Abstract
Plants, algae and cyanobacteria need to regulate photosynthetic light harvesting in response to the constantly changing light environment. Rapid adjustments are required to maintain fitness because of a tradeoff between efficient solar energy conversion and photoprotection. The xanthophyll cycle, in which the carotenoid pigment violaxanthin is reversibly converted into zeaxanthin, is ubiquitous among green algae and plants and is necessary for the regulation of light harvesting, protection from oxidative stress, and adaptation to different light conditions1,2. Violaxanthin de-epoxidase (VDE) is the key enzyme responsible for zeaxanthin synthesis from violaxanthin under excess light. Here we show that the CVDE gene from the model green alga Chlamydomonas reinhardtii encodes an atypical VDE. This protein is not homologous to the VDE found in plants and is instead related to a lycopene cyclase from photosynthetic bacteria3. Unlike the plant-type VDE that is located in the thylakoid lumen, the Chlamydomonas CVDE protein is located on the stromal side of the thylakoid membrane. Phylogenetic analysis suggests that CVDE evolved from an ancient de-epoxidase that was present in the common ancestor of green algae and plants, providing evidence of unexpected diversity in photoprotection in the green lineage.
Photosynthetic organisms are subjected to a large dynamic range of light intensities, which can vary rapidly due to canopy shading, passing clouds, or sunflecks, as well as on a daily or seasonal basis. To allow optimal photosynthesis at low light intensities and to avoid photo-oxidative damage due to the formation of reactive oxygen species (ROS) under excess light, photosynthetic organisms have evolved the ability to regulate light harvesting. Under excess light, photosynthetic light harvesting is regulated by nonphotochemical quenching (NPQ) mechanisms that are responsible for dissipating excess absorbed light as heat4–7. The major and most intensively investigated component of NPQ is called qE, which is turned on and off on the time scale of seconds to minutes. qE depends on acidification of the thylakoid lumen upon formation of high ΔpH across the thylakoid membrane in excess light8. In plants, this results in two important changes that facilitate qE: conformational changes of light-harvesting complex proteins by protonation and the activation of a lumen-localized violaxanthin (Vio) de-epoxidase (VDE) enzyme. VDE catalyzes the conversion of Vio to zeaxanthin (Zea) via the intermediate antheraxanthin (Anthera). Zea and Anthera (xanthophylls with a de-epoxidized 3-hydroxy β-ring end group) are the major xanthophyll pigments that are involved in qE in plants. Zea epoxidase converts Zea back to Vio in limiting light. Together, these light intensity-dependent interconversions are known as the xanthophyll cycle (Fig. 1a). Xanthophyll de-epoxidation occurs in almost all photosynthetic eukaryotes, although it contributes to qE and other NPQ mechanisms to different extents in different organisms9–11. In green algae and plants, Zea also plays important roles in photoprotection as an antioxidant that directly quenches singlet oxygen and triplet chlorophyll species12–14.
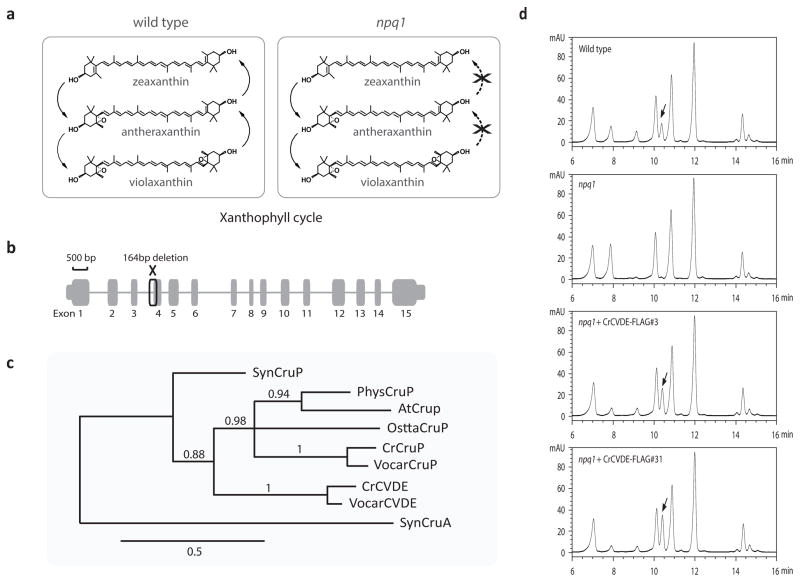
Figure 1. Molecular analysis and complementation of npq1 mutation in Chlamydomonas.
a, Xanthophyll cycle reactions. The de-epoxidation of violaxanthin to zeaxanthin via antheraxanthin is defective in the npq1 mutant. b, Schematic showing the Cre04.g221550 (CrCVDE) gene model and the 164-bp deletion in the npq1 mutant allele. c, Phylogenetic analysis of CVDE and CruP proteins. Syn, Synechococcus sp. strain PCC7002; Phys, Physcomitrella patens; At, Arabidopsis thaliana; Ostta, Ostreococcus tauri; Cr, Chlamydomonas reinhardtii; Vocar, Volvox carteri. d, HPLC phenotype of wild type, npq1, and two independent complemented lines. Arrows denote the Zea peak resulting from CVDE activity.
Mutants defective in the xanthophyll cycle and qE have been identified in the unicellular green alga Chlamydomonas reinhardtii and the model plant Arabidopsis thaliana15,16. The npq1 mutants are defective in VDE activity and are unable to convert Vio to Anthera and Zea in high light (Fig. 1a and d). Although the Arabidopsis npq1 mutant was shown to affect the VDE gene16, the molecular basis of the Chlamydomonas npq1 mutant has been mysterious, because the Chlamydomonas genome lacks an obvious ortholog of the VDE gene found in plants and other algae. Furthermore, VDE activity is not inhibited by dithiothreitol (DTT) in Chlamydomonas cells11, unlike in plants, indicating that Chlamydomonas most likely employs a novel type of VDE.
The Chlamydomonas npq1 mutation had been previously mapped to linkage group IV17. By fine mapping, we localized the npq1 mutation to a small region containing 13 gene models as candidate genes. One of these gene models (Cre04.g221550) encodes a putative FAD-dependent oxidoreductase with a predicted chloroplast transit peptide. Genomic polymerase chain reaction (PCR) analysis showed that there was a 164 bp deletion in the npq1 allele (Fig. 1b, Supplementary Fig. 1) of this gene. Introducing a Cre04.g221550 genomic clone into the npq1 mutant strain restored Zea synthesis in high light (Fig. 1d). Interestingly, some rescued lines accumulated higher levels of Zea than the wild type (Fig. 1c), which correlated with higher accumulation of the protein encoded by Cre04.g221550 (Supplementary Fig. 2). From the results of these experiments, it is clear that the Zea deficiency of npq1 is caused by the loss of Cre04.g221550 function.
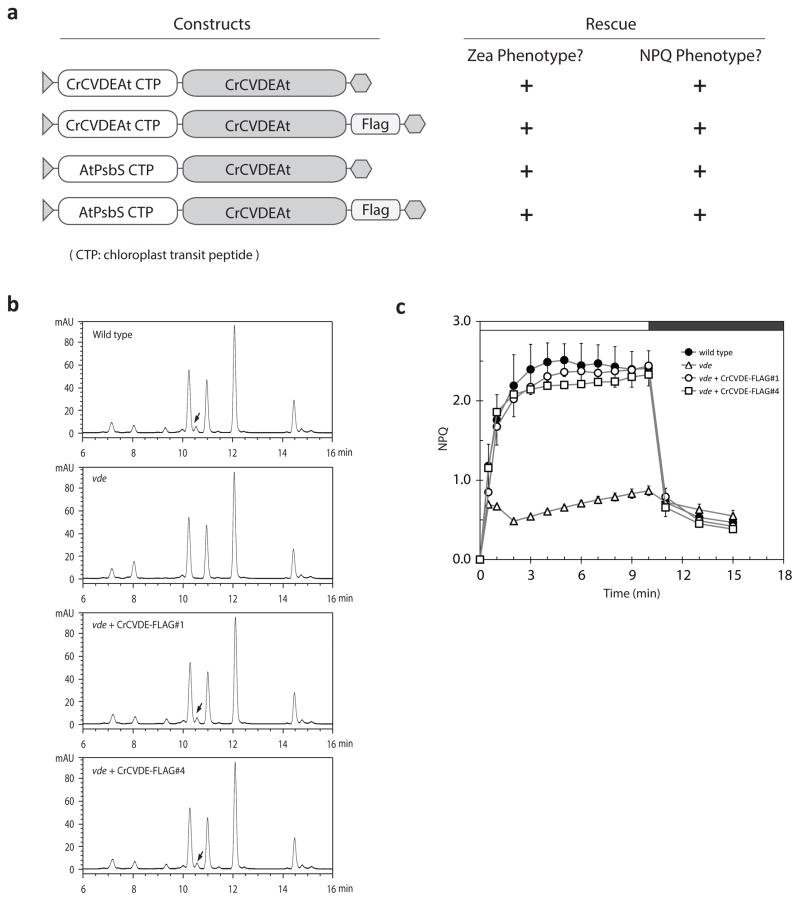
To determine if Cre04.g221550 actually encodes a protein with VDE activity, we tested if this gene could complement the Arabidopsis npq1 mutation (here called vde1), which is known to disrupt the endogenous plant-type VDE gene16. To ensure proper expression and chloroplast targeting of the Cre04.g221550 protein, we codon-optimized the Cre04.g221550 gene sequence for Arabidopsis, either with a sequence encoding its native, amino-terminal chloroplast transit peptide or the chloroplast transit peptide from the Arabidopsis PsbS protein, and with or without a carboxyl-terminal FLAG epitope tag (Fig. 2a). Arabidopsis vde1 lines expressing each of the four versions of Cre04.g221550 displayed excess-light-induced Zea synthesis and NPQ phenotypes similar to wild-type plants (Fig. 2b,c), showing that the Cre04.g221550 gene indeed encodes a functional, evolutionarily distinct VDE enzyme. Based on the presence of homologs of Cre04.g221550 in sequenced green algae of the class Chlorophyceae, we designate this gene as Chlorophycean VDE (CVDE) to distinguish it from the plant-type VDE gene.
Figure 2. Functional complementation of Arabidopsis vde1 mutant by expression of the Chlamydomonas CVDE protein.
a, Constructs used for transformation of the Arabidopsis vde1 mutant and their ability to complement the zeaxanthin accumulation and NPQ phenotypes. “+” indicates successful rescue of the phenotype. b, HPLC phenotypes of wild type, vde1 mutant, and two complemented lines. Arrows denote the Zea peak resulting from CVDE activity (or plant-type VDE activity in the wild type). c, NPQ induction and relaxation of wild type, vde1 mutant, and two independent complemented lines. White bar above graph indicates illumination with 1250 μmol photons m−2 sec−1; black bar indicates darkness (with only very weak measuring light). Error bars represent standard deviation (n=5).
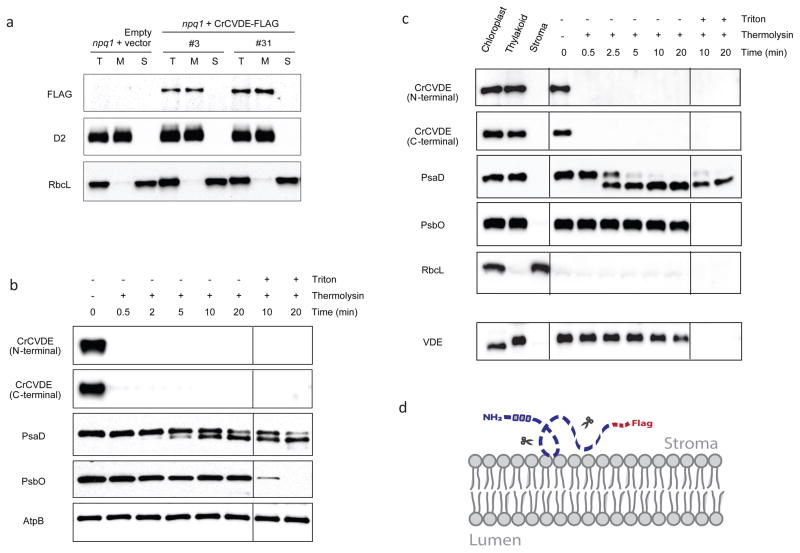
Plant-type VDE is localized in the thylakoid lumen and associates with the thylakoid membrane, where it catalyzes the de-epoxidation reaction on membrane-associated Vio. We used lines of both the Chlamydomonas npq1 mutant and the Arabidopsis vde1 mutant complemented with a carboxyl-terminal FLAG-tagged version of the Chlamydomonas CVDE (CrCVDE) protein to determine its localization. The functional carboxyl-terminal tagging demonstrated that this modification does not impair CrCVDE enzyme activity (Fig. 2b, c). Using either a polyclonal antibody raised against an N-terminal 15 amino acid peptide of mature CrCVDE or a commercial antibody raised against the FLAG epitope, we detected the CrCVDE protein at a molecular mass of 87 kDa (Fig. 3), which is the predicted size of the mature protein after cleavage of the chloroplast transit peptide. As expected, the CrCVDE protein is associated with the thylakoid membrane in both Chlamydomonas and Arabidopsis (Fig. 3a, b, c). To determine the topology of CrCVDE, we performed a limited proteolysis experiment with isolated thylakoid membranes from both Chlamydomonas and Arabidopsis complemented lines. Thermolysin treatment resulted in complete cleavage of the CrCVDE protein, even more rapidly than the cleavage of the stroma-exposed PsaD subunit of photosystem I, which was quickly digested to a thermolysin-resistant fragment (Fig. 3b, c). In contrast, the PsbO subunit of photosystem II, located in the thylakoid lumen, was completely resistant to thermolysin unless the membrane was solubilized with detergent (Fig. 3b, c). In Arabidopsis, the lumen-localized plant-type VDE protein (in the vde1 mutant complemented with a FLAG-tagged version of the Arabidopsis VDE gene) was not affected unless the membrane was solubilized with detergent (Fig. 3c). These results strongly suggest that the epitope-tagged CrCVDE protein is located on the stromal side of the thylakoid membrane when expressed in either Chlamydomonas or in Arabidopsis (Fig. 3d), which differs from the plant-type VDE that is located in the thylakoid lumen (Fig. 3c). The stroma-exposed location of CrCVDE was further supported by the presence of an FAD-binding domain in the mature CrCVDE protein (FAD is present in the stroma but not the thylakoid lumen). Salt wash assays indicated that CrCVDE is peripherally associated with the membrane and could be extracted by NaSCN (Supplementary Fig. 3).
Figure 3. Subcellular localization of CrCVDE proteins expressed in Chlamydomonas and Arabidopsis.
a, Immunoblot analysis of total cell (T), membrane (M), and soluble (S) fractions of Chlamydomonas strains. The FLAG-tagged CrCVDE protein is detected in the membrane fraction and not in the soluble fraction in two independent transformants. Subcellular markers: D2 for membrane fraction and RbcL for soluble fraction. b, Protease protection assay of isolated intact thylakoids from Chlamydomonas complemented lines. Isolated thylakoids were treated with thermolysin in the presence and absence of Triton X-100. Aliquots were removed at the specified times, samples were separated by SDS-PAGE, and immunodetection was performed with specified antibodies. Thermolysin-resistant Atpβ was used as a loading control. The FLAG-tagged CrCVDE protein was probed with both the N-terminal epitope antibody and the C-terminal FLAG antibody. Subcellular markers: PsaD for stroma-exposed membrane protein, PsbO for thylakoid lumen. c, Immunoblot analysis and protease protection assay of the FLAG-tagged CrCVDE protein expressed in the Arabidopsis vde1 mutant. Left panel: the CrCVDE protein is detected in the thylakoid membrane fraction and not in the soluble stroma fraction in Arabidopsis. Subcellular markers: PsaD for stroma-exposed membrane protein, PsbO for thylakoid lumen, and RbcL for stroma. Right panel: protease protection assay of isolated thylakoids from Arabidopsis complemented lines. RbcL was not present in the thylakoid fraction. Lower panel: the location of the plant-type VDE in the thylakoid lumen was confirmed by analysis of a transformant expressing the FLAG-tagged Arabidopsis VDE protein in the vde1 mutant. The migration of the VDE protein in the chloroplast fraction is altered by the comigration of a protein that is absent from the thylakoid fraction. d, Proposed topology of CrCVDE in both Chlamydomonas and Arabidopsis.
The in vivo substrate of VDE, Vio, is free in the membrane lipid phase rather than bound to pigment proteins2,18. Therefore, one possible explanation of functional replacement of plant-type VDE in Arabidopsis by CrCVDE is that substrate Vio molecules are accessible to enzymes on either side of the thylakoid membrane (i.e., in the thylakoid lumen or in the stroma of the chloroplast). This is likely, because addition of partially purified plant-type VDE from spinach to the stromal side of thylakoids isolated from the Arabidopsis vde1 mutant rescued the mutant phenotype in vitro19. Similar to plant-type VDE, the CVDE activity of intact Chlamydomonas cells was inhibited by the uncoupler nigericin (Supplementary Fig. 4), indicating that the activation of this stromal enzyme also requires the buildup of a large pH gradient in excess light. Plant-type VDE requires ascorbate to catalyze the de-epoxidation reaction, but at this time it is not clear what other substrates are required for CVDE activity.
The evolutionary origins of plant-type VDE and CVDE are clearly distinct. CVDE is a homolog of CruP and CruA (Fig. 1c and Supplementary Fig. 5). CruA is known to be involved in bacterial carotenoid biosynthesis as a lycopene cyclase3, whereas CruP is a paralog of CruA. We note that the proposed carotenoid cyclase20 and de-epoxidase reaction mechanisms are similar (Supplementary Fig. 6), suggesting that a de-epoxidase enzyme could evolve from a cyclase. Our demonstration that CrCVDE has VDE activity suggests that its paralog CruP, which is widely distributed in oxygenic photosynthetic organisms21, might also be a de-epoxidase. Based on the observation that cruP mutants or overexpressors of Arabidopsis accumulate more or less β-carotene-5,6-epoxide (an oxidized derivative of β-carotene), respectively, when challenged by stress21, we hypothesize that CruP is a β-carotene-5,6-epoxide de-epoxidase. CVDE and CruP homologs are present in Chlamydomonas and its multicellular relative Volvox carteri, but only CruP homologs can be found in Ostreococcus tauri, Arabidopsis thaliana, and Physcomitrella patens. Phylogenetic analysis strongly suggests that CVDE evolved by duplication of CruP in the ancestor of green algae and plants and that CVDE has been selectively lost in some clades of the Viridiplantae (Fig. 1c). This might explain some previous observations of DTT-resistant VDE activity in green algae 9–11, however the limited numbers of genomes sequenced within this clade prohibits any further speculation about the distribution or origin of CVDE-related xanthophyll cycling.
The evolutionary history of algae (and plants) is complicated by endosymbiosis and horizontal gene transfer events. We showed that a novel de-epoxidase from a green algal group is functional in a land plant, despite their evolutionary separation by over 700 million years22. Therefore it may be possible to mix and match the regulatory components of light harvesting from different clades of photosynthetic organisms to effectively tune photosynthetic efficiency and increase photosynthetic productivity.
METHODS
Genetic mapping and PCR analysis
The fine mapping of the npq1 mutation was done by scoring PCR-based markers on selected tetrad mutant progeny derived from a cross between npq1 (137c strain background) and the polymorphic wild-type strain S1D2 (CC-2090). Markers were designed based on information in Kathir et al.23 and the marker list from David Stern available at www.chlamy.org. To identify the mutation in the CVDE gene, genomic DNA PCR was performed with a series of primer pairs that collectively span the entire gene, and the PCR products were sequenced for comparison between the wild type and the npq1 mutant. The primers that resulted in different length products between wild type and npq1 were RMD345 (5′-CTTGGCGGAAGCAGAGTATGGC-3′) and RMD346 (5′-CGGCCTCCCTTCATCCCTCCCAC-3′).
Phylogenetic analysis
CVDE homologs and CruP homologs were identified by searching via BlastP and tBlastN against the sequenced proteome and genome database, respectively, with an evalue cutoff of 1e−90.The potential chloroplast transit peptides for CVDE homologs or CruP homologs were predicted by aligning respective homologs from organisms with or without chloroplasts using the Clustal Omega program (version1.2.1; http://www.ebi.ac.uk/Tools/msa/clustalo/). The predicted mature proteins were aligned using Clustal Omega and BoxShade (version 3.21; www.ch.embnet.org/software/BOX_form.html). The phylogenetic tree was constructed at Phylogeny.fr (http://phylogeny.lirmm.fr/phylo_cgi/advanced.cgi) with Gblocks for alignment curation, PhyML for construction of Phylogenetic tree, and Tree Dyn for visualization of phylogenetic tree.
Complementation of Chlamydomonas npq1 mutant
For complementation of npq1, an 11.5-kb EcoRV/NotI fragment of BAC clone 33B9 containing the CVDE gene was subcloned into the pBC1 vector24 to generate pCVDEg. For complementation of npq1 with a carboxyl-terminal FLAG-tagged version of the CVDE protein, the 1.4 kb SbfI/BglII fragment of pCVDEg containing the 3′ terminus of the CVDE gene was subcloned into the pUC19-BglII vector to generate pUC19-BglII-pCVDE. The 0.4 kb NcoI/BglII fragment of pUC19-BglII-pCVDE was then replaced by a synthesized version (Integrated DNA Technologies, Inc.), which contains a carboxyl-terminal FLAG-tag linked with the CVDE protein through two glycines to generate plasmid pUC19-BglII-pCVDE-FLAG. The 1.4 kb SbfI/BglII fragment of pUC19-BglII-pCVDE-FLAG was then ligated into pCVDEg double-digested with same enzymes to generate pCVDEg-FLAG. Both pCVDEg and pCVDEg-FLAG were separately transformed into the npq1 mutant using the glass bead method as described previously25. The positive transformants were selected on paromomycin and then screened for zeaxanthin accumulation after high light exposure by HPLC as previously described26.
Complementation of Arabidopsis vde1 mutation by Chlamydomonas CVDE
The predicted protein sequences of Chlamydomonas CVDE were retrieved from both Phytozome at http://www.phytozome.net (protein ID: Cre04.g221550.t1.2) and the Joint Genome Institute at http://genome.jgi-psf.org/Chlre4/Chlre4.home.html (protein ID: 522089). The predicted CVDE protein sequences were confirmed by comparing against each other and against the cDNA consensus obtained from UCSC/UCLA genome browser at http://genomes.mcdb.ucla.edu. The CDS of the CrCVDEAt gene was then codon-optimized and synthesized for Arabidopsis nuclear/cytoplasmic expression (GenScript). The synthetic CrCVDEAt gene was subcloned into the Gateway vector pDONR221, and a FLAG-tag was added right before the stop codon by ‘Round-the-horn’ site-directed mutagenesis (http://openwetware.org/wiki/%27Round-the-horn_site-directed_mutagenesis). Sequence encoding the Arabidopsis PSBS transit peptide (first 54 amino acids) was amplified to replace the predicted native CrCVDE transit peptide (first 56 amino acids) in versions of each construct using gene SOEing27. The CrCVDEAt gene and the FLAG-tagged CrCVDEAt gene were subcloned into the pEarleyGate100 vector28 and transformed into the Arabidopsis vde1 mutant16 using the floral dip method29. As a positive control, a vector containing a FLAG-tagged version of the Arabidopsis VDE1 gene30 was also transformed. The transformants were selected on Murashige and Skoog plates containing 20 μg/mL glufosinate ammonium, screened for NPQ capacity with the IMAGING-PAM M-series (Heinz Walz), measured for NPQ induction with an FMS2 fluorometer (Hansatech Instruments) as previously described31, and assayed for the accumulation of zeaxanthin after high light exposure by HPLC as described26.
Chlamydomonas cell fractionation
Chlamydomonas cells were grown photoheterotrophically in TAP medium32 to medium logarithmic phase (approximately 5 × 106 cells mL−1) and harvested by centrifugation at 3,000g for 5 min. Cells were resuspended in PBS buffer to a density of 2 × 108 cells mL−1 and broken by FastPrep-24 (MP Biomedicals, Solon, OH) with lysing matrix J at a speed of 4.0 m/sec for 40 sec. Total membrane and total supernatant were separated by centrifugation at 20,000g, 4°C for 10 min. Total membranes were washed three times before being resuspended with 1 × PBS buffer containing 100 μM phenylmethylsulfonyl fluoride (PMSF). Samples were then subjected to immunoblot analysis as described below.
Chlamydonas and Arabidopsis thylakoid isolation
The Chlamydomonas thylakoid were isolated by a modification of the flotation procedure described previously33. The Chlamydomonas cells were grown in 400 mL TAP under low light and harvested at mid-logarithmic growth phase. The cell pellet was resuspended in 20 mL of 25 mM HEPES (pH 7.5), 0.3 M sucrose, 10 mM CaCl2, 10 mM MgCl2 with protease inhibitors. The cells were broken by passing the resuspended cells through a chilled French pressure cell, and the homogenate was centrifuged at 18,000 rpm for 10 min. The supernatant was discarded and the pellet was gently resuspended with a paintbrush in 5 mL of 5 mM HEPES (pH 7.5), 1.8 M sucrose, 10 mM CaCl2, 10 mM MgCl2. The resuspension was carefully transferred into a clear tube for SW41 rotor and topped with 6 mL of 5 mM HEPES (pH 7.5), 0.5 M sucrose, 10 mM CaCl2, 10 mM MgCl2. The tubes were centrifuged at 38,000 rpm (SW41, 4°C) for 1 hour. The membrane layer at the interface of two solutions was carefully transferred to a 1.5 mL eppendorf tube containing 1 m: of 25 mM HEPES (pH 7.5), 0.3 M sucrose, 10 mM CaCl2, 10 mM MgCl2.
Fresh Arabidopsis rosette leaves were harvested from 4-week-old plants grown in controlled conditions of 14 h light, 22°C/10 h dark, 23°C, with a light intensity of 150 μmol photons m−2 s−1 and stored on ice. The Arabidopsis thylakoids were isolated from the leaves as previously described34.
Protease protection assay
Thylakoids were resuspended in 0.3 M sorbitol, 2.5 mM EDTA 5 mM MgCl2, 0.5% (w/v) BSA, 20 mM HEPES (pH 7.6) at 0.3 nmol chlorophyll a per mL. The reaction was started by the addition of thermolysin (EMD Millipore) at a final concentration of 20 μg mL−1 to 400 μL thylakoids preparation. The reaction was stopped by transferring 60 μL to a tube containing 6 μL of 500 mM EDTA at six different time points: 0, 0.5, 2, 5, 10, 20 min. The tubes were votexed immediately, and 66 μL of 2X sample buffer was added.
CVDE antibody generation and immunoblot analysis
The polyclonal antibody recognizing CrCVDE was generated in rabbits against an epitope located near the N-terminus of the protein sequence of CrCVDE (CLRNQKHEPEKKGPK), and the resulting crude serum was affinity purified (ProSci Inc., Poway, CA). Polyclonal antibodies against D2, PsbO, PsaD, and RbcL were obtained from Agrisera (Sweden) and FLAG antibody was from Thermo Fisher Scientific. Protein samples were solubilized with 2× solubilization buffer (500 mM Tris-HCl (pH 6.8), 7% SDS, 20% glycerol (v/v), 2 M urea, 10% β-mercaptoethanol (v/v)) by pipetting up and down several times before incubation at room temperature for 30 min. For immunoblot analysis of CVDE, protein samples were separated with NuPAGE Novex 3–8% Tris-Acetate mini gels (Life Technologies, Carlsbad, CA). For immunoblot analysis of all other proteins, protein samples were separated with Novex 10–20% Tris-Glycine mini gels (Life Technologies, Carlsbad, CA). A total of 5 × 105 cells was loaded per lane for Chlamydomonas samples, and a total of 1.5 μg chlorophyll was loaded per lane for Arabidopsis samples. Proteins were then transferred to nitrocellulose membranes, blocked with 5% nonfat dry milk, and blotted with specific polyclonal antibodies. The signals were detected by Supersignal West Femto Chemiluminescent substrate detection system (Thermo Scientific).
Chlamydomonas cell fractionation, Chlamydomonas and Arabidopsis thylakoid preparations, protease protection assays, and western experiments were successfully repeated three times.
Polypeptide extraction from thylakoid
Freshly isolated thylakoids were resuspended at 0.5 mg chlorophyll/ml in thylakoid resuspension buffer (0.3M sorbitol, 2.5 mM EDTA, 5 mM MgCl2, 0.5% (wt/vol) BSA, 20 mM HEPES (pH 7.6)) containing 2 M NaBr, or 0.1 M Na2CO3, or 2 M NaSCN, or no additive. After incubation on ice for 30 min, the membrane and the supernatant fraction were separated by centrifugation at 20,000g, 4°C for 10 min. The membrane fractions were washed three times before being resuspended with 1 × PBS buffer containing 1mM PMSF. The supernatants were precipitated in 80% acetone and centrifuged at 20,000g, 4°C for 10 min to collect pellets. The pellets were then resuspended with 1 × PBS buffer containing 1 mM PMSF. The membrane and supernatant fraction were subsequently subjected to immunoblot analysis.
Supplementary Material
Acknowledgments
We thank José García-Cerdán and Robbie Calderon for helpful discussion of Chlamydomonas subcellular localization. This work was supported by the U.S. Department of Energy, Office of Science, Basic Energy Sciences, Chemical Sciences, Geosciences, and Biosciences Division under field work proposal 449B. K.K.N. is an investigator of the Howard Hughes Medical Institute and the Gordon and Betty Moore Foundation (through Grant GBMF3070).
Footnotes
Author contributions
Z.L., G.P., R.M.D., Y.B., W.A., S.Y.Y., and L.L. performed research; Z.L., G.P., R.M.D., and K.K.N. designed research; Z.L., G.P., and K.K.N. analysed data and wrote the paper; all authors discussed the results and commented on the manuscript.
Competing interests
The authors declare no competing financial interests.
References
- 1.Demmig-Adams B. Carotenoids and photoprotection in plants: A role for the xanthophyll zeaxanthin. Biochim Biophys Acta. 1990;1020:1–24. [Google Scholar]
- 2.Jahns P, Latowski D, Strzalka K. Mechanism and regulation of the violaxanthin cycle: The role of antenna proteins and membrane lipids. Biochim Biophys Acta. 2009;1787:3–14. doi: 10.1016/j.bbabio.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 3.Maresca JA, Graham JE, Wu M, Eisen JA, Bryant DA. Identification of a fourth family of lycopene cyclases in photosynthetic bacteria. Proc Natl Acad Sci USA. 2007;104:11784–11789. doi: 10.1073/pnas.0702984104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Niyogi KK. PHOTOPROTECTION REVISITED: Genetic and Molecular Approaches. Ann Rev Plant Physiol Plant Mol Biol. 1999;50:333–359. doi: 10.1146/annurev.arplant.50.1.333. [DOI] [PubMed] [Google Scholar]
- 5.Müller P, Li XP, Niyogi KK. Non-photochemical quenching. A response to excess light energy. Plant Physiol. 2001;125:1558–1566. doi: 10.1104/pp.125.4.1558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ruban AV, Johnson MP, Duffy CDP. The photoprotective molecular switch in the photosystem II antenna. Biochim Biophys Acta. 2012;1817:167–181. doi: 10.1016/j.bbabio.2011.04.007. [DOI] [PubMed] [Google Scholar]
- 7.Niyogi KK, Truong TB. Evolution of flexible non-photochemical quenching mechanisms that regulate light harvesting in oxygenic photosynthesis. Curr Op Plant Biol. 2013;16:307–314. doi: 10.1016/j.pbi.2013.03.011. [DOI] [PubMed] [Google Scholar]
- 8.Briantais JM, Vernotte C, Picaud M, Krause GH. A quantitative study of the slow decline of chlorophyll a fluorescence in isolated chloroplasts. Biochim Biophys Acta. 1979;548:128–138. doi: 10.1016/0005-2728(79)90193-2. [DOI] [PubMed] [Google Scholar]
- 9.Casper-Lindley C, Björkman O. Fluorescence quenching in four unicellular algae with different light-harvesting and xanthophyll-cycle pigments. Photosynth Res. 1998;56:277–289. [Google Scholar]
- 10.Lunch CK, et al. The xanthophyll cycle and NPQ in diverse desert and aquatic green algae. Photosynth Res. 2013;115:139–151. doi: 10.1007/s11120-013-9846-x. [DOI] [PubMed] [Google Scholar]
- 11.Quaas T, et al. Non-photochemical quenching and xanthophyll cycle activities in six green algal species suggest mechanistic differences in the process of excess energy dissipation. J Plant Physiol. 2015;172:92–103. doi: 10.1016/j.jplph.2014.07.023. [DOI] [PubMed] [Google Scholar]
- 12.Baroli I, Do AD, Yamane T, Niyogi KK. Zeaxanthin accumulation in the absence of a functional xanthophyll cycle protects Chlamydomonas reinhardtii from photooxidative stress. Plant Cell. 2003;15:992–1008. doi: 10.1105/tpc.010405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baroli I, Niyogi KK. Molecular genetics of xanthophyll-dependent photoprotection in green algae and plants. Phil Trans R Soc Lond B: Biol Sci. 2000;355:1385–1394. doi: 10.1098/rstb.2000.0700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Havaux M, Niyogi KK. The violaxanthin cycle protects plants from photooxidative damage by more than one mechanism. Proc Natl Acad Sci USA. 1999;96:8762–8767. doi: 10.1073/pnas.96.15.8762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niyogi KK, Björkman O, Grossman AR. Chlamydomonas xanthophyll cycle mutants identified by video imaging of chlorophyll fluorescence quenching. Plant Cell. 1997;9:1369–1380. doi: 10.1105/tpc.9.8.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Niyogi KK, Grossman AR, Björkman O. Arabidopsis mutants define a central role for the xanthophyll cycle in the regulation of photosynthetic energy conversion. Plant Cell. 1998;10:1121–1134. doi: 10.1105/tpc.10.7.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Anwaruzzaman M, et al. Genomic analysis of mutants affecting xanthophyll biosynthesis and regulation of photosynthetic light harvesting in Chlamydomonas reinhardtii. Photosynth Res. 2004;82:265–276. doi: 10.1007/s11120-004-2439-y. [DOI] [PubMed] [Google Scholar]
- 18.Yamamoto HY. In: Photoprotection, Photoinhibition, Gene Regulation, and Environment Vol. 21 Advances in Photosynthesis and Respiration. Demmig-Adams Barbara, Adams William W, III, Mattoo Autar K., editors. Ch 1. Springer; Netherlands: 2006. pp. 1–10. [Google Scholar]
- 19.Macko S, Wehner A, Jahns P. Comparison of violaxanthin de-epoxidation from the stroma and lumen sides of isolated thylakoid membranes from Arabidopsis: implications for the mechanism of de-epoxidation. Planta. 2002;216:309–314. doi: 10.1007/s00425-002-0848-8. [DOI] [PubMed] [Google Scholar]
- 20.Britton G. Later reactions of carotenoid biosynthesis. Pure Appl Chem. 1976;47:223–236. [Google Scholar]
- 21.Bradbury LMT, et al. Lycopene cyclase paralog CruP protects against reactive oxygen species in oxygenic photosynthetic organisms. Proc Natl Acad Sci USA. 2012;109:E1888–E1897. doi: 10.1073/pnas.1206002109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Leliaert F, et al. Phylogeny and molecular evolution of the green algae. Crit Rev Plant Sci. 2012;31:1–46. [Google Scholar]
- 23.Kathir P, et al. Molecular Map of the Chlamydomonas reinhardtii nuclear genome. Eukaryotic Cell. 2003;2:362–379. doi: 10.1128/EC.2.2.362-379.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Dent RM, et al. Large-scale insertional mutagenesis of Chlamydomonas supports phylogenomic functional prediction of photosynthetic genes and analysis of classical acetate-requiring mutants. Plant J. 2015;82:337–351. doi: 10.1111/tpj.12806. [DOI] [PubMed] [Google Scholar]
- 25.Dent RM, Haglund CM, Chin BL, Kobayashi MC, Niyogi KK. Functional genomics of eukaryotic photosynthesis using insertional mutagenesis of Chlamydomonas reinhardtii. Plant Physiol. 2005;137:545–556. doi: 10.1104/pp.104.055244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Müller-Moulé P, Conklin PL, Niyogi KK. Ascorbate deficiency can limit violaxanthin de-epoxidase activity in vivo. Plant Physiol. 2002;128:970–977. doi: 10.1104/pp.010924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Horton RM, Cai ZL, Ho SN, Pease LR. Gene splicing by overlap extension: tailor-made genes using the polymerase chain reaction. Biotechniques. 1990;8:528–535. [PubMed] [Google Scholar]
- 28.Earley KW, et al. Gateway-compatible vectors for plant functional genomics and proteomics. Plant J. 2006;45:616–629. doi: 10.1111/j.1365-313X.2005.02617.x. [DOI] [PubMed] [Google Scholar]
- 29.Zhang X, Henriques R, Lin SS, Niu QW, Chua NH. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nature Protocols. 2006;1:641–646. doi: 10.1038/nprot.2006.97. [DOI] [PubMed] [Google Scholar]
- 30.Leonelli L, Erickson E, Lyska D, Niyogi KK. Transient expression in Nicotiana benthamiana for rapid functional analysis of genes involved in non-photochemical quenching and carotenoid biosynthesis. Plant J. 2016 doi: 10.1111/tpj.13268. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brooks M, Niyogi K. In: Chloroplast Research in Arabidopsis Vol. 775 Methods in Molecular Biology. Paul Jarvis R, editor. Ch 16. Humana Press; 2011. pp. 299–310. [DOI] [PubMed] [Google Scholar]
- 32.Harris EH. The Chlamydomonas sourcebook. A comprehensive guide to biology and laboratory use. Academic Press; 1989. [DOI] [PubMed] [Google Scholar]
- 33.Chua NH, Bennoun P. Thylakoid membrane polypeptides of Chlamydomonas reinhardtii: wild-type and mutant strains deficient in photosystem II reaction center. Proc Natl Acad Sci USA. 1975;72:2175–2179. doi: 10.1073/pnas.72.6.2175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Brooks MD, Sylak-Glassman EJ, Fleming GR, Niyogi KK. A thioredoxin-like/β-propeller protein maintains the efficiency of light harvesting in Arabidopsis. Proc Natl Acad Sci USA. 2013;110:E2733–E2740. doi: 10.1073/pnas.1305443110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.