationship between affection status and eigenvectors 1, 2, and 4. Following the widely accepted standard for interpreting GWAS (Pe’er et al. 2008), significance was established for SNPs with p < 5e−8. Regional association plots were generated with the LocusExplorer software (Dadaev et al. 2016). Details of the publicly available data sets that were used in the preparation of the figure are described elsewhere (Al Olama et al. 2015). Linkage disequilibrium (LD) estimates between markers originated from 1000 Genomes Project phase 3 data (May 2013 release; 1000 Genomes Consortium et al. 2012). Human genome version 19 coordinates are given throughout.
Identifying independent signals in regions of association
Because of the potential for linkage disequilibrium between markers, we analyzed each region containing multiple significant SNPs with forward stepwise regression and elastic net to identify SNPs with an independent contribution to the signal at each region. Markers were selected from each region with GWAS p < 0.01 (to reduce the number of simultaneously analyzed markers); all were within 500 kb of the significant flanking markers in the region. For the forward selection analysis, the ‘step’ function in R (base package) was employed for model development which included study site assignment, and the first four eigenvectors estimated during quality control as covariates. Although stepwise selection is commonly used in GWAS, there are several problems that compromise its value for high-dimensional data: the resulting regression coefficients are biased away from zero, it does not adequately handle correlated markers, and there is an inflated risk of capitalizing on chance features of the data. For these reasons, we also used the elastic-net method, which is designed to select variables of interest by addressing the high dimensionality of the data set and correlated markers. The ‘glmnet’ package in R (Friedman et al. 2010; Tibshirani et al. 2012) was used for the elastic-net analysis; study site and the PCA eigenvectors were retained in all models. We selected SNPs from the elastic-net analysis that had non-zero coefficients after tuning α and λ penalties by tenfold cross validation (×100) with the ‘caret’ R package.
Comparison of familial prostate cancer outcomes with previously associated SNPs
We compared the ORs estimated for SNPs previously reported to be associated with prostate cancer with the ORs estimated for this study. We used the following statistic to test for a significant departure between the ORs of the two studies being compared:
where OR1 is the previously reported OR for an SNP, OR2 is the estimated OR from the current study for that SNP, and SE1 and SE2 are the standard errors of the ORs. The statistic Z follows a normal distribution. A low p value indicates a substantial departure between the two estimated ORs. Positive Z scores indicate a stronger effect size in the previous study, and a negative Z score indicates a stronger effect size in the familial cohort.
Investigation of a high OR SNP
The SNP rs138042437 (referred to as kgp28696802 in the imputed data set), which had a high OR in the GWAS, was followed up to assess the segregation of the rare allele with prostate cancer. The frequency of the SNP was estimated from genotype data for all available cases and controls from the PRACTICAL Consortium (members of the consortium can be found in the Supplemental Table 1) to gauge effect size in cases not ascertained on the basis of their family history of prostate cancer. Sanger sequencing of additional relatives for rs138042437 used Qiagen Multiplex Master Mix following the Qiagen protocol with a 64 degree annealing temperature using forward primer [gatcagagtggtccagaatgg] and reverse primer [ggagaagacagagactgaagaagg], which produced a product size of 492 bases.
A test of over-transmission of the rare allele at rs138042437 to affected relatives assumes a null hypothesis of random segregation and that the number of pedigrees where specified individuals share a rare variant is the sum of independent Bernoulli trials with differing probabilities of success depending on the configuration of observed carriers in each pedigree. The exact distribution of the statistic was computed with the probability of success for each Bernoulli trial (pedigree) defined as the probability of each configuration of carriers in the pedigree as determined by RVsharing program [RVsharing R software package (Bureau et al. 2014)], conditional on the proband carrying the variant. The RVsharing program computes the probability that specified carriers of a rare variant (assumed to be <1 %) in the pedigree share the variant, assuming that the variant has entered the pedigree only once (Bureau et al.2014). The probability mass function (pmf) for the number of sharing cases was calculated for each pedigree. The pmf for the total number of sharing cases summed over all pedigrees was then computed simply by taking the convolution of the pedigree pmfs.
Haplotypes at the 8q24.21 locus were estimated from imputed genotypes at 8q24.21 using the SHAPEIT2 software (Delaneau et al. 2011). Haplotypes were also estimated from genotypes derived from targeted sequencing data of the 8q24.21 locus in an independent set of 56 prostate cancer cases, also ascertained from ICPCG member sites but not included in the GWAS data set. Haplotypes were estimated from the targeted sequencing data to identify the presence of other rare or novel variants that might not be present in the imputed markers. Targeted sequencing data capture used a targeted Agilent SureSelect Custom capture library designed to collect all coding and non-repetitive non-coding bases in the 8q24.21 locus. Sequence data were generated following the protocol for Ilumina HiSeq 101 Cycle Paired End Sequencing (v4) until an average read depth of 100 was achieved for intended capture bases. Reads were aligned with BWA v0.7.12 (Li and Durbin 2010), variants were called with GATK v3.5 (McKenna et al. 2010), and variant annotation was produced by Annovar (2015-06-17) (Wang et al. 2010).
Results
All prostate cancer
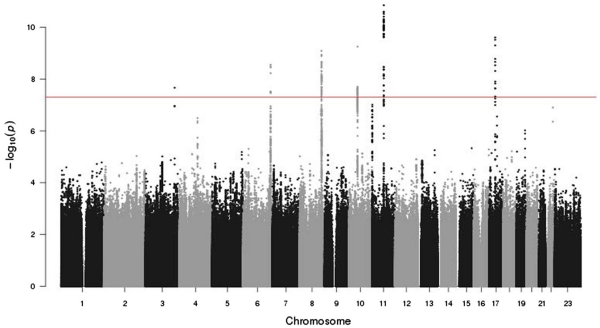
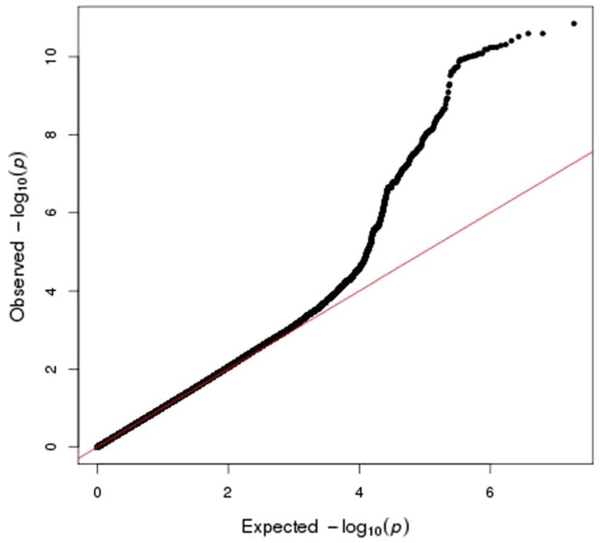
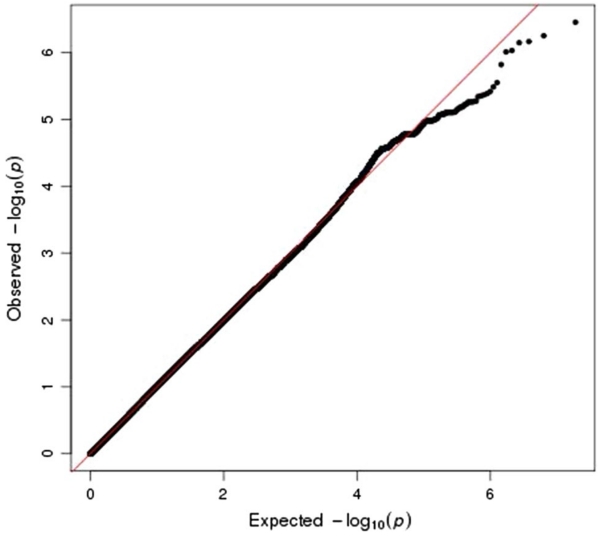
Figure 1 shows the Manhattan plot of the GWAS results. Figure 2 shows a QQ plot for all the GWAS statistics. The QQ plot shows a slight departure of the distribution of test statistics from the expected distribution (λ = 1.08) that is likely an artifact of imputation and not indicative of population stratification between cases and controls (since no inflation was apparent in the observed genotype data, λ = 0.98). The GWAS identified multiple significant SNPs at six regions, including 3q26.31, 6q24.3, 8q24.21, 10q11.23, 11q13.3, and 17q12. Table 2 reports the most significant SNP from each of these regions, and Supplemental Table 2 shows the 135 significant SNPs (p < 5e−8) that occurred in the GWAS.
Fig. 1.
Manhattan plot of genome-wide association of 2511 familial prostate cancer cases and 1382 controls
Fig. 2.
QQ plot of genome-wide association results from 2511 familial prostate cancer cases and 1382 controls using observed genotypes from the Illumina 5M SNP platform
Table 2.
Most significant SNP in six regions from the genome-wide association of 2511 prostate cancer cases and 1382 controls
| Chromosome | Base-pair position (hg19) |
Accession ID | Minor allele |
Minor allele frequency |
Odds ratio | Lower 95 % CI |
Upper 95 % CI |
p value |
|---|---|---|---|---|---|---|---|---|
| 3 | 170,074,517 | kgp1923988 | C | 0.192 | 1.41 | 1.25 | 1.59 | 3.4e–08 |
| 6 | 160,842,537 | rs3123636 | C | 0.356 | 1.37 | 1.23 | 1.51 | 2.8e–09 |
| 8 | 128,077,146 | kgp9645322 | A | 0.044 | 2.39 | 1.81 | 3.16 | 8.2e–10 |
| 10 | 51,549,496 | rs10763567 | A | 0.489 | 1.31 | 1.19 | 1.44 | 5.6e–10 |
| 11 | 69,012,244 | rs12270641 | T | 0.450 | 1.40 | 1.27 | 1.55 | 1.4e–11 |
| 17 | 36,102,381 | rs11651052 | A | 0.435 | 1.37 | 1.24 | 1.51 | 2.5e–10 |
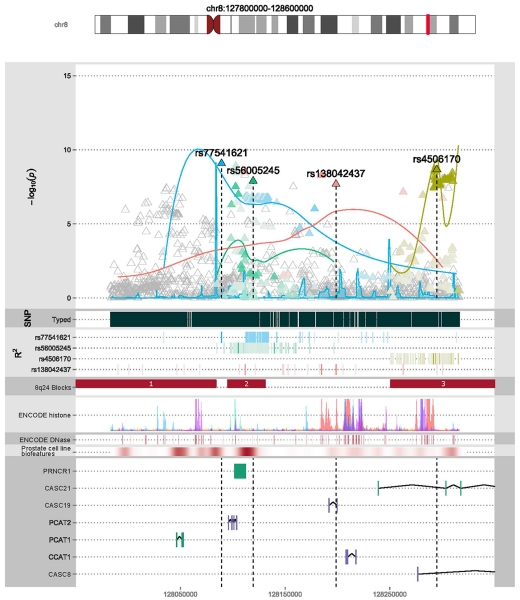
For the regions with multiple significant SNPs, a forward selection and elastic-net analysis in each region identified which SNPs within the region contributed an independent signal. The analyzed regions were 6q25.3, 8q24.21, 10q11.23, 11q13.3, and 17q12. The 3q26.31 region had only one significant SNP and was not investigated further. SNPs contributing independent signals in these regions were all previously published and validated, or proximal and in high LD with associated SNPs. Table 3 shows the markers included by these two selection approaches and the nearest previously reported SNP, if different. The overlap of the forward selection and elastic-net analysis at each region retained only one SNP in the final model for each region, with the exception of the 8q24.21 region, which retained nine SNPs, four of which were also significant in the GWAS. A benefit of elastic net is that it allows highly correlated SNPs to be selected in a model, whereas forward selection might select only one. This makes sense when it is statistically impossible to distinguish among highly correlated SNPs, any of which could be a functional SNP, or any of which could be highly correlated with a functional SNP. Elastic-net achieve this by imposing a penalty on the absolute values of the SNP regression coefficients, which shrinks the unimportant coefficients to zero, while also shrinking the larger coefficients. This avoids the overly optimistic results from forward selection while causing the coefficients from elastic net to be closer to zero than those from forward selection, as illustrated in Table 3. Figure 3 shows the locations of significant markers from the GWAS that were retained by the variance reduction methods applied to genotype data in the region for the 8q24.21 locus as well as the approximate LD structure of the region.
Table 3.
Results for forward selection and elastic-net analyses for SNPs selected by both methods
| Cytogenetic band |
Nearest genomic featureA |
Accession number (bp position, hgl9) |
Accession number for equivalent previously published SNP (bp position, hgl9) |
Forward selection co-efficient (standard error) |
Elastic-net co-efficient |
|---|---|---|---|---|---|
| 6q25.3 | SLC22A3 | *rs3123636 (160.84 Mb) | ars9364554 (160.83 Mb) | 0.28 (0.05) | 0.005 |
| 8q24.21 | Intergenic (10 kb upstream of PCAT2) | *rs77541621 (128.08 Mb) | brs6983561 (128.10 Mb) | 0.68 (0.15) | 0.103 |
| PRNCR1 | *rs56005245 (128.11 Mb) | crsl6901979 (128.12 Mb) | 0.39 (0.07) | 0.036 | |
| Intergenic, 50Kb downstream of PRNCR1, 50-kb upstream of CASC19 |
rs2392725 (128.16 Mb) | Unreported | −0.29 (0.05) | −0.018 | |
| CASC19 | *rsl38042437 (128.208 Mb) | drsl88140481 (128.19 Mb) | 2.21 (0.46) | 0.143 | |
| 400 bases downstream of CASC19 | rs79797202 (128.210 Mb) | Unreported | −0.47 (0.14) | −0.016 | |
| Intergenic, 10-kb downstream of CASC19 | rsl44940621 (128.215 Mb) | Unreported | 0.88 (0.27) | 0.096 | |
| CASC8/CASC21 | *rs4506170 (128.32 Mb) | ers620861 (128.34 Mb) | 0.31 (0.07) | 0.027 | |
| CASC8 | rsl2682374 (128.41 Mb) | frs6983267 (128.41 Mb) | −0.19 (0.05) | −0.004 | |
| Intergenic, 20-kb downstream of CASC8 | rs7841251 (128.52 Mb) | grs1447295 (128.48 Mb) | −0.26 (0.07) | −0.012 | |
| 10q11.23 | TIMM23 | *rsl0763567 (51.54 Mb) | grsl0993994 (51.49 Mb) | −0.27 (0.05) | – |
| rs72795883 (51.57 Mb) | – | 0.26 | |||
| 11ql3.3 | Intergenic (75-kb upstream of MYEOV) | *rsl2270641 (69.01 Mb) | ars7931342 (68.99 Mb) | 0.33 (0.05) | 0.024 |
| 17q12 | HNF1B | *rs11651052 (36.10Mb) | hrs4430796 (36.10 Mb) | 0.31 (0.05) | 0.044 |
SNP was significant in the genome-wide association of all prostate cancer cases versus controls
References:
Refseq annotation names
Fig. 3.
Genomic features within the 8q24.21 region. The top plot shows the location of the plotted region on chromosome 8 with respect to cytogenetic bands. The x-axis of all subsequent plots is base-pair position, denoted along the bottom of the figure. For the main plot, the y-axis is −log10 (p value) from the GWAS. Triangles represent SNPs tested in the GWAS. Significant SNPs with an independent signal in the variance reduction analyses are labeled, have outlined triangles, and are assigned different colored shading; the colored shading is also applied to other SNPs in the region in high linkage disequilibrium (LD), where darker color indicates higher LD. The curved lines are normalized LD curves and indicate the general strength and position of LD between the labeled SNPs and other SNPs in the region. The next plot shows the density of genotyped SNPs in the region, followed by a heat map of r2 estimates between labeled SNPs and other SNPs in the region, where darker shading indicates higher LD. The next plot shows the position of established LD blocks in the 8q24.21 region (Al Olama et al. 2009). The next plot shows ENCODE histone modification binding sites with colors representing different tracks using the same color scheme as the UCSC genome browser (https://genome.ucsc.edu), and the height represents strength of the signal. The next plot shows a heat map of ENCODE DNase binding sites, with darker color representing higher sensitivity. The next plot shows a heat map of expression in a prostate cancer cell line, with darker color representing higher expression. Finally, the genomic features in the region are given (RefSeq ID)
The SNP rs138042437 (p = 1.7e−8, OR = 13.3, 95 % confidence interval [5.4, 32.7], minor allele G, MAF = 0.015) exhibited the highest estimated effect size of any marker in the GWAS and is in high LD (r2 = 1) with previously reported SNP rs188140481 (Gudmundsson et al. 2012) (p = 5e−9, OR 9.4, 95 % confidence interval [4.4, 19.9], minor allele = A, MAF = 0.016), and SNP rs183373024 (p = 2.9e−9, OR = 7.6, 95=% confidence interval [3.9, 14.9], minor allele = G, MAF = 0.017). For rs138042437, familial cases in this study had MAF = 2.2 %, compared to 0.2 % for controls. Population estimates of global minor allele frequency are 0.4 % in 1000 Genomes Project SNP annotation (1000 Genomes Project Consortium et al. 2012) and 0.2 % in dbSNP annotation (Sherry et al. 2001). The MAF of the rare allele for rs138042437 in the prostate cancer case–control data set from the PRACTICAL Consortium was 2.2 % (out of 22,898 alleles) for cases and 1.1 % (out of 23,054 total alleles) for controls [OR = 1.90, 95 % confidence interval (1.71, 2.12)]. The comparatively high frequency of the rare allele at rs138042437 in the much larger set of controls from the PRACTICAL Consortium data set than controls in this study indicates that the estimated allele frequency in the controls is most likely too low, and hence, the magnitude of the estimated effect size for the marker is likely over-stated in this data set.
Known predisposition genes
We also scanned for association signals at SNPs within known predisposition genes for prostate cancer, BRCA1 (Agalliu et al. 2009), BRCA2 (Edwards et al. 2003), and HOXB13 (Ewing et al. 2012). Association at BRCA1/2 genes was modest (most extreme p = 1e−3). For HOXB13, the SNP rs138213197 (p = 2.8e−7; OR = 4.7), representing the G84E mutation, had a similar estimated effect size to a recent meta-analysis of prostate cancer in 120,617 men (OR = 4.5, 95 % confidence interval [3.28, 6.20]; Huang and Cai 2014).
Comparison of effect size for significant SNPs observed in non-familial data sets
Ninety-one of the 103 SNPs previously reported and validated as significantly associated with prostate cancer risk in GWAS of cases and controls not ascertained on the basis of family history (summarized in Al Olama et al. 2014), and were available for analysis. The Z statistic compared the estimated ORs between the current study and the original report of a significant association for each SNP. Supplemental Figure 2 shows all of the outcomes of the comparisons of ORs for 91 of the SNPs previously associated with prostate cancer. For the large majority of SNPs, the effect size (OR) did not differ significantly between the originally reported estimate and the one observed for the familial prostate cancer cases vs. controls comparisons. Only three SNPs had a significantly higher estimated risk among the familial cases compared to controls: rs7584330 at 2q37 in MLPH [OR = 1.26, previously reported OR = 1.06(Al Olama et al. 2014); Z = −2.9], rs9364554 at 6q25 in SLC22A3 [OR = 1.35, previously reported OR = 1.28 (Eeles et al. 2008), Z = −3.5], and rs7931342 at 11q13 near MYEOV [OR 1.38, previously reported OR = −1.27 (Eeles et al. 2008), =Z = −2.7]. However, the outcomes of these comparisons appear to fall within an expected distribution that would be achieved by chance, suggesting that increased ORs may likely represent false positives, and also suggesting that most of the risk SNPs detected in population-based GWAS apply to our study with cased enriched for prostate cancer family history.
Aggressive prostate cancer
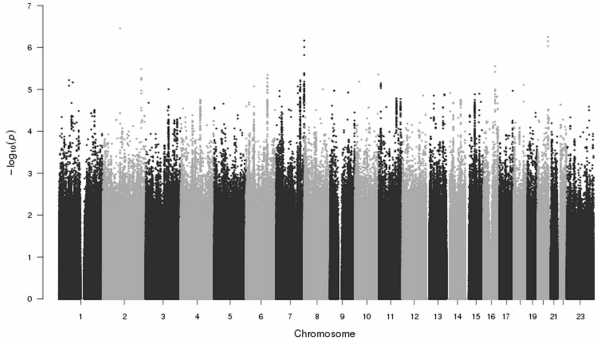
A Manhattan plot showing the outcomes from the GWAS of aggressive cases (n 1394) and less-aggressive cases (n = 1096) appears in Fig. 4, and Fig. 5 shows the corresponding QQ plot (λ = 1.08). No markers were significant (p < 5e−8) in this analysis. Similarly, none of the SNPs previously associated with aggressive disease were significant in this GWAS; Specifically, SNPs rs2735839 (chr19:51.36 Mb; p = 0.110; Helfand et al. 2015), rs11672691 (chr19:41.99 Mb; p = 0.020; Al Olama et al. 2013), rs11704416 (chr22:40.44 Mb; p 0.017; Al Olama et al. 2013), rs35148638 (chr5:86.61 Mb; p = 0.33; Berndt et al. 2015), and rs78943174 (chr3:175.25 Mb; p = 0.34; Berndt et al. 2015).
Fig. 4.
Genome-wide association of 1394 aggressive versus 1096 non-aggressive familial prostate cancer cases
Fig. 5.
QQ plot of genome-wide association results from 1394 aggressive versus 1096 non-aggressive familial prostate cancer cases
Testing for segregation of rs138042437 in relatives
The GWAS identified 112 carriers of the rare allele for rs138042437 (107 cases and 5 controls). Given the evidence of association of SNP rs188140481 (in high LD with rs138042437; r2 = 1.0) with prostate cancer presented in a previous GWAS study (Gudmundsson et al. 2012), the very high estimated effect size of the variants in this data, and the availability of already sampled affected relatives, 116 affected relatives of 62 rare-allele carriers in the GWAS cases were tested for the presence of the rare allele; 78 additional affected carriers and 38 non-carriers were observed. The 116 affected relatives included 79 siblings (56 carriers), 3 half-siblings (3 carriers), 9 avunculars (5 carriers), 20 cousins (13 carriers), and 5 more distant relatives (1 carrier). The exact p value for the observed sharing calculated from the convolution pmf was 8.5e−11. For reference, the mean and standard deviation of the distribution were 43.9 and 5.0, respectively, which, using the normal approximation would give p = 6e−12. The extreme p value indicates strong evidence for segregation of the rare allele in affected relatives of index carriers.
Haplotypes were estimated for 1083 imputed markers falling within 10 kb of three significant markers in high LD at the 8q24.21 locus [rs183373024, rs188140481, and rs138042437 (r2 = 1.0, MAFs < 1.7)], defined as 128.094–128.218 kb. All 112 risk-allele carriers at rs138042437 (107 cases and 5 controls) also carried the risk allele at rs183373024 and rs188140481. An identical haplotype was estimated for 98 cases and 5 controls (92 % of risk-allele carriers) that contained the rare allele of all three SNPs and no other rare variants (MAFs < 2 %). The complete haplotype is given in Supplemental Table 3. Haplotypes were also estimated from genotypes of 56 prostate cancer cases (in an independent study of cases ascertained from ICPCG member sites and not included in the GWAS data set) with targeted sequencing data available at the 8q24 region to test for the presence of other rare variants not captured in the imputed genotype data that may be detectable with targeted sequence data. Seven carriers of the risk alleles at rs183373024, rs188140481, and rs138042437 were identified among the 56 cases who shared an identical estimated haplotype spanning 128,103,679–128,223,746 bp on chromosome 8 that did not contain any other rare or novel variants. Two of 112 carriers observed in the GWAS data set (both were cases), 3 of 116 additionally sampled affected relative carriers, and 1 of 7 sequenced case carriers were homozygous for the rare allele. No determination of the presence of a deletion in the homozygous carriers could be made from available data.
In terms of aggressiveness, the three SNPs on the haplotype rs183373024, rs188140481, and rs138042437 had high effect sizes in the GWAS of all prostate cancer (ORs ≥ 7.6) and low effect sizes in the aggressive GWAS (ORs = 0.9). The investigation of clinical variables that accompanied the GWAS samples did not distinguish affected risk-allele carriers at rs138042437 from non-carrier cases in the ICPCG data set. Affected carriers were not different with respect to age at diagnosis (average age for carriers = 60.45 years, non-carriers = 60.35; t test p = 0.9), ICPCG aggressiveness status (aggressive carriers = 37 %, aggressive non-carriers = 35 %; X2 p = 0.7), or Gleason score (high = 7+, moderate or low <7; high Gleason carriers = 63 %, high Gleason non-carriers = 60 %; X2 p = 0.6).
Discussion
This GWAS of familial prostate cancer cases and matched controls independently detected significantly associated SNPs in six regions, all of which had been previously associated with prostate cancer risk in earlier GWAS and confirmation studies. Of particular interest, however, is that SNPs rs183373024, rs188140481, and rs138042437 at 8q24.21 exhibited very high-risk estimates in this study (ORs ≥ 7.6). SNP rs188140481 was previously associated with prostate cancer in a GWAS conducted on 1795 Icelanders, where SNPs were imputed from the whole genome data (Gudmundsson et al. 2012). Two of the SNPs occur in long non-coding RNAs, CASC19 (rs138042437), and PRNCR1 (rs183373024), which are a predominant feature of the 8q24.21 region (Xiang et al. 2015; Han et al. 2016). The rare alleles of all three SNPs were observed in 107 familial cases in this GWAS population, and 62 cases of which had 116 additionally sampled affected relatives available who were subsequently genotyped; the variant was observed in 78 additional relatives (p = 8.5e−11), indicating strong evidence for co-segregation of the variants with prostate cancer in these families.
A consistent haplotype of three rare SNPs was observed among the majority of risk-allele carriers in the GWAS cohort at the 8q24.21 locus, rs183373024, rs188140481, and rs130842437 in LD block 2 of the 8q24.21 region (Al Olama et al. 2009). A consistent haplotype was also identified for seven risk-allele carriers for the three SNPs in a set of 56 additional familial prostate cancer cases with targeted sequencing data available for the 8q24.21 region. The fact that no additional novel variants were detected among the set of variants on the haplotype estimated from the targeted sequence data indicates either high fidelity of the imputation strategy, or low fidelity of targeted sequencing, in terms of capturing the diversity of the region. It may be possible that further rare variation exists on the risk haplotype that is not observable through either high-density SNP genotyping or targeted sequencing, but may be observable with the application of the whole genome sequencing of carriers.
It is not clear which of the three variants on the risk haplotype conveys an alteration in risk for prostate cancer, either singly or in combination. However, a recent investigation of ENCODE ChipSeq data using the FunciSNP R package identified that rs183373024 occurs at a FoxA1 binding site (Hazelett et al. 2013), which could plausibly indicate regulation of genomic features in the region. Furthermore, SNP rs183373024 is in the long non-coding RNA PRNCR1, upregulation of which has recently been shown to enhance cellular proliferation in colon cancer (Yang et al. 2016). The SNP rs138042437 resides within a POLR2A binding site indicating that this SNP may impact transcription of nearby genomic features (Chen et al. 2015). Despite the fact that biological activity is not currently well understood for most long non-coding RNAs, some progress is being made. For instance, another long non-coding RNA in the 8q24.21 region, CARLo-5 (CCAT1 locus), has recently been shown through functional investigation to affect cell-cycle regulation and tumor development (Kim et al. 2014) indicating that long non-coding RNAs can have substantial impact on currently cryptic biological processes.
No significant differences were observed when considering clinical variables (i.e., age at diagnosis, ICPCG aggressiveness status, and Gleason score) for carriers and non-carriers of the risk alleles at rs138042437, and the three SNPs on the risk haplotype did not show even nominal evidence of association in the case–case aggressiveness GWAS comparisons. Furthermore, no significant differences were observed in a previous investigation that reported results by age at diagnosis and aggressiveness status for carriers and non-carriers of rs188140481 in the Icelandic population (Gudmundsson et al. 2012). Taken together, these observations suggest that the haplotype is not characterized as a marker for more aggressive disease.
Of particular note, the three SNPs on the haplotype exhibited large estimated effect sizes in the GWAS (ORs ≥ 7.6), larger in magnitude than the recently discovered predisposition gene for prostate cancer, HOXB13 (rs138213197; OR = 4.7; p = 2.8e−7), which was characterized as a prostate cancer predisposition gene on the basis of demonstrated segregation of a deleterious variant in prostate cancer pedigrees (Huang 2012).
Comparisons of the estimated ORs between previously reported prostate cancer risk SNPs and the estimates from this familial cases vs. unaffected control data set did not have detectable differences for the majority of SNPs tested, indicating similar risks for the majority of known prostate cancer associated SNPs in this study population. The GWAS of familial aggressive cases did not reveal any novel significant findings and did not statistically confirm SNPs previously associated with aggressive disease, possibly indicating a lack of statistical power for the aggressive phenotype in this data set.
The major strengths of a GWAS, including familial cases, are the ability to detect SNPs with larger effect sizes using a modest sample size and the availability of previously sampled affected relatives who can be subsequently analyzed to test for segregation. Here, the strengths of this design were sufficient to compensate for the lower statistical power of the design compared to larger case–control data sets. One way to interpret the findings of the study is to consider that if functional validation of the three candidate SNPs is achieved, which appears at least probable in the context of other functional investigations of the 8q24.21 region, the study will have at least partially explained prostate cancer susceptibility in 48 families with multiple affected men. Fine-mapping of this region in a large multiethnic cohort will add further insight on these variants/haplotypes and help to determine the most likely causal variants. In summary, the strong evidence for co-segregation of the risk haplotype with prostate cancer further characterizes the haplotype as a predisposition locus for prostate cancer.
Supplementary Material
Acknowledgments
This research was supported by the National Cancer Institute Grant U01 CA 89600 (Support for the ICPCG), and a grant from the Center for Inherited Disease Research supported the genotyping aspect of the study (contract no. HHSN268201200008I; PI Cannon-Albright). The Geneva Coordinating Center at the University of Washington conducted quality control aspects of the study and submitted public facing data to the database of Genotypes and Phenotypes (dbGaP) at the National Center for Biotechnology Information. The authors acknowledge the Keith and Susan Warshaw Fund, the Maurice Warshaw Fund, the C. Scott Watkins Fund, and the Tennity Family Fund. The FHCRC portion of the study was supported by NIH grants R01 CA080122, R01 CA056678, and R01 CA092579, Fred Hutchinson Cancer Research Center and the Prostate Cancer Foundation. Funding for the iCOGS infrastructure (PRACTICAL data) came from the European Community’s Seventh Framework Programme under grant agreement no 223175 (HEALTH-F2-2009-223175) (COGS), Cancer Research UK (C1287/A10118, C1287/A 10710, C12292/A11174, C1281/A12014, C5047/A8384, C5047/A15007, C5047/A10692, C8197/A16565), the National Institutes of Health (CA128978) and Post-Cancer GWAS initiative (1U19 CA148537, 1U19 CA148065, and 1U19 CA148112—the GAME-ON initiative), the Department of Defence (W81XWH-10-1-0341), the Canadian Institutes of Health Research (CIHR) for the CIHR Team in Familial Risks of Breast Cancer, Komen Foundation for the Cure, the Breast Cancer Research Foundation, and the Ovarian Cancer Research Fund. The Biomedical Research Centre at the Institute of Cancer Research (UK) acknowledges the support of the National Institute of Health Research, the Royal Marsden NHS Foundation Trust, and Prostate Cancer UK.
Footnotes
Members from the Prostate Cancer Association Group to Investigate Cancer Associated Alterations in the Genome (PRACTICAL) consortium are provided in the Supplemental Material. Information of the consortium can be found at http://practical.ccge.medschl.cam.ac.uk/.
Members from the International Consortium for Prostate Cancer Genetics can be found at https://www.icpcg.org/?q=centers.
Electronic supplementary material The online version of this article (doi:10.1007/s00439-016-1690-6) contains supplementary material, which is available to authorized users.
References
- 1000 Genomes Project Consortium. Abecasis GR, Auton A, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agalliu I, Gern R, Leanza S, Burk RD. Associations of high-grade prostate cancer with BRCA1 and BRCA2 founder mutations. Clin Cancer Res. 2009;15:1112–1120. doi: 10.1158/1078-0432.CCR-08-1822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Olama A, Dadaev T, Hazelett DJ, Li Q, Leongamornlert D, Saunders EJ, Stephens S, Cieza-Borrella C, Whitmore I, Benlloch Garcia S, Giles GG, Southey MC, Fitzgerald L, Gronberg H, Wiklund F, Aly M, Henderson BE, Schumacher F, Haiman CA, Schleutker J, Wahlfors T, Tammela TL, Nordestgaard BG, Key TJ, Travis RC, Neal DE, Donovan JL, Hamdy FC, Pharoah P, Pashayan N, Khaw KT, Stanford JL, Thibodeau SN, Mcdonnell SK, Schaid DJ, Maier C, Vogel W, Luedeke M, Herkommer K, Kibel AS, Cybulski C, Wokolorczyk D, Kluzniak W, Cannon-Albright L, Brenner H, Butterbach K, Arndt V, Park JY, Sellers T, Lin HY, Slavov C, Kaneva R, Mitev V, Batra J, Clements JA, Spurdle A, Teixeira MR, Paulo P, Maia S, Pandha H, Michael A, Kierzek A, Govindasami K, Guy M, Lophatonanon A, Muir K, Viñuela A, Brown AA, PRACTICAL Consortium. COGS-CRUK GWAS-ELLIPSE (Part of GAME-ON) Initiative. Australian Prostate Cancer BioResource. UK Genetic Prostate Cancer Study Collaborators. UK ProtecT Study Collaborators. Freedman M, Conti DV, Easton D, Coetzee GA, Eeles RA, Kote-Jarai Z. Multiple novel prostate cancer susceptibility signals identified by fine-mapping of known risk loci among Europeans. Hum Mol Genet. 2015;24:5589–5602. doi: 10.1093/hmg/ddv203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Olama AA, Kote-Jarai Z, Giles GG, Guy M, Morrison J, Severi G, Leongamornlert DA, Tymrakiewicz M, Jhavar S, Saunders E, Hopper JL, Southey MC, Muir KR, English DR, Dearnaley DP, Ardern-Jones AT, Hall AL, O’Brien LT, Wilkinson RA, Sawyer E, Lophatananon A, UK Genetic Prostate Cancer Study Collaborators. British Association of Urological Surgeons’ Section of Oncology. UK Prostate testing for cancer and Treatment study (ProtecT Study) Collaborators. Horwich A, Huddart RA, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper C, Donovan JL, Hamdy FC, Neal DE, Eeles RA, Easton DF. Multiple loci on 8q24.21 associated with prostate cancer susceptibility. Nat Genet. 2009;41:1058–1060. doi: 10.1038/ng.452. [DOI] [PubMed] [Google Scholar]
- Al Olama AA, Kote-Jarai Z, Schumacher FR, Wiklund F, Berndt SI, Benlloch S, Giles GG, Severi G, Neal DE, Hamdy FC, Donovan JL, Hunter DJ, Henderson BE, Thun MJ, Gaziano M, Giovannucci EL, Siddiq A, Travis RC, Cox DG, Canzian F, Riboli E, Key TJ, Andriole G, Albanes D, Hayes RB, Schleutker J, Auvinen A, Tammela TL, Weischer M, Stanford JL, Ostrander EA, Cybulski C, Lubinski J, Thibodeau SN, Schaid DJ, Sorensen KD, Batra J, Clements JA, Chambers S, Aitken J, Gardiner RA, Maier C, Vogel W, Dörk T, Brenner H, Habuchi T, Ingles S, John EM, Dickinson JL, Cannon-Albright L, Teixeira MR, Kaneva R, Zhang HW, Lu YJ, Park JY, Cooney KA, Muir KR, Leongamornlert DA, Saunders E, Tymrakiewicz M, Mahmud N, Guy M, Govindasami K, O’Brien LT, Wilkinson RA, Hall AL, Sawyer EJ, Dadaev T, Morrison J, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Dudderidge T, Ogden C, Cooper CS, Lophatonanon A, Southey MC, Hopper JL, English D, Virtamo J, Le Marchand L, Campa D, Kaaks R, Lindstrom S, Diver WR, Gapstur S, Yeager M, Cox A, Stern MC, Corral R, Aly M, Isaacs W, Adolfsson J, Xu J, Zheng SL, Wahlfors T, Taari K, Kujala P, Klarskov P, Nordestgaard BG, Røder MA, Frikke-Schmidt R, Bojesen SE, FitzGerald LM, Kolb S, Kwon EM, Karyadi DM, Orntoft TF, Borre M, Rinckleb A, Luedeke M, Herkommer K, Meyer A, Serth J, Marthick JR, Patterson B, Wokolorczyk D, Spurdle A, Lose F, McDonnell SK, Joshi AD, Shahabi A, Pinto P, Santos J, Ray A, Sellers TA, Lin HY, Stephenson RA, Teerlink C, Muller H, Rothenbacher D, Tsuchiya N, Narita S, Cao GW, Slavov C, Mitev V, UK Genetic Prostate Cancer Study Collaborators. British Association of Urological Surgeons’ Section of Oncology. UK ProtecT Study Collaborators. Australian Prostate Cancer Bioresource. PRACTICAL Consortium. Chanock S, Gronberg H, Haiman CA, Kraft P, Easton DF, Eeles RA. A meta-analysis of genome-wide association studies to identify prostate cancer susceptibility loci associated with aggressive and non-aggressive disease. Hum Mol Genet. 2013;22:408–415. doi: 10.1093/hmg/dds425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al Olama AA, Kote-Jarai Z, Berndt SI, Conti DV, Schumacher F, Han Y, Benlloch S, Hazelett DJ, Wang Z, Saunders E, Leongamornlert D, Lindstrom S, Jugurnauth-Little S, Dadaev T, Tymrakiewicz M, Stram DO, Rand K, Wan P, Stram A, Sheng X, Pooler LC, Park K, Xia L, Tyrer J, Kolonel LN, Le Marchand L, Hoover RN, Machiela MJ, Yeager M, Burdette L, Chung CC, Hutchinson A, Yu K, Goh C, Ahmed M, Govindasami K, Guy M, Tammela TL, Auvinen A, Wahlfors T, Schleutker J, Visakorpi T, Leinonen KA, Xu J, Aly M, Donovan J, Travis RC, Key TJ, Siddiq A, Canzian F, Khaw KT, Takahashi A, Kubo M, Pharoah P, Pashayan N, Weischer M, Nordestgaard BG, Nielsen SF, Klarskov P, Røder MA, Iversen P, Thibodeau SN, McDonnell SK, Schaid DJ, Stanford JL, Kolb S, Holt S, Knudsen B, Coll AH, Gapstur SM, Diver WR, Stevens VL, Maier C, Luedeke M, Herkommer K, Rinckleb AE, Strom SS, Pettaway C, Yeboah ED, Tettey Y, Biritwum RB, Adjei AA, Tay E, Truelove A, Niwa S, Chokkalingam AP, Cannon-Albright L, Cybulski C, Wokolorczyk D, Kluźniak W, Park J, Sellers T, Lin HY, Isaacs WB, Partin AW, Brenner H, Dieffenbach AK, Stegmaier C, Chen C, Giovannucci EL, Ma J, Stampfer M, Penney KL, Mucci L, John EM, Ingles SA, Kittles RA, Murphy AB, Pandha H, Michael A, Kierzek AM, Blot W, Signorello LB, Zheng W, Albanes D, Virtamo J, Weinstein S, Nemesure B, Carpten J, Leske C, Wu SY, Hennis A, Kibel AS, Rybicki BA, Neslund-Dudas C, Hsing AW, Chu L, Goodman PJ, Klein EA, Zheng SL, Batra J, Clements J, Spurdle A, Teixeira MR, Paulo P, Maia S, Slavov C, Kaneva R, Mitev V, Witte JS, Casey G, Gillanders EM, Seminara D, Riboli E, Hamdy FC, Coetzee GA, Li Q, Freedman ML, Hunter DJ, Muir K, Gronberg H, Neal DE, Southey M, Giles GG, Severi G, Breast and Prostate Cancer Cohort Consortium (BPC3) PRACTICAL (Prostate Cancer Association Group to Investigate Cancer-Associated Alterations in the Genome) Consortium. COGS (Collaborative Oncological Gene-environment Study) Consortium. GAME-ON/ELLIPSE Consortium. Cook MB, Nakagawa H, Wiklund F, Kraft P, Chanock SJ, Henderson BE, Easton DF, Eeles RA, Haiman CA. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat Genet. 2014;46:1103–1109. doi: 10.1038/ng.3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amundadottir LT, Sulem P, Gudmundsson J, Helgason A, Baker A, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Cazier JB, Sainz J, Jakobsdottir M, Kostic J, Magnusdottir DN, Ghosh S, Agnarsson K, Birgisdottir B, Le Roux L, Olafsdottir A, Blondal T, Andresdottir M, Gretarsdottir OS, Bergthorsson JT, Gudbjartsson D, Gylfason A, Thorleifsson G, Manolescu A, Kristjansson K, Geirsson G, Isaksson H, Douglas J, Johansson JE, Bälter K, Wiklund F, Montie JE, Yu X, Suarez BK, Ober C, Cooney KA, Gronberg H, Catalona WJ, Einarsson GV, Barkardottir RB, Gulcher JR, Kong A, Thorsteinsdottir U, Stefansson K. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- Berndt SI, Wang Z, Yeager M, Alavanja MC, Albanes D, Amundadottir L, Andriole G, Beane Freeman L, Campa D, Cancel-Tassin G, Canzian F, Cornu JN, Cussenot O, Diver WR, Gapstur SM, Grönberg H, Haiman CA, Henderson B, Hutchinson A, Hunter DJ, Key TJ, Kolb S, Koutros S, Kraft P, Le Marchand L, Lindström S, Machiela MJ, Ostrander EA, Riboli E, Schumacher F, Siddiq A, Stanford JL, Stevens VL, Travis RC, Tsilidis KK, Virtamo J, Weinstein S, Wilkund F, Xu J, Lilly Zheng S, Yu K, Wheeler W, Zhang H, African Ancestry Prostate Cancer GWAS Consortium. Sampson J, Black A, Jacobs K, Hoover RN, Tucker M, Chanock SJ. Two susceptibility loci identified for prostate cancer aggressiveness. Nat Commun. 2015;6:6889. doi: 10.1038/ncomms7889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bureau A, Younkin SG, Parker MM, Bailey-Wilson JE, Marazita ML, Murray JC, Mangold E, Albacha-Hejazi H, Beaty TH, Ruczinski I. Inferring rare disease risk variants based on exact probabilities of sharing by multiple affected relatives. Bioinformatics. 2014;30:2189–2196. doi: 10.1093/bioinformatics/btu198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Yu H, Wang J, Zhang Z, Gao Z, Chen Z, Lu Y, Liu W, Jiang D, Zheng SL, Wei G, Isaacs WB, Feng J, Xu J. Systematic enrichment analysis of potentially functional regions for 103 prostate cancer risk-associated loci. Prostate. 2015;75:1264–1276. doi: 10.1002/pros.23008. [DOI] [PubMed] [Google Scholar]
- Christensen GB, Camp NJ, Farnham JM, Cannon-Albright LA. Genome-wide linkage analysis for aggressive prostate cancer in Utah high-risk pedigrees. Prostate. 2007;67:605–613. doi: 10.1002/pros.20554. [DOI] [PubMed] [Google Scholar]
- Dadaev T, Leongamornlert DA, Saunders EJ, Eeles R, Kote-Jarai Z. Locus Explorer: a user-friendly tool for integrated visualisation of genetic association data and biological annotations. Bioinformatics. 2016;32:949–951. doi: 10.1093/bioinformatics/btv690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delaneau O, Marchini J, Zagury J-F. A linear complexity phasing method for thousands of genomes. Nat Methods. 2011;9:179–181. doi: 10.1038/nmeth.1785. [DOI] [PubMed] [Google Scholar]
- Duggan D, Zheng SL, Knowlton M, Benitez D, Dimitrov L, Wiklund F, Robbins C, Isaacs SD, Cheng Y, Li G, Sun J, Chang BL, Marovich L, Wiley KE, Bälter K, Stattin P, Adami HO, Gielzak M, Yan G, Sauvageot J, Liu W, Kim JW, Bleecker ER, Meyers DA, Trock BJ, Partin AW, Walsh PC, Isaacs WB, Grönberg H, Xu J, Carpten JD. Two genome-wide association studies of aggressive prostate cancer implicate putative prostate tumor suppressor gene DAB2IP. J Natl Cancer Inst. 2007;99:1836–1844. doi: 10.1093/jnci/djm250. [DOI] [PubMed] [Google Scholar]
- Edwards SM, Kote-Jarai Z, Meitz J, Hamoudi R, Hope Q, Osin P, Jackson R, Southgate C, Singh R, Falconer A, Dearnaley DP, Ardern-Jones A, Murkin A, Dowe A, Kelly J, Williams S, Oram R, Stevens M, Teare DM, Ponder BA, Gayther SA, Easton DF, Eeles RA. Two percent of men with early-onset prostate cancer harbor germline mutations in the BRCA2 gene. Am J Hum Genet. 2003;72:1–12. doi: 10.1086/345310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eeles RA, Kote-Jarai Z, Giles GG, Olama AA, Guy M, Jugurnauth SK, Mulholland S, Leongamornlert DA, Edwards SM, Morrison J, Field HI, Southey MC, Severi G, Donovan JL, Hamdy FC, Dearnaley DP, Muir KR, Smith C, Bagnato M, Ardern-Jones AT, Hall AL, O’Brien LT, Gehr-Swain BN, Wilkinson RA, Cox A, Lewis S, Brown PM, Jhavar SG, Tymrakiewicz M, Lophatananon A, Bryant SL, UK Genetic Prostate Cancer Study Collaborators. British Association of Urological Surgeons’ Section of Oncology. UK ProtecT Study Collaborators. Horwich A, Huddart RA, Khoo VS, Parker CC, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Fisher C, Jamieson C, Cooper CS, English DR, Hopper JL, Neal DE, Easton DF. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- Eeles RA, Al Olama AA, Benlloch S, Saunders EJ, Leongamornlert DA, Tymrakiewicz M, Ghoussaini M, Luccarini C, Dennis J, Jugurnauth-Little S, Dadaev T, Neal DE, Hamdy FC, Donovan JL, Muir K, Giles GG, Severi G, Wiklund F, Gronberg H, Haiman CA, Schumacher F, Henderson BE, Le Marchand L, Lindstrom S, Kraft P, Hunter DJ, Gapstur S, Chanock SJ, Berndt SI, Albanes D, Andriole G, Schleutker J, Weischer M, Canzian F, Riboli E, Key TJ, Travis RC, Campa D, Ingles SA, John EM, Hayes RB, Pharoah PD, Pashayan N, Khaw KT, Stanford JL, Ostrander EA, Signorello LB, Thibodeau SN, Schaid D, Maier C, Vogel W, Kibel AS, Cybulski C, Lubinski J, Cannon-Albright L, Brenner H, Park JY, Kaneva R, Batra J, Spurdle AB, Clements JA, Teixeira MR, Dicks E, Lee A, Dunning AM, Baynes C, Conroy D, Maranian MJ, Ahmed S, Govindasami K, Guy M, Wilkinson RA, Sawyer EJ, Morgan A, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As NJ, Woodhouse CJ, Thompson A, Dudderidge T, Ogden C, Cooper CS, Lophatananon A, Cox A, Southey MC, Hopper JL, English DR, Aly M, Adolfsson J, Xu J, Zheng SL, Yeager M, Kaaks R, Diver WR, Gaudet MM, Stern MC, Corral R, Joshi AD, Shahabi A, Wahlfors T, Tammela TL, Auvinen A, Virtamo J, Klarskov P, Nordestgaard BG, Røder MA, Nielsen SF, Bojesen SE, Siddiq A, Fitzgerald LM, Kolb S, Kwon EM, Karyadi DM, Blot WJ, Zheng W, Cai Q, McDonnell SK, Rinckleb AE, Drake B, Colditz G, Wokolorczyk D, Stephenson RA, Teerlink C, Muller H, Rothenbacher D, Sellers TA, Lin HY, Slavov C, Mitev V, Lose F, Srinivasan S, Maia S, Paulo P, Lange E, Cooney KA, Antoniou AC, Vincent D, Bacot F, Tessier DC, COGS-Cancer Research UK GWAS-ELLIPSE (part of GAME-ON) Initiative. Australian Prostate Cancer Bioresource. UK Genetic Prostate Cancer Study Collaborators. British Association of Urological Surgeons’ Section of Oncology. UK ProtecT (Prostate testing for cancer and Treatment) Study Collaborators. PRACTICAL (Prostate Cancer Association Group to Investigate Cancer-Associated Alterations in the Genome) Consortium. Kote-Jarai Z, Easton DF. Identification of 23 new prostate cancer susceptibility loci using the iCOGS custom genotyping array. Nat Genet. 2013;45:385–391. doi: 10.1038/ng.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, Wiley KE, Isaacs SD, Johng D, Wang Y, Bizon C, Yan G, Gielzak M, Partin AW, Shanmugam V, Izatt T, Sinari S, Craig DW, Zheng SL, Walsh PC, Montie JE, Xu J, Carpten JD, Isaacs WB, Cooney KA. Germline mutations in HOXB13 and prostate-cancer risk. N Engl J Med. 2012;366:141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman J, Hastie T, Tibshirani R. Regularization paths for generalized linear mixed models via coordinate descent. J Stat Softw. 2010;33:1–22. [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Manolescu A, Amundadottir LT, Gudbjartsson D, Helgason A, Rafnar T, Bergthorsson JT, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Xu J, Blondal T, Kostic J, Sun J, Ghosh S, Stacey SN, Mouy M, Saemundsdottir J, Backman VM, Kristjansson K, Tres A, Partin AW, Albers-Akkers MT, Godino-Ivan Marcos J, Walsh PC, Swinkels DW, Navarrete S, Isaacs SD, Aben KK, Graif T, Cashy J, Ruiz-Echarri M, Wiley KE, Suarez BK, Witjes JA, Frigge M, Ober C, Jonsson E, Einarsson GV, Mayordomo JI, Kiemeney LA, Isaacs WB, Catalona WJ, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Genome-wide association study identifies a second prostate cancer susceptibility variant at 8q24.21. Nat Genet. 2007a;39:631–637. doi: 10.1038/ng1999. [DOI] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Steinthorsdottir V, Bergthorsson JT, Thorleifsson G, Manolescu A, Rafnar T, Gudbjartsson D, Agnarsson BA, Baker A, Sigurdsson A, Benediktsdottir KR, Jakobsdottir M, Blondal T, Stacey SN, Helgason A, Gunnarsdottir S, Olafsdottir A, Kristinsson KT, Birgisdottir B, Ghosh S, Thorlacius S, Magnusdottir D, Stefansdottir G, Kristjansson K, Bagger Y, Wilensky RL, Reilly MP, Morris AD, Kimber CH, Adeyemo A, Chen Y, Zhou J, So WY, Tong PC, Ng MC, Hansen T, Andersen G, Borch-Johnsen K, Jorgensen T, Tres A, Fuertes F, Ruiz-Echarri M, Asin L, Saez B, van Boven E, Klaver S, Swinkels DW, Aben KK, Graif T, Cashy J, Suarez BK, van Vierssen Trip O, Frigge ML, Ober C, Hofker MH, Wijmenga C, Christiansen C, Rader DJ, Palmer CN, Rotimi C, Chan JC, Pedersen O, Sigurdsson G, Benediktsson R, Jonsson E, Einarsson GV, Mayordomo JI, Catalona WJ, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Two variants on chromosome 17 confer prostate cancer risk, and the one in TCF2 protects against type 2 diabetes. Nat Genet. 2007b;39:977–983. doi: 10.1038/ng2062. [DOI] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Rafnar T, Bergthorsson JT, Manolescu A, Gudbjartsson D, Agnarsson BA, Sigurdsson A, Benediktsdottir KR, Blondal T, Jakobsdottir M, Stacey SN, Kostic J, Kristinsson KT, Birgisdottir B, Ghosh S, Magnusdottir DN, Thorlacius S, Thorleifsson G, Zheng SL, Sun J, Chang BL, Elmore JB, Breyer JP, McReynolds KM, Bradley KM, Yaspan BL, Wiklund F, Stattin P, Lindström S, Adami HO, McDonnell SK, Schaid DJ, Cunningham JM, Wang L, Cerhan JR, St Sauver JL, Isaacs SD, Wiley KE, Partin AW, Walsh PC, Polo S, Ruiz-Echarri M, Navarrete S, Fuertes F, Saez B, Godino J, Weijerman PC, Swinkels DW, Aben KK, Witjes JA, Suarez BK, Helfand BT, Frigge ML, Kristjansson K, Ober C, Jonsson E, Einarsson GV, Xu J, Gronberg H, Smith JR, Thibodeau SN, Isaacs WB, Catalona WJ, Mayordomo JI, Kiemeney LA, Barkardottir RB, Gulcher JR, Thorsteinsdottir U, Kong A, Stefansson K. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Gudbjartsson DF, Blondal T, Gylfason A, Agnarsson BA, Benediktsdottir KR, Magnusdottir DN, Orlygsdottir G, Jakobsdottir M, Stacey SN, Sigurdsson A, Wahlfors T, Tammela T, Breyer JP, McReynolds KM, Bradley KM, Saez B, Godino J, Navarrete S, Fuertes F, Murillo L, Polo E, Aben KK, van Oort IM, Suarez BK, Helfand BT, Kan D, Zanon C, Frigge ML, Kristjansson K, Gulcher JR, Einarsson GV, Jonsson E, Catalona WJ, Mayordomo JI, Kiemeney LA, Smith JR, Schleutker J, Barkardottir RB, Kong A, Thorsteinsdottir U, Rafnar T, Stefansson K. Genome-wide association and replication studies identify four variants associated with prostate cancer susceptibility. Nat Genet. 2009;41:1122–1126. doi: 10.1038/ng.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gudmundsson J, Sulem P, Gudbjartsson DF, Masson G, Agnarsson BA, Benediktsdottir KR, Sigurdsson A, Magnusson OT, Gudjonsson SA, Magnusdottir DN, Johannsdottir H, Helgadottir HT, Stacey SN, Jonasdottir A, Olafsdottir SB, Thorleifsson G, Jonasson JG, Tryggvadottir L, Navarrete S, Fuertes F, Helfand BT, Hu Q, Csiki IE, Mates IN, Jinga V, Aben KK, van Oort IM, Vermeulen SH, Donovan JL, Hamdy FC, Ng CF, Chiu PK, Lau KM, Ng MC, Gulcher JR, Kong A, Catalona WJ, Mayordomo JI, Einarsson GV, Barkardottir RB, Jonsson E, Mates D, Neal DE, Kiemeney LA, Thorsteinsdottir U, Rafnar T, Stefansson K. A study based on whole-genome sequencing yields a rare variant at 8q24 associated with prostate cancer. Nat Genet. 2012;44:1326–1329. doi: 10.1038/ng.2437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Patterson N, Freedman ML, Myers SR, Pike MC, Waliszewska A, Neubauer J, Tandon A, Schirmer C, McDonald GJ, Greenway SC, Stram DO, Le Marchand L, Kolonel LN, Frasco M, Wong D, Pooler LC, Ardlie K, Oakley-Girvan I, Whittemore AS, Cooney KA, John EM, Ingles SA, Altshuler D, Henderson BE, Reich D. Multiple regions within 8q24.21 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haiman CA, Chen GK, Blot WJ, Strom SS, Berndt SI, Kittles RA, Rybicki BA, Isaacs WB, Ingles SA, Stanford JL, Diver WR, Witte JS, Hsing AW, Nemesure B, Rebbeck TR, Cooney KA, Xu J, Kibel AS, Hu JJ, John EM, Gueye SM, Watya S, Signorello LB, Hayes RB, Wang Z, Yeboah E, Tettey Y, Cai Q, Kolb S, Ostrander EA, Zeigler-Johnson C, Yamamura Y, Neslund-Dudas C, Haslag-Minoff J, Wu W, Thomas V, Allen GO, Murphy A, Chang BL, Zheng SL, Leske MC, Wu SY, Ray AM, Hennis AJ, Thun MJ, Carpten J, Casey G, Carter EN, Duarte ER, Xia LY, Sheng X, Wan P, Pooler LC, Cheng I, Monroe KR, Schumacher F, Le Marchand L, Kolonel LN, Chanock SJ, Van Den Berg D, Stram DO, Henderson BE. Genome-wide association study of prostate cancer in men of African ancestry identifies a susceptibility locus at 17q21. Nat Genet. 2011;43:570–573. doi: 10.1038/ng.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han Y, Rand KA, Hazelett DJ, Ingles SA, Kittles RA, Strom SS, Rybicki BA, Nemesure B, Isaacs WB, Stanford JL, Zheng W, Schumacher FR, Berndt SI, Wang Z, Xu J, Rohland N, Reich D, Tandon A, Pasaniuc B, Allen A, Quinque D, Mallick S, Notani D, Rosenfeld MG, Jayani RS, Kolb S, Gapstur SM, Stevens VL, Pettaway CA, Yeboah ED, Tettey Y, Biritwum RB, Adjei AA, Tay E, Truelove A, Niwa S, Chokkalingam AP, John EM, Murphy AB, Signorello LB, Carpten J, Leske MC, Wu SY, Hennis AJ, Neslund-Dudas C, Hsing AW, Chu L, Goodman PJ, Klein EA, Zheng SL, Witte JS, Casey G, Lubwama A, Pooler LC, Sheng X, Coetzee GA, Cook MB, Chanock SJ, Stram DO, Watya S, Blot WJ, Conti DV, Henderson BE, Haiman CA. Prostate cancer susceptibility in men of African ancestry at 8q24. J Natl Cancer Inst. 2016;108:djv431. doi: 10.1093/jnci/djv431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hazelett DJ, Coetzee SG, Coetzee GA. A rare variant, which destroys a FoxA1 site at 8q24, is associated with prostate cancer risk. Cell Cycle. 2013;12:379–380. doi: 10.4161/cc.23201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Helfand BT, Roehl KA, Cooper PR, McGuire BB, Fitzgerald LM, Cancel-Tassin G, Cornu JN, Bauer S, Van Blarigan EL, Chen X, Duggan D, Ostrander EA, Gwo-Shu M, Zhang ZF, Chang SC, Jeong S, Fontham ET, Smith G, Mohler JL, Berndt SI, McDonnell SK, Kittles R, Rybicki BA, Freedman M, Kantoff PW, Pomerantz M, Breyer JP, Smith JR, Rebbeck TR, Mercola D, Isaacs WB, Wiklund F, Cussenot O, Thibodeau SN, Schaid DJ, Cannon-Albright L, Cooney KA, Chanock SJ, Stanford JL, Chan JM, Witte J, Xu J, Bensen JT, Taylor JA, Catalona WJ. Associations of prostate cancer risk variants with disease aggressiveness: results of the NCI-SPORE Genetics Working Group analysis of 18,343 cases. Hum Genet. 2015;134:439–450. doi: 10.1007/s00439-015-1534-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howie B, Machini J. Genotype imputation for genome-wide association studies. PLoS Genet. 2010;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Cai B. G84E mutation in HOXB13 is firmly associated with prostate cancer risk: a meta-analysis. Tumour Biol. 2014;35:1177–1182. doi: 10.1007/s13277-013-1157-5. [DOI] [PubMed] [Google Scholar]
- Kim T, Cui R, Jeon YJ, Lee JH, Lee JH, Sim H, Park JK, et al. Long-range interaction and correlation between MYC enhancer and oncogenic long noncoding RNA CARL-o5. Proc Nat Acad Sci. 2014;111:4173–4178. doi: 10.1073/pnas.1400350111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kote-Jarai Z, Easton DF, Stanford JL, Ostrander EA, Schleutker J, Ingles SA, Schaid D, Thibodeau S, Dörk T, Neal D, Donovan J, Hamdy F, Cox A, Maier C, Vogel W, Guy M, Muir K, Lophatananon A, Kedda MA, Spurdle A, Steginga S, John EM, Giles G, Hopper J, Chappuis PO, Hutter P, Foulkes WD, Hamel N, Salinas CA, Koopmeiners JS, Karyadi DM, Johanneson B, Wahlfors T, Tammela TL, Stern MC, Corral R, McDonnell SK, Schürmann P, Meyer A, Kuefer R, Leongamornlert DA, Tymrakiewicz M, Liu JF, O’Mara T, Gardiner RA, Aitken J, Joshi AD, Severi G, English DR, Southey M, Edwards SM, Al Olama AA, PRACTICAL Consortium. Eeles RA. Multiple novel prostate cancer predisposition loci confirmed by an international study: the PRACTICAL Consortium. Cancer Epidemiol Biomark Prev. 2008;17:2052–2061. doi: 10.1158/1055-9965.EPI-08-0317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kote-Jarai Z, Al Olama AA, Giles GG, Severi G, Schleutker J, Weischer M, Campa D, Riboli E, Key T, Gronberg H, Hunter DJ, Kraft P, Thun MJ, Ingles S, Chanock S, Albanes D, Hayes RB, Neal DE, Hamdy FC, Donovan JL, Pharoah P, Schumacher F, Henderson BE, Stanford JL, Ostrander EA, Sorensen KD, Dörk T, Andriole G, Dickinson JL, Cybulski C, Lubinski J, Spurdle A, Clements JA, Chambers S, Aitken J, Gardiner RA, Thibodeau SN, Schaid D, John EM, Maier C, Vogel W, Cooney KA, Park JY, Cannon-Albright L, Brenner H, Habuchi T, Zhang HW, Lu YJ, Kaneva R, Muir K, Benlloch S, Leongamornlert DA, Saunders EJ, Tymrakiewicz M, Mahmud N, Guy M, O’Brien LT, Wilkinson RA, Hall AL, Sawyer EJ, Dadaev T, Morrison J, Dearnaley DP, Horwich A, Huddart RA, Khoo VS, Parker CC, Van As N, Woodhouse CJ, Thompson A, Christmas T, Ogden C, Cooper CS, Lophatonanon A, Southey MC, Hopper JL, English DR, Wahlfors T, Tammela TL, Klarskov P, Nordestgaard BG, Røder MA, Tybjærg-Hansen A, Bojesen SE, Travis R, Canzian F, Kaaks R, Wiklund F, Aly M, Lindstrom S, Diver WR, Gapstur S, Stern MC, Corral R, Virtamo J, Cox A, Haiman CA, Le Marchand L, Fitzgerald L, Kolb S, Kwon EM, Karyadi DM, Orntoft TF, Borre M, Meyer A, Serth J, Yeager M, Berndt SI, Marthick JR, Patterson B, Wokolorczyk D, Batra J, Lose F, McDonnell SK, Joshi AD, Shahabi A, Rinckleb AE, Ray A, Sellers TA, Lin HY, Stephenson RA, Farnham J, Muller H, Rothenbacher D, Tsuchiya N, Narita S, Cao GW, Slavov C, Mitev V, Easton DF, Eeles RA, UK Genetic Prostate Cancer Study Collaborators. British Association of Urological Surgeons’ Section of Oncology. UK ProtecT Study Collaborators. The Australian Prostate Cancer BioResource. PRACTICAL Consortium Seven prostate cancer susceptibility loci identified by a multi-stage genome-wide association study. Nat Genet. 2011;10(43):785–791. doi: 10.1038/ng.882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurie CC, Doheny KF, Mirel DB, Pugh EW, Bierut LJ, Bhangale T, Boehm F, Caporaso NE, Cornelis MC, Edenberg HJ, Gabriel SB, Harris EL, Hu FB, Jacobs KB, Kraft P, Landi MT, Lumley T, Manolio TA, McHugh C, Painter I, Paschall J, Rice JP, Rice KM, Zheng X, Weir BS. Quality control and quality assurance in genotypic data for genome-wide association studies. Genet Epidemiol. 2010;34:591–602. doi: 10.1002/gepi.20516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Durbin R. Fast and accurate long-read alignment with Burrows–Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, DePristo MA. The genome analysis toolkit: a mapreduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe’er I, Yelensky R, Altshuler D, Daly MJ. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008;32:381–385. doi: 10.1002/gepi.20303. [DOI] [PubMed] [Google Scholar]
- Price AL, Zaitlen NA, Reich D, Patterson N. New approaches to population stratification in genome-wide association studies. Nat Revi Genet. 2010;11:459–463. doi: 10.1038/nrg2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC. PLINK: a toolset for whole-genome association and population based linkage analysis. Am J Human Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salinas CA, Kwon E, Carlson CS, Koopmeiners JS, Feng Z, Karyadi DM, Ostrander EA, Stanford JL. Multiple independent genetic variants in the 8q24.21 region are associated with prostate cancer risk. Cancer Epidemiol Biomark Prev. 2008;17:1203–1213. doi: 10.1158/1055-9965.EPI-07-2811. [DOI] [PubMed] [Google Scholar]
- Schaid DJ, McDonnell SK, Zarfas KE, Cunningham JM, Hebbring S, Thibodeau SN, Eeles RA, Easton DF, Foulkes WD, Simard J, Giles GG, Hopper JL, Mahle L, Moller P, Badzioch M, Bishop DT, Evans C, Edwards S, Meitz J, Bullock S, Hope Q, Guy M, Hsieh CL, Halpern J, Balise RR, Oakley-Girvan I, Whittemore AS, Xu J, Dimitrov L, Chang BL, Adams TS, Turner AR, Meyers DA, Friedrichsen DM, Deutsch K, Kolb S, Janer M, Hood L, Ostrander EA, Stanford JL, Ewing CM, Gielzak M, Isaacs SD, Walsh PC, Wiley KE, Isaacs WB, Lange EM, Ho LA, BeebeDimmer JL, Wood DP, Cooney KA, Seminara D, Ikonen T, Baffoe-Bonnie A, Fredriksson H, Matikainen MP, Tammela TL, Bailey-Wilson J, Schleutker J, Maier C, Herkommer K, Hoegel JJ, Vogel W, Paiss T, Wiklund F, Emanuelsson M, Stenman E, Jonsson BA, Grönberg H, Camp NJ, Farnham J, Cannon-Albright LA, Catalona WJ, Suarez BK, Roehl KA. Pooled genome linkage scan of aggressive prostate cancer: results from the International Consortium for Prostate Cancer Genetics. Hum Genet. 2006;120:471–485. doi: 10.1007/s00439-006-0219-9. [DOI] [PubMed] [Google Scholar]
- Schumacher FR, Berndt SI, Siddiq A, Jacobs KB, Wang Z, Lindstrom S, Stevens VL, Chen C, Mondul AM, Travis RC, Stram DO, Eeles RA, Easton DF, Giles G, Hopper JL, Neal DE, Hamdy FC, Donovan JL, Muir K, Al Olama AA, Kote-Jarai Z, Guy M, Severi G, Grönberg H, Isaacs WB, Karlsson R, Wiklund F, Xu J, Allen NE, Andriole GL, Barricarte A, Boeing H, Bueno-de-Mesquita HB, Crawford ED, Diver WR, Gonzalez CA, Gaziano JM, Giovannucci EL, Johansson M, Le Marchand L, Ma J, Sieri S, Stattin P, Stampfer MJ, Tjonneland A, Vineis P, Virtamo J, Vogel U, Weinstein SJ, Yeager M, Thun MJ, Kolonel LN, Henderson BE, Albanes D, Hayes RB, Feigelson HS, Riboli E, Hunter DJ, Chanock SJ, Haiman CA, Kraft P. Genome-wide association study identifies new prostate cancer susceptibility loci. Hum Mol Genet. 2011;20:3867–3875. doi: 10.1093/hmg/ddr295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun J, Zheng SL, Wiklund F, Isaacs SD, Purcell LD, Gao Z, Hsu FC, Kim ST, Liu W, Zhu Y, Stattin P, Adami HO, Wiley KE, Dimitrov L, Sun J, Li T, Turner AR, Adams TS, Adolfsson J, Johansson JE, Lowey J, Trock BJ, Partin AW, Walsh PC, Trent JM, Duggan D, Carpten J, Chang BL, Grönberg H, Isaacs WB, Xu J. Evidence for two independent prostate cancer risk-associated loci in the HNF1B gene at 17q12. Nat Genet. 2008;40:1153–1155. doi: 10.1038/ng.214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomas G, Jacobs KB, Yeager M, Kraft P, Wacholder S, Orr N, Yu K, Chatterjee N, Welch R, Hutchinson A, Crenshaw A, CancelTassin G, Staats BJ, Wang Z, Gonzalez-Bosquet J, Fang J, Deng X, Berndt SI, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cussenot O, Valeri A, Andriole GL, Crawford ED, Tucker M, Gerhard DS, Fraumeni JF, Jr, Hoover R, Hayes RB, Hunter DJ, Chanock SJ. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- Tibshirani R, Bien J, Friedman J, Hastie T, Simon NT, Taylor J, Tibshirani R. Strong rules for discarding predictors in Lassotype problems. J R Stat Soc. 2012;74:245–266. doi: 10.1111/j.1467-9868.2011.01004.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Res. 2010;38:e164. doi: 10.1093/nar/gkq603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiang JF, Yang L, Chen LL. The long noncoding RNA regulation at the MYC locus. Curr Opin Genet Dev. 2015;33:41–48. doi: 10.1016/j.gde.2015.07.001. [DOI] [PubMed] [Google Scholar]
- Yang L, Qiu M, Xu Y, Wang J, Zheng Y, Li M, Xu L, Yin R. Upregulation of long non-coding RNA PRNCR1 in colorectal cancer promotes cell proliferation and cell cycle progression. Oncol Rep. 2016;35:318–324. doi: 10.3892/or.2015.4364. [DOI] [PubMed] [Google Scholar]
- Yeager M, Orr N, Hayes RB, Jacobs KB, Kraft P, Wacholder S, Minichiello MJ, Fearnhead P, Yu K, Chatterjee N, Wang Z, Welch R, Staats MJ, Calle EE, Feigelson HS, Thun MJ, Rodriguez C, Albanes D, Virtamo J, Weinstein S, Schumacher FR, Giovannucci E, Willett WC, Cancel-Tassin G, Cussenot O, Valeri A, Andriole GL, Gelmann EP, Tucker M, Gerhard DS, Fraumeni JF, Hoover R, Hunter DJ, Chanock SJ, Thomas G. Genomewide association study of prostate cancer identifies a second risk locus at 8q24.21. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.