Sargent et al. identify the E3 ubiquitin ligase PEX2 as the causative agent of mammalian pexophagy. During amino acid starvation, PEX2 expression increases to ubiquitinate peroxisomal membrane proteins and signal peroxisome degradation by autophagy.
Abstract
Peroxisomes are metabolic organelles necessary for anabolic and catabolic lipid reactions whose numbers are highly dynamic based on the metabolic need of the cells. One mechanism to regulate peroxisome numbers is through an autophagic process called pexophagy. In mammalian cells, ubiquitination of peroxisomal membrane proteins signals pexophagy; however, the E3 ligase responsible for mediating ubiquitination is not known. Here, we report that the peroxisomal E3 ubiquitin ligase peroxin 2 (PEX2) is the causative agent for mammalian pexophagy. Expression of PEX2 leads to gross ubiquitination of peroxisomes and degradation of peroxisomes in an NBR1-dependent autophagic process. We identify PEX5 and PMP70 as substrates of PEX2 that are ubiquitinated during amino acid starvation. We also find that PEX2 expression is up-regulated during both amino acid starvation and rapamycin treatment, suggesting that the mTORC1 pathway regulates pexophagy by regulating PEX2 expression levels. Finally, we validate our findings in vivo using an animal model.
Introduction
Organelle quality control is essential for the proper function of cells. Failure to remove damaged organelles is detrimental to the cell and is the basis of various diseases (Lynch-Day et al., 2012). For an effective organelle quality-control system, cells must be able to distinguish between aberrant and normal organelles and efficiently target the damaged organelles for removal while minimizing contact with the healthy organelles. Thus, a key hallmark of any quality control system is a mechanism that allows for a high degree of target selectivity.
Autophagy is vital in organelle quality control. It is the cellular process of degrading large cytoplasmic components such as large protein complexes, protein aggregates, invasive organisms, and organelles in the lysosome (Yang and Klionsky, 2010). Macroautophagy, hereafter referred to as autophagy, is the cellular process where cytoplasmic material is delivered to the lysosome via a double-membrane sequestering vesicle called the autophagosome (Kirisako et al., 1999). Autophagy can be a selective process where specific cytoplasmic components are targeted to the autophagosome by autophagy receptor proteins (Khaminets et al., 2016). Selective autophagy is described by a general mechanism where substrates are sequentially (a) designated for degradation, (b) targeted by autophagy receptor proteins, (c) recruited to the nascent autophagosome, and (d) degraded by the lysosome. The latter two steps of this process appear to be conserved for all substrates, but the mechanism by which various substrates are designated and targeted by autophagy receptor proteins conveys the discriminatory nature of selective autophagy and remains an active field of research.
One of these substrates, the peroxisome, is highly regulated by autophagy to meet the metabolic needs of the cell (Nordgren et al., 2013). Peroxisome numbers can be rapidly reduced in response to various cellular stress conditions, including amino acid starvation (Hara-Kuge and Fujiki, 2008), oxidative stress (Zhang et al., 2015), and hypoxia (Walter et al., 2014). Currently, it is thought that ubiquitination of a peroxisomal membrane protein acts as the signal to designate peroxisomes for degradation (Kim et al., 2008). Ubiquitination of the peroxisome surface leads to the recruitment of at least two ubiquitin-binding autophagy receptors, NBR1 and p62, which target the ubiquitinated peroxisomes to autophagosomes for degradation (Deosaran et al., 2013; Walter et al., 2014; Yamashita et al., 2014). Of the several peroxisomal proteins identified to be involved in pexophagy (Brown et al., 2014; Nazarko et al., 2014; Yamashita et al., 2014; Jiang et al., 2015), only the peroxisomal matrix protein receptor peroxin 5 (PEX5) has been shown to be ubiquitinated in pexophagy (Nordgren et al., 2015; Zhang et al., 2015). Currently, it is not known whether other peroxisomal proteins are ubiquitinated to signal pexophagy. Similarly, the E3 ubiquitin ligase mediating ubiquitination and the regulatory mechanism for pexophagy remain elusive.
To understand the mechanisms by which peroxisomes are designated for pexophagy, the E3 ubiquitin ligase required for this process needs to be identified. In this paper, we begin our investigation with the hypothesis that one or more of the peroxisomal E3 ubiquitin ligases PEX2, PEX10, and PEX12 may be required for pexophagy. These three RING-finger E3 ubiquitin ligases form a transmembrane complex that has active sites located at the outer surface of the peroxisomal membrane (El Magraoui et al., 2012). These ligases are primarily believed to function together in the PEX5 import cycle, the mechanism by which peroxisomal matrix proteins are shuttled and imported to the matrix of the peroxisome (Platta et al., 2007). Here, for the first time, we show that PEX2, but not PEX10 or PEX12, acts as the E3 ubiquitin ligase to selectively ubiquitinate peroxisomal membrane proteins to designate peroxisomes for autophagy-mediated degradation during amino acid starvation conditions.
Results
Overexpression of PEX2 leads to peroxisome degradation
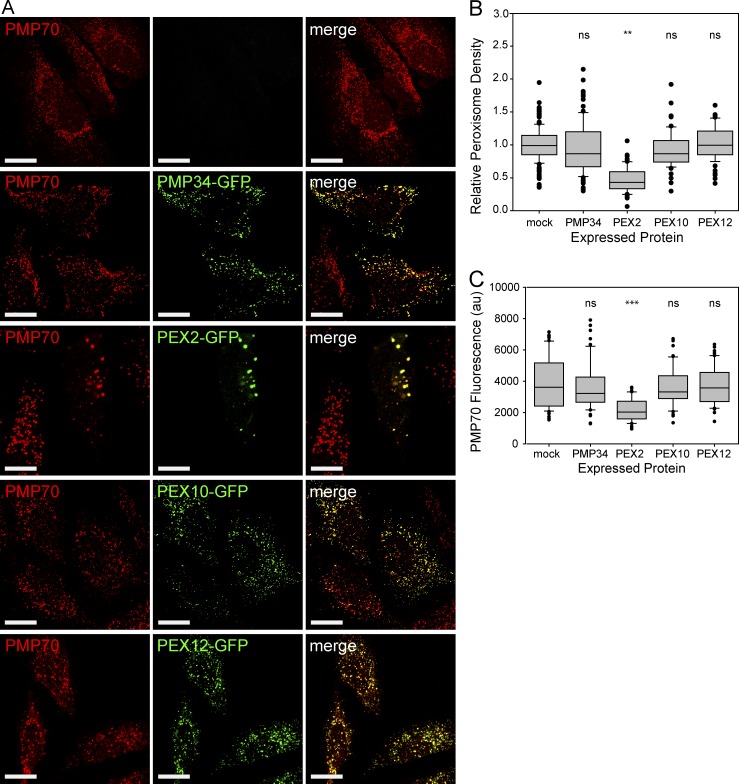
To study the potential role of the peroxisomal E3 ubiquitin ligases in pexophagy, we examined whether overexpression of the GFP-tagged peroxisomal E3 ubiquitin ligases can induce pexophagy in a mammalian cell line, HeLa cells. The peroxisomal membrane protein PMP34-GFP was also expressed as a control. After 48-h expression, immunofluorescent staining was performed against the peroxisomal membrane protein peroxisomal membrane protein 70 (PMP70) and imaged using confocal fluorescence microscopy (Fig. 1 A and Fig. S1). Quantification of the number of PMP70-positive puncta structures within the entire cell (peroxisome density) showed a decrease in peroxisome density in cells expressing PEX2-GFP, but not PMP34-GFP, PEX10-GFP, or PEX12-GFP (Fig. 1 B).
Figure 1.
Overexpression of the E3 ubiquitin ligase PEX2 decreases the number of peroxisomes. (A) HeLa cells were transfected with PMP34 or with one of the peroxisomal E3 ubiquitin ligases (PEX2, PEX10, and PEX12) fused to GFP. The cells were grown for 48 h, then fixed and stained for the peroxisomal membrane protein PMP70. Bars, 20 µm. (B) Box plot of the peroxisomal density of cells expressing the constructs in A. The peroxisomal density was calculated by quantifying the number of PMP70 puncta structures from Z-stack confocal images of the entire cell by and dividing by the cell volume. (C) Box plot of the total PMP70 fluorescence intensity from the entire cell. Both B and C are quantification of 150 cells from three independent trials. **, P < 0.01; ***, P < 0.001; ns (not significant), P > 0.05. In B and C, boxes show the 25th, 50th, and 75th percentiles, and lines show one-way standard deviations. Points represent all cells that did not fall within one standard deviation.
In some cells expressing PEX2-GFP, the PMP70-positive structures appeared as clusters of peroxisomes that were not observed in any of the other conditions. We have previously shown that peroxisome clustering preceded degradation when the autophagy receptor NBR1 was overexpressed (Deosaran et al., 2013); thus, the observed clustering of peroxisomes in cells expressing PEX2 is likely part of the degradation process. Because quantification of peroxisome density does not distinguish between loss of peroxisomes and clustering of peroxisomes, we also quantified the total fluorescence of the PMP70 puncta in these cells. The sum of the intensities of all PMP70 fluorescent voxels contained within each PMP70-positive puncta was quantified for all cells (Fig. 1 C). A rank-sum test was used to demonstrate that there is a significant reduction in PMP70 fluorescent signal in PEX2-GFP–expressing cells, but not in cells expressing GFP-tagged PMP34, PEX10, or PEX12.
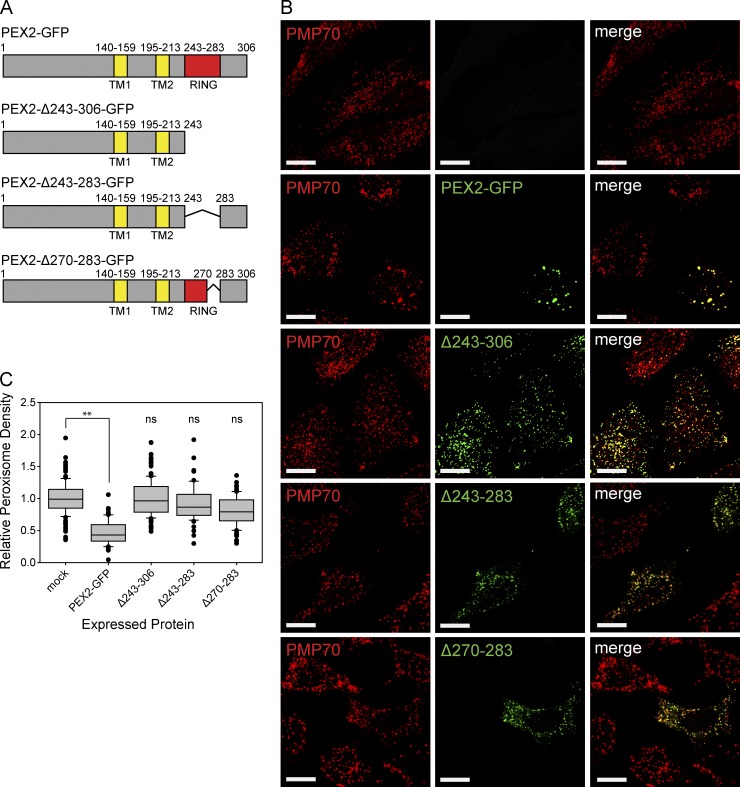
To test whether the E3 ubiquitin ligase activity of PEX2 was required to induce peroxisome loss, we constructed mutants of PEX2-GFP–containing deletions within its RING-domain active site (Fig. 2 A). The PEX2-Δ243-306-GFP construct deletes the C terminus of the protein, the PEX2-Δ243-283-GFP construct deletes the RING-finger domain, and the PEX2-Δ270-283-GFP construct deletes a region that is thought, by homology, to be critical in E2 binding (Deshaies and Joazeiro, 2009). When expressed in cells, we found that none of the PEX2-GFP deletion mutants caused a significant loss of peroxisomes, suggesting that the E3 ligase activity is required for PEX2-mediated peroxisome loss (Fig. 2, B and C).
Figure 2.
PEX2-mediated peroxisome loss requires the RING-finger domain. (A) Schematic of PEX2 and its deletion mutant constructs. RING, RING-finger domain; TM, transmembrane domain. Number above the image represents residue number. (B) HeLa cells were transfected with various PEX2-GFP constructs in A and fixed and stained for the peroxisomal membrane protein PMP70 after 48 h. Bars, 20 µm. (C) Box plots of the peroxisomal density of cells expressing the constructs in B. Plotted are a total of 150 cells from three independent trials. **, P < 0.01; ns (not significant), P > 0.05. Boxes show the 25th, 50th, and 75th percentiles, and lines show one-way standard deviations. Points represent all cells that did not fall within one standard deviation.
PEX2-induced peroxisome loss is mediated by autophagy and requires NBR1
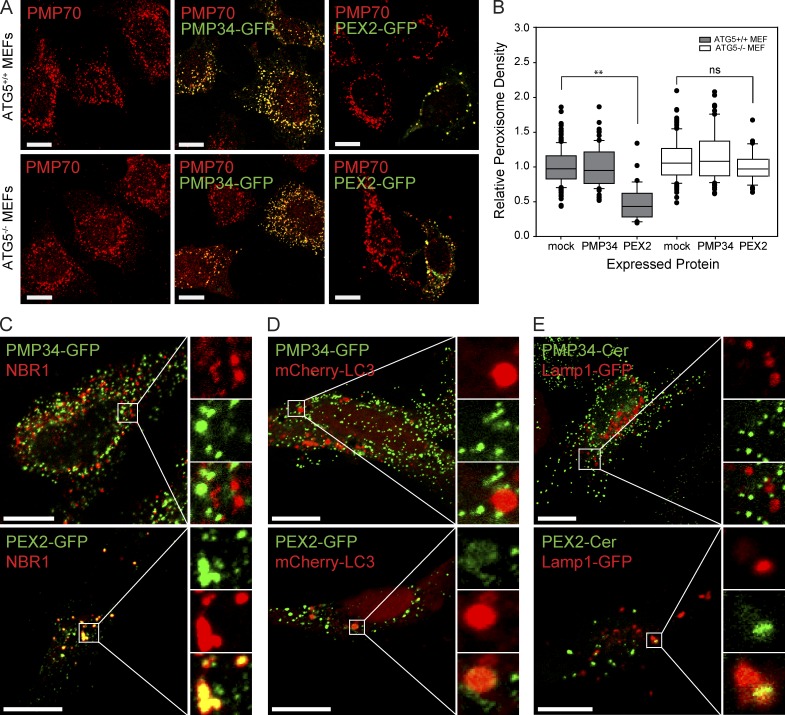
To investigate whether PEX2-mediated peroxisome loss was an autophagic process, we expressed PEX2-GFP in mouse embryonic fibroblast (MEF) cells incapable of autophagy-related gene 5 (ATG5)–dependent autophagy (Kuma et al., 2004). In the wild-type ATG5+/+ MEF cell line, expression of PEX2 caused a phenotype similar to that observed in HeLa cells, with peroxisome clustering and visible reduction in peroxisome numbers (Fig. 3 A). Quantification of peroxisomes in the PEX2-GFP expressing wild-type MEF cells showed a significant decrease in the peroxisomal density compared with mock transfected (mock) and PMP34-GFP expressing cells (Fig. 3 B). In the ATG5−/− MEF cell line, although some clustering of peroxisomes was observed, PEX2-GFP expression did not cause a significant decrease in peroxisomes (Fig. 3, A and B). This suggests that autophagy is required for the PEX2-mediated loss of peroxisomes. To further validate that PEX2 overexpression induces pexophagy, we examined the localization of three pexophagy markers during PEX2 expression: the peroxisomal autophagy receptor NBR1 (Deosaran et al., 2013; Walter et al., 2014; Yamashita et al., 2014), the autophagosomal marker LC3, and the lysosomal marker Lamp1 (Klionsky et al., 2016). Consistent with our finding that PEX2-GFP induces pexophagy, most of the detectable endogenous NBR1 in the cell was found colocalized with the clustered peroxisomes in cells expressing PEX2-GFP, but not in cells expressing PMP34-GFP (Fig. 3 C). Similarly, coexpressed LC3 and Lamp1 were also found colocalized with peroxisomes in multiple locations in each cell expressing PEX2-GFP, but not in PMP34-GFP–expressing cells (Fig. 3, D and E).
Figure 3.
PEX2-mediated peroxisome loss is an autophagic process. (A) ATG5+/+ and ATG5−/− MEF cells were mock transfected, transfected with PMP34-GFP, or transfected with PEX2-GFP as indicated. 48 h later, the cells were fixed, stained, and imaged for the peroxisomal membrane protein PMP70. Bars, 20 µm. (B) Box plot of the peroxisomal density of cells in A. Plotted are a total of 150 cells from three independent trials. **, P < 0.01; ns, not significant. boxes show the 25th, 50th, and 75th percentiles, and lines show one-way standard deviations. Points represent all cells that did not fall within one standard deviation. (C) Immunofluorescent image of HeLa cells transfected with PMP34-GFP or PEX2-GFP (green) and probed for endogenous NBR1 (red). Panels on the right show enlargement of the region of interest (white box). The channels are shown individually with the merge as indicated. (D) To detect autophagosomes, cells were cotransfected with mCherry-LC3. (E) To detect lysosomes, cells were cotransfected with Lamp1-GFP. Bars, 20 µm. The region of interest (white box) is magnified and each channel is shown separately along with the merged image on the right side in C–E. The pseudocolor used for each probe in the images is indicated by the color of the name in each panel.
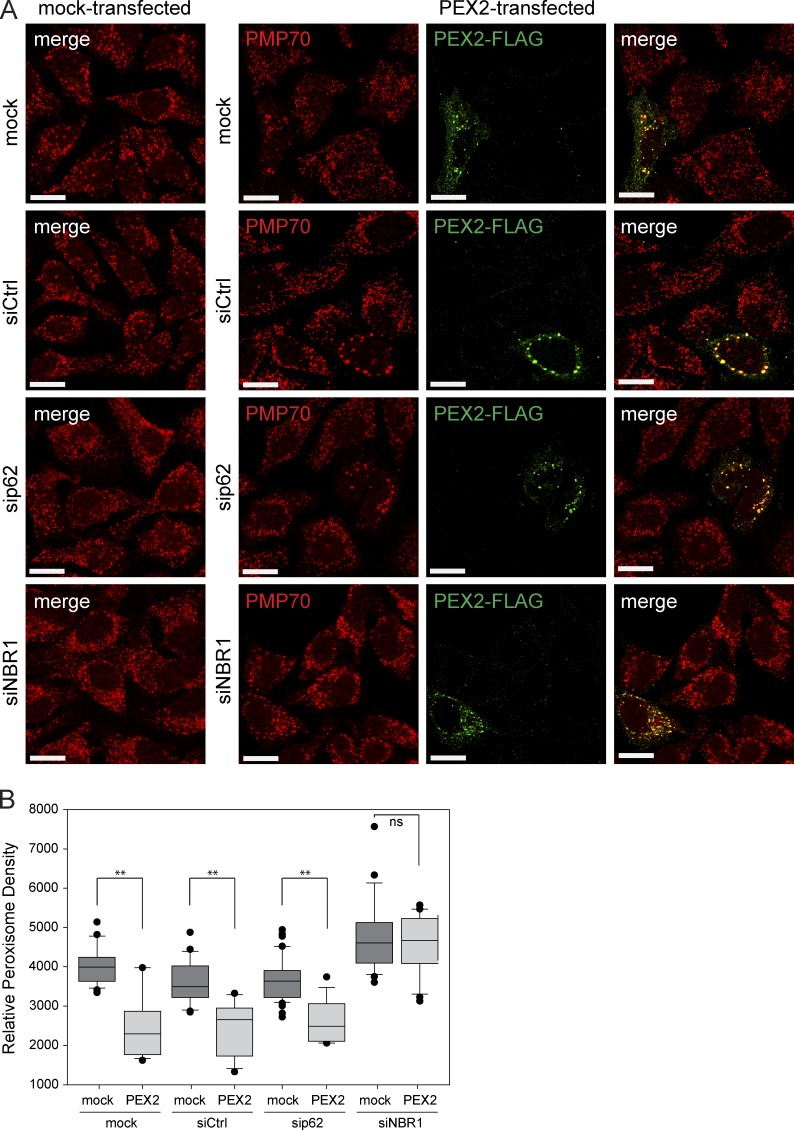
Previously, we have shown that NBR1 is the main autophagy receptor for pexophagy and that p62 acts as an aide to NBR1 (Deosaran et al., 2013). To investigate the contributions of NBR1 and p62 to PEX2-mediated pexophagy, we silenced the expression of each receptor and quantified peroxisome density. In mock-treated cells, cells transfected with nontargeting siRNA or cells depleted of p62, peroxisome loss was observed upon PEX2-FLAG expression (Fig. 4, A and B). However, in cells depleted of NBR1 expression, peroxisome loss was not observed upon PEX2-FLAG expression. Altogether, these findings support the idea that PEX2-mediated peroxisome loss is an autophagic process that requires the autophagy receptor NBR1.
Figure 4.
PEX2-mediated pexophagy requires NBR1. (A) Representative images of HeLa cells treated with nontargeting siRNA (siCtrl), or with siRNA against p62 or NBR1. 48 h after siRNA transfection, these cells were transfected with PEX2-FLAG, and 24 h afterward they were fixed and stained for the peroxisomal membrane protein PMP70 and with an antibody against the FLAG epitope. “mock” and “mock-transfected” are cells treated with transfection agent, but not with siRNA or plasmids, respectively. Bars, 20 µm. (B) Box plot of the PMP70 puncta density of cells in A. Plotted are a total of 50 cells from three independent trials. **, P < 0.01; ns (not significant), P > 0.05. Boxes show the 25th, 50th, and 75th percentiles, and lines show one-way standard deviations. Points represent all cells that did not fall within one standard deviation.
Peroxisomes are selectively degraded by autophagy during amino acid starvation
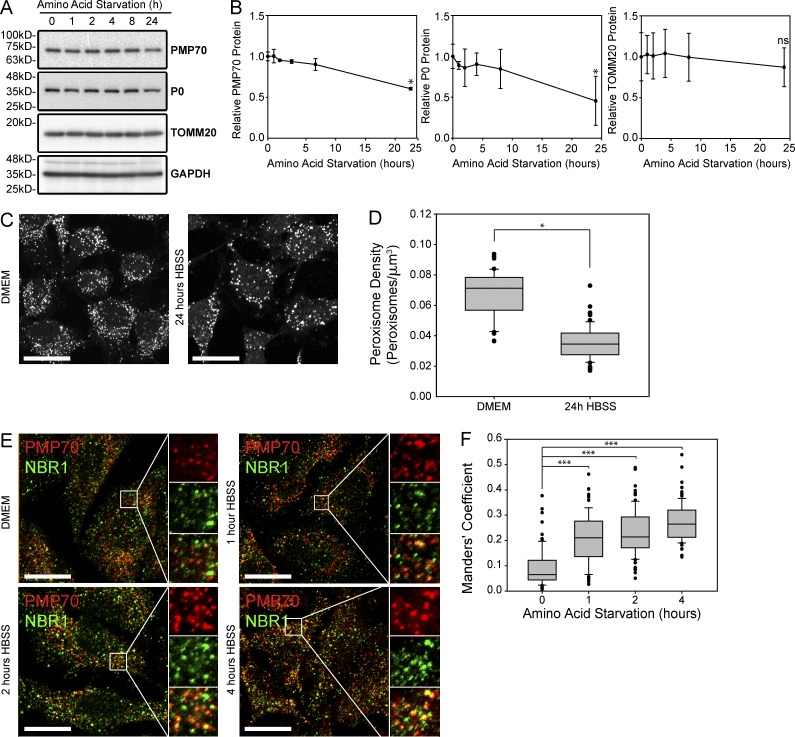
It has previously been shown that peroxisomes are degraded during low–amino acid conditions (Hara-Kuge and Fujiki, 2008; Jiang et al., 2015). To demonstrate that the degradation of peroxisomes is caused by selective pexophagy, but not bulk degradation, we characterized the loss of peroxisomes during low–amino acid conditions using three different assays. First, we examined the protein levels of various organelles. During amino acid starvation, both peroxisomes and ribosomes have been shown to be degraded by autophagy whereas other organelles such as mitochondria are initially protected (Hara-Kuge and Fujiki, 2008; Kraft et al., 2008; Gomes et al., 2011; Rambold et al., 2011; Jiang et al., 2015), suggesting a selective process. To confirm, HeLa cells were grown in HBSS, a medium that is absent of amino acids but contains sufficient glucose for cell survival over a 24-h period. The cell lysates were prepared at various time points and analyzed by Western blotting, probing for the peroxisome marker PMP70, the ribosomal component P0 (RPLP0), the mitochondria marker TOMM20, and the cytosolic marker GAPDH (Fig. 5 A). Quantifying these proteins demonstrated a significant decrease in both PMP70 and P0 but no significant change in the mitochondrial protein TOMM20 after 24 h of amino acid starvation in HBSS (Fig. 5 B).
Figure 5.
Peroxisomes are selectively degraded during amino acid starvation. (A) Immunoblot of HeLa cell lysates incubated in HBSS between 0 and 24 h as indicated. Shown are blots probed for the peroxisomal marker PMP70, the ribosomal marker PO, the mitochondrial marker TOMM20, and the cytosolic marker GAPDH. (B) Line graphs of the quantification of PMP70, PO, and TOMM20 blots relative to GAPDH and normalized against 0 h. Shown is the mean ± standard deviation from three independent experiments. (C) Immunofluorescent image of PMP70-stained HeLa cells grown in regular growth media (DMEM) or in HBSS media for 24 h. Bars, 20 µm. (D) Box plots of the peroxisomal density from 150 cells from three trials as shown in C. (E) HeLa cells were grown in either the growth media DMEM or the amino acid depletion media HBSS for time indicated and stained for PMP70 (red) and NBR1 (green). Panels on the right show enlargement of the region of interest (white box). The channels are shown individually with the merge as indicated. Bar, 20 µm. (F) Box plot of Manders’ coefficients showing the percentage of NBR1 that is colocalized with PMP70-positive puncta. Plotted are 150 cells from three trials. *, P < 0.05; ***, P < 0.001; ns (not significant), P > 0.05. Boxes show the 25th, 50th, and 75th percentiles, and lines show one-way standard deviations. Points represent all cells that did not fall within one standard deviation.
Next, we quantified the number of peroxisomes from confocal fluorescent images of cells immunostained for the endogenous peroxisomal protein PMP70 to visualize the loss of peroxisomes. After 24 h of amino acid starvation, we found a decreased number of PMP70 puncta structures compared with cells grown in DMEM, indicating a loss of peroxisomes (Fig. 5, C and D). Finally, to implicate pexophagy in the process, we examined whether NBR1 was localized to peroxisomes during amino acid starvation. Here, we quantified the degree of NBR1 colocalization on PMP70 structures using Manders’ coefficient of NBR1 in confocal fluorescent images of cells incubated in HBSS for up to 4 h (Fig. 5 E). We found that colocalization increased significantly with continued amino acid starvation, suggesting that NBR1 was being rapidly recruited to the peroxisomes (Fig. 5 F). Together, these results suggest that amino acid starvation induces selective peroxisome degradation via pexophagy.
PEX2 is required for both steady-state and amino acid starvation–induced pexophagy
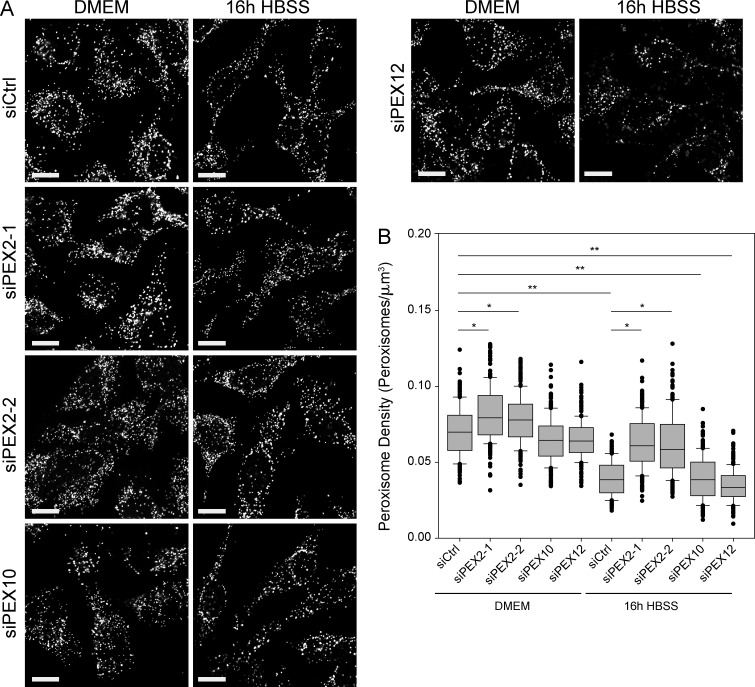
To determine whether any of the peroxisomal E3 ligases were involved in the activation of pexophagy during amino acid starvation, HeLa cells were depleted of PEX2, PEX10, or PEX12 before treatment with HBSS media. Knockdown of each E3 ubiquitin ligase was validated via quantitative PCR to confirm that the siRNA constructs were specific (Fig. S2). In cells transfected with nontargeting siRNA (siCtrl), amino acid starvation led to a significant decrease in the peroxisome density (Fig. 6, A and B, compare siCtrl-DMEM to siCtrl-HBSS). Similarly, a significant decrease in peroxisome density was observed in cells depleted of either PEX10 or PEX12 expression during HBSS treatment. However, when PEX2 expression was knocked down by siRNA, no significant loss of peroxisome density was observed during amino acid starvation. To demonstrate that this phenotype was not caused by potential off-target effects of the siRNA, a second siRNA against PEX2 (siPEX2-2) was used (Fig. 6, A and B). Additionally, we performed complementation studies using a construct resistant to the siRNA (PEX2-siR-FLAG) to show that the phenotype could be rescued (Fig. S3, A and B).
Figure 6.
PEX2 is required for pexophagy during both amino acid starvation and basal turnover. (A) HeLa cells were transfected with nontargeting siRNA (siCtrl) or with siRNA against PEX2, PEX10, or PEX12 for 48 h, and then grown in either regular DMEM media or amino acid starved in HBSS media for an additional 16 h. Shown are the representative fluorescent images of cells immunostained for PMP70. Bars, 20 µm. (B) Box plot of the peroxisomal density of cells after amino acid depletion. Plotted are a total of 150 cells from three independent trials. Only those with P < 0.05 when compared with either siCtrl DMEM or siCtrl HBSS are indicated. *, P < 0.05; **, P < 0.01. Boxes show the 25th, 50th, and 75th percentiles, and lines show one-way standard deviations. Points represent all cells that did not fall within one standard deviation.
Interestingly, we also observed a significant increase in peroxisome density in cells depleted of PEX2 expression grown in DMEM (Fig. 6, A and B). Because peroxisomes in cultured cells are regularly turned over by pexophagy, and have a half-life of ∼2–3 d (Ivashchenko et al., 2011), PEX2 may be involved during the basal turnover of peroxisomes as well as during amino acid starvation.
PEX5 and PMP70 are ubiquitinated by PEX2 during amino acid starvation
PEX5 is a known substrate of the peroxisomal E3 ubiquitin ligases PEX2 and PEX12 (Platta et al., 2009), and recently, PEX5 has been shown to be ubiquitinated during oxidative-stress–induced pexophagy (Zhang et al., 2015), as well as in pexophagy induced by defects in the matrix protein import cycle (Nordgren et al., 2015). To determine whether PEX5 is ubiquitinated during amino acid starvation and whether PEX2 was required for the process, we examined PEX5 ubiquitination in wild-type cells and cells depleted of PEX2. Additionally, because it is not known whether PEX5 is the sole peroxisomal protein ubiquitinated during pexophagy, we also examined PMP70, as it is the most abundant peroxisomal membrane protein (Hashimoto et al., 1986; Hartl and Just, 1987).
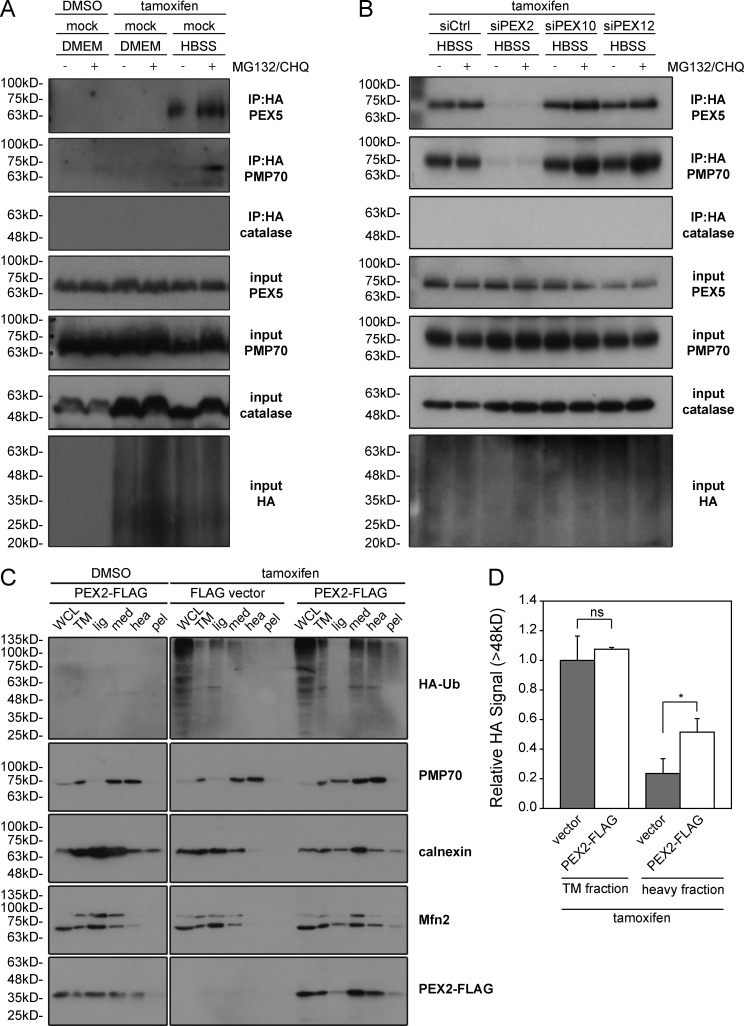
To detect ubiquitination, we used a HEK293 cell line stably expressing HA-tagged ubiquitin (HA-Ub) under a tamoxifen-inducible promoter. After 24 h of HA-Ub induction, these cells were subjected to 4 h of amino acid starvation. The cells were also treated with the proteasomal inhibitor MG132 to prevent proteasomal degradation and the lysosomal inhibitor chloroquine to prevent peroxisome degradation. To detect ubiquitinated peroxisomal proteins, HA-Ub was enriched by immunoprecipitation and probed for PEX5 and PMP70 and for the matrix protein catalase as a negative control. In cells grown in HBSS, PEX5 and PMP70 were both found to be ubiquitinated but were not ubiquitinated in cells grown in DMEM (Fig. 7 A).
Figure 7.
PEX5 and PMP70 are ubiquitinated by PEX2. (A) HEK293 cells stably expressing a HA-ubiquitin (HA-Ub) construct under the control of a tamoxifen-inducible promoter were treated with either DMSO or tamoxifen for 24 h. 4 h before lysis, cells were depleted of amino acids in HBSS media, either in the absence or presence of proteasomal inhibitor MG132 (10 µM) and the lysosomal inhibitor chloroquine (10 nM). Ubiquitinated proteins were immunoprecipitated with an antibody against the FLAG epitope and lysates (input) and immunoprecipitation (IP:HA) were blotted for PMP70, PEX5, catalase, and HA as indicated. (B) As in A, but before induction of HA-Ub, cells were mock treated or transfected with nontargeting siRNA (siCtrl) or siRNA against PEX2, PEX10, or PEX12. 4 h before lysis, cells were treated grown in HBSS media, either in the presence or absence of the proteasomal inhibitor MG132, and analyzed as in A. (C) Peroxisomes isolated from cells expressing PEX2-FLAG show an increase in ubiquitinated proteins. The HA-Ub HEK293 cell line was transfected with either an empty FLAG vector or PEX2-FLAG. 4 h before lysis, the cells were treated with the proteasomal inhibitor MG132 (10 µM). After treatment, cell lysates were collected (WCL), the total membrane (TM) fraction was separated (TM), and the TM fraction was further separated into light (lig), medium (med), heavy (hea), and pellet (pel) fractions. Each fraction was then analyzed by immunoblot using antibodies against HA, PMP70 (peroxisome membrane), Mfn2 (mitochondrial outer membrane), calnexin (ER membrane), and FLAG as indicated. (D) Bar graph showing the relative HA signal >48 kD from the total membrane fraction (TM) and the heavy fraction (heavy) lanes from three independent experiments. Means and standard deviations are shown. *, P < 0.05; ns (not significant), P > 0.05.
To determine whether any of the peroxisomal E3 ubiquitin ligases were required for this process, we depleted the cells of PEX2, PEX10, and PEX12 individually in the HEK293–HA-Ub cells. Here, we found that only the loss of PEX2 expression resulted in the loss of PEX5 and PMP70 ubiquitination, whereas both peroxisomal membrane proteins were found to be ubiquitinated in cells depleted of either PEX10 or PEX12 (Fig. 7 B). This suggests that PEX2 is solely responsible for PMP70 and PEX5 ubiquitination during amino acid starvation.
To further validate that PEX2 is responsible for the increased ubiquitination of peroxisomal membrane proteins, we examined whether overexpression of PEX2 resulted in an increase in ubiquitinated proteins on peroxisomes. To determine whether peroxisomal proteins were ubiquitinated, peroxisomes were isolated from the HEK293–HA-Ub stable cell line, which had been transiently transfected with either empty FLAG vector or PEX2-FLAG. Membranes were first isolated by centrifugation, and then fractionated into light, medium, heavy, and pellet fractions by density centrifugation. We found peroxisomes (PMP70) to be enriched in both the medium and heavy fractions, whereas the ER (calnexin) and mitochondria (MFN2) were enriched in only the medium fraction (Fig. 7 C). In both the medium and heavy fractions, we detected multiple HA-Ub bands in cells expressing PEX2-FLAG, but not in the cells transfected with empty vector, suggesting the expression of PEX2-FLAG causes an increase in the ubiquitination of peroxisomal membrane proteins (Fig. 7, C and D). These multiple ubiquitin signals further suggest that the PEX2 E3 ubiquitin ligase activity may be promiscuous against multiple peroxisomal membrane proteins.
PEX2 protein levels are regulated by mTOR
When we examined the expression levels of transiently transfected peroxisomal E3 ligases, we noticed that the expression of PEX2 was consistently lower than that of the other peroxisomal E3 ligases even though all three constructs used the same expression vector (Fig. S4 A). However, we found that PEX2 expression increased upon inhibiting the proteasome with MG132, suggesting that the low expression of PEX2 may be caused by proteasomal turnover (Fig. S4, B and C). Together, these results suggested that PEX2 might be regulated posttranslationally, at the level of protein degradation.
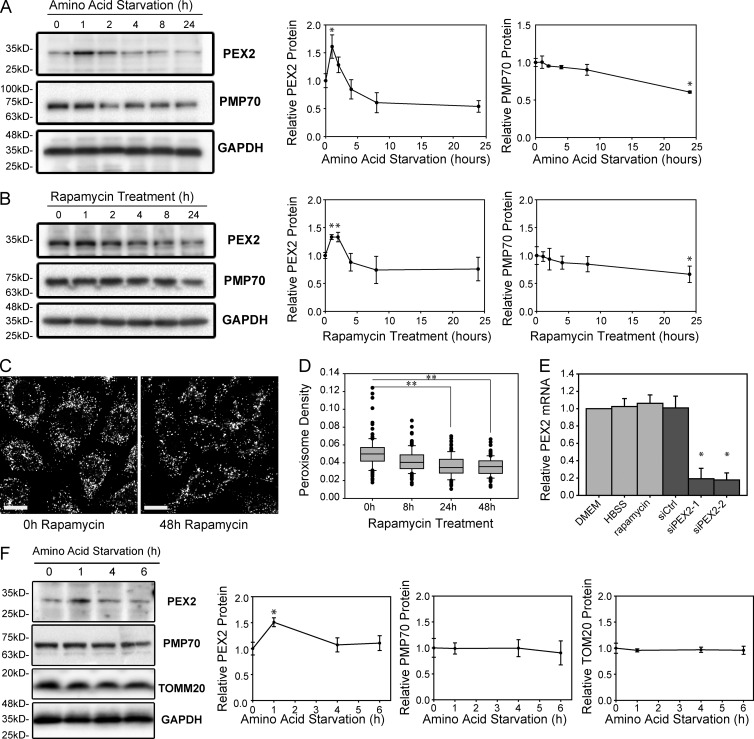
Because the overexpression of PEX2 induced pexophagy (Fig. 1, A and B), we next asked whether regulation of PEX2 expression may be a mechanism of regulating pexophagy. To address this hypothesis, we examined whether PEX2 expression levels were regulated during amino acid starvation conditions by analyzing PEX2 expression in HeLa cells grown in HBSS between 1 and 24 h. During these conditions, we found that PEX2 expression was biphasic, rapidly increasing during the first 2 h of amino acid starvation before returning to the basal level (Fig. 8 A). As before, PMP70 levels gradually decreased and a significant difference was observed after 24 h of amino acid starvation.
Figure 8.
PEX2 protein levels increase but mRNA is unchanged during amino acid starvation or mTORC1 inhibition. (A) HeLa cells were depleted of amino acids for 0–24 h as indicated in HBSS and analyzed by immunoblotting for PEX2, PMP70, and GAPDH. Line graphs show the amount of each protein relative to GAPDH and normalized to the start of HBSS treatment (0 h). n = 3. (B) HeLa cells were treated with 5 µM rapamycin for 0–24 h as indicated to inhibit mTOR and cell lysates were analyzed by immunoblotting for PEX2, PMP70, and GAPDH. Line graphs show the amount of each protein relative to GAPDH and normalized to 0 h. n = 3. (C) Immunofluorescent images of PMP70 in HeLa cells treated with 5 µM rapamycin for 0–48 h. Bars, 20 µm. (D) Box plot of the peroxisomal density of cells in (C). Plotted are a total of 150 cells from three independent trials. Boxes show the 25th, 50th, and 75th percentiles, and lines show one-way standard deviations. Points represent all cells that did not fall within one standard deviation. (E) Quantitative PCR of PEX2 mRNA in HeLa cells grown in DMEM, HBSS, or 5 µM rapamycin for 1 h. Also shown is the PEX2 mRNA level in cells treated with a nontargeting siRNA (siCtrl) or treated with two different siRNA constructs against PEX2. The means and standard deviations of three independent trials are shown. (F) Western blots of PEX2, PMP70, TOMM20, and GAPDH from primary rat hepatocytes after 0, 1, 4, or 6 h treatment in HBSS media. The line graphs represent the protein levels of PEX2, PMP70 and TOMM20 relative to GAPDH loading control. Each point represents the mean of three different trials with standard deviations shown. *, P < 0.05; **, P < 0.01.
Because amino acid starvation activates autophagy through the inhibition of mTORC1 (Kim et al., 2011), we next asked whether the mTORC1 signaling pathway was involved in the regulation of PEX2. To address this question, PEX2 expression levels were examined in HeLa cells treated with the mTORC1 inhibitor rapamycin. Similar to what was observed during amino acid starvation, we observed biphasic regulation of PEX2 during mTOR inhibition and a significant decrease in PMP70 after 24 h, suggesting that the mTOR signaling pathway regulates PEX2 expression (Fig. 8 B). To further validate the role of mTORC1 in pexophagy, we quantified the peroxisome density in HeLa cells treated with rapamycin for 0–48 h and found that peroxisomes decreased upon rapamycin treatment (Fig. 8, C and D). To ensure that autophagy was being up-regulated during amino acid starvation and rapamycin treatment, we examined the processing of LC3. During either treatment, PEX2 up-regulation was synchronized with the conversion of LC3 to its lipidated form (Fig. S5, A and B).
To determine whether PEX2 was regulated at the level of transcription or mRNA stability, we examined the PEX2 mRNA levels using quantitative PCR. Although PEX2 mRNA levels were significantly affected by siRNA treatment, we were unable to observe a significant change in PEX2 mRNA levels during amino acid starvation or rapamycin treatment (Fig. 8 E). Together with the observation that PEX2 appears to be rapidly degraded by the proteasome (Fig. S4 D), we concluded that PEX2 regulation during amino acid starvation or the inhibition of mTORC1 occurs posttranscriptionally.
Finally, we examined whether the regulation of PEX2 expression could be replicated in primary cells. Primary rat liver hepatocytes were depleted of amino acids in HBSS for 0–6 h, and their lysates were blotted for PEX2, PMP70, TOMM20, and GAPDH (Fig. 8 F). The ratio of PEX2, PMP70, or TOMM20 to GAPDH protein levels was quantified from three trials and graphed. We were able to confirm that PEX2 protein levels are significantly increased in just 1 h after starvation. Altogether, our findings strongly suggest a novel role for PEX2 in amino acid starvation–induced autophagic destruction of peroxisomes.
Peroxisomes are degraded and PEX2 protein levels are elevated during protein restriction in vivo
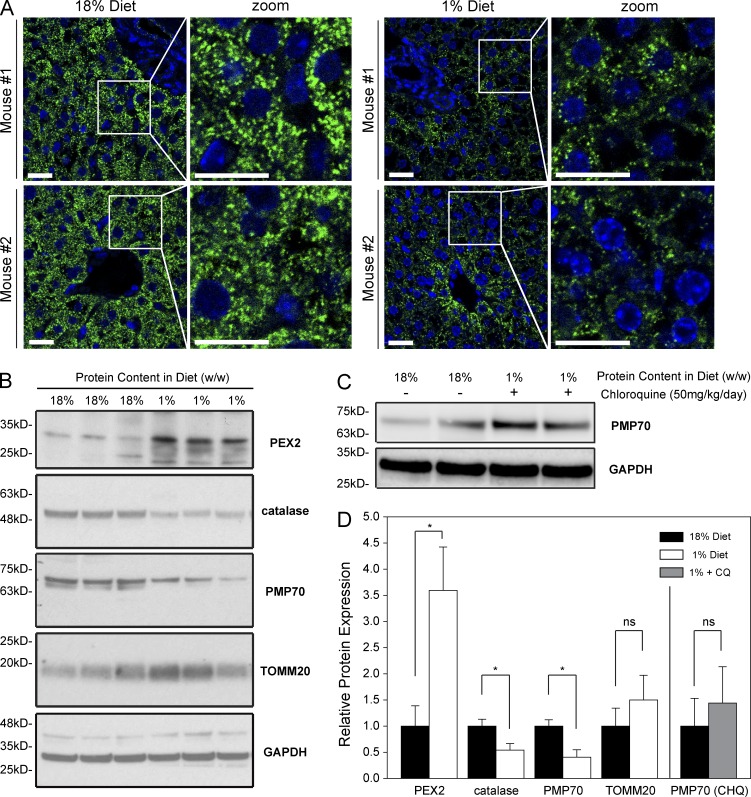
Although several studies have shown that acute amino acid starvation can induce pexophagy in cultured cells, it is not known whether a similar phenotype is observed in vivo. Recently, we described the loss of peroxisomes in rats fed a low protein diet (Van Zutphen et al., 2016). To determine whether amino acid starvation can induce pexophagy in vivo, we adapted the rat model to study mice fed a protein-restricted diet. Newly weaned mice were protein restricted for 12 d by free feeding with a 1% protein chow (wt/wt) compared with 18% protein (wt/wt) for the control animals. To determine whether the low protein diet led to peroxisome loss in mouse livers, we performed immunofluorescence microscopy on thin sections of the liver and immunoblotting analysis of liver lysates. We first examined changes in peroxisome density via immunofluorescent staining of PMP70. In the livers of the protein-restricted animals, the peroxisome density was visibly decreased compared with the control animals (Fig. 9 A). To quantify the observed loss of peroxisomes, we performed immunoblotting analysis of liver lysates. The peroxisomal membrane protein PMP70 was significantly decreased in the protein-restricted livers compared with control (Fig. 9, B–D). However, we did not observe a decrease in the mitochondrial membrane protein TOMM20, in agreement with what has been previously reported in cultured cells (Blackstone and Chang, 2011; Gomes et al., 2011; Rambold et al., 2011).
Figure 9.
Peroxisomes are degraded and PEX2 protein levels elevated in a mouse model of amino acid starvation. (A) Mice were fed a control diet containing 18% protein (wt/wt) or a protein-restriction diet containing 1% protein (wt/wt). Representative images of fluorescent immunohistochemistry images thin slices of the livers stained with PMP70 (green) or DAPI (blue) as a marker of the nucleus. Images from two different mice for each diet conditions are shown. Bars, 20 µm. (B) Whole-liver lysates from three different mice for each condition were separated by SDS-PAGE and blotted for PEX2, catalase, PMP70, TOMM20, and GAPDH. (C) Immunoblot of liver lysate of 1% diet animals treated with chloroquine compared with control. (D) Bar graphs represent the relative expression of PMP70/GAPDH ratio in B and C normalized to the 18% diet control animals. Means and standard deviations are shown. *, P < 0.05; **, P < 0.01; ns (not significant), P > 0.05.
We also examined PEX2 expression levels in the mouse liver. To determine whether PEX2 expression increased during low–amino acid conditions, mouse liver lysates were analyzed by SDS-PAGE immunoblot analysis. Similar to what was observed in cultured cells (Fig. 8), we found that PEX2 levels were elevated in the protein restricted animals compared with their control littermates fed a normal diet (Fig. 9, B and D). This suggested that the loss of peroxisomes in the low protein diet animals may be induced by PEX2-induced pexophagy.
To confirm that the loss of peroxisomes is caused by pexophagy, we treated the animals with chloroquine, a lysosomotyroic drug that inhibits autophagy by preventing the lysosomal degradation of autophagosomes. When protein-restricted animals were treated with chloroquine, there was no decrease in peroxisomal levels, and instead, an increase in PMP70 was observed (Fig. 9, C and D), analogous to the increase in peroxisome levels observed during NBR1 or PEX2 depletion (Figs. 4 B and 6 B). This suggests that the loss of peroxisomes in the livers of protein-restricted animals was caused by pexophagy and not a decrease in peroxisome biogenesis.
Discussion
The peroxisomal E3 ubiquitin ligase PEX2 has previously been described to play a critical role in peroxisome assembly and was the first gene to be associated with Zellweger spectrum disorder, a collection of autosomal-recessive disorders affecting the biogenesis of peroxisomes (Dodt and Gould, 1996; Faust and Hatten, 1997). During peroxisome biogenesis, PEX2 is required to ubiquitinate the membrane-bound form of the matrix protein receptor PEX5 in order for it to be removed from the membrane (Okumoto et al., 1997; Matsumura et al., 2000; Shimura et al., 2000; Platta et al., 2007; Prestele et al., 2010). Here, we describe an additional role for PEX2 as a regulator of peroxisome turnover. We show that during cellular stress, such as amino acid starvation, PEX2 expression increases and at least two peroxisomal membrane proteins are ubiquitinated to designate the peroxisomes for autophagic degradation.
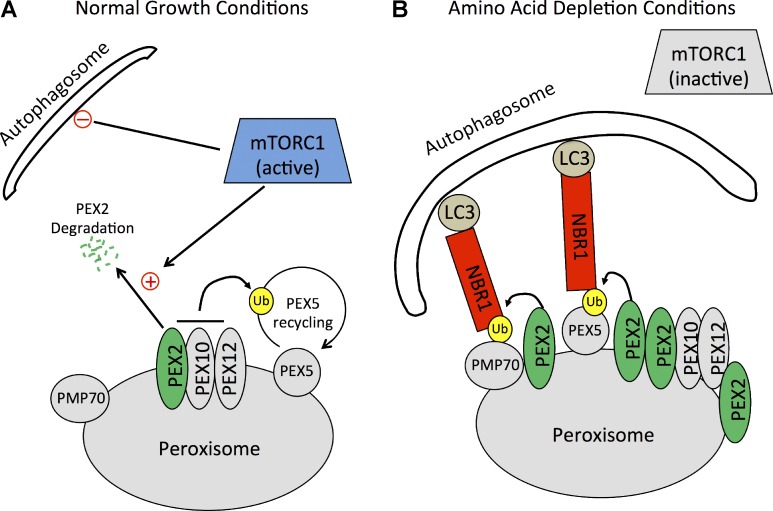
Based on our findings, we suggest a potential mechanism by which peroxisomes numbers are regulated during amino acid starvation (Fig. 10). During normal growth conditions, PEX2 expression is regulated by mTORC1 to maintain PEX2 at an expression level high enough to form the E3 complex with PEX10 and PEX12 for PEX5 recycling but low enough to prevent unintended pexophagy. Although the mechanism by which mTORC1 regulates PEX2 expression is unclear, we hypothesize that mTORC1 accelerates PEX2 degradation by the proteasome (Fig. 10 A). This hypothesis is supported by the recent finding that mTORC1 enhances proteasome activity of specific proteins (Zhang et al., 2014). During low–amino acid conditions, the inhibition of mTOR results in the stabilization of PEX2 at the peroxisome membrane and an increase in the ubiquitination of peroxisomal membrane proteins such as PEX5 and PMP70 at the peroxisome membrane. Ubiquitination of the peroxisome surface designates the peroxisome for degradation, which recruits the autophagy receptor NBR1 to target peroxisomes to autophagosomes for degradation (Fig. 10 B).
Figure 10.
A model of pexophagy activation during amino acid starvation. (A) In cells grown in normal growth media, mTORC1 maintains a low PEX2 expression level by promoting PEX2 degradation. A low level of PEX2 is maintained to allow for ubiquitination (Ub) of PEX5 for its recycling from the peroxisomal membrane. mTORC1 also inhibits the activation of autophagy. (B) During amino acid depletion conditions, mTORC1 is inactivated, releasing its inhibition of both autophagy and the expression levels of PEX2. Autophagosome formation increases, and PEX2 levels are elevated. The increase in PEX2 on peroxisomes results in increased ubiquitination of at least two peroxisomal proteins, PEX5 and PMP70, which allows for the recruitment of the autophagy receptor NBR1. NBR1 then targets these ubiquitinated peroxisomes to autophagosomes for degradation.
One important corollary of our model is that PEX5 is not the only substrate ubiquitinated by PEX2. Ubiquitinated PEX5 has previously been predicted to be a signal for pexophagy (Kim et al., 2008; Deosaran et al., 2013; Brown et al., 2014), but it was not until recently that ubiquitinated PEX5 was shown to induce pexophagy (Nordgren et al., 2015; Zhang et al., 2015). Nordgren et al. (2015) demonstrated that PEX5 molecules tagged with a bulky C-terminal tag are inefficiently removed from the membrane and are subsequently monoubiquitinated to signal pexophagy, whereas Zhang et al. (2015) showed that oxidative stress can target ataxia-telangiectasia mutated to peroxisomes, leading to phosphorylation and subsequent ubiquitination of PEX5 to promote pexophagy. Here, we show that PEX5 ubiquitination may not be the only signal for pexophagy during amino acid starvation, as we find that PMP70 is also ubiquitinated by PEX2 during amino acid starvation (Fig. 7 A). Furthermore, our data suggest that other peroxisomal membrane proteins may also be ubiquitinated, as multiple ubiquitination bands can be observed during PEX2 overexpression (Fig. 7 B). Although we do not know whether the ubiquitination of PMP70 or other PMPs can induce pexophagy at this time, given that expression of a ubiquitin motif on the surface of peroxisomes is sufficient to induce pexophagy (Kim et al., 2008), we suggest that PEX5 is not the only peroxisomal membrane protein to be ubiquitinated by PEX2 to signal pexophagy.
Recently, Zhang et al. (2015) reported that the depletion of any of the peroxisomal E3 ligases prevented PEX5 ubiquitination during oxidative stress, suggesting that all three ligases may play a role in oxidative-stress–induced pexophagy. In contrast, we found that PEX2 knockdown prevented both PEX5 ubiquitination and pexophagy, whereas knockdown of PEX10 or PEX12 did not affect PEX5 ubiquitination or pexophagy during amino acid starvation (Fig. 6, A and B; and Fig. 7, A and B). Furthermore, overexpression of PEX2 without coexpression of PEX10 or PEX12 was sufficient to induce ubiquitination of the peroxisome membrane (Fig. 7 C) and pexophagy (Fig. 1, A and B). It is not clear at this time what role PEX10 and PEX12 play in amino acid starvation induced pexophagy but it appears that pexophagy induction may be mediated by distinct mechanisms depending on the stimulus.
Finally, our work suggests a possible mechanistic insight into the pathogenesis of a common form of severe malnutrition in young children called kwashiorkor. Kwashiorkor is characterized by edema and disturbed hepatic metabolism, illustrated by steatosis and hypoglycemia (Grover and Ee, 2009). Interestingly, postmortem EM studies on the livers of kwashiorkor patients showed a decrease in peroxisomes (Brooks et al., 1992), similar to what we observed in our mouse model. Recently, we have demonstrated that prolonged protein restriction in young rats resulted in peroxisome loss and disruption in liver metabolic function (Van Zutphen et al., 2016). Here, our work uncovers a possible cellular mechanism by which peroxisomes are lost during severe malnutrition. In our mouse model, we find that peroxisome numbers decrease while PEX2 protein levels are elevated and that the loss in peroxisome numbers is mediated by autophagy. However, it still needs to be determined whether the loss of peroxisomes plays a direct role in the other impairments in hepatic metabolism. Further studies are currently being performed to explain a possible relationship between the loss of the metabolic organelle and liver pathogenesis during severe malnutrition.
Materials and methods
Plasmids and siRNA constructs
PMP34-GFP, mCherry-LC3, and Lamp1-GFP have all been previously described (Deosaran et al., 2013) HA-Ub has been previously described (Coyaud et al., 2015). PEX2-GFP, PEX10-GFP, PEX12-GFP, PEX2-Δ243-306-GFP, PEX2-Δ243-283-GFP, PEX2-Δ270-283-GFP, PEX2-CFP, PEX2-FLAG, PEX2-siR-FLAG, and PEX2-FlpIn-FLAG were synthesized for this work. For these constructs, open reading frames were sourced from SIDNET (Hospital for Sick Children, Toronto, Canada), primers were sourced from Sigma-Aldrich, and constructs were sequenced by The Center for Applied Genomics (Toronto, Canada). A complete description of the construction of all plasmids used in this study is available upon request (siCtrl, 5′-AAUAAGGCUAUGAAGAGAUAC-3′; siPEX2-1, 5′-GCUAGUUUGGUCCCAGUUU-3′; siPEX2-2, 5′-GAAGAACGAUGCUAUGAUU-3′; siPEX10, 5′-UCACUUAUUUGGACGGGAUUUC-3′; siPEX12, 5′-UGUUGCCUUAUCCCUGUCUA-3′; sip62, 5′-GCAUUGAAGUUGAUAUCGAUTT-3′; and siNBR1, 5′-GAACGUAUACUUCCCAUUGUU-3′).
Antibodies
Rabbit polyclonal α-PMP70 was purchased from Abcam. Rabbit polyclonal α-NBR1 was a gift from I. Dikic (Goethe Univerity, Frankfurt, Germany). Human polyclonal α-P0 was a gift from A. Palazzo (University of Toronto, Toronto, Canada). Mouse monoclonal α-TOMM20 was purchased from Santa Cruz Biotechnology, Inc. Mouse monoclonal α-Mfn2 was purchased from Cedarlane. Rabbit monoclonal α-calnexin was a gift from D. Williams (University of Toronto, Toronto, Canada). Mouse monoclonal α-HA was purchased from Covance. Rabbit polyclonal α-Atg12 was purchased from Cell Signaling Technology. Rabbit polyclonal α-PEX14 was purchased from Millipore. Rabbit polyclonal α-PEX2 was generated from peptide by GenScript. Rabbit polyclonal α-PEX2 was purchased from Abcam. Rabbit polyclonal α-PEX5 was generated by GenScript. Rabbit polyclonal α-catalase was purchased from EMD Millipore. Rabbit monoclonal α-4E-BP was a gift from J. Brumell (University of Toronto, Toronto, Canada). Mouse monoclonal α-FLAG was purchased from Sigma-Aldrich.
HRP-conjugated goat polyclonal α-mouse secondary and HRP-conjugated goat polyclonal α-rabbit secondary was purchased from Cedarlane. HRP-conjugated mouse monoclonal α-GAPDH was purchased from Abcam. Fluorescent goat polyclonal α-rabbit Alexa Fluor 568 secondary antibody and goat polyclonal α-mouse Alexa Fluor 488 secondary antibody were purchased from Invitrogen.
Cell culture
HeLa cells were purchased from ATCC (CCL-2) and were cultured in DMEM (SH3008101; HyClone) supplemented with 10% FBS (Invitrogen) and 6 mM l-glutamine (Invitrogen). ATG5+/+ and ATG5−/− MEFs were donated by J. Brumell (University of Toronto, Toronto, Canada) and cultured in DMEM supplemented with 10% FBS and 6 mM l-glutamine. Stably transfected HA-Ub HEK293 cells donated by B. Raught (University of Toronto, Toronto, Canada) and cultured in DMEM supplemented with 10% FBS, 6 mM l-glutamine, and G418. HBSS was purchased from Lonza (10-527F) and contains 1 g/l glucose. All cells were cultured in 37°C in humidified air conditions containing 5% CO2.
siRNA and plasmid transfections
In all experiments where siRNA was transfected, cells were transfected twice for 8 h and allowed to recover for 16 h. Lipofectamine-2000 or Lipofectamine-3000 (Invitrogen) was used for plasmid transfections. Lipofectamine-2000 was used for siRNA transfection. In experiments where cells were pretransfected with siRNA, cells were transfected with plasmids 24 h after the last siRNA treatment and 24–48 h before fixation or lysis. The media was changed 8 h after transfection.
Microscopy and data analysis
All fluorescent imaging was performed on an LSM710 (ZEISS) with a 63×/1.4 Plan-Apochromat oil objective using Argon2, HeNe, and HeNe2 lasers and Zen software (ZEISS). Z-stack images were acquired at thicknesses of between 0.60 µm and 1.00 µm. Live-cell imaging was performed at 37°C in CO2-independent medium (Thermo Fisher Scientific) containing FBS, leupeptin, and E-64 as specified. For all treatments within a particular experiment, image acquisitions were obtained on the same day using the same microscope settings. The images were acquired in 1,024 × 1,024 pixels at the depth of 12 bits. For visual presentation, only the brightness was adjusted.
Box plots were used to represent data from multiple experiments under the same heading. In all box plots, boxes span from the 25th percentile to the 75th percentile with the median shown. One-way standard deviations are plotted as lines, and any data points beyond one standard deviation are shown. Line graphs were used to represent data from multiple experiments under the same time point. In all line graphs, points represent the mean and standard deviations are shown. Bar graphs were used to represent data from multiple experiments under the same heading. In all bar graphs, the bar represents the mean, and standard deviations are shown.
When comparing data points, a normality test was performed to determine whether distributions were normal. For normal distributions, comparisons were made using Student’s t test. For nonnormal distributions, comparisons were made using the Mann–Whitney U test. Only the Manders’ coefficient of NBR1 on peroxisomes (Fig. 5 F) was determined to be nonnormal. Manders’ coefficient of PMP70 structures that contained NBR1 was calculated using the Volocity software (v.6.3; PerkinElmer).
Peroxisome density
Z-stack confocal images were used to determine peroxisome density using Volocity software (PerkinElmer). For quantification of peroxisome density in HeLa and MEF cells, the number of PMP70-stained punctate structures was determined by Volocity via the FindSpots function and divided by the volume of each cell to obtain the number of peroxisome per cubic micrometer. The mean peroxisome density was then normalized to the wild-type controls. At least 50 cells were analyzed for each trial, and at least three trials were performed for each condition as indicated in each figure.
Total PMP70 fluorescence
To determine the total fluorescent intensity of PMP70 within a cell, the fluorescent signal from the entire cell was obtained and subtracted from background from images taken with a fully opened pinhole. All treatments were prepared and imaged at the same time using the same settings. At least 50 cells were analyzed for each trial, and at least three trials were performed.
Immunoblotting
For cell lysates, cells were lysed with 100 mM Tris, pH 9.0, containing 1% SDS and Halt protease inhibitor cocktail (Thermo Fisher Scientific), and the lysate was heated at 95°C with vortexing for 15 min. For animal lysates, protein lysates were prepared by lysing cells in RIPA buffer, and samples were run on SDS polyacrylamide gel and transferred to nitrocellulose membrane. For pull-downs, cells were lysed in 1% SDS, diluted tenfold in 1% NP-40, pH 9, incubated with primary antibody and protein G Sepharose overnight, washed three times in 1% NP-40, and boiled in 1% SDS to release antibody from the beads.
Protein concentrations were determined by BCA assay (EMD Millipore). Equivalent sample amounts were subjected to SDS-PAGE. Protein was transferred to 0.45-µm BioTrace PVDF membrane (Pall) and probed with the appropriate primary and HRP-conjugated secondary antibodies following standard protocols.
Blots were developed using Luminata Crescendo (EMD Millipore) or with ECL Plus or ECL Advance reagents (GE Healthcare) or Super Signal West Dura-Extended Duration Substrate (Thermo Fisher Scientific). Proteins were detected on either a ChemiDoc (Bio-Rad Laboratories) or using a FluorChem E detector purchased from Protein Simple.
Immunofluorescence
For immunofluorescence, cells were fixed using 3.7% paraformaldehyde (Electron Microscopy Sciences) in PBS for 15 min and permeabilized using 0.1% Triton X-100 (Thermo Fisher Scientific) in PBS for 15 min, followed by incubation with the appropriate primary and secondary antibodies for 2 h and 1 h, respectively. Alexa Fluor 488 signal was acquired using a 488-nm argon laser with a 493- to 565-nm bandpass filter, and Alexa Fluor 561 signals were acquired using a 561-nm diode laser with a 600- to 700-nm bandpass filter.
Immunoprecipitation
HEK293 cells stably transfected with HA-Ub plasmid under the control of a tamoxifen promoter were treated with 10 µm tamoxifen for 24 h to induce the expression of HA-Ub. Cells were washed in PBS and lysed in 500 µl lysis buffer containing 1% NP-40, 1% SDS, 10 mM Tris-HCl buffered to pH 9, 133 mM NaCl, 1 mM EDTA, and protease inhibitor cocktail. The lysis buffer either did or did not contain 10 nM MG132 and 10 nM NEM as shown in Fig. 7 (A and B). Lysates were boiled for 15 min, cleared via centrifugation, and diluted to 5 ml in the same lysis buffer without SDS. Lysates were then incubated with primary antibody and protein G Sepharose overnight. Beads were then washed three times using the same lysis buffer, and proteins were eluted by boiling in SDS-containing sample buffer for 15 min.
Quantitative PCR
RNA was collected and isolated using the Promega RNA SV Isolation kit. A cDNA library was synthesized by first-strand synthesis using the Applied Biosystems High-Capacity cDNA Archive kit. Quantitative PCR was performed on the Applied Biosystems StepOne Real Time PCR System using TaqMan Fast Advanced Master Mix and FAM-labeled probes. Quantities of RNA transcripts are shown relative to Abt1 loading control.
Peroxisome purification
Peroxisomes were purified using the Peroxisome Isolation kit (PEROX1; Sigma-Aldrich) as described by the manufacturer. HA-Ub stably transfected HEK cells were simultaneously treated with 10 µM tamoxifen and transfected with plasmid. 8 h after transfection, the media was changed to remove lipofectamine and fresh tamoxifen was added. 24 h after transfection, cells were trypsinized, washed, and resuspended in Peroxisome Extraction Buffer (Sigma-Aldrich). The Sigma-Aldrich PEROX1 protocol was then followed, with the exceptions that a 5-mm metal cell homogenizer was used to mechanically lyse the cells and the final fractions were pelleted at 200,000 g and resuspended in 100 mM Tris, pH 9.0, containing 1% SDS. Two 15-cm dishes of cells were used for each treatment.
Rat liver hepatocyte extraction
Rat liver hepatocytes were isolated by a two-step perfusion method using collagenase as described by Moshage et al. (1990). In brief, the liver was first perfused through portal vein, with the dissected inferior vena cava used as the outflow port. Perfusion was performed with Ca2+-free Krebs-Ringer Hepes buffer, pH 7.4, maintained at 37°C (10 min; flow rate = 25 ml/min), followed by perfusion with Mg2+-free Krebs-Ringer Hepes buffer containing Ca2+ (5.7 mmol/l) and collagenase type I (0.12–0.16 U/ml, 10 min, flow rate 8 ml/min; Sigma-Aldrich). The liver was then removed and placed in the same buffer containing 1% BSA (Sigma-Aldrich) without collagenase. Hepatocytes were then released by gentle touch of the liver and filtered through 60-mesh sterile nylon gauze. Cells were washed three times with HBSS at 50 g for 5 min, and the supernatant was discarded. The final cell pellet was resuspended and cultured in William’s E medium at 37°C and 5% CO2.
Mouse studies
Immediately after weaning, male C57BL/6 mice (Charles River) were placed on either a control diet (18% protein; Envigo) or a low-protein diet (1% protein; Envigo) for a period of 12 d. Protein-restricted animals were also given carbohydrates (42 g/l with 55% fructose and 45% sucrose by weight) dissolved in their drinking water to ensure they met their caloric requirement. For chloroquine treatment, animals on the low-protein diet were administered with chloroquine (50 mg/kg/day; Alfa Aesar) daily by intraperitoneal injection starting on the first day of diet. All animals had ad libitum access to food and were housed in Lab Animal Services at the Hospital for Sick Children (Toronto, Canada). Animals were sacrificed by cervical dislocation, and liver tissue was snap frozen after harvest and stored at −80°C until analysis or fixed in 10% neutral buffered formalin and embedded in paraffin. 4-µm-thick tissue sections were deparaffinized in three changes of xylene and rehydrated in a series of alcohol. Antigen retrieval was done through digestion with 0.01% trypsin (10 min at 37°C) followed by microwave heating in 10 mM sodium citrate buffer, pH 6.0, for PMP70. To block nonspecific binding sites, sections were treated with blocking buffer containing 10% goat serum, 3% BSA in TBS, pH 7.4, with 0.1% Tween 20, and stained with rabbit polyclonal antibody against PMP70 (1:2,000) overnight at 4°C. Goat anti–rabbit Alexa Fluor 488 antibody (1:1,000; A-11011; Invitrogen) was used for visualization of PMP70. Nuclei were stained with DAPI. All samples were prepared and imaged at the same time using the microscopy settings as described in the Microscopy and data anaylsis section.
For immunoblot analysis, the frozen liver tissues were homogenized in tissue lysis buffer (FNN0071; Thermo Fisher Scientific) with protease inhibitor (P2714; Sigma-Aldrich), and 20 ng protein was separated on a gradient gel (Bolt 4–12% Bis-Tris Plus, NW04120Box; Thermo Fisher Scientific) and transferred to a PVDF membrane (iBlot Transfer stack, IB4010-02; Thermo Fisher Scientific). The membranes were then incubated with primary antibody as indicated overnight at 4°C followed by the complementing HRP secondary antibody. Super Signal West Dura-Extended Duration Substrate (Thermo Fisher Scientific) was used to detect the immunocomplexes, which were then visualized using a LI-COR Biosciences Odyssey FC imaging system.
Online supplemental material
Fig. S1 shows the same HeLa cell expressing PEX2-GFP at two different brightness settings to demonstrate that the apparent loss of peroxisomes is not caused by a morphological change. Fig. S2 shows the mRNA levels of the peroxisomal E3 ligases as determined by quantitative PCR to show that the siRNA constructs used are specific to their target genes. Fig. S3 shows that the PEX2-siR-FLAG construct can complement siPEX2-1. Fig. S4 shows that PEX2 protein expression is stabilized by the inhibition of proteasomes. Fig. S5 shows that the endogenous level of PEX2 regulation occurs on a similar timescale as LC3 during both nitrogen starvation and rapamycin treatment. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201511034/DC1.
Supplementary Material
Acknowledgments
We are grateful to Ms. Rong Hua and Dr. Yuqing Wang for critical reading of the manuscript, Ms. Rong Hua for discussion on Manders’ colocalization coefficient calculation, and Drs. Brian Raught and John Brumell for providing the HA-Ub stable HEK293 cell line and ATG5 knockout MEF, respectively.
This work is supported by an operating grant from the Canadian Institutes of Health Research (to P.K. Kim), the Ontario Early Researcher Award (to P.K. Kim), and the North American Society for Pediatric Gastroenterology, Hepatology, and Nutrition Young Investigator Development Award (to R. Bandsma).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- ATG
- autophagy-related gene
- HA-Ub
- HA-tagged ubiquitin
- HEK
- human embryonic kidney
- MEF
- mouse embryonic fibroblast
- NBR1
- neighbor of BRCA1
- PEX
- peroxin
- PMP
- peroxisomal membrane protein
References
- Blackstone C., and Chang C.-R.. 2011. Mitochondria unite to survive. Nat. Cell Biol. 13:521–522. 10.1038/ncb0511-521 [DOI] [PubMed] [Google Scholar]
- Brooks S.E., Goldon M.H., and Taylor E.. 1992. Hepatic ultrastructure in children with protein-energy malnutrition. West Indian Med. J. 41:139–145. [PubMed] [Google Scholar]
- Brown A.I., Kim P.K., and Rutenberg A.D.. 2014. PEX5 and ubiquitin dynamics on mammalian peroxisome membranes. PLOS Comput. Biol. 10:e1003426 10.1371/journal.pcbi.1003426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyaud E., Mis M., Laurent E.M.N., Dunham W.H., Couzens A.L., Robitaille M., Gingras A.C., Angers S., and Raught B.. 2015. BioID-based identification of Skp Cullin F-box (SCF)β-TrCP1/2 E3 ligase substrates. Mol. Cell. Proteomics. 14:1781–1795. 10.1074/mcp.M114.045658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deosaran E., Larsen K.B., Hua R., Sargent G., Wang Y., Kim S., Lamark T., Jauregui M., Law K., Lippincott-Schwartz J., et al. 2013. NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 126:939–952. 10.1242/jcs.114819 [DOI] [PubMed] [Google Scholar]
- Deshaies R.J., and Joazeiro C.A.. 2009. RING domain E3 ubiquitin ligases. Annu. Rev. Biochem. 78:399–434. 10.1146/annurev.biochem.78.101807.093809 [DOI] [PubMed] [Google Scholar]
- Dodt G., and Gould S.J.. 1996. Multiple PEX genes are required for proper subcellular distribution and stability of Pex5p, the PTS1 receptor: evidence that PTS1 protein import is mediated by a cycling receptor. J. Cell Biol. 135:1763–1774. 10.1083/jcb.135.6.1763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Magraoui F., Bäumer B.E., Platta H.W., Baumann J.S., Girzalsky W., and Erdmann R.. 2012. The RING-type ubiquitin ligases Pex2p, Pex10p and Pex12p form a heteromeric complex that displays enhanced activity in an ubiquitin conjugating enzyme-selective manner. FEBS J. 279:2060–2070. 10.1111/j.1742-4658.2012.08591.x [DOI] [PubMed] [Google Scholar]
- Faust P.L., and Hatten M.E.. 1997. Targeted deletion of the PEX2 peroxisome assembly gene in mice provides a model for Zellweger syndrome, a human neuronal migration disorder. J. Cell Biol. 139:1293–1305. 10.1083/jcb.139.5.1293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomes L.C., Di Benedetto G., and Scorrano L.. 2011. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat. Cell Biol. 13:589–598. 10.1038/ncb2220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grover Z., and Ee L.C.. 2009. Protein energy malnutrition. Pediatr. Clin. North Am. 56:1055–1068. 10.1016/j.pcl.2009.07.001 [DOI] [PubMed] [Google Scholar]
- Hara-Kuge S., and Fujiki Y.. 2008. The peroxin Pex14p is involved in LC3-dependent degradation of mammalian peroxisomes. Exp. Cell Res. 314:3531–3541. 10.1016/j.yexcr.2008.09.015 [DOI] [PubMed] [Google Scholar]
- Hartl F.-U., and Just W.W.. 1987. Integral membrane polypeptides of rat liver peroxisomes : response to different metabolic states. Arch Biochem Biophys. 255:109–119. http:/dx.doi.org/10.1016/0003-9861(87)90300-6 [DOI] [PubMed] [Google Scholar]
- Hashimoto T., Kuwabara T., and Usuda N.. 1986. Purification of membrane polypeptides of rat liver peroxisomes. J Biochem. 100:301–310. [DOI] [PubMed] [Google Scholar]
- Ivashchenko O., Van Veldhoven P.P., Brees C., Ho Y.-S., Terlecky S.R., and Fransen M.. 2011. Intraperoxisomal redox balance in mammalian cells: oxidative stress and interorganellar cross-talk. Mol. Biol. Cell. 22:1440–1451. 10.1091/mbc.E10-11-0919 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang L., Hara-Kuge S., Yamashita S., and Fujiki Y.. 2015. Peroxin Pex14p is the key component for coordinated autophagic degradation of mammalian peroxisomes by direct binding to LC3-II. Genes Cells. 20:36–49. 10.1111/gtc.12198 [DOI] [PubMed] [Google Scholar]
- Khaminets A., Behl C., and Dikic I.. 2016. Ubiquitin-dependent and independent signals in selective autophagy. Trends Cell Biol. 26:6–16. 10.1016/j.tcb.2015.08.010 [DOI] [PubMed] [Google Scholar]
- Kim J., Kundu M., Viollet B., and Guan K.L.. 2011. AMPK and mTOR regulate autophagy through direct phosphorylation of Ulk1. Nat. Cell Biol. 13:132–141. 10.1038/ncb2152 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim P.K., Hailey D.W., Mullen R.T., and Lippincott-Schwartz J.. 2008. Ubiquitin signals autophagic degradation of cytosolic proteins and peroxisomes. Proc. Natl. Acad. Sci. USA. 105:20567–20574. 10.1073/pnas.0810611105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirisako T., Baba M., Ishihara N., Miyazawa K., Ohsumi M., Yoshimori T., Noda T., and Ohsumi Y.. 1999. Formation process of autophagosome is traced with Apg8/Aut7p in yeast. J. Cell Biol. 147:435–446. 10.1083/jcb.147.2.435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klionsky D.J., Abdelmohsen K., Abe A., Abedin J., Abeliovich H., Arozena A.A., Adachi H., Adams M., Adams P.D., Adeli K., et al. 2016. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy. 12:1–22. 10.1080/15548627.2015.1100356 [DOI] [PMC free article] [PubMed]
- Kraft C., Deplazes A., Sohrmann M., and Peter M.. 2008. Mature ribosomes are selectively degraded upon starvation by an autophagy pathway requiring the Ubp3p/Bre5p ubiquitin protease. Nat. Cell Biol. 10:602–610. 10.1038/ncb1723 [DOI] [PubMed] [Google Scholar]
- Kuma A., Hatano M., Matsui M., Yamamoto A., Nakaya H., Yoshimori T., Ohsumi Y., Tokuhisa T., and Mizushima N.. 2004. The role of autophagy during the early neonatal starvation period. Nature. 432:1032–1036. 10.1038/nature03029 [DOI] [PubMed] [Google Scholar]
- Lynch-Day M.A., Mao K., Wang K., Zhao M., and Klionsky D.J.. 2012. The role of autophagy in Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2:a009357 10.1101/cshperspect.a009357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumura T., Otera H., and Fujiki Y.. 2000. Disruption of the interaction of the longer isoform of Pex5p, Pex5pL, with Pex7p abolishes peroxisome targeting signal type 2 protein import in mammals. Study with a novel Pex5-impaired Chinese hamster ovary cell mutant. J. Biol. Chem. 275:21715–21721. 10.1074/jbc.M000721200 [DOI] [PubMed] [Google Scholar]
- Moshage H., Casini A., and Lieber C.S.. 1990. Acetaldehyde selectively stimulates collagen production in cultured rat liver fat-storing cells but not in hepatocytes. Hepatology. 12:511–518. 10.1002/hep.1840120311 [DOI] [PubMed] [Google Scholar]
- Nazarko T.Y., Ozeki K., Till A., Ramakrishnan G., Lotfi P., Yan M., and Subramani S.. 2014. Peroxisomal Atg37 binds Atg30 or palmitoyl-CoA to regulate phagophore formation during pexophagy. J. Cell Biol. 204:541–557. 10.1083/jcb.201307050 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordgren M., Wang B., Apanasets O., and Fransen M.. 2013. Peroxisome degradation in mammals: mechanisms of action, recent advances, and perspectives. Front. Physiol. 4:145 10.3389/fphys.2013.00145 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordgren M., Francisco T., Lismont C., Hennebel L., Brees C., Wang B., Van Veldhoven P.P., Azevedo J.E., and Fransen M.. 2015. Export-deficient monoubiquitinated PEX5 triggers peroxisome removal in SV40 large T antigen-transformed mouse embryonic fibroblasts. Autophagy. 11:1326–1340. 10.1080/15548627.2015.1061846 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumoto K., Bogaki A., Tateishi K., Tsukamoto T., and Osumi T.. 1997. Isolation and characterization of peroxisome-deficient Chinese hamster ovary cell mutants representing human complementation group III. Exp. Cell Res. 233:11–20. 10.1006/excr.1997.3552 [DOI] [PubMed] [Google Scholar]
- Platta H.W., El Magraoui F., Schlee D., Grunau S., Girzalsky W., and Erdmann R.. 2007. Ubiquitination of the peroxisomal import receptor Pex5p is required for its recycling. J. Cell Biol. 177:197–204. 10.1083/jcb.200611012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platta H.W., El Magraoui F., Bäumer B.E., Schlee D., Girzalsky W., and Erdmann R.. 2009. Pex2 and pex12 function as protein-ubiquitin ligases in peroxisomal protein import. Mol. Cell. Biol. 29:5505–5516. 10.1128/MCB.00388-09 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestele J., Hierl G., Scherling C., Hetkamp S., Schwechheimer C., Isono E., Weckwerth W., Wanner G., and Gietl C.. 2010. Different functions of the C3HC4 zinc RING finger peroxins PEX10, PEX2, and PEX12 in peroxisome formation and matrix protein import. Proc. Natl. Acad. Sci. USA. 107:14915–14920. 10.1073/pnas.1009174107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rambold A.S., Kostelecky B., Elia N., and Lippincott-Schwartz J.. 2011. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc. Natl. Acad. Sci. USA. 108:10190–10195. 10.1073/pnas.1107402108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimura H., Hattori N., Kubo S., Mizuno Y., Asakawa S., Minoshima S., Shimizu N., Iwai K., Chiba T., Tanaka K., and Suzuki T.. 2000. Familial Parkinson disease gene product, parkin, is a ubiquitin-protein ligase. Nat. Genet. 25:302–305. 10.1038/77060 [DOI] [PubMed] [Google Scholar]
- Van Zutphen T., Ciapaite J., Bloks V.W., Ackereley C., Gerding A., Jurdzinski A., De Moraes R.A., Zhang L., Wolters J.C., Bischoff R., et al. 2016. Malnutrition-associated liver steatosis and ATP depletion is caused by peroxisomal and mitochondrial dysfunction. J. Hepatol. In press. 10.1016/j.jhep.2016.05.046 [DOI] [PubMed]
- Walter K.M., Schönenberger M.J., Trötzmüller M., Horn M., Elsässer H.-P., Moser A.B., Lucas M.S., Schwarz T., Gerber P.A., Faust P.L., et al. 2014. Hif-2α promotes degradation of mammalian peroxisomes by selective autophagy. Cell Metab. 20:882–897. 10.1016/j.cmet.2014.09.017 [DOI] [PubMed] [Google Scholar]
- Yamashita S., Abe K., Tatemichi Y., and Fujiki Y.. 2014. The membrane peroxin PEX3 induces peroxisome-ubiquitination-linked pexophagy. Autophagy. 10:1549–1564. 10.4161/auto.29329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z., and Klionsky D.J.. 2010. Mammalian autophagy: core molecular machinery and signaling regulation. Curr. Opin. Cell Biol. 22:124–131. 10.1016/j.ceb.2009.11.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Tripathi D.N., Jing J., Alexander A., Kim J., Powell R.T., Dere R., Tait-Mulder J., Lee J.-H., Paull T.T., et al. 2015. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol. 17:1259–1269. 10.1038/ncb3230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y., Nicholatos J., Dreier J.R., Ricoult S.J.H., Widenmaier S.B., Hotamisligil G.S., Kwiatkowski D.J., and Manning B.D.. 2014. Coordinated regulation of protein synthesis and degradation by mTORC1. Nature. 513:440–443. 10.1038/nature13492 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.