Abstract
Nuc‐ErbB3 an alternative transcript from the ErbB3 locus binds to a specific DNA motif and associates with Schwann cell chromatin. Here we generated a nuc‐ErbB3 knockin mouse that lacks nuc‐ErbB3 expression in the nucleus without affecting the neuregulin‐ErbB3 receptor signaling. Nuc‐ErbB3 knockin mice exhibit hypermyelination and aberrant myelination at the paranodal region. This phenotype is attributed to de‐repression of myelination associated gene transcription following loss of nuc‐ErbB3 and histone H3K27me3 promoter occupancy. Nuc‐ErbB3 knockin mice exhibit reduced association of H3K27me3 with myelination‐associated gene promoters and increased RNA Pol‐II rate of transcription of these genes. In addition, nuc‐ErbB3 directly regulates levels of H3K27me3 in Schwann cells. Nuc‐ErbB3 knockin mice exhibit significant decrease of histone H3K27me3 methyltransferase (HMT) activity and reduced levels of H3K27me3. Collectively, nuc‐ErbB3 is a master transcriptional repressor, which regulates HMT activity to establish a repressive chromatin landscape on promoters of genes during peripheral myelination. GLIA 2016;64:977–992
Keywords: PNS myelination, chromatin, transcriptional repression
Main Points
Nuc‐ErbB3 knock‐in mice exhibit peripheral hypermyelination.
Nuc‐ErbB3 regulates total levels of H3K27me3 and HMT activity.
Nuc‐ErbB3 induces transcriptional repression of myelination associated genes.
Introduction
The ErbB receptor tyrosine kinase family is comprised of four members: EGF receptor (ErbB1), ErbB2, ErbB3, and ErbB4 (Citri et al., 2003). ErbB3, which lacks intrinsic kinase activity is expressed throughout the Schwann cell lineage and forms heterodimers with ErbB2 (Citri et al., 2003). In the peripheral nervous system (PNS), the ligand for ErbB3 that induces signaling activation is Neuregulin‐1 (NRG1) (Meyer et al., 1997). Signaling through the neuregulin/ErbB2–3 axis has been implicated in the regulation of Schwann cell myelination (Garratt et al., 2000a; Michailov et al., 2004; Taveggia et al., 2005), migration and axonal sorting (Yamauchi et al., 2008). In cultured Schwann cells NRG1 activates mitogen‐activated protein kinase (MAPK) and PI3K pathways (Ogata et al., 2006). Activation of PI3K and its downstream target Akt are crucial for the trophic, proliferative, and differentiation responses of Schwann cells to NRG1 (Maurel and Salzer, 2000). Two other signals that also regulate Schwann cell differentiation, laminin in the extracellular matrix and cAMP‐dependent pathways, were recently implicated in NRG1‐dependent signaling (Arthur‐Farraj et al., 2011; Monje et al., 2006; Yu et al., 2005). Finally, signaling though the G‐protein coupled receptor Gpr126 has been shown to regulate Schwann cell differentiation and myelination (Mogha et al., 2013).
Various transcription factors also regulate Schwann cell differentiation and myelination including Egr2/Krox20, Oct‐6, Sox10, and Brn2 (Britsch et al., 2001; Jaegle et al., 2003, 1996; Ryu et al., 2007; Topilko et al., 1994). Among them, Egr2 is the major transcriptional modulator of Schwann cell myelination, which is regulated by soluble and membrane‐bound neuregulin (Murphy et al., 1996; Svaren and Meijer, 2008; Taveggia et al., 2005). Recent studies implicate histone deacetylases (HDACs) as global regulators of Schwann cell gene expression through the regulation of NF‐κB, Sox10, and Egr2 (Chen et al., 2011; Jacob et al., 2011). Although it is generally thought that histone deacetylation results in gene repression, in Schwann cells the role of HDACs is to activate transcription rather than repress it. Recently, histone H3K27m3 and the polycomb repressive complex 2 (PRC2) were shown to regulate maintenance of peripheral myelination through gene repression (Ma et al., 2015). Moreover, the member of PRC2 Enhancer of Zeste Homolog 2 (Ezh2) has been shown to regulate Schwann cell differentiation (Heinen et al., 2012). However, a global mechanism for the regulation of chromatin mediated transcriptional repression during peripheral myelination has not been established so far.
Recently, we discovered nuc‐ErbB3, a variant of ErbB3 that is expressed in the nucleus of Schwann cells and transcribed through alternative transcription initiation (Adilakshmi et al., 2011). Here we ablated nuc‐ErbB3 from the nucleus by generating a knockin mouse (KI), which carries a point mutation at the nuclear localization signal (NLS). This genetic manipulation does not affect the expression or function of the full‐length ErbB3 receptor tyrosine kinase. Computational analysis of in vivo ChIP data demonstrates that nuc‐ErbB3 regulates transcription through (1) co‐occupancy with H3K27me3 on promoter regions of myelination‐associated genes that contain a specific nuc‐ErbB3 DNA binding motif and (2) association with H3K27me3 enriched promoters without specific DNA motif binding. In both instances, loss of nuc‐ErbB3 from the nucleus of Schwann cells is accompanied by deassociation of the repressive histone mark H3K27me3 from the nuc‐ErbB3 bound promoters resulting in increased rate of RNA Pol‐II transcription of these genes. Transcriptional de‐repression in nuc‐ErbB3 KI mice results in peripheral hypermyelination and aberrant myelination, which is more prominent at the paranodal region.
The H3K27 histone methyltransferase (HMT) Ezh2 (enhancer of zeste 2) is a Polycomb group protein that catalyzes the tri‐methylation of lysine 27 on histone H3 (H3K27me3) to impose epigenetic gene silencing and transcriptional repression (Lee et al., 2006; Plath et al., 2003; Wu and Zhang, 2011). We show here the novel role of nuc‐ErbB3 in regulating histone H3K27 tri‐methylation. Nuc‐ErbB3 KI Schwann cells exhibit significant decrease of histone H3K27me3 HMT activity and reduced total levels of H3K27me3. In addition, overexpression of nuc‐ErbB3 in Schwann cells results in significantly elevated levels of HeK27me3. Collectively, we show for the first time that nuc‐ErbB3 is a master transcriptional repressor, which regulates histone HMT activity to establish a repressive chromatin landscape on promoters of genes during peripheral myelination.
Materials and Methods
Primary Cell Culture and DRG Explants
Mouse dorsal root ganglia explants and Schwann cell cultures were cultured as described earlier (Ness et al., 2013). Schwann cell purity was confirmed to be 100% using Sox10 and S100 staining (representative S100 staining in Fig. 2, Supporting Information). For activation of ErbB2/B3 signaling, cells were starved overnight and incubated in 20 ng/mL beta1‐heregulin for 10 min. Transfections of mouse DRG explants with Brn2 construct (Origene) were conducted using TransIT‐Neural Transfection reagent (Mirus) according to the manufacturer's protocol.
Figure 2.
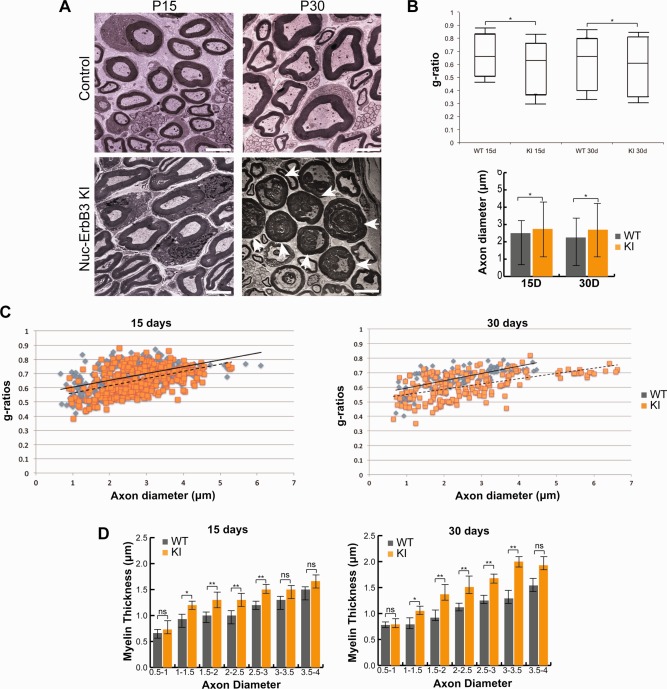
Nuc‐ErbB3 KI mice exhibit peripheral hypermyelination. (A) Electron microscopy of P15 and P30 sciatic nerves from WT and nuc‐ErbB3 KI mice shows hypermyelination in KI mice. Arrowheads at P30 show examples of severely hypermyelinated axons with condensed axoplasm in nuc‐ErbB3 KI mice. Magnification: ×4,000. (B) Quantification of g‐ratios from at least 300 myelinated axons from each time interval shows significant decrease in g‐ratios of nuc‐ErbB3 KI mice at P15 and P30. At P15 and P30 KI mice exhibit increased axonal diameter of the myelinated axons. The results are presented as average ± s.d. and significance was calculated using a paired Student's t‐test (*:P < 0.05, n = 4 mice, n.s: not significant). (C) Quantification of the g‐ratios versus axon diameters of myelinated axons between WT and nuc‐ErbB3 KI mice shows lower distribution of g‐ratios in KI mice. (D) Binning analysis of myelin thickness of group of axons with the indicated diameters. Nerves of WT and nuc‐ErbB3 KI mice at P15, and P30 were analyzed. The results are presented as average ± s.d. and significance was calculated with two‐way Anova (*:P < 0.05, **:P < 0.001, n.s.: not significant). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Animal Use and Care
To obtain a point mutated nuc‐ErbB3 KI mouse strain, the mutation was inserted in the near region of exon 27 using a floxed neomycin selection cassette (about 500 bp). The cassette was inserted in a region devoid of predicted transcription factor binding sites of the Pa2g4 downstream gene. The inserted mutation was introduced into the gene of interest using homologous recombination in embryonic stem cells. This technology enables to express the mutated form of ErbB3 and replace the wild‐type form of the protein under the control of the ErbB3 endogenous gene promoter without altering potential regulatory elements of this gene. Generation of the nuc‐ErbB3 KI mice was performed by GenoWay on a C57/B6 background. C57/B6 WT mice were obtained from Jackson Labs. Animals were maintained according to the NIH Guide for the Care and Use of Laboratory Animals. Nuc‐ErbB3 KI congenic mice were maintained on a C57/B6 background and all in vivo experiments used age‐matched control animals. All animal protocols were approved by the Institutional Animal Care and Use Committee of the Weis Center for Research, Geisinger Clinic.
Transmission Electron Microscopy and Data Analysis
Isolated sciatic nerves were fixed by perfusion with 3% gluteraldehyde in phosphate buffer pH 7.4 at 4°C. Nerves were then postfixed in 2% osmium tetroxide in 100 mM phosphate buffer, dehydrated in ethanol and embedded in Epox812 (Fullam, Latham, NY). For ultrastructural analysis, ultrathin sections were cut, placed on copper grids, stained with lead citrate and uranyl magnesium acetate, and examined in Jeol (Peabody, MA) JEM‐1200EX transmission/scanning electron microscope. To calculate g‐ratios, we used EM photomicrographs from sciatic nerves of three WT and three nuc‐ErbB3 KI mice for each time interval and measured ∼300 axons from each group. The results are presented as average ± s.d. For binning analysis of myelination, axons were divided into groups based on their diameters and the myelin thickness was calculated by subtracting the axonal diameter from the total diameter of the myelinated fiber. Results are presented as average ± s.d.
Three‐Dimensional‐EM Reconstruction Using Serial Blockface SEM and Serial Ion Ablation SEM
Serial blockface scanning EM imaging was performed at Renovo Neural Inc (Cleveland, OH) using standard protocols. Briefly, tissue preparation and imaging were performed as previously described (Ohno et al., 2011) using the staining methods described by Deerinck et al. (West et al., 2010). Sample blocks of approx 0.5 × 0.5 mm were examined in a Zeiss Sigma VP scanning EM fitted with a Gatan 3View in‐chamber ultramicrotome and using a Gatan low‐kV backscattered electron detector. At low magnification, regions of interest with longitudinally oriented axons were identified and image stacks covering 60 µm wide and 150–200 µm in length generated. Images were collected at 2.25 kV, x3.5 K with pixel size of 6 nm (x,y) using the 30 μm aperture in high current mode. Two stacks, of 250 and 500 slices were generated with 100 nm slice thicknesses. Myelin was segmented based on density thresholding, downsampled to 100 nm voxels and 3D surface visualizations generated using the ImageJ 3D Viewer plugin. Single slice views of axons were generated automatically from multiple slices using an algorithm that assembled multiple ROIs obtained along a user‐defined path that followed the axon centerline through the stack (x, y, z coordinates). Video sequences were generated in ImageJ and converted to standard codecs using winFF software.
Immunocytochemistry
Cells were fixed with 4% formaldehyde and permeabilized with 0.1% Triton‐X100 or methanol. The following primary antibodies were used: MBP (Covance), neurofilament (Encor Biotechnology), Oct‐6 (Santa Cruz), ErbB3, and Ki67 (Cell Signaling), S100 (Dako) and p75NGFR (Abcam). The cultures were then washed and incubated with the respective AlexaFluor‐488 or rhodamine‐conjugated secondary antibody (Jackson Immunochemicals). Cultures were co‐stained with 4′6‐diamidino‐2‐phenylindole or Hoescht dye. Images were captured with a Zeiss Axiovert inverted microscope equipped with Apotome and a high‐resolution digital camera.
5‐Bromodeoxyuridine and FLICA Labeling
To detect cell proliferation, cells were incubated with 5‐bromodeoxyuridine (BrdU) for 90 min, fixed with 70% ethanol in 50 mM glycine, pH 2, and BrdU immunostaining was done according to manufacturer's directions (Roche). To detect caspase activity, cells were incubated with the FLICA substrate for 1 h and stained with propidium iodide according to manufacturer's directions (Immunochemistry Technologies, LLC).
Protein Immunoprecipitation, Western Blotting, and Sandwich ELISA
For immunoprecipitation studies, cultures were lysed in 1× RIPA buffer containing phosphatase and protease inhibitors (Sigma). Lysates were cleared by centrifugation and precleared for nonspecific binding by rotating with beads alone before primary antibodies against ErbB3 (Santa Cruz) were added to the cleared lysates and rotated overnight at 4°C. TrueBlot antirabbit IgG beads (eBioscience) were added to the lysates and immunoprecipitates were washed, solubilized with 2× SDS buffer, boiled, loaded on 4–12% SDS–PAGE gels (NuPAGE) and processed as described below. Cytosolic extracts from rat Schwann cells expressing V5‐tagged nuc‐ErbB3 constructs (described below) and anti‐V5 magnetic beads (MBL) were used to coimmunoprecipitate interacting cytosolic partners. Precipitates were isolated and run 1 cm into a 4‐12% Novex gel (Life Technologies), stained with Commassie blue, and bands were analyzed by nanoLcMS/MS mass spectrometry (Alphalyse).
For total cell lysates, cultures were lysed in 1× SDS lysis buffer containing phosphatase and protease inihibitors (Sigma). Sciatic nerves were frozen in liquid nitrogen and homogenized in 1× SDS lysis buffer. Samples were separated on 4–12% SDS–PAGE gels (NuPAGE), transferred onto nitrocellulose and immunoblotted. The following primary antibodies were used: ErbB3, P‐ErbB3, Brn2, total histone H3 (Cell Signaling), Egr2, MBP (Covance), P‐Tyr 4G10 (Millipore), Oct‐6 (Origene), Actin, Pa2G4 (Sigma), neuregulin‐CRD (AB_10672412), and neuregulin‐HBD (AB_2154679) (UC Davis/NIH NeuroMab Facility), pan‐neurofilament (Covance), FLAG (a generous gift from the Wang lab), and V5 (Life Technologies). Secondary antibodies were horseradish peroxidase‐conjugated goat antimouse and goat antirabbit (Cell Signaling) and donkey antigoat (Jackson Immunolabs). Semi‐quantitative analysis was performed with FluorChem SP analytical software. Statistical significance was determined by two‐tailed Student's t‐test.
Electrophoretic Mobility Shift Assay (EMSA)
EMSA was completed according to the User Manual guidelines from Panomics with minor modifications. Complimentary transcription factor (TF) oligonucleotides for the nuc‐ErbB3 binding motif (5′‐TNNGNNNNCNNCCAGAGNCNNN‐3′) and scrambled oligonucleotides (5′‐CNNTNNNNANNAGGCCANANNN‐3′) were synthesized, labeled with biotin and annealed. Cold oligonucleotides for the nuc‐ErbB3 binding motif were synthesized and annealed. To perform EMSA, samples were prepared with 0.3 μg nuc‐ErbB3 recombinant protein or 1 μg of nuclear extract, 1 μg Poly d(I‐C), 4× binding buffer (10 mM HEPES pH 7.9, 100 mM KCl, 4 mM DTT, 0.5% Triton X‐100, and 2.5% Glycerol), nuclease‐free water and biotin‐labeled TF probe (20 fmol). A negative control sample was prepared without recombinant protein. To perform the cold assay, the sample was incubated at room temperature for 15 min with 7‐fold excess of the cold TF probe (1.4 μg) prior to the addition of the biotin‐labeled TF probe. For the scrambled assay, the biotin‐labeled TF probe was replaced with the scrambled probe (200 ng). The binding reaction with the TF probe was performed at room temperature for 30 min. For the supershift assay, 2 μg anti‐V5 antibody was added to the reaction and incubated an additional 15 min at room temperature. Protein‐DNA complexes were then separated through gel electrophoresis on a 6% nondenaturing polyacrylamide gel using 0.5× tris‐borate‐EDTA buffer (TBE) at 4°C. Transfer was performed on a Pall Biodyne B nylon membrane using an electroblotting device with chilled 0.5× TBE then fixed using by UV crosslinking. The membrane was washed, blocked, incubated with Strep‐Alkaline Phosphate followed by CDP‐Star according the protocol for a nonisotopic detection of biotinylated DNA probes (Ambion Bright Star Biodetect Kit) or Strep‐HRP conjugate followed by ECL (Chemiluminescent Nucleic Acid Detection Module). The membrane was then exposed to Classic Blue Autoradiography film BX for 5 min and developed.
Generation of Recombinant nuc‐ErbB3
Expression and purification of the Fc‐nuc‐ErbB3 fusion protein was performed by GeneScript USA Inc. Nuc‐ErbB3 protein was transiently expressed as Fc‐nuc‐ErbB3 fusion with an artificial signal peptide in 293 cells. Fc‐nuc‐ErbB3 fusion protein was purified by one‐step affinity purification from culture supernatant using a HiTrap rProtein A HP column. Samples collected during cell culture and after purification were analyzed by SDS‐PAGE and Western blot.
Generation of nucErbB3 Constructs, Transfections, and Nuclear Extracts Analysis
The coding region of the nuc‐ErbB3 was amplified from the GeneScript construct (described above) and subcloned into the pcDNA 3.1/V5‐His expression vector (Life Technologies). The nucErbB3 clone containing the mutated NLS sequence was then produced using the QuikChange Site Directed Mutagenesis kit (Agilent) according to manufacturer's protocol. Nucleofection (Amaxa) of rat Schwann cells were conducted using manufacturer's protocol and nuclear extracts were isolated using the Nuclear Complex Co‐IP kit (Active Motif). The total nuclear fraction was used for EMSA (described above).
Determination of Transcription Factor Binding Motifs
Specific DNA binding motifs were predicted for nuc‐ErbB3 using data from our previously performed genome‐wide chromatin immunoprecipitation (ChIP)–chip analysis on nuc‐ErbB3 using the rat ChIP 385K promoter set from NimbleGen. For each of the 589 genes for which nuc‐ErbB3 binding was detected the nearest instance of a ChIP‐chip peak to the transcription start site was located, resulting in 410 unique genomic locations, with peak scores ranging from 0.62 to 4.14. For the analysis 75 peaks were selected with scores equal or greater than 2.0. The average length of these 75 peak sequences was 370 bp. Peak sequences were downloaded from UCSC genome browser (Kent et al., 2002) (build rn4 (Kent et al., 2002)), with low complexity regions masked using the built‐in repeat masking algorithm in the UCSC genome browser and Tandem Repeat Finder (Benson, 1999). De novo motif discovery was performed using the command‐line version of WebMotifs (Romer et al., 2007). The set of 75 peak sequences was used in three different motif discovery programs: AlignACE (Hughes et al., 2000), Weeder (Pavesi et al., 2004) and MDscan (Liu et al., 2002). Over‐represented motifs were identified by these programs in the peak sequences relative to a background distribution, which later was compiled from the entire 15,912 promoter sequences used in the ChIP array, and masked using the same protocol as for peak sequences. To objectively evaluate the significance of motifs found by the three programs, statistical Z‐scores were calculated for each predicted motif against randomized controls. For this purpose 50 control sets of 75 sequences, each 370 bp long, were randomly selected from the genome‐wide background sequences, and fed into the three motif discovery programs.
Searching the nuc‐ErbB3 Motif Against Genome‐Wide Promoter Regions
Variants of the nuc‐ErbB3 DNA binding motif (Fig. 6A) were searched against peak sequences from ChIP‐chip analysis to identify specific genes that are potentially regulated by nuc‐ErbB3. To compile the peak sequences, the set of 589 genes previously reported that have nuc‐ErbB3‐interacting promoters (Adilakshmi et al., 2011) were clustered at 90% sequence identity to a nonredundant set of 478 genes using CD‐HIT (Fu et al., 2012). All peaks were collected for each gene from the ChIP‐chip data, resulting in 486 peak sequences. Motifs search was performed using regular expressions against both forward and reverse complement strands of DNA.
Figure 6.
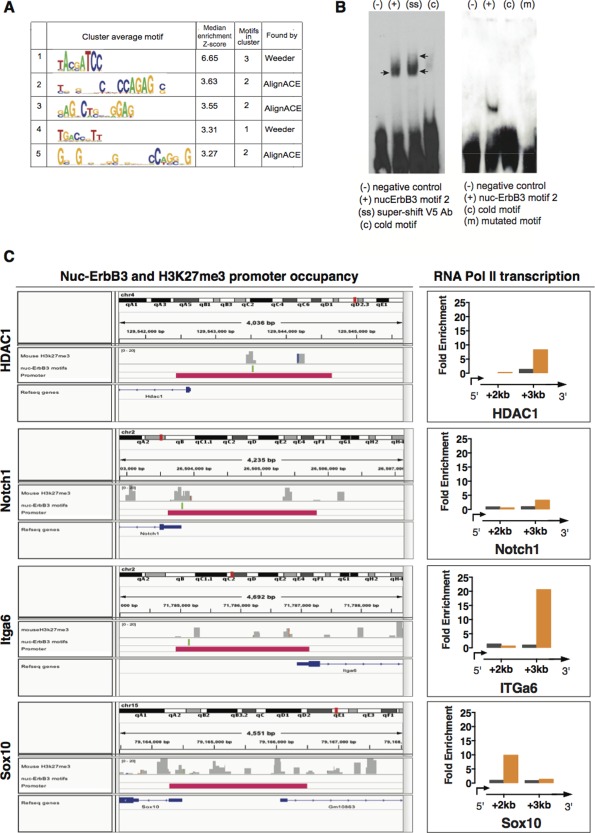
(A) Nuc‐ErbB3 binding motifs were located from ChIP‐chip peak data using the program WebMotifs, which combines three de novo motif discovery programs: AlignACE, Weeder and MDscan. 5 groups of motifs were identified. (B) EMSA with nuclear extracts from rat Schwann cells transfected with nuc‐ErbB3‐V5 show specific binding to motif 2. Addition of anti‐V5 antibody results in a supershift (arrows). Cold (nonbiotinylated) oligo competes and prevents binding of motif 2 to nuc‐ErbB3 (left panel). EMSA with recombinant nuc‐ErbB3 shows specific binding to motif 2. Cold (nonbiotinylated) oligo competes and prevents binding of motif 2 to nuc‐ErbB3, while mutation of the core sequence of motif 2 abrogates binding to recombinant nuc‐ErbB3 (right panel). (C) Computational analysis of nuc‐ErbB3 and H3K27me3 promoter occupancy from in vivo sciatic nerve ChIP data. Co‐occurrence of nuc‐ErbB3 binding motifs and chromatin regions associated with H3K27me3 were discovered in the promoters of HDAC1, Notch1 and Itga6 while Sox10 promoter contains only H3K27me3 binding sites. Sonicated chromatin isolated from P30 sciatic nerves of WT or nuc‐ErbB3 KI mice was immunoprecipitated with antibodies against RNA Polymerase II (Millipore). Fold enrichment over input is presented for HDAC, Notch1, Itga6, and Sox10. Localization of RNA Pol II greater than 1 kb downstream of the TSS identifies active transcription and increased levels at the nuc‐ErbB3 KI samples indicate elevated transcriptional rate following loss of nuc‐ErbB3 from the nucleus. Genes with nuc‐ErbB3 and H3K27me3 co‐occupancy on their promoters (e.g., HDAC1, Itga6, and Notch1) are transcriptionally repressed since they exhibit increased rate of transcription (derepression) in the absence of nuc‐ErbB3 in KI mice. The same effect on transcription was observed for Sox10 verifying that the association of nuc‐ErbB3 with Sox10 promoter, which occurs in the context of the repressive histone mark H3K27me3 without direct DNA binding, results in transcriptional repression. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Real‐Time qPCR
Total RNA from WT and nuc‐ErbB3 KI sciatic nerves was extracted using Trizol and treated with RNase‐free DNAse I. cDNA was synthesized using the High Capacity RNA to cDNA kit (Applied Biosystems). Quantitative PCR for neuregulin isoforms was performed using PowerSYBR Green Mastermix (Life Technologies) and the following primers, adapted for mouse sequence from published rat Nrg1 isoform primers (Liu et al., 2011):
Nrg1Type3‐specific primers: forward‐GGACCCCTGAGGTGAGAACA, reverse‐ CAGTCGTGGATGTAGATGTGGTT)
Nrg1‐EGF domain: forward‐ACCAGCCATCTCATAAAGTGTGC, reverse‐ TTGAGGGGTTTGACAGGTCC
GAPDH primers (Qiagen) were used for normalization and samples were quantified using the comparative C T method and fold change was calculated with WT samples (n = 4).
H3K27 ELISA and HMT Activity Assay
Total levels of trimethyl Lys27 on histone H3 was quantified using a H3K27me3 ELISA (Active Motif). Briefly, acid extracted histones from WT and KI mouse Schwann cells were prepared using manufacturer's directions and protein concentrations were calculated using a Bradford Assay (Biorad). Equal amounts of histone extracts were added to the plate and the sandwich ELISA assay was conducted following the manufacturer's protocol.
Histone methyltransferase activity was quantified in WT and KI nuclear extracts from mouse Schwann cells. Nuclear extracts were isolated using Abcam's Nuclear Extraction Kit and protein concentrations were calculated using a Bradford Assay (Biorad). Equal amounts of nuclear extract protein were analyzed using Histone Methyltransferase H3K27 Activity Quantification Assay Kit (Abcam) and following the manufacturer's protocol. Results were normalized to WT activity levels and HMT activity is represented as a percentage of WT activity.
Chromatin immunoprecipitation‐Quantitative PCR Array
For ChIP experiments, mouse Schwann cells were cross‐linked in 1% formaldehyde, isolated, and sonicated on ice in ChIP lysis buffer (Epitect ChIP OneDay kit) for 10 s ON and 90 s OFF for 5 cycles using a Misonix Sonicator 3000. Chromatin was isolated from sciatic nerves from WT or nuc‐ErbB3 KI mice as previously described (Adilakshmi et al., 2012). Immunoprecipitations were carried out on precleared lysates using Epitect ChIP OneDay kit (Qiagen) according to manufacturers directions. The following antibodies were used: ErbB3, total histone H3 ChIP grade antibody (Cell Signaling), H3K27me3 ChIP grade antibody, RNA Polymerase II ChIP grade antibody (Millipore). Purified DNA was mixed with RT2 SYBR green PCR Mastermix (Qiagen) and Epitect ChIP qPCR arrays were performed in a StepOnePlus real‐time cycler (ABI) according to manufacturers directions. Threshold fluorescence was manually defined across plates using the Positive PCR Control (PPC). IP C t values were normalized to input C t values and background (normalized IgG control C t) was subtracted for the normalized IP (ΔΔCt[IP]) value, and the linear conversion (2(−ΔΔCt[IP])) was calculated to determine Fold Enrichment, according to manufacturers directions. Fold difference for each gene was calculated as Fold Enrichment KI/Fold Enrichment WT. Data analysis and quality control assessment was conducted for all PCR arrays using Excel templates from Qiagen.
For Western blotting following chromatin immunoprecipitation, the IP sample was reversed cross‐linked, sample loading buffer with DTT was added and samples were run on a 4–12% gel and immunoblotted as described above.
Statistical Analysis
Statistical analysis for protein expression was conducted with unpaired Student's two‐tailed t tests with significance declared at P < 0.05, as described in the text. Paired Student's two‐tailed t test was used for statistical analysis of myelin internode numbers in explant cultures and g‐ratios in EM sections of sciatic nerves. Binning analysis of myelin thickness was calculated with two‐way Anova and the results are presented as average ± s.d.
Results
Generation and Characterization of the nuc‐ErbB3 Knockin Mouse
To reveal the role of nuc‐ErbB3 in vivo we generated a nuc‐ErbB3 knockin mouse (GenOway). The targeting strategy included the introduction of a floxed neomycin selection cassette that generated a point mutation in exon 27 at the nuclear localization signal (NLS) sequence of nuc‐ErbB3. The NLS sequence within exon 27 is SRSRSRS (Adilakshmi et al., 2011). We mutated the NLS to SRSRSAS, which rendered the NLS non functional. The cassette was introduced in intron 26, 50 bp downstream of exon 26 in a region devoid of transcription factor binding sites to minimize potential deregulation of the Pa2g4 downstream gene promoter (Fig. 1A). The mutant strain was generated by homologous recombination in embryonic stem (ES) cells derived from a 129Sv strain. ES cells were then injected into C57BL/6J blastocysts. Chimeric mice were bred to Cre deleter mice to remove the Neomycin selection cassette and thus create the Neo‐excised heterozygote mutated KI line (Fig. 1A).
Figure 1.
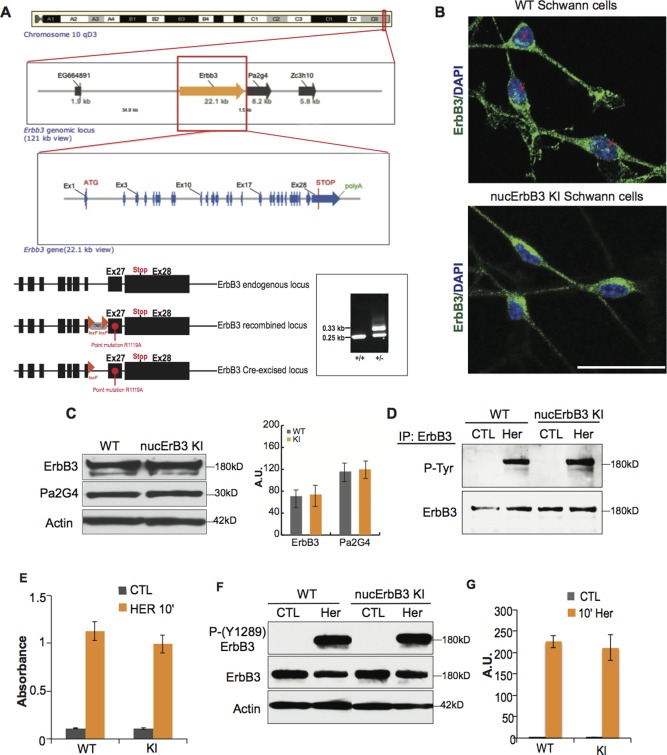
Nuc‐ErbB3 KI mice show lack of nuc‐ErbB3 localization in the nucleus without affecting the ErbB3 receptor signaling. (A) Genomic organization of the ErbB3 locus on chromosome 10 showing the presence of Pa2g4 1.5 kb downstream of ErbB3. A floxed Neomycin selection cassette carrying the R1119A mutation at the NLS motif of exon 27 was recombined in the ErbB3 endogenous locus to generate targeted nuc‐ErbB3 chimeric mice. Crossing the nuc‐ErbB3 chimeras with Cre‐expressing mice results in deletion of the neomycin cassette and generation of the nuc‐ErbB3 KI mice. (B) Nuc‐ErbB3 KI Schwann cells show complete lack of nuc‐ErbB3 localization in the nucleus, as compared with nuclear localization in WT Schwann cells (arrows indicate positive nuclear stain). The images correspond to maximum projection view from Confocal z‐stacks at 20× magnification (ErbB3: green, DAPI: blue). (C) Equal expression of ErbB3 and Pa2G4 in sciatic nerve lysates of nuc‐ErbB3 KI and WT mice. Actin was used to demonstrate equal protein loading. Densitometric quantification from three independent experiments presented as average ± s.d. (D) ErbB3 immunoprecipitation from WT and nuc‐ErbB3 KI Schwann cell lysates with or without the addition of 20 ng/mL beta1‐heregulin followed by total phospho‐tyrosine Western blotting shows equal phosphorylation of ErbB3 in WT and KI Schwann cells. Total ErbB3 was used to demonstrate equal precipitation. (E) Nuc‐ErbB3 KI and WT Schwann cells exhibit equal pan phospho‐tyrosine activation of ErbB3 receptor after addition of 20 ng/mL beta1‐heregulin for 10min using an ErbB3 pan phospho‐Tyr ELISA (Cell Signaling Technologies). Results are expressed as average ± s.d. from three independent experiments. (F) Incubation of WT and nuc‐ErbB3 KI Schwann cells with 10 ng/mL beta1‐heregulin for 10 min results in equal phosphorylation of Y1289 of the ErbB3 receptor. (G) Densitometric analysis of three independent experiments shows equal phosphorylation of Y1280 of ErbB3 in WT and KI Schwann cells. Results are expressed as average ± s.d. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Our targeting strategy results in complete ablation of nuc‐ErbB3 expression from the nucleus of Schwann cells (Fig. 1B). An Ensembl database search of the ErbB3 locus revealed that Pa2G4 gene is located 1.5 kb downstream of ErbB3 and the Pa2G4 promoter is within the 3′‐end of ErbB3 including exon 27. We verified that our targeting strategy does not affect the expression of Pa2G4 or the expression of the full‐length ErbB3 receptor in nuc‐ErbB3 KI Schwann cells (Fig. 1C). To examine if the neuregulin‐induced activation of the ErbB3 receptor is affected in the nuc‐ErbB3 KI Schwann cells, we immunoprecipitated full‐length ErbB3 from WT and KI Schwann cells after activation with beta1‐heregulin and examined the phosphorylation state of ErbB3. Full‐length ErbB3 is equally activated by beta1‐heregulin in KI Schwann cells as compared with WT Schwann cells (Fig. 1D). To confirm that nuc‐ErbB3 retention in the cytosol does not contribute to an increase in total phospho‐Tyr ErbB3 activity, a phospho‐ErbB3 sandwich ELISA revealed equal phospho‐Tyr activation of ErbB3 in nuc‐ErbB3 KI mouse Schwann cells and WT Schwan cells after 10 min beta1‐heregulin treatment (Fig. 1E). Western blotting of the same lysates with an antibody against Y1289 of ErbB3, which resides within a YXXM motif and participates in signaling to PI3K (Kim et al., 1994) verified the equal activation of the full‐length ErbB3 receptor in WT and KI mouse Schwann cells (Fig. 1F,G). Nuc‐ErbB3 KI and WT Schwann cells exhibit equal activation of Erk1/2 and Akt signaling pathways as well as equal levels of proliferation and apoptosis (Fig. 1A,B, Supporting Information). KI67‐positive proliferating Schwann cells in 3 d sciatic nerves were less than 0.1% in both WT and nuc‐ErbB3 KI nerves. Finally, nuc‐ErbB3 is not expressed in axons and neuregulin‐1 isoforms do not change in expression in nuc‐ErbB3 KI axons (Fig. 1C–E, Supporting Information).
Nuc‐ErbB3 KI Mice Exhibit Peripheral Hypermyelination
Neuregulin I Type III‐induced activation of ErbB2/3 is an instructive signal for peripheral myelination and regulates the thickness of peripheral myelin (Garratt et al., 2000b; Taveggia et al., 2005). We analyzed myelination in nuc‐ErbB3 KI mice and demonstrate that ablation of nuc‐ErbB3 from the nucleus of Schwann cells results in hypermyelination and increased axonal diameter (Fig. 2A,B) even though KI mice exhibit equal ErbB3 receptor activation as compared with WT mice (Fig. 1C–G). Myelin sheaths were thicker at P15 and P30 (Fig. 2A) and in some instances severe hypermyelination at P30 results in compressed axoplasm of some axons (Fig. 2A arrowheads). The g‐ratio at these time intervals is significantly lower in nuc‐ErbB3 KI mice (Fig. 2B,C) (P < 0.05) and the diameter of myelinated axons is also significantly greater at P15 and P30 (Fig. 2B,C). Increased myelin thickness was particularly pronounced in axons with diameters between 1 and 3.5 microns at P15 and P30 (Fig. 2D). We conclude from these data that nuc‐ErbB3 regulates the initial extent of peripheral myelination and loss of nuc‐ErbB3 results in acute pronounced hypermyelination in the presence of normal neuregulin‐ErbB3 signaling.
Hypermyelination in nuc‐ErbB3 KI Mice Results in Aberrant Myelin Structures
To verify that the myelin pathology was consistent along the length of the axons in sciatic nerves of P30 nuc‐ErbB3 KI mice and does not represent focal abnormalities, we performed 3D‐EM reconstruction using Serial Blockface SEM and Serial Ion Ablation SEM (Renovo Neural). Images were collected at 2.25 kV, x3.5 K with pixel size of 6 nm (x, y) using the 30 μm aperture in high current mode. Two stacks, of 250 and 500 slices were generated with 100 nm slice thicknesses. 3D reconstruction of nuc‐ErbB3 KI sciatic nerve axons identified numerous irregular myelin foldings (tomaculi). The aberrant myelin infoldings and outfoldings were elaborate at the node (Fig.3A–H), while most were continuous along the myelin internode and not restricted to the nodal region (Fig. 3I,J, arrows). Sciatic nerves from WT litermates show that the nodal axoplasm is open and continuous and the myelin outfoldings are not as pronounced as in nuc‐ErbB3 nerves. 3D view of the compact myelin shows lack of aberrant myelin outfoldings along the axon (Fig. 3K–N).
Figure 3.
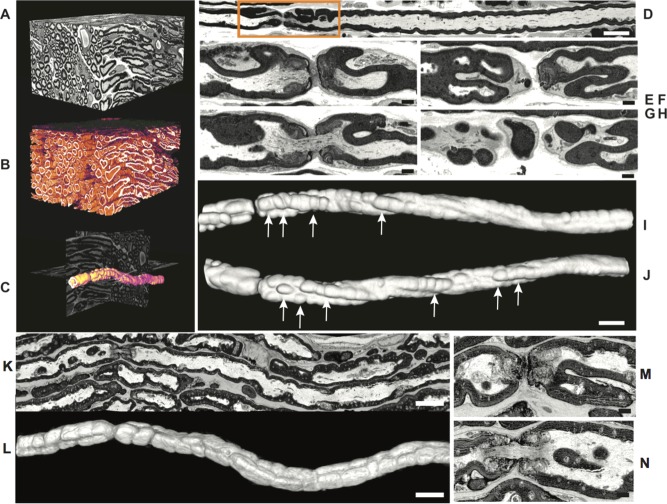
Serial blockface scanning EM analysis of nuc‐ErbB3 KI sciatic nerves shows aberrant myelination. (A–J) Three‐dimensional reconstruction of image stack (120 × 80 × 60 μm). Intensity‐based segmentation of myelinated fibers identified many with irregular myelin foldings (B, false color scale). Myelin foldings at nodes are elaborate as illustrated for a representative axon from this set (C, shown in orange on orthogonal slices). A single slice view (D) indicates myelin outfoldings are prominent around the node. In single slices (E–H, from box in D), myelin folds appear isolated, but in 3D view of the compact myelin (I, same orientation as D and J rotated through 180°) aberrant myelin foldings were continuous along the internode and not restricted to the nodal region (arrows). (K–N) Single slice view of control (WT) sciatic nerves shows that the nodal axoplasm is open and continuous and the perinodal crenulation and myelin outpocketing is not as pronounced as in nuc‐ErbB3 nerves. Three‐dimensional view of the compact myelin along the axon shows lack of aberrant myelin foldings along the axon (L). Scale bars A–D, K, 5 μm; E–J, L–N 1 μm). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Loss of nuc‐ErbB3 Leads to Increased Initiation of Myelination
To understand the role of nuc‐ErbB3 in the regulation of peripheral myelination we utilized the in vitro dorsal root ganglion (DRG) explant culture system. In culture the number of Schwann cells that initiate myelination is low compared with the total number of Schwann cells that populate an explant culture. DRG explants from nuc‐ErbB3 KI mice exhibited a significant increase in myelin internodes formed by 12 days after the initiation of myelination by ascorbate addition (Fig. 4A,B). The cultures maintained a higher number of myelin internodes through 4 weeks of ascorbate treatment (Fig. 4B). After 4 weeks, nuc‐ErbB3 KI explant cultures have significantly increased Egr2, Brn2, and MBP protein levels as compared with control, but Oct‐6 expression levels remained equal to control cultures (Fig. 4C,D). qPCR using RNA from 4 week nuc‐ErbB3 KI and WT explant cultures shows significant increase of the steady–state levels of Egr2 mRNA, while Brn2 and Oct6 show no significant differences (Fig. 2, Supporting Information). These results suggest that loss of nuc‐ErbB3 creates a more permissive environment for the initiation of myelination in DRG cultures.
Figure 4.
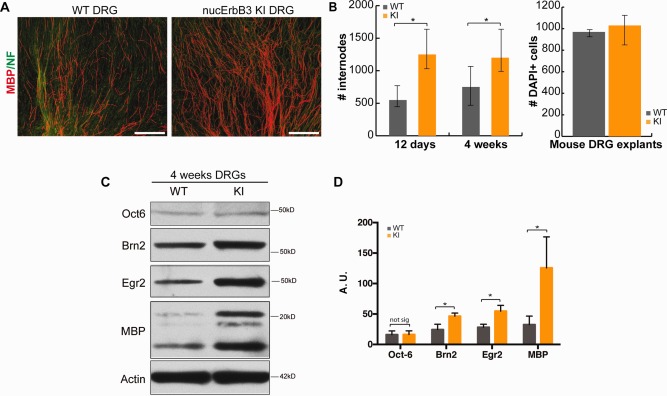
Loss of nuc‐ErbB3 leads to increased initiation of myelination in mouse DRG explants and increased Brn2, Egr2 and MBP expression. (A) Myelinated mouse DRG explants from WT or nuc‐ErbB3 KI mice were immunostained for MBP and Neurofilament. Representative photomicrographs of myelination at 12 days are shown. (B) The numbers of myelin internodes were significantly increased in nuc‐ErbB3 KI explants at 12 days (*P = 0.025, n = 4, paired Student's t‐test) and 4wk (*P = 0.007, n = 6, paired Student's t‐test), while the number of DAPI positive cells in both sets of cultures was not significantly different. (C and D) Protein lysates from 4 week explants were analyzed for Oct6, Brn2, Egr2, and MBP with actin as a loading control. Expression was significantly upregulated in nuc‐ErbB3 KI cultures (*P = 0.0003 (Brn2), *P = 0.01 (Egr2), *P = 0.004 (MBP), n = 8, Student's t‐test, error bars ± s.d.). Oct6 protein expression did not show significant change. [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Nuc‐ErbB3 Regulates Histone H3K27 Methyltransferase Activity and Total Levels of H3K27me3
Proper levels of myelination in the PNS are achieved and maintained through terminal differentiation of Schwann cells. This is accompanied by widespread transcriptional repression of genes that negatively affect the myelination program (Jessen and Mirsky, 2008). Since the hypermyelination and increased initiation of myelination seen in the KI nerves and Schwann cell cultures suggest loss of transcriptional repression, we hypothesized that nuc‐ErbB3 may regulate epigenetic marks to induce transcriptional repression. From the known histone marks, H3K27me3 is associated with transcriptional repression in a cell type‐specific manner (Boyer et al., 2006; Lee et al., 2006; Mikkelsen et al., 2007).
Analysis of total HeK27me3 levels in cultured WT and KI Schwann cells reveals that KI Schwann cells have significantly less H3K27me3 (Fig. 5A). Transient overexpression of nuc‐ErbB3 in Schwann cells significantly increases levels of H3K27me3 after 24 h (Fig. 5B). Transient knockdown of nuc‐ErbB3 through siRNA induced a slight but not significant decrease in H3K27me3 levels (Fig. 5B). To determine if nuc‐ErbB3 regulates HMT activity in order to affect the levels of the repressive histone mark H3K27me3, we performed an HMT activity assay in WT and nuc‐ErbB3 KI Schwann cells. We demonstrate that nuc‐ErbB3 KI Schwann cells have significantly reduced HMT activity as compared with WT Schwann cells (Fig. 5C).
Figure 5.
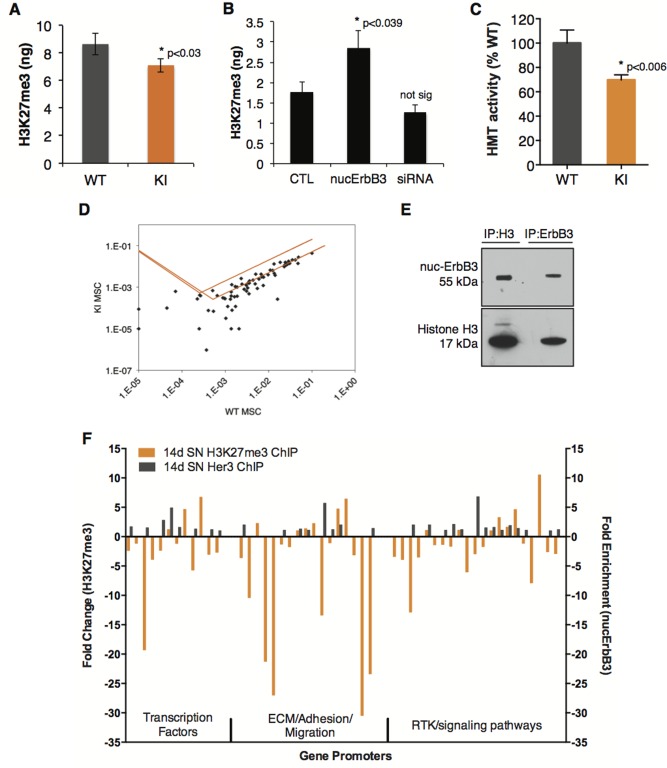
Nuc‐ErbB3 regulates total levels of H3K27me3 and HMT activity and associates with specific set of promoters in conjunction with H3K27me3. (A) Equal protein amounts of acid‐extracted histones from WT and nuc‐ErbB3 mouse Schwann cells were analyzed with a tri‐methyl H3K27 ELISA kit (Active Motif). The total amount of H3K27me3 in nuc‐ErbB3 KI mouse Schwann cells is significantly decreased compared with WT cells (n = 4, P = 0.03). (B) Rat Schwann cells were transfected with a nuc‐ErbB3 expression vector (24 h) or nuc‐ErbB3‐specific siRNA (48 h). Total amount of H3K27me3 in the cells following transfection was analyzed as in (A). Overexpression of nuc‐ErbB3 significantly increased tri‐methylation of H3K27 (n = 3, P = 0.039) while knockdown of nuc‐ErbB3 expression did not significantly change H3K27me3 levels. (C) Nuclear extracts from WT and nucErbB3 KI Schwann cells were prepared and histone methyltransferase (HMT) activity was quantified. HMT activity in nuc‐ErbB3 KI cells was normalized to WT cells (% WT activity) and was significantly decreased as compared with WT (n = 3, P = 0.006). (D) Sonicated chromatin fractions from WT and nuc‐ErbB3 KI Schwann cells were immunoprecipitated with H3K27me3‐specific antibody (Millipore) and the purified DNA was analyzed with Epitect ChIP real time PCR arrays (Qiagen) to quantify H3K27me3 marked gene promoters. Fold enrichment (normalized to input and control IgG fractions) was calculated and fold difference nuc‐ErbB3 KI versus WT is shown as a scatter plot, with the orange lines indicating the two‐fold threshold. (E) Sonicated chromatin fractions from P14 WT and KI sciatic nerves were immunoprecipitated with antibodies against total histone H3 or ErbB3 (Cell Signaling). Nuc‐ErbB3 and total histone H3 are coimmunoprecipitated from sciatic nerve chromatin. (F) H3K27me3 and nuc‐ErbB3 were immunoprecipitated from P14 WT sciatic nerve chromatin and purified DNA was analyzed with Epitect ChIP real time PCR arrays (Qiagen) as in (D). Input normalized IP values (adjusted to control IgG background) are presented as Fold Enrichment (2− ΔΔCt). Nuc‐ErbB3 enrichment at gene promoters was compared with H3K27me3 association with the same promoters. For the nuc‐ErbB3 ChIP, all genes analyzed that had >One‐fold increase are displayed on the graph (gray bars). For the H3K27me3 ChIP, all genes with >two‐fold change in nuc‐ErbB3 KI sciatic nerves and/or had nuc‐ErbB3 binding are displayed (orange bars). [Color figure can be viewed in the online issue, which is available at wileyonlinelibrary.com.]
Nuc‐ErbB3 Associates with Specific Gene Promoters in Conjunction with H3K27me3
We previously demonstrated that nuc‐ErB3 associates with promoter regions of genes expressed in cultured rat Schwann cells through genome‐wide ChIP‐chip analysis (Adilakshmi et al., 2011). To determine if the loss of nuc‐ErbB3 affects the association of Schwann cell promoters with repressive histone marks, we performed ChIP‐qPCR analysis of WT and nuc‐ErbB3 KI mouse Schwann cells using an antibody against tri‐methylated histone H3 K27 (H3K27me3). We used an 84‐gene promoter array (Qiagen) to compare promoter association with H3K27me3 in KI and WT mouse Schwann cells (MSCs). Loss of nuc‐ErbB3 results in 31 of 84 genes exhibiting a >2‐fold decrease in association with H3K27me3 as compared with WT MSCs (Fig. 5D), which indicates that nuc‐ErbB3 mediates the interaction of specific gene promoters with the repressive histone mark H3K27me3.
Next we investigated whether nuc‐ErbB3 interacts directly with the promoters with reduced H3K27me3 in nuc‐ErbB3 KI sciatic nerves. We confirmed that the ErbB3 antibody (Cell Signaling) can immunoprecipitate nuc‐ErbB3 associated with chromatin by performing an in vivo ChIP on P14 sciatic nerves from WT mice and analyzed immunoprecipitated proteins by Western blotting. Chromatin immunoprecipitation with a total histone H3 antibody or ErbB3 antibody pulls down both proteins, respectively (Fig. 5E). We then performed ChIP‐qPCR on chromatin samples from P14 WT mouse sciatic nerves using the ErbB3 antibody (Cell Signaling) and H3K27me3 using the same 84 promoter array as above (Qiagen). Promoters enriched over input (>1) after immunoprecipitation with ErbB3 antibody were compared with the promoters that had >2‐fold loss of repressive histone H3K27me3 in nuc‐ErbB3 KI sciatic nerves (Fig. 5F). Nuc‐ErbB3 was enriched at 1/3 of gene promoters represented in the array, and 45% of these promoters also showed a >2‐fold decrease in association with H3K27me3 in nuc‐ErbB3 KI MSCs (Fig. 5F, Table S1, Supporting Information). This indicates that nuc‐ErbB3 forms epigenetic repressive complexes on specific gene promoters in conjunction with the repressive histone mark H3K27me3.
Prediction and Identification of nuc‐ErbB3 DNA Binding Motif
To understand how nuc‐ErbB3 binds to specific gene promoters we analyzed our previously published genome‐wide ChIP‐chip peak data (Adilakshmi et al., 2011) using the program WebMotifs (Romer et al., 2007), which combines three de novo motif discovery programs: AlignACE (Hughes et al., 2000), Weeder (Pavesi et al., 2004) and MDscan (Liu et al., 2002) in order to discover specific nuc‐ErbB3 DNA binding motifs. Of the 323 motifs predicted [MDScan (10), AlignACE (301) and Weeder (12)], 313 were considered insignificant (Z‐score <3) and the remaining 10 motifs were clustered into five groups (Fig. 6A).
Next we synthesized 5 biotinylated oligonucleotides corresponding to the five groups of motifs to perform EMSAs with recombinant nuc‐ErbB3 protein or nuc‐ErbB3‐V5 tagged transfected Schwann cell nuclear extracts. Electrophoretic mobility shift assay (EMSA) shows that nuc‐ErbB3 binds specifically to motif 2, which shows a modest supershift with the addition of anti‐V5 antibody (Fig. 6B). Similarly, the protein was transiently expressed in Hek 293 cells as an Fc‐Nuc‐ErbB3 fusion protein and purified (GeneScript). EMSA shows that recombinant nuc‐ErbB3 binds specifically to motif 2, while a cold (not biotinylated) motif 2‐oligonucleotide or a mutated motif 2 show no binding (Fig. 6B). Mutation of the core sequence of motif 2 inhibits the binding of nuc‐ErbB3, which suggests that this is the specific DNA binding motif of nuc‐ErbB3 (Fig. 6 B).
Co‐Occurrence of nucErbB3 Binding Motifs and Chromatin Regions Associated with K27‐Trimethylated Histone H3
A position‐specific weight matrix (PWM), representing the nucleotide preferences of the binding motif, was generated based on the WebLogo. We used two different PWMs, one specifying the 17 positions described in the WebLogo, and a shorter matrix with the rightmost 11 positions, which captures a region with stronger nucleotide preferences. The promoter sequences of 31 genes selected for their association with nucErbB3 were collected from the mm10 assembly of the mouse genome, based on coordinates extracted from the refGene.txt file from UCSC (http://hgdownload.cse.ucsc.edu/goldenPath/mm10/database/). The promoter region was defined as that from −2000 to +200 nucleotides with respect to the annotated transcription start site of a transcript. Motif locations were identified on each promoter using the two PWMs and the program MOODS (Korhonen et al., 2009). In total, 31 motifs were found in 20 different promoters.
To examine the association of nucErbB3 motifs with genomic regions containing histone H3‐K27 trimethylation, we queried the NCBI GEO Datasets database (http://www.ncbi.nlm.nih.gov/) for mouse H3K27 trimethylation in peripheral nerve, and downloaded the SRX815105 data of the GSM1567362 entry. The extracted fastq file was aligned to the mm10 assembly of the mouse genome using Bowtie (Langmead et al., 2009), and the aligned reads, together with the predicted motifs, were visualized with the Integrative Genomics Viewer (http://www.broadinstitute.org/igv/). We demonstrate that in the case of HDAC1, Itga6, and Notch1 nuc‐ErbB3 exhibits promoter co‐occupancy with H3K27me3 (Fig. 6C). In total, 60% of the gene promoters identified to interact with nuc‐ErbB3 from the in vivo ChIP analysis show co‐occupancy with H3K27me3. Even though Sox10 promoter associates with nuc‐ErbB3 in our in vivo ChIP, it does not contain nuc‐ErbB3 DNA binding sites. However, H3K27me3 shows multiple binding sites on Sox10 promoter (Fig. 6C). It is possible that nuc‐ErbB3 associates with Sox10 promoter in the context of an epigenetic repressive complex with H3K27me3 since it regulates both total levels of H3K27me3 and HMT activity (Fig. 5A–C).
Nuc‐ErbB3 Regulates the RNA Pol II Rate of Transcription of Genes in Association with the Repressive Histone Mark H3K27me3
To discover if the presence of nuc‐ErbB3 on gene promoters in association with H3K27me3 results in functional regulation of gene transcription we performed in vivo RNA Pol II ChIP using sciatic nerve chromatin from P14 nuc‐ErbB3 KI and WT mice. RNAPol II ChIP consists of the detection of RNA polymerase II within the coding region of the genes, which allows the quantification of actual transcriptional rates of target genes (Sandoval et al., 2004). Genes with nuc‐ErbB3 and H3K27me3 co‐occupancy on their promoters (e.g., HDAC1, Itga6, and Notch1) are transcriptionally repressed since they exhibit increased rate of transcription (derepression) in the absence of nuc‐ErbB3 in KI mice (Fig. 6C). The same effect on transcription was observed for Sox10 verifying that the association of nuc‐ErbB3 with Sox10 promoter, which occurs in the context of the repressive histone mark H3K27me3 without direct DNA binding, results in transcriptional repression (Fig. 6C).
Discussion
The onset and maintenance of myelination in the PNS depends on a tightly modulated transcriptional program that involves Sox10, Egr2, Oct‐6, Brn2, NF‐κB (Jaegle et al., 2003, 1996; Kuhlbrodt et al., 1998; Nickols et al., 2003; Topilko et al., 1994) and the regulatory role of HDAC1 and HDAC2 (Jacob et al., 2011). In addition, Neuregulin‐1 (NRG1) signaling through the ErbB3 receptor tyrosine kinase has an instructive role for Schwann cell myelination (Taveggia et al., 2005) and a crucial role for the regulation of myelin thickness (Michailov et al., 2004). Recently, the role of histone H3K27 methylation was explored through the generation of a Schwann cell specific knock‐out of Eed, which is a subunit of polycomb repressive complex 2 (PRC2) that catalyzes the methylation of H3K27 (Ma et al., 2015). On a separate study, the role of the histone methyltransferase (HMT), enhancer of zeste 2 (Ezh2) has been explored (Heinen et al., 2012). Suppression of Ezh2 expression in cultured rat Schwann cells resulted in reduced length of Schwann cell processes and incomplete Schwann cell maturation. These studies showcase the significance of epigenetic repressive complexes on peripheral myelination. However, how levels of H3K27me3 and HMT activity are regulated in Schwann cells and means to modulate HMT activity have not been discovered so far.
Here we developed a KI mouse to study the role of nuc‐ErbB3 in the PNS. Nuc‐ErbB3 KI mice lack localization of nuc‐ErbB3 in the nucleus due to a point mutation at the nuclear localization signal. The classical ErbB3/NRG1 signaling axis is intact in nuc‐ErbB3 KI mice and the ErbB3 receptor is phosphorylated at equal levels with WT mice. Nuc‐ErbB3 KI mice exhibit peripheral hypermyelination and aberrant myelin structures (myelin outfoldings and infoldings). The hypermyelination phenotype is accompanied by increased expression of Egr2, Brn2, and MBP in KI Schwann cell DRG cocultures, which are key mediators of the transition from promyelinating Schwann cells to myelinating Schwann cells (Garratt et al., 2000b; Jaegle et al., 2003; Topilko et al., 1994). Epigenetic mechanisms that have recently been shown to regulate Schwann cell differentiation include the histone modifying enzymes histone deacetylases 1 and 2 (HDAC1/2) and the BAF complexes (BRG1 associated factors), chromatin remodeling ATPases that reposition nucleosomes. HDAC2 synergizes with Sox10 to activate transcription (Chen et al., 2011; Jacob et al., 2011) and BRG1 complexes with NF‐kB (Limpert et al., 2013) or Sox10 (Weider et al., 2012) to drive the transcription of Oct‐6 and Egr2. Loss of Brg1 or HDAC2 inhibits Schwann cell myelination. In the case of nuc‐ErbB3 KI Schwann cell DRG cocultures, the increase in Egr2, Brn2, and MBP expression suggests either a targeted derepression of gene transcription or de novo increase of transcription or translation of these genes.
To clarify the molecular mechanism of nuc‐ErbB3 function we discovered that nuc‐ErbB3 regulates the total levels of the repressive histone mark H3K27me3. Nuc‐ErbB3 exerts this epigenetic function through direct regulation of HMT activity. In vivo ChIP using sciatic nerve chromatin revealed that nuc‐ErbB3 associates with H3K27me3‐bound promoters and loss of nuc‐ErbB3 in KI mice results in reduced association of the repressive histone mark H3K27me3 with specific promoters. We have previously shown that nuc‐ErbB3 associates with chromatin in a genome‐wide manner (Adilakshmi et al., 2011) and here we discovered that nuc‐ErbB3 binds DNA either directly on a specific DNA motif, or in conjunction with H3K27me3. Both cases result in significant decrease of the RNA Pol II rate of transcription of the corresponding genes suggesting a role of nuc‐ErbB3 as transcriptional repressor during peripheral myelination. The small number of genes identified by our analysis suggests the possible presence of a redundant mechanism of repression or the existence of other repressive histone marks regulating gene transcription. It is also possible that the function of histone deacetylases within repressive complexes, such as the NuRD complex (Chen et al., 2011; Hung et al., 2012; Jacob et al., 2011) mediate transcriptional repression of genes during peripheral myelination.
In a recent study Ma et al., investigated the function of H3K27me3 in peripheral nerve by creating a Schwann cell specific deletion of Eed, an integral component of the PRC2. Eed knockout mice did not have any early postnatal defect in myelin structure but exhibited abnormal gene expression and progressively they developed peripheral hypermyelination. In the case of nuc‐ErbB3 KI mice we detected early postnatal hypermyelination, abnormal gene expression and reduced HMT activity and H3K27me3 expression. The difference in the timing of hypermyelination suggests that nuc‐ErbB3 is not an integral component of the epigenetic repressive complexes but an important regulator of epigenetic repressive marks on gene promoters. Since epigenetic control of gene expression is vital for cellular homeostasis, we believe that loss of nuc‐ErbB3 is gradually compensated by other regulatory mechanisms.
The in vivo nuc‐ErbB3 phenotype is different from the inhibition of myelination produced by acute in vitro siRNA knockdown of nuc‐ErbB3 expression in DRG cultures (Adilakshmi et al., 2011). The hypermyelination phenotype is the outcome of the chronic depletion of nuc‐ErbB3 expression from the nucleus during embryonic development and differentiation in vivo, while acute knockdown of nuc‐ErbB3 expression in adult Schwann cells results in inhibition of myelination. The apparent discrepancy is not surprising since it has been recently described that in vitro siRNA‐mediated and in vivo genetically modified phenotypes could be completely opposite (Hall et al., 2013). In addition, acute siRNA knockdown of nuc‐ErbB3 does not have a significant effect on the levels of H3K27me3 suggesting a different effect on gene transcription and eventually phenotypic expression. It is possible that the acute knockdown of nuc‐ErbB3 expression affects its affinity for direct DNA binding e.g., on the promoters of Ezrin and HMGB1 without interfering with the regulation of repressive histone marks. We are currently trying to determine the underlying differences between acute and chronic depletion of nuc‐ErbB3 from the nucleus of Schwann cells and how possible interacting proteins regulate the different outcomes.
We have shown that nuc‐ErbB3 regulates the transcription of Ezrin and HMGB1 promoters (Adilakshmi et al., 2011). A motif search of these two promoters shows that HMGB1 contains a nuc‐ErbB3 motif while Ezrin promoter does not have a nuc‐ErbB3 motif. We discovered cooccupancy of nuc‐ErbB3 and H3K27me3 on HMGB1 promoter while Ezrin promoter does not associate with nuc‐ErbB3 in the context of H3K27me3. In addition, secondary structure prediction of nuc‐ErbB3 shows that nuc‐ErbB3 is a helix‐loop‐helix transcription factor that suggests both direct (motif‐based) and indirect DNA binding and regulation. We believe that the inhibition of Ezrin promoter is the result of an indirect regulation, while HMGB1 is repressed through nuc‐ErbB3/H3K27me3 promoter co‐occupancy.
Finally, it has been shown that epigenetic repression of gene promoters occurs through specific binding of transcription factors to Ezh2 and recruitment of the PRC2 complex on targeted promoter regions (Tien et al., 2015) or through transcription factor interaction with long noncoding RNAs, which specifically determines the recruitment and function of PRC2 (Sarma et al., 2014). Both mechanisms result in the presence of the repressive histone mark H3K27me3 on gene promoters and subsequently in transcriptional repression. Since nuc‐ErbB3 regulates both the total levels of H3K27me3 and the HMT activity of PRC2 we hypothesize that the role of nuc‐ErbB3 is to specifically recruit and regulate the function of PRC2 on promoter regions during myelination.
Our novel discovery for the role of nuc‐ErbB3 explains how expression of the repressive histone mark H3K27me3 and activity of HMT are regulated in Schwann cells, which enriches the picture of epigenetic repressive mechanisms in the modulation of peripheral myelination.
Supporting information
Supporting Information
Supporting Information
Supporting Information
Supporting Information
Acknowledgment
The authors thank Renovo Neural Inc. for the 3D‐EM reconstruction service and John Shaw and Charles Schworer at Geisinger Clinic for their technical assistance with transmission electron microscopy. They would also like to thank John Zepecki for cell quantifications and all members of the Tapinos Lab for reviewing the manuscript. All animal work was performed according to institutional guidelines.
References
- Adilakshmi T, Ness‐Myers J, Madrid‐Aliste C, Fiser A, Tapinos N. 2011. A nuclear variant of ErbB3 receptor tyrosine kinase regulates ezrin distribution and Schwann cell myelination. J Neurosci 31:5106–5119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adilakshmi T, Sudol I, Tapinos N. 2012. Combinatorial action of miRNAs regulates transcriptional and post‐transcriptional gene silencing following in vivo PNS injury. PLoS One 7:e39674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arthur‐Farraj P, Wanek K, Hantke J, Davis CM, Jayakar A, Parkinson DB, Mirsky R, Jessen KR. 2011. Mouse schwann cells need both NRG1 and cyclic AMP to myelinate. Glia 59:720–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benson G. 1999. Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27:573–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Plath K, Zeitlinger J, Brambrink T, Medeiros LA, Lee TI, Levine SS, Wernig M, Tajonar A, Ray MK, Bell GW, Otte AP, Vidal M, Gifford DK, Young RA, Jaenisch R. 2006. Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441:349–353. [DOI] [PubMed] [Google Scholar]
- Britsch S, Goerich DE, Riethmacher D, Peirano RI, Rossner M, Nave KA, Birchmeier C, Wegner M. 2001. The transcription factor Sox10 is a key regulator of peripheral glial development. Genes Dev 15:66–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang H, Yoon SO, Xu X, Hottiger MO, Svaren J, Nave KA, Kim HA, Olson EN, Lu QR. 2011. HDAC‐mediated deacetylation of NF‐kappaB is critical for Schwann cell myelination. Nat Neurosci 14:437–441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Citri A, Skaria KB, Yarden Y. 2003. The deaf and the dumb: the biology of ErbB‐2 and ErbB‐3. Exp Cell Res 284:54–65. [DOI] [PubMed] [Google Scholar]
- Fu LM, Niu BF, Zhu ZW, Wu ST, Li WZ. 2012. CD‐HIT: Accelerated for clustering the next‐generation sequencing data. Bioinformatics 28:3150–3152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garratt AN, Britsch S, Birchmeier C. 2000a. Neuregulin, a factor with many functions in the life of a schwann cell. Bioessays 22:987–996. [DOI] [PubMed] [Google Scholar]
- Garratt AN, Voiculescu O, Topilko P, Charnay P, Birchmeier C. 2000b. A dual role of erbB2 in myelination and in expansion of the schwann cell precursor pool. J Cell Biol 148:1035–1046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall EA, Keighren M, Ford MJ, Davey T, Jarman AP, Smith LB, Jackson IJ, Mill P. 2013. Acute versus chronic loss of mammalian Azi1/Cep131 results in distinct ciliary phenotypes. PLoS Genet 9:e1003928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinen A, Tzekova N, Graffmann N, Torres KJ, Uhrberg M, Hartung HP, Kury P. 2012. Histone methyltransferase enhancer of zeste homolog 2 regulates Schwann cell differentiation. Glia 60:1696–1708. [DOI] [PubMed] [Google Scholar]
- Hughes JD, Estep PW, Tavazoie S, Church GM. 2000. Computational identification of cis‐regulatory elements associated with groups of functionally related genes in Saccharomyces cerevisiae. J Mol Biol 296:1205–1214. [DOI] [PubMed] [Google Scholar]
- Hung H, Kohnken R, Svaren J. 2012. The nucleosome remodeling and deacetylase chromatin remodeling (NuRD) complex is required for peripheral nerve myelination. J Neurosci 32:1517–1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacob C, Christen CN, Pereira JA, Somandin C, Baggiolini A, Lotscher P, Ozcelik M, Tricaud N, Meijer D, Yamaguchi T, Matthias P, Suter U. 2011. HDAC1 and HDAC2 control the transcriptional program of myelination and the survival of Schwann cells. Nat Neurosci 14:429–436. [DOI] [PubMed] [Google Scholar]
- Jaegle M, Ghazvini M, Mandemakers W, Piirsoo M, Driegen S, Levavasseur F, Raghoenath S, Grosveld F, Meijer D. 2003. The POU proteins Brn‐2 and Oct‐6 share important functions in Schwann cell development. Genes Dev 17:1380–1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaegle M, Mandemakers W, Broos L, Zwart R, Karis A, Visser P, Grosveld F, Meijer D. 1996. The POU factor Oct‐6 and Schwann cell differentiation. Science 273:507–510. [DOI] [PubMed] [Google Scholar]
- Jessen KR, Mirsky R. 2008. Negative regulation of myelination: relevance for development, injury, and demyelinating disease. Glia 56:1552–1565. [DOI] [PubMed] [Google Scholar]
- Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. 2002. The human genome browser at UCSC. Genome Res 12:996–1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HH, Sierke SL, Koland JG. 1994. Epidermal growth factor‐dependent association of phosphatidylinositol 3‐kinase with the erbB3 gene product. J Biol Chem 269:24747–24755. [PubMed] [Google Scholar]
- Korhonen J, Martinmaki P, Pizzi C, Rastas P, Ukkonen E. 2009. MOODS: fast search for position weight matrix matches in DNA sequences. Bioinformatics 25:3181–3182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuhlbrodt K, Herbarth B, Sock E, Hermans‐Borgmeyer I, Wegner M. 1998. Sox10, a novel transcriptional modulator in glial cells. J Neurosci 18:237–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. 2009. Ultrafast and memory‐efficient alignment of short DNA sequences to the human genome. Genome Biol 10:R25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee TI, Jenner RG, Boyer LA, Guenther MG, Levine SS, Kumar RM, Chevalier B, Johnstone SE, Cole MF, Isono K, Koseki H, Fuchikami T, Abe K, Murray HL, Zucker JP, Yuan B, Bell GW, Herbolsheimer E, Hannett NM, Sun K, Odom DT, Otte AP, Volkert TL, Bartel DP, Melton DA, Gifford DK, Jaenisch R, Young RA. 2006. Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125:301–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Limpert AS, Bai S, Narayan M, Wu J, Yoon SO, Carter BD, Lu QR. 2013. NF‐kappaB forms a complex with the chromatin remodeler BRG1 to regulate Schwann cell differentiation. J Neurosci 33:2388–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X, Bates R, Yin DM, Shen C, Wang F, Su N, Kirov SA, Luo Y, Wang JZ, Xiong WC, Mei L. 2011. Specific regulation of NRG1 isoform expression by neuronal activity. J Neurosci 31:8491–8501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu XS, Brutlag DL, Liu JS. 2002. An algorithm for finding protein‐DNA binding sites with applications to chromatin‐immunoprecipitation microarray experiments. Nat Biotechnol 20:835–839. [DOI] [PubMed] [Google Scholar]
- Ma KH, Hung HA, Srinivasan R, Xie H, Orkin SH, Svaren J. 2015. Regulation of peripheral nerve myelin maintenance by gene repression through polycomb repressive complex 2. J Neurosci 35:8640–8652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurel P, Salzer JL. 2000. Axonal regulation of Schwann cell proliferation and survival and the initial events of myelination requires PI 3‐kinase activity. J Neurosci 20:4635–4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer D, Yamaai T, Garratt A, Riethmacher‐Sonnenberg E, Kane D, Theill LE, Birchmeier C. 1997. Isoform‐specific expression and function of neuregulin. Development 124:3575–3586. [DOI] [PubMed] [Google Scholar]
- Michailov GV, Sereda MW, Brinkmann BG, Fischer TM, Haug B, Birchmeier C, Role L, Lai C, Schwab MH, Nave KA. 2004. Axonal neuregulin‐1 regulates myelin sheath thickness. Science 304:700–703. [DOI] [PubMed] [Google Scholar]
- Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, Lee W, Mendenhall E, O'Donovan A, Presser A, Russ C, Xie X, Meissner A, Wernig M, Jaenisch R, Nusbaum C, Lander ES, Bernstein BE. 2007. Genome‐wide maps of chromatin state in pluripotent and lineage‐committed cells. Nature 448:553–560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogha A, Benesh AE, Patra C, Engel FB, Schoneberg T, Liebscher I, Monk KR. 2013. Gpr126 functions in Schwann cells to control differentiation and myelination via G‐protein activation. J Neurosci 33:17976–17985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monje PV, Bartlett Bunge M, Wood PM. 2006. Cyclic AMP synergistically enhances neuregulin‐dependent ERK and Akt activation and cell cycle progression in Schwann cells. Glia 53:649–659. [DOI] [PubMed] [Google Scholar]
- Murphy P, Topilko P, Schneider‐Maunoury S, Seitanidou T, Baron‐Van Evercooren A, Charnay P. 1996. The regulation of Krox‐20 expression reveals important steps in the control of peripheral glial cell development. Development 122:2847–2857. [DOI] [PubMed] [Google Scholar]
- Ness JK, Snyder KM, Tapinos N. 2013. Lck tyrosine kinase mediates beta1‐integrin signalling to regulate Schwann cell migration and myelination. Nat Commun 4:1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickols JC, Valentine W, Kanwal S, Carter BD. 2003. Activation of the transcription factor NF‐kappaB in Schwann cells is required for peripheral myelin formation. Nat Neurosci 6:161–167. [DOI] [PubMed] [Google Scholar]
- Ogata T, Yamamoto S, Nakamura K, Tanaka S. 2006. Signaling axis in schwann cell proliferation and differentiation. Mol Neurobiol 33:51–62. [DOI] [PubMed] [Google Scholar]
- Ohno N, Kidd GJ, Mahad D, Kiryu‐Seo S, Avishai A, Komuro H, Trapp BD. 2011. Myelination and axonal electrical activity modulate the distribution and motility of mitochondria at CNS nodes of Ranvier. J Neurosci 31:7249–7258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavesi G, Mauri G, Pesole G. 2004. In silico representation and discovery of transcription factor binding sites. Briefings Bioinf 5:217–236. [DOI] [PubMed] [Google Scholar]
- Plath K, Fang J, Mlynarczyk‐Evans SK, Cao R, Worringer KA, Wang H, de la Cruz CC, Otte AP, Panning B, Zhang Y. 2003. Role of histone H3 lysine 27 methylation in X inactivation. Science 300:131–135. [DOI] [PubMed] [Google Scholar]
- Romer KA, Kayombya GR, Fraenkel E. 2007. WebMOTIFS: automated discovery, filtering and scoring of DNA sequence motifs using multiple programs and Bayesian approaches. Nucleic Acids Res 35:W217–W220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryu EJ, Wang JY, Le N, Baloh RH, Gustin JA, Schmidt RE, Milbrandt J. 2007. Misexpression of Pou3f1 results in peripheral nerve hypomyelination and axonal loss. J Neurosci 27:11552–11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandoval J, Rodriguez JL, Tur G, Serviddio G, Pereda J, Boukaba A, Sastre J, Torres L, Franco L, Lopez‐Rodas G. 2004. RNAPol‐ChIP: a novel application of chromatin immunoprecipitation to the analysis of real‐time gene transcription. Nucleic Acids Res 32:e88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarma K, Cifuentes‐Rojas C, Ergun A, Del Rosario A, Jeon Y, White F, Sadreyev R, Lee JT. 2014. ATRX directs binding of PRC2 to Xist RNA and Polycomb targets. Cell 159:869–883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Svaren J, Meijer D. 2008. The molecular machinery of myelin gene transcription in Schwann cells. Glia 56:1541–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taveggia C, Zanazzi G, Petrylak A, Yano H, Rosenbluth J, Einheber S, Xu X, Esper RM, Loeb JA, Shrager P, Chao MV, Falls DL, Role L, Salzer JL. 2005. Neuregulin‐1 type III determines the ensheathment fate of axons. Neuron 47:681–694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tien CL, Jones A, Wang H, Gerigk M, Nozell S, Chang C. 2015. Snail2/Slug cooperates with Polycomb repressive complex 2 (PRC2) to regulate neural crest development. Development 142:722–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topilko P, Schneider‐Maunoury S, Levi G, Baron‐Van Evercooren A, Chennoufi AB, Seitanidou T, Babinet C, Charnay P. 1994. Krox‐20 controls myelination in the peripheral nervous system. Nature 371:796–799. [DOI] [PubMed] [Google Scholar]
- Weider M, Kuspert M, Bischof M, Vogl MR, Hornig J, Loy K, Kosian T, Muller J, Hillgartner S, Tamm ER, Metzger D, Wegner M. 2012. Chromatin‐remodeling factor Brg1 is required for Schwann cell differentiation and myelination. Dev Cell 23:193–201. [DOI] [PubMed] [Google Scholar]
- West JB, Fu Z, Deerinck TJ, Mackey MR, Obayashi JT, Ellisman MH. 2010. Structure‐function studies of blood and air capillaries in chicken lung using 3D electron microscopy. Respir Physiol Neurobiol 170:202–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu SC, Zhang Y. 2011. Cyclin‐dependent kinase 1 (CDK1)‐mediated phosphorylation of enhancer of zeste 2 (Ezh2) regulates its stability. J Biol Chem 286:28511–28519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi J, Miyamoto Y, Chan JR, Tanoue A. 2008. ErbB2 directly activates the exchange factor Dock7 to promote Schwann cell migration. J Cell Biol 181:351–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu WM, Feltri ML, Wrabetz L, Strickland S, Chen ZL. 2005. Schwann cell‐specific ablation of laminin gamma1 causes apoptosis and prevents proliferation. J Neurosci 25:4463–4472. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supporting Information
Supporting Information
Supporting Information
Supporting Information


