Organogenesis is regulated by specialized clusters of cells, the signaling centers. Their molecular regulation is well known, but the cellular behaviors involved remain ill defined. Here, Ahtiainen et al. use live imaging of embryonic mouse teeth to unveil cellular mechanisms driving epithelial invagination.
Abstract
During organogenesis, cell fate specification and patterning are regulated by signaling centers, specialized clusters of morphogen-expressing cells. In many organs, initiation of development is marked by bud formation, but the cellular mechanisms involved are ill defined. Here, we use the mouse incisor tooth as a model to study budding morphogenesis. We show that a group of nonproliferative epithelial cells emerges in the early tooth primordium and identify these cells as a signaling center. Confocal live imaging of tissue explants revealed that although these cells reorganize dynamically, they do not reenter the cell cycle or contribute to the growing tooth bud. Instead, budding is driven by proliferation of the neighboring cells. We demonstrate that the activity of the ectodysplasin/Edar/nuclear factor κB pathway is restricted to the signaling center, and its inactivation leads to fewer quiescent cells and a smaller bud. These data functionally link the signaling center size to organ size and imply that the early signaling center is a prerequisite for budding morphogenesis.
Introduction
Cellular behaviors, including cell proliferation, movement, and differentiation, are regulated in part by morphogens during embryonic development. In many organs, signaling molecules are produced by groups of specialized cells, signaling centers that control cell fates and cellular behaviors in the surrounding tissue. Well-defined examples include the apical ectodermal ridge and zone of polarizing activity of the developing limb, the isthmus regulating midbrain and hindbrain regionalization, the notochord that patterns the neural tube and the somatic mesoderm, and the enamel knot that instructs tooth morphogenesis (Jernvall and Thesleff, 2000; Partanen, 2007; Towers et al., 2012). These signaling centers share features: they are composed of a relatively small number of cells that regulate cell fates and behaviors by secreted factors, including sonic hedgehog (Shh) and members of the fibroblast growth factor (Fgf), bone morphogenetic protein (Bmp), and Wnt families. The signals expressed by the signaling centers are well characterized, but less is known about the origins and fate of signaling center cells and the cellular behaviors they regulate.
Ectodermal organs such as hair follicles, feathers, and mammary glands are initiated as a thickening of the epithelium called a placode (Pispa and Thesleff, 2003). Soon after, the underlying mesenchymal cells condense, and the epithelium invaginates to produce a bud (Jernvall and Thesleff, 2000; Schmidt-Ullrich and Paus, 2005). Like other ectodermal appendages, teeth form through epithelial–mesenchymal interactions and proceed via similar early stages (Jussila and Thesleff, 2012). In mice, tooth morphogenesis is initiated around embryonic day 11 (E11) when a horseshoe-shaped epithelial thickening, the dental lamina, appears in both jaws. Gene expression analyses indicate that the continuous dental lamina resolves into two separate domains in each jaw half, the incisor and the molar placodes, between E11 and E12 (Biggs and Mikkola, 2014). Genes that are initially expressed along the entire dental lamina and later become confined to incisor and molar primordia include Foxi3, Pitx2, and Sox2 (St Amand et al., 2000; Juuri et al., 2013; Shirokova et al., 2013). In addition, several signaling molecules such as Shh, Wnt10a/b, and Bmp2 are expressed in a more restricted manner (Dassule and McMahon, 1998; Keränen et al., 1998), suggesting the presence of a specific signaling center within the tooth placodes (Dassule and McMahon, 1998; Jernvall and Thesleff, 2000). However, the exact identity and function of this signaling center has remained obscure.
After the placode stage, tooth morphogenesis proceeds through bud, cap, and bell stages before hard tissue mineralization (Tummers and Thesleff, 2009). Immediately before the transition from the bud to the cap stage, a well-defined signaling center called the primary enamel knot (EK) forms at the tip of the bud and exhibits restricted expression of Shh, multiple members of the Bmp, Fgf, and Wnt families, and the cyclin-dependent kinase inhibitor gene p21 (Jernvall et al., 1998; Sarkar and Sharpe, 1999; Jernvall and Thesleff, 2000). In addition, expression of Edar, the receptor of the Tnf-like ligand ectodysplasin (Eda), is restricted to the EK (Tucker et al., 2000; Laurikkala et al., 2001). The EK is essential in controlling later stages of folding and patterning of the dental epithelium, where it stimulates proliferation of the mesenchyme and the surrounding epithelial cells, whereas cells within the EK remain nonproliferative (Jernvall and Thesleff, 2000; Jussila and Thesleff, 2012).
The molecular regulation of early tooth morphogenesis has been studied extensively, and a large number of mouse mutants displaying early developmental arrest have been described (Bei, 2009; Jussila and Thesleff, 2012; Biggs and Mikkola, 2014; Lan et al., 2014). However, the cellular behaviors contributing to dental morphogenesis are just beginning to be understood. Here, we use the mandibular incisor as a model to study the formation and function of the poorly characterized early signaling center. With the aid of Fucci cell cycle indicator mice and live laser scanning confocal tissue microscopy, we have recently shown that the cellular mechanisms driving hair placode formation include a switch to G1 phase and centripetal cell migration (Ahtiainen et al., 2014). The current study shows that in tooth placodes only a subset of cells become quiescent and form an early signaling center. These cells exhibit migratory behavior whereby they condense and form the mature signaling center. The signaling center cells remain nonproliferative, whereas a burst of cell proliferation in the neighboring dental epithelial cells leads to bud formation. We show that Eda/Edar/NF-κB signaling regulates the number of signaling center cells and thereby tooth bud size. Finally, our findings indicate that the EK is not derived from the early signaling center cells, but rather it forms de novo at the tip of the tooth bud.
Results
A subset of epithelial cells remain in G1 phase throughout embryonic mouse incisor placode and bud morphogenesis
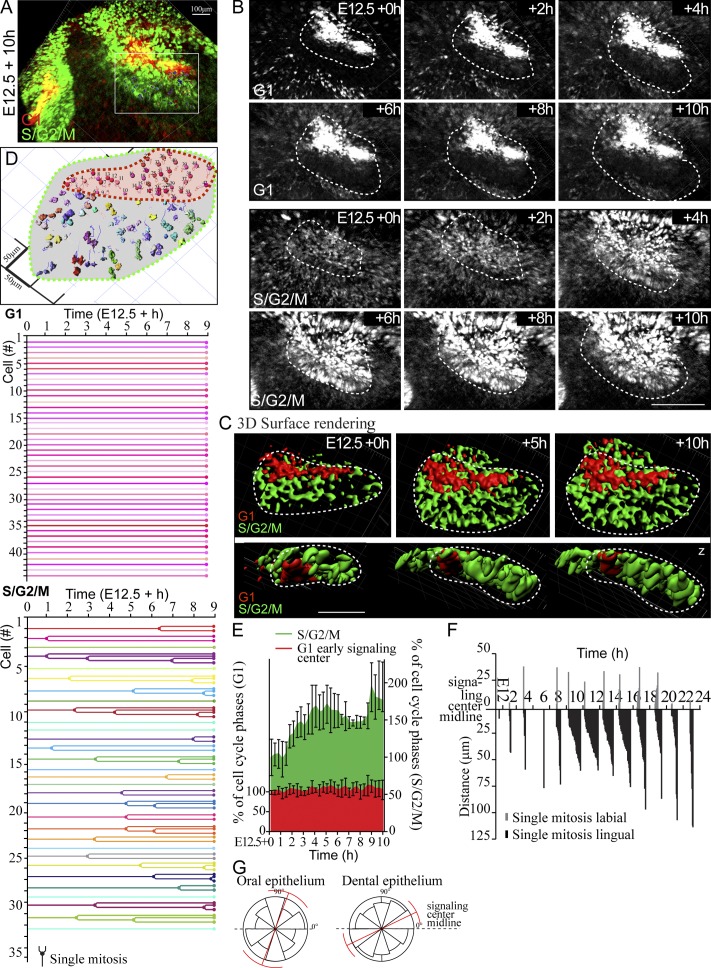
To understand cellular mechanisms in early tooth morphogenesis, we analyzed cell cycle dynamics in developing mandibular incisors before the onset of morphological development (E10.5), at dental lamina stage (E11.0–11.5), placode stage (E12.0–E12.5), and early (E13.0) and late (E13.5) bud stage. Fluorescent cell cycle indicator (Fucci) reporter mice (Sakaue-Sawano et al., 2008) were used to monitor cell cycle status. At E10.5 and E11.0, G1/G0 (hereafter G1) and S/G2/M phase cells (identified by nuclear red and green fluorescence, respectively) were evenly distributed throughout the mandible (Fig. S1). At E11.5, however, a clear clustering of G1 cells was observed; the G1 focus seemed to coincide with the dental lamina. Between E11.5 and E12.0, the G1 foci in the prospective incisor and molar region were connected initially, but by E12.5 they were fully split into two separate G1 foci (Fig. S1).
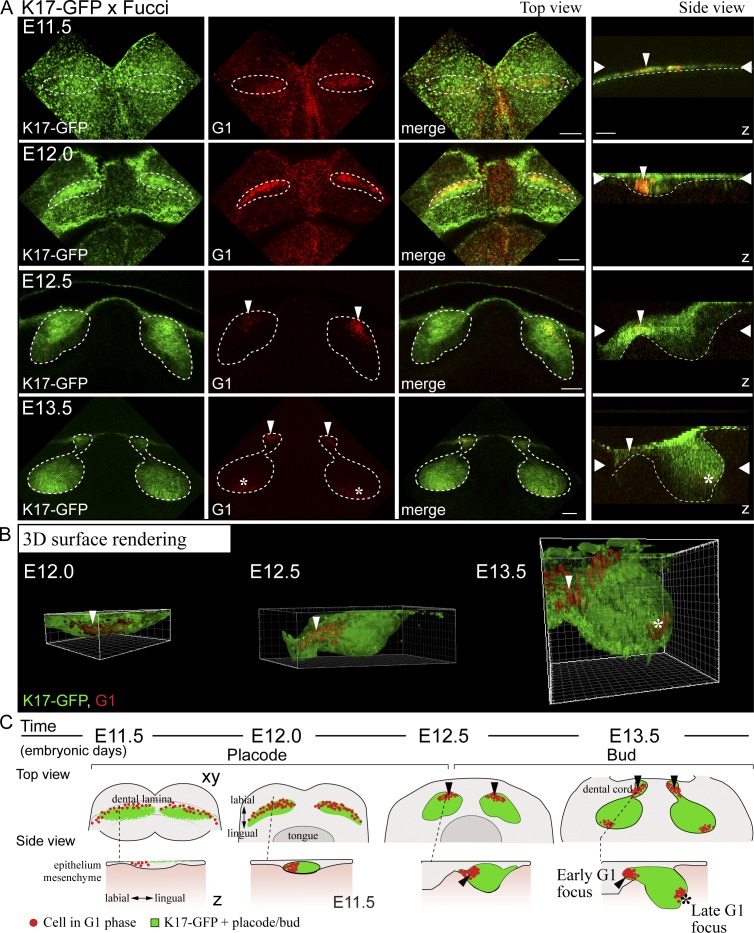
To assess the location and identity of the G1 foci, we analyzed these cells in the developing incisors of transgenic mice expressing Keratin17-GFP (K17-GFP), a marker for the epithelium of ectodermal appendages (McGowan and Coulombe, 1998). K17-GFP was initially expressed in the periderm throughout the oral epithelium (Fig. 1 A). At E12.0, K17-GFP expression became more restricted to the forming placode, and a loose band of nonproliferating Fucci G1 cells was seen in the labial part of this area. At E12.5, the G1 cells had condensed into a restricted area closer to the mandibular midline (Fig. 1, A and B). At the late bud stage (E13.5), two G1 foci were evident in the incisors: one in the dental cord (junction between the bud and oral epithelium) and a new distinct cluster of cells at the tip of the bud (Fig. 1, A and B). This G1 focus corresponds to the known location of the emerging EK (Jernvall et al., 1998; Munne et al., 2009). A schematic representation illustrates the G1 cell populations in incisor morphogenesis (Fig. 1 C). To conclude, there are distinct subpopulations of epithelial cells in the forming incisor that exhibit cell cycle cessation in a specific spatiotemporal pattern.
Figure 1.
A subset of epithelial cells are nonproliferative throughout incisor placode and bud morphogenesis. Confocal fluorescence images of mouse embryonic mandible expressing keratin 17-GFP (green) and fluorescent cell cycle indicator for G1 cell cycle phase (red). (top view) Each figure is an optical section in xy plane, and the dotted line represents the prospective tooth area. (side view) Each figure is a projection in z plane, the epithelial border is delineated with a dotted line, and wide arrowheads represent the position of the xy-section in the z-axis. (A) K17-GFP was initially expressed throughout the oral epithelium in the superficial peridermal layer. At E11.5, the dental lamina appeared as a thickening of the epithelium. There was a diffuse stripe of nonproliferating Fucci G1 cells appearing along the dental lamina (marked by thin arrowhead in z-projection). K17-GFP expression became restricted to the forming placode cells between E12.0 and 12.5, and there was specific subset of cells in G1 in the placode, localized to the labial part of the thickening (arrowhead). At E12.5, invagination of the tooth bud began, and G1 cell focus became restricted at the anterior part of the bud. At E13.5, the early G1 foci remained in the dental cord, and new distinct G1 foci appeared in the tip of the bud (asterisk). Bars, 100 µm. (B) 3D surface rendering of the forming incisor tooth placode/bud shows the initial G1 focus at E12.0 and 12.5 (thin arrowhead) in the anterior part of the bud, while a new G1 focus appeared at the tip of the bud (asterisk). (C) A schematic representation of the G1 cell populations in the developing incisor tooth placode/bud. The early G1 focus in the anterior part of the forming tooth bud is separate from the G1 focus corresponding with the EK.
The early G1 cell population colocalizes with signaling center markers
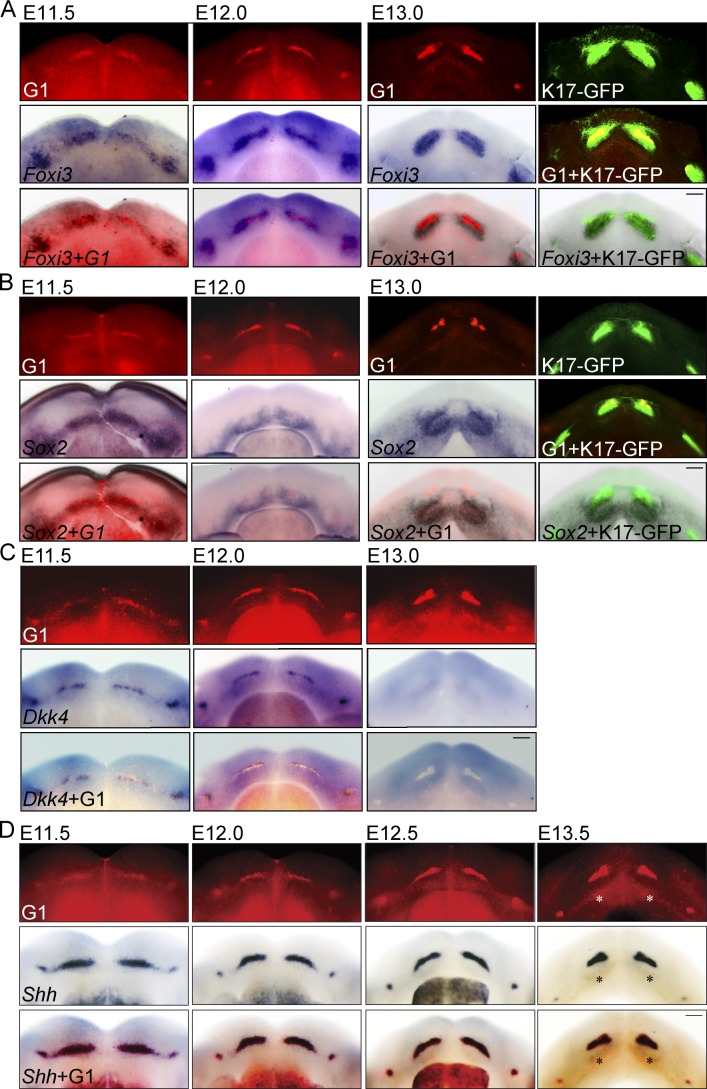
To characterize the G1 cell populations, mandibles were first imaged by fluorescence microscopy and then subjected to whole-mount in situ hybridization for known dental epithelial markers. Foxi3 is expressed early in the dental lamina and later at placode and bud stages; its expression spans the entire thickened dental epithelium but is absent from the oral epithelium (Shirokova et al., 2013). Dkk4 is expressed as a narrow stripe along the prospective dental epithelium from E10.5 onward (Bazzi et al., 2007; Fliniaux et al., 2008). Sox2 is initially expressed in the dental lamina during E12 and becomes confined to incisor and molar regions (Juuri et al., 2013). Shh is expressed in the dental lamina and a subset of epithelial cells throughout early tooth morphogenesis and marks a putative early signaling center (Dassule and McMahon, 1998; Keränen et al., 1998; Hovorakova et al., 2011).
We found that the labial G1 population coincided with Foxi3 expression in the dental lamina at E11.5 and later in the forming placode and bud (Fig. 2 B). However, the Foxi3 expression domain was broader at all time points and colocalized with K17-GFP positivity (E12.0–E13.0; Fig. 2 A). In contrast, Sox2-expressing cells colocalized only partially with the early G1 population and K17-GFP–positive cells and mainly resided lingual to them (Fig. 2 B and Fig. S2, A and B). Dkk4 colocalized with the G1 population but was down-regulated at E13.0, whereas the G1 focus still persisted (Fig. 2 C). Interestingly, Shh expression colocalized with the G1 cell population at all observed time points (E11.5, 12.0, 12.5, and 13.5; Fig. 2 D), identifying this subset of cells as an early epithelial signaling center.
Figure 2.
G1 cell population domain coincides with Shh expression in placodes and buds. Fucci G1 cell cycle transgene expression in control or K17-GFP background was analyzed with fluorescence microscopy. The same specimens were then analyzed by whole-mount in situ hybridization with probes specific for Foxi3, Sox2, Dkk4, or Shh. (A) The early G1 population coincided with, but was more restricted than, the Foxi3 expression domain in the dental lamina at E11.5 and later in the forming placode and bud. Foxi3 expression colocalized with K17-GFP positivity. (B) Early G1 population and K17-GFP–positive cells partially colocalized, but mostly resided buccally to Sox2-expressing cells. (C) Dkk4 colocalized with the G1 population; however, by E13.0 Dkk4 expression was already down-regulated, whereas the G1 focus persisted in the anterior part of the incisor bud. (D) Shh expression colocalized with the G1 cell population at all observed time points (E11.5, 12.5, 12.0, and 13.5; emerging enamel knot marked with an asterisk). This identifies the early G1 cell population as an early signaling center characterized by cell cycle cessation and signaling center marker expression. Bars, 100 µm.
The number of G1 cells stays constant through placode and bud stages
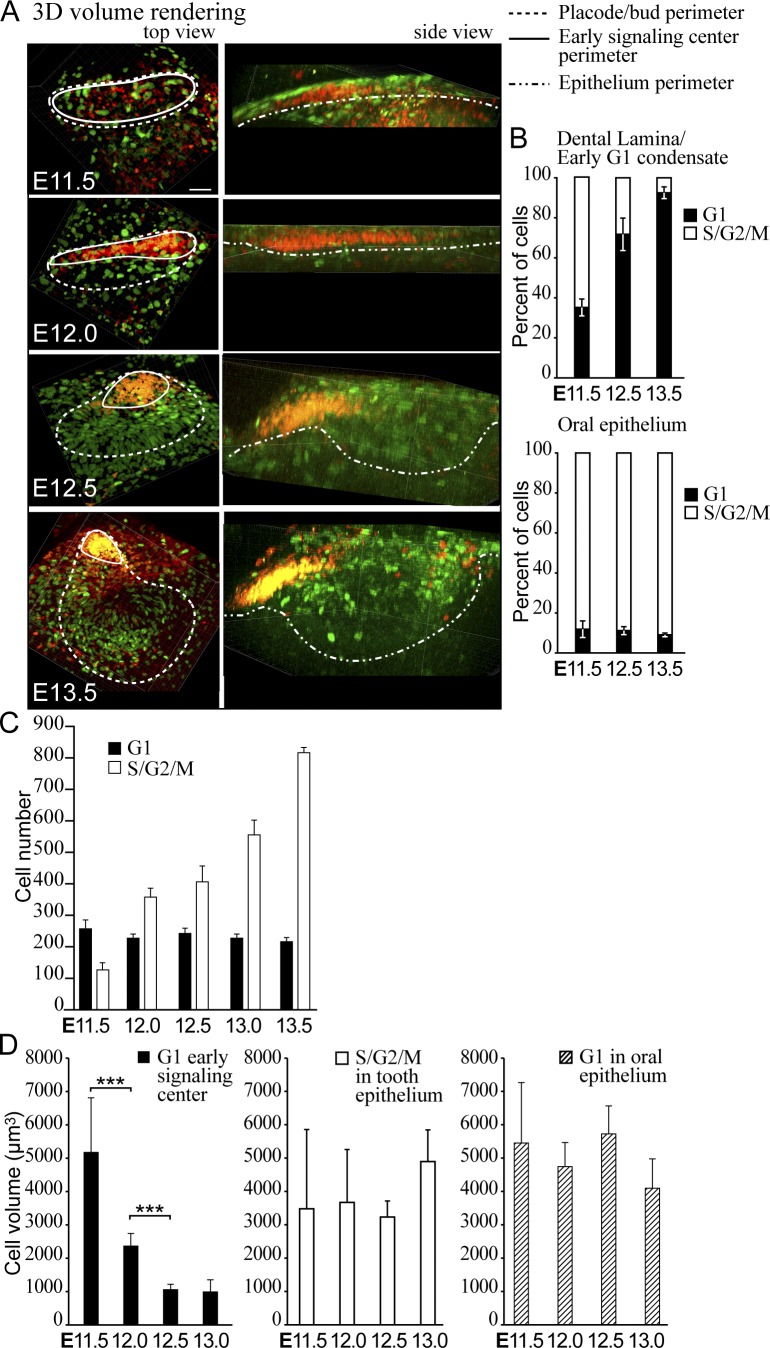
The early focus of G1 cells was restricted to a progressively smaller area, whose shape changed from a long narrow stripe into a more triangular shape in whole-mount view (Fig. 2) and also in 3D volume renderings from fluorescence confocal z-stacks (Fig. 3 A and Video 1). To understand the mechanisms of this phenomenon, we assayed the cell number and volume in the G1 population.
Figure 3.
G1 cells compact, but their number stays constant during budding morphogenesis. (A) 3D volume renderings of fluorescence confocal images of whole-mount samples show the contribution of all G1 (red) and S/G2/M (green) cells at different stages of incisor bud formation. Top view: dotted line, placode/bud volume (quantification of cell number in C); solid line, representative volumes of the G1 signaling center foci (quantifications of cell percentages/numbers in B and C). Side view: epithelium volume is delineated with a dotted line. Bar, 50 µm. (B) Quantification of the percentage of cells in G1 and S/G2/M stages in the oral epithelium not contributing to the incisor showed a low number of nonproliferating cells throughout morphogenesis. In contrast to 38 ± 7.7% cells in the dental lamina, at E12.5 and later, the majority of cells in the early condensate area were in G1 (ncells oral epithelium = 350 and ncells lamina/condensate = 700). (C) Quantification of the number of cells in G1 and S/G2/M phase in the placode/bud showed a constant number of G1 cells, whereas the number of proliferating cells increased (nplacodes = 8; data shown are means). (D) Quantification of cell volume in the early signaling center and oral epithelium showed a significant reduction of cell volume over time in the G1 early signaling center population but not in the S/G2/M population or in G1 cells elsewhere in the oral epithelium (Student’s t test, ***, P < 0.001; nplacodes/time point = 8; data shown are means ± SD).
G1 cells remained in the front part of the developing bud, whereas proliferating cells appeared on the lingual side as shown by Fucci S/G2/M reporter (Fig. 3 A). This was also seen by incorporation of 5-ethynyl-2′-deoxyurinide (EdU) nucleoside analog delivered via in vivo injections of pregnant K17-GFP;Fucci G1 dams at E11.5, 12.5, and 13.5 (Fig. S4 A). Quantification of percentage of cells in G1 in the condensate area showed a specific increase in the proportion of G1 cells, whereas in the oral epithelium most of the cells remained proliferative (Fig. 3 B). However, the absolute number of G1 cells in the early signaling center remained constant throughout morphogenesis, whereas the number of S/G2/M cells adjacent to the early signaling center increased significantly (Fig. 3 C).
Because reduction in G1 cell number cannot explain the restriction of the early signaling center area, we next measured cell volume changes during placode formation. The cell volume in the early signaling center decreased significantly with time, whereas the volume of S/G2/M cells in the tooth epithelium and G1 cells in the oral epithelium stayed constant (Fig. 3 D and Fig. S3), showing that the early signaling center cells become more compact as the early signaling center changes shape.
Differential cell migration contributes to early signaling center condensation
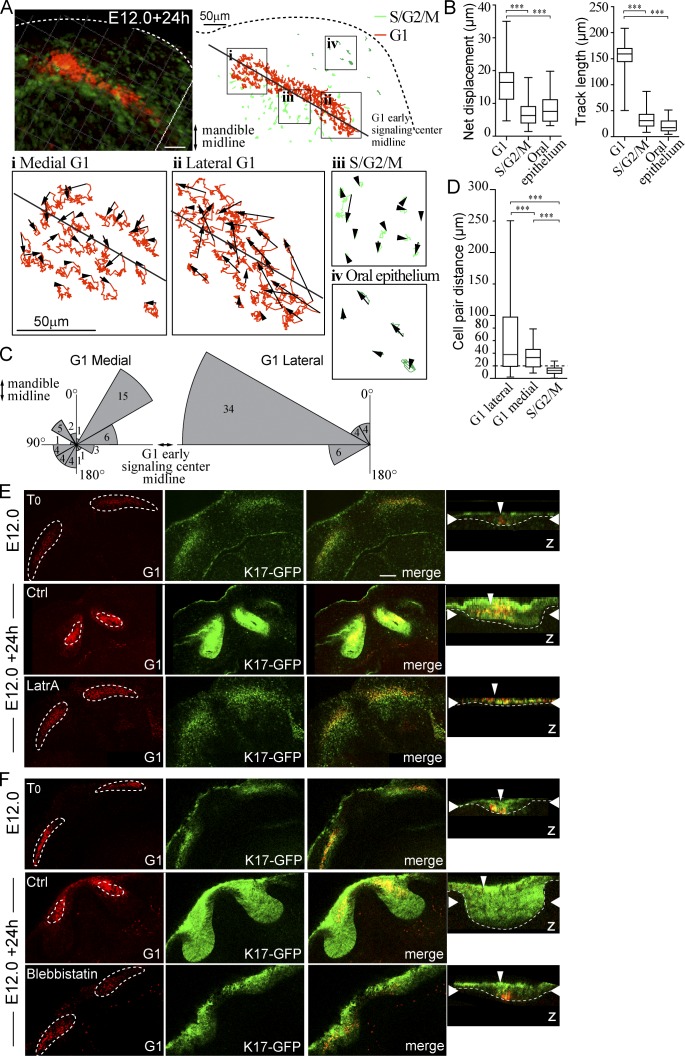
To study the cellular behaviors that mediate early signaling center compaction during placode morphogenesis, we used Fucci whole-mount explant cultures and live confocal fluorescence microscopy. In E12.0 mandibles, the incisor G1 focus is distinguishable as a narrow stripe. These mandibles were dissected and imaged for 24 h, and cell movement was tracked in four distinct cell populations: the medial and lateral G1 signaling center cells, the adjacent S/G2/M population, and cells of the oral epithelium not contributing to tooth formation. The cells in the lateral part of the G1 signaling center migrated directionally toward the medial midline along the dental lamina (Fig. 4 A). In contrast, the cells in the proximal part of the area showed smaller overall displacement. The S/G2/M placodal cells directly adjacent to the G1 subpopulation and oral epithelial cells outside the placode area stayed localized (Fig. 4 A). Quantification revealed that both the net displacement and track length were significantly higher in G1 early signaling center cells compared with the adjacent S/G2/M and oral epithelial cells (Fig. 4 B). The G1 cells moved up to 35 µm from their original position, whereas the other two cell populations moved on average less than their own circumference, thus staying nonmotile (Fig. 4 B). Analysis of the angle of movement of G1 cells showed that the lateral G1 cells, but not those close to the mandibular midline, displayed directional movement toward the midline (Fig. 4 C).
Figure 4.
Differential cell migration contributes to early signaling center condensation. Live confocal fluorescence microscopy of Fucci transgenes in mandibular whole-mount explant was used to characterize cell movement during placode formation. Explants were imaged at E12.0 + 24 h, and movement of placode cells, adjacent S/G2/M population, and cells of the oral epithelium was tracked. (A) Cell tracks in a representative incisor in four locations: (i) medial G1, (ii) lateral G1, (iii) S/G2/M in close proximity to the early signaling center, and (iv) S/G2/M in the oral epithelium. (B) Quantification of net displacement and track length showed significantly higher values in G1 early signaling center cells (medial and lateral populations together) compared with the adjacent S/G2/M and oral epithelial cells (Student’s t test, ***, P < 0.001; nareas = 5 and ncells = 30; data shown are means ± SD). The early signaling center G1 cells exhibited higher motility with respect to other epithelial cell populations. (C) G1 cell movement angles showed persistent directional movement by the lateral cells toward the mandible midline. Medial G1 cells, in contrast, moved toward the center of the early signaling center. (D) Movement of G1 cell pairs within the forming signaling center and S/G2/M cells in the placode were studied. Cells were followed by live imaging for 24 h (E12.0 onward), and cell pairs, initially ≤15 µm distance from each other, were traced (ncell pairs = 32 in nplacodes = 3). The pairwise comparison revealed that placode cells switch their neighbors frequently in both medial and lateral populations (Student’s t test, ***, P < 0.001; mean cell pair distances = 37.7, 25.2, and 12.6, respectively). The lateral G1 pairs end at longer distances apart from each other. In contrast, S/G2/M cells retained their original neighbors. Box and whisker plot represented as minimum, 25th percentile, median, 75th percentile, and maximum values for each dataset. (E) Latrunculin A (8 µM) was applied to K17-GFP;Fucci G1 explants at E12.0 followed by 24-h culture. Initially G1 cells were distributed along the dental lamina. After 24-h control treatment, G1 cells were observed in a condensed area focus in the anterior part of the K17-GFP positive bud, similar to E13.0 incisors (compare to Fig. S1). In latrunculin A-treated explants, morphogenesis was arrested at placode stage: the signaling center did not change shape, and no invagination of the epithelium was seen. (F) As with LatrA treatment, early signaling center G1 cells stayed dispersed in samples treated with blebbistatin (100 µM), and budding morphogenesis was abrogated. Narrow vertical arrowheads indicate the position of the early signaling center and wide horizontal arrowheads show the xy optical section position in the orthogonal section. Bars, 50 µm.
To distinguish whether the observed cell movement results from displacement of the entire placode cell population by contraction or active migration of individual cells, we studied the movement of cell pairs with respect to each other. We traced cell pairs that were initially within close proximity and analyzed their distances in 3D 20 h later. This pairwise comparison revealed that the S/G2/M cells mostly retained their original neighbors (Fig. 4 D). However, G1 signaling center cells did switch their partners frequently in both medial and lateral populations, the lateral G1 pairs ending furthest apart from each other (Fig. 4 D).
We also inhibited cell movement by actin cytoskeleton disrupting agent latrunculin A (LatrA) or myosin II inhibitor blebbistatin. Inhibitors were applied at E12.0 (Fig. 4, E and F), and explants were cultured for 24 h. In contrast to control medium, both LatrA and blebbistatin prevented compaction of the G1 signaling center, which remained as a stripe similar to stage E12.0 and halted epithelial morphogenesis at placode stage (Fig. 4, E and F). Collectively, these data support directional cell migration as a mechanism for compaction of the early signaling center.
Early signaling center cells remain in G1, whereas budding morphogenesis is driven by cell proliferation in the adjacent lingual cell population
To evaluate the contribution of cell proliferation to tooth budding morphogenesis, we first followed budding morphogenesis in the very initial stages by live imaging from E12.0 to E12.0 + 22 h (Fig. S4, B and C; and Video 2). Live imaging indicated that the early signaling center cells remained in G1, whereas a wave of cell divisions occurred in the thickened placode starting at +6 h. These cells were adjacent to the G1 area on the lingual side and to a small extent on the labial side. A lower number of cytokinesis events was seen throughout the oral epithelium (Fig. S4, B and C; and Video 2).
For more detailed analysis, individual cells were traced during the extensive phase of bud growth (E12.5 + 10 h; Video 3). The G1 cells remained localized in the dental cord, close to the oral surface, in their original position and number (Fig. 5, A and B). In contrast, the S/G2/M proliferation wave in the adjacent cell population led to epithelial invagination and formation of the bud (Fig. 5, A–C). Next, we tracked individual cells, both G1 cells in the early signaling center and the proliferating S/G2/M population, to see if and when they proceeded into mitosis. None of the followed G1 cells proceeded to S/G2/M during the time lapse; rather, they stayed localized in the early signaling center (Fig. 5 D) and thus did not contribute to the invaginating tooth bud. In contrast, the cells originally in S/G2/M phase remained in this cell cycle phase and divided during the time lapse. Their progeny contributed to the growing bud but did not exhibit obvious migratory behavior. Quantification of G1 and S/G2/M cells from confocal fluorescence live imaging samples showed an increase in the bud-forming population, whereas the number of the early signaling center cells stayed constant (Fig. 5 E). When the kinetics of the mitoses was scored, most mitotic activity was found lingual to the early signaling center (Fig. 5 F). There was no evident preference in mitotic orientation with respect to the longitudinal midline of the early signaling center (Fig. 5 G). Thus we conclude that the G1 cells of the early signaling center cells do not reenter the cell cycle and stay localized close to the oral epithelium, in the dental cord. Instead, budding morphogenesis is driven by proliferation of their epithelial neighbors.
Figure 5.
Early signaling center G1 cells do not contribute to the tooth bud. Cell division kinetics and single-cell fates in the developing incisor were analyzed with the aid of Fucci transgenes in explants imaged at E12.5 + 10 h. (A) Volume rendering of the end point of the time lapse of E12.5 + 10 h. (B) Still images of the time lapse (volume rending of separate channels for G1 and S/G2/M). G1 cells remained condensed and localized in the dental cord. In contrast, G2/S/M nuclei appeared in the adjacent cell population, and the number of proliferating cells consecutively increased as the bud invaginated. (C) Surface rendering of the bud at different stages of invagination. G1 early signaling center stayed localized whereas the cells adjacent to it proliferated and gave rise to the bud. (D) Tracing the contribution of individual G1 and S/G2/M cells originating from various positions of the forming bud. None of the early signaling center G1 cells (n = 44) progressed into S/G2/M. Of the 31 traced S/G2/M cells, 21 proceeded to cytokinesis and divided during the time lapse. The daughter cells contributed to the growing bud locally, and the cells did not exhibit apparent migratory behavior. (E) Quantification of cells from confocal fluorescence live imaging samples. Cell number at t = 0 h was adjusted to 100%. There was an increase in the bud-forming population, whereas the number of the early signaling center cells stayed constant (nplacodes = 8; data shown are means ± SD). (F) Plot of kinetics of cell proliferation. Most mitotic activity was found in the lingual cell population adjacent to the early signaling center, where a wave of mitoses started at E12.0 + 6 h, and highest mitotic activity was seen in the next 10–18 h. (G) There was no clear preference in the mitotic orientation in the forming bud (nmitoses = 100, nexplants = 5). Bars: (A and B) 100 µm; (C) 50 µm. Error bars indicate ±SD.
The enamel knot is not a derivative of the early signaling center but is induced de novo in the posterior part of the bud
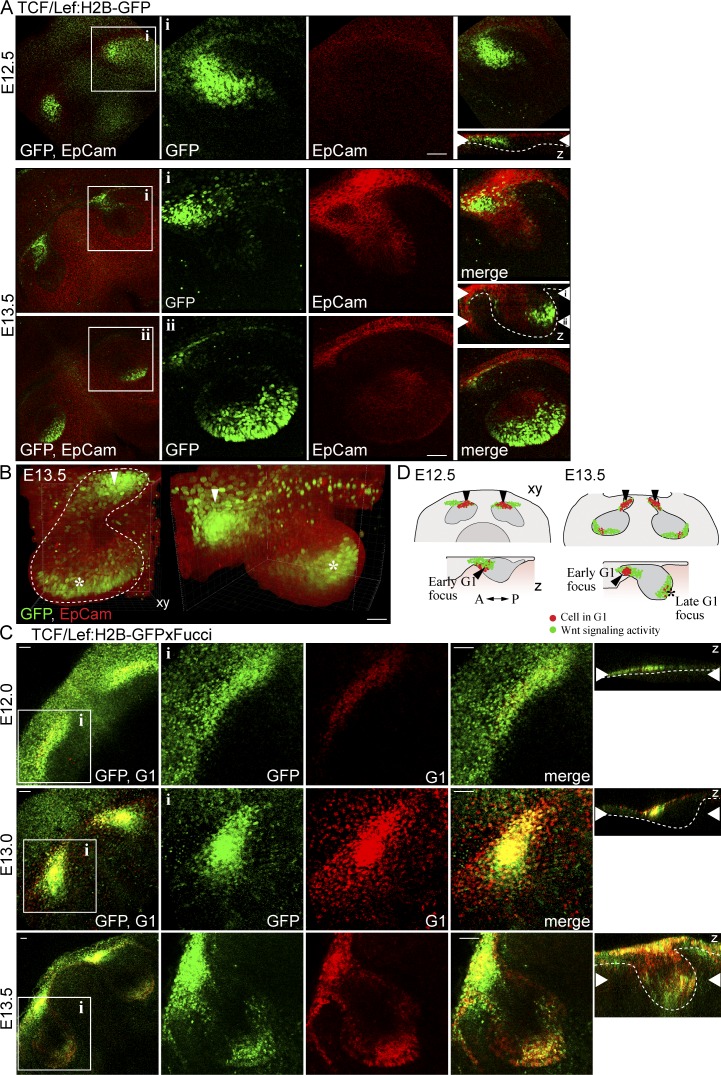
Our analysis of fixed samples showed that the two signaling centers, the early signaling center and the EK, represent two spatially separate regions of G1 cells. The signals expressed in the two signaling centers are largely the same (Jernvall and Thesleff, 2000), but it is not known whether there is a clonal relationship between the cells of the two signaling centers. The EK displays high Wnt/β-catenin signaling activity (Lammi et al., 2004; Liu et al., 2008). To gain further insight into EK induction and the origin of EK cells, we used the Tcf/Lef:H2B-GFP mouse model (Ferrer-Vaquer et al., 2010) that expresses nuclear histone 2B-GFP fusion protein as a marker of canonical Wnt signaling activity.
We first analyzed the Tcf/Lef:H2B-GFP reporter expression in fixed samples, costained with the epithelial marker EpCam. At E12.5, GFP-expressing cells were observed at the labial part of the incisor (Fig. 6 A). At E13.5, a separate area of GFP-positive cells was detected in the posterior part of the bud (Fig. 6, A and B). The GFP-positive regions corresponded with the early signaling center and the EK (Fig. 6, C and D).
Figure 6.
The early and late G1 foci coincide with the Wnt reporter expression. (A) Analysis of Tcf/Lef:H2B-GFP reporter expression (nuclear GFP green) in fixed E12.5 and E13.5 whole-mount explants immunostained with an epithelial membrane marker EpCam to visualize the epithelial tissue (red). At E12.5, the Wnt reporter was expressed at the anterior part of the bud, and at E13.5 a new area of GFP-positive cells was seen at the tip of the bud. (B) 3D volume rendering showed the two separate areas of GFP positivity in the E13.5 tooth bud (early expression marked with an arrowhead and late expression with an asterisk). (C) The Tcf/Lef:H2B-GFP together with Fucci G1 showed colocalization in the early signaling center cells at E12.0, although GFP positivity was more widespread along the placode. At E13.0, the Tcf/Lef:H2B-GFP positive domain displayed a triangular shape similar to the G1 focus. At E13.5, a new G1 focus (the enamel knot) was apparent at the tip of the bud. (D) A schematic representation of the different cell populations in the forming bud. Bars: (A) 50 µm; (C) 40 µm.
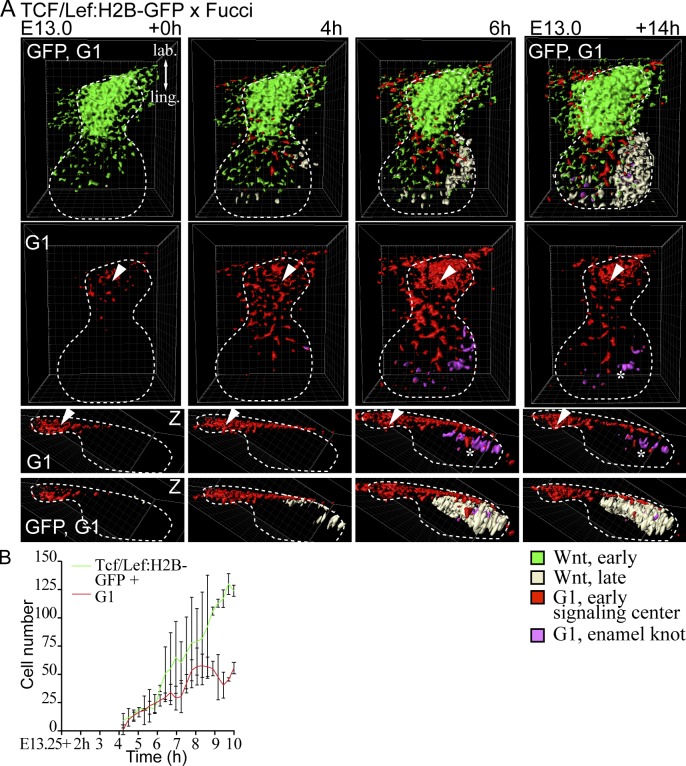
Next, we imaged E13.0 Tcf/Lef:H2B-GFP;Fucci-G1 tissues up to 14 h. Initially, only one G1 focus, the early signaling center, was observed, and it overlapped with the Wnt reporter expression (Fig. 7 A). At E13.0 + 4 h, a novel cluster of Tcf/Lef:H2B-GFP cells began to emerge at the lingual part of the bud. Soon after, the first G1 cells appeared within the Wnt reporter–positive cluster. After 14 h of culture, the new signaling center (G1 focus) had further expanded. Wnt signaling activity coincided with the G1 cells but was more widespread (Fig. 7 B and Video 4). Quantification of GFP-positive and G1 cells showed an increase in number of both, but there was a higher increase in the number GFP-positive cells (Fig. 7 B).
Figure 7.
The enamel knot appears de novo at the tip of the incisor bud. Live imaging was used to study the spatiotemporal dynamics of the early and late signaling centers using Tcf/Lef:H2B-GFP and Fucci G1 reporters. (A) The explant was live imaged from E13.0 + 14 h. In still images of the time lapse with 3D surface rendering, GFP (bright green) was initially seen in the labial part of the growing bud in the same area with the G1 early signaling center (red). At E13.0 + 6 h, new GFP-positive cells appeared at the tip of the bud (light yellow), and consecutively G1 cells appeared (magenta) in the area forming the EK. The G1 early signaling center is marked with an arrowhead and the EK with an asterisk. Bar, 50 µm. (B) Quantification of representative live imaging samples. GFP-positive and G1 cells showed initially the same number, but there was a higher increase in the number of GFP-positive cells (nbuds = 3; data shown are means ± SD).
In conclusion, live imaging of E12.5–E13.5 explants (Figs. 5 and 7) indicated that the early signaling center cells stay close to the oral surface and do not contribute to the EK. Instead, the EK appears de novo at the tip of the mature bud, as the bud cells transition into G1 phase.
The size of the early signaling center correlates with the size of the tooth bud
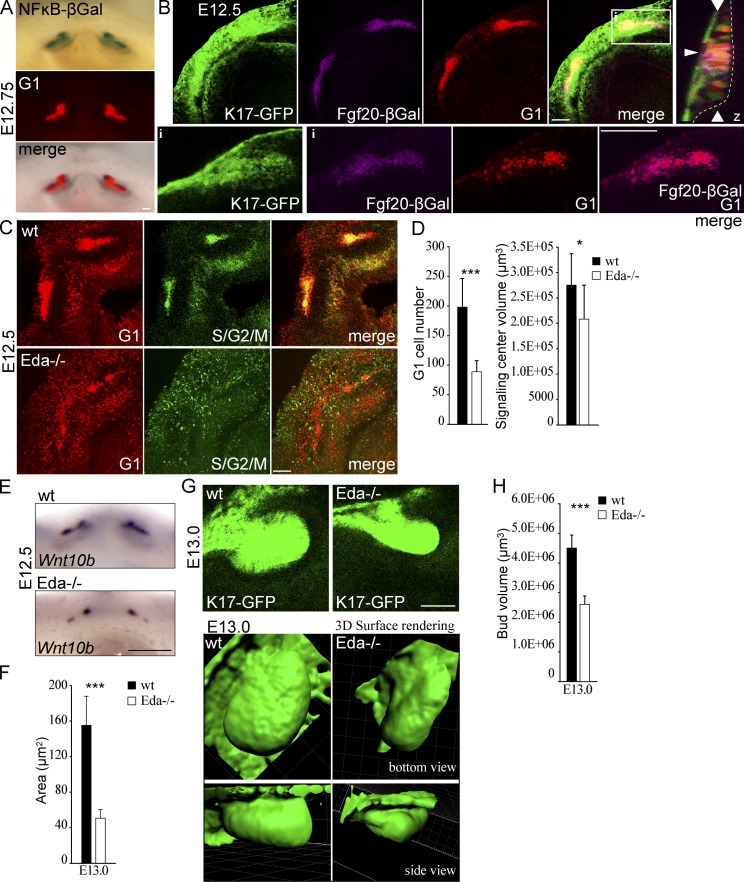
The Eda/Edar/NF-κB pathway is an important regulator of tooth size and shape: Eda-null mice (Tabby mice) have small molar buds that give rise to teeth of reduced size (Grüneberg, 1965; Pispa et al., 1999). Edar/NF-κB activity is confined to the EK, and in its absence, the EK is substantially smaller (Pispa et al., 1999; Ohazama et al., 2004; Harjunmaa et al., 2014). These findings prompted us to analyze the function of Eda in the early signaling center. Expression of an NFκB-LacZ reporter transgene, known to fully depend on Eda signaling activity in embryonic teeth (Pispa et al., 2008), was first detected at ~E11.0 in the incisor region (Fig. S5) in a manner reminiscent of the G1 focus (Fig. S1). The same was evident also at later stages (Fig. S5). Unfortunately, the expression level of the NFκB-LacZ reporter was too low to allow reliable analysis by immunofluorescence, but overlay of Fucci-G1 expression with the whole-mount X-gal–stained E12.5/12.75 explants revealed that the Eda/Edar activity colocalized with the early signaling center (Figs. 8 A and S5 B). In addition, we used the expression of Fgf20, a target gene of Eda whose expression level is greatly reduced in developing Eda-null teeth (Häärä et al., 2012), as a proxy for Eda signaling activity. In Fgf20βGal mice, the LacZ gene has been inserted into the Fgf20 locus (Huh et al., 2012). β-Galactosidase staining was limited to the G1 early signaling center cells in Fgf20βGal/+;K17-GFP;Fucci-G1 embryos, further confirming that Eda signaling activity is localized to these cells (Fig. 8 B).
Figure 8.
The reduced G1 cell number in the early signaling center of Eda null incisor placodes correlates with a smaller bud size. NF-κB reporter and Fgf20βGal mice were used as reporters for Eda/NF-κB signaling activity in developing incisors. (A) Whole-mount X-gal staining of an E12.75 mandible shows colocalization of NF-κB reporter activity with the Fucci G1 population. (B) Maximum-intensity projection and a single optical section (inset) of Fgf20βGal/+;Fucci G1;K17-GFP E12.5 mouse mandible. βGal staining (magenta) was confined to G1 (red) early signaling center cells (merged image of βGal and G1 only in the inset), whereas K17-GFP was expressed throughout the bud. (inset) Narrow horizontal arrowhead indicates the position of the early signaling center and wide vertical arrowheads show the xy optical section position in the orthogonal section. (C) Expression analysis of Fucci transgenes in Eda-null mandibles at E12.5 revealed that the early G1 focus often displayed a bipartite structure and was less coherent compared with the control signaling center (maximum-intensity projection). (D) Quantitation of the early signaling center G1 cell number and overall volume showed a significant decrease in the number of cells and also a decrease in the overall volume of the signaling center (Student’s t test, ***, P < 0.001; *, P < 0.05; n = 5 wt mice and n = 7 Eda−/− mice). (E) Similarly to the reduction seen in the Fucci G1 cell population, the Wnt10b signaling domain was reduced in E12.5 Eda−/− samples. (F) Quantification of the size of the Wnt10b expression domain in mutant versus control embryos (Student’s t test, ***, P < 0.001; n = 10 ctrl mice and n = 9 Eda−/− mice). (G) Comparison of bud shape (based on K17-GFP expression) showed a flatter shape in Eda-null embryos compared with control at E13.0. (H) Bud volume at E13.0 was reduced in the absence of Eda signaling (Student’s t test, ***, P < 0.01; n = 8 wt mice and n = 6 Eda−/− mice). All data shown are mean ± SD. Bars: (A and B) 100 µm; (C and E) 50 µm.
Next we studied the effect of the loss of Eda signaling pathway on the early signaling center G1 cell population. At E12.5, the G1 foci were less prominent in Eda-null embryos and often showed a bipartite structure (Fig. 8 C). Quantification showed a significant decrease in the number of G1 cells and in overall volume of the signaling center (Fig. 8 D). In addition, the expression domain of signaling center marker Wnt10b was reduced in Eda−/− incisors compared with controls (Fig. 8, E and F). Comparison of the bud shape and volume at E13.0 revealed that incisor buds of Eda-null embryos were smaller in size and protruded less into the mesenchyme than in wild-type (wt) littermates (Fig. 8, G and H). In conclusion, the size of the early signaling center was reduced by loss of Eda, and this correlated with the smaller size of the tooth bud.
Discussion
Here we have dissected the cellular events driving epithelial morphogenesis in early tooth development, thereby providing insights into the mechanisms driving epithelial invagination. The presence of an early signaling center has been postulated in placode-stage teeth based on the shared molecular signals with the EK, the later-forming and well-characterized signaling center (Jernvall and Thesleff, 2000). However, the exact identity, formation, and functions of the early signaling center have remained obscure. Here we show that there is a molecularly and functionally discernible cell population within the tooth placode that is characterized by cell cycle cessation and differential cell migration. The early signaling center regulates tooth budding morphogenesis through cell proliferation of the neighboring cells in the placode. Yet the signaling center remains quiescent throughout early tooth development; the cells remain in the dental cord that later on becomes thinner and ultimately disappears (Jussila and Thesleff, 2012). We propose that this early signaling center be called the initiation knot.
In several contexts, cell cycle arrest is important for signaling center fate and function; the mammalian ventral node and notochord are relatively quiescent (Bellomo et al., 1996), as are the most central cells of the isthmic organizer in the midbrain/hindbrain border (Trokovic et al., 2005). The latter express cyclin-dependent kinase inhibitor p21, a known regulator of G1 cell cycle arrest, and display reduced proliferation (Trokovic et al., 2005). In nascent hair follicles, the entire placode appears to act as a signaling center. Initiation of hair placode formation is marked by G1 entry, and cells remain in G1 throughout placodogenesis (Ahtiainen et al., 2014).
The early signaling center shares several features with the primary EK. Like the early signaling center, the EK shows a distinct lack of cell proliferation as indicated by absence of BrdU incorporation (Jernvall et al., 1994). Furthermore, p21 is focally expressed in the EK (Jernvall et al., 1998), as well as in a subset of tooth placode cells (Keränen et al., 1998), likely representing the G1 population identified in this study. Although comparison of published in situ hybridization data from different studies is somewhat challenging, it seems plausible that the gene expression signature of the early signaling center and the EK is largely shared. In addition to p21, shared genes include Shh, Wnt10a/b, Fgf20, Bmp2, Lrp4, and Dkk4 (Vaahtokari et al., 1996; Åberg et al., 1997; Dassule and McMahon, 1998; Keränen et al., 1998; Fliniaux et al., 2008; Liu et al., 2008; Ohazama et al., 2008; Häärä et al., 2012). This molecular similarity raises the possibility that the EK could be derived from the early signaling center cells: the early signaling center cells could be “pushed down” from their original position as the tooth bud grows. Alternatively, the early signaling center cells could exit the G1 phase and contribute to bud growth by reentering the cell cycle. We show here, however, that neither is the case. The early signaling center cells stay in G1 phase and remain in the dental cord throughout budding morphogenesis. Thus, the two signaling centers are distinct from each other, and the EK arises de novo in the tip of the growing bud at E13.5 (Jernvall and Thesleff, 2000).
It has been suggested that the early Shh+ focus, i.e., the early signaling center, represents a rudimentary, deciduous incisor (Hovorakova et al., 2011). Our data do not support this interpretation, but rather establish that the early signaling center is an integral part of the developing tooth. We propose that the early signaling center functions to regulate budding morphogenesis via several signaling pathways. First, deficiency in Eda signaling leads to a smaller incisor tooth bud (Miard et al., 1999; this study), similar to the molar bud (Pispa et al., 1999). NF-κB reporter expression is restricted to the signaling centers: it first appears in the early signaling center of the incisors and somewhat later in molars, but is not present in the intervening domain at any time point, nor in the proliferating cells of the placode. Our study shows that the number of G1 cells in the early signaling center, as well as the expression domain of a signaling center marker Wnt10b, are significantly reduced in Eda-null embryos. Given that Eda/Edar regulates the expression of multiple signaling center genes (Wnt10a/b, Fgf20, and Shh; Pummila et al., 2007; Zhang et al., 2009; Häärä et al., 2012; Voutilainen et al., 2012), it is likely that the diminished bud size is caused by a combinatorial effect of reduced expression of multiple Eda target genes. Also, the EK is smaller in Eda-null mice (Pispa et al., 1999; Harjunmaa et al., 2014), suggesting that the function of Eda is conserved in the two signaling centers. Second, we show that Shh expression correlates with the G1 cells, whereas the cells of the invaginating epithelium are in S/G2/M phase and do not express Shh. Conditional deletion of Shh, leading to loss of expression at around E12.5, leads to a flat, poorly invaginated tooth bud (Dassule et al., 2000). Furthermore, application of Shh-releasing beads on E11.5 mandibles results in ectopic epithelial invaginations, suggesting that Shh drives epithelial cell proliferation locally (Hardcastle et al., 1998). However, it is apparent that in addition to growth factors from the early signaling center, several mesenchymal cues regulate epithelial cell proliferation and bud growth, including activin A (Ferguson et al., 1998) and agonists of the epithelial Fgfr2b (De Moerlooze et al., 2000; Hosokawa et al., 2009; Veistinen et al., 2009). Mesenchymal signals may directly impact the early signaling center, such as mesenchymal Bmp4, which is required for maintenance of epithelial Shh expression (Fujimori et al., 2010).
What then causes the switch to G1 in incisor early signaling center cells, and is it sufficient for changing cell fate? Although the Eda/NF-κB pathway plays an important role, a signaling center still forms even in the absence of Eda/NF-κB signaling. Our previous study in developing skin (Ahtiainen et al., 2014) showed that in addition to Eda, stimulation of canonical Wnt activity resulted in G1 switch. This likely also happens in the tooth: forced epithelial activation of Wnt/β-cat signaling induces the formation of multiple signaling centers (and ectopic teeth) in the oral cavity (Järvinen et al., 2006; Liu et al., 2008), and conversely K14-Cre–mediated epithelial deletion of β-catenin leads to loss of Shh expression and a developmental arrest at the placode–bud transition (Liu et al., 2008). Further studies will be required to clarify the role of the canonical Wnt pathway in the induction of the early signaling center.
We show here that the nonproliferative early signaling center cells show differential migratory behavior toward the mandibular midline, leading to a change in the signaling center shape from a wide stripe to a compact triangle. The molecular mechanisms governing the directional movement remain to be identified. We suggest that this phenomenon is important to ensure that a single, large incisor forms, as opposed to two or three incisors typical to many mammals. Several genetic and ex vivo manipulations have shown the propensity of the incisor signaling center to disintegrate. Inactivation of mesenchymal β-catenin, leading to reduced mesenchymal Bmp4 expression, results in splitting of the incisor placode into two domains, giving rise to two adjacent incisors (Fujimori et al., 2010). Similarly, ectopic application of either Bmp antagonist noggin or exogenous activin A causes splitting of the epithelial Shh domain ex vivo (Munne et al., 2010). The disintegrated small placodes gave rise to smaller incisors, suggesting that the large mouse incisor signaling center is required for correct incisor size (Munne et al., 2010).
What then is the ultimate fate of the early signaling center cells? Studies on the EK have shown that at least some of the EK cells are cleared by apoptosis and also autophagy (Vaahtokari et al., 1996; Yang et al., 2013), and this could serve as the silencing of the signaling center. The same likely applies to the early signaling center, as apoptotic cells have been noted previously (Munne et al., 2009) in a position that correlates well with the G1 population identified in this study. There is reason to believe that attenuation of Wnt signaling is required for the clearance of the signaling center. In mice lacking the Wnt/Bmp inhibitor Ectodin/Sostdc1/Wise, no apoptosis was detected, but instead ectopic Wnt reporter expression was apparent in the early signaling center area at E13–E14, and a supernumerary bud was induced, leading to formation of an extra incisor in the adult (Munne et al., 2009). This phenotype was rescued by reducing the dosage of Wnt coreceptors Lrp5 and Lrp6 (Ahn et al., 2013). A highly similar incisor phenotype is also caused by forced epithelial activation of NF-κB (Blackburn et al., 2015), which in the context of the hair placode signaling center stimulates Wnt signaling (Zhang et al., 2009). Hypothetically, excess Wnt signaling may prolong the activity of the early signaling center, thereby driving the budding of a second, lingually positioned incisor in these mouse models.
In conclusion, we show that growth of the tooth bud is steered by proliferation of the cell population adjacent to the nonproliferating early signaling center, the initiation knot. Directional cell migration shapes the early signaling center, which may be important in determining the number of incisors. Eda/Edar/NF-κB signaling plays a role in defining the number of G1 cells and consequently regulates the size of the growing bud. Although we did not address the role of the early signaling center in developing molars, the colocalization of Shh and other signals with the G1 focus strongly suggests that similar mechanisms account for molar budding as well. However, current evidence suggests that somewhat different mechanisms of bud formation are used in other ectodermal organs. Cell proliferation seems to have a minor contribution in the epithelial morphogenesis of developing mammary glands until the primary sprout forms. A lower mitotic index compared with the adjacent epidermal epithelium and near-absence of BrdU incorporation has been reported up to late bud stage (Balinsky, 1950; Lee et al., 2011). In contrast, as with teeth, hair budding is thought to be achieved through cell proliferation, but whether all or only a subset of placode cells reenter the cell cycle is still unclear (Biggs and Mikkola, 2014). Shh, expressed at the leading edge of the hair bud, is also a key signal driving epithelial cell proliferation in hair follicles (Mill et al., 2003). The extent of the similarities in cellular mechanisms of budding morphogenesis of different ectodermal appendages is the focus of future studies.
Materials and methods
Animals and preparation and culture of embryonic skin
All mouse studies were approved by the National Animal Experiment Board. Transgenic mouse lines used in this study have been described earlier: [mK17 5′]-GFP mice express GFP under the Keratin 17 promoter (Bianchi et al., 2005); Fucci mice are bitransgenic mice expressing nuclear red (mKO-Ctd1) and nuclear green (mAZ-Gem) in G1 and S/G2/M phases, respectively (Sakaue-Sawano et al., 2008); Tcf/Lef:H2B-GFP mice express nuclear GFP as a marker of canonical Wnt signaling activity (#013752; The Jackson Laboratory; Ferrer-Vaquer et al., 2010); transgenic NF-κB reporter mice express β-galactosidase under an NF-κB–responsive element (Bhakar et al., 2002); FGF20βGal mice harbor an Fgf20-β-galactosidase (β-Gal) knock-in allele (Huh et al., 2012); and Eda-null (Tabby) mice were obtained from the Jackson Laboratory (#000314). All embryos were staged according to limb and other external morphological criteria.
Embryonic mandibles were dissected from E10.5–E13.5 embryos and cultured in Trowell-type tissue culture as described previously (Närhi and Thesleff, 2010). The culture medium consisted of DMEM/F12 (1:1) without phenol red supplemented with 50 U/ml penicillin, 50 µg/ml streptomycin, 10% FCS, and 15 mM Hepes (Gibco). For inhibition of actin dynamics, latrunculin A (8 µM) or blebbistatin (100 µM; Sigma-Aldrich) was added to the growth medium.
Whole-mount immunofluorescence, X-gal staining, and in situ hybridization
For whole-mount fluorescence studies, tissues were fixed for 2 h or overnight in 4% PFA and/or permeabilized with 0.1% Triton X-100 in PBS for a minimum of 1 h and washed with PBS. Nuclei were counterstained with Hoechst 33342. The epithelial cell layer was identified with a polyclonal EpCam antibody (1:500, CD326; BD) together with Alexa Fluor 568–conjugated secondary antibody and βGal with polyclonal rabbit antibody (1:1,500; MP Biomedicals) together with Alexa Fluor 647–conjugated secondary antibody (BD and Invitrogen). Tissues were mounted with Vectashield (Vector Laboratories). All results represent at least three independent experiments, and the number of samples for each analysis is presented in the figure legends. Whole-mount X-gal staining was performed as described previously (Pispa et al., 2008). Whole-mount in situ hybridizations with digoxigenin-labeled probes for Foxi3, Sox2, Dkk4, Shh, and Wnt10b were performed as described previously (Wang and Shackleford, 1996; Fliniaux et al., 2008; Juuri et al., 2013; Shirokova et al., 2013). Fluorescent imaging was done with a ZEISS SteREO Lumar.V12 microscope, NeoLumar S 0.8×/WD 80-mm objective, and ZEISS Axiocam MRm3 CCD camera and X-gal staining with a SZX9 stereomicroscope (Olympus) and ColorView IIIu CCD camera (Olympus). The Wnt10b expression domain in whole-mount specimens was quantified from mutants and littermate wt or age-matched wt controls with ImageJ software (National Institutes of Health).
Fluorescence confocal microscopy and time-lapse imaging
For 3D time-lapse imaging studies, dissected tissues were allowed to recover for 2 h. The explants were imaged as described previously (Ahtiainen et al., 2014) with a Leica Biosystems TCS SP5 microscope and HC PL APO 10×/0.4 (air) objective and, for fixed samples, in addition HCX PL APO 20×/0.7 Imm Corr (water, glycerol, oil) Lbd.bl and HCX APO 63×/1.30 Corr (glycerol) CS 21 objectives. Images of fixed samples were acquired as z-stacks of 3-µm optical sections or xzy-stacks of 1.5-µm optical sections. Images were acquired as z-stacks of 3-µm optical sections acquired at 20-min intervals for all time lapses. Lack of pyknotic nuclei and frequency of mitoses in every acquired z-stack, as visualized directly with the aid of the Fucci reporter, confirmed good tissue health. For determination of cell cycle status and cell quantification, only cells that were distinctly identified as either G1 or S/G2/M were scored. All results represent at least three independent experiments.
Quantitative and statistical analyses of experimental data
Images were analyzed and quantitative measurements performed with Imaris 7.2.1 (Bitplane) and ImageJ software. Images were processed for presentation with Photoshop CS5 and Illustrator CS5 software (Adobe Systems). Statistical analysis and further graphing were done with Prism 5 (Graphpad Software), PAST (http://folk.uio.no/ohammer/past/; Hammer et al., 2001), and SPSS Statistics (IBM) software. Differences between groups in cell volume and proliferation analyses were assessed with the parametric Student’s t test. For cell shape and condensation studies, measurements were done as described previously (Ahtiainen et al., 2014). In brief, all measurements were done in three dimensions from confocal optical stacks. For cell tracking purposes, to minimize the global motion of cells caused by tissue deformation or growth, the tissues were allowed to recover after dissection and were then mounted on a solid support. They did not, therefore, show significant amounts of rotational distortion, random vibration, or distortion in arbitrary planes, but rather a constant even movement in the yz plane (initial flattening and following growth in a plane perpendicular to the epithelial sheet) or a similar movement in the xy plane. To remove this background in cell tracking experiments, global movement of the tissue was subtracted from the movement of the cell population under observation in Imaris: at least four stationary points were defined as reference points (at least 100 µm away from the target cells), and global movement was corrected according to these.
For cell track plots, at least 10 representative cells from at least three placodes were selected, and cell movement was plotted on axes representing displacement length in micrometers and relative angle. Cell movement (the angle from 0° to 359° at which the cell moved with respect to the mandible medial axis used as the 0/180° line of reference) was measured for individual cells and represented on a polar plot. For 4D cell migration tracking, track length and cell net displacement were analyzed. Box-and-whiskers plots represent minimum, 25th percentile, median, 75th percentile, and maximum values for each dataset.
For analysis of cell volume, the G1 condensate volume, S/G2/M total volume in tooth epithelium, or equal volume in the oral epithelium was masked, individual nuclei were counted, and mean single-cell volume was defined as total volume divided by cell number in the respective volume.
Sox2 immunofluorescence staining
For whole-mount immunofluorescence staining, tissues were fixed for 2 h in 4% PFA, permeabilized, and incubated with polyclonal goat anti-Sox2 (1:100; Santa Cruz Biotechnology, Inc.) and Alexa Fluor 647–conjugated secondary antibody (Invitrogen) overnight in 0.3% Triton X-100, 0.5% BSA, and 5% normal donkey serum in PBS. Tissues were mounted with Vectashield.
EdU in vivo injection
For the in vivo proliferation assay, pregnant K17-GFP;Fucci G1 dams were injected at E11.5, 12.5, and 13.5 with 25 mg/kg EdU (Invitrogen) in 7.5 mg/ml sterile saline solution 2 h before sacrifice. Tissues were fixed and processed as whole mount (time points E11.5 and 12.5) or embedded in paraffin (E13.5). EdU detection was done from whole-mount samples or frontal paraffin sections as described in the Click-iT EdU Alexa Fluor 674 Imaging kit protocol (Invitrogen).
Individual cell size measurements
Whole-mount immunofluorescence samples stained for E-cadherin (polyclonal rat antibody, 1:300; Invitrogen) were used to visualize cell perimeters, and nuclei were visualized with Hoechst 33342 staining. For measurements of individual cell sizes, high-resolution images in both xy- and z-axis were acquired as z-stacks (xy-scan) and xzy-stacks (vertical scanning) with a 63× high numerical aperture HCX APO 63×/1.30 Corr (glycerol) CS 21 objective. From these optical stacks, individual G1 cell sizes were defined by one of two methods: (a) cell area was measured at the center of each cell inspected from consecutive optical sections in 3D, and the section that was within equal distance from cell perimeters in horizontal, vertical, and depth measurements was chosen; or (b) cell volume was defined by measuring height (major axis), width (vertical axis), and depth (minor axis) and defining the ellipsoid volume as π/6 × (major × minor × vertical).
Online supplemental material
Fig. S1 shows the distribution of Fucci cell cycle indicator transgene-expressing cells in the developing mouse mandible: maximum-intensity projections of confocal fluorescence microscopy z-stacks of Fucci cell cycle reporter embryonic mouse mandible whole mounts at different stages of embryonic development from E10.0 to E13.5 (related to Fig. 1). Fig. S2 shows that Sox2-positive cells reside lingually to the early signaling center. Immunofluorescence staining of Sox2 in E12.5 Fucci G1 whole-mount samples confirmed that Sox2-positive cells are distinct from the early signaling center G1 cells and reside on the lingual side of the forming epithelial bud (related to Fig. 2). Fig. S3 shows cell condensation in the early signaling center: high-resolution imaging of E-cadherin–stained incisors showing reduction in G1 cell size from E11.5 to E12.5 (related to Fig. 3). Fig. S4 shows that early signaling center cells remain in G1, whereas budding morphogenesis takes place through local cell proliferation in the adjacent lingual cell population. Incorporation of EdU nucleoside analog delivered via in vivo injections of pregnant K17-GFP;Fucci G1 dams at E11.5, E12.5, and E13.5 shows G1 cells in the front part of the developing bud, whereas proliferating cells appeared on the lingual side. Still images from time lapse of Fucci E12.0 + 22 h mandible show mitoses highlighted in the bud adjacent to the G1 area on the lingual side (related to Figs. 4 and 5). Fig. S5 shows NF-κB reporter expression in the prospective incisor region. Whole-mount X-gal staining of mandibles at E11.0–E12.5 (related to Fig. 8). Video 1 shows the contribution of cells in different cell cycle phases to the forming incisor at different stages of development. 3D surface renderings show the contribution of G1 and S/G2/M cells at E11.5, E12.0, E12.5, and E13.5 (related to Fig. 3). Video 2 shows that the S/G2/M switch is initiated in the cell population adjacent to the early signaling center. Confocal microcopy time lapse of an E12.0 Fucci mandible imaged for 22 h; individual S/G2/M cells that go into cytokinesis are highlighted (related to Fig. 5). Video 3 shows that proliferating cells next to the G1 early signaling center form the incisor tooth bud. Confocal microscopy time lapse of an E12.5 Fucci mandible imaged for 10 h; 3D volume rendering overview is followed by a closeup of the left incisor, in which individual G1 and S/G2/M cell divisions have been followed (related to Fig. 5). Video 4 shows visualization of the early signaling center and the emergence of the enamel knot by Fucci G1 and TCF/Lef:H2B-GFP reporter. Confocal time-lapse microscopy of an E13.0 TCF/Lef:H2B-GFP, Fucci G1 mandible imaged for 14 h. The cells in the early signaling center are both G1 and GFP positive and remain in the labial part, whereas the EK is initiated at the posterior part of the developing bud (related to Fig. 7). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201512074/DC1.
Supplementary Material
Acknowledgments
We thank Ms. Raija Savolainen and Ms. Riikka Santalahti for technical assistance. We thank Jukka Jernvall and Jacqueline Moustakas-Verho for critical reading of the manuscript and for Fucci mice provided by the Institute of Physical and Chemical Research BioResource Center through the National Bio-Resource Project of the Ministry of Education, Culture, Sports, Science and Technology, Ibaraki, Japan. Imaging was conducted at the Light Microscopy Unit of the Institute of Biotechnology, University of Helsinki.
This work was financially supported by the Academy of Finland, the Jane and Aatos Erkko Foundation, the Sigrid Jusélius Foundation, and Biocentrum Helsinki.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- E
- embryonic day
- EdU
- 5-ethynyl-2′-deoxyurinide
- EK
- enamel knot
- wt
- wild type
References
- Åberg T., Wozney J., and Thesleff I.. 1997. Expression patterns of bone morphogenetic proteins (Bmps) in the developing mouse tooth suggest roles in morphogenesis and cell differentiation. Dev. Dyn. 210:383–396. [DOI] [PubMed] [Google Scholar]
- Ahn Y., Sims C., Logue J.M., Weatherbee S.D., and Krumlauf R.. 2013. Lrp4 and Wise interplay controls the formation and patterning of mammary and other skin appendage placodes by modulating Wnt signaling. Development. 140:583–593. 10.1242/dev.085118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahtiainen L., Lefebvre S., Lindfors P.H., Renvoisé E., Shirokova V., Vartiainen M.K., Thesleff I., and Mikkola M.L.. 2014. Directional cell migration, but not proliferation, drives hair placode morphogenesis. Dev. Cell. 28:588–602. 10.1016/j.devcel.2014.02.003 [DOI] [PubMed] [Google Scholar]
- Balinsky B.I. 1950. On the prenatal growth of the mammary gland rudiment in the mouse. J. Anat. 84:227–235. [PMC free article] [PubMed] [Google Scholar]
- Bazzi H., Fantauzzo K.A., Richardson G.D., Jahoda C.A., and Christiano A.M.. 2007. The Wnt inhibitor, Dickkopf 4, is induced by canonical Wnt signaling during ectodermal appendage morphogenesis. Dev. Biol. 305:498–507. 10.1016/j.ydbio.2007.02.035 [DOI] [PubMed] [Google Scholar]
- Bei M. 2009. Molecular genetics of tooth development. Curr. Opin. Genet. Dev. 19:504–510. 10.1016/j.gde.2009.09.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bellomo D., Lander A., Harragan I., and Brown N.A.. 1996. Cell proliferation in mammalian gastrulation: The ventral node and notochord are relatively quiescent. Dev. Dyn. 205:471–485. [DOI] [PubMed] [Google Scholar]
- Bhakar A.L., Tannis L.L., Zeindler C., Russo M.P., Jobin C., Park D.S., MacPherson S., and Barker P.A.. 2002. Constitutive nuclear factor-kappa B activity is required for central neuron survival. J. Neurosci. 22:8466–8475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bianchi N., Depianto D., McGowan K., Gu C., and Coulombe P.A.. 2005. Exploiting the keratin 17 gene promoter to visualize live cells in epithelial appendages of mice. Mol. Cell. Biol. 25:7249–7259. 10.1128/MCB.25.16.7249-7259.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biggs L.C., and Mikkola M.L.. 2014. Early inductive events in ectodermal appendage morphogenesis. Semin. Cell Dev. Biol. 25-26:11–21. 10.1016/j.semcdb.2014.01.007 [DOI] [PubMed] [Google Scholar]
- Blackburn J., Kawasaki K., Porntaveetus T., Kawasaki M., Otsuka-Tanaka Y., Miake Y., Ota M.S., Watanabe M., Hishinuma M., Nomoto T., et al. . 2015. Excess NF-κB induces ectopic odontogenesis in embryonic incisor epithelium. J. Dent. Res. 94:121–128. 10.1177/0022034514556707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dassule H.R., and McMahon A.P.. 1998. Analysis of epithelial-mesenchymal interactions in the initial morphogenesis of the mammalian tooth. Dev. Biol. 202:215–227. 10.1006/dbio.1998.8992 [DOI] [PubMed] [Google Scholar]
- Dassule H.R., Lewis P., Bei M., Maas R., and McMahon A.P.. 2000. Sonic hedgehog regulates growth and morphogenesis of the tooth. Development. 127:4775–4785. [DOI] [PubMed] [Google Scholar]
- De Moerlooze L., Spencer-Dene B., Revest J.M., Hajihosseini M., Rosewell I., and Dickson C.. 2000. An important role for the IIIb isoform of fibroblast growth factor receptor 2 (FGFR2) in mesenchymal-epithelial signalling during mouse organogenesis. Development. 127:483–492. [DOI] [PubMed] [Google Scholar]
- Ferguson C.A., Tucker A.S., Christensen L., Lau A.L., Matzuk M.M., and Sharpe P.T.. 1998. Activin is an essential early mesenchymal signal in tooth development that is required for patterning of the murine dentition. Genes Dev. 12:2636–2649. 10.1101/gad.12.16.2636 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrer-Vaquer A., Piliszek A., Tian G., Aho R.J., Dufort D., and Hadjantonakis A.K.. 2010. A sensitive and bright single-cell resolution live imaging reporter of Wnt/β-catenin signaling in the mouse. BMC Dev. Biol. 10:121 10.1186/1471-213X-10-121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliniaux I., Mikkola M.L., Lefebvre S., and Thesleff I.. 2008. Identification of dkk4 as a target of Eda-A1/Edar pathway reveals an unexpected role of ectodysplasin as inhibitor of Wnt signalling in ectodermal placodes. Dev. Biol. 320:60–71. 10.1016/j.ydbio.2008.04.023 [DOI] [PubMed] [Google Scholar]
- Fujimori S., Novak H., Weissenböck M., Jussila M., Gonçalves A., Zeller R., Galloway J., Thesleff I., and Hartmann C.. 2010. Wnt/β-catenin signaling in the dental mesenchyme regulates incisor development by regulating Bmp4. Dev. Biol. 348:97–106. 10.1016/j.ydbio.2010.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüneberg H. 1965. Genes and genotypes affecting the teeth of the mouse. J. Embryol. Exp. Morphol. 14:137–159. [PubMed] [Google Scholar]
- Häärä O., Harjunmaa E., Lindfors P.H., Huh S.H., Fliniaux I., Åberg T., Jernvall J., Ornitz D.M., Mikkola M.L., and Thesleff I.. 2012. Ectodysplasin regulates activator-inhibitor balance in murine tooth development through Fgf20 signaling. Development. 139:3189–3199. 10.1242/dev.079558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer Ø., Harper D.A.T., and Ryan P.D.. 2001. PAST: Paleontological statistics software package for education and data analysis. Palaeontol. Electronica. 4:9. [Google Scholar]
- Hardcastle Z., Mo R., Hui C.C., and Sharpe P.T.. 1998. The Shh signalling pathway in tooth development: Defects in Gli2 and Gli3 mutants. Development. 125:2803–2811. [DOI] [PubMed] [Google Scholar]
- Harjunmaa E., Seidel K., Häkkinen T., Renvoisé E., Corfe I.J., Kallonen A., Zhang Z.Q., Evans A.R., Mikkola M.L., Salazar-Ciudad I., et al. . 2014. Replaying evolutionary transitions from the dental fossil record. Nature. 512:44–48. 10.1038/nature13613 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hosokawa R., Deng X., Takamori K., Xu X., Urata M., Bringas P. Jr., and Chai Y.. 2009. Epithelial-specific requirement of FGFR2 signaling during tooth and palate development. J. Exp. Zoolog. B Mol. Dev. Evol. 312B:343–350. 10.1002/jez.b.21274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hovorakova M., Prochazka J., Lesot H., Smrckova L., Churava S., Boran T., Kozmik Z., Klein O., Peterkova R., and Peterka M.. 2011. Shh expression in a rudimentary tooth offers new insights into development of the mouse incisor. J. Exp. Zoolog. B Mol. Dev. Evol. 316:347–358. 10.1002/jez.b.21408 [DOI] [PubMed] [Google Scholar]
- Huh S.H., Jones J., Warchol M.E., and Ornitz D.M.. 2012. Differentiation of the lateral compartment of the cochlea requires a temporally restricted FGF20 signal. PLoS Biol. 10:e1001231 10.1371/journal.pbio.1001231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Järvinen E., Salazar-Ciudad I., Birchmeier W., Taketo M.M., Jernvall J., and Thesleff I.. 2006. Continuous tooth generation in mouse is induced by activated epithelial Wnt/beta-catenin signaling. Proc. Natl. Acad. Sci. USA. 103:18627–18632. 10.1073/pnas.0607289103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jernvall J., and Thesleff I.. 2000. Reiterative signaling and patterning during mammalian tooth morphogenesis. Mech. Dev. 92:19–29. 10.1016/S0925-4773(99)00322-6 [DOI] [PubMed] [Google Scholar]
- Jernvall J., Kettunen P., Karavanova I., Martin L.B., and Thesleff I.. 1994. Evidence for the role of the enamel knot as a control center in mammalian tooth cusp formation: Non-dividing cells express growth stimulating Fgf-4 gene. Int. J. Dev. Biol. 38:463–469. [PubMed] [Google Scholar]
- Jernvall J., Åberg T., Kettunen P., Keränen S., and Thesleff I.. 1998. The life history of an embryonic signaling center: BMP-4 induces p21 and is associated with apoptosis in the mouse tooth enamel knot. Development. 125:161–169. [DOI] [PubMed] [Google Scholar]
- Jussila M., and Thesleff I.. 2012. Signaling networks regulating tooth organogenesis and regeneration, and the specification of dental mesenchymal and epithelial cell lineages. Cold Spring Harb. Perspect. Biol. 4:a008425 10.1101/cshperspect.a008425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juuri E., Jussila M., Seidel K., Holmes S., Wu P., Richman J., Heikinheimo K., Chuong C.M., Arnold K., Hochedlinger K., et al. . 2013. Sox2 marks epithelial competence to generate teeth in mammals and reptiles. Development. 140:1424–1432. 10.1242/dev.089599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keränen S.V., Aberg T., Kettunen P., Thesleff I., and Jernvall J.. 1998. Association of developmental regulatory genes with the development of different molar tooth shapes in two species of rodents. Dev. Genes Evol. 208:477–486. 10.1007/s004270050206 [DOI] [PubMed] [Google Scholar]
- Lammi L., Arte S., Somer M., Jarvinen H., Lahermo P., Thesleff I., Pirinen S., and Nieminen P.. 2004. Mutations in AXIN2 cause familial tooth agenesis and predispose to colorectal cancer. Am. J. Hum. Genet. 74:1043–1050. 10.1086/386293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lan Y., Jia S., and Jiang R.. 2014. Molecular patterning of the mammalian dentition. Semin. Cell Dev. Biol. 25-26:61–70. 10.1016/j.semcdb.2013.12.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laurikkala J., Mikkola M., Mustonen T., Åberg T., Koppinen P., Pispa J., Nieminen P., Galceran J., Grosschedl R., and Thesleff I.. 2001. TNF signaling via the ligand-receptor pair ectodysplasin and edar controls the function of epithelial signaling centers and is regulated by Wnt and activin during tooth organogenesis. Dev. Biol. 229:443–455. 10.1006/dbio.2000.9955 [DOI] [PubMed] [Google Scholar]
- Lee M.Y., Racine V., Jagadpramana P., Sun L., Yu W., Du T., Spencer-Dene B., Rubin N., Le L., Ndiaye D., et al. . 2011. Ectodermal influx and cell hypertrophy provide early growth for all murine mammary rudiments, and are differentially regulated among them by Gli3. PLoS One. 6:e26242 10.1371/journal.pone.0026242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu F., Chu E.Y., Watt B., Zhang Y., Gallant N.M., Andl T., Yang S.H., Lu M.M., Piccolo S., Schmidt-Ullrich R., et al. . 2008. Wnt/beta-catenin signaling directs multiple stages of tooth morphogenesis. Dev. Biol. 313:210–224. 10.1016/j.ydbio.2007.10.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGowan K.M., and Coulombe P.A.. 1998. Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J. Cell Biol. 143:469–486. 10.1083/jcb.143.2.469 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miard S., Peterková R., Vonesch J.L., Peterka M., Ruch J.V., and Lesot H.. 1999. Alterations in the incisor development in the Tabby mouse. Int. J. Dev. Biol. 43:517–529. [PubMed] [Google Scholar]
- Mill P., Mo R., Fu H., Grachtchouk M., Kim P.C., Dlugosz A.A., and Hui C.C.. 2003. Sonic hedgehog-dependent activation of Gli2 is essential for embryonic hair follicle development. Genes Dev. 17:282–294. 10.1101/gad.1038103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Munne P.M., Tummers M., Järvinen E., Thesleff I., and Jernvall J.. 2009. Tinkering with the inductive mesenchyme: Sostdc1 uncovers the role of dental mesenchyme in limiting tooth induction. Development. 136:393–402. 10.1242/dev.025064 [DOI] [PubMed] [Google Scholar]
- Munne P.M., Felszeghy S., Jussila M., Suomalainen M., Thesleff I., and Jernvall J.. 2010. Splitting placodes: Effects of bone morphogenetic protein and Activin on the patterning and identity of mouse incisors. Evol. Dev. 12:383–392. 10.1111/j.1525-142X.2010.00425.x [DOI] [PubMed] [Google Scholar]
- Närhi K., and Thesleff I.. 2010. Explant culture of embryonic craniofacial tissues: Analyzing effects of signaling molecules on gene expression. Methods Mol. Biol. 666:253–267. 10.1007/978-1-60761-820-1_16 [DOI] [PubMed] [Google Scholar]
- Ohazama A., Hu Y., Schmidt-Ullrich R., Cao Y., Scheidereit C., Karin M., and Sharpe P.T.. 2004. A dual role for Ikk alpha in tooth development. Dev. Cell. 6:219–227. 10.1016/S1534-5807(04)00024-3 [DOI] [PubMed] [Google Scholar]
- Ohazama A., Johnson E.B., Ota M.S., Choi H.Y., Porntaveetus T., Oommen S., Itoh N., Eto K., Gritli-Linde A., Herz J., and Sharpe P.T.. 2008. Lrp4 modulates extracellular integration of cell signaling pathways in development. PLoS One. 3:e4092 10.1371/journal.pone.0004092 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Partanen J. 2007. FGF signalling pathways in development of the midbrain and anterior hindbrain. J. Neurochem. 101:1185–1193. 10.1111/j.1471-4159.2007.04463.x [DOI] [PubMed] [Google Scholar]
- Pispa J., and Thesleff I.. 2003. Mechanisms of ectodermal organogenesis. Dev. Biol. 262:195–205. 10.1016/S0012-1606(03)00325-7 [DOI] [PubMed] [Google Scholar]
- Pispa J., Jung H.S., Jernvall J., Kettunen P., Mustonen T., Tabata M.J., Kere J., and Thesleff I.. 1999. Cusp patterning defect in Tabby mouse teeth and its partial rescue by FGF. Dev. Biol. 216:521–534. 10.1006/dbio.1999.9514 [DOI] [PubMed] [Google Scholar]
- Pispa J., Pummila M., Barker P.A., Thesleff I., and Mikkola M.L.. 2008. Edar and Troy signalling pathways act redundantly to regulate initiation of hair follicle development. Hum. Mol. Genet. 17:3380–3391. 10.1093/hmg/ddn232 [DOI] [PubMed] [Google Scholar]
- Pummila M., Fliniaux I., Jaatinen R., James M.J., Laurikkala J., Schneider P., Thesleff I., and Mikkola M.L.. 2007. Ectodysplasin has a dual role in ectodermal organogenesis: Inhibition of Bmp activity and induction of Shh expression. Development. 134:117–125. 10.1242/dev.02708 [DOI] [PubMed] [Google Scholar]
- Sakaue-Sawano A., Kurokawa H., Morimura T., Hanyu A., Hama H., Osawa H., Kashiwagi S., Fukami K., Miyata T., Miyoshi H., et al. . 2008. Visualizing spatiotemporal dynamics of multicellular cell-cycle progression. Cell. 132:487–498. 10.1016/j.cell.2007.12.033 [DOI] [PubMed] [Google Scholar]
- Sarkar L., and Sharpe P.T.. 1999. Expression of Wnt signalling pathway genes during tooth development. Mech. Dev. 85:197–200. 10.1016/S0925-4773(99)00095-7 [DOI] [PubMed] [Google Scholar]
- Schmidt-Ullrich R., and Paus R.. 2005. Molecular principles of hair follicle induction and morphogenesis. BioEssays. 27:247–261. 10.1002/bies.20184 [DOI] [PubMed] [Google Scholar]
- Shirokova V., Jussila M., Hytönen M.K., Perälä N., Drögemüller C., Leeb T., Lohi H., Sainio K., Thesleff I., and Mikkola M.L.. 2013. Expression of Foxi3 is regulated by ectodysplasin in skin appendage placodes. Dev. Dyn. 242:593–603. 10.1002/dvdy.23952 [DOI] [PubMed] [Google Scholar]
- St Amand T.R., Zhang Y., Semina E.V., Zhao X., Hu Y., Nguyen L., Murray J.C., and Chen Y.. 2000. Antagonistic signals between BMP4 and FGF8 define the expression of Pitx1 and Pitx2 in mouse tooth-forming anlage. Dev. Biol. 217:323–332. 10.1006/dbio.1999.9547 [DOI] [PubMed] [Google Scholar]
- Towers M., Wolpert L., and Tickle C.. 2012. Gradients of signalling in the developing limb. Curr. Opin. Cell Biol. 24:181–187. 10.1016/j.ceb.2011.11.005 [DOI] [PubMed] [Google Scholar]
- Trokovic R., Jukkola T., Saarimäki J., Peltopuro P., Naserke T., Weisenhorn D.M., Trokovic N., Wurst W., and Partanen J.. 2005. Fgfr1-dependent boundary cells between developing mid- and hindbrain. Dev. Biol. 278:428–439. 10.1016/j.ydbio.2004.11.024 [DOI] [PubMed] [Google Scholar]
- Tucker A.S., Headon D.J., Schneider P., Ferguson B.M., Overbeek P., Tschopp J., and Sharpe P.T.. 2000. Edar/Eda interactions regulate enamel knot formation in tooth morphogenesis. Development. 127:4691–4700. [DOI] [PubMed] [Google Scholar]
- Tummers M., and Thesleff I.. 2009. The importance of signal pathway modulation in all aspects of tooth development. J. Exp. Zoolog. B Mol. Dev. Evol. 312B:309–319. 10.1002/jez.b.21280 [DOI] [PubMed] [Google Scholar]
- Vaahtokari A., Åberg T., and Thesleff I.. 1996. Apoptosis in the developing tooth: Association with an embryonic signaling center and suppression by EGF and FGF-4. Development. 122:121–129. [DOI] [PubMed] [Google Scholar]
- Veistinen L., Åberg T., and Rice D.P.. 2009. Convergent signalling through Fgfr2 regulates divergent craniofacial morphogenesis. J. Exp. Zoolog. B Mol. Dev. Evol. 312B:351–360. 10.1002/jez.b.21276 [DOI] [PubMed] [Google Scholar]
- Voutilainen M., Lindfors P.H., Lefebvre S., Ahtiainen L., Fliniaux I., Rysti E., Murtoniemi M., Schneider P., Schmidt-Ullrich R., and Mikkola M.L.. 2012. Ectodysplasin regulates hormone-independent mammary ductal morphogenesis via NF-κB. Proc. Natl. Acad. Sci. USA. 109:5744–5749. 10.1073/pnas.1110627109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J., and Shackleford G.M.. 1996. Murine Wnt10a and Wnt10b: Cloning and expression in developing limbs, face and skin of embryos and in adults. Oncogene. 13:1537–1544. [PubMed] [Google Scholar]
- Yang J.W., Zhu L.X., Yuan G.H., Chen Y.X., Zhang L., Zhang L., and Chen Z.. 2013. Autophagy appears during the development of the mouse lower first molar. Histochem. Cell Biol. 139:109–118. 10.1007/s00418-012-1016-2 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Tomann P., Andl T., Gallant N.M., Huelsken J., Jerchow B., Birchmeier W., Paus R., Piccolo S., Mikkola M.L., et al. . 2009. Reciprocal requirements for EDA/EDAR/NF-kappaB and Wnt/beta-catenin signaling pathways in hair follicle induction. Dev. Cell. 17:49–61. 10.1016/j.devcel.2009.05.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.