Abstract
Nasopharyngeal carcinoma (NPC) is one of the most common malignancies of the head and neck. It arises from the nasopharynx epithelium and is associated with high morbidity and mortality. Long non‐coding RNA (lncRNA) have been reported to regulate gene interaction and play critical roles in carcinogenesis and progression. LncRNA‐ROR, a recently identified lncRNA, has been shown to be involved in initiation, progression and metastasis of several tumors, including hepatocellular carcinoma, breast cancer and glioma. However, whether lncRNA‐ROR is associated with the progression of NPC remains unknown. Resistance to radiotherapy and chemotherapy is the primary cause of NPC patients’ death. In this study, we found that lncRNA‐ROR was significantly upregulated in NPC tissues compared with normal tissues. Next, our study proved that lncRNA‐ROR was highly associated with the proliferation, metastasis and apoptosis of NPC. The enrichment of lncRNA‐ROR played a critucal functional role in chemoresistance. The mechanism by which NPC resists chemotherapy might be that lncRNA‐ROR suppress p53 signal pathway. Taken together, these data suggested that lncRNA‐ROR played an important role in the progression of NPC; thereby it might become a therapeutic target and reduce chemoresistance for NPC.
Keywords: Chemoresistance, lncRNA‐ROR, metastasis, nasopharyngeal carcinoma, proliferation
Nasopharyngeal carcinoma (NPC) is one of the most common malignancies of the head and neck.1 It is a unique disease with significantly different risk factors, pathogenesis, clinical behaviors and treatment options than other head and neck cancers.2 NPC arises in the surface epithelium of the nasopharynx and is associated with a high frequency of neck lymph node metastasis and distant metastases.3 Distinguished by its unique clinical and pathologic characteristics, NPC is sensitive to radiotherapy and chemotherapy.4 However, the majority of patients appeared radioresistance and chemoresistance. Some patients present with damage to adjacent normal tissues as a result of radiation.5 Although some progress has been researched, the molecular mechanisms of radioresistance and chemoresistance are poorly understood. Therefore, it is of great clinical value to further reveal the molecular mechanisms of NPC carcinogenesis and develop effective therapeutic strategies.
Long non‐coding RNA (lncRNA) are non‐protein coding transcripts that are longer than 200 nucleotides.6 Resent research demonstrates that lncRNAs are involved in multilevel regulation of gene expression including the epigenetic, transcriptional and post‐transcriptional levels.7 Dysregulated lncRNA expression levels characterize the entire spectrum of disease, and aberrant lncRNA function drives cancer through the disruption of cell processes, typically by facilitating transcriptional regulation.8 It has been found that numbers of lncRNA play pivotal roles in the cancer‐related gene regulatory system, and the disorder of their expression can promote cancer cell proliferation, invasion and metastasis.9 However, the expression and function of lncRNA in NPC have not been studied intensively.
The lncRNA‐ROR is emerging as an important player in reprogramming differentiated cells turn into pluripotent stem cells.10 The LncRNA‐ROR gene is 2.6 kb in length and consists of four exons.11 Recently it has been reported to be upregulated and promote the progression of hepatocellular carcinoma, breast cancer, mammary stem cells and glioma.12, 13, 14 For example, lncRNA–ROR is a hypoxia‐responsive lncRNA which is functionally linked to hypoxia signaling in HCC through miR‐145–HIF‐1 signaling module. LncRNA‐ROR has a key role in regulating breast cancer by preventing the activation of cellular stress pathways including the p53 response.11 Whereas the implications of lncRNA‐ROR have been studied in many tumors, the role of lncRNA‐ROR in NPC proliferation and metastasis remains unexplored.
In the present study, we found that the expression of lncRNA‐ROR increased in tumor tissues compared with non‐cancerous tissues. Inhibition of lncRNA‐ROR suppressed the proliferation and migration but improved the apoptosis of CNE2 cells. The expression of lncRNA‐ROR was correlated with the concentration gradient of cis‐Dichlorodiamineplatinum (II) (DDP) in CNE2. In addition, down‐expression of lncRNA‐ROR improved the expression of p53, especially after treatment with DDP. These results suggested that lncRNA‐ROR can regulate the proliferation, migration and chemoresistance of NPC.
Materials and Methods
Specimens and cell lines
Nasopharyngeal carcinoma tissue specimens were obtained from the Affiliated Hospital of Nantong University, China. The patients were diagnosed using histopathological evidence. Samples were frozen in liquid nitrogen immediately after surgical removal. The non‐cancer tissues were provided by patients who had nasal operations. All human tissues were collected using the protocols approved by the Ethics Committee of the Affiliated Hospital of Nantong University.
All the cell lines (CNE2, CNE1, 5‐8F and 6‐18B) from our laboratory were cultured in RPMI medium 1640 (Gibco BRL, Grand Island, NY, USA) supplemented with 10% FBS (Gibco BRL, Grand Island, NY, USA). 5‐8F and 6‐10B cells were cultured in completed medium with 100 U/mL penicillin and 100 μg/mL streptomycin (Shanghai Genebase Gen‐Tech, Shanghai, China). The immortalized normal nasopharyngeal epithelial cell line NP69 was cultured in Keratinocyte‐SFM medium supplemented with epidermal growth factor (Invitrogen, Carlsbad, NM, USA). All cells were cultured at 37°C under 5% CO2.
Quantitative real‐time PCR
LncRNA‐ROR expression in NPC compared with normal cells and tissues was measured with SYBR quantitative real‐time PCR (qRT‐PCR). Total RNA was isolated from cancerous/non‐cancerous specimens or cell lines using TRIzol Reagent (Invitrogen, Shanghai, China) according to the manufacturer's instructions. LncRNA‐ROR cDNA was generated using the Reverse Transcription System Kit. qRT‐PCR reactions were performed using an ABI7500 System and SYBR Green PCR Master Mix (Takara, Dalian, China). The primers were designed and synthesized by Guangzhou RiboBio (Guangzhou, China). The primer sequences for lncRNA‐ROR were 5′‐CTCCAGCTATGCAGACCACTC‐3′ (forward) and 5′‐GTGACGCCTGACCTGTTGAC‐3′ (reverse). Each assay was performed in triplicate, and the average was calculated. LncRNA‐ROR expression levels were normalized to GAPDH.
Western blot analysis
Proteins were measured by western blot analysis. Total protein was extracted from transfected cells using the RIPA lysis buffer (Beyotime Institute of Biotechnology, Nantong, China). For western blot analysis, equal amounts of protein samples were separated by 10% SDS‐PAGE and transferred to PVDF membranes (Millipore, Billerica, MA, USA). Membranes were incubated with the primary antibodies. The antibodies used were as follows: anti‐p53 (Abcam, UK; 1:2000 dilution), anti‐p‐ERK (Santa Cruz Biotechnology, USA; 1:500 dilution), anti‐ERK (Santa Cruz Biotechnology, USA; 1:500 dilution) and mouse anti‐β‐actin (Santa Cruz Biotechnology, USA; 1:1,000 dilution). Blots were then incubated with goat anti‐rabbit or anti‐mouse secondary antibody conjugated to HRP (Santa Cruz Biotechnology, USA; 1:1000 dilution) and visualized by enhanced chemiluminescence (Cell Signaling Technology, Danfoss, USA). The values are representative of at least three independent experiments.
Small interfering RNA and transfection
CNE2 cells were transfected with either 50nMsiRNA targeting ROR or scrambled negative controls (RiboBio, Guangzhou, Shanghai, China). Transfections were performed using Lipofectamine 2000 Reagent (Invitrogen) following the manufacturer's protocol. Following culture for a further 48 h, total RNA and cellular protein lysates were collected and used for reverse transcription‐quantitative PCR (qRT‐PCR) and western blot analysis, respectively.
Cell viability assay (Cell Counting Kit‐8)
Cells were seeded into 96‐well plates, then transfected with siRNA ROR or scrambled negative controls and were cultured for 6, 24, 48, 72 and 96 h. Cell viability was assessed by Cell Counting Kit‐8 assay (Beyotime Institute of Biotechnology, Shanghai, China). The absorbance of each well was read on a microplate reader (F‐2500 florescence spectrophotometer; Hitachi, Tokyo, Japan) at 450 nm. All of the experiments were independently repeated at least three times.
Flow cytometric analysis of proliferation and apoptosis
CNE2 cells, transiently transfected with siRNA ROR, were harvested 48 h after transfection by trypsinization. After double staining with an Annexin V‐FITC Apoptosis Detection Kit (BD Biosciences, Oxford, UK). The apoptotic cells were detected by flow cytometry. All samples were assayed in triplicate.
At 48 h after transfection, the adhered cells were obtained by trypsinization and pooled with the flating cells and centrifuged at 300 g for 5 min. Then, the cells were ‘stained with a’ Cell Cycle Detection Kit (KeyGene, Holland) according to the manufacturer's instructions. Finally, the samples were analyzed using the FACSCalibur Flow Cytometer (BD Bioscience, USA) and BD CellQuest software (BD Bioscience, USA).
Wound healing assay
Cells were seeded in 6‐well plates in normal cell growth media and incubated to confluence. An artificial homogeneous wound was created on the monolayer using a sterilized 200‐μL micropipette tip when the cells reached approximately 90% density. The medium was changed to 1640 containing 2% FBS. Duplicate wells for each condition were examined, and each experiment was repeated in triplicate.
Matrigel invasion and migration assays
For the transwell migration assays, CNE2 cells were plated in a serum‐free medium in the top chamber with a non‐coated membrane (24‐well insert; 8‐μm pore size; Millipore) and a medium supplemented with 10% serum was in the lower chamber. The Matrigel invasion assay was done using the BD BioCoat Matrigel Invasion Chamber (pore size, 8 mm, 24‐well; BD Biosciences, CA, USA) following the manufacturer's protocol. After 48 h, the bottom of the chamber insert was stained with methanol and 0.1% crystal violet, imaged, and counted using an IX70 inverted microscope. Each Matrigel invasion assay was conducted in at least three replicates.
Statistical analysis
Data are presented as mean ± SE; the Student's t‐test was used for assessing the difference between individual groups and P ≤ 0.05 was considered statistically significant.
Results
Overexpression of lncRNA‐ROR in human nasopharyngeal carcinoma tissues and cell lines
Recent data have indicated that lncRNA‐ROR is overexpressed in several cancers and is critical in tumor progression (e.g in proliferation, migration and invasion).11, 14, 15 First, we assessed the lncRNA‐ROR expression level in clinical specimens by qRT‐PCR. In a collection of six pairs of randomly chosen samples of NPC, we found that the lncRNA‐ROR level was higher in cancerous tissues compared with normal tissues (Fig. 1a). To confirm the overexpression of lncRNA‐ROR in NPC, we then measured the expression of lncRNA‐ROR in NPC cell lines. In agreement with the data above, lncRNA‐ROR expressed a higher level in NPC cell lines (CNE2, CNE1, 5‐8F and 6‐10B) than in nasopharyngeal epithelia cell NP69 (Fig. 1b). Taken together, these findings suggest that lncRNA‐ROR was upregulated in NPC.
Figure 1.
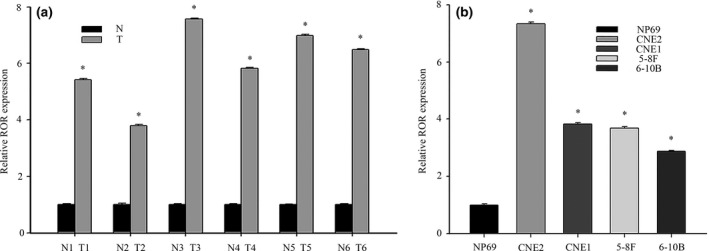
Aberrant expression of lncRNA‐ROR in human nasopharyngeal carcinoma (NPC) tissue samples and cell lines. (a) The expression levels of lncRNA‐ROR in NPC and non‐cancer tissue were measured by qRT‐PCR. T (human NPC tissues), N (non‐cancer tissue). (b) qRT‐PCR assays for lncRNA‐ROR expression in NPC cell lines (CNE2, CNE1, 5‐8F and 6‐10B). Relative levels compared with the nasopharyngeal epithelia cell NP69 for qRT‐PCR. Data are mean ± SE. *P < 0.05.
LncRNA‐ROR promotes nasopharyngeal carcinoma cells proliferation by regulating the cell cycle
Of these NPC cell lines, the highest expression level of lncRNA‐ROR was found in CNE2. Herein, CNE2 was selected to study the function of lncRNA‐ROR. The proliferation function of lncRNA‐ROR in CNE2 was examined first. Three different siRNA targeted to lncRNA‐ROR was designed and the effect on lncRNA‐ROR was confirmed by qRT‐PCR (Fig. 2a). As we expected, compared with the scrambled negative controls, the intracellular level of lncRNA‐ROR was degraded after transcription with siRNA ROR. In addition, siRNA‐1 was chosen as the most effective siRNA. Accordingly, the proliferation of transfected CNE2 cells was measured by CCK‐8 assay. As shown in Figure 2(b), cells transfected with siRNA‐1 showed weak proliferation ability (Fig. 2b). Furthermore, flow cytometry was carried out to evaluate the effect of lncRNA‐ROR on cell proliferation, and the results showed that lncRNA‐ROR had significant effects on the cell cycle (Fig. 2c). Inhibition of the expression of ROR increased the G1 phase population (from 59.91% to 70.11%), and at the same time decreased the S and G2 phase population (from 40.01% to 29.89%) in the CNE2 cells (Fig. 2c). Western blot was then carried out to prove that lncRNA‐ROR promotes NPC cell proliferation by regulating the cell cycle. After knockdown of lncRNA‐ROR, the expression of PCNA (Proliferating cell nuclear antigen) and cyclin A were clearly downregulated compared with scrambled siRNA (Fig. 2d). Furthermore, we examined the effects of lncRNA‐ROR on the apoptosis of CNE2 cells transfected with siRNA‐1 by flow cytometry. The results showed that transfection with siRNA‐1 increased the total apoptosis rate of CNE2 cells from 10.81% to 20.57% compared to scrambled siRNA (Fig. 3a). These results suggest that lncRNA‐ROR can reduce the apoptosis of CNE2 and promote cell proliferation as a tumor regulator through altering the cell cycle pathway in NPC.
Figure 2.
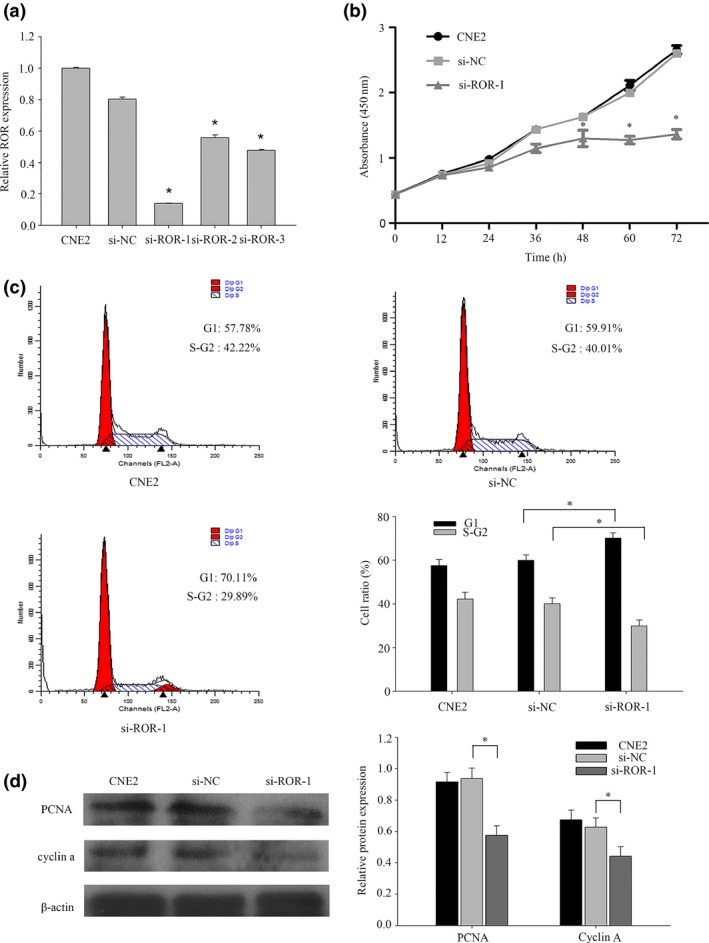
LncRNA‐ROR promotes nasopharyngeal carcinoma (NPC) cell proliferation by regulating the cell cycle. (a) CNE2 cells were transfected with no siRNA (CNE2), si‐NC (scrambled siRNA) and siRNA ROR. After 48 h, total RNA from CNE2 cells was isolated and qRT‐PCR for lncRNA‐ROR was performed. (b) CCK8 assays were used to determine the viability of siRNA ROR transfected CNE2 cells. (c) Flow cytometry assays were performed to analyze the cell cycle progression when CNE2 cells were transfected with siRNA ROR 24 h later. (d) The expression of Proliferating Cell Nuclear Antigen and cyclin A were detected by western blot analysis. Data are mean ± SE. The same experiments were performed in triplicate. *P < 0.05.
Figure 3.
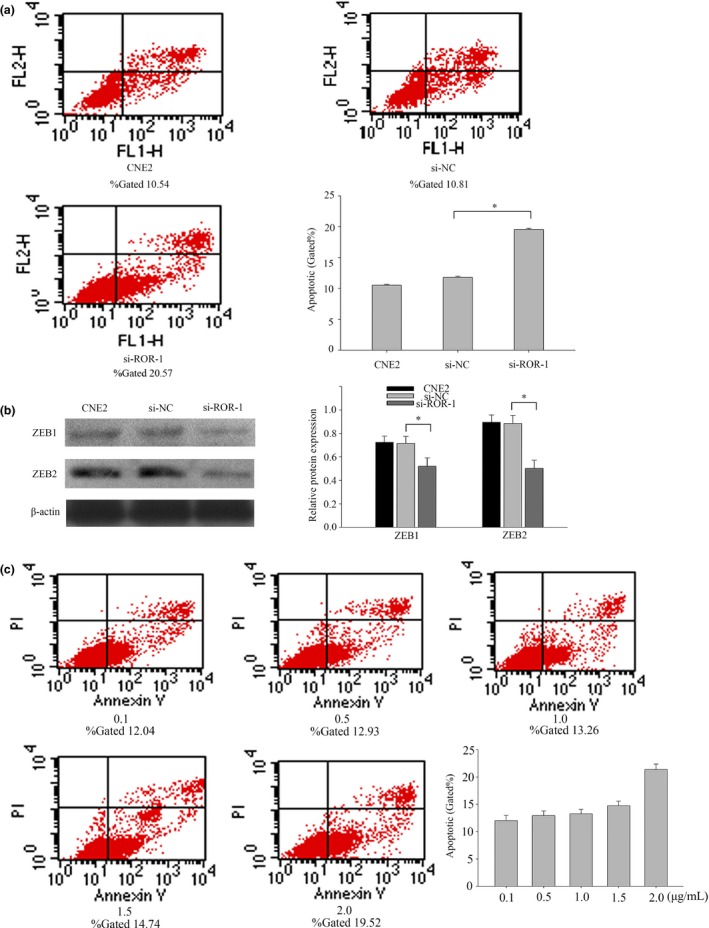
Knockdown of lncRNA‐ROR promotes nasopharyngeal carcinoma (NPC) cell apoptosis and increases the expression of epithelial–mesenchymal transition (EMT) inducers. (a) Flow cytometry assays were performed to analyze the cell apoptosis when CNE2 cells were transfected with siRNA ROR 48 h later. (b) The expression of ZEB1 and ZEB2 were detected by western blot analysis. ZEB1 and ZEB2 are EMT inducers. (c) Flow cytometry assays were performed to analyze the cell apoptosis when CNE2 cells cultured with different concentrations of DDP. Data are mean ± SE. The same experiments were performed in triplicate.*P < 0.01.
LncRNA‐ROR promotes nasopharyngeal carcinoma migration and invasion by influencing the epithelial–mesenchymal transition program
Based on the different expression of lncRNA‐ROR, we proposed that this lncRNA might play an important role in regulating cell migration and invasion of NPC cells. To test this hypothesis, cell migration and invasion assays were performed in CNE2 cells transfected with siRNA‐1 and a negative control.The wound healing assay showed that cell migration was inhibited in ROR‐knockdown NPC cells compared to the negative control (Fig. 4a). Moreover, the transwell assay indicated a significant reduction in cell migration and invasion ability after siRNA‐1 transfection into CNE2 cells (Fig. 4b). Taken together, these results suggest that lncRNA‐ROR may act as a tumor promoter through promoting cell migration and invasion. To verify the mechanism of invasion, we then examined the epithelial and mesenchymal markers using western blotting. After transfection with siRNA‐1, the epithelial markers E‐cadherin were upregulated in CNE2 cells, whereas the mesenchymal markers vimentin and N‐cadherin were significantly downregulated (Fig. 4c). Thus, molecular parameters suggested that lncRNA‐ROR may be able to drive the CNE2 cells from an epithelial to mesenchymal status. After transfection with siRNA‐1, the expressions of several epithelial–mesenchymal transition (EMT) inducers (ZEB1 and ZEB2) were downregulated (Fig. 3b). Together, these results suggest that lncRNA‐ROR may be a novel EMT inducer, and that it has a role in NPC progression.
Figure 4.
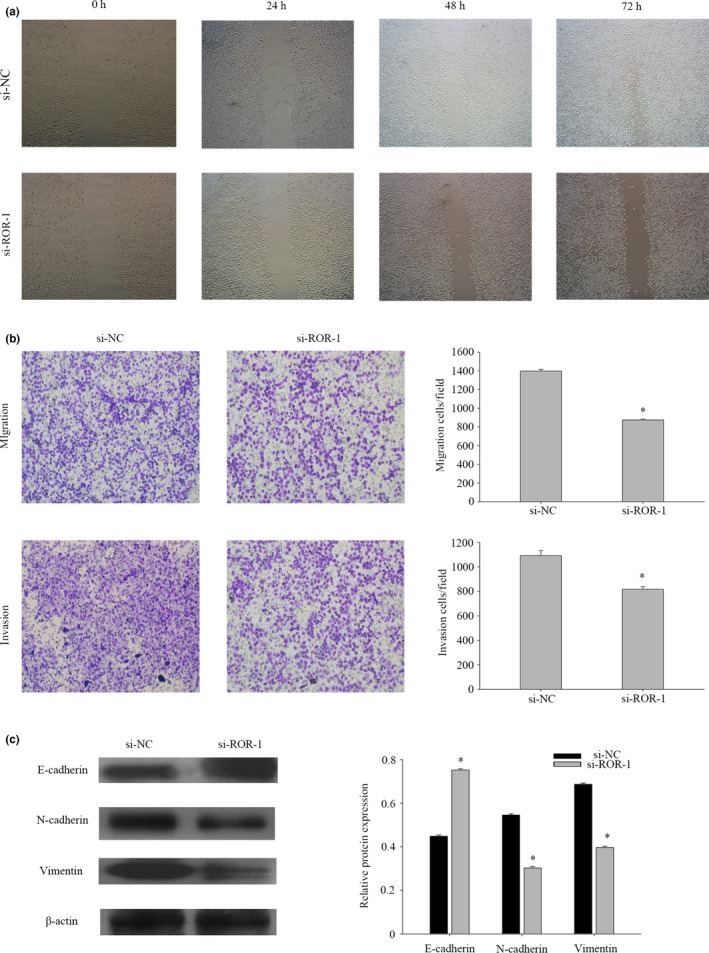
LncRNA‐ROR promotes nasopharyngeal carcinoma (NPC) migration and invasion by influencing the epithelial–mesenchymal transition (EMT) program. (a) Wound‐healing assay to assess the effect of lncRNA‐ROR on cell mobility in CNE2 cells. (b) The effect of lncRNA‐ROR on migration or invasion ability was measured by transwell assays. The data represent the mean number of cells per field and are presented as the means ± SEM. *P < 0.01. All the experiments were repeated three times. (c) The protein expression of E‐cadherin, N‐cadherin and vimentin in CNE2 cells was analyzed by western blot. The same experiments were performed in triplicate. Data are mean ± SE. *P < 0.01.
LncRNA‐ROR promotes nasopharyngeal carcinoma chemotherapy resistance ability by p53 pathways
Some published studies have shown that lncRNA‐ROR regulates many different kinds of cancer cells, especially in DNA damage.11, 16 We speculate that lncRNA‐ROR can regulate the NPC cells with chemotherapy. First, NPC cells were treated with DDP at 2.0 μg/mL for 24 h. The expression of ROR was then extracted. The results indicated that the expression levels of lncRNA‐ROR in four different kinds of NPC cells treated with DDP were rising in varying degrees(Fig. 5a). Then CNE2 was treated with DDP concentration gradient and the expression of lncRNA‐ROR was highest in 2.0 μg/mL (Fig. 5b). DDP‐induced cell death was revealed in a dose‐dependent manner (Fig. 3c). qRT‐PCR suggested that lncRNA‐ROR could regulate chemotherapy. Cells were treated with DDP at 2 μg/mL and then transfected with siRNA‐1 or negative control. The proliferation of transfected CNE2 cells was measured using a CCK‐8 assay (Fig. 5c). We found that lncRNA‐ROR can promote the proliferation of CNE2 cells with DDP. CNE2 cells were first treated by DDP then transfected with negative control or siRNA‐1 at the indicated concentrations for 24 h before harvesting for flow cytometry assays (Fig. 5d). Inhibition of the expression of lncRNA‐ROR significantly reduced cells’ ability to resist chemotherapeutic drugs. The evidence indicates that p53 and p21 translational regulation after chemotherapy may play an important role in chemoresistance. Herein, we explored the relationship between lncRNA‐ROR and p53. CNE2 cells were first transfected with negative control or siRNA‐1 and then treated with DDP at 2 μg/mL for 24 h before harvesting for western blot (Fig. 5e). Of interest, knockdown of ROR leads to an increase in the activation of p53 pathways. We show that lncRNA‐ROR functions as a negative regulator of p53.
Figure 5.
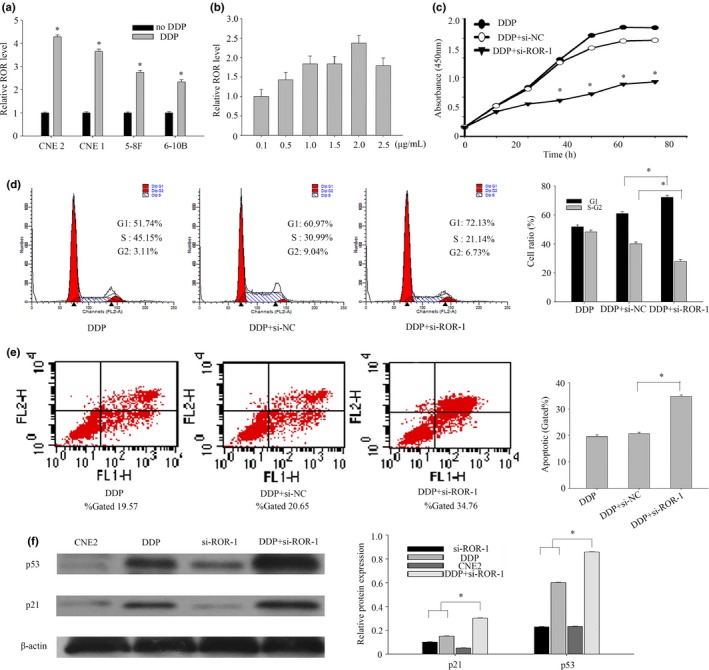
LncRNA‐ROR can improve the chemoresistance of nasopharyngeal carcinoma (NPC). (a) The expression levels of lncRNA‐ROR in treated CNE2 were measured by qRT‐PCR. (b) qRT‐PCR assays for lncRNA‐ROR expression in CNE2 cultured with DDP concentration gradient. (c) CCK8 assays were used to determine the viability of siRNA‐1 transfected CNE2 cells with DDP. (d) Flow cytometry assays were performed to analyze the cell cycle progression. CNE2 cells were first treated by DDP then transfected with negative control or siRNA‐1 at the indicated concentrations for 24 h before harvesting for flow cytometry assays. (e) Flow cytometry assays were performed to analyze the cell cycle progression. CNE2 cells were first treated by DDP then transfected with negative control or siRNA‐1 at the indicated concentrations for 24 h before harvesting for apoptosis. (f) CNE2 cells were transfected with control siRNA or siRNA‐1 for 6 h, treated with DDP in 2.0 μg/mL. The cells were harvested for western blot at the indicated time points after DDP treatment. The same experiments were performed in triplicate. Data are mean ± SE. *P < 0.01.
Discussion
Mounting evidence indicates that eukaryotic transcriptomes and genomes are not the simple substrates of protein‐coding gene transcription that they were once thought to be, but rather exhibit extensive non‐coding RNA (ncRNA) expression.17 In the past decade, the discovery of numerous lncRNA has dramatically altered our understanding of the biology of complex diseases, including cancers. They have emerged as an important player in cancer biology. Increasing evidence suggests that lncRNA may function as oncogenes or tumor suppressors. Hundreds of lncRNA have been found frequently dysregulated in cancer cells.18 For example, some results have demonstrated that high lncRNA‐HOTAIR expression is associated with NPC progression and predicts poor patient prognosis.19 LncRNA‐ROR has been identified to play a role in promoting survival in iPSC and ESC by preventing the activation of cellular stress pathways.20 Recently, some genomewide association studies have identified lncRNA‐ROR as a risk locus for several other cancers, including breast cancer, hepatocellular carcinoma, and glioma.12, 13, 14 For example, lncRNA‐ROR can influence the metastasis of breast cancer by EMT and regulate the chemothresistance of hepatocellular carcinoma by p53.13, 15
Inspired by these lines of evidence, we investigated lncRNA‐ROR expression in NPC and considered its clinical significance in the present study. We found that lncRNA‐ROR expression is significantly upregulated in NPC tissues compared with normal tissues and cells. This strongly suggests the possibility that lncRNA‐ROR can be used as a potential therapeutic target to NPC. It also suggests that its overexpression may be an important factor for NPC progression. Moreover, suppressed lncRNA‐ROR expression reduced the proliferation ability of CNE2 cells. Flow cytometry and western blot indicated that lncRNA‐ROR may regulate the proliferation ability by influencing cell cycles. Refractoriness to apoptosis is believed to be a possible cause of cancer formation. We found that knockdown of lncRNA‐ROR can also increase NPC cell apoptosis. These data suggest that lncRNA‐ROR contributes to NPC development through regulation of cell proliferation and apoptosis. We have demonstrated that lncRNA‐ROR is a critical positive regulator of EMT. EMT has been recognized as an important process in normal organ development, as well as in human pathology, including cancer progression.21, 22 Increasing evidence indicates that EMT is a crucial step for tumor infiltration and metastasis, and it has been generally accepted that EMT is a key mechanism underlying metastasis in many types of cancer. The wound healing assay and transwell invasion assay showed that cell migration was inhibited in ROR‐knockdown NPC cells compared to the negative controls. It potentiated NPC cell migration and invasion in vitro. This suggested that ROR might promote tumor metastasis through regulation of tumor cell adhesion and mobility.
Previous studies have revealed that lncRNA‐ROR contributes to various cellular events in many cancers, such as facilitated cell proliferation and chemoresistance.11, 16 Distinguished by NPC‐unique clinical and pathologic characteristics, NPC is highly chemoradiotherapy sensitive.23 However, there are still some NPC patients’ performance tolerating chemotherapy and serious complications were occured in others after radiotherapy and chemotherapy.5 Improving the efficacy of radiotherapy and chemotherapy is very important. Some research has reported that p53 regulates the expression of lncRNA upon DNA damage.24 This study demonstrated that lncRNA‐ROR is a unique member of p53 negative regulators because lncRNA‐ROR is capable of suppressing the cellular p53 level after DNA damage; this ROR‐mediated p53 repression occurs, in part, through translation regulation. Culturing NPC cells with DDP after inhibition of lncRNA‐ROR greatly reduces the proliferation and improves the apoptosis. Along with these findings, our study provides further evidence that non‐coding RNA, particularly lncRNA‐ROR in this case, are important interplayers in the p53 mediated tumor suppression network.
In summary, we show that lncRNA‐ROR has functions in modulating the proliferation, migration and chemoresistance of NPC. It may be critical for the biological processes in NPC. (proliferation, migration and chemoresistance). It is more important is that lncRNA‐ROR could be a crucial therapy resistance site, and could improve patients’ quality of life. Therefore, this study extends our knowledge on the oncogenesis of NPC.
Disclosure Statement
The authors have no conflict of interest to declare.
Acknowledgments
This study was supported by grants from the Chinese National Natural Science Foundation (nos. 81172841, 81202368 and 81471603), the China Postdoctoral Science Foundation (2013M541708); the “333 Natural Science Foundation” of Jiangsu Grant (BRA2013286); the Jiangsu Provincial Health Department (Z201005); an innovative project for Nantong University postgraduate students (13025043); the Jiangsu Province's Outstanding Medical Academic Leader Program (LJ201136); and the Natural Science Foundation of Jiangsu Province (SBK2015022581).
Cancer Sci 107 (2016) 1215–1222
Funding Information
Chinese National Natural Science Foundation (nos. 81172841, 81202368 and 81471603); China Postdoctoral Science Foundation (2013M541708); “333 Natural Science Foundation” of Jiangsu Grant (BRA2013286); Jiangsu Provincial Health Department (Z201005); Nantong University Postgraduate Students (13025043); Jiangsu Province's Outstanding Medical Academic Leader Program (LJ201136); Natural Science Foundation of Jiangsu Province (SBK2015022581).
References
- 1. Gong Z, Zhang S, Zeng Z et al LOC401317, a p53‐regulated long non‐coding RNA, inhibits cell proliferation and induces apoptosis in the nasopharyngeal carcinoma cell line HNE2. PLoS ONE 2014; 9: e110674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bo H, Gong Z, Zhang W et al Upregulated long non‐coding RNA AFAP1‐AS1 expression is associated with progression and poor prognosis of nasopharyngeal carcinoma. Oncotarget 2015; 6: 20404–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chua ML, Wee JT, Hui EP, Chan AT. Nasopharyngeal carcinoma. Lancet 2015; 7(3): 388–93. [Google Scholar]
- 4. Blanchard P, Lee A, Marguet S et al Chemotherapy and radiotherapy in nasopharyngeal carcinoma: an update of the MAC‐NPC meta‐analysis. Lancet Oncol 2015; 16: 645–55. [DOI] [PubMed] [Google Scholar]
- 5. Wang Q, Fan H, Liu Y et al Curcumin enhances the radiosensitivity in nasopharyngeal carcinoma cells involving the reversal of differentially expressed long non‐coding RNAs. Int J Oncol 2014; 44: 858–64. [DOI] [PubMed] [Google Scholar]
- 6. Zhao W, Luo J, Jiao S. Comprehensive characterization of cancer subtype associated long non‐coding RNAs and their clinical implications. Sci Rep 2014; 4: 6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bergmann JH, Spector DL. Long non‐coding RNAs: modulators of nuclear structure and function. Curr Opin Cell Biol 2014; 26: 10–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. George J, Patel T. Noncoding RNA as therapeutic targets for hepatocellular carcinoma. Semin Liver Dis 2015; 35: 63–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ling H, Vincent K, Pichler M et al Junk DNA and the long non‐coding RNA twist in cancer genetics. Oncogene 2015; 34: 5003–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Wang Y, Xu Z, Jiang J et al Endogenous miRNA sponge lincRNA‐RoR regulates Oct4, Nanog, and Sox2 in human embryonic stem cell self‐renewal. Dev Cell 2013; 25: 69–80. [DOI] [PubMed] [Google Scholar]
- 11. Zhang A, Zhou N, Huang J et al The human long non‐coding RNA‐RoR is a p53 repressor in response to DNA damage. Cell Res 2013; 23: 340–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Feng S, Yao J, Chen Y et al Expression and functional role of reprogramming‐related long noncoding RNA (lincRNA‐ROR) in glioma. J Mol Neurosci 2015; 56: 623–30. [DOI] [PubMed] [Google Scholar]
- 13. Hou P, Zhao Y, Li Z et al LincRNA‐ROR induces epithelial‐to‐mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis 2014; 5: e1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Takahashi K, Yan IK, Haga H, Patel T. Modulation of hypoxia‐signaling pathways by extracellular linc‐RoR. J Cell Sci 2014; 127: 1585–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Takahashi K, Yan IK, Kogure T, Haga H, Patel T. Extracellular vesicle‐mediated transfer of long non‐coding RNA ROR modulates chemosensitivity in human hepatocellular cancer. FEBS Open Bio 2014; 4: 458–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Huang J, Zhou N, Watabe K et al Long non‐coding RNA UCA1 promotes breast tumor growth by suppression of p27 (Kip1). Cell Death Dis 2014; 5: e1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wilusz JE, Sunwoo H, Spector DL. Long noncoding RNAs: functional surprises from the RNA world. Genes Dev 2009; 23: 1494–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Silva A, Bullock M, Calin G. The clinical relevance of long non‐coding RNAs in cancer. Cancers 2015; 7: 2169–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nie Y, Liu X, Qu S, Song E, Zou H, Gong C. Long non‐coding RNA HOTAIR is an independent prognostic marker for nasopharyngeal carcinoma progression and survival. Cancer Sci 2013; 104: 458–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Okita K, Ichisaka T, Yamanaka S. Generation of germline‐competent induced pluripotent stem cells. Nature 2007; 448: 313–7. [DOI] [PubMed] [Google Scholar]
- 21. Thiery JP. Epithelial–mesenchymal transitions in development and pathologies. Curr Opin Cell Biol 2003; 15: 740–6. [DOI] [PubMed] [Google Scholar]
- 22. Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial–mesenchymal transitions in development and disease. Cell 2009; 139: 871–90. [DOI] [PubMed] [Google Scholar]
- 23. Jin C, Yan B, Lu Q, Lin Y, Ma L. The role of MALAT1/miR‐1/slug axis on radioresistance in nasopharyngeal carcinoma. Tumour Biol 2016; 37: 4025–33. [DOI] [PubMed] [Google Scholar]
- 24. Sanchez Y, Segura V, Marin‐Bejar O et al Genome‐wide analysis of the human p53 transcriptional network unveils a lncRNA tumour suppressor signature. Nat Commun 2014; 5: 5812. [DOI] [PMC free article] [PubMed] [Google Scholar]


