Pichon et al. describe a method to visualize translation of single endogenous mRNPs in live cells and provide evidence for specialized translation factories, as well as measurements of translation elongation rate, ribosome loading, and movements of single polysomes.
Abstract
Translation is an essential step in gene expression. In this study, we used an improved SunTag system to label nascent proteins and image translation of single messenger ribonucleoproteins (mRNPs) in human cells. Using a dedicated reporter RNA, we observe that translation of single mRNPs stochastically turns on and off while they diffuse through the cytoplasm. We further measure a ribosome density of 1.3 per kilobase and an elongation rate of 13–18 amino acids per second. Tagging the endogenous POLR2A gene revealed similar elongation rates and ribosomal densities and that nearly all messenger RNAs (mRNAs) are engaged in translation. Remarkably, tagging of the heavy chain of dynein 1 (DYNC1H1) shows this mRNA accumulates in foci containing three to seven RNA molecules. These foci are translation sites and thus represent specialized translation factories. We also observe that DYNC1H1 polysomes are actively transported by motors, which may deliver the mature protein at appropriate cellular locations. The SunTag should be broadly applicable to study translational regulation in live single cells.
Introduction
Translation is a fundamental step in gene expression, for which the importance is nicely demonstrated by the thousands of microRNAs that fine-tune expression of a large fraction of mRNAs (Lewis et al., 2005; Baek et al., 2008; Selbach et al., 2008; Hafner et al., 2010). Translational regulation not only provides a quantitative control of protein levels, but it also determines when and where a protein is produced (Sonenberg and Hinnebusch, 2009; Liu et al., 2016). In particular, proteins can be produced locally, in specific subcellular compartments, and this process is intimately linked to subcellular mRNA localization (Martin and Ephrussi, 2009; Jung et al., 2014; Buxbaum et al., 2015). Although many mRNAs localize randomly throughout the cytoplasm, some are highly enriched in particular cytoplasmic areas. Such specific RNA localization occurs in many organisms from bacteria to humans and plays important roles in a variety of cellular processes. Although localized mRNAs are believed to be translated locally, RNA localization can also be the result of other processes. For instance, mRNA can accumulate in processing bodies (P-bodies) for storage or degradation (Cougot et al., 2004; Pillai et al., 2005), and likewise, stress granules are believed to function as mRNA-protective and sorting centers when translation is globally repressed (Kedersha et al., 2000; Mollet et al., 2008; Decker and Parker, 2012). Recent large-scale studies in Drosophila melanogaster embryos and in human cell lines have revealed that a large number of mRNAs can localize in specific subcellular areas (Lécuyer et al., 2007; Battich et al., 2013; Wilk et al., 2016). These studies have also shown many unexpected localization patterns. Whether these patterns are related to local protein synthesis currently remains a mystery.
Although mRNA localization suggests local translation, spatial translational regulation is a distinct process that provides an additional layer of control. A well-known example is that of nanos mRNA in Drosophila oocytes (Gavis and Lehmann, 1994). This mRNA is weakly enriched at the posterior pole of oocytes, with only 4% of the total mRNAs being localized there (Bergsten and Gavis, 1999). However, a tight spatial control of protein synthesis prevents translation of nonlocalized mRNAs, such that the Nanos protein is only produced at the posterior pole. Thus, the spatial control of translation is an important process in itself.
Several microscopy methods have been devised to study translation at the level of single cells (Chao et al., 2012). FRAP and local protein photoconversion can be used to visualize newly translated proteins, but are limited by the time required for chromophore maturation. The translating RNA imaging by coat protein knock-off biosensor identifies single mRNAs that were never engaged in translation (Halstead et al., 2015), but it does not provide information on translation itself. In this study, we developed a strategy to directly image the translational activity of single mRNPs in live cells. Our approach makes use of the SunTag system (Tanenbaum et al., 2014), which allows visualization of single molecules of proteins. It is inspired from the MS2 and Laci technology that image single RNA and DNA loci, respectively, using a repeated tag (Robinett et al., 1996; Bertrand et al., 1998). The SunTag uses a recombinant single-chain variable fragment (scFv) antibody that recognizes a peptide from the yeast Gcn4 protein. The protein of interest carries up to 24 tandem repeats of the peptide epitope, whereas the scFv is fused to superfolder GFP (sfGFP). In vivo binding of the scFv-sfGFP to the epitope yields up to 24 molecules of sfGFP per protein, and this is sufficiently bright to visualize single molecules of proteins in wide-field microscopy. We show that the SunTag can also be used to visualize nascent proteins while still being translated. It provides a powerful tool to image translation of endogenous mRNAs in live cells and at the levels of single molecules.
Results
An improved SunTag system to visualize translation of single mRNPs
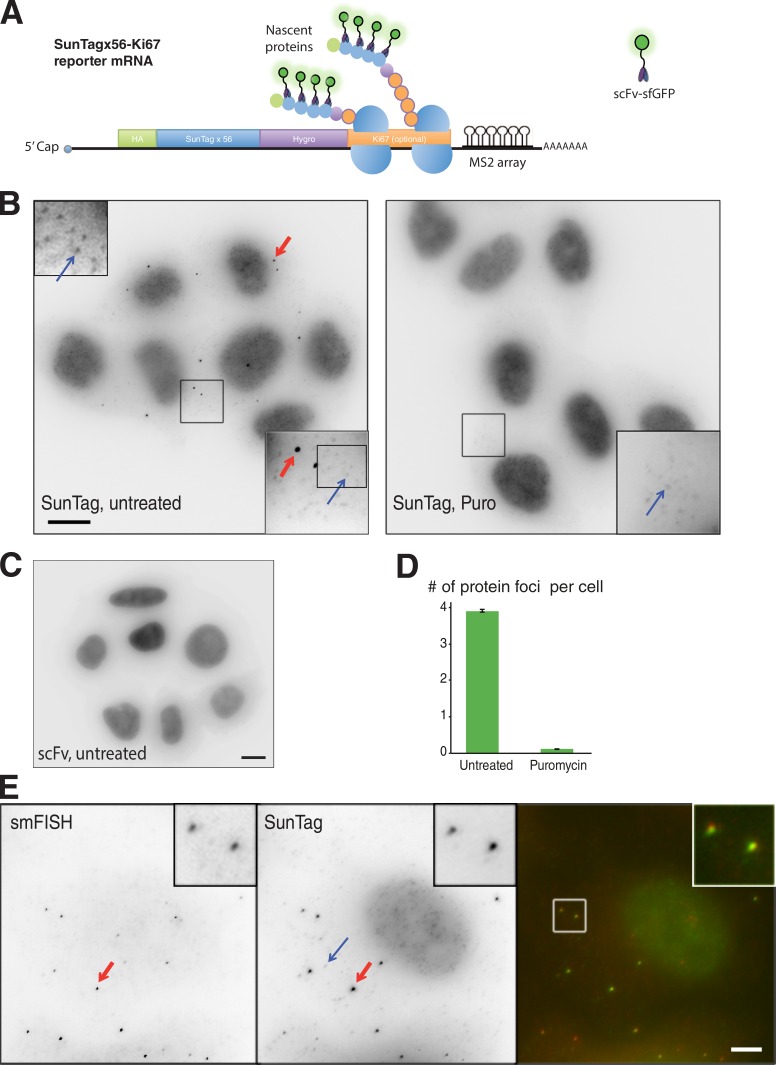
The SunTag system relies on the binding of a fluorescent antibody to a peptide epitope (Tanenbaum et al., 2014), and this epitope thus becomes immediately detectable upon binding of the antibody. We reasoned that this property could be used to image nascent proteins during their translation, by inserting the SunTag at their N termini. The original SunTag has up to 24 repetitions of the peptide epitope. To improve the sensitivity of the system, we generated a new repeat of 32 epitopes and combined it with the original SunTagx24 to obtain a repeat of 56 copies. We inserted the SunTagx56 at the N terminus of a hygromycin selectable gene and further fused the resulting construct to the mouse Ki67 cDNA, which encodes a 3,177-aa-long nuclear protein (Fig. 1 A). We reasoned that this would increase the number of ribosomes loaded on the reporter mRNA, thereby generating a stronger signal. We additionally introduced the MINX intron into the construct (Zillmann et al., 1988) to allow assembly of the exon junction complex and promote export and translation of the resulting mRNA (Le Hir et al., 2016). The polyA signal of the reporter was derived from the herpes simplex virus thymidine kinase gene, and we further added an array of MS2 stem-loops in the 3′ UTR to visualize single mRNAs in live cells with a fluorescent version of the MS2 coat protein (MCP; Fusco et al., 2003). The resulting construct was termed SunTagx56-Ki67.
Figure 1.
Imaging translation of single mRNAs with the SunTag system. (A) Schematic of the Suntagx56-Ki67 reporter mRNA. (B) Effect of puromycin (Puro) on the brighter protein foci labeled by the scFv-sfGFP. Panels represent microscopy images of HeLa cells stably expressing the scFv-sfGFP and the SunTagx56-Ki67 mRNA and untreated (left) or treated with puromycin (right). The blue arrow indicates a spot corresponding to a single molecule of SunTagx56-Ki67 protein, whereas the red arrow points to a brighter protein foci. (bottom right inset) A zoom of the boxed area (10 × 10 µm). (top left inset) An additional zoom of the first inset to show single molecules of proteins. Bar, 10 µm. (C) Expression of the scFV-sfGFP alone does not generate foci but a homogenous signal. The microscopy image is taken from HeLa cells stably transduced with the scFv retroviral vector (the parental cells of the clone shown in B). Bar, 10 µm. (D) Quantification of the number of brighter protein foci per cell (±2 SD; mean of four experiments; >160 cells counted in each condition). (E) Colocalization of the bright protein foci labeled by the scFv-sfGFP, with the SunTagx56-Ki67 mRNAs. Panels represent microscopy images of cells stably expressing the scFv-sfGFP and the SunTagx56-Ki67 mRNA and labeled with smFISH probes recognizing the SunTag-Hygro sequences. Right panel: color overlay of the smFISH image (red, left panel) and the SunTag signal (green, middle panel). The blue arrow indicates a spot corresponding to a single molecule of SunTagx56-Ki67 protein (middle panel), whereas the red arrow points to a brighter protein foci that colocalizes with the SunTagx56-Ki67 mRNA. Inset: a zoom of the boxed area (4 × 4 µm). Bar, 4 µm.
Imaging single polysomes of SunTagx56-Ki67
We generated stable clones of HeLa cells that expressed both this reporter and a nuclear version of the scFv-sfGFP. We hypothesized that this would decrease background signals, first by reducing the levels of free scFv-sfGFP in the cytoplasm and second by transporting the full-length SunTagx56-Ki67 protein to the nucleus. Clones expressing both the scFv-sfGFP and the SunTagx56-Ki67 reporter displayed numerous small dots corresponding to single proteins, as well as brighter protein foci (Fig. 1 B). In contrast, clones expressing only the scFv-sfGFP displayed a diffuse signal as previously reported (Tanenbaum et al., 2014; Fig. 1 C). Interestingly, the brighter protein foci of the SunTagx56-Ki67 clones disappeared after a brief treatment with the translational inhibitor puromycin, whereas single molecules of proteins were still visible (Fig. 1, B and D). This suggested that the brighter foci were translation sites. In agreement, single-molecule FISH (smFISH; Femino et al., 1998) with SunTag-hygromycin probes revealed that these brighter foci colocalized with single mRNAs (>95% of the time, n > 700; Fig. 1 E and Fig. S1). These brighter protein foci thus corresponded to nascent protein chains translated from a single polysome of the reporter mRNA.
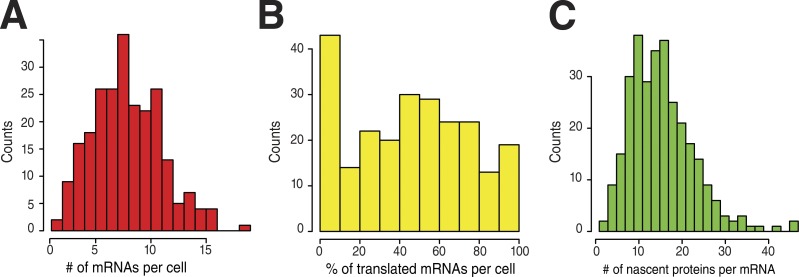
The ability to detect single proteins and single polysomes allowed us to quantify translation at the level of single mRNAs. On average, we detected 8.4 mRNA per cell with 47% of them engaged in translation, but this fraction varied greatly from cell to cell (Fig. 2, A and B). We further measured that the mean nascent protein foci were as bright as 16 single proteins (±2.6 on replicate counts), corresponding to a mean density of 1.3 ribosome per kilobase (see Materials and methods). The nascent protein foci also showed a relatively wide intensity distribution (Fig. 2 C), with the first and last quartiles having means of 8.3 and 27 nascent proteins, respectively. This suggested that different mRNA molecules had different abilities to be translated or that their translation rate varied over time.
Figure 2.
Single-cell, single-molecule quantification of gene expression of the SunTagx56-Ki67 reporter. Panels represent histograms of the number of mRNAs per cell (A; n = 238 cells), the percentage of translated mRNAs (B; n = 238 cells), and the number of nascent proteins per translated mRNA (C; n = 300 mRNAs).
Translation of single SunTagx56-Ki67 mRNPs alternates between active and inactive states
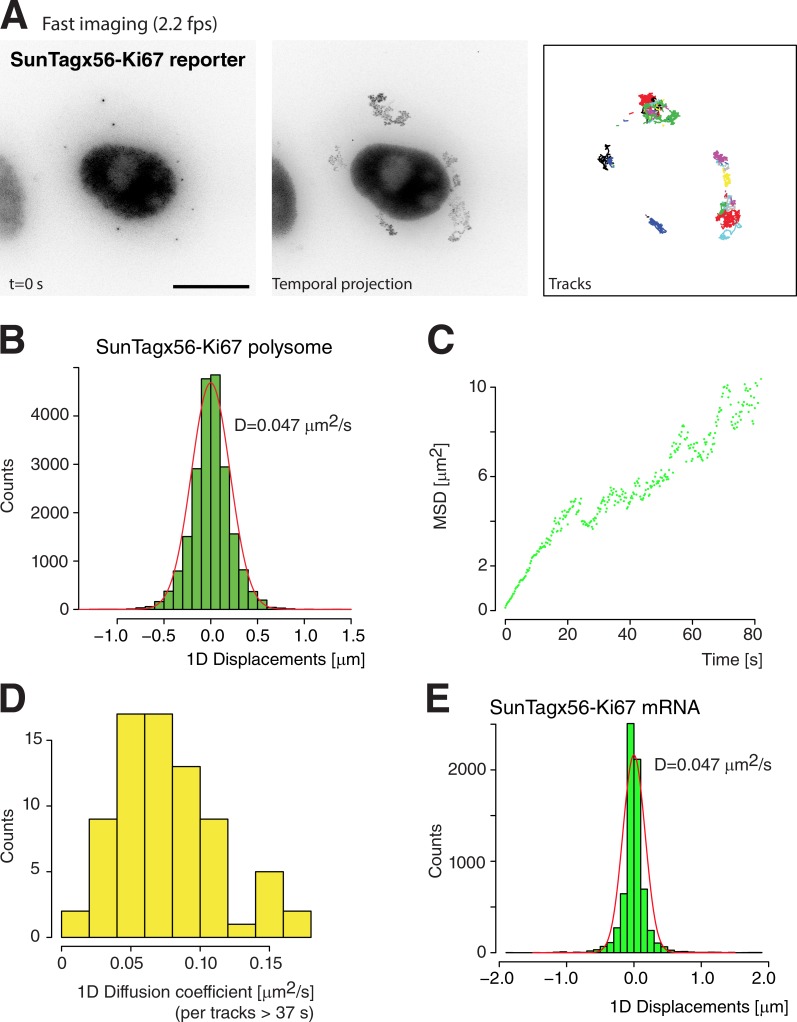
To gain more insights into the dynamics of translation, we imaged live cells. We first imaged mRNAs and polysomes at the same time using cells stably expressing an nls-MCP-TagRFPt fusion in addition to the scFv-sfGFP (Video 1). This demonstrated the feasibility of the approach and the possibility to simultaneously visualize both translated and untranslated mRNAs. However, the relatively poor quality of the red signal and its rapid bleaching made the experiment technically challenging. Most of the following experiments were thus performed in absence of MCP fusions, by imaging polysomes in a single color with the scFv-sfGFP. We acquired 3D image stacks for 45 min at a rate of six images per minute (Fig. 3 A and Video 2). Individual nascent protein foci corresponding to single polysomes remained often detectable for tens of minutes, although their intensity could vary (Fig. 3 B). Interestingly, the intensity of a small number of protein foci gradually decreased for ∼1 to 2 min before they became undetectable (Fig. 3 and Video 3). This suggested that translation of these mRNPs had stopped, thereby resulting in the gradual release of the nascent proteins from the mRNP until it no longer contained any ribosomes. In addition, we could also detect particles appearing, as well as particles disappearing and reappearing again (Fig. 3 B and Videos 4 and 5), thus indicating that translation of single mRNPs alternated between active and inactive states. The number of translating mRNPs in the cytoplasm as well as their rapid diffusion made the long-term tracking of single polysomes difficult, and this precluded rigorous quantifications of the on and off time of single mRNPs. However, the gradual decrease of intensity occurring when a single polysome turned off could be used to provide an estimate of the ribosome elongation rate. To analyze these data, we implemented a mathematical model assuming a uniform ribosome distribution, a constant ribosome velocity, and a release of nascent proteins occurring immediately after completion of their synthesis (see Materials and methods). Individual “ribosome runoff” curves are shown in Fig. 3 C, and the fit to the model indicated an elongation rate of 13.2 aa/s (Fig. 3 D). To get additional insights into the dynamics of translation, we performed FRAP experiments of nascent protein foci (Fig. 3, E and F). It was previously shown that binding of the scFv-sfGFP to its target is stable in vivo, with a t1/2 dissociation rate between 5 and 10 min (Tanenbaum et al., 2014). Recovery of single translating mRNPs was complete in 3.5 min. Thus, FRAP recovery indicated synthesis and release of the tagged Ki67 protein rather than exchange of the scFv antibody on its target. To model the data, we used the same model as for ribosome runoff analysis (see Materials and methods). Fitting the FRAP data yielded a ribosome elongation rate of 18 aa/s, in reasonable agreement with the rate measured from the ribosomal runoff experiments. These rates are 2.5–3 times faster than genome-wide estimates based on runoff experiments using the translational inhibitor harringtonine (Ingolia et al., 2011). Given that foci of nascent proteins were as bright as 16 individual proteins, the FRAP data implied that ribosomes initiated a mean of once every 13 s.
Figure 3.
Stochastic translational activity of single SunTagx56-Ki67 polysomes. (A) Tracks of single SunTagx56-Ki67 polysomes, imaged for 45 min at a rate of 0.1 frames/s (fps). Left panel: image of the SunTag signal at the start of the video; middle panel: temporal maximal intensity projection of the video; right panel: identified tracks, and each track is color-coded. Bar, 10 µm. (B) Quantification of the intensity of individual polysomes over time. Each color is a single polysome from the tracks of A. (C and D) Measurements of translation elongation rate from ribosome run-off curves. In C, the intensity of individual polysomes was measured over time, and tracks were aligned by the last image in which the polysome was detected. In D, the mean curve (n = 10) and a fit to a model describing ribosomes running off the mRNA (gray). (E and F) Measurements of translation elongation rate by FRAP. Individual polysomes were photobleached, and fluorescence recovery was measured over time. The curves in E represent the recovery of individual polysomes, and the curve in F is the mean (n = 13), with a fit to the model in gray. a.u., arbitrary units.
Translating SunTagx56-Ki67 mRNPs diffuse through the cytoplasm
The live-cell movies described in the previous section showed that the nascent protein foci moved rapidly throughout the cytoplasm. To obtain quantitative insights, we acquired 3D stacks at a higher frame rate (2.2 images/s for 4 min; Fig. 4 A and Video 6), which allowed tracking of the translating mRNPs. The histogram of displacements between two consecutive frames revealed a single population with a diffusion coefficient of 0.047 µm2/s (Fig. 4 B). We also calculated the mean square displacement (MSD) as a function of time by averaging individual particles. This indicated that diffusion was normal between 1 and 20 s, with a coefficient of 0.034 µm2/s (Fig. 4 C and Fig. S2 A), in reasonable agreement with the aforementioned estimate. These values were also in range with previous measurements of ribosome movements by spt-PALM (0.1 µm2/s; Katz et al., 2016). We then calculated individual diffusion coefficients for the subset of particles that could be tracked for at least 75 time points (i.e., traces longer than 36 s; n = 75 particles). This yielded a wide distribution of diffusion coefficient (0.042 ± 0.021 µm2/s; Fig. 4 D), indicating substantial differences between individual translating mRNAs (see Materials and methods for details). To test whether this heterogeneity correlated with the ribosome load, we plotted this diffusion coefficient as a function of particle brightness. A weak negative correlation was obtained (Fig. S2 B), indicating that the ribosome load had only a small effect on diffusion. This was further confirmed by measuring the diffusion of mRNAs using the nls-MCP-TagRFPt (Fig. 4 E). We focused on cells in which most of the mRNAs were not translated as assessed by the lack of scFv foci, and we observed that the SunTagx56-Ki67 mRNPs diffused at the same rate as SunTagx56-Ki67 polysomes. Again, this suggested that the ribosome load had little effect on the mRNP diffusion rate.
Figure 4.
Diffusion of single SunTagx56-Ki67 polysomes and mRNAs. (A) Legend as in Fig. 3 A, except that cells were imaged at 4 frames/s (fps) for 2 min. Bar, 10 µm. (B–D) Quantification of the diffusion rates of SunTagx56-Ki67 polysomes. In B, the graph is a histogram of 1D displacements measured between two consecutive video frames (n = 957). In C, the graph represents the mean MSD as a function of time, for the same 957 particles. In D, the histogram represents the diffusion coefficient of individual polysomes, measured for tracks longer than 36 s (n = 75). (E) Quantification of the diffusion rates of SunTagx56-Ki67 mRNAs. Legend as in Fig. 2 B, except that the SunTagx56-Ki67 mRNA were tracked using nls-MCP-TagRFPt (n = 2,021). Only cells with a majority of untranslated SunTagx56-Ki67 mRNA were analyzed.
Visualizing translation of the large subunit of RNA polymerase II
Next, we attempted to visualize translation of endogenous mRNAs and focused first on the housekeeping gene POLR2A, which codes for the large subunit of RNA polymerase II. As in the case of Ki67, we reasoned that the length of its coding region (1,970 aa) would facilitate visualization of the nascent proteins. We generated a CRISPR tagging cassette by fusing a puromycin selectable marker upstream of the SunTagx56 repeat, with a P2A peptide in between them (Fig. 5 A). This leads to a physical separation of the puromycin N-acetyltransferase from the SunTag and thus ensured that the selectable marker was fully functional. The resulting cassette was introduced into a repair construct carrying POLR2A homology arms, such that the Puro-P2A-SunTagx56 was in frame with both the natural ATG codon and the POLR2A coding sequence. This cassette was then transfected together with guide RNAs and the Cas9 nickase (Ran et al., 2013) in HeLa cells expressing either a cytoplasmic or nuclear version of the scFv-sfGFP fusion. Heterozygous recombinant clones were identified by genomic PCR and observed by fluorescence microscopy. We obtained the best signals with the cytoplasmic version of scFv-sfGFP and performed subsequent experiments with these clones. As in the case of the Ki67 reporter mRNA, the scFv-sfGFP detected single proteins as well as brighter protein foci (Fig. S3 A). The single proteins localized in the cytoplasm and not the nucleus, indicating that the SunTagx56 cassette disrupted the normal biogenesis pathway of RNA polymerase II (Boulon et al., 2010). Nevertheless, the brighter protein foci labeled by the scFv-sfGFP colocalized with smFISH probes detecting the RNA sequences of POLR2A (Fig. S3 B) and Puro-SunTagx56 (Fig. 5 B). Colocalization analysis revealed that 98% of the protein foci associated with single RNAs detected with the Puro-SunTagx56 probes (±1%, three replicate counts with n > 800; note that this is not the case for the POLR2A probes, as they also detected the untagged mRNA from the wild-type allele). In addition, the brighter protein foci disappeared after a brief exposure to puromycin, whereas single proteins were still visible (Fig. S3 A). This demonstrated that the bright scFv-sfGFP foci corresponded to nascent proteins being translated from a single mRNA and thus represented single POLR2A polysomes.
Figure 5.
Imaging translation of single molecules of endogenous POLR2A mRNAs. (A) Schematic of the recombination cassette used to edit the POLR2A gene. (B) Colocalization of the bright protein foci labeled by the scFv-sfGFP with tagged POLR2A mRNAs. Panels represent microscopy images of cells stably expressing the scFv-sfGFP and containing the SunTagx56-POLR2A allele and labeled with smFISH probes recognizing the SunTag-Puro sequences. (right) Color overlay of the smFISH image (red, left) and the SunTag signal (green, middle). (inset) Zoom of the boxed area (4.5 × 4.5 µm). Bar, 4.5 µm. (C) Single-cell, single-molecule quantification of POLR2A gene expression. Legend as in Fig. 2 (n = 94 cells for the left and middle panels; n = 326 mRNAs for the right panel). (D) FRAP recovery curves of SunTagx56-POLR2A polysomes. The curve is the mean of the bleaching of 16 single polysomes, with a fit to the model in gray. (E and F) Diffusion of POLR2A polysomes. Legend as in Fig. 4. Data are from 5,888 particles. a.u., arbitrary units; fps, frames per second.
Quantification of the tagged POLR2A mRNA indicated that cells contained a mean of 33 mRNAs (±3 on replicate counts), with a unimodal distribution (Fig. 5 C, left). Interestingly, and in contrast to what was observed with the Ki67 reporter RNA, nearly all of the tagged POLR2A mRNAs appeared to be actively engaged in translation (mean of 91%), with little cell-to-cell variation (Fig. 5 C, middle). Image quantification indicated that the nascent protein foci were as bright as 12 individual proteins (Fig. 5 C, right). This corresponded to a density of 1.3 ribosome/kb, similar to the one observed for the SunTagx56-Ki67 mRNA. The frequency of initiation events thus appears to be similar for these two mRNAs, as long as they are engaged in translation. FRAP analysis of translating POLR2A mRNPs indicated that recovery occurred in ∼3.5 min (Fig. 5 D), which corresponded to an elongation rate of 13.8 aa/s, similar to what was found for the SunTagx56-Ki67 reporter. Finally, analysis of the movements of translating POLR2A mRNPs revealed that these were diffusing at a rate similar to the one of the SunTagx56-Ki67 reporter (Fig. 5, E and F; Fig. S2 A; and Video 7). Again, individual particles displayed widely varying diffusion constants (Fig. 5 F, right).
Dynein heavy chain mRNAs accumulate in cytoplasmic blobs, where they are translated
We then focused on the heavy chain of dynein 1 (DYNC1H1), the main minus end–directed motor in the cytoplasm of human cells. smFISH revealed that this mRNA was not randomly distributed throughout the cytoplasm but accumulated in foci (hereafter referred to as “blobs”), although single, isolated mRNA molecules were also present (Fig. 6 A). To confirm this localization pattern, we used a HeLa cell line containing a bacterial artificial chromosome (BAC) bearing the entire DYNC1H1 gene with a GFP tag inserted in frame at its C terminus (Poser et al., 2008). smFISH with a probe set recognizing the GFP tag showed that the exogenous mRNA also accumulated in blobs, confirming the specificity of the DYNC1H1 smFISH probes (Fig. 6 B). Interestingly, we could detect similar DYNC1H1 RNA blobs in all of the rodent and human cells that we analyzed: NIH3T3 cells (Fig. 6 C), U2OS, HEK, and primary neurons (not depicted), demonstrating the generality of this observation. Image quantification indicated that, depending on the cell line, cells contained a mean of 1.8 to 9.5 blobs, with three to seven mRNA molecules per blob (Fig. 6 D). Overall, 20–45% of DYNC1H1 mRNA accumulated in blobs. We then tested whether these blobs colocalized with P-bodies or stress-granule markers. P-bodies are small cytoplasmic structures known to degrade and store untranslated mRNAs (Cougot et al., 2004; Decker and Parker, 2012), whereas stress granules form only in stressed cells and are made of untranslated mRNPs (Decker and Parker, 2012). In unstressed cells, the stress-granule marker G3BP did not accumulate in the DYNC1H1 mRNA blobs (Fig. S4 A), and a P-body marker also labeled distinct structures (Fig. S4 B).
Figure 6.
Dynein 1 heavy chain mRNA accumulates in blobs that are translation sites. (A) Localization of endogenous DYNC1H1 mRNAs in HeLa cells. Image displays the smFISH signal of HeLa cells labeled with probes recognizing DYNC1H1 mRNAs. Blue arrow: single RNA molecules; red arrow: RNA blob. The green dashed outline represents the position of the nucleus. Bar, 6 µm. (B) Localization of BAC-tagged DYNC1H1 mRNAs in HeLa cells. Legend as in A, except that the probes labeled the GFP sequences. Left panel: smFISH images; right panel: GFP signal. Bar, 6 µm.(C) Localization of endogenous DYNC1H1 mRNAs in NIH3T3 cells. Legend as in A. Bar, 10 µm. (D) Quantification of the number of blobs per cell (left graph), the number of DYNC1H1 mRNAs per blob (middle graph), and the fraction of DYNC1H1 mRNAs present in blobs (right graph). 3T3: NIH3T3 mouse cells (n = 60 cells); BAC: HeLa cells expressing the DYNC1H1 BAC (n = 220 cells); HeLa: HeLa cells expressing the SunTagx32-DYNC1H1 allele (n = 385 cells). The error bars represent the SD of the mean between experimental replicates (n = 3). (E) Colocalization among the bright protein foci labeled by the scFv-sfGFP, the endogenous DYNCH1 mRNAs, and the tagged DYNC1H1 mRNAs. Panels represent microscopy images of cells stably expressing the scFv-sfGFP and containing the SunTagx32-DYNC1H1 allele. Left panel: signal of the scFv; middle left panel: signal from smFISH probes recognizing the DYNC1H1 sequences; middle right panel: signal from smFISH probes recognizing the SunTag-Puro sequences; right panel: color overlay of the scFv signal (green), the endogenous DYNC1H1 mRNAs (red), and the tagged DYNC1H1 mRNA (green). (inset) A 2.5 × 2.5-µm zoom of the boxed area. Bar, 10 µm. (F) Percentage of translated SunTagx32-DYNC1H1 mRNAs, for mRNAs localizing in blobs or being single mRNAs (n = 260 cells). The error bars represent the SD of the mean between experimental replicates (n = 3). (G) Effect of inhibition of translation initiation on the accumulation of DYNC1H1 in blobs. (left) Number of DYNC1H1 polysome per cell, in cells untreated or incubated with Torin1. (right) Fraction of DYNCH1 mRNA in blobs, in cells untreated or incubated with Torin1. The error bars represent the SD of the mean between experimental replicates (n = 4).
In the cell line with the BAC expressing GFP-tagged DYN1CH1, the GFP-tagged dynein subunit accumulated throughout the cytoplasm and was not enriched in the mRNA blobs (Fig. 6 B). However, GFP has a maturation rate of ∼15 min, and we cannot determine where the protein is translated. To address this question, we tagged the endogenous dynein subunit at its N terminus with the Puro-P2A-SunTag cassette as done above for POLR2A. We obtained positive clones with both the SunTagx56 and the SunTagx32 cassette and did the following experiment with the SunTagx32. Heterozygous clones were confirmed by genomic PCR, and, in these clones, the scFv-sfGFP labeled single molecules of proteins as well as brighter protein foci that colocalized with mRNAs detected with oligonucleotide probes hybridizing to the Puro-SunTagx32 sequences (Fig. 6 E). The brighter protein foci were no longer detected after a brief treatment with puromycin (Fig. S5 A), confirming that they corresponded to nascent proteins still associated to translating mRNPs. Quantifications of the intensities of the nascent protein foci indicated that the DYNC1H1 mRNA associated with a mean of 34 proteins (Fig. S5 B). This corresponded to a mean density of 2.1 ribosomes/kb, higher than for the Ki67 reporter and the endogenous POLR2A mRNA. We then determined whether the tagged allele was properly targeted to the DYNC1H1 mRNA blobs. To this end, we performed a two-color smFISH experiment using probes labeling either the endogenous DYNC1H1 sequences or the Puro-SunTagx32 sequences (Fig. 6 E). We counted a total of 27 DYNC1H1 mRNAs per cell, including 7 from the SunTagx32 allele. We found that 20% of the wild-type mRNA and 13% of the tagged allele accumulated in blobs. This indicated that the SunTagx32-DYNC1H1 mRNAs were targeted to the mRNA blobs, albeit at a lower efficiency than the untagged allele. Blobs thus contained a mixture of tagged and untagged mRNAs: a mean of 0.6 molecule of tagged mRNA for 3 molecules of untagged mRNAs. We then asked whether DYNC1H1 mRNAs were translated in blobs. To this end, we used the triple-labeling experiment (scFv, DYNC1H1 mRNAs, and Puro-SunTagx32 mRNAs) to computationally separate mRNAs into blob or single categories. Then, the fraction of translated mRNAs in each category was computed (Fig. 6 F). This analysis was possible because blobs contained at most one molecule of tagged RNA, and it indicated that >90% of the blob mRNAs were translated, against 70% of the single. We also found that the intensity of translation foci was 20% higher when present in blob. Thus, SunTagx32-DYNC1H1 mRNAs were translated slightly more frequently and more efficiently when present in blob.
Next, we investigated whether the formation of blobs was dependent on translation. We treated cells with Torin1, a mammalian target of rapamycin inhibitor that results in a general decrease in translation initiation (approximately two- to threefold; Thoreen et al., 2012). In agreement, a threefold decrease in the number of DYNC1H1 polysomes was observed (Fig. 6 G, left panel). The fraction of DYNC1H1 mRNAs accumulating in blobs concomitantly decreased from 20 to 9% (Fig. 6 G, right panel), thus indicating that formation of the mRNA blobs depends on translation, possibly through the nascent protein chains.
DYNC1H1 polysomes display rapid rectilinear motions
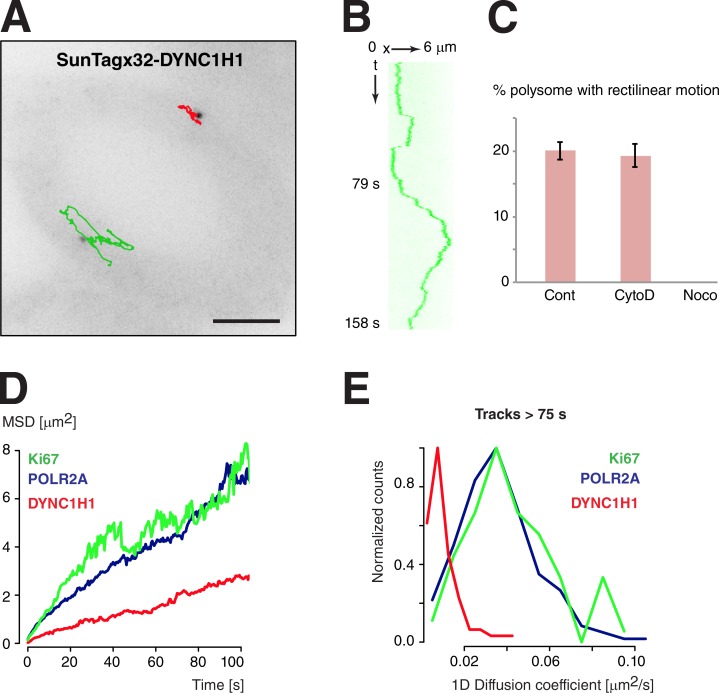
The cytoplasmic distribution of DYNC1H1 mRNAs is peculiar, as these mRNAs accumulate in multicopy structures where they can be translated. To test whether this spatial distribution could be correlated with particular movements or diffusion properties of the corresponding polysomes, we imaged live cells containing the SunTagx32-DYNC1H1 allele. Remarkably, this showed that DYNC1H1 polysomes displayed rapid rectilinear motion, suggestive of motor-dependent movements (Fig. 7, A and B; and Video 8). In a 4-min time window, 19% of the particles displayed a rapid rectilinear motion, with the median speed of the particles being 1 µm/s and the median distance traveled during a run being 2.3 µm. These values are similar to what has been reported for the directional movements of localized mRNPs in cell lines (Fusco et al., 2003). These rapid movements were dependent on an intact microtubule network and not on actin (Fig. 7 C and Video 9). This suggested that they resulted from an active process involving molecular motors.
Figure 7.
Microtubule-dependent movements of Dynein 1 heavy chain polysomes. (A) Movements of tagged DYNC1H1 polysomes. Legend as in Fig. 4. Bar, 6 µm. (B) Kymograph showing rapid rectilinear motions of the green particle of A. (C) Percentage of DYNC1H1 polysome displaying rapid rectilinear motion over a 4-min time period, in untreated cells (Cont) or in cells treated with cytochalasin D (CytoD) or nocodazole (Noco). The error bars represent the SD of the mean between experimental replicates (n = 3). (D and E) Comparison of the diffusion of DYNC1H1 polysomes with that of POL2RA and Ki67. Legend as in Fig. 4 (C and D). Data of DYNC1H1 are from 176 particles.
In contrast to this active transport, diffusive movement of DYNC1H1 polysomes was five times slower than that of Ki67 and POLR2A polysomes (0.01 vs. 0.05 µm2/s; Fig. 7 D and Fig. S2 A). In addition, calculation of diffusion coefficients for single DYNC1H1 polysomes revealed that many were nearly immobile for tens of seconds, suggesting that they may be anchored on some cellular structures (Fig. 7 E). Collectively, these results suggested that translating DYNC1H1 mRNPs alternate between cycles of diffusion, anchoring and motor-dependent transport. Because blobs contained at most one molecule of tagged mRNA, it was not possible to determine whether specific movements were associated with mRNAs localized in blobs or occurring as single molecules. Nevertheless, the peculiar localization of DYNC1H1 mRNAs correlated with specific movement properties of these polysomes.
Discussion
The SunTag as a tool to monitor translation of single endogenous mRNPs
In this study, we show that by labeling a peptide epitope right after its synthesis, the SunTag system allows us to visualize nascent proteins that are still being translated by the ribosome. The SunTag thus provides a convenient assay to monitor translation of single mRNPs in living cells. The use of a larger SunTag repeat, as we did in this study, facilitates visualization of newly translated proteins. Likewise, fusing it to a long reporter protein increases the signal of nascent proteins. However, these improvements are not absolutely necessary, as we show that it is possible to visualize translation sites using a short hygromycin protein reporter (Fig. S5 D), as well as using shorter SunTag repeats (Fig. 6). We demonstrate that combining the SunTag with genome editing allows us to visualize translation of endogenous mRNAs and thus in a context that keeps all the regulatory sequences that control mRNA transport and localization. We thus believe that this technology holds great promise to visualize and measure translation in live cells and at the level of single mRNPs. It will provide a unique view of this fundamental process, and, in particular, it should be very informative to analyze the spatial and temporal regulation of translation, as well as to better understand to the cell-to-cell variability of gene expression. Although this manuscript was under consideration, other studies showed the use of the SunTag to monitor translation in live cells (Wang et al., 2016; Wu et al., 2016; Yan et al., 2016). Our work further extends these studies by showing that combining the SunTag with genome editing allows us to image translation of endogenous mRNAs.
Translation of single mRNA alternates between active and inactive states
Translation of the SunTagx56-Ki67 reporter appears discontinuous: single mRNAs appear to switch between translated and untranslated states. The reasons for this phenomenon are at present unclear. It could be that these switches are regulated. Alternatively, they may be related to the stochastic behavior often observed at the level of single molecules (Sanchez and Golding, 2013). Much evidence has shown that during transcription initiation, the stochastic binding of transcription factors on a single promoter DNA molecule often results in stochastic switching of promoters between active and inactive states (Sanchez and Golding, 2013). Likewise, it may be possible that single mRNAs randomly switch between translationally competent and incompetent states. Translation initiation requires the assembly of a large macromolecular complex comprising many initiation factors, and this complex may have a finite t1/2 and may spontaneously dissociate within the cell, thereby turning off the mRNP until the initiation complex reassembles. Interestingly, we observed that nearly all of the POLR2A mRNAs are actively engaged in translation. This mRNA codes for an abundant housekeeping protein and may thus be an optimal substrate for translation. This suggests that the switch between active and inactive states may be somehow regulated by the mRNA sequence. In the future, it will be interesting to analyze mRNAs regulated at the translational level.
Polysomes diffuse rapidly through the cytoplasm of human cells
The ability to visualize polysomes in live cells allowed us to compare the mobility of translated and untranslated mRNAs. Polysomes were tracked using the scFv, whereas untranslated mRNAs were tracked using nls-MCP-tagRFPt. We found that translation had little effect on mRNA mobility, which may be because the diffusion coefficient varies with the volume of the particle and thus with the cubic root of its mass. An eightfold increase in the mass thus translates into only a twofold decrease in diffusion coefficient. If one assumes that ribosomes are twice as compact as a random coil mRNA of similar mass and that the ribosomal density is 1.3/kb, then translation is expected to decrease diffusion by 1.3-fold. In contrast to the small effect of translation, diffusion coefficients of individual polysomes varied over almost an order of magnitude, even when coding for the same protein. The reasons for this heterogeneity are at present unclear. It could be due to the location of the mRNP in the cell, the cellular components that associate with it, or other features of the cellular environment.
A fraction of dynein heavy chain is synthesized in translation factories
The SunTagx56-Ki67 and the endogenous POLR2A mRNAs occur as isolated single molecules that are dispersed throughout the cytoplasm. In contrast, 20–50% of the DYNC1H1 mRNAs concentrate in blobs that contain multiple mRNAs (a mean of three to seven). We show that translation DYNC1H1 mRNAs occur on both isolated molecules and blobs, although it appears slightly more frequent and efficient in blobs. The translation of DYNC1H1 mRNAs in blobs indicate that the blobs represent specialized translation factories. Such structures have been hypothesized before (Chang et al., 2006), but the use of the SunTag allows to provide a direct proof of their existence. The function of such factories is currently elusive. One possibility would be that they help produce the mature protein complex. Dyneins are large macromolecular complexes of ∼2 MD, which also require dedicated factors for their assembly (Carter et al., 2016). They are composed of two heavy chains, two intermediate chains, two light intermediate chains, and multiple light chains. The dynein heavy chain itself is a large polypeptide that contains several structural domains, including an AAA+ motor domain. Folding of dynein heavy chain and assembly of the dynein complex may thus be favored by the concentration of the nascent chains in dedicated factories that may contain the required chaperone and assembly factors. It is, however, interesting to note that the mRNAs coding for other dynein subunits do not localize in the DYNC1H1 blobs (unpublished data). Future studies will be thus required to understand the functions of the DYNC1H1 translation factories.
Large-scale RNA localization studies in Drosophila have revealed that numerous mRNAs accumulate in cytoplasmic foci (Lécuyer et al., 2007; Wilk et al., 2016). In the future, it will be interesting to determine whether these correspond to translation factories or to sites of accumulation of untranslated mRNA, as in the case of P-bodies. It would also be interesting to determine whether the accumulation of mRNAs in foci is linked to a specific transport pathway that may help to deliver them when and where they are needed. Motor-dependent movements of foci containing untranslated mRNAs may help to transport them to their future translation site, as proposed for instance in the case of neuronal P-bodies (Cougot et al., 2008; Zeitelhofer et al., 2008). In this study, we show that polysomes can also be actively transported, and an interesting speculation would be that motor-dependent movements of dynein heavy chain polysomes may help to deliver newly synthesized dyneins to their site of action. Another possibility could be that this active transport may be required to counterbalance the particularly slow diffusion of this mRNP (0.008 µm2/s, or 50 h to explore a cellular area of 1,600 µm2; Fig. S2 A; Fisher and Cooper, 1967). Although future studies will be needed to explore the function of motor-dependent movements of polysomes, this phenomenon reveals a new facet of RNA metabolism.
Materials and methods
Cells
HeLa cells were maintained in DMEM supplemented with 10% FBS, 10 U/ml penicillin/streptomycin and 2.9 mg/ml glutamine in a humidified CO2 incubator at 37°C. Cells were transfected with JetPrime (Polyplus) and selected on 150 µg/ml hygromycin or 0.25 µg/ml puromycin. For each stable cell line, several individual clones were picked and screened by smFISH with sets of fluorescent oligonucleotide probes against the integrated sequence. For CRISPR recombination, clones were additionally analyzed by genomic PCR using amplicons specific for either the nonrecombined or the recombined allele.
The scFv-GB1-sfGFP and scFv-GB1-sfGFP-NLS (plasmids 60907 and 60906; Addgene; Tanenbaum et al., 2014) are referred in the text to as scFv-sfGFP. These plasmids were introduced into cells by retroviral infection. HEK293T cells were transiently transfected with a cocktail of plasmids coding for retroviral components and producing the genomic scFv-sfGFP retroviral RNAs. Viral particles were collected and used to infect recipient HeLa cells, which were then sorted by FACS. Only lowly expressing cells were selected. The phage-Ubc-nls-MCP-tagRFPt was introduced into selected cells by retroviral infection following the same procedure. For translational inhibition, cells were treated with puromycin at 100 µg/ml for 30 min. Cells were treated with nocodazole, cytochalasin D, and Torin1 at 10 µg/ml for 1 h, 5 µM for 1 h, and 250 nM for 2 h, respectively.
Plasmids
Sequences of the plasmids are available upon request. The arrays of MS2 stem-loops consisted of a repeat of 132 MS2 binding sites that was generated by gene-synthesis techniques. It is composed of 33 unique stem-loops that were designed to minimize their similarities, and this sequence was multimerized four times. The size of the tag is 2.9 kb, and its sequence is listed in Table S1.
The repeat of 56 SunTag sequence was composed of the original SunTagx24 (plasmid 60910; Addgene; Tanenbaum et al., 2014), which was fused to a new SunTagx32 sequence. This new repeat was designed to minimize the similarities between individual repeat at the nucleotide level while keeping the same protein sequence. The SunTagx32 sequence was cloned by gene-synthesis techniques. The constructs used in this study were assembled using a mixture of gene synthesis, Gateway reactions, Gibson assembly, and traditional cloning techniques. The sequences are available in Table S1.
The nls-MCP-TagRFPt plasmid expresses a nuclear version of MCP that fluoresces in red, and it was generated from pHAGE-Ubc-nls-HA-MCP-YFP (plasmid 31320; Addgene) by replacing GFP with TagRFPt by traditional cloning techniques.
Genomic PCR
Genomic DNA was prepared with GenElute Mammalian Genomic DNA Miniprep (Sigma-Aldrich) and analyzed by PCR with Platinum Taq DNA Polymerase (Invitrogen). The sequences of oligonucleotides were as follows: POLR2A-wt forward, 5′-TTTACCCACGACTCTGGCTC-3′ and POLR2A-wt reverse, 5′-TGCTCTTGAAGGTAGGGTCC-3′; Puro-reverse, 5′-GGTGACCCGCTCGATGTG-3′; DYNC1H1-wt forward, 5′-GGTAGCTGTTCTCAGTAGGT-3′ and DYNC1H1-wt reverse, 5′-CCTACAACAGTGATGCTCGC-3′; and SunTag reverse, 5′-TACCCTTCTTCAGTCTGGCG-3′.
Heterozygous clones were further characterized by microscopy. The SunTagx56-POLR2A proteins localized in the cytoplasm and were thus nonfunctional. In contrast, live imaging of the SunTagx32-DYNC1H1 cell line showed that single molecules of protein displayed rapid rectilinear motions over long distances, suggesting that the tagged dyneine subunit was fully functional.
In situ hybridization
Cells were grown on glass coverslips (0.17 mm), washed with PBS, fixed in 4% PFA for 20 min, and permeabilized in 70% ethanol overnight at 4°C. Cells were hybridized as previously described (Fusco et al., 2003), except that the probes were sets of unlabeled oligonucleotides hybridizing against the target RNA and additionally contained a common supplementary sequence that was preannealed to a fluorescent oligonucleotide probe termed the FLAP (Tsanov et al., 2016). We used sets of 24 oligonucleotides to detect the POLR2A mRNA, 36 for the DYNC1H1 mRNA (60 for the triple-staining experiment), 36 for the SunTag-Puro, and 48 for the SunTag-Hygro sequences. Sequences of the probes are available in Table S1. The GFP probes hybridized to the GFP-IRES-Neo sequences of the LAP tag present in the BAC (Poser et al., 2008) and were a set of 40 oligonucleotides, each 40 nt long and conjugated to three or four Cy3 molecules (Femino et al., 1998). Slides were mounted in Vectashield with DAPI (Vector Laboratories).
Image acquisition in fixed cells
Fixed cells were imaged at room temperature on an Axioimager Z1 wide-field microscope (63×, NA 1.4; ZEISS) equipped with an sCMOs Zyla 4 0.2 camera (Andor Technology) and controlled by MetaMorph (Universal Imaging). 3D image stacks were collected with a Z-spacing of 0.3 µm. Figures were prepared with ImageJ (National Institutes of Health), Photoshop (Adobe Systems), and Illustrator (Adobe Systems), and graphs were generated with R.
P-bodies were labeled with a mouse monoclonal antibody raised against S6K and that is known to recognize GE-1/Helds (sc-8418; Santa Cruz Biotechnology, Inc.; Kedersha and Anderson, 2007). The secondary anti-mouse antibody was coupled to FITC. Slides were mounted in Vectashield with DAPI (Vector Laboratories).
Image acquisition in live cells
Cells were plated on 25-mm-diameter coverslips (0.17-mm thick) in nonfluorescent media (DMEM gfp-2 of Evrogen complemented with rutin and 10% FCS). Coverslips were mounted in a temperature-controlled chamber (37°C) with 5% CO2 and imaged on an inverted OMXv3 Deltavision microscope (GE Healthcare) in time-lapse mode. A 100×, NA 1.4 objective was used, with an intermediate 2× lens and an Evolve 512 × 512 EMCCD camera (Photometrics). For fast imaging, we imaged at a rate of 2.2 stacks/s for 2 min, using stacks of 10 planes with a Z-spacing of 0.6 µm. This spacing allowed accurate point spread function determination without excessive oversampling. For slow imaging, we collected stacks of 14 planes with a Z-spacing of 0.6 µm and at a frame rate of one stack every 10 s. The power of illuminating light and the exposure time were set to the lowest values that still allowed visualization of single molecules of proteins (laser at 1% of full power, exposure of 10 ms/plane). This minimized bleaching and maximized the number of frames that was collected.
FRAP
FRAP was performed on a confocal microscope (Meta LSM780; ZEISS) with a 63×, 1.4 NA objective, at 37°C and in DMEM gfp2 complemented with rutin and 10% FCS. The translation foci labeled by the scFv-sfGFP were bleached at 488 nm in a circle of 1.3-µm diameter at full laser power for 300 ms. Recoveries were measured by making stacks in 3D at a rate of one stack every 2 s (one stack was made of five slices in Z, 0.4 µm apart). Recoveries were analyzed by defining a tracking area in the 4D image stack and by measuring the total intensity of a cube of 3 × 3 pixels centered on the brightest pixel in the tracking area (Boireau et al., 2007). Background was removed, intensities at each time point were corrected for bleaching by dividing them by the total cell fluorescence, and these values were normalized to the fluorescence intensity before the bleach. Finally, we eliminated the first 15 s after recovery to remove the contribution of diffusing scFv-sfGFP molecules, either free or labeling single molecules of tagged proteins.
Ribosome runoff experiments
Live-cell videos were acquired at a rate of one image every 10 s on an OMXv3 Deltavision (GE Healthcare; slow imaging conditions; see Image acquisition in live cells). Single SunTagx56-Ki67 polysomes were then tracked using the FIJI plugin TrackMate, and their intensity over time was quantitated (see Analysis of live-cell videos). Polysomes that turned off during the video were identified visually, and their tracks were aligned by the last image in which the polysome was visible. The intensities of a polysome over time were then normalized by dividing them by the median intensity of the polysome over the track. The mean normalized intensities of different polysomes over time were then fit to the model described in the section Modeling of the FRAP and ribosome runoff experiments.
Modeling of the FRAP and ribosome runoff experiments
Fit of the recovery curve was done with a linear model that assumes that ribosomes are distributed homogenously along the mRNA and move at a constant speed, as done previously for modeling transcription (Boireau et al., 2007). The model further assumes that proteins diffuse away immediately after completion of their synthesis. The ribosome runoff and the FRAP experiments yield curves that are symmetric with respect to time, and the calculations are thus presented only for the ribosome runoff experiments. The fit yields three parameters: the intensity before translation turns off (I0), the time at which translation turns off (t0), and the elongation rate (v). The curve is composed of two parts. In the first part, ribosomes are located on both the SunTag repeat and the downstream coding region, whereas in the second part, the last ribosome has passed the SunTag, and ribosomes are all located on the downstream coding region. During this second period, there is no synthesis of SunTag repetition while ribosomes terminate translation at a constant rate. This part thus translates into a linear segment, for which the slope depends on the elongation rate. During the first period, ribosomes similarly terminate transcription, but at the same time, ribosomes located on the SunTag repeat generate a new signal, which is proportional to the number of ribosomes on the repeat. In turn, this number decreases linearly with time because ribosomes move to the 3′ end at a constant rate. Assuming that the length of the SunTag repeat is L1 and that the length of the downstream region is L2, then a formalization of the model yields the following equation for I(t), the intensity at time t, when t′ < 0, then I(t′) = I0 and when 0 < t′ < L1/v, then
when L1/v < t′< (L1+L2)/v, then
and when (L1+L2)/v < t′, then I(t′) = 0, with t′ = t − t0.
Analysis of smFISH images
The numbers of mRNA per cell were counted manually for Figs. 2 and 5. To quantify single mRNA molecules and mRNA blobs in Fig. 6, we used an automated pipeline. Cell segmentation was performed with CellCognition (Held et al., 2010), using DAPI for nuclear segmentation and the smFISH images for cellular segmentation with a watershed method. Fluorescent spots were detected with FISH-quant (Mueller et al., 2013), with the connected components methods after manual thresholding of Laplacian of Gaussian filtered image. To separate individual mRNA molecules and blob, a threshold on their raw intensity was applied. This threshold was defined as 1.5, the median intensity of all the detected spots. This value was determined by comparing DYNC1H1 images with images obtained from an mRNA that does not make blobs (e.g., CRM1) and by selecting the value that maximized the differences in the number of blobs between the two genes. Blobs inside the nucleus were excluded to avoid transcription sites in the analysis.
To quantify the number of mRNAs per blob, we first calculated the intensities of single mRNA molecules. Images around individual mRNAs were cropped and used to calculate a mean image of all individual mRNAs. The resulting mean image was fit with a 3D Gaussian function and the integrated intensity above background estimated. Each individual RNA blob was analyzed in a similar fashion (cropping and Gaussian fitting). To infer how many individual mRNA molecules are present in a particular blob, we divided its integrated intensity by the integrated intensity of the mean individual mRNA molecules.
Analysis of SunTag images in fixed cells
The percentage of translated mRNA per cell were counted manually for Figs. 2 and 4, by counting mRNAs molecules that colocalized with a SunTag foci. We used a semiautomated pipeline to quantify the number of nascent protein chains per translating mRNAs. Translation foci and individual proteins spots labeled by the SunTag were defined manually in 3D stacks in a homemade Matlab interface. Images of individual proteins were cropped ±3 pixels in XY and ±1 pixel in Z, with respect to the pixel with maximum intensity. An averaged image of the individual proteins was then calculated using at least 15 individual proteins in the vicinity of a translation foci. The image of each translation foci was cropped to have similar dimensions, and a maximum intensity projection along Z was performed. The resulting images were fit with a 2D Gaussian function, and the integrated intensity above background was calculated. The number of proteins per translation foci was estimated as the ratio of the integrated intensity of the translation foci to the intensity of the averaged image of single proteins. To calculate the density of ribosomes, we divided the number of nascent protein per mRNA by Lnorm, the normalized cDNA length. If the length of the SunTag coding region is L1, and the length of the coding region after the SunTag is L2, Lnorm is L1 × 0.5 + L2. The reason is that ribosomes located on the SunTag repeat are less bright than those located after the repeat, because only a fraction of the repeat has been translated. If we assume that ribosomes are uniformly distributed along the length of the coding region, then the mean location of the ribosomes that are still on the SunTag repeat is at half the length of the repeat. This means that their mean brightness is half of the brightness of the ribosomes located after the SunTag. A formal calculation has been done in Boireau et al. (2007) in the case of RNA polymerases transcribing an MS2 repeat, but can be equally applied to ribosomes translating a SunTag repeat. The length, in nucleotides, of the various coding sequences is: 2,380 for the SunTagx32; 4,123 for the SunTagx56; 1,023 for hygromycin; 660 for puromycin; 9,531 for Ki67; 5,910 for POLR2A; and 13,938 for DYNC1H1.
Analysis of live-cell videos
Videos were projected along Z using pixels of maximal intensity and corrected for photobleaching using the histogram matching method in ImageJ (National Institutes of Health). Individual spots were identified and tracked with the TrackMate plugin in ImageJ, using the DoG detector, subpixel localization, a blob diameter of 0.8 µm, and a threshold manually adapted for each video. For fast imaging conditions, tracks were reconstructed using the simple LAP tracker option, using a maximal linking distance of 1.5 µm, a gap-closing distance of 1 µm, and a maximal frame gap of 2.
Tracks were imported and analyzed in R. Instant 1D displacements between frames were calculated along the x and y axis, and the resulting histograms were fitted to a Gaussian function, for which variance is directly proportional to the diffusion coefficient (D). The values obtained for the x and y displacements were treated independently and within 1% for the SunTagx56-Ki67 and SunTag-POLR2A polysomes and within 7% for the SunTagx32-DYNC1H1. We also calculated a mean MSD as a function of time, by aligning all tracks at their start and averaging the resulting 2D displacements. The segment of the resulting curves comprised between 0 and 18 s was fit to a linear model, and the slope was used to calculate D. These values were approximately twofold lower than the ones calculated from instant displacements, which may be a result of errors in pointing accuracy for the instant displacements (mean instant displacement was between 100 and 300 nm) or to a subdiffusive behavior for the particles. We also analyzed single individual tracks, for tracks longer than 36 s. For each track, we calculated all instant 1D displacements in x and y, pooled them, and estimated D for each polysome from the variance of the resulting data (>150 data points). We then calculated the SD of individual diffusion coefficients. If all particles were diffusing identically, they should have the same D, and the measured SD between individual particles should be D/(nbp)0.5, with nbp being the number of data points used to calculate D for each track (in this study, 150). For the three polysomes analyzed in this study (Ki67, POLR2A, and DYNC1H1), the measured SD was 5–10-fold higher than expected, indicating that particles are heterogeneous in terms of diffusional properties.
To quantify the fluctuation of polysomes intensity over time, the tracking data were used to locate the particles in the videos. The videos were bleach corrected, projected along Z using the pixels of maximal intensities, and cropped around the spot location at each time point. The resulting images were then fit to 2D Gaussian, and particle intensity was measured from the integrated intensity above background.
Online supplemental material
Fig. S1 shows colocalization of the bright protein foci labeled by the scFv-sfGFP with the SunTagx56-Ki67 mRNAs. Fig. S2 shows diffusion of polysomes and correlation with the number of nascent proteins. Fig. S3 shows characterization of the SunTagx56-POLR2A polysomes. Fig. S4 shows characterization of DYNC1H1 RNA blobs. Fig. S5 shows characterization of DYNC1H1 polysomes. Table S1 contains the sequence of the oligo probes used for the smFISH experiments, as well as the sequence of the SunTagx32, SunTagx56, and MS2 × 132 tags. Video 1 shows fast imaging of the SunTagx56-Ki67 reporter (2.2 stacks/s; each Z stack with 10 slices 0.6 µm apart), accelerated four times. Video 2 shows slow imaging of the SunTagx56-Ki67 reporter (1 stack per 10 s), accelerated 160 times. Video 3 shows slow imaging of the SunTagx56-Ki67 reporter (1 stack per 10 s), accelerated 160 times. Video 4 shows slow imaging of the SunTagx56-Ki67 reporter (1 stack per 10 s), accelerated 80 times. Video 5 shows slow imaging of the SunTagx56-Ki67 reporter (1 stack per 10 s), accelerated 80 times. Video 6 shows fast imaging of the SunTagx56-Ki67 reporter (2.2 stacks/s), accelerated four times. Video 7 shows fast imaging of the SunTagx56-POLR2A gene (2.2 stacks/s), accelerated four times. Video 8 shows fast imaging of the SunTagx32-DYNC1H1 gene (2.2 stacks/s), accelerated four times. Video 9 shows the same as Video 8, except that the cells were treated with cytochalasin D (5 µM for 1 h). Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.201605024/DC1.
Supplementary Material
Acknowledgments
We thank D. Fisher and A. Camas for the gift of a Gateway mouse Ki67 donor vector, R. Vale for the gift of the SunTag plasmids, I. Poser and T. Hyman for the gift of the DYNC1H1 BAC-GFP cell line, and the Montpellier RIO Imaging facility for help with the imaging experiments.
A. Safieddine is supported by a Ministère de l'Éducation Nationale de la Recherche et de Technologie fellowship. This work was supported by the Site de Recherche Intégrée sur le Cancer (SIRIC) Montpellier Cancer (grant INCa-DGOS-INSERM 6045) and Fondation de la Recherche Medicale and Agence Nationale de la Recherche HI-FISH grants to E. Basyuk.
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- BAC
- bacterial artificial chromosome
- MCP
- MS2 coat protein
- MSD
- mean square displacement
- P-body
- processing body
- scFv
- single-chain variable fragment
- sfGFP
- superfolder GFP
- smFISH
- single-molecule FISH
References
- Baek D., Villén J., Shin C., Camargo F.D., Gygi S.P., and Bartel D.P.. 2008. The impact of microRNAs on protein output. Nature. 455:64–71. 10.1038/nature07242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battich N., Stoeger T., and Pelkmans L.. 2013. Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat. Methods. 10:1127–1133. 10.1038/nmeth.2657 [DOI] [PubMed] [Google Scholar]
- Bergsten S.E., and Gavis E.R.. 1999. Role for mRNA localization in translational activation but not spatial restriction of nanos RNA. Development. 126:659–669. [DOI] [PubMed] [Google Scholar]
- Bertrand E., Chartrand P., Schaefer M., Shenoy S.M., Singer R.H., and Long R.M.. 1998. Localization of ASH1 mRNA particles in living yeast. Mol. Cell. 2:437–445. 10.1016/S1097-2765(00)80143-4 [DOI] [PubMed] [Google Scholar]
- Boireau S., Maiuri P., Basyuk E., de la Mata M., Knezevich A., Pradet-Balade B., Bäcker V., Kornblihtt A., Marcello A., and Bertrand E.. 2007. The transcriptional cycle of HIV-1 in real-time and live cells. J. Cell Biol. 179:291–304. 10.1083/jcb.200706018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulon S., Pradet-Balade B., Verheggen C., Molle D., Boireau S., Georgieva M., Azzag K., Robert M.C., Ahmad Y., Neel H., et al. 2010. HSP90 and its R2TP/Prefoldin-like cochaperone are involved in the cytoplasmic assembly of RNA polymerase II. Mol. Cell. 39:912–924. 10.1016/j.molcel.2010.08.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buxbaum A.R., Yoon Y.J., Singer R.H., and Park H.Y.. 2015. Single-molecule insights into mRNA dynamics in neurons. Trends Cell Biol. 25:468–475. 10.1016/j.tcb.2015.05.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter A.P., Diamant A.G., and Urnavicius L.. 2016. How dynein and dynactin transport cargos: a structural perspective. Curr. Opin. Struct. Biol. 37:62–70. 10.1016/j.sbi.2015.12.003 [DOI] [PubMed] [Google Scholar]
- Chang L., Shav-Tal Y., Trcek T., Singer R.H., and Goldman R.D.. 2006. Assembling an intermediate filament network by dynamic cotranslation. J. Cell Biol. 172:747–758. 10.1083/jcb.200511033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao J.A., Yoon Y.J., and Singer R.H.. 2012. Imaging translation in single cells using fluorescent microscopy. Cold Spring Harb. Perspect. Biol. 4:a012310 10.1101/cshperspect.a012310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Babajko S., and Séraphin B.. 2004. Cytoplasmic foci are sites of mRNA decay in human cells. J. Cell Biol. 165:31–40. 10.1083/jcb.200309008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cougot N., Bhattacharyya S.N., Tapia-Arancibia L., Bordonné R., Filipowicz W., Bertrand E., and Rage F.. 2008. Dendrites of mammalian neurons contain specialized P-body-like structures that respond to neuronal activation. J. Neurosci. 28:13793–13804. 10.1523/JNEUROSCI.4155-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Decker C.J., and Parker R.. 2012. P-bodies and stress granules: possible roles in the control of translation and mRNA degradation. Cold Spring Harb. Perspect. Biol. 4:a012286 10.1101/cshperspect.a012286 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Femino A.M., Fay F.S., Fogarty K., and Singer R.H.. 1998. Visualization of single RNA transcripts in situ. Science. 280:585–590. 10.1126/science.280.5363.585 [DOI] [PubMed] [Google Scholar]
- Fisher H.W., and Cooper T.W.. 1967. Electron microscope studies of the microvilli of HeLa cells. J. Cell Biol. 34:569–576. 10.1083/jcb.34.2.569 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fusco D., Accornero N., Lavoie B., Shenoy S.M., Blanchard J.M., Singer R.H., and Bertrand E.. 2003. Single mRNA molecules demonstrate probabilistic movement in living mammalian cells. Curr. Biol. 13:161–167. 10.1016/S0960-9822(02)01436-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavis E.R., and Lehmann R.. 1994. Translational regulation of nanos by RNA localization. Nature. 369:315–318. 10.1038/369315a0 [DOI] [PubMed] [Google Scholar]
- Hafner M., Landthaler M., Burger L., Khorshid M., Hausser J., Berninger P., Rothballer A., Ascano M. Jr., Jungkamp A.C., Munschauer M., et al. 2010. Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell. 141:129–141. 10.1016/j.cell.2010.03.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halstead J.M., Lionnet T., Wilbertz J.H., Wippich F., Ephrussi A., Singer R.H., and Chao J.A.. 2015. An RNA biosensor for imaging the first round of translation from single cells to living animals. Science. 347:1367–1371. 10.1126/science.aaa3380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Held M., Schmitz M.H., Fischer B., Walter T., Neumann B., Olma M.H., Peter M., Ellenberg J., and Gerlich D.W.. 2010. CellCognition: time-resolved phenotype annotation in high-throughput live cell imaging. Nat. Methods. 7:747–754. 10.1038/nmeth.1486 [DOI] [PubMed] [Google Scholar]
- Ingolia N.T., Lareau L.F., and Weissman J.S.. 2011. Ribosome profiling of mouse embryonic stem cells reveals the complexity and dynamics of mammalian proteomes. Cell. 147:789–802. 10.1016/j.cell.2011.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung H., Gkogkas C.G., Sonenberg N., and Holt C.E.. 2014. Remote control of gene function by local translation. Cell. 157:26–40. 10.1016/j.cell.2014.03.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Z.B., English B.P., Lionnet T., Yoon Y.J., Monnier N., Ovryn B., Bathe M., and Singer R.H.. 2016. Mapping translation ‘hot-spots’ in live cells by tracking single molecules of mRNA and ribosomes. eLife. 5:e10415 10.7554/eLife.10415 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kedersha N., and Anderson P.. 2007. Mammalian stress granules and processing bodies. Methods Enzymol. 431:61–81. 10.1016/S0076-6879(07)31005-7 [DOI] [PubMed] [Google Scholar]
- Kedersha N., Cho M.R., Li W., Yacono P.W., Chen S., Gilks N., Golan D.E., and Anderson P.. 2000. Dynamic shuttling of TIA-1 accompanies the recruitment of mRNA to mammalian stress granules. J. Cell Biol. 151:1257–1268. 10.1083/jcb.151.6.1257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lécuyer E., Yoshida H., Parthasarathy N., Alm C., Babak T., Cerovina T., Hughes T.R., Tomancak P., and Krause H.M.. 2007. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell. 131:174–187. 10.1016/j.cell.2007.08.003 [DOI] [PubMed] [Google Scholar]
- Le Hir H., Saulière J., and Wang Z.. 2016. The exon junction complex as a node of post-transcriptional networks. Nat. Rev. Mol. Cell Biol. 17:41–54. 10.1038/nrm.2015.7 [DOI] [PubMed] [Google Scholar]
- Lewis B.P., Burge C.B., and Bartel D.P.. 2005. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 120:15–20. 10.1016/j.cell.2004.12.035 [DOI] [PubMed] [Google Scholar]
- Liu Y., Beyer A., and Aebersold R.. 2016. On the dependency of cellular protein levels on mRNA abundance. Cell. 165:535–550. 10.1016/j.cell.2016.03.014 [DOI] [PubMed] [Google Scholar]
- Martin K.C., and Ephrussi A.. 2009. mRNA localization: gene expression in the spatial dimension. Cell. 136:719–730. 10.1016/j.cell.2009.01.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollet S., Cougot N., Wilczynska A., Dautry F., Kress M., Bertrand E., and Weil D.. 2008. Translationally repressed mRNA transiently cycles through stress granules during stress. Mol. Biol. Cell. 19:4469–4479. 10.1091/mbc.E08-05-0499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mueller F., Senecal A., Tantale K., Marie-Nelly H., Ly N., Collin O., Basyuk E., Bertrand E., Darzacq X., and Zimmer C.. 2013. FISH-quant: automatic counting of transcripts in 3D FISH images. Nat. Methods. 10:277–278. 10.1038/nmeth.2406 [DOI] [PubMed] [Google Scholar]
- Pillai R.S., Bhattacharyya S.N., Artus C.G., Zoller T., Cougot N., Basyuk E., Bertrand E., and Filipowicz W.. 2005. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 309:1573–1576. 10.1126/science.1115079 [DOI] [PubMed] [Google Scholar]
- Poser I., Sarov M., Hutchins J.R., Hériché J.K., Toyoda Y., Pozniakovsky A., Weigl D., Nitzsche A., Hegemann B., Bird A.W., et al. 2008. BAC TransgeneOmics: a high-throughput method for exploration of protein function in mammals. Nat. Methods. 5:409–415. 10.1038/nmeth.1199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ran F.A., Hsu P.D., Lin C.Y., Gootenberg J.S., Konermann S., Trevino A.E., Scott D.A., Inoue A., Matoba S., Zhang Y., and Zhang F.. 2013. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 154:1380–1389. (published erratum appears in Cell 2013. 155:479-480) 10.1016/j.cell.2013.08.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinett C.C., Straight A., Li G., Willhelm C., Sudlow G., Murray A., and Belmont A.S.. 1996. In vivo localization of DNA sequences and visualization of large-scale chromatin organization using lac operator/repressor recognition. J. Cell Biol. 135:1685–1700. 10.1083/jcb.135.6.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez A., and Golding I.. 2013. Genetic determinants and cellular constraints in noisy gene expression. Science. 342:1188–1193. 10.1126/science.1242975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selbach M., Schwanhäusser B., Thierfelder N., Fang Z., Khanin R., and Rajewsky N.. 2008. Widespread changes in protein synthesis induced by microRNAs. Nature. 455:58–63. 10.1038/nature07228 [DOI] [PubMed] [Google Scholar]
- Sonenberg N., and Hinnebusch A.G.. 2009. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 136:731–745. 10.1016/j.cell.2009.01.042 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanenbaum M.E., Gilbert L.A., Qi L.S., Weissman J.S., and Vale R.D.. 2014. A protein-tagging system for signal amplification in gene expression and fluorescence imaging. Cell. 159:635–646. 10.1016/j.cell.2014.09.039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoreen C.C., Chantranupong L., Keys H.R., Wang T., Gray N.S., and Sabatini D.M.. 2012. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature. 485:109–113. 10.1038/nature11083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsanov N., Samacoits A., Chouaib R., Traboulsi A., Gostan T., Weber C., Zimmer C., Zibara K., Walter T., Marion P., et al. 2016. smiFISH and FISH-quant - a flexible single RNA detection approach with super-resolution capability. Nucleic Acids Res. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C., Han B., Zhou R., and Zhuang X.. 2016. Real-time imaging of translation on single mRNA transcript in live cells. Cell. 165:990–1001. 10.1016/j.cell.2016.04.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilk R., Hu J., Blotsky D., and Krause H.M.. 2016. Diverse and pervasive subcellular distributions for both coding and long noncoding RNAs. Genes Dev. 30:594–609. 10.1101/gad.276931.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu B., Eliscovich C., Yoon Y.J., and Singer R.H.. 2016. Translation dynamics of single mRNAs in live cells and neurons. Science. 352:1430–1435. 10.1126/science.aaf1084 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan X., Hoek T.A., Vale R.D., and Tanenbaum M.E.. 2016. Dynamics of translation of single mRNA molecules in vivo. Cell. 165:976–989. 10.1016/j.cell.2016.04.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitelhofer M., Karra D., Macchi P., Tolino M., Thomas S., Schwarz M., Kiebler M., and Dahm R.. 2008. Dynamic interaction between P-bodies and transport ribonucleoprotein particles in dendrites of mature hippocampal neurons. J. Neurosci. 28:7555–7562. 10.1523/JNEUROSCI.0104-08.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zillmann M., Zapp M.L., and Berget S.M.. 1988. Gel electrophoretic isolation of splicing complexes containing U1 small nuclear ribonucleoprotein particles. Mol. Cell. Biol. 8:814–821. 10.1128/MCB.8.2.814 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.