Abstract
We have assessed whether HLA immunogenicity as defined by differences in donor–recipient HLA amino‐acid sequence (amino‐acid mismatch score, AMS; and eplet mismatch score, EpMS) and physicochemical properties (electrostatic mismatch score, EMS) enables prediction of allosensitization to HLA, and also prediction of the risk of an individual donor–recipient HLA mismatch to induce donor‐specific antibody (DSA). HLA antibody screening was undertaken using single‐antigen beads in 131 kidney transplant recipients returning to the transplant waiting list following first graft failure. The effect of AMS, EpMS, and EMS on the development of allosensitization (calculated reaction frequency [cRF]) and DSA was determined. Multivariate analyses, adjusting for time on the waiting list, maintenance on immunosuppression after transplant failure, and graft nephrectomy, showed that AMS (odds ratio [OR]: 1.44 per 10 units, 95% CI: 1.02–2.10, p = 0.04) and EMS (OR: 1.27 per 10 units, 95% CI: 1.02–1.62, p = 0.04) were independently associated with the risk of developing sensitization to HLA (cRF > 15%). AMS, EpMS, and EMS were independently associated with the development of HLA‐DR and HLA‐DQ DSA, but only EMS correlated with the risk of HLA‐A and ‐B DSA development. Differences in donor–recipient HLA amino‐acid sequence and physicochemical properties enable better assessment of the risk of HLA‐specific sensitization than conventional HLA matching.
Keywords: translational research/science, histocompatibility, kidney transplantation/nephrology, alloantibody, alloantigen, major histocompatibility complex (MHC), organ allocation, retransplantation
Short abstract
This study applies the Cambridge HLA Immunogenicity Algorithm to assess alloantibody responses after a failed kidney transplant and provides strong evidence that amino acid sequence and physicochemical analyses of donor–recipient HLA immunogenicity enables prediction of HLA class I and II donor‐specific alloantibody development and offers additional value to that of conventional HLA mismatch grade for predicting overall HLA‐specific sensitization to the potential donor pool.
Abbreviations
- AMS
amino acid mismatch score
- cRF
calculated reaction frequency
- DSA
donor‐specific antibody
- EMS
electrostatic mismatch score
- EpMS
eplet mismatch score
Introduction
Many countries operate deceased donor kidney allocation schemes that aim to ensure equity of access to transplantation, while minimizing the number of donor HLA mismatches to reduce the risk of graft rejection. The diversity of HLA types is such that while poorly HLA matched grafts can usually be avoided, most (>80%) recipients receive grafts with one or more HLA mismatches. Inevitably, many grafts eventually fail and this is often associated with the development of antibodies against mismatched donor HLA. If repeat transplantation is undertaken, it is usually necessary to avoid donor HLA mismatches against which the patient is sensitized, a requirement that markedly limits access to transplantation.
It was generally assumed that the breadth of sensitization following a failed transplant increased with the number of donor HLA mismatches, although the precise relationship had not been examined. We recently showed that the risk of allosensitization following failure of a first renal transplant increases incrementally with the number of mismatches at individual HLA‐A, ‐B, ‐C, ‐DR, and –DQ loci 1. In this study, mismatches were based on HLA specificities and the number of donor mismatches within each locus was enumerated as 0, 1, or 2. However, all HLA mismatches within a given locus were considered to have equal relevance to allosensitization and no account was taken of potential differences in immunogenicity according to donor HLA mismatch and recipient HLA type.
Recent studies, by our group 2, 3, 4 and others 5, 6, 7, 8, have shown that HLA alloantigen immunogenicity can be more accurately assessed by evaluating differences in the number and location of amino acid (AA) mismatches at continuous and discontinuous (eplet) positions, as well as their physicochemical properties. In these approaches, interlocus (HLA‐A, ‐B, ‐C, or HLA‐DRB1/3/4/5) or intralocus (HLA‐DQA1/DQB1) AA sequence subtraction is performed on the assumption that a polymorphic AA residue at a given sequence position within a donor HLA can be considered nonimmunogenic if it is expressed on the recipient HLA molecules. In the present study we sought to determine whether donor HLA immunogenicity as defined by differences in the number of amino acid mismatches as well as their physicochemical properties enables better prediction of the development of HLA‐specific antibodies in patients listed for repeat renal transplantation.
Methods
Patients and HLA‐specific antibody screening
The patient population studied and the antibody screening protocol used have been described in detail previously 1. Briefly, the study cohort comprised 131 consecutive patients (87 males, 44 females, median age 38) who received a primary kidney allograft between 1995 and 2010, and returned to the Cambridge kidney transplant waiting list following failure of their graft during this time period (56 patients [43%] underwent transplant nephrectomy). Of the 131 patients, 66 (50.4%) continued to receive immunosuppression after return to the waiting list (36 patients received a single agent [prednisolone in all but 4 patients] and 30 received multiple immunosuppressive agents [mostly a calcineurin inhibitor and prednisolone]). During the period when recipients received their primary kidney transplant, organ allocation favored HLA matching, particularly at the HLA‐DR locus. Whereas only 11% of the recipient cohort received a donor kidney transplant with 0–1 HLA‐A, ‐B, and ‐C mismatches, 49% received a graft with 0–1 HLA‐DR mismatch. Antibody screening was undertaken at the time of (and prior to) the first transplant, after return to the transplant waiting list following graft failure, and at 3‐monthly intervals while remaining on the list for retransplantation. Screening was undertaken using Luminex single antigen beads with mean fluorescence intensity (MFI) cut‐off thresholds of 2000 and 8000 to identify the presence of donor‐specific antibodies (DSA) and to allow determination of the calculated reaction frequency (cRF) against a panel of 10 000 consecutive UK organ donors 9. For each patient, cRF was determined for HLA class I loci (HLA‐A, ‐B, ‐C), for HLA class II loci (HLA‐DRB1/3/4/5 and HLA‐DQ), and for HLA class I and II loci combined. Multiple sera for each patient listed for retransplantation were examined and the peak reactive serum was identified as that showing the highest cRF within a median (standard deviation [SD]) follow‐up period since first transplantation of 2539 (1605) days. Patient sera may exhibit high reactivity to HLA (high cRF) due to the presence of multiple alloantibodies or due to a limited number of alloantibodies directed against broadly reactive public epitopes; such analyses were beyond the scope of this study.
Determination of HLA amino acid mismatch score (AMS), electrostatic mismatch score (EMS), and eplet mismatch score (EpMS)
The amino acid mismatch score (AMS) for each mismatched donor HLA was determined by performing inter‐ and intralocus amino acid sequence comparisons between the donor HLA and the recipient HLA class I or class II type using a previously described computer algorithm 3, 4. Similarly, the electrostatic mismatch score (EMS) for each mismatched donor HLA was calculated as the sum of the differences in isoelectric point for each mismatched amino acid (identified above, 3, 4). For each patient, the total AMS and the total EMS were calculated by summing the AMS or the EMS for each mismatched HLA present on the kidney donor HLA type. The computer algorithm is freely available for download (http://www.hlaimmunogenicity.org/download/Cambridge_HLA_Class_I_Immunogenicity_Algorithm.xls and http://www.hlaimmunogenicity.org/download/Cambridge_HLA_Class_II_Immunogenicity_Algorithm.xls).
The eplet mismatch score (EpMS) between kidney donor and recipient HLA class I and class II types was determined using the HLAMatchmakerTM computer algorithm 6, 8.
Statistical methods
Study population characteristics and descriptive statistics for this patient cohort have been detailed previously 1. A univariate exploratory analysis incorporating HLA immunogenicity variables was performed and is presented in Table S1. Logistic regression was used to perform univariate and multivariate analyses to explore the association of conventional HLA mismatch grade, HLA immunogenicity scores, and clinical variables, with the risk of developing posttransplant failure HLA‐specific sensitization (cRF > 15%) and with the risk of becoming highly sensitized (cRF ≥ 85%). To examine for an independent effect of HLA immunogenicity scores on posttransplant sensitization, adjusting for the effect of conventional HLA mismatch grade, and to account for potential collinearity between these variables, linear regression was used to de‐correlate AMS, EpMS, or EMS from HLA mismatch grade before inclusion into the models. The p‐values were taken from likelihood ratio tests. For the DSA analyses, logistic regression models were used to investigate the association between the development of DSA responses (at MFI levels of >2000 and >8000) and clinical and HLA immunogenicity explanatory variables. Initially, each explanatory variable was modelled separately; further models investigated the additional value in incorporating AMS, EpMS, or EMS into models including dual immunosuppression while on the waiting list, length of time on the waiting list, and allograft nephrectomy (DSA analyses consider individual donor–recipient HLA mismatches and, therefore, correction for conventional HLA match grade is not applicable). For presentation, AMS, EpMS, and EMS were grouped, but for regression models, the absolute value was used. Statistical significance was assessed using likelihood ratio tests at 5% significance level. Due to the inherent correlation between HLA immunogenicity scores, AMS, EpMS, or EMS were included separately into the multivariate models. All analyses were performed in R (R Foundation for Statistical Computing, Vienna, Austria) 10.
Results
Antibody screening of the 131 patients comprising the study cohort showed that before transplantation, 16.0% of patients were sensitized (cRF > 15%) and 3.8% were highly sensitized (cRF ≥ 85%) to HLA. While on the waiting list for repeat kidney transplantation, 67.9% became sensitized and 49.6% became highly sensitized to HLA. As reported previously, the level of sensitization in this cohort increased incrementally with the number of donor HLA mismatches of their failed transplant, and all HLA loci assessed (HLA‐A, ‐B, ‐C, ‐DRB1, ‐DRB3/4/5, and DQB1) contributed independently to sensitization (adjusted for pretransplant sensitization), although the contribution of HLA‐C locus mismatches was less pronounced. Sensitization was also independently associated with length of time on the waiting list for repeat transplantation and with maintenance of dual‐therapy immunosuppression 1.
In the present study we examined the association between HLA‐specific antibody formation and the immunogenicity of donor HLA mismatches as determined by the AMS, EpMS, and the EMS between donor and recipient HLA molecules. The mean (SD) AMS, EpMS, and EMS for HLA class I was 20 (11.1), 17 (9.4), and 31 (20.8), respectively; the mean (SD) AMS, EpMS, and EMS for HLA‐DR (‐DRB1 and ‐DRB3/4/5) was 5 (7.2), 8 (10.0), and 7 (9.3), respectively; and the mean (SD) AMS, EpMS, and EMS for HLA‐DQ (‐DQA1 and ‐DQB1) was 11 (15.4), 12 (13.9), and 15 (22.8), respectively.
Influence of donor HLA immunogenicity on development of posttransplant HLA‐specific sensitization (expressed as cRF)
An exploratory univariate analysis was undertaken to determine whether the immunogenicity of donor HLA mismatches expressed by the failed kidney transplant, as assessed by AMS, EpMS, and EMS, was associated with subsequent sensitization detected on analysis of peak reactive sera while patients were on the list for repeat transplantation. For this analysis, cRF levels were categorized into four bands (0–15%, 16–50%, 51–84%, and 85–100%). As shown in Figure 1, sensitization to HLA class I, HLA class II, and overall HLA class I and class II increased with increasing AMS (odds ratio [OR] on overall cRF > 15%: 1.40, 95% CI: 1.16–1.71 per 10 unit increase of AMS, p < 0.001), EpMS (OR on overall cRF > 15%: 1.36, 95% CI: 1.13–1.64 per 10 unit increase of EpMS, p < 0.001) or EMS (OR on overall cRF > 15%: 1.27, 95% CI: 1.11–1.45 per 10 unit increase of EMS, p < 0.001).
Figure 1.

Association between the immunogenicity of first transplant donor HLA mismatches and posttransplant HLA ‐specific sensitization expressed as calculated reaction frequency ( cRF ). HLA‐specific alloantibodies were detected using single‐antigen HLA beads (mean fluorescence intensity cut‐off threshold of 2000); the likelihood of identifying an antibody‐compatible organ donor (cRF) was determined by comparing individual patient HLA‐specific antibody profiles with the HLA types of 10 000 consecutive UK deceased organ donors. Panel (A) shows peak cRF levels while on the waiting list attributable to antibodies against HLA‐A, ‐B, and ‐C considered collectively according to the immunogenicity of donor HLA class I mismatches expressed by the failed kidney transplant, as assessed by amino acid mismatch score (AMS), eplet mismatch score (EpMS), and electrostatic mismatch score (EMS). Panel (B) shows peak cRF levels while on the waiting list attributable to antibodies against HLA‐DRB1, ‐DRB3/4/5 and ‐DQ, considered collectively according to the immunogenicity of donor HLA class II mismatches present on the failed kidney transplant, as assessed by AMS, EpMS and EMS. Panel (C) shows peak cRF levels while on the waiting list attributable to antibodies against HLA class I and class II considered collectively according to the immunogenicity of donor HLA class I and class II mismatches present on the failed kidney transplant, as assessed by AMS, EpMS, and EMS. Patients were categorized according to the likelihood of identifying an antibody‐compatible organ donor as cRF 0–15%, cRF 16–50%, cRF 51–84%, and cRF 85–100%. Patients were grouped in quantiles of the variable of interest (AMS, EpMS, or EMS) and within each group the number of patients is shown.
Subsequently, multivariate logistic regression was used to adjust for the effect on sensitization of the length of time on the waiting list and of maintenance of dual‐therapy immunosuppression while on the waiting list for retransplantation. The analysis was also controlled for the inherent correlation between conventional HLA mismatch grade (0, 1, or 2 HLA mismatches per locus) and HLA immunogenicity scores, using linear regression to de‐correlate the AMS, EpMS, or EMS from the number of donor HLA mismatches present on the failed kidney transplant. As shown in Table 1, donor HLA immunogenicity as assessed by AMS, EpMS, and EMS was independently associated with the risk of developing posttransplant HLA class I and class II specific antibodies (cRF 16–100%), providing additional predictive value to that of conventional HLA mismatch grade. HLA mismatch grade (OR: 1.29, 95% CI: 1.07–1.59, p = 0.01), dual agent immunosuppression (OR: 0.28, 95% CI: 0.08–0.81, p = 0.03), and time on the waiting list (OR: 1.35, 95% CI: 1.13–1.67, p = 0.002) were all associated with the risk of a patient becoming highly sensitized (cRF ≥ 85%), whereas mismatched amino acids (AMM), EpMS, and EMS had no independent effect.
Table 1.
Multivariate analysis: influence of donor HLA immunogenicity on the development of posttransplant HLA class I and class II specific antibodies (expressed as calculated reaction frequency [cRF])
| Variable | Odds ratio (95% CI) on developing HLA‐specific sensitization (cRF 16–100%) | Odds ratio (95% CI) on becoming highly sensitized (cRF 85–100%) | ||
|---|---|---|---|---|
| OR (95% CI) | p‐value | OR (95% CI) | p‐value | |
| AMS (per 10 AA MM)a | 1.44 (1.02, 2.10) | 0.04 | 1.22 (0.91, 1.65) | 0.18 |
| HLA (per MM) | 1.29 (1.05, 1.62) | 0.02 | 1.29 (1.07, 1.59) | 0.01 |
| Dual agent immunosuppression | 0.42 (0.16, 1.11) | 0.08 | 0.28 (0.08, 0.81) | 0.03 |
| Time on the waiting list (per year) | 1.54 (1.21, 2.07) | 0.001 | 1.35 (1.13, 1.67) | 0.002 |
| EpMS (per 10 eplet MM)a | 1.41 (1.00, 2.05) | 0.05 | 1.26 (0.94, 1.71) | 0.13 |
| HLA (per MM) | 1.39 (1.05, 1.63) | 0.02 | 1.30 (1.07, 1.59) | 0.01 |
| Dual agent immunosuppression | 0.39 (0.15, 1.04) | 0.06 | 0.26 (0.08, 0.77) | 0.02 |
| Time on the waiting list (per year) | 1.51 (1.19, 2.01) | 0.002 | 1.34 (1.11, 1.65) | 0.003 |
| EMS (per 10 units)a | 1.27 (1.02, 1.62) | 0.04 | 1.13 (0.94, 1.37) | 0.20 |
| HLA (per MM) | 1.30 (1.05, 1.64) | 0.02 | 1.30 (1.07, 1.59) | 0.01 |
| Dual agent immunosuppression | 0.40 (0.15, 1.04) | 0.06 | 0.27 (0.08, 0.77) | 0.02 |
| Time on the waiting list (per year) | 1.54 (1.19, 2.07) | 0.002 | 1.34 (1.11, 1.65) | 0.003 |
A minority of this patient cohort had low level HLA‐specific sensitization before transplantation; adjustment for pretransplant sensitization levels was performed and did not change significantly the results of these analyses.
Statistically significant values are indicated in bold font.
AMS, amino acid mismatch score; EMS, electrostatic mismatch score; EpMS, eplet mismatch score; MM, mismatches.
Linear regression was used to de‐correlate AMS, EpMS, or EMS from HLA mismatch grade before inclusion into the multivariate models.
We also examined the effect of donor HLA immunogenicity scores on the risk of developing sensitization to HLA‐A, ‐B, ‐C; HLA‐DR (‐DRB1 and ‐DRB3/4/5); and HLA‐DQ. Multivariate analyses showed that AMS, EpMS, and EMS were independently associated with the risk of developing HLA class I (cRF > 15% and cRF ≥ 85%) and HLA‐DQ specific antibodies (cRF > 15%), whereas HLA‐DR mismatch grade correlated with locus‐specific sensitization with an additional effect attributable to HLA‐DR EMS for high (≥85%) HLA‐DR specific cRF (Table S2).
Influence of donor HLA immunogenicity on development of posttransplant DSA
We next sought to determine the factors associated with the development of DSA against the HLA mismatches present on the failed renal allograft. For this analysis, all donor–recipient HLA mismatches for the entire study cohort (n = 671) were pooled and analyzed together. While on the waiting list for retransplantation, 40 patients developed DSA against HLA class I, 4 against HLA class II, and 31 against both HLA class I and II. Overall, DSA was detected against 235 of the 671 (35%) donor–recipient HLA mismatches with a median (SD) MFI of 8071 (5129). DSA responses against HLA‐C mismatches were infrequent (16.8%) and not associated with donor HLA‐C alloantigen immunogenicity. Univariate logistic regression analysis (Figure 2A) focusing on HLA‐A and ‐B DSA responses showed that the EMS, but not AMS or EpMS, of a donor HLA correlated with the likelihood of an antibody response. Multivariate analyses, adjusting for length of time on the waiting list, maintenance on dual‐therapy immunosuppression, and for nephrectomy, confirmed that EMS was independently associated with HLA‐A and ‐B DSA development (for DSA MFI > 2000, OR: 1.81, 95% CI: 1.16–2.86, p = 0.01 per 10 EMS units; and for DSA MFI > 8000, OR: 1.62, 95% CI: 1.01–2.59, p = 0.04 per 10 EMS units; Table 2). Multivariate logistic regression analyses of HLA Class II DSA responses showed that all three HLA immunogenicity scores were independently associated with the development of HLA‐DR (at MFI > 2000 and >8000) and HLA‐DQ DSA (Table 2 and Figures 2B and C) and no differences in the predictive power of AMS, EpMS, or EMS were observed.
Figure 2.
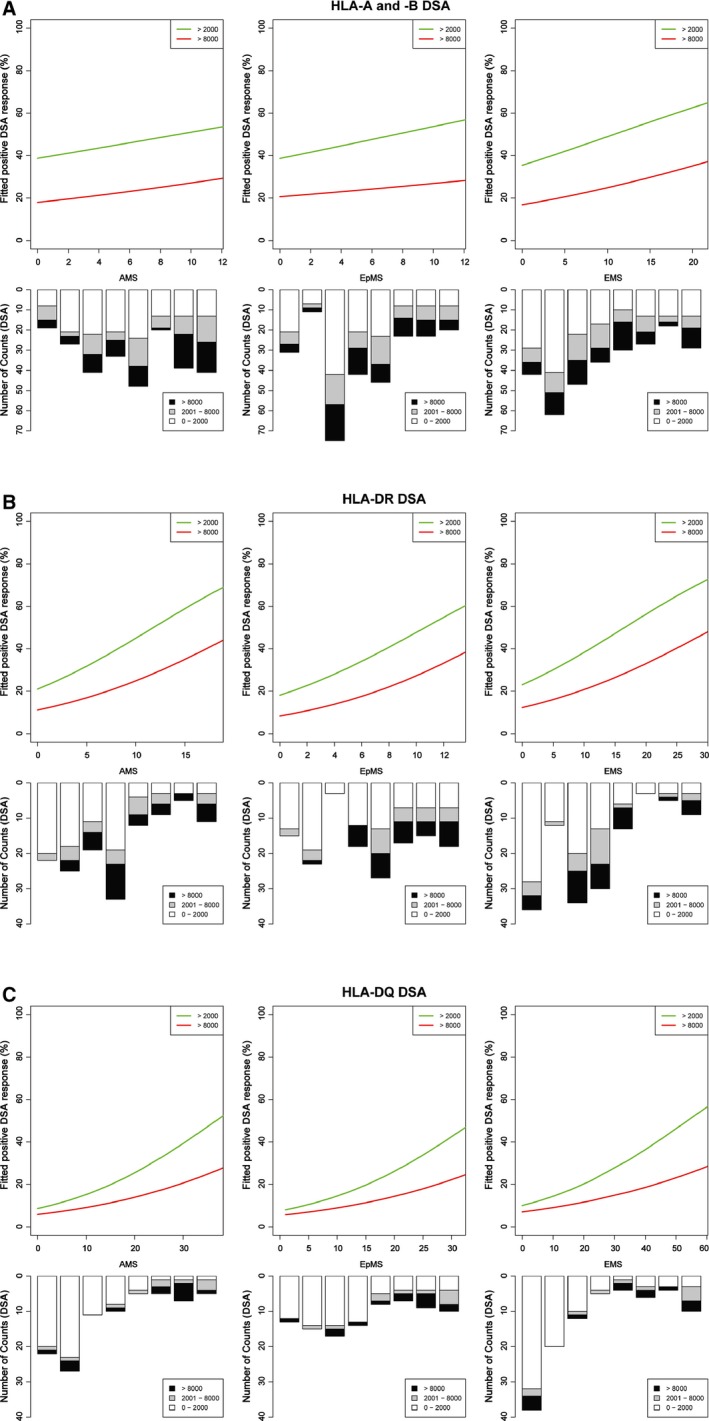
Logistic regression analyses of the relationship between the immunogenicity of donor HLA mismatches and development of posttransplant donor‐specific antibodies ( DSA ). Development of alloantibodies against donor HLA mismatches expressed by the failed kidney transplant were detected using single‐antigen HLA bead analysis of sera obtained following transplant failure (using mean fluorescence intensity [MFI] cut‐off thresholds of 2000 and 8000 to reflect increasing levels of DSA). Panels (A), (B), and (C) show the fitted logistic regression curves (green line for DSA with MFI > 2000 and red line for DSA with MFI > 8000) for HLA‐A and ‐B, HLA‐DRB1/3/4/5, and HLA‐DQ DSA, respectively. For the regression models absolute values were used, but for presentation amino acid mismatch score (AMS), eplet mismatch score (EpMS), and electrostatic mismatch score (EMS) were grouped and the number of DSA and MFI levels within each group is shown.
Table 2.
Multivariate analysis: influence of donor HLA immunogenicity on development of posttransplant donor‐specific antibodies (DSA)
| Variable | Odds ratio (95% CI) on developing HLA donor‐specific antibodies (MFI > 2000) | Odds ratio (95% CI) on developing high level HLA donor‐specific antibodies (MFI > 8000) | ||
|---|---|---|---|---|
| OR (95% CI) | p‐value | OR (95% CI) | p‐value | |
| HLA‐A and –B | ||||
| AMS (per 10 AA MM) | 2.02 (1.01, 4.12) | 0.05 | 1.72 (0.81, 3.65) | 0.16 |
| Dual agent immunosuppression | 0.36 (0.14, 0.83) | 0.02 | 0.29 (0.09, 0.79) | 0.02 |
| Time on the waiting list (per year) | 1.31 (1.16, 1.48) | <0.001 | 1.02 (0.90, 1.15) | 0.75 |
| Nephrectomy | 2.27 (1.27, 4.15) | 0.006 | 1.19 (0.64, 2.26) | 0.59 |
| EpMS (per 10 eplet MM) | 2.04 (0.90, 4.69) | 0.09 | 1.44 (0.59, 3.46) | 0.42 |
| Dual agent immunosuppression | 0.35 (0.14, 0.82) | 0.02 | 0.29 (0.09, 0.79) | 0.02 |
| Time on the waiting list (per year) | 1.30 (1.15, 1.47) | <0.001 | 1.01 (0.90, 1.14) | 0.81 |
| Nephrectomy | 2.20 (1.23, 4.02) | 0.009 | 1.17 (0.63, 2.22) | 0.62 |
| EMS (per 10 units) | 1.81 (1.16, 2.86) | 0.01 | 1.62 (1.01, 2.59) | 0.04 |
| Dual agent immunosuppression | 0.34 (0.13, 0.80) | 0.02 | 0.28 (0.09, 0.76) | 0.02 |
| Time on the waiting list (year) | 1.29 (1.15, 1.47) | <0.001 | 1.00 (0.90, 1.14) | 0.87 |
| Nephrectomy | 2.15 (1.20, 3.95) | 0.01 | 1.13 (0.60, 2.15) | 0.71 |
| HLA‐DRB1/3/4/5 | ||||
| AMS (per 10 AA MM) | 5.42 (2.23, 15.01) | <0.001 | 4.02 (1.65, 10.94) | 0.003 |
| Dual agent immunosuppression | 0.05 (0.01, 0.21) | <0.001 | N/Aa | – |
| Time on the waiting list (per year) | 1.00 (0.83, 1.19) | 0.96 | 0.93 (0.75, 1.15) | 0.53 |
| EpMS (per 10 eplet MM) | 6.30 (2.30, 19.30) | <0.001 | 6.97 (2.24, 25.58) | 0.002 |
| Dual agent immunosuppression | 0.06 (0.01, 0.23) | <0.001 | N/Aa | – |
| Time on the waiting list (per year) | 0.98 (0.82, 1.17) | 0.83 | 0.93 (0.75, 1.16) | 0.54 |
| EMS (per 10 units) | 2.77 (1.52, 5.52) | 0.002 | 2.37 (1.32, 4.68) | 0.006 |
| Dual agent immunosuppression | 0.06 (0.01, 0.24) | <0.001 | N/Aa | – |
| Time on the waiting list (per year) | 0.94 (0.79, 1.11) | 0.50 | 0.89 (0.72, 1.09) | 0.28 |
| HLA‐DQ | ||||
| AMS (per 10 AA MM) | 1.79 (1.19, 2.71) | 0.005 | 1.49 (0.92, 2.47) | 0.11 |
| Dual agent immunosuppression | 0.18 (0.01, 1.10) | 0.12 | 0.29 (0.01, 1.93) | 0.28 |
| Time on the waiting list (per year) | 0.91 (0.70, 1.15) | 0.43 | 0.82 (0.58, 1.09) | 0.20 |
| EpMS (per 10 eplet MM) | 1.99 (1.20, 3.47) | 0.011 | 1.59 (0.86, 3.08) | 0.15 |
| Dual agent immunosuppression | 0.17 (0.01, 1.00) | 0.10 | 0.28 (0.01,1.78) | 0.25 |
| Time on the waiting list (per year) | 0.91 (0.71, 1.15) | 0.45 | 0.82 (0.58, 1.10) | 0.21 |
| EMS (per 10 units) | 1.46 (1.14, 1.90) | 0.003 | 1.26 (0.93, 1.70) | 0.14 |
| Dual agent immunosuppression | 0.17 (0.01, 1.01) | 0.11 | 0.27 (0.01, 1.72) | 0.24 |
| Time on the waiting list (per year) | 0.89 (0.69, 1.14) | 0.37 | 0.81 (0.57, 1.09) | 0.20 |
Statistically significant values are indicated in bold font.
AA, amino acid; AMS, amino acid mismatch score; DSA, donor‐specific antibodies; EMS, electrostatic mismatch score; EpMS, eplet mismatch score; MFI, mean fluorescence intensity; MM, mismatches.
HLA‐DR DSA in patients on dual agent immunosuppression had MFI values below 8000.
Discussion
The risk of allosensitization following failure of a first renal transplant increases incrementally with the number of mismatches at individual HLA‐A, ‐B, ‐C, ‐DR, and ‐DQ loci 1. However, this simple numerical approach to assessing HLA mismatch grade takes no account of differences in donor HLA immunogenicity according to recipient HLA type and this is likely to have an important influence on the alloimmune response. Knowledge of HLA structure, along with the ability to characterize alloantibody specificities in patient sera using single antigen bead technology, now allows the potential impact of differences between donor and recipient HLA molecules to be determined, with a view to developing improved strategies for kidney allocation.
In the present study we examined three different approaches for assessment of HLA class I and class II immunogenicity. These ranged from simply enumerating the number of AMM between donor and recipient HLA, to counting the number of polymorphic surface accessible amino acid residues at discontinuous positions of donor HLA that cluster together to form a potential epitope (EpMS), to assessing the physicochemical disparity between the side chains of mismatched amino acids between donor and recipient HLA (EMS). The principal finding was that assessment of donor HLA immunogenicity based on AMS, EpMS, or EMS offers additional value to that of conventional HLA mismatch grade for predicting sensitization to HLA in patients awaiting retransplantation after a failed first kidney transplant. Moreover, donor HLA‐DR and ‐DQ alloantigens with high AMS, EpMS, or EMS were more likely to induce DSA responses, which in the case of HLA‐DR were more likely to be of high level (MFI > 8000). Importantly, donor HLA EMS, but not AMS or EpMS, predicted the development of DSA (at MFI > 2000 and >8000) against HLA‐A and ‐B mismatches.
Following kidney transplantation, DSA development against both HLA class I and class II alloantigens is an important risk factor for subsequent chronic humoral rejection and allograft failure 11, 12, 13, 14. Humoral responses against HLA class II are frequent and commonly involve HLA‐DQ specific antibodies 15, 16. Our study suggests that the risk of developing both HLA‐DR and ‐DQ DSA can be predicted by accounting for the immunogenicity of donor HLA class II mismatches. Our findings agree with recent reports from Wiebe et al demonstrating that high donor HLA‐DR and ‐DQ immunogenicity, as assessed by high epitope (eplet) load, increases the risk of DSA development and of subsequent kidney graft failure 17, 18. We did not, however, demonstrate an advantage in using an eplet approach to assess HLA immunogenicity over simply enumerating the number of amino acid polymorphisms between donor and recipient HLA molecules. AMS and EpMS both reflect differences in amino acid sequence between donor and recipient HLA mismatches and while aiding prediction of immunogenicity of a particular HLA mismatch, they do not take into account the physicochemical properties of the amino acid polymorphisms involved. The specificity and affinity of antibody binding to target antigen is strongly influenced by electrostatic interactions, and these are determined by the number and polar charges of amino acid side chains 2, 19. EMS integrates information on the number of mismatched amino acids and the differences in electrostatic charges of their side chains between donor and recipient HLA class I and class II molecules. Our results show that this additional information improves the ability to predict the development of an alloantibody response against a given HLA mismatch.
While the present study clearly shows that prediction of HLA immunogenicity based on information derived from polymorphic amino acids on donor HLA and their physicochemical properties is superior to the traditional approach of assigning equal weight to all HLA mismatches within a particular locus, there are some limitations to our study. First, we analyzed alloantibody responses after kidney transplant failure and our findings would be strengthened if they were confirmed in patients with functioning grafts. This would require access to data from a prospective posttransplant alloantibody monitoring program with long‐term follow‐up, which is not currently widely available. Second, our analysis is strengthened by quantitative analyses of DSA development based on MFI cut‐off levels of >2000 and >8000. However, even though we routinely treat sera with EDTA to overcome the prozone phenomenon 20, 21, we acknowledge that titration studies would have provided further evidence on alloantibody strength 22. Moreover, HLA‐DP type was not routinely performed during the period of the study, so we were unable to consider its influence on allosensitization, and it is apparent that many patients become sensitized to HLA‐DP after transplant failure 23. There is, however, no a priori reason why amino acid comparison after intralocus subtraction for HLA‐DP should not predict allosensitization since HLA‐DP is structurally very similar to HLA‐DR and ‐DQ 24. As described previously 1, the patient cohort in the present study was moderately well matched particularly for HLA‐DR and ‐DQ. While the size of the study cohort was sufficient to demonstrate the additional influence of AMS, EpMS, and EMS over simply counting mismatched HLA specificities, it did not allow in‐depth analysis of HLA‐DQ immunogenicity, because of the limited number of mismatched HLA‐DQ specificities within the study cohort. Finally, we have previously shown that transplant nephrectomy did not have an independent effect on overall sensitization to HLA when withdrawal of immunosuppression was taken into account 1. However, the present study showed that transplant nephrectomy was independently associated with DSA development against donor HLA‐A and ‐B alloantigens, suggesting that these alloantibodies may be absorbed to an extent by the graft and become more apparent after its removal. A similar effect for DSA against HLA class II was not demonstrated and, as explained above, this may be due to the relatively limited number of HLA class II mismatches in this patient cohort.
In conclusion, our findings demonstrate a clear relationship between the immunogenicity of donor HLA class I and class II mismatches and the development of HLA‐specific antibodies after graft failure and relisting for transplantation. HLA antibodies severely limit the chance of finding an antibody‐compatible donor kidney for patients requiring retransplantation and HLA matching is, therefore, particularly important in recipients who are likely to require repeat transplantation in the future. While the traditional approach to HLA matching, based on counting the number of mismatched HLA specificities has merit, our findings show that more sophisticated approaches to determining HLA compatibility improve assessment of HLA immunogenicity and consideration should be given to incorporating them into HLA matching algorithms. Eurotransplant has implemented the use of HLAMatchmaker to identify antibody‐compatible donors for patients who are already highly sensitized 25, 26. The present study supports the incorporation of such approaches to HLA matching for allocation of deceased donor kidneys to first‐time recipients. Although further validation is required, our findings suggest that information on the electrostatic charge of polymorphic amino acids in mismatched HLA alleles (EMS) should be introduced into HLA matching algorithms, as it improves prediction of DSA development and HLA‐specific sensitization. Such approaches to HLA matching are also more permissive than simply aiming to avoid as many HLA mismatches as possible, because they identify acceptable HLA mismatches that are likely to be of low immunogenicity, thereby increasing the number of deceased donors that might be considered a suitable HLA match for a given recipient.
Disclaimer
The views expressed are those of the authors and not necessarily those of the NHS, the NIHR, the Department of Health, or NHSBT.
Disclosure
The authors of this manuscript have no conflicts of interest to disclose as described by the American Journal of Transplantation.
Supporting information
Table S1: Exploratory analysis of explanatory variables and posttransplant sensitization (expressed as calculated reaction frequency–cRF; mean fluorescence intensity threshold of >2000).
Table S2: Multivariate analysis: influence of donor HLA immunogenicity on the development of posttransplant HLA class I, HLA‐DRB1/3/4/5, and HLA‐DQ specific antibodies (expressed as calculated reaction frequency–cRF; mean fluorescence intensity threshold of >2000).
Acknowledgments
This study was supported by the Cambridge National Institute for Health Research Biomedical Research Centre and the NIHR Blood and Transplant Research Unit in Organ Donation and Transplantation at the University of Cambridge in collaboration with Newcastle University and in partnership with National Health Service Blood and Transplant (NHSBT). V.K. was supported by an Academy of Medical Sciences Grant and an Evelyn Trust Grant. D.H.M. was supported by an RCSEng Research Fellowship.
Kosmoliaptsis V, Mallon DH, Chen Y, Bolton EM, Bradley JA & Taylor CJ. Alloantibody Responses After Renal Transplant Failure Can Be Better Predicted by Donor–Recipient HLA Amino Acid Sequence and Physicochemical Disparities Than Conventional HLA Matching. Am J Transplant 2016; 16: 2139–2147
References
- 1. Kosmoliaptsis V, Gjorgjimajkoska O, Sharples LD, et al. Impact of donor mismatches at individual HLA‐A, ‐B, ‐C, ‐DR, and ‐DQ loci on the development of HLA‐specific antibodies in patients listed for repeat renal transplantation. Kidney Int 2014; 50: 540–544. [DOI] [PubMed] [Google Scholar]
- 2. Kosmoliaptsis V, Dafforn TR, Chaudhry AN, Halsall DJ, Bradley JA, Taylor CJ. High‐resolution, three‐dimensional modeling of human leukocyte antigen class I structure and surface electrostatic potential reveals the molecular basis for alloantibody binding epitopes. Hum Immunol 2011; 72: 1049–1059. [DOI] [PubMed] [Google Scholar]
- 3. Kosmoliaptsis V, Sharples LD, Chaudhry AN, Halsall DJ, Bradley JA, Taylor CJ. Predicting HLA class II alloantigen immunogenicity from the number and physiochemical properties of amino acid polymorphisms. Transplantation 2011; 91: 183–190. [DOI] [PubMed] [Google Scholar]
- 4. Kosmoliaptsis V, Chaudhry AN, Sharples LD, et al. Predicting HLA Class I alloantigen immunogenicity from the number and physiochemical properties of amino acid polymorphisms. Transplantation 2009; 88: 791–798. [DOI] [PubMed] [Google Scholar]
- 5. Duquesnoy RJ. Antibody‐reactive epitope determination with HLAMatchmaker and its clinical applications. Tissue Antigens 2011; 77: 525–534. [DOI] [PubMed] [Google Scholar]
- 6. Duquesnoy RJ. A structurally based approach to determine HLA compatibility at the humoral immune level. Hum Immunol 2006; 67: 847–862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Tambur AR, Claas FH. HLA epitopes as viewed by antibodies: What is it all about? Am J Transplant 2015; 15: 1148–1154. [DOI] [PubMed] [Google Scholar]
- 8. Duquesnoy RJ, Askar M. HLAMatchmaker: A molecularly based algorithm for histocompatibility determination. V. Eplet matching for HLA‐DR, HLA‐DQ, and HLA‐DP. Hum Immunol 2007; 68: 12–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Howell WM, Harmer A, Briggs D, et al. British Society for Histocompatibility & Immunogenetics and British Transplantation Society guidelines for the detection and characterisation of clinically relevant antibodies in allotransplantation. Int J Immunogenet 2010; 37: 435–437. [DOI] [PubMed] [Google Scholar]
- 10. R Core Team . R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2013. Available from: http://www.R-project.org/. [Google Scholar]
- 11. Lefaucheur C, Viglietti D, Bentlejewski C, et al. IgG donor‐specific anti‐human HLA antibody subclasses and kidney allograft antibody‐mediated injury. J Am Soc Nephrol 2016; 27: 293–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lee PC, Zhu L, Terasaki PI, Everly MJ. HLA‐specific antibodies developed in the first year posttransplant are predictive of chronic rejection and renal graft loss. Transplantation 2009; 88: 568–574. [DOI] [PubMed] [Google Scholar]
- 13. Mao Q, Terasaki PI, Cai J, et al. Extremely high association between appearance of HLA antibodies and failure of kidney grafts in a five‐year longitudinal study. Am J Transplant 2007; 7: 864–871. [DOI] [PubMed] [Google Scholar]
- 14. Mehra NK, Siddiqui J, Baranwal A, Goswami S, Kaur G. Clinical relevance of antibody development in renal transplantation. Ann N Y Acad Sci 2013; 1283: 30–42. [DOI] [PubMed] [Google Scholar]
- 15. Willicombe M, Brookes P, Sergeant R, et al. De novo DQ donor‐specific antibodies are associated with a significant risk of antibody‐mediated rejection and transplant glomerulopathy. Transplantation 2012; 94: 172–177. [DOI] [PubMed] [Google Scholar]
- 16. DeVos JM, Gaber AO, Knight RJ, et al. Donor‐specific HLA‐DQ antibodies may contribute to poor graft outcome after renal transplantation. Kidney Int 2012; 82: 598–604. [DOI] [PubMed] [Google Scholar]
- 17. Wiebe C, Nevins TE, Robiner WN, Thomas W, Matas AJ, Nickerson PW. The synergistic effect of Class II HLA epitope‐mismatch and nonadherence on acute rejection and graft survival. Am J Transplant 2015; 15: 2197–2202. [DOI] [PubMed] [Google Scholar]
- 18. Wiebe C, Pochinco D, Blydt‐Hansen TD, et al. Class II HLA epitope matching‐A strategy to minimize de novo donor‐specific antibody development and improve outcomes. Am J Transplant 2013; 13: 3114–3122. [DOI] [PubMed] [Google Scholar]
- 19. Mallon DH, Bradley JA, Winn PJ, Taylor CJ, Kosmoliaptsis V. Three‐dimensional structural modelling and calculation of electrostatic potentials of HLA Bw4 and Bw6 epitopes to explain the molecular basis for alloantibody binding: Toward predicting HLA antigenicity and immunogenicity. Transplantation 2015; 99: 385–390. [DOI] [PubMed] [Google Scholar]
- 20. Kosmoliaptsis V, Bradley JA, Peacock S, Chaudhry AN, Taylor CJ. Detection of immunoglobulin G human leukocyte antigen‐specific alloantibodies in renal transplant patients using single‐antigen‐beads is compromised by the presence of immunoglobulin M human leukocyte antigen‐specific alloantibodies. Transplantation 2009; 87: 813–820. [DOI] [PubMed] [Google Scholar]
- 21. Schnaidt M, Weinstock C, Jurisic M, Schmid‐Horch B, Ender A, Wernet D. HLA antibody specification using single‐antigen beads—a technical solution for the prozone effect. Transplantation 2011; 92: 510–515. [DOI] [PubMed] [Google Scholar]
- 22. Tambur AR, Herrera ND, Haarberg KM, et al. Assessing antibody strength: Comparison of MFI, C1q, and titer information. Am J Transplant 2015; 15: 2421–2430. [DOI] [PubMed] [Google Scholar]
- 23. Jolly EC, Key T, Rasheed H, et al. Preformed donor HLA‐DP‐specific antibodies mediate acute and chronic antibody‐mediated rejection following renal transplantation. Am J Transplant 2012; 12: 2845–2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tambur AR, Buckingham M, McDonald L, Luo X. Development of donor‐specific and non‐donor‐specific HLA‐DP antibodies post‐transplant: The role of epitope sharing and epitope matching. Clin Transpl 2006; 399–404. [PubMed] [Google Scholar]
- 25. Heidt S, Witvliet MD, Haasnoot GW, Claas FH. The 25th anniversary of the Eurotransplant Acceptable Mismatch program for highly sensitized patients. Transpl Immunol 2015; 33: 51–57. [DOI] [PubMed] [Google Scholar]
- 26. Claas FH, Doxiadis II. Management of the highly sensitized patient. Curr Opin Immunol 2009; 21: 569–572. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1: Exploratory analysis of explanatory variables and posttransplant sensitization (expressed as calculated reaction frequency–cRF; mean fluorescence intensity threshold of >2000).
Table S2: Multivariate analysis: influence of donor HLA immunogenicity on the development of posttransplant HLA class I, HLA‐DRB1/3/4/5, and HLA‐DQ specific antibodies (expressed as calculated reaction frequency–cRF; mean fluorescence intensity threshold of >2000).


