Invadopodia are membrane protrusions used by cancer cells to remodel and invade the extracellular matrix. Here, Rajadurai et al. show that the lipid phosphatase SHIP2 recruits the Ena/VASP-family actin regulatory protein Mena to stabilize invadopodia membrane protrusions and promote cell invasion.
Abstract
Invadopodia are specialized membrane protrusions that support degradation of extracellular matrix (ECM) by cancer cells, allowing invasion and metastatic spread. Although early stages of invadopodia assembly have been elucidated, little is known about maturation of invadopodia into structures competent for ECM proteolysis. The localized conversion of phosphatidylinositol(3,4,5)-triphosphate and accumulation of phosphatidylinositol(3,4)-bisphosphate at invadopodia is a key determinant for invadopodia maturation. Here we investigate the role of the 5′-inositol phosphatase, SHIP2, and reveal an unexpected scaffold function of SHIP2 as a prerequisite for invadopodia-mediated ECM degradation. Through biochemical and structure-function analyses, we identify specific interactions between SHIP2 and Mena, an Ena/VASP-family actin regulatory protein. We demonstrate that SHIP2 recruits Mena, but not VASP, to invadopodia and that disruption of SHIP2–Mena interaction in cancer cells leads to attenuated capacity for ECM degradation and invasion in vitro, as well as reduced metastasis in vivo. Together, these findings identify SHIP2 as a key modulator of carcinoma invasiveness and a target for metastatic disease.
Introduction
Normal epithelium is separated from the underlying stroma by a specialized layer of ECM, the basement membrane (BM). During localized invasion and metastasis, invasive carcinoma cells break through this barrier, commonly by proteolytic remodeling of the BM, and penetrate into the interstitial matrix of the stroma (Hoshino et al., 2013). The acquired ability of carcinoma cells to proteolytically remodel the ECM is often supported by their capacity to form invadopodia, which are dynamic, actin cytoskeleton–supported membrane protrusions that function as sites for intracellular trafficking and secretion of matrix metalloproteases (MMPs; Murphy and Courtneidge, 2011; Hoshino et al., 2013). Upon BM perforation, invadopodia are converted into larger pseudopodia structures, allowing carcinoma cells to transmigrate through the BM and invade into the stroma, thus initiating the process of metastasis to distant organs (Schoumacher et al., 2010).
Invadopodia biogenesis is triggered through the oncogenic activity or activation of multiple cell surface receptors, whose signals converge on downstream regulatory signaling molecules involved in cytoskeletal organization. Of these, class I phosphoinositide-3-kinase (PI3K), an enzyme that phosphorylates the D3 position of the inositol ring of phosphatidylinositol(4,5)-bisphosphate (PtdIns(4,5)P2) to produce PtdIns(3,4,5)-triphosphate (PtdIns(3,4,5)P3), has recently emerged as a critical regulator of invadopodia (Hoshino et al., 2012). Inhibition of PI3K activity or sequestration of D3 phosphoinositides attenuates invadopodia formation, whereas a constitutively active p110 subunit of PI3K enhances invadopodia-mediated ECM degradation (Yamaguchi et al., 2011). Mechanistically, enrichment of PtdIns(3,4,5)P3 at sites of invadopodia initiation coincides with recruitment of regulators of the Arp2/3 actin nucleation complex, cortactin and N-WASP. Together these suffice for initiation of invadopodia assembly through enhanced nucleation of branched actin filaments (Sharma et al., 2013). In contrast, invadopodia maturation into proteolytically active structures (comprising membrane protrusions), coupled with targeted trafficking of MMPs, requires local accumulation of phosphoinositide, PtdIns(3,4)P2 (Sharma et al., 2013). Dephosphorylation of PtdIns(3,4,5)P3 at the D5 position of the inositol ring by 5′-inositol phosphatases, including SHIP2, yields PtdIns(3,4)P2 (Ooms et al., 2009). Localized accumulation of PtdIns(3,4)P2 at nascent invadopodia leads to recruitment of several effector proteins, including the Tks4/Tks5 family of adaptors that are believed to sustain invadopodia maturation through regulation of further nucleation of actin filaments and targeted delivery of MT1-MMP (Sharma et al., 2013). Although sustained Arp2/3-mediated branched actin filament nucleation at nascent membrane protrusions has emerged as a crucial regulatory step for invadopodia formation, little is known about how subsequent actin filament elongation contributes to the maturation process. In this respect, Mena and VASP, members of the enabled (Ena)/vasodilator-stimulated phosphoprotein (VASP) family involved in actin filament elongation, localize to invadopodia, and overexpression of an invasion-associated isoform of Mena (MenaINV) can prolong invadopodia lifetime (Philippar et al., 2008; Schoumacher et al., 2010). However, mechanisms for recruitment or the potential functional redundancy of Mena and VASP for invadopodia biogenesis remain unknown.
Here, we investigate the role of the 5′-inositol phosphatase, SHIP2, in maturation of invadopodia. Our analyses reveal that in addition to its lipid phosphatase activity, SHIP2 functions as a scaffold critical for recruitment of Mena to invadopodia. Uncoupling SHIP2–Mena interactions in cancer cells leads to decreased stability of invadopodia, resulting in attenuated ECM degradation in vitro and metastatic capacity in vivo. Together, these findings provide new insight into molecular mechanisms underlying invadopodia maturation into proteolytically active structures and highlight the importance of an unexpected scaffold function of SHIP2 as a key modulator of cell invasion and a potential therapeutic target for metastatic disease.
Results
SHIP2 regulation of invadopodia requires an intact proline-rich sequence and its phosphatase activity
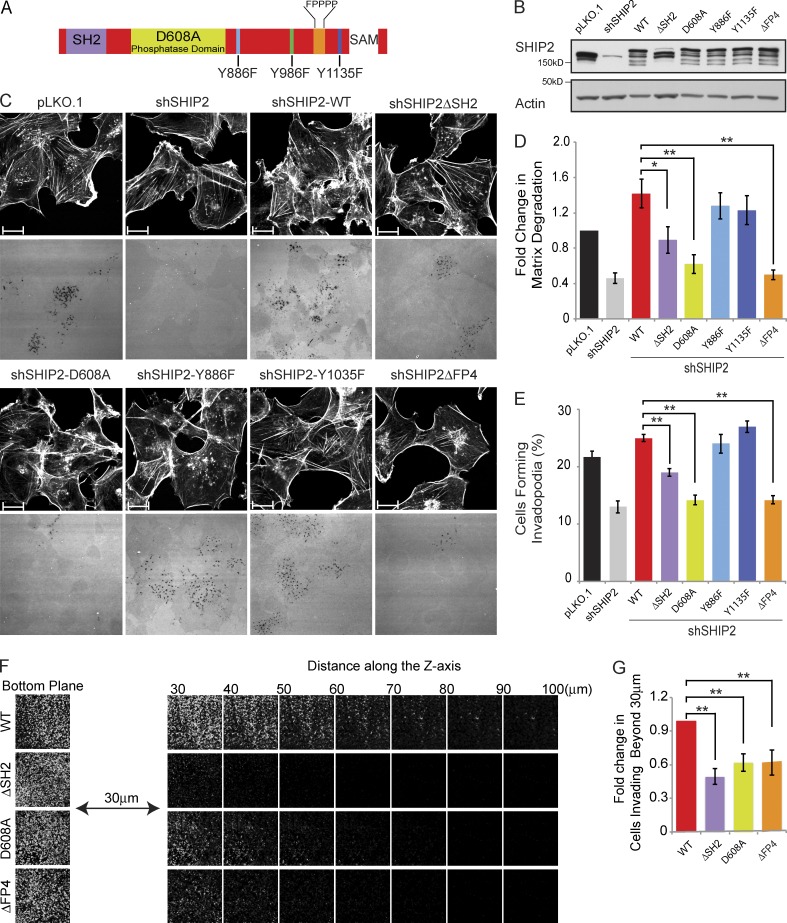
Invadopodia-related functions of SHIP2 were supported by knockdown experiments and were attributed to its enzymatic activity using a small molecule inhibitor (AS1949490; Sharma et al., 2013). However, no detailed structure–function analyses have been performed, and additional roles for SHIP2 have not been explored. To identify protein interaction modules and partners of SHIP2 that contribute to invadopodia maturation, we evaluated a panel of SHIP2 mutants for their ability to form invadopodia competent to degrade the ECM. The panel of SHIP2 mutants lack specific domains (SH2 and proline-rich sequence [PRS]) and contain amino acid substitutions of specific tyrosine residues (Y886 and Y1135) implicated in cell invasion when tyrosine phosphorylated in response to growth factor stimulation (Johnson et al., 2013), as well as a catalytically inactive SHIP2 mutant (D608A; Ooms et al., 2009; Fig. 1 A).
Figure 1.
Proteolytically active invadopodia formation requires both the phosphatase activity and the scaffolding functions of SHIP2. (A) Schematic diagram of SHIP2 depicts its SH2 domain, inositol phosphatase domain, SAM domain, proline-rich region, and several phosphorylated tyrosine residues (Y886, Y996, and Y1135). (B) MDA-MB-231 cells stably expressing SHIP2 shRNA (shSHIP2-C) were transduced with retroviral vectors containing a panel of shRNA-resistant SHIP2 mutants. Upon selection for stable expression of SHIP2 mutants, cell lysates were separated by SDS-PAGE and probed as indicated to assess the protein level of SHIP2 mutants. (C) MDA-MB-231 cells stably expressing SHIP2 shRNA were transduced with retroviral vectors containing a panel of shRNA-resistant SHIP2 mutants. Upon selection for stable expression of SHIP2 mutants, cells were cultured on fluorescently labeled gelatin, and abilities of different SHIP2 mutants to rescue SHIP2 knockdown-induced invadopodia formation (percentage of cells positive for F-actin punctate overlaying matrix degradation area) and ECM degradation (mean matrix degradation area per fields of view) phenotypes were compared. (D) ECM degradation by cells expressing different SHIP2 mutants was quantified and expressed as the fold change of control (pLKO.1). (E) Percentage of cells that form proteolytically active invadopodia in cells expressing different SHIP2 mutants was quantified in comparison to control (pLKO.1). (F) SHIP2-depleted MDA-MB-231 cells rescued with shRNA-resistant wild-type SHIP2 or SHIP2 mutants (ΔSH2, D608A, and ΔFP4) were subjected to an inverted invasion assay. Representative confocal z-stack from each condition depicts relative cell invasion. (G) For quantification of cell invasion, five confocal z-stacks per condition per experiment were acquired. Fluorescence intensity of the planes within each z-stack was measured using Cell Profiler software. Cells invading to and past 30 µm inside the matrix were quantified as a percentage of total cell population. Experiments were repeated four times. All quantified data indicate the mean values ± SE from at least four independent experiments. *, P < 0.05; **, P < 0.01. Bars, 10 µm.
Stable depletion of SHIP2 in invasive MDA-MB-231 breast cancer cells results in 35% fewer cells with proteolytically active invadopodia (Fig. S1, A and B) and a 54% decrease in overall matrix degradation capacity (Fig. S1, A and C). However, SHIP2 depletion does not affect the ability of MDA-MB-231 cells to form ventrally located, cortactin-positive, F-actin–rich punctate structures (Fig. S1, D and E). Consistent with the decrease in invadopodia-mediated ECM degradation, SHIP2 depletion results in a 36% decrease in cell invasion through 3D matrix (Fig. S1, F and G), although no noticeable differences are observed in 2D cell migration parameters, including speed of migration or persistence (Video 1 and Fig. S2). Collectively, these data argue that the contribution of SHIP2 to cell invasion is predominantly mediated through its invadopodia-mediated matrix remodeling capacity.
Populations of SHIP2-depleted cells stably expressing each SHIP2 mutant (Fig. 1 B) were established and assayed for their ability to form proteolytically active invadopodia and for overall ECM degradation capacity. Reexpression of wild-type (WT) SHIP2 rescued both invadopodia formation (114% of pLKO.1) and ECM degradation (139% of pLKO.1) to levels similar to or greater than those of control cells expressing endogenous SHIP2 (Fig. 1, C–E). The ability of SHIP2-depleted cells to form invadopodia and degrade the ECM was also rescued by expression of SHIP2-Y886F and SHIP2-Y1135F mutants, indicating that phosphorylation of these tyrosine residues is not required for SHIP2-dependent invadopodia-mediated ECM degradation (Fig. 1, C–E).
In contrast, several SHIP2 mutants displayed decreased ability to rescue invadopodia formation and ECM degradation after SHIP2 knockdown. The catalytically inactive SHIP2 mutant (D608A; Ooms et al., 2009) failed to rescue invadopodia formation (65% of pLKO.1) and ECM degradation (59% of pLKO.1) to levels of WT SHIP2 (Fig. 1, C–E). This is consistent with the ability of SHIP2 to catalyze conversion of PtdIns(3,4,5)P3 to PtdIns(3,4)P2, which is required for recruitment of the Tks5 scaffold protein and stabilization of the branched actin nucleation complex, consisting of Arp2/3, N-WASP, and cortactin (Sharma et al., 2013). Similarly, a moderate but consistent decrease in ability to rescue invadopodia formation (87.5% of pLKO.1) and ECM degradation (86% of pLKO.1) was observed for the SHIP2ΔSH2 mutant (Fig. 1, C–E). Several SHIP2 SH2 domain–interacting proteins, including p130Cas, localize to invadopodia (Prasad et al., 2001; Alexander et al., 2008), supporting that optimal recruitment of SHIP2 to invadopodia may be mediated by its SH2 domain.
Most strikingly, a SHIP2 mutant that retains phosphatase activity and an intact SH2 domain, but lacks a C-terminal PRS (SHIP2ΔPPPDFPPPP, referred to as SHIP2ΔFP4), showed decreased ability to rescue invadopodia formation (65% of pLKO.1) and overall ECM degradation (49% of pLKO.1) to levels similar to the phosphatase-inactive mutant (SHIP2-D608A; Fig. 1, C–E). The SHIP2ΔFP4 mutant localized to invadopodia (Fig. S3, A and B), indicating no requirement for the PRS (PPPDFPPPP) for SHIP2 recruitment, instead supporting a role for the PRS in invadopodia maturation and ECM degradation. Consistent with their inability to rescue invadopodia-mediated matrix remodeling, the SHIP2ΔSH2, SHIP2-D608A, and SHIP2ΔFP4 mutants showed reduced ability to rescue cell invasion (50%, 62%, and 63% of WT-SHIP2, respectively; Fig. 1, F and G).
SHIP2 recruitment to invadopodia requires an intact SH2 domain
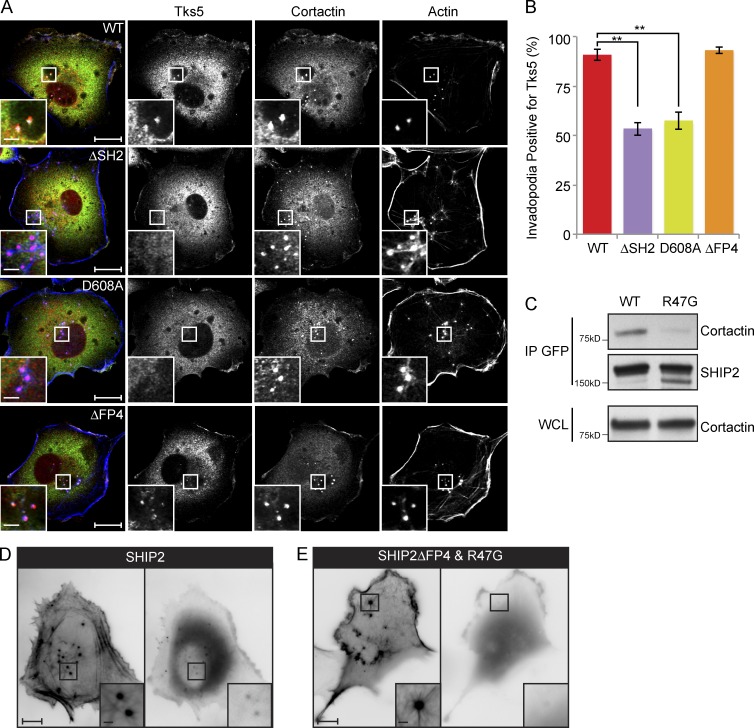
Invadopodia are composed of multiple cytoskeletal, trafficking, and scaffold proteins. Among these, Tks5 has emerged as a scaffold protein required for invadopodia stability and functional integrity (Murphy and Courtneidge, 2011). To assess whether Tks5 recruitment to invadopodia is altered in cells expressing SHIP2 mutants with diminished capacity to rescue matrix degradation (SHIP2ΔSH2, SHIP2-D608A, and SHIP2ΔFP4), we performed quantification of Tks5 content in ventrally located membrane protrusions that stained positive for the invadopodia marker proteins cortactin and F-actin. Whereas the majority of invadopodia (91%) from MDA-MB231-shSHIP2 cells rescued with WT-SHIP2 scored positive for Tks5, only 54% and 57% of invadopodia from cells rescued with SHIP2ΔSH2 and SHIP2-D608A, respectively, were positive for Tks5 (Fig. 2, A and B). Recruitment of Tks5 to nascent invadopodia is thought to be mediated via its N-terminally located PtdIns(3,4)P2-binding Phox-homology (PX) domain. Given that SHIP2 catalytic activity generates PtdIns(3,4)P2 from PtdIns(3,4,5)P3, it is feasible that reduced recruitment of Tks5 to invadopodia sites in cells expressing the catalytically inactive mutant of SHIP2 (SHIP2-D608A) is caused by decreased production of PtdIns(3,4)P2.
Figure 2.
SHIP2 is recruited to invadopodia via its SH2 domain, possibly through an interaction with cortactin. (A) SHIP2-depleted MDA-MB-231 cells rescued with shRNA-resistant wild-type SHIP2 or SHIP2 mutants (ΔSH2, D608A, and ΔFP4) were immunostained for invadopodia markers cortactin and Tks5, and confocal sections were acquired at the ventral surface. (B) As in A, MDA-MB-231-shSHIP2 cells rescued with WT-SHIP2, SHIP2 ΔSH2, SHIP2-D608A, or SHIP2ΔFP4 were immunostained for F-actin (phalloidin), cortactin, and Tks5. Percentage of F-actin– (phalloidin) and cortactin-positive punctae at the ventral surface of cells that were also positive for Tks5 was quantified based on three independent experiments. (C) Cell lysates were isolated from MDA-MB-231 cells transiently transfected with GFP-SHIP2 or GFP-SHIP2ΔFP4-R47G. WT-SHIP2 or SHIP2ΔFP4-R47G was immunoprecipitated using GFP antibody, and immunoprecipitates were separated by SDS-PAGE and probed as indicated. (D) MDA-MB-231 cells were transiently transfected with tagRFP-LifeAct and GFP-SHIP2 and subjected to time-lapse video microscopy analysis. Single frame depicts SHIP2 localization with respect to actin punctae. (E) MDA-MB-231 cells were transiently transfected with tagRFP-LifeAct and GFP-SHIP2ΔFP4-R47G and subjected to time-lapse video microscopy analysis. Single frame illustrates that GFP-SHIP2ΔFP4-R47G failed to localize to actin punctae. All quantified data indicate the mean values ± SE from at least three independent experiments. **, P < 0.01. Bars: 10 µm; (inset) 2 µm.
Our data indicate that the SHIP2ΔSH2 mutant that retains catalytic activity has reduced ability to promote recruitment of Tks5 to nascent invadopodia. Because SHIP2 localizes to the cytosol and translocates to the plasma membrane in response to extracellular stimuli through interactions involving its SH2 domain (Pesesse et al., 2001; Prasad et al., 2001; Ishihara et al., 2002; Smith et al., 2010), we rationalized that reduced recruitment of Tks5 to nascent invadopodia may be caused by impaired localization of SHIP2ΔSH2. In agreement with this, a SHIP2 mutant, SHIP2ΔFP4-R47G, that renders the SH2 domain unable to bind phosphotyrosine (R47G; Prasad et al., 2009) and is unable to engage with Mena tetramers bound to endogenous wild-type SHIP2 (SHIP2ΔFP4) fails to localize to ventral F-actin–positive protrusions (Video 2 and Fig. 2 D). However, a SHIP2ΔFP4 mutant with an intact SH2 domain still localizes to ventral F-actin–positive protrusions, demonstrating requirement for an intact SH2 domain for SHIP2 recruitment (Fig. S3, A and B). Collectively, these data establish that a functional SH2 domain is critical for localization of SHIP2 to invadopodia sites.
Cortactin precedes SHIP2 recruitment to nascent membrane protrusions at sites of invadopodia (Sharma et al., 2013), and cortactin is tyrosine phosphorylated in response to multiple receptor and nonreceptor tyrosine kinases (Ammer and Weed, 2008; Weaver, 2008). To test whether cortactin recruits SHIP2 to nascent invadopodia, we performed coimmunoprecipitation of endogenous cortactin with exogenously expressed GFP-SHIP2 or GFP-SHIP2ΔSH2 in MDA231 cells. An interaction was observed between endogenous cortactin with exogenous SHIP2, which is dependent on the integrity of the SHIP2 SH2 domain (Fig. 2 C). Collectively, these data support the ability of tyrosine-phosphorylated cortactin to recruit SHIP2 to nascent invadopodia via its SH2 domain.
SHIP2 interacts with Mena and VASP
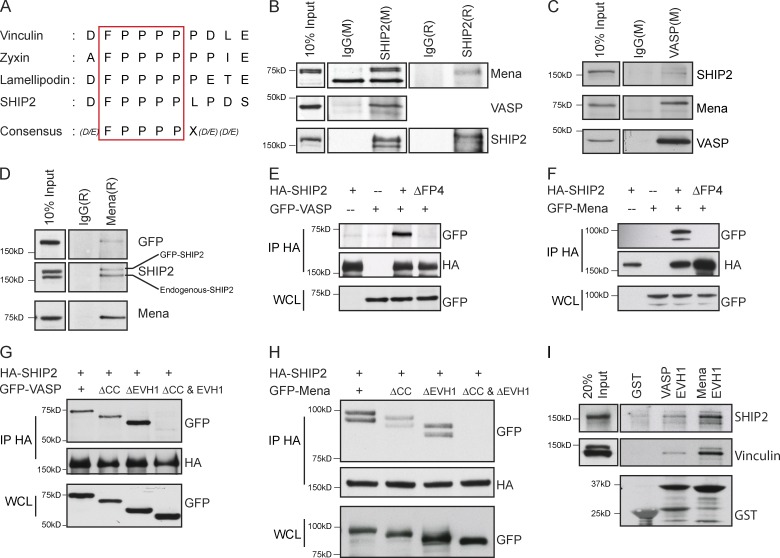
To identify interaction partners for the SHIP2 PRS required for invadopodia maturation, we first tested known binding partners of SHIP2 PRS. The SH3 domains of the endocytic adaptor protein, intersectin, associates with the PPPDFPPPP motif of SHIP2 (Nakatsu et al., 2010). However, depletion of intersectin failed to abrogate invadopodia formation or ECM degradation (Fig. S4), suggesting that SHIP2-dependent invadopodia formation and ECM degradation are mediated by other binding partners of its PRS motif. Intriguingly, the PPPDFPPPP motif of SHIP2 harbors a putative FPPPP (FP4) consensus sequence for the EVH1 domain of the Ena/VASP family of proteins (Fig. 3 A; Carl et al., 1999; Ball et al., 2000; Bear and Gertler, 2009). Ena/VASP proteins elongate actin filaments and possess actin barbed-end anticapping properties (Bear et al., 2002; Barzik et al., 2005; Hansen and Mullins, 2010; Breitsprecher et al., 2011), hence playing a significant role in cellular processes dependent on actin filament dynamics, particularly at the leading edge of lamellipodia and tips of filopodia (Krause et al., 2002, 2003; Murphy and Courtneidge, 2011; Carmona et al., 2016). In addition, two members of the Ena/VASP protein family, Mena and VASP, localize at invadopodia sites (Philippar et al., 2008; Schoumacher et al., 2010). To determine whether Mena or VASP interacts with SHIP2 in MDA-MB-231 cells, we performed reciprocal immunoprecipitations of SHIP2 and Ena/VASP proteins from cell lysates. We found that both Mena and VASP coimmunoprecipitate with endogenous SHIP2 (Fig. 3, B–D) as well as exogenously expressed WT-SHIP2 (Fig. 3, E and F), but not with an exogenously expressed SHIP2ΔFP4 mutant (Fig. 3, E and F). Under these conditions, interaction between SHIP2 and a third member of the Ena/VASP family of proteins, EVL, was not detected, likely because of low endogenous levels of EVL in MDA-MB-231 cells compared with Mena and VASP (not depicted).
Figure 3.
SHIP2 interacts with the EVH1 domains of Mena and VASP via the FPPPP motif. (A) Schematic diagram depicts the SHIP2 consensus motif for the Mena and VASP EVH1 domains and corresponding sequences of the other known interaction partners of Mena and VASP (Zyxin, Lamellipodin, and Vinculin). (B) SHIP2 was immunoprecipitated from MDA-MB-231 cell lysates with either mouse (M) or rabbit (R) SHIP2 antibodies, or respective IgG controls, and immunoprecipitates were separated by SDS-PAGE and probed for Mena and VASP. (C) Coimmunoprecipitation was performed from MDA-MB-231 cell lysates with a mouse VASP antibody or mouse IgG control, and immunoprecipitates were probed for SHIP2, Mena, and VASP. (D) Coimmunoprecipitation was performed from MDA-MB-231 cell lysates expressing GFP-SHIP2 using a rabbit Mena antibody or rabbit IgG control, and immunoprecipitates were probed for SHIP2, Mena, and GFP. (E) HEK293 cells were transiently cotransfected with HA-SHIP2 or HA-SHIP2ΔFP4 and GFP-VASP as indicated, SHIP2 was immunoprecipitated from the cell lysates with an anti-HA antibody, and immunoprecipitates were probed for the presence of GFP-VASP. (F) As in D, HA-SHIP2 or HA-SHIP2ΔFP4 and GFP-Mena were cotransfected as indicated and immunoprecipitated with an anti-HA antibody, and immunoprecipitates were probed for the presence of GFP-Mena. (G) HEK293 cells were transiently cotransfected with HA-SHIP2 and mutants of GFP-VASP (WT, VASPΔCC, VASPΔEVH1, and VASPΔCC and ΔEVH1) as indicated, SHIP2 was immunoprecipitated from the cell lysates with anti-HA antibody, and immunoprecipitates were probed for the presence of GFP-VASP. (H) As in G, HA-SHIP2 and mutants of GFP-Mena (WT, MenaΔCC, MenaΔEVH1, and MenaΔCC and ΔEVH1) were cotransfected as indicated and immunoprecipitated with an anti-HA antibody, and immunoprecipitates were probed for the presence of GFP-Mena. (I) GST, GST-VASP-EVH1, and GST-Mena-EVH1 were coupled to glutathione-Sepharose beads and used to pull down interaction partners from the lysates of MDA-MB-231 cells. Resulting samples were probed for Vinculin and SHIP2 as indicated.
To establish whether the EVH1 domains of Mena and VASP interact with the FP4 motif in SHIP2, we created MenaΔEVH1 and VASPΔEVH1 mutants and analyzed their ability to coimmunoprecipitate with SHIP2. Surprisingly, when overexpressed, both MenaΔEVH1 and VASPΔEVH1 coimmunoprecipitated with SHIP2 (Fig. 3, G and H). Because both Mena and VASP are able to tetramerize through their C-terminal coiled-coil domains (Loureiro et al., 2002; Bear and Gertler, 2009), we hypothesized that MenaΔEVH1 and VASPΔEVH1 could form mixed tetramers with endogenous Mena and VASP, respectively, allowing indirect interaction with WT-SHIP2. In agreement with this hypothesis, we established that simultaneous deletion of the coiled-coil and EVH1 domains from Mena and VASP abrogates binding with SHIP2, whereas loss of either domain alone is insufficient to disrupt the interactions (Fig. 3, G and H). To establish whether the EVH1 domains alone are sufficient for the interactions between SHIP2 and Mena/VASP, we used GST-fused EVH1 domains of VASP and Mena as baits to capture binding proteins from the cell lysates. In these experiments, both SHIP2 and vinculin (another known binding partner of Mena/VASP proteins) were efficiently captured, further demonstrating that EVH1 domains of Mena and VASP are sufficient for interaction with SHIP2 (Fig. 3 I). Collectively, our results reveal that SHIP2 interacts with Mena and VASP via an EVH1-binding site in SHIP2. These findings indicate that in addition to its role in generating PI(3,4)P2, SHIP2 may promote invadopodia maturation through its scaffolding functions, that is, through physical recruitment of Mena and/or VASP.
SHIP2 recruits Mena, but not VASP, to invadopodia to confer functionality
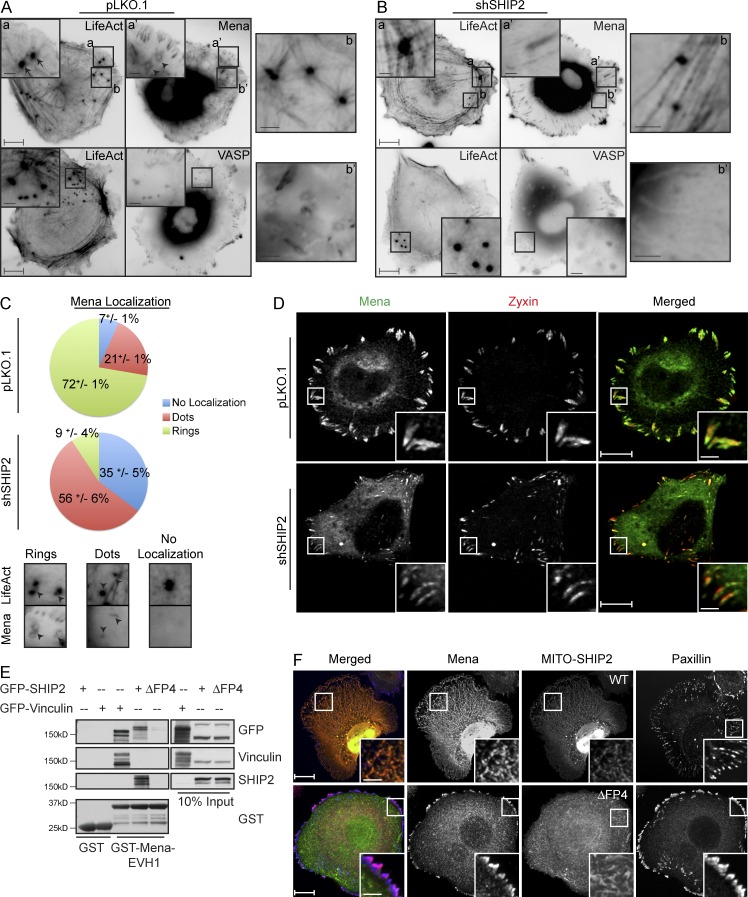
When plated on fluorescent gelatin matrix, MDA-MB-231 cells form invadopodia that actively degrade ECM. The F-actin–rich cores of these structures can be identified during time-lapse live cell imaging using LifeAct fluorescent probe (Fig. S3 C and Video 3). Using this time-lapse microscopy approach, we observed that localization of VASP and Mena differs within invadopodia. Whereas VASP localizes to F-actin–rich cores of protrusions, Mena is targeted to rings overlapping the F-actin– and Tks5-rich cores (Fig. 4 A and Videos 4 and 5). Despite differential localization of Mena and VASP, F-actin– and Tks5-rich cores of protrusions are readily detected by live cell imaging, but the ring-like pattern of Mena localization is often lost during the process of fixation and immunofluorescence staining, indicating that it may be transient in nature. As a result, only a minority of fixed invadopodia structures retain the Mena localization to rings at the distal end of F-actin– and cortactin-positive cores (Fig. S5, A and B).
Figure 4.
SHIP2 recruits Mena, but not VASP, to promote proteolytically active invadopodia formation. (A) MDA-MB-231-pLKO.1 cells were transiently transfected with tagRFP-LifeAct and GFP-Mena or GFP-VASP and subjected to time-lapse video microscopy. Single frame illustrates the differential localization of Mena and VASP with respect to filamentous actin at invadopodia protrusions. (B) As in A, MDA-MB-231-shSHIP2 cells were transiently transfected with tagRFP-LifeAct and GFP-Mena or GFP-VASP and subjected to time-lapse video microscopy. Single frame illustrates that, under SHIP2-depleted conditions, Mena localizes to focal adhesions but not to filamentous actin at invadopodia protrusions, whereas VASP still localizes to filamentous actin at invadopodia protrusions. (C) GFP-Mena and tagRFP-LifeAct were transiently coexpressed in SHIP2-depleted MDA-MB-231 cells and subjected to the time-lapse imaging. Three different localization patterns of Mena relative to F-actin punctae (tagRFP-LifeAct) are observed. The relative distribution of Mena localization pattern under SHIP2-depleted condition compared with control is quantified and depicted as a pie chart. Quantification addresses percentage of invadopodia from 36 cells during the course of the video across three independent experiments. (D) MDA-MB-231-pLKO.1 and MDA-MB-231-shSHIP2 cells were immunostained with Zyxin and Mena, and confocal images were acquired at the ventral surface. (E) GST and GST-Mena-EVH1 were coupled to glutathione-Sepharose beads and used to pull down interaction partners from the lysates of HEK293 cells transiently transfected with GFP-Vinculin, GFP-SHIP2, or GFP-SHIP2ΔFP4. Resulting samples were probed for GFP, Vinculin, and SHIP2 as indicated. (F) Mitochondria-targeted SHIP2 (Mito-SHIP2) or Mito-SHIP2ΔFP4 was transiently coexpressed together with mCherry-Mena in SKBr3 cells and immunostained as indicated to establish the localization of Mena with respect to SHIP2 and focal adhesions. All quantified data indicate the mean values ± SE from at least three independent experiments. Bars: 10 µm; (inset) 2 µm.
To establish whether SHIP2 is required for localization of Mena or VASP to invadopodia, we examined their localization after the knockdown of SHIP2. Under SHIP2-depleted conditions, actin- and cortactin-rich invadopodia-like protrusions form but fail to efficiently degrade matrix (Fig. S1, D and E). Although SHIP2 depletion had no effect on the stability of Mena or VASP (Fig. S5 D), it significantly reduced the localization of Mena at rings overlapping the F-actin core of invadopodia-like protrusions (9% of all invadopodia in SHIP2-depleted cells compared with 72% in control cells; Fig. 4, A–C; and Videos 6 and 7). A separate pool of Mena was still observed at focal adhesions, indicating that Mena localization to other sites in the cell is not abrogated (Videos 8 and 9 and Fig. 4 D). In contrast, SHIP2 depletion had no effect on VASP localization to F-actin cores of invadopodia-like protrusions (Fig. 4, A and B), supporting the hypothesis that proteins other than SHIP2 recruit VASP to these F-actin–rich protrusions. Hence, the distinct pattern of Mena localization at invadopodia compared with VASP may reflect a differential dependence on SHIP2 for recruitment of Mena.
The ability of Mena and SHIP2 to colocalize at the ventral cell surfaces, where invadopodia and focal adhesions form, was also evaluated by time-lapse total internal reflection fluorescent microscopy analysis. Mena was initially observed predominantly at the focal adhesions, whereas SHIP2 was mainly targeted to distinct punctate structures (Fig. S5 C). Analysis of time-lapse images revealed that over time, Mena accumulates within punctate SHIP2-positive regions, consistent with SHIP2 recruiting Mena to these ventral cell protrusions (Fig. S5 C).
To establish whether SHIP2 is sufficient to recruit Mena to specific subcellular locations, we targeted SHIP2 to mitochondria and examined its ability to redirect recruitment of Mena. In these experiments, we used WT-SHIP2 and SHIP2ΔFP4 fused with the mitochondria-targeting sequence (presequence) of Su9 from ATPase subunit 9 of Neurospora crassa (Frederick et al., 2008). Localization of both proteins at mitochondria was established by costaining with a mitochondrial dye (MitoTracker red; Fig. S5 E). When coexpressed with mitochondria-targeted wild-type SHIP2 (Mito-SHIP2), Mena readily accumulated at the mitochondria, whereas it failed to localize to mitochondria in cells expressing Mito-SHIP2ΔFP4 and instead predominantly localized to focal adhesions (Fig. 4 F). In contrast, VASP, when coexpressed with Mito-SHIP2, failed to localize to mitochondria (Fig. S5 F). These observations are consistent with the biochemical evidence that the EVH1 domain of Mena is sufficient and necessary for interaction of Mena via the FP4 motif of SHIP2 (Fig. 4 E). Together, these data support that the FP4 motif of SHIP2 is required and sufficient for recruitment of Mena to specific SHIP2-enriched subcellular locations such as invadopodia.
SHIP2-mediated recruitment of Mena stabilizes invadopodia protrusions
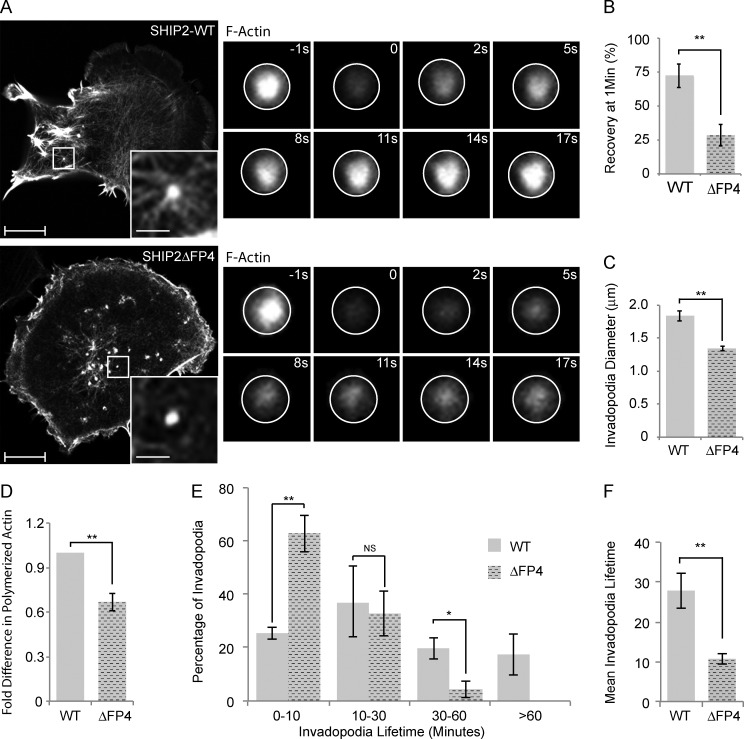
To understand the functional consequences of SHIP2-dependent recruitment of Mena to invadopodia, we performed a series of short (1 frame/5 s for 5 min) time-lapse video microscopy analyses of the dynamics of invadopodia formation in SHIP2-depleted cells. Expression of either WT-SHIP2 or SHIP2ΔFP4 supported formation of ventral F-actin–rich protrusions. However, in cells expressing the SHIP2ΔFP4 mutant, many of the newly formed F-actin–rich protrusions were transient in nature, breaking into smaller protrusions and rapidly disappearing (Video 10), whereas cells expressing WT-SHIP2 formed stable F-actin–rich protrusions (Video 10). Because Mena is an actin filament elongation factor, we assessed F-actin content. Overall F-actin content in MDA-MB-231-shSHIP2 cells rescued with SHIP2ΔFP4 was 33% lower compared with cells rescued with WT-SHIP2 (Fig. 5 D). Taking into account that Mena forms tetramers and functions as an actin filament elongation factor that clusters filaments at barbed ends (Krause et al., 2003), we hypothesized that inefficient recruitment of Mena to invadopodia precursors diminishes elongation of actin filaments required to support membrane protrusion. To address this possibility, cells expressing LifeAct, a fluorescent probe that binds to filamentous but not monomeric actin, were subjected to FRAP analysis, using fluorescence recovery as a proportional readout for actin filament elongation. SHIP2ΔFP4-expressing cells exhibited a 60% decrease in the rate of recovery after photobleaching (28% recovery after 50 s) compared with WT-SHIP2 cells (72% recovery after 50 s; Fig. 5, A and B). Hence, our data indicate that failure to recruit Mena leads to delayed elongation of actin filaments within nascent invadopodia. Moreover, the actin-rich invadopodia-like protrusions that formed in SHIP2ΔFP4-expressing cells were smaller in diameter (27%) than those that formed in WT-SHIP2–expressing cells (Fig. 5 C), consistent with defective assembly of the actin network within invadopodia precursors.
Figure 5.
SHIP2 recruits Mena to regulate the actin network within the invadopodia membrane protrusions, hence invadopodia stability. (A) SHIP2-knockdown cells rescued with either WT-SHIP2 or SHIP2ΔFP4 were transiently transfected with tagRFP-LifeAct and subjected to FRAP analysis on F-actin–rich protrusions. Time series of WT-SHIP2 and SHIP2ΔFP4 rescued cells illustrate the delay in fluorescence recovery. (B) FRAP analysis was performed as described in A, and fluorescence recovery of 30 invadopodia protrusions acquired from three independent experiments from WT-SHIP2– and SHIP2ΔFP4-rescued cells was quantified 50 s after photobleaching. (C) SHIP2-knockdown cells rescued with either WT-SHIP2 or SHIP2ΔFP4 were transiently transfected with tagRFP-LifeAct and subjected to live time-lapse imaging, and the diameter of at least 200 invadopodia-like protrusions from three independent experiments were measured. (D) Filamentous actin content was measured from SHIP2-knockdown cells rescued with either WT-SHIP2 or SHIP2ΔFP4 and expressed as fold change. (E) SHIP2-knockdown cells rescued with either WT-SHIP2 or SHIP2ΔFP4 were transiently transfected with tagRFP-LifeAct and subjected to time-lapse video microscopy. Invadopodia lifetime was analyzed for 120 invadopodia from cells rescued with WT-SHIP2 and 85 invadopodia from cells rescued with SHIP2ΔFP4 and expressed as binned data. (F) As in E, mean lifetime was calculated for invadopodia formed in cells rescued with WT-SHIP2 and SHIP2ΔFP4. Invadopodia with lifetimes of less than 1 hour were considered for this analysis. All quantified data indicate the mean values ± SE from at least three independent experiments. *, P < 0.05; **, P < 0.01; NS, not statistically significant. Bars: 10 µm; (inset) 2 µm.
To determine whether delayed elongation of actin filaments at nascent protrusions results in shorter invadopodia lifetime, we performed time-lapse imaging (1 frame/min for 1 h) of MDA-MB-231-shSHIP2 cells transiently expressing LifeAct and rescued with either WT-SHIP2 or SHIP2ΔFP4. Analysis of time-lapse imaging revealed that SHIP2ΔFP4 promotes invadopodia that have shorter lifetimes compared with WT-SHIP2. Whereas 18% of F-actin protrusions that formed in MDA-MB-231-shSHIP2 cells rescued with WT-SHIP2 displayed a lifetime of >1 h, all F-actin protrusions formed by cells rescued with SHIP2ΔFP4 displayed a lifetime of <1 h (Fig. 5 E), and the majority (63%) of invadopodia in SHIP2ΔFP4 cells had a lifetime of 0–10 min (Fig. 5, E and F). Collectively, these data demonstrate that SHIP2-mediated recruitment of Mena contributes to actin filament elongation at invadopodia sites and consequently increases stability of these structures.
Mena, but not VASP, contributes to invadopodia-dependent ECM proteolysis and cell invasion
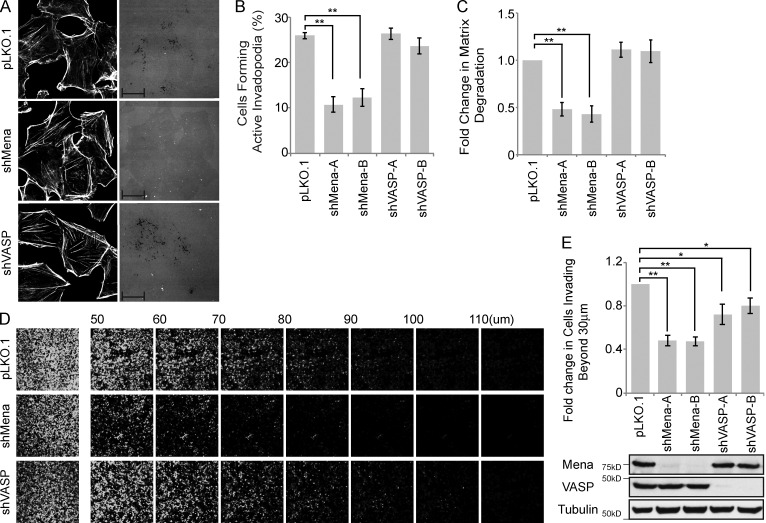
Given that the SHIP2ΔFP4 mutant is unable to rescue invadopodia-mediated ECM degradation and that SHIP2 depletion abrogates localization of Mena, but not VASP, to nascent invadopodia protrusions, we hypothesized that Mena but not VASP may play a critical role downstream from SHIP2 in invadopodia maturation and ECM degradation. To test this possibility, using lentiviral transduction, we established stable cell populations of MDA-MB-231 cells expressing two different shRNA duplexes targeting Mena or VASP. Stable depletion of Mena led to a significant decrease in formation of proteolytically active invadopodia (43% of control; Fig. 6, A and B), ECM degradation (44% of control; Fig. 6, A and C), and cell invasion (47% of control; Fig. 6, D and E), whereas depletion of VASP resulted in a modest decrease in cell invasion (retaining 75% of control; Fig. 6, D and E) but had no significant effect on invadopodia formation (Fig. 6, A–C). Collectively, these data support that Mena, but not VASP, functions downstream from SHIP2 in formation of proteolytically competent invadopodia, resulting in increased invasion of the extracellular matrix.
Figure 6.
Mena, but not VASP, promotes proteolytically active invadopodia formation and cell invasion. (A–C) MDA-MB-231 cells were stably transduced with pLKO.1 or two individual shRNA duplexes targeting Mena or VASP. (A) Cells were seeded on fluorescently labeled gelatin and stained with phalloidin (F-actin), and confocal images were acquired. (B) The percentage of VASP- or Mena-knockdown cells that formed proteolytically active invadopodia was quantified in comparison to control cells. (C) Amounts of ECM degradation by VASP- or Mena-knockdown cells were quantified as fold change compared with control cells. (D) MDA-MB-231 cells stably transduced with pLKO.1 or individual shRNA duplexes targeting VASP or Mena were subjected to an inverted invasion assay. Representative confocal z-stacks depict relative cell invasion. (E) For quantification of cell invasion, five confocal z-stacks per condition per experiment were acquired. Fluorescence intensity of the planes within each z-stack was measured using Cell Profiler software. Cells invading to and past 30 µm inside the matrix were quantified as percentage of total cell population. Experiments were repeated four times. Western blot reveals the relative protein levels of VASP and Mena between different MDA-MB-231 cells stably transduced with pLKO.1 and individual shRNA duplexes targeting VASP or Mena. All quantified data indicate the mean values ± SE from at least three independent experiments. *, P < 0.05; **, P < 0.01. Bars: 10 µm; (inset) 2 µm.
Ring-like localization of Mena contributes to proteolytic activity
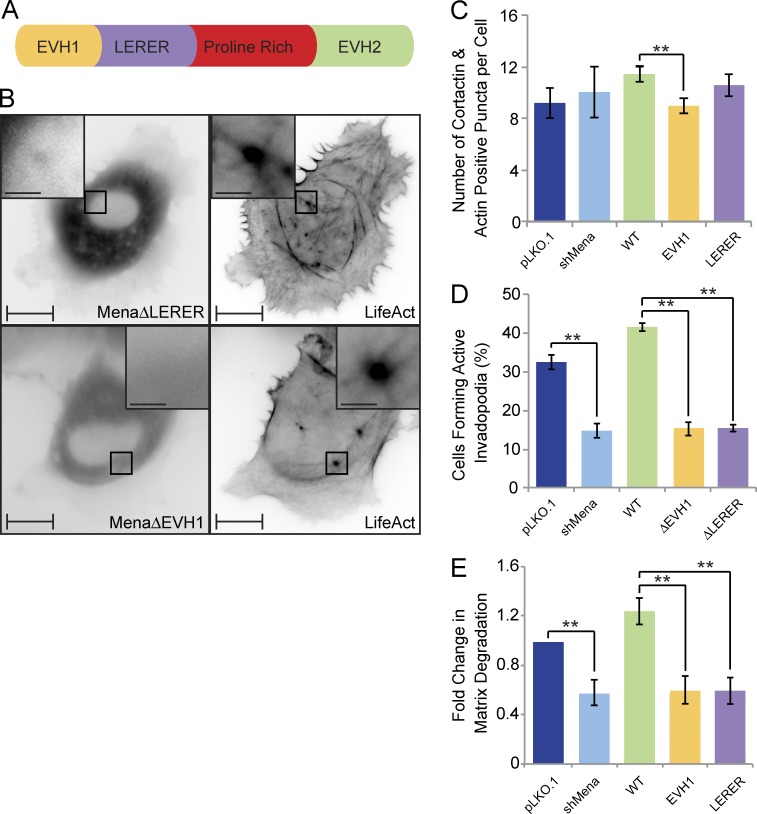
All Ena/VASP proteins have similar domain organization and share several interaction partners; however, Mena has features that are not found in VASP and hence may be required for invadopodia-related functions. Among these are LERER repeats located between the EVH1 domain and the proline-rich sequence of Mena (Fig. 7 A). To determine which domains within Mena contribute to its targeting to invadopodia and invadopodia maturation into ECM-degrading structures, we generated Mena mutant proteins lacking either the EVH1 domain (MenaΔEVH1) or LERER repeats (MenaΔLERER) and examined their localization at the ventral F-actin–rich punctae. When expressed in Mena-depleted MDA-MB-231 cells, WT-Mena is enriched in a ring surrounding the distal ends of F-actin cores. In contrast, MenaΔEVH1 failed to localize to the F-actin punctae (Fig. 7 B), and MenaΔLERER failed to target to ring-like structures and was found predominantly at the F-actin core in a pattern reminiscent of VASP localization (Fig. 7 B). Collectively, these data support that the EVH1 domain is required for recruitment of Mena to F-actin–positive membrane protrusions, whereas LERER repeats are required for retention of Mena at the distal ends of the F-actin cores.
Figure 7.
Mena is recruited to invadopodia via its EVH1 domain and requires the LERER sequences for its ring-like localization and proteolytic activity. (A) Schematic diagram of Mena depicts its EVH1 domain, LERER repeats, proline-rich region, and the EVH2 domain. (B) MDA-MB-231-shMena cells were transiently transfected with tagRFP-LifeAct and GFP-MenaΔEVH1 or GFP-MenaΔLERER and subjected to time-lapse video microscopy. Single frame illustrates GFP-MenaΔEVH1 and GFP-MenaΔLERER localization with respect to F-actin cores of invadopodia protrusions. (C) MDA-MB-231-pLKO.1, MDA-MB-231-shMena, and MDA-MB-231-shMena cells transiently rescued with WT-Mena, MenaΔEVH1, or MenaΔLERER were seeded on unconjugated gelatin matrix and immunostained for invadopodia markers (F-actin and cortactin), and confocal images were acquired at the ventral cell surface. The number of F-actin– and cortactin-positive protrusions per cell was quantified. (D) MDA-MB-231-pLKO.1, MDA-MB-231-shMena, and MDA-MB-231-shMena cells transiently rescued with WT-Mena, MenaΔEVH1, or MenaΔLERER were seeded on fluorescent gelatin matrix and immunostained with phalloidin (F-actin), confocal images were acquired at the ventral cell surface, and the percentage of cells that formed proteolytically active invadopodia (positive for F-actin punctate overlaying matrix degradation area) was quantified. (E) As in D, invadopodia assay was performed, and the amount of ECM degradation (mean matrix degradation area per fields of view) by MDA-MB-231-pLKO.1, MDA-MB-231-shMena, and MDA-MB-231-shMena cells transiently rescued with WT-Mena, MenaΔEVH1, or MenaΔLERER was quantified as fold change compared with control (pLKO.1) cells. All quantified data indicate the mean values ± SE from at least three independent experiments. **, P < 0.01. Bars: 10 µm; (inset) 2 µm.
To determine the functional consequences of differential localization of Mena within invadopodia (ring-like vs. punctate), we rescued MDA-MB-231-shMena cells with WT-Mena, MenaΔEVH1, or MenaΔLERER and assayed transfected cells for their ability to form proteolytically active invadopodia and degrade ECM. Stable depletion of Mena in invasive MDA-MB-231 breast cancer cells resulted in no significant difference in the number of F-actin– and cortactin-positive punctae (Fig. 7 C), although the punctae per se appear smaller (not depicted). However, loss of Mena led to a 52% reduction in the number of cells with proteolytically active invadopodia (Fig. 7 D) and a 42% decrease in overall matrix degradation capacity (Fig. 7 E). In support of this result, reexpression of WT-Mena rescued both invadopodia formation (≈129% of control) and ECM degradation (≈126% of control) to levels similar to or greater than those observed in control cells expressing endogenous Mena (Fig. 7, D and E). Reexpression of neither MenaΔEVH1 nor MenaΔLERER fully rescued invadopodia formation (47% and 46% of control, respectively) and ECM degradation (60% and 61% of control, respectively; Fig. 7, D and E). MenaΔLERER failed to rescue invadopodia-mediated matrix degradation despite recruitment to the F-actin cores of nascent invadopodia, implying a role for the LERER repeats in MMP secretion. Collectively, these data support that the LERER repeats retain Mena at the distal ends of the F-actin–rich cores of invadopodia and promote recruitment of MT1-MMP to invadopodia sites.
SHIP2 phosphatase activity and proline-rich sequence are required for cell invasion in vivo
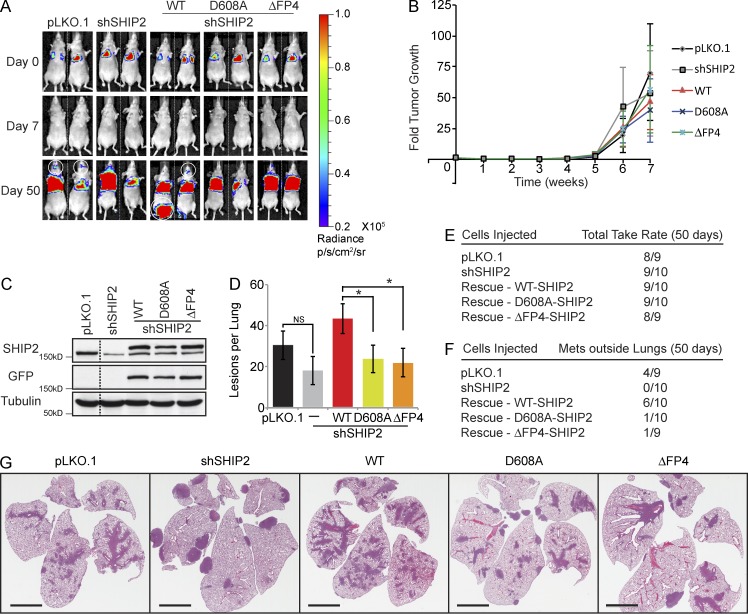
To assess the in vivo relevance of SHIP2–Mena interaction for cancer cell invasion and metastasis, we used the bone-specific metastatic variant of MDA-MB-231 cells (1833-TR) stably tagged with a luciferase reporter (Kang et al., 2003) that allows visualization of tumor growth. Stable populations of 1833-TR cells expressing SHIP2 shRNA or vector were established, and SHIP2-depleted cells were rescued to similar levels of expression with WT-SHIP2, the catalytic inactive SHIP2 mutant (SHIP2-D608A), or SHIP2 uncoupled from Mena (SHIP2ΔFP4; Fig. 8 C). All of the resulting cell lines were able to colonize the lungs when injected in the tail vain of nude mice, with detectable tumor growth starting 5 wk after injection (Fig. 8, A and B). In all cases, ∼90% of mice developed lung tumors with similar growth rates (Fig. 8, B and E). Microscopic examination of lung lesions by hematoxylin and eosin staining revealed that although total tumor volume was similar across all conditions, numbers and gross morphology of the lesions were distinct (Fig. 8, D and G). Specifically, mice injected with SHIP2-depleted cells and rescued with SHIP2-D608A or SHIP2ΔFP4 developed fewer lung lesions (46%) and a preference for growth in peripheral capillaries compared with mice injected with cells rescued with WT-SHIP2 (Fig. 8, D and G).
Figure 8.
SHIP2 phosphatase activity and capacity to interact with Mena are required for cell invasion in vivo. (A) Mice injected (tail vein) with a MDA-MB-231-1833-TR bone metastatic variant expressing vector (pLKO.1), SHIP2 shRNA, or SHIP2 shRNA plus GFP-SHIP2-WT, GFP-SHIP2-D608A, or GFP-SHIP2ΔFP4 were imaged to detect luciferase activity. Bioluminescence images of two representative mice per condition acquired immediately and 1 and 7 wk after injection are depicted. (B) Tumor growth within lungs, for mice injected with cells as indicated in A, was quantified and expressed as fold change of initial inoculum on day 0. (C) SDS-PAGE followed by Western blot analysis depicts protein levels of SHIP2 between MDA-MB-231-1833-TR cells transduced with pLKO.1 and shSHIP2 as well as exogenous rescue levels of GFP-SHIP2-WT, GFP-SHIP2-D608A, and GFP-SHIP2ΔFP4. (D) Number of lung lesions per mouse was quantified based on hematoxylin and eosin staining of serial step sections of paraffin embedded tissue. (E) Table summarizes the cells injected via tail vein and their respective take rate for tumor growth at the lungs. (F) Table summarizes the cells injected via tail vein and their respective tumor spread and growth rate outside lungs. (G) Representative hematoxylin and eosin staining of a single section of all five lobes of mouse lung shows characteristic lesions among different conditions. All quantified data indicate the mean values ± SE. *, P < 0.05; NS, not statistically significant. Bars, 4 mm.
Although incidence and growth rate of tumors in lungs were similar across all conditions, differences were observed in the ability of the rescued cell lines to escape the lungs and colonize other organs, predominantly the bones (Fig. 8, A and F). Whereas ∼40% of mice injected with 1883-TR-pLKO.1 cells developed bone lesions, mice injected with SHIP2-depleted cells had no detectable lesions (Fig. 8 F). Reexpression of WT-SHIP2 resulted in an increase of bone lesion incidence to ∼60%, whereas expression of SHIP2-D608A or SHIP2ΔFP4 resulted in only ∼10% of injected mice developing bone lesions (Fig. 8 F), consistent with decreased potential of these SHIP2 mutants to support invadopodia formation and ECM degradation. Hence, collectively, our data demonstrate a requirement for SHIP2 for cell invasion and metastasis in vivo that requires both intact phosphatase activity and its function as a molecular scaffold to recruit Mena.
Discussion
Elevated expression of the 5′-inositol phosphatase SHIP2 is observed in aggressive cancers and is associated with increased invasive capacity (Prasad et al., 2008; Fu et al., 2013, 2014; Yang et al., 2014), yet its role in cell invasion is still poorly understood. Here we identify a critical regulatory function for SHIP2 in maturation of invadopodia into proteolytically active structures that enhance invasive capacity in 3D cultures in vitro and in vivo. This was achieved by analyzing invadopodia formation and maturation and cancer cell invasion under conditions in which SHIP2 levels were depleted and rescued with SHIP2 mutants. These multiple approaches yielded quantitative, complementary results that support a model wherein cancer cell invasion requires both the lipid phosphatase activity of SHIP2 and an unexpected role for SHIP2 as a scaffold protein that recruits the actin regulatory protein Mena to invadopodia. The localization of a SHIP2/Mena complex to invadopodia is dependent on the SHIP2 SH2 domain and its ability to couple with tyrosine-phosphorylated cortactin, a key protein in invadopodia formation. SHIP2-dependent recruitment of Mena to cortactin within nascent invadopodia is required for invadopodia maturation and recruitment of matrix metalloproteases, which is required for matrix degradation and cell invasion.
SHIP2 uses the PI3K product phosphoinositide, PtdIns(3,4,5)P3, as a high-affinity substrate to yield PtdIns(3,4)P2. Nascent invadopodia (early membrane protrusion) are predominantly enriched in PtdIns(3,4,5)P3, which suffices to recruit the regulators of the Arp2/3 actin nucleation complex, cortactin and N-WASP, and hence trigger the nucleation of actin filaments at sites of invadopodia initiation. A local switch in phosphoinositide composition from PtdIns(3,4,5)P3 to PtdIns(3,4)P2 at nascent invadopodia provides docking sites for the Tks4/Tks5 family of scaffolding proteins (Sharma et al., 2013). Recruitment of Tks5 to PtdIns(3,4)P2-enriched invadopodia membranes via its PX domain anchors the Arp2/3 complex via interactions with Nck and N-WASP and, as a result, promotes nucleation of actin filaments that sustain membrane protrusion and subsequent delivery of metalloprotease MT1-MMP to invadopodia (Stylli et al., 2009).
This process is consistent with our observations that SHIP2 is indispensable for invadopodia stability and maturation and SHIP2 catalytic activity contributes to the recruitment and anchoring of Tks5. In SHIP2-depleted cells rescued with a lipid phosphatase inactive mutant of SHIP2 (SHIP2-D608A), a significant percentage of nascent cortactin- and F-actin–positive invadopodia are negative for Tks5 (43% compared with only 9% rescued with WT-SHIP2; Fig. 2). Cortactin accumulation precedes Tks5 recruitment to nascent invadopodia, and tyrosine phosphorylation of cortactin is required for invadopodia maturation (Magalhaes et al., 2011; Beaty et al., 2013; Sharma et al., 2013). We identified a specific interaction between SHIP2 and cortactin that is dependent on tyrosine phosphorylation of cortactin and an intact SH2 domain of SHIP2, implying a role for cortactin in recruiting SHIP2 to nascent invadopodia. In support of this, a SHIP2 mutant lacking a functional SH2 domain (SHIP2ΔFP4-R47G) is not recruited to nascent F-actin–positive invadopodia protrusions (Figs. 2 and S3; Video 2), consistent with decreased conversion of PtdIns(3,4,5)P3 into PtdIns(3,4)P2 and decreased recruitment of Tks5 (Fig. 2).
Structure–function analyses revealed that SHIP2 contains a consensus binding sequence for the EVH1 domain of the Ena/VASP family of actin elongation factors (Bear and Gertler, 2009). The actin filament elongation factors, Ena/VASP family proteins Mena and VASP, are known to localize at invadopodia, yet modes of their recruitment to invadopodia had not been elucidated (Philippar et al., 2008; Schoumacher et al., 2010). A SHIP2 mutant lacking the Ena/VASP consensus binding sequence (SHIP2ΔFP4) fails to induce invadopodia capable of ECM degradation (Fig. 1). Moreover, we demonstrate a direct interaction between SHIP2 and Mena/VASP and provide evidence that SHIP2 localizes Mena but not VASP to invadopodia (Figs. 3 and 4). Depletion of SHIP2 decreases Mena, but not VASP, localization within invadopodia, and a mitochondria-targeted SHIP2 recruits Mena, but not VASP, to mitochondria (Fig. 4). These results support a specific role for Mena downstream from SHIP2 in invadopodia biogenesis. Invadopodia protrusions formed in cells expressing SHIP2ΔFP4 were smaller in diameter than those of cells expressing WT-SHIP2, perhaps because of impaired capacity to elongate actin filaments that sustain membrane protrusion (Fig. 5). Because the ability of both Mena and VASP to bind SHIP2 relies on their EVH1 domains, differential affinity of the Mena and VASP EVH1 domains for the FP4 consensus sequence within SHIP2 may be a likely explanation that calls for further investigation. In support of this, the tumor suppressor protein, Tes, shows selective interaction with Mena over the VASP EVH1 domain through the Tes LIM domain (Boëda et al., 2007; Small, 2008).
Localization of Mena and VASP at invadopodia and their main function as actin elongation factors pose the question of their functional redundancy during invadopodia biogenesis. In our experiments, depletion of Mena, but not VASP, abrogated invadopodia-mediated matrix remodeling (Fig. 6). Nonetheless, depletion of VASP was associated with a less pronounced reduction in cell-invasive potential than depletion of Mena. This observation may reflect involvement of VASP in alternative protease-independent mechanisms of invasion.
Increased expression of Mena is found in high-risk preneoplastic lesions and correlates with augmented tumor invasiveness and poor prognosis in breast cancer patients (Gertler and Condeelis, 2011; Oudin et al., 2016a). Precise mechanistic functions of Mena in targeted delivery of vesicular MMPs to nascent invadopodia sites remain unclear. One model argues that Mena recruitment increases elongation of actin filaments within invadopodia structures, leading to membrane protrusion that in turn serves as a signal to trigger targeted delivery of MMPs. Another possibility is that the ability of Mena to couple targeted delivery of MMPs to nascent invadopodia protrusions is determined by the presence of additional sequence features that are not found in VASP, such as the LERER repeats between the EVH1 domain and the PRS. In support of this, the integrin αVβ3 complex, like VASP, localizes to invadopodia as punctae, whereas the α5β1 complex localizes as a ring overlapping the punctae (Mueller et al., 1999; Branch et al., 2012). Intriguingly, the ring-like distribution of Mena within invadopodia is reminiscent of the adhesion rings described by Branch et al. (2012). Formation of these adhesion rings is highly dependent on integrin activity and correlates with targeted delivery of MT1-MMP and capacity to proteolytically remodel ECM (Branch et al., 2012). Mena can bind the cytoplasmic tails of α5 integrin via LERER repeats and activate inside-out α5β1 integrin signaling (Gupton and Gertler, 2010; Oudin et al., 2016b). Mena-mediated activation of the α5β1 integrin complex promotes exocytosis initiated by the exocyst complex containing Exo70 and VAMP7, both of which can regulate MMP secretion at invadopodia (Sakurai-Yageta et al., 2008; Steffen et al., 2008; Gupton and Gertler, 2010; Monteiro et al., 2013). In view of these findings, the ring pattern of Mena localization within invadopodia may reflect its potential requirement for inside-out activation of α5β1 integrin complexes, coupling vesicular trafficking of MMPs to invadopodia protrusions.
Mena mRNA has been reported to undergo alternative splicing events, resulting in expression of multiple tissue-specific protein isoforms that can elicit differential biological responses (Gertler and Condeelis, 2011). The most characterized Mena isoforms are MenaINV and MenaΔv6. MDA-MB-231 invasive breast cancer cells express a specific Mena isoform that lacks exon 6 (MenaΔv6). Expression of this isoform is associated with increased cell motility (Di Modugno et al., 2012). Therefore, it is possible that, in MDA-MB-231 cells, MenaΔv6 cooperates with SHIP2 to promote invadopodia-mediated matrix remodeling and cell invasion. In addition, circulating tumor cells, micrometastatic lesions, and the invasive front of bulk primary tumor also express another isoform of Mena, MenaINV, which includes a 19-aa exon (Goswami et al., 2009; Gertler and Condeelis, 2011; Oudin et al., 2016a). Expression of MenaINV has only been observed in vivo, implying its regulation by the tumor microenvironment (Gertler and Condeelis, 2011). Depletion of SHIP2 had no effect on the ability of MDA-MB-231 cells to colonize lungs in nude mice upon tail vein injection but decreased the ability of tumor cells to subsequently escape the lungs, enter the bloodstream, and colonize distant organs (mainly bone). These findings support a role for SHIP2 in cell invasion in vivo (Fig. 8). Moreover, whereas expression of WT-SHIP2 in SHIP2-depleted MDA-MB-231 cells rescued this metastatic phenotype, expression of SHIP2 uncoupled from Mena failed to rescue metastasis to the bones (Fig. 8). This may be caused by the inability of SHIP2ΔFP4 to cooperate with Mena or MenaINV and trigger invadopodia-mediated ECM remodeling. In agreement with this rationale, overexpression of MenaINV prolongs invadopodia lifetime in vitro and promotes distant metastasis in vivo (Philippar et al., 2008).
Collectively, our findings reveal a dual role of SHIP2 in coupling nascent actin-rich membrane protrusions to targeted MMP secretion, thus leading to maturation of invadopodia into structures competent for ECM degradation. First, SHIP2 promotes PtdIns(3,4)P2-mediated recruitment of Tks5 that anchors the Arp2/3 actin nucleation complex. Second, it directly recruits Mena to concomitantly elongate newly nucleated actin filaments, thereby regulating the architecture of the actin network that sustains membrane protrusion. Therefore, pharmacological disruption of SHIP2–Mena interaction may hold a therapeutic potential for metastatic disease. In summary, our work provides novel insights into the molecular mechanisms governing invadopodia biogenesis and identifies new avenues for specific therapeutic targeting of SHIP2 invadopodia-related functions.
Materials and methods
Cell culture and transfection
HEK293, SKBr3, and MDA-MB-231 cells were maintained in DMEM containing 10% FBS. Transient transfections of HEK293 cells were performed using Lipofectamine/Plus reagent, and transfections of MDA-MB-231 and SKBr3 cells were performed using Lipofectamine 2000, according to the manufacturer’s instructions (Invitrogen). The following shRNA sequences (Thermo Fisher Scientific) were used to establish stable SHIP2 depletion: shSHIP2-A (5′-CCGGGCAGCTCAATGCCTTTGACATCTCGAGTGTCAAAGGCATTGAGCTGCTTTTTG-3′), shSHIP2-B (5′-CCGGCCTGAACTACATCAGCAGGAACTCGAGTTCCTGCTGATGTAGTTCAGGTTTTTG-3′), and shSHIP2-C (5′-CCGGCCGGATTCTGTGGAAATCCTACTCGAGTAGGATTTCCACAGAATCCGGTTTTTG-3′). Cells stably expressing shSHIP2-C were used to establish stable rescue lines.
Antibodies and reagents
Commercial antibodies to SHIP2, Mena, and VASP (rabbit) were obtained from Cell Signaling Technology; SHIP2 and VASP (mouse) from Santa Cruz Biotechnology, Inc.; vinculin, actin, and tubulin from Sigma-Aldrich; GFP antibody, phalloidin Alexa Fluor 546, and Alexa Fluor 488– and 555–conjugated secondary antibodies from Thermo Fisher Scientific; and HA.11 monoclonal antibody from Covance. HA-SHIP2 and GFP-SHIP2 were gifts from C.A. Mitchell (Monash University, Clayton, Australia) and K. Kaibuchi (Nagoya University, Nagoya, Japan), respectively. GFP-VASP and GFP-Mena were gifts from F. Gertler (Massachusetts Institute of Technology, Cambridge, MA). pLifeAct-TagRFP was purchased from Ibidi. Point mutants of pEGFP-N3-SHIP2 were generated by site-directed mutagenesis using a kit from Stratagene (Agilent Technologies). Deletion mutants of pEGFP-N3-SHIP2, GFP-VASP, and GFP-Mena were generated by PCR amplification. mCherry-Mena and pLXSN-SHIP2 WT and mutants were generated by subcloning. Mitochondria targeting vector (MITO-GFP) was generated by subcloning the presequence of Su9 from ATPase subunit 9 of N. crassa into pGFP-C2 vector (Frederick et al., 2008). MITO-SHIP2 was generated by PCR amplification of SHIP2 and insertion into the EcoRI and XhoI sites of the MITO-GFP vector. SHIP2 deletion mutants lacking aa 18–108 (SHIP2ΔSH2) and 1,048–1,059 (SHIP2ΔFP4) were generated by PCR.
Immunoprecipitation and Western blotting
Cells were harvested in lysis buffer (150 mM NaCl, 20 nM Tris-HCl, 1 mM EDTA, 1 mM EGTA, 1% Triton X-100, and 1% sodium deoxycholate, pH 7.4) supplemented with 1 mM PMSF, 1 mM sodium vanadate, 1 mM sodium fluoride, 10 µg/ml aprotinin, and 10 µg/ml leupeptin. For immunoprecipitation, lysates were incubated with the indicated antibodies for 2 h at 4°C with gentle rotation followed by the addition of 30 µl of a 50% slurry of either protein A–Sepharose or protein G–Sepharose (GE Healthcare) and an additional hour of gentle rotation before the collection of immune complexes. The resulting immune complexes were washed three times with lysis buffer, eluted with SDS sample buffer, and resolved by SDS-PAGE. Bound proteins were visualized using an ECL detection kit (GE Healthcare) as described previously (Rajadurai et al., 2012).
GST pull-down assay
Cell lysates were precleared with glutathione-Sepharose beads for 1 h at 4°C, and 5 µg fusion protein was immobilized on 25 µl glutathione-Sepharose beads at RT. MDA-MB-231 cell lysates were incubated with glutathione-Sepharose beads loaded with GST alone, GST-VASP-EVH1, or GST-Mena-EVH1 for 2 h at 4°C. The resulting samples were washed three times with lysis buffer, eluted with SDS sample buffer, resolved by SDS-PAGE, and subjected to Western blotting.
Inverted invasion assay
Inverted invasion assays were performed as described previously (Caswell et al., 2007). In brief, modified Boyden chambers with 8-µm membrane pore size were coated with a mixture of Matrigel (BD) and acid-purified rat-tail collagen (Gibco) at final concentrations of 2.5 mg/ml of each for 2 h at 37°C. Inserts were then inverted, and 106 cells were seeded directly onto the opposite surface of the filter and allowed to adhere for 4 h at 37°C and 5% CO2. After 4 h, filters were inverted, and the top compartments of the chambers were filled with DMEM plus 10% FBS, and the bottom compartments with plain DMEM. Cells were allowed to invade through the matrix for 72 h and were stained with Calcein-AM (Invitrogen) according to the manufacturer’s instructions; cells that did not enter the matrix were removed using cotton swabs. Invading cells were visualized by confocal microscopy, and serial confocal sections (z-stacks) were acquired at 10-µm intervals. The fluorescence intensity reflecting the number of invading cells was then measured for each section using Cell Profiler software (Stöter et al., 2013), and cells invading into the matrix at least 30 µm and beyond were quantified as the percentage of total cell population within the matrix.
Fluorescent gelatin degradation assay
Glass coverslips were coated with poly-d-lysine for 20 min, washed three times with PBS, incubated for 20 min in 0.5% glutaraldehyde (Sigma-Aldrich), and washed three times with PBS. Fluorescein-conjugated gelatin (Invitrogen) was diluted to 20 µg/ml in 0.1% unconjugated gelatin (STEMCELL Technologies), and the resulting mixture was incubated on coverslips at 37°C for 1 h. Coverslips were then washed three times with PBS and quenched in 5 mg/ml sodium borohydride. Coverslips were washed again three times with PBS and sterilized in 70% ethanol for 20 min. Finally, coverslips were washed with DMEM, and 5 × 104 cells were seeded and incubated at 37°C and 5% CO2 for 24 h, unless specified otherwise. Cells were fixed with 4% PFA and stained according to accepted immunofluorescence protocol.
Live cell imaging
Live cell imaging was performed as described previously (Rajadurai et al., 2012) using an Axiovert 200 microscope (ZEISS) and Apochromat 1.4-NA 100× objective. In brief, cells grown on the 35-mm glass-bottom dishes (Ibidi) coated with either fluorescently labeled or nonconjugated gelatin were positioned on the motorized stage on the inverted microscope Axiovert 200M (ZEISS), equipped with a 100× planApo 1.4-NA objective, AxioCam HRM (ZEISS) digital camera, and a small transparent environmental chamber Climabox (ZEISS) with 5% CO2 at 37°C. The microscope was controlled by the AxioVision LE software (ZEISS). Images were collected at set time intervals and analyzed using the MetaMorph software.
FRAP
Images were acquired using a 63×, 1.7-NA oil-immersion objective on a 710 laser scanning confocal microscope (ZEISS). Cells were kept in a heated (37°C) CO2 (5%) chamber throughout the experiment. The 561-nm laser line was used to excite RFP. Photobleaching was achieved by 100% laser illumination of a region of interest. Low-intensity illumination (2%) was used to record fluorescence before and after bleaching for a total time period of 5 min. Mean pixel intensity was evaluated using ZEN2009/2011 software. Data were corrected for background fluctuations and bleaching artifacts and normalized to prebleaching intensity. The recovered fluorescence intensity was calculated as follows: (IP − I0)/(II − I0), where II is the initial intensity measured before bleaching, I0 is the immediate fluorescence intensity after bleaching, and IP is the postbleaching values of the recovery intensities. Fluorescence recovery was calculated 60 s after photobleaching for graphical representation.
F-actin measurement
F-Actin was quantified as previously described (Diakonova et al., 2002). In brief, 7 × 105 cells per well were plated in six-well dishes. After 5 h, cells were fixed in 4% PFA for 20 min, permeabilized with 0.1% Triton-100 for 20 min, and stained with 250 nM rhodamine phalloidin in PHEM buffer for 30 min in the dark. After three washes with PBS, cells were scraped off with 700 µl methanol and extracted for 24 h at −20°C. Extracted rhodamine-phalloidin was measured at 540 nm using a spectrofluorometer.
Tail vein injection and metastatic tumor growth in vivo
Female nude mice (6–8 wk old) were injected into the tail vein with 105 MDA-MB-231-1833-TR cells expressing pLKO.1 vector, shRNA against SHIP2, and, in addition to SHIP2 shRNA, exogenous GFP-SHIP2-WT, GFP-SHIP2-D608A, or GFP-SHIP2ΔFP4. Luciferase activity in the lungs was confirmed immediately after injection using an IVIS 100 (Caliper Life Sciences) bioluminescence imaging system as previously described (Knight et al., 2013). Mice were imaged once a week thereafter, and luciferase activity was monitored to detect tumor growth in the lungs and distant metastasis to other organs. Luminescence signal within the lungs was measured and quantified as fold change of initial inoculum as described in Mourskaia et al. (2012). Mice were sacrificed 8 wk after injection, and distant metastasis to other organs, including bones and lymph nodes, was evaluated. Tissues with tumorigenic/metastatic lesions (lungs, mandibles, vertebrae, sternum, and tibia/fibula) were collected at necropsy for histological analysis. Tissue samples were fixed in either 10% formalin (lungs and lymph nodes) or 4% PFA (bones), embedded in paraffin, and sectioned (50-µm step sections). Tissue sections were stained with hematoxylin and eosin and imaged using an Aperio-XT slide scanner (Aperio). The number of lesions throughout the lung (step sections) was quantified. All animal experiments were performed in accordance with the guidelines of the McGill University animal ethics committee and the Canadian Council on Animal Care as approved by the facility animal care committee (protocol #5562).
Statistical analyses
All statistical analyses were performed using Microsoft Excel. Graphed data represent the mean values ± standard error (SE) from at least three independent experiments. Two-tailed, paired Student’s t test was used to determine the statistical significance unless otherwise specified.
Online supplemental material
Fig. S1 shows the ability of SHIP2-depleted MDA-MB-231 cells to form functional invadopodia and invade the ECM. Fig. S2 shows the migratory capacity of SHIP2-depleted MDA-MB-231 cells. Fig. S3 shows the localization of various SHIP2 mutants to invadopodia. Fig. S4 shows that intersectin is dispensable for functional invadopodia formation. Fig. S5 shows that whereas VASP localizes to the invadopodia core, Mena localizes as a ring at the distal tip of invadopodia core. It also shows that the differential localization of Mena and VASP is caused by the ability of SHIP2 to recruit Mena, but not VASP, to invadopodia. Video 1 shows that SHIP2 depletion does not affect random cell migration in MDA-MB-231 cells. Video 2 shows that SHIP2ΔFP4-R47G fails to localize to actin-rich invadopodia protrusions. Video 3 shows matrix degradation under F-actin–rich membrane protrusions in MDA-MB-231 cells. Video 4 shows the localization of Mena in a ring around the actin core within invadopodia. Video 5 shows the localization of Mena in a ring around the Tks5 core within invadopodia. Video 6 shows that depletion of SHIP2 affects Mena localization with respect to F-actin core of invadopodia. Video 7 shows that depletion of SHIP2 affects Mena localization with respect to Tks5 core of invadopodia. Video 8 shows that depletion of SHIP2 does not affect Mena localization to Paxillin-positive focal adhesions. Video 9 shows that depletion of SHIP2 does not affect Mena localization to Zyxin-positive focal adhesions. Video 10 shows that a SHIP2 mutant uncoupled from Mena destabilizes actin-rich membrane protrusions at the ventral surface of MDA-MB-231 cells. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201501003/DC1.
Supplementary Material
Acknowledgments
We thank members of the Park laboratory for their helpful comments on the manuscript. We thank Dr. Claire Brown and Dr. Erika Tse-Luen Wee in the Life Sciences Imaging Core for help with live cell imaging and quantification, and Ken McDonald in the FACS Core for his help with cell sorting.
This research was supported by fellowships to C.V. Rajadurai from the Canadian Institutes of Health Research, C.D.H. Ratcliffe and P.P. Coelho from the Fonds de Recherche du Québec - Santé, K. Zaoui from the Susan G. Komen for the Cure, and S. Havrylov from the Canadian Institutes of Health Research/Fonds de Recherche du Québec - Santé training grant in cancer research FRN53888 to the McGill Integrated Cancer Research Training Program. This work was supported by a Foundation operating grant to M. Park from the Canadian Institutes of Health Research (242529). M. Park holds the Diane and Sal Guerrera Chair in Cancer Genetics.
The authors declare no competing financial interests.
Author contributions: C.V. Rajadurai conceived the ideas and hypothesis, designed and performed the majority of the experiments, analyzed the data, and contributed to the writing of the manuscript. S. Havrylov designed the pipeline for automated quantification of matrix degradation and cell invasion and contributed to the writing of the manuscript. P.P. Coelho tracked videos, quantified 2D cell migration, and participated in revisions of the manuscript. C.D.H. Ratcliffe tracked videos, quantified invadopodia lifetime, and participated in revisions of the manuscript. K. Zaoui provided technical assistance with live cell imaging experiments and performed FRAP analysis. B.H. Huang participated in revisions of the manuscript. A. Monast and N. Chughtai provided assistance with animal experiments. V. Sangwan provided assistance with manuscript preparation. F.B. Gertler provided reagents and feedback on experiment design, interpretation of the data, and revision of the written manuscript. P.M. Siegel provided feedback on experiment design and interpretation of the data. M. Park coordinated the study, provided feedback in experiment design and interpretation of the data, and contributed to the writing and revision of the manuscript.
Footnotes
Abbreviations used:
- BM
- basement membrane
- MMP
- matrix metalloprotease
- P2
- bisphosphate
- P3
- triphosphate
- PI3K
- phosphoinositide-3-kinase
- PRS
- proline-rich sequence
- PtdIns
- phosphatidylinositol
- SE
- standard error
- WT
- wild-type
References
- Alexander N.R., Branch K.M., Parekh A., Clark E.S., Iwueke I.C., Guelcher S.A., and Weaver A.M.. 2008. Extracellular matrix rigidity promotes invadopodia activity. Curr. Biol. 18:1295–1299. 10.1016/j.cub.2008.07.090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammer A.G., and Weed S.A.. 2008. Cortactin branches out: Roles in regulating protrusive actin dynamics. Cell Motil. Cytoskeleton. 65:687–707. 10.1002/cm.20296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball L.J., Kühne R., Hoffmann B., Häfner A., Schmieder P., Volkmer-Engert R., Hof M., Wahl M., Schneider-Mergener J., Walter U., et al. 2000. Dual epitope recognition by the VASP EVH1 domain modulates polyproline ligand specificity and binding affinity. EMBO J. 19:4903–4914. 10.1093/emboj/19.18.4903 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzik M., Kotova T.I., Higgs H.N., Hazelwood L., Hanein D., Gertler F.B., and Schafer D.A.. 2005. Ena/VASP proteins enhance actin polymerization in the presence of barbed end capping proteins. J. Biol. Chem. 280:28653–28662. 10.1074/jbc.M503957200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J.E., and Gertler F.B.. 2009. Ena/VASP: Towards resolving a pointed controversy at the barbed end. J. Cell Sci. 122:1947–1953. 10.1242/jcs.038125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bear J.E., Svitkina T.M., Krause M., Schafer D.A., Loureiro J.J., Strasser G.A., Maly I.V., Chaga O.Y., Cooper J.A., Borisy G.G., and Gertler F.B.. 2002. Antagonism between Ena/VASP proteins and actin filament capping regulates fibroblast motility. Cell. 109:509–521. 10.1016/S0092-8674(02)00731-6 [DOI] [PubMed] [Google Scholar]
- Beaty B.T., Sharma V.P., Bravo-Cordero J.J., Simpson M.A., Eddy R.J., Koleske A.J., and Condeelis J.. 2013. β1 integrin regulates Arg to promote invadopodial maturation and matrix degradation. Mol. Biol. Cell. 24:1661–1675: S1–S11. 10.1091/mbc.E12-12-0908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boëda B., Briggs D.C., Higgins T., Garvalov B.K., Fadden A.J., McDonald N.Q., and Way M.. 2007. Tes, a specific Mena interacting partner, breaks the rules for EVH1 binding. Mol. Cell. 28:1071–1082. 10.1016/j.molcel.2007.10.033 [DOI] [PubMed] [Google Scholar]
- Branch K.M., Hoshino D., and Weaver A.M.. 2012. Adhesion rings surround invadopodia and promote maturation. Biol. Open. 1:711–722. 10.1242/bio.20121867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitsprecher D., Kiesewetter A.K., Linkner J., Vinzenz M., Stradal T.E.B., Small J.V., Curth U., Dickinson R.B., and Faix J.. 2011. Molecular mechanism of Ena/VASP-mediated actin-filament elongation. EMBO J. 30:456–467. 10.1038/emboj.2010.348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carl U.D., Pollmann M., Orr E., Gertlere F.B., Chakraborty T., and Wehland J.. 1999. Aromatic and basic residues within the EVH1 domain of VASP specify its interaction with proline-rich ligands. Curr. Biol. 9:715–718. 10.1016/S0960-9822(99)80315-7 [DOI] [PubMed] [Google Scholar]
- Carmona G., Perera U., Gillett C., Naba A., Law A.-L., Sharma V.P., Wang J., Wyckoff J., Balsamo M., Mosis F., et al. 2016. Lamellipodin promotes invasive 3D cancer cell migration via regulated interactions with Ena/VASP and SCAR/WAVE. Oncogene. In press. 10.1038/onc.2016.47 [DOI] [PMC free article] [PubMed]
- Caswell P.T., Spence H.J., Parsons M., White D.P., Clark K., Cheng K.W., Mills G.B., Humphries M.J., Messent A.J., Anderson K.I., et al. 2007. Rab25 associates with α5β1 integrin to promote invasive migration in 3D microenvironments. Dev. Cell. 13:496–510. 10.1016/j.devcel.2007.08.012 [DOI] [PubMed] [Google Scholar]
- Diakonova M., Bokoch G., and Swanson J.A.. 2002. Dynamics of cytoskeletal proteins during Fcgamma receptor-mediated phagocytosis in macrophages. Mol. Biol. Cell. 13:402–411. 10.1091/mbc.01-05-0273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Modugno F., Iapicca P., Boudreau A., Mottolese M., Terrenato I., Perracchio L., Carstens R.P., Santoni A., Bissell M.J., and Nisticò P.. 2012. Splicing program of human MENA produces a previously undescribed isoform associated with invasive, mesenchymal-like breast tumors. Proc. Natl. Acad. Sci. USA. 109:19280–19285. 10.1073/pnas.1214394109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frederick R.L., Okamoto K., and Shaw J.M.. 2008. Multiple pathways influence mitochondrial inheritance in budding yeast. Genetics. 178:825–837. 10.1534/genetics.107.083055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu C.-H., Lin R.-J., Yu J., Chang W.-W., Liao G.-S., Chang W.-Y., Tseng L.-M., Tsai Y.-F., Yu J.-C., and Yu A.L.. 2014. A novel oncogenic role of inositol phosphatase SHIP2 in ER-negative breast cancer stem cells: involvement of JNK/vimentin activation. Stem Cells. 32:2048–2060. 10.1002/stem.1735 [DOI] [PubMed] [Google Scholar]
- Fu M., Fan W., Pu X., Ni H., Zhang W., Chang F., Gong L., Xiong L., Wang J., and Gu X.. 2013. Elevated expression of SHIP2 correlates with poor prognosis in non-small cell lung cancer. Int. J. Clin. Exp. Pathol. 6:2185–2191. [PMC free article] [PubMed] [Google Scholar]
- Gertler F., and Condeelis J.. 2011. Metastasis: Tumor cells becoming MENAcing. Trends Cell Biol. 21:81–90. 10.1016/j.tcb.2010.10.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goswami S., Philippar U., Sun D., Patsialou A., Avraham J., Wang W., Di Modugno F., Nistico P., Gertler F.B., and Condeelis J.S.. 2009. Identification of invasion specific splice variants of the cytoskeletal protein Mena present in mammary tumor cells during invasion in vivo. Clin. Exp. Metastasis. 26:153–159. 10.1007/s10585-008-9225-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupton S.L., and Gertler F.B.. 2010. Integrin signaling switches the cytoskeletal and exocytic machinery that drives neuritogenesis. Dev. Cell. 18:725–736. 10.1016/j.devcel.2010.02.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen S.D., and Mullins R.D.. 2010. VASP is a processive actin polymerase that requires monomeric actin for barbed end association. J. Cell Biol. 191:571–584. 10.1083/jcb.201003014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino D., Jourquin J., Emmons S.W., Miller T., Goldgof M., Costello K., Tyson D.R., Brown B., Lu Y., Prasad N.K., et al. 2012. Network analysis of the focal adhesion to invadopodia transition identifies a PI3K-PKCα invasive signaling axis. Sci. Signal. 5:ra66–ra66. 10.1126/scisignal.2002964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshino D., Branch K.M., and Weaver A.M.. 2013. Signaling inputs to invadopodia and podosomes. J. Cell Sci. 126:2979–2989. 10.1242/jcs.079475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara H., Sasaoka T., Ishiki M., Wada T., Hori H., Kagawa S., and Kobayashi M.. 2002. Membrane localization of Src homology 2-containing inositol 5′-phosphatase 2 via Shc association is required for the negative regulation of insulin signaling in Rat1 fibroblasts overexpressing insulin receptors. Mol. Endocrinol. 16:2371–2381. 10.1210/me.2002-0083 [DOI] [PubMed] [Google Scholar]
- Johnson H., Lescarbeau R.S., Gutierrez J.A., and White F.M.. 2013. Phosphotyrosine profiling of NSCLC cells in response to EGF and HGF reveals network specific mediators of invasion. J. Proteome Res. 12:1856–1867. 10.1021/pr301192t [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang Y., Siegel P.M., Shu W., Drobnjak M., Kakonen S.M., Cordón-Cardo C., Guise T.A., and Massagué J.. 2003. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 3:537–549. 10.1016/S1535-6108(03)00132-6 [DOI] [PubMed] [Google Scholar]
- Knight J.F., Lesurf R., Zhao H., Pinnaduwage D., Davis R.R., Saleh S.M.I., Zuo D., Naujokas M.A., Chughtai N., Herschkowitz J.I., et al. 2013. Met synergizes with p53 loss to induce mammary tumors that possess features of claudin-low breast cancer. Proc. Natl. Acad. Sci. USA. 110:E1301–E1310. 10.1073/pnas.1210353110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krause M., Bear J.E., Loureiro J.J., and Gertler F.B.. 2002. The Ena/VASP enigma. J. Cell Sci. 115:4721–4726. 10.1242/jcs.00218 [DOI] [PubMed] [Google Scholar]
- Krause M., Dent E.W., Bear J.E., Loureiro J.J., and Gertler F.B.. 2003. Ena/VASP proteins: Regulators of the actin cytoskeleton and cell migration. Annu. Rev. Cell Dev. Biol. 19:541–564. 10.1146/annurev.cellbio.19.050103.103356 [DOI] [PubMed] [Google Scholar]
- Loureiro J.J., Rubinson D.A., Bear J.E., Baltus G.A., Kwiatkowski A.V., and Gertler F.B.. 2002. Critical roles of phosphorylation and actin binding motifs, but not the central proline-rich region, for Ena/vasodilator-stimulated phosphoprotein (VASP) function during cell migration. Mol. Biol. Cell. 13:2533–2546. 10.1091/mbc.E01-10-0102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magalhaes M.A.O., Larson D.R., Mader C.C., Bravo-Cordero J.J., Gil-Henn H., Oser M., Chen X., Koleske A.J., and Condeelis J.. 2011. Cortactin phosphorylation regulates cell invasion through a pH-dependent pathway. J. Cell Biol. 195:903–920. 10.1083/jcb.201103045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monteiro P., Rossé C., Castro-Castro A., Irondelle M., Lagoutte E., Paul-Gilloteaux P., Desnos C., Formstecher E., Darchen F., Perrais D., et al. 2013. Endosomal WASH and exocyst complexes control exocytosis of MT1-MMP at invadopodia. J. Cell Biol. 203:1063–1079. 10.1083/jcb.201306162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mourskaia A.A., Amir E., Dong Z., Tiedemann K., Cory S., Omeroglu A., Bertos N., Ouellet V., Clemons M., Scheffer G.L., et al. 2012. ABCC5 supports osteoclast formation and promotes breast cancer metastasis to bone. Breast Cancer Res. 14:R149 10.1186/bcr3361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller S.C., Ghersi G., Akiyama S.K., Sang Q.X.A., Howard L., Pineiro-Sanchez M., Nakahara H., Yeh Y., and Chen W.T.. 1999. A novel protease-docking function of integrin at invadopodia. J. Biol. Chem. 274:24947–24952. 10.1074/jbc.274.35.24947 [DOI] [PubMed] [Google Scholar]
- Murphy D.A., and Courtneidge S.A.. 2011. The ‘ins’ and ‘outs’ of invadopodia: Characteristics, formation and function. Nat. Rev. Mol. Cell Biol. 12:413–426. 10.1038/nrm3141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakatsu F., Perera R.M., Lucast L., Zoncu R., Domin J., Gertler F.B., Toomre D., and De Camilli P.. 2010. The inositol 5-phosphatase SHIP2 regulates endocytic clathrin-coated pit dynamics. J. Cell Biol. 190:307–315. 10.1083/jcb.201005018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ooms L.M., Horan K.A., Rahman P., Seaton G., Gurung R., Kethesparan D.S., and Mitchell C.A.. 2009. The role of the inositol polyphosphate 5-phosphatases in cellular function and human disease. Biochem. J. 419:29–49. 10.1042/BJ20081673 [DOI] [PubMed] [Google Scholar]
- Oudin M.J., Hughes S.K., Rohani N., Moufarrej M.N., Jones J.G., Condeelis J.S., Lauffenburger D.A., and Gertler F.B.. 2016a Characterization of the expression of the pro-metastatic MenaINV isoform during breast tumor progression. Clin. Exp. Metastasis. 33:249–261. 10.1007/s10585-015-9775-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oudin M.J., Jonas O., Kosciuk T., Broye L.C., Guido B.C., Wyckoff J., Riquelme D., Lamar J.M., Asokan S.B., Whittaker C., et al. 2016b Tumor cell-driven extracellular matrix remodeling drives haptotaxis during metastatic progression. Cancer Discov. 6:516–531. 10.1158/2159-8290.CD-15-1183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pesesse X., Dewaste V., De Smedt F., Laffargue M., Giuriato S., Moreau C., Payrastre B., and Erneux C.. 2001. The Src homology 2 domain containing inositol 5-phosphatase SHIP2 is recruited to the epidermal growth factor (EGF) receptor and dephosphorylates phosphatidylinositol 3,4,5-trisphosphate in EGF-stimulated COS-7 cells. J. Biol. Chem. 276:28348–28355. 10.1074/jbc.M103537200 [DOI] [PubMed] [Google Scholar]
- Philippar U., Roussos E.T., Oser M., Yamaguchi H., Kim H.-D., Giampieri S., Wang Y., Goswami S., Wyckoff J.B., Lauffenburger D.A., et al. 2008. A Mena invasion isoform potentiates EGF-induced carcinoma cell invasion and metastasis. Dev. Cell. 15:813–828. 10.1016/j.devcel.2008.09.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad N., Topping R.S., and Decker S.J.. 2001. SH2-containing inositol 5′-phosphatase SHIP2 associates with the p130(Cas) adapter protein and regulates cellular adhesion and spreading. Mol. Cell. Biol. 21:1416–1428. 10.1128/MCB.21.4.1416-1428.2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prasad N.K., Tandon M., Badve S., Snyder P.W., and Nakshatri H.. 2008. Phosphoinositol phosphatase SHIP2 promotes cancer development and metastasis coupled with alterations in EGF receptor turnover. Carcinogenesis. 29:25–34. 10.1093/carcin/bgm213 [DOI] [PubMed] [Google Scholar]
- Prasad N.K., Werner M.E., and Decker S.J.. 2009. Specific tyrosine phosphorylations mediate signal-dependent stimulation of SHIP2 inositol phosphatase activity, while the SH2 domain confers an inhibitory effect to maintain the basal activity. Biochemistry. 48:6285–6287. 10.1021/bi900492d [DOI] [PubMed] [Google Scholar]
- Rajadurai C.V., Havrylov S., Zaoui K., Vaillancourt R., Stuible M., Naujokas M., Zuo D., Tremblay M.L., and Park M.. 2012. Met receptor tyrosine kinase signals through a cortactin-Gab1 scaffold complex, to mediate invadopodia. J. Cell Sci. 125:2940–2953. 10.1242/jcs.100834 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakurai-Yageta M., Recchi C., Le Dez G., Sibarita J.B., Daviet L., Camonis J., D’Souza-Schorey C., and Chavrier P.. 2008. The interaction of IQGAP1 with the exocyst complex is required for tumor cell invasion downstream of Cdc42 and RhoA. J. Cell Biol. 181:985–998. 10.1083/jcb.200709076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoumacher M., Goldman R.D., Louvard D., and Vignjevic D.M.. 2010. Actin, microtubules, and vimentin intermediate filaments cooperate for elongation of invadopodia. J. Cell Biol. 189:541–556. 10.1083/jcb.200909113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma V.P., Eddy R., Entenberg D., Kai M., Gertler F.B., and Condeelis J.. 2013. Tks5 and SHIP2 regulate invadopodium maturation, but not initiation, in breast carcinoma cells. Curr. Biol. 23:2079–2089. 10.1016/j.cub.2013.08.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small J.V. 2008. Facing up to Mena: Tes(ting) times for EVH1 domains. Nat. Cell Biol. 10:118–120. 10.1038/ncb0208-118 [DOI] [PubMed] [Google Scholar]
- Smith K., Humphreys D., Hume P.J., and Koronakis V.. 2010. Enteropathogenic Escherichia coli recruits the cellular inositol phosphatase SHIP2 to regulate actin-pedestal formation. Cell Host Microbe. 7:13–24. 10.1016/j.chom.2009.12.004 [DOI] [PubMed] [Google Scholar]
- Steffen A., Le Dez G., Poincloux R., Recchi C., Nassoy P., Rottner K., Galli T., and Chavrier P.. 2008. MT1-MMP-dependent invasion is regulated by TI-VAMP/VAMP7. Curr. Biol. 18:926–931. 10.1016/j.cub.2008.05.044 [DOI] [PubMed] [Google Scholar]
- Stöter M., Niederlein A., Barsacchi R., Meyenhofer F., Brandl H., and Bickle M.. 2013. CellProfiler and KNIME: Open source tools for high content screening. In Methods in Molecular Biology. Humana Press, Totowa, NJ: 105–122. 10.1007/978-1-62703-311-4_8 [DOI] [PubMed] [Google Scholar]
- Stylli S.S., Stacey T.T., Verhagen A.M., Xu S.S., Pass I., Courtneidge S.A., and Lock P.. 2009. Nck adaptor proteins link Tks5 to invadopodia actin regulation and ECM degradation. J. Cell Sci. 122:2727–2740. 10.1242/jcs.046680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver A.M. 2008. Cortactin in tumor invasiveness. Cancer Lett. 265:157–166. 10.1016/j.canlet.2008.02.066 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi H., Yoshida S., Muroi E., Yoshida N., Kawamura M., Kouchi Z., Nakamura Y., Sakai R., and Fukami K.. 2011. Phosphoinositide 3-kinase signaling pathway mediated by p110α regulates invadopodia formation. J. Cell Biol. 193:1275–1288. 10.1083/jcb.201009126 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Fu M., Ding Y., Weng Y., Fan W., Pu X., Ge Z., Zhan F., Ni H., Zhang W., et al. 2014. High SHIP2 expression indicates poor survival in colorectal cancer. Dis. Markers. 2014:218968 10.1155/2014/218968 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.