Abstract
Development of myopia is associated with large-scale changes in ocular tissue gene expression. Although differential expression of coding genes underlying development of myopia has been a subject of intense investigation, the role of non-coding genes such as microRNAs in the development of myopia is largely unknown. In this study, we explored myopia-associated miRNA expression profiles in the retina and sclera of C57Bl/6J mice with experimentally induced myopia using microarray technology. We found a total of 53 differentially expressed miRNAs in the retina and no differences in miRNA expression in the sclera of C57BL/6J mice after 10 days of visual form deprivation, which induced -6.93 ± 2.44 D (p < 0.000001, n = 12) of myopia. We also identified their putative mRNA targets among mRNAs found to be differentially expressed in myopic retina and potential signaling pathways involved in the development of form-deprivation myopia using miRNA-mRNA interaction network analysis. Analysis of myopia-associated signaling pathways revealed that myopic response to visual form deprivation in the retina is regulated by a small number of highly integrated signaling pathways. Our findings highlighted that changes in microRNA expression are involved in the regulation of refractive eye development and predicted how they may be involved in the development of myopia by regulating retinal gene expression.
Introduction
Myopia is the most common vision disorder and a leading cause of visual impairment worldwide [1]. The prevalence of myopia has increased from 25% to 44% of the adult population in the United States in the last 30 years [2], and reached more than 80% of young adults in many parts of East Asia [3, 4]. In addition to its direct impact on visual acuity and quality of life, myopia is a major risk factor for potentially blinding ocular disorders such as cataract, glaucoma, retinal detachment, and myopic maculopathy, and represents one of the leading causes of blindness [5–7].
Postnatal refractive eye development is a tightly coordinated process, which is regulated by visual input [8]. Visual experience modulates ocular growth during early postnatal period and drives it towards sharp vision (i.e., perfect match between the eye’s optical power and its axial length) in a process called emmetropization [9]. During emmetropization, optical defocus evokes a signaling cascade that originates in the retina, propagates through other ocular tissues (i.e., retinal pigment epithelium and choroid), and results in scleral wall remodeling with increased eye growth. This signaling is associated with large-scale changes in gene expression in all ocular tissues, which was extensively studied at the mRNA level in several animal models of myopia [10–17]. Although these studies established that modulation of gene expression plays an important role in refractive eye development, non-coding transcriptome changes underlying refractive eye development have been largely unexplored.
MicroRNAs (miRNAs) are small non-coding RNAs that direct post-transcriptional regulation of gene expression by either facilitating degradation of their target mRNAs or suppressing mRNA translation [18–20]. MiRNAs often serve as nodes in signaling networks and modulate many cell activities, including cell proliferation, cell differentiation, metabolism, and synaptic function [21–24]. The potential influence of miRNAs on various biological processes is immense, as at least 50% of all coding genes are thought to be regulated by 1193 annotated miRNAs in mice and 1881 miRNAs in humans [22, 23, 25]. Insights into the full range of biologic functions of miRNAs are recent, and their involvement in disease has generated significant interest due to strong potential for therapeutic development [24, 26, 27].
MiRNA transcriptomes have been profiled in several ocular tissues [28] and miRNAs have been found to be expressed in the cornea, lens, retina, retinal pigment epithelium (RPE), and sclera of both embryonic and adult eyes [29–36]. A growing number of studies have shown that miRNAs play key roles in regulating both eye development and eye diseases [32, 34, 37–55]. For example, miR-96, miR-183, miR-1, and miR-133 have been implicated in retinitis pigmentosa [49], while miR-31, miR-150, and miR-184 have been associated with choroidal neovascularization [50] and diabetic retinopathy [51]. Mutations in miR-184 cause EDICT syndrome, familial keratoconus with cataract, and sporadic keratoconus [52, 53]. Mutations in the binding site of miR-328 within 3’-UTR of a myopia-causing gene Pax6 were also shown to be associated with high myopia in a Chinese cohort [54, 55], suggesting that miRNAs play an important role in refractive eye development as well.
In the current study, we performed large-scale miRNA expression profiling in a mouse model of form-deprivation myopia and discovered extensive changes in miRNA expression in the myopic retina. The identified differentially expressed miRNAs were used to perform analysis of the miRNA-mRNA interaction networks in the myopic retina using a database of mRNAs differentially expressed in the myopic eye. This analysis revealed multiple mRNA targets for differentially expressed miRNAs and putative signaling pathways involved in the development of myopia.
Materials and Methods
Animals
C57BL/6J mice were obtained from the Jackson Laboratory (Bar Harbor, ME) and were maintained as an in-house breeding colony. C57BL/6J mice were recently shown not to have Rd8 mutation causing retinal degeneration in C57BL/6N mice [56]; however, C57BL/6J mice are known to have a relatively high incidence of microphthalmia, which affects from 4.4% to 10% of animals [57, 58]. Therefore, animals were screened for the presence of microphthalmia and other ophthalmic abnormalities such as corneal opacities and anterior polar cataracts often associated with this condition [59]. Each animal was first examined visually, which is often sufficient to identify animals with microphthalmia or cataract. If no anomalies are identified after visual examination, eyes were examined under the dissecting microscope and using handmade slit lamp. This was followed by the examination of the pupil image with the photorefractor. Animals found to have microphthalmia, corneal opacities or cataract were removed from the study (~10% in our colony). All procedures adhered to the ARVO Statement for the Use of Animals in Ophthalmic and Vision Research and were approved by the Columbia University Institutional Animal Care and Use Committee.
Induction of Form-Deprivation Myopia
Visual form deprivation (VFD) was induced in 12 mice by applying a unilateral frosted hemispherical plastic diffusers over the right eyes as previously described [60]. Briefly, frosted hemispherical plastic diffusers were hand-made using caps from 0.2 ml PCR tubes (Molecular BioProducts, San Diego, CA) and rings made from medical tape (inner diameter 6 mm; outer diameter 8 mm). A cap was frosted with fine sandpaper and attached to a ring with Loctite™ Super Glue (Henkel Consumer Adhesives, Avon, OH). On the first day of the experiment (P24), animals were anesthetized via intraperitoneal injection of ketamine (90 mg/kg) and xylazine (10 mg/kg), and diffusers were attached to the skin surrounding the right eye with several stitches using size 5–0 ETHILON™ microsurgical sutures (Ethicon, Somerville, NJ) and reinforced with Vetbond™ glue (3M Animal Care Products, St. Paul, MN). The contralateral untreated left eyes were used as control. Toenails were covered with adhesive tape to prevent mice from removing the diffusers. Animals recovered on a warming pad and were then housed in transparent plastic cages for the duration of the experiment (10 days). A control group comprised of 8 age-matched C57BL/6J mice was maintained under the same experimental conditions as experimental group, but without VFD.
RNA Extraction
After 10 days of visual form deprivation, mice were euthanized following the approved experimental protocol. Both myopic and control eyes were enucleated, cleaned by removing surrounding tissues and the crystalline lens, retina and sclera were dissected, snap-frozen in liquid nitrogen and stored in RNAlater®-ICE (Life Technologies, Grand Island, NY). In order to obtain sufficient amount of tissue for RNA isolation, the retinas or scleras from three myopic or control eyes were pooled for RNA extraction. Three replicates (3 eyes per replicate) were processed in parallel. Total RNA was isolated from the retina and sclera samples using mirVanaTM miRNA isolation kit (Life Technologies, Grand Island, NY) according to the manufacturer’s instructions. RNA concentration was measured using NanoDrop 8000 spectrophotometer (Thermo Scientific, Wilmington, DE). The quality of the total RNA was assessed using Agilent RNA 6000 Nano kit and 2100 Agilent Bioanalyzer (Agilent Technologies, Santa Clara, CA) following the manufacturer’s instructions.
MicroRNA expression profiling and microarray data analysis
MiRNA expression profiling was carried out at the Microarray Core Facility of the Duke University Institute for Genome Sciences and Policy (IGSP) using Agilent Mouse microRNA Microarray (release 15.0) (Agilent Technologies, Santa Clara, CA). MiRNA labeling, hybridization and washing were carried out according to the manufacturer’s recommendations. Following hybridization, microarrays were scanned on a DNA microarray scanner (Agilent G2565BA) and features were extracted using the Agilent Feature Extraction (AFE) image analysis tool (version A.9.5.3) with default protocols and settings. Gene expression data were analyzed using Partek Genomics Suit 6.6. Data were adjusted to bring the minimal signal to 0.5, normalized using quantile normalization procedure, and log2-transformed. This was followed by the removal of absent features and outliers. The normalized data were then analyzed using ANOVA to identify the differences in miRNA expression levels between myopic and control eyes. Differentially expressed miRNAs were identified using an FDR-adjusted P-value threshold of 0.05 and a cutoff of 2-fold change in expression. Differential expression was calculated as fold change (FC, myopic samples vs control).
Analysis of miRNA-mRNA Signaling Pathways
To identify biologically relevant miRNA-regulated genetic networks, differentially expressed miRNAs were analyzed using QIAGEN’s Ingenuity Pathway Analysis (IPA®) software and database (QIAGEN, Redwood City, CA). Putative mRNA targets were identified for the differentially expressed miRNAs, and then the mRNAs were filtered using published datasets of mRNAs differentially expressed in the myopic retina [10, 13, 14, 61, 62]. Mouse orthologs of mRNAs found to be differentially expressed in the species other than mouse were identified using Ensembl Compara [63]. The list of miRNAs and their associated target mRNAs were then subjected to core functional analysis in IPA® that uses ~1.5 million microRNA targeting interactions from sources including miRecords, TarBase, TargetScan, Ingenuity Expert findings and Ingenuity ExpertAssist findings to identify the miRNA-mRNA target relationships. Specifically, TargetScanMouse 6.2 [64], which was integrated with the latest versions of miRBase [65] and RefSeq [66, 67] databases, was used to identify mRNA targets. The stringency level for miRNA-mRNA interactions was set at “High (predicted)” or “Experimentally Observed”. “High (predicted)” confidence level was assigned to the relationships between a highly conserved microRNA and at least one conserved site on the targeted mRNA with the total TargetScan context score of -0.4 or less, which indicated that the microRNA was predicted to repress the expression of its mRNA target to at least 40% of the "normal" level. The “Experimentally Observed” targeting interactions were high quality manually curated miRNA-mRNA interactions with documented experimental support for each interaction. The homolog adjustment was applied to all miRNAs, which represent the miRNA families that share the same seed region, and IPA® mouse miRNA cluster symbols were used in the network images for each miRNA. Significant and enriched networks and Gene Ontology categories were obtained using the Fisher's exact test with a P-value threshold of 0.05.
Results
MicroRNA expression profiling in the mouse form-deprivation myopia
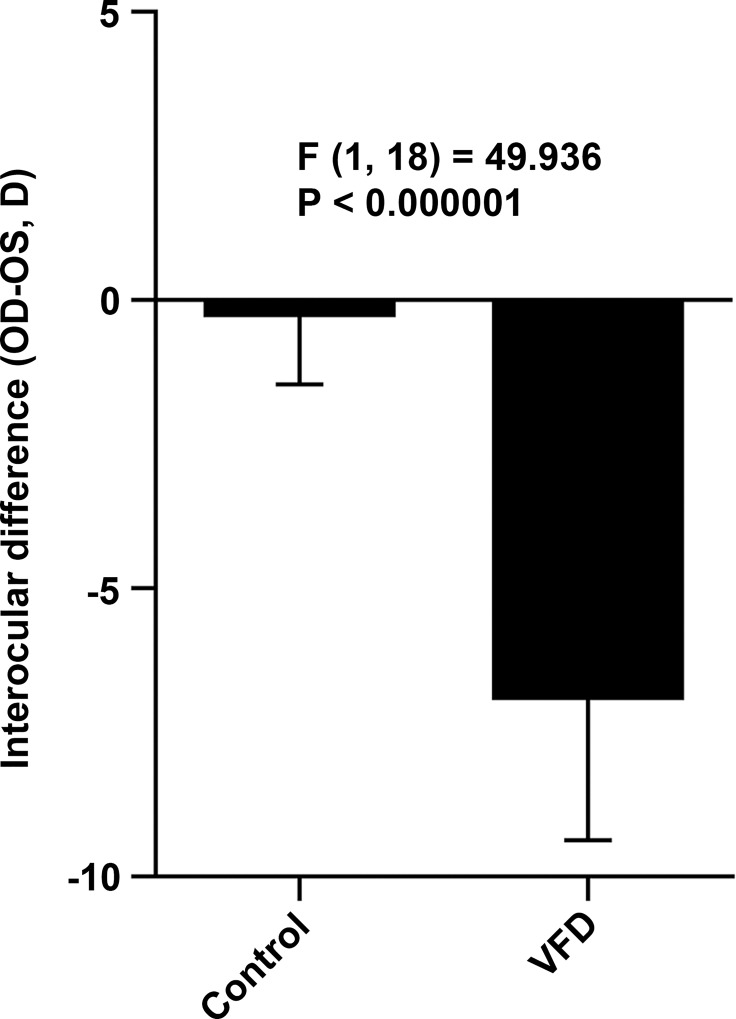
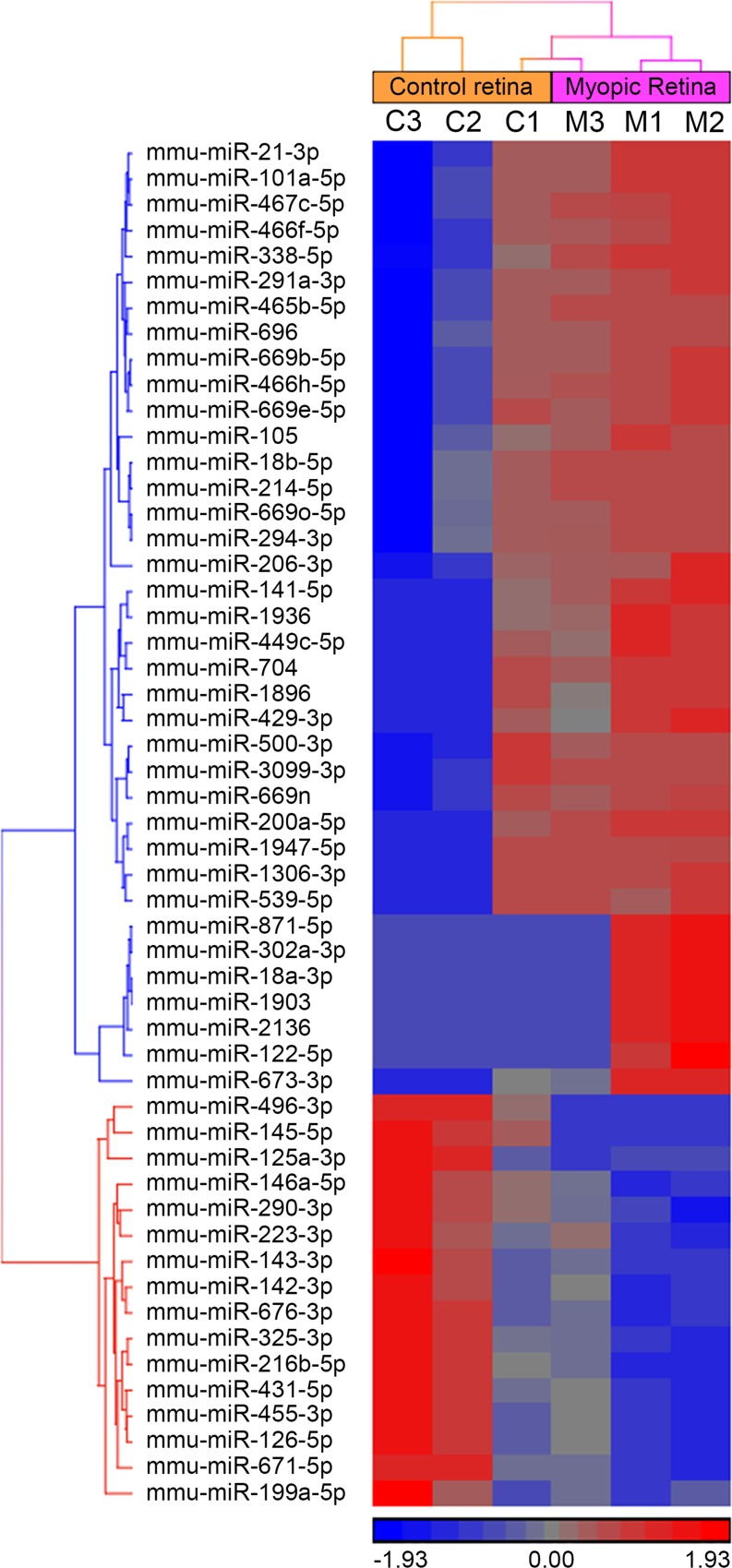
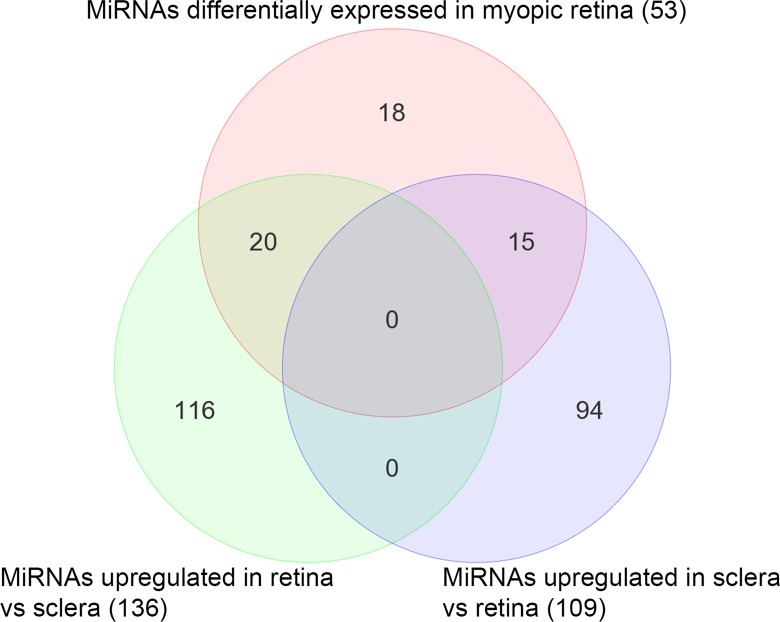
To examine potential involvement of miRNAs in the development of myopia, we analyzed miRNA expression in a mouse model of myopia. Twelve P24 C57BL/6J mice were subjected to monocular visual form deprivation (VFD). After 10 days of VFD, we detected a myopic shift in refraction in the deprived eyes of -6.93 ± 2.44 D (p < 0.000001, n = 12) relative to the control eyes (Fig 1). The difference in the interocular difference (OD-OS) between the VFD group and control group was also statistically significant (F(1, 18) = 49.936, p < 0.000001). A large-scale miRNA expression profiling was performed in the retina and sclera of the VFD mice using Agilent mouse microRNA microarrays which contained 627 mature mouse and 39 mouse viral miRNAs. This profiling revealed that a total of 53 miRNAs were differentially expressed (FC ≥ 2.0; FDR-adjusted p < 0.05) in the myopic retina compared to the contralateral control retina, whereas no differentially expressed miRNAs were identified in the sclera (Fig 2; Table 1). Thirty seven out of the 53 miRNAs were up-regulated and 16 out of the 53 miRNAs were down-regulated in the myopic retina (Fig 2; Table 1). Analysis of differential expression of these 53 miRNAs in the retina versus sclera revealed that 18 miRNAs were equally expressed in both retina and sclera, whereas 20 miRNAs were upregulated in the retina versus sclera and 15 were upregulated in the sclera versus retina (Fig 3; S1 Fig; S1 Table). Although the majority of the differentially expressed miRNAs originated from different miRNA clusters, mmu-miR-429-3p and mmu-miR-200a-5p belonged to the same cluster (MID < 5 kb) on chromosome 4 and were both up-regulated in myopic retina, while mmu-miR-145-5p and mmu-miR-143-3p localized within the same cluster (MID < 5 kb) on chromosome 18 and were both down-regulated in myopic retina (Table 1) [68]. Several miRNAs exhibited more than 10-fold change in expression in the myopic retina (Table 1), including mmu-miR-1947-5p (FC = 31.5, p = 1.47 × 10−04), mmu-miR-200a-5p (FC = 18.8, p = 9.46 × 10−05), mmu-miR-141-5p (FC = 13.9, p = 4.75 × 10−06), mmu-miR-465b-5p (FC = 12.8, p = 5.93 × 10−04), mmu-miR-214-5p (FC = 12.6, p = 8.27 × 10−03), mmu-miR-1936 (FC = 12.3, p = 9.56 × 10−06), mmu-miR-466f-5p (FC = 11.5, p = 3.85 × 10−03), mmu-miR-669o-5p (FC = 10.9, p = 2.18 × 10−03), mmu-miR-18b-5p (FC = 10.1, p = 1.79 × 10−03), and mmu-miR-145-5p (FC = -10.5, p = 8.87 × 10−09). These data suggest that development of form-deprivation myopia is associated with large-scale changes in miRNA expression in the retina.
Fig 1. Form-deprivation myopia in C57BL/6J mice.
Form-deprivation myopia was induced in C57BL/6J mice by applying a diffuser to the right eye of P24 animals. Ten days of visual form deprivation induced -6.93 ± 2.44 D (p < 0.000001, n = 12) of myopia in the right eyes compared to the contralateral control eyes.
Fig 2. Hierarchical cluster analysis of 53 miRNAs differentially expressed in the myopic retina versus control retina.
Logarithmic values (base 2) of Agilent total gene signal for differentially expressed miRNAs (cutoff: FC > 2, FDR-adjusted p-value < 0.05) were quantile normalized, shifted to mean of zero, scaled to standard deviation of 1.0 and subjected to hierarchical clustering using Euclidean dissimilarity and average linkage. The color scale indicates transcript abundance relative to the mean of zero: red identifies an increase in relative miRNA abundance; blue identifies a decrease in relative miRNA abundance. Columns show individual samples, whereas rows show individual miRNAs. Control samples c1, c2, and c3 correspond to myopic samples m1, m2, and m3 respectively. The “co-clustering” of the control sample c1 and myopic sample m3 resulted from the clustering algorithm that was used to generate the cluster and reflects individual differences in gene expression as well as differences in the myopic response to visual form deprivation between animals. It appears that the myopic response in the animals comprising experimental group 1 (samples c1 and m1) was the weakest among the 3 groups. However, the relationship between control sample c1 and the corresponding myopic sample m1 follows the same pattern as in the other two experimental groups even though samples c1 and m1 fall within the same color scheme. Consistent up- or down-regulation of the specific miRNAs in the myopic eyes versus corresponding control eyes across all samples is reflected by the low p-values shown in Table 1.
Table 1. MiRNAs differentially expressed in myopic retina versus control retina.
| miRBase ID | miRNA name | miRNA cluster | Fold change | P-value |
|---|---|---|---|---|
| MIMAT0009413 | mmu-miR-1947-5p | miR-1947-5p (miRNAs w/seed GGACGAG) | 31.5 | 1.47 × 10−04 |
| MIMAT0004619 | mmu-miR-200a-5p | miR-200a-5p (and other miRNAs w/seed AUCUUAC) | 18.8 | 9.46 × 10−05 |
| MIMAT0004533 | mmu-miR-141-5p | miR-141-5p (and other miRNAs w/seed AUCUUCC) | 13.9 | 4.75 × 10−06 |
| MIMAT0004871 | mmu-miR-465b-5p | miR-465b-5p (and other miRNAs w/seed AUUUAGA) | 12.8 | 5.93 × 10−04 |
| MIMAT0004664 | mmu-miR-214-5p | miR-214-5p (miRNAs w/seed GCCUGUC) | 12.6 | 8.27 × 10−03 |
| MIMAT0009400 | mmu-miR-1936 | miR-1936 (miRNAs w/seed AACUGAC) | 12.3 | 9.56 × 10−06 |
| MIMAT0004881 | mmu-miR-466f-5p | miR-466f-5p (miRNAs w/seed ACGUGUG) | 11.5 | 3.85 × 10−03 |
| MIMAT0009421 | mmu-miR-669o-5p | miR-669o-5p (miRNAs w/seed AGUUGUG) | 10.9 | 2.18 × 10−03 |
| MIMAT0004858 | mmu-miR-18b-5p | miR-18a-5p (and other miRNAs w/seed AAGGUGC) | 10.1 | 1.79 × 10−03 |
| MIMAT0009411 | mmu-miR-1306-3p | miR-1306-3p (miRNAs w/seed CGUUGGC) | 9.3 | 3.67 × 10−03 |
| MIMAT0000368 | mmu-miR-291a-3p | miR-291a-3p (and other miRNAs w/seed AAGUGCU) | 9.2 | 3.22 × 10−04 |
| MIMAT0003483 | mmu-miR-696 | miR-696 (miRNAs w/seed CGUGUGC) | 9.0 | 1.28 × 10−03 |
| MIMAT0004628 | mmu-miR-21-3p | miR-21-3p (and other miRNAs w/seed AACAGCA) | 8.9 | 8.01 × 10−05 |
| MIMAT0004824 | mmu-miR-673-3p | miR-673-3p (and other miRNAs w/seed CCGGGGC) | 8.4 | 1.65 × 10−05 |
| MIMAT0001537 | mmu-miR-429-3p | miR-200b-3p (and other miRNAs w/seed AAUACUG) | 7.8 | 2.05 × 10−03 |
| MIMAT0011212 | mmu-miR-2136 | miR-2136 (miRNAs w/seed UGGGUGU) | 6.3 | 5.02 × 10−04 |
| MIMAT0004841 | mmu-miR-871-5p | miR-743a-5p (and other miRNAs w/seed AUUCAGA) | 5.8 | 5.51 × 10−04 |
| MIMAT0004526 | mmu-miR-101a-5p | miR-101a-5p (miRNAs w/seed CAGUUAU) | 5.1 | 1.88 × 10−04 |
| MIMAT0007873 | mmu-miR-1896 | miR-1896 (miRNAs w/seed UCUCUGA) | 4.9 | 1.66 × 10−03 |
| MIMAT0004885 | mmu-miR-467c-5p | miR-467c-5p (and other miRNAs w/seed AAGUGCG) | 4.6 | 1.68 × 10−04 |
| MIMAT0004884 | mmu-miR-466h-5p | miR-669m-5p (and other miRNAs w/seed GUGUGCA) | 4.3 | 1.29 × 10−03 |
| MIMAT0003476 | mmu-miR-669b-5p | miR-669b-5p (miRNAs w/seed GUUUUGU) | 4.3 | 1.02 × 10−03 |
| MIMAT0005853 | mmu-miR-669e-5p | miR-331-5p (and other miRNAs w/seed GUCUUGU) | 4.2 | 2.41 × 10−03 |
| MIMAT0004856 | mmu-miR-105 | miR-105 (miRNAs w/seed CAAGUGC) | 4.2 | 1.83 × 10−04 |
| MIMAT0003494 | mmu-miR-704 | miR-704 (miRNAs w/seed GACAUGU) | 4.0 | 3.00 × 10−04 |
| MIMAT0000372 | mmu-miR-294-3p | miR-291a-3p (and other miRNAs w/seed AAGUGCU) | 3.9 | 3.79 × 10−03 |
| MIMAT0004626 | mmu-miR-18a-3p | miR-18a-3p (and other miRNAs w/seed CUGCCCU) | 3.9 | 6.01 × 10−04 |
| MIMAT0003169 | mmu-miR-539-5p | miR-539-5p (miRNAs w/seed GAGAAAU) | 3.5 | 3.47 × 10−04 |
| MIMAT0009427 | mmu-miR-669n | miR-5010-3p (and other miRNAs w/seed UUUGUGU) | 3.3 | 7.22 × 10−03 |
| MIMAT0004647 | mmu-miR-338-5p | miR-338-5p (miRNAs w/seed ACAAUAU) | 3.2 | 4.09 × 10−04 |
| MIMAT0003460 | mmu-miR-449c-5p | miR-34a-5p (and other miRNAs w/seed GGCAGUG) | 2.6 | 1.41 × 10−04 |
| MIMAT0000239 | mmu-miR-206-3p | miR-1-3p (and other miRNAs w/seed GGAAUGU) | 2.5 | 4.14 × 10−04 |
| MIMAT0007868 | mmu-miR-1903 | miR-1903 (and other miRNAs w/seed CUUCUUC) | 2.4 | 5.95 × 10−04 |
| MIMAT0000380 | mmu-miR-302a-3p | miR-291a-3p (and other miRNAs w/seed AAGUGCU) | 2.4 | 5.27 × 10−04 |
| MIMAT0014816 | mmu-miR-3099-3p | miR-3099 (and other miRNAs w/seed AGGCUAG) | 2.4 | 7.47 × 10−03 |
| MIMAT0003507 | mmu-miR-500-3p | miR-501-3p (and other miRNAs w/seed AUGCACC) | 2.2 | 4.14 × 10−03 |
| MIMAT0000246 | mmu-miR-122-5p | miR-122-5p (miRNAs w/seed GGAGUGU) | 2.0 | 8.09 × 10−04 |
| MIMAT0000247 | mmu-miR-143-3p | miR-143-3p (and other miRNAs w/seed GAGAUGA) | -2.0 | 1.43 × 10−03 |
| MIMAT0003738 | mmu-miR-496-3p | miR-503-3p (and other miRNAs w/seed GAGUAUU) | -2.1 | 8.01 × 10−10 |
| MIMAT0001418 | mmu-miR-431-5p | miR-431-5p (and other miRNAs w/seed GUCUUGC) | -2.1 | 7.76 × 10−04 |
| MIMAT0000137 | mmu-miR-126-5p | miR-126a-5p (and other miRNAs w/seed AUUAUUA) | -2.3 | 3.67 × 10−03 |
| MIMAT0004572 | mmu-miR-290-3p | miR-467a-5p (and other miRNAs w/seed AAGUGCC) | -2.4 | 6.43 × 10−03 |
| MIMAT0003731 | mmu-miR-671-5p | miR-671-5p (miRNAs w/seed GGAAGCC) | -2.6 | 2.30 × 10−03 |
| MIMAT0003729 | mmu-miR-216b-5p | miR-216b-5p (miRNAs w/seed AAUCUCU) | -2.6 | 1.51 × 10−05 |
| MIMAT0000665 | mmu-miR-223-3p | miR-223-3p (miRNAs w/seed GUCAGUU) | -2.7 | 4.84 × 10−04 |
| MIMAT0003782 | mmu-miR-676-3p | miR-676 (and other miRNAs w/seed CGUCCUG) | -2.7 | 9.35 × 10−04 |
| MIMAT0004640 | mmu-miR-325-3p | miR-325-3p (miRNAs w/seed UUAUUGA) | -2.8 | 5.59 × 10−04 |
| MIMAT0003742 | mmu-miR-455-3p | miR-455-3p (miRNAs w/seed CAGUCCA) | -3.1 | 1.74 × 10−03 |
| MIMAT0000229 | mmu-miR-199a-5p | miR-199a-5p (and other miRNAs w/seed CCAGUGU) | -3.2 | 6.65 × 10−03 |
| MIMAT0000158 | mmu-miR-146a-5p | miR-146a-5p (and other miRNAs w/seed GAGAACU) | -3.2 | 2.70 × 10−05 |
| MIMAT0000155 | mmu-miR-142-3p | miR-142-3p (and other miRNAs w/seed GUAGUGU) | -3.3 | 2.13 × 10−03 |
| MIMAT0004528 | mmu-miR-125a-3p | miR-125a-3p (miRNAs w/seed CAGGUGA) | -3.4 | 1.21 × 10−03 |
| MIMAT0000157 | mmu-miR-145-5p | miR-145-5p (and other miRNAs w/seed UCCAGUU) | -10.5 | 8.87 × 10−09 |
Fig 3. Overlap between miRNAs differentially expressed in the myopic retina and miRNAs differentially expressed in the retina versus sclera.
Venn diagram shows overlap between 53 miRNAs, which were differentially expressed in the myopic retina, 136 miRNAs, which were up-regulated in the retina versus sclera, and 109 miRNAs, which were up-regulated in the sclera versus retina. Eighteen differential miRNAs were equally expressed in both retina and sclera, 20 differential miRNAs were up-regulated in the retina versus sclera and 15 differential miRNAs were down-regulated in the retina versus sclera.
Identification of putative target mRNAs for the miRNAs differentially expressed in myopic retina
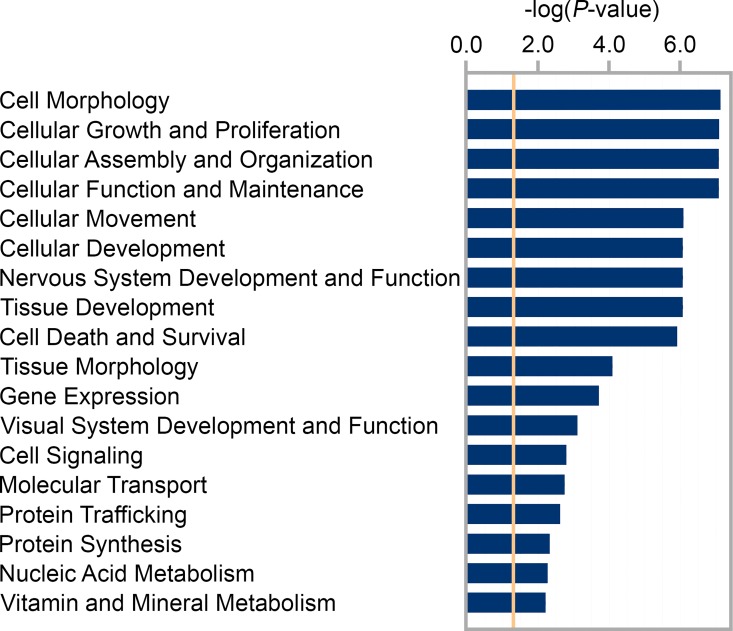
In order to explore potential signaling pathways regulated by the identified differentially expressed miRNAs during the development of form-deprivation myopia in mice, we performed miRNA-mRNA interaction network analyses using a custom database of mRNAs differentially expressed in the retina upon induction of experimental myopia (S2 Table) [10, 13, 14, 61, 62] using IPA®. This analysis identified a total of 135 mRNA targets for 21 out of 53 differentially expressed miRNAs (Table 2), whereas no target mRNAs were found for the remaining 32 miRNAs among 611 mRNAs, which were found to be differentially expressed in the myopic retina (S3 Table). Gene ontology analysis of the target mRNAs revealed that these mRNAs encode proteins primarily involved in cellular growth, proliferation, nervous and visual systems development (Fig 4). The miRNA-mRNA networks within these gene ontology categories were characterized by complex combinatorial interactions with one miRNA targeting multiple mRNAs and one mRNA often being targeted by several miRNAs. For example, mmu-miR-145-5p, which was strongly down-regulated in myopic retina (FC = -10.5, p = 8.87 × 10−09), targeted 25 mRNAs (the largest number among all 21 miRNAs) (Table 2); while mmu-miR-429-3p (FC = 7.8, p = 2.05 × 10−03), mmu-miR-143-3p (FC = -2.0, p = 1.43 × 10−03), mmu-miR-223-3p (FC = -2.7, p = 4.84 × 10−04) and mmu-miR-146a-5p (FC = -3.2, p = 2.70 × 10−05) targeted 17, 17, 16 and 14 mRNAs respectively. The average number of mRNAs targeted by one miRNA was 9.0 ± 6.2. On the other hand, each mRNA was targeted by only 1.4 ± 0.7 miRNAs and maximum by 4 miRNAs.
Table 2. MiRNAs and their target mRNAs differentially expressed in myopic retina versus control retina.
| miRNA name | miRNA fold change | Target mRNA name and fold change |
|---|---|---|
| mmu-miR-18b-5p | 10.1 | BRWD3 (-2.5); NOS1 (-2.0); CTGF (-2.0); NXT2 (-1.7); HLF (-1.6); TMEM2 (-1.6) |
| mmu-miR-1306-3p | 9.3 | DUSP4 (-2.5) |
| mmu-miR-291a-3p | 9.2 | CNOT6 (-2.2); RAB11A (-2.0); CUL3 (-1.7); CLIP4 (-1.6); HLF (-1.6); BAZ1A (-1.6); ZC3H13 (-1.2); ZFP91 (-1.0) |
| mmu-miR-429-3p | 7.8 | ZEB2 (-3.5); ATP11B (-2.7); PRKAR1A (-2.6); BRWD3 (-2.5); RIMS2 (-1.9); RBFOX2 (-1.8); SENP5 (-1.8); ARIH1 (-1.7); MAPK9 (-1.7); COMMD3 (-1.7); HLF (-1.6); SLC30A5 (-1.6); TAOK3 (-1.6); ETV5 (-1.5); NEGR1 (-1.5); MPRIP (-1.2); MAP2 (-1.2) |
| mmu-miR-539-5p | 3.5 | RAB11A (-2.0); SENP5 (-1.8); WNK1 (-1.8); NPL (-1.7); ZSWIM5 (-1.7); MAP2 (-1.2) |
| mmu-miR-449c-5p | 2.6 | NOS1 (-2.0); RBFOX2 (-1.8); MRPL17 (-1.7); COPS7B (-1.7); ZSWIM5 (-1.7); TCF12 (-1.6) |
| mmu-miR-206-3p | 2.5 | CNOT6 (-2.2); MAN1C1 (-1.8); ARIH1 (-1.7); NXT2 (-1.7); RICTOR (-1.7); TAOK3 (-1.6); FAM101B (-1.6); KLHL5 (-1.5); ZFP91 (-1.0) |
| mmu-miR-1903 | 2.4 | ASCL1 (-1.6) |
| mmu-miR-500-3p | 2.2 | OLFM4 (-3.5) |
| mmu-miR-122-5p | 2.0 | DUSP4 (-2.5); UBAP2 (-1.9); NEGR1 (-1.5) |
| mmu-miR-143-3p | -2.0 | FUT4 (1.2); COX18 (1.2); CHST10 (1.2); BCL2 (1.2); PPP2R3A (1.3); GABARAPL1 (1.3); DCLK1 (1.3); HTR7 (1.3); LBH (1.3); GXYLT1 (1.3); ADD3 (1.5); ZNF275 (1.5); MAP1B (1.6); DCX (1.6); PRKCE (1.8); RNF165 (1.8); TPM3 (1.9) |
| mmu-miR-496-3p | -2.1 | CSRNP3 (1.2); CNOT2 (1.2); AEBP2 (1.2); INSIG1 (1.3); PRKCE (1.8); NT5C2 (6.3) |
| mmu-miR-431-5p | -2.1 | PTPRF (1.1); AKAP12 (1.3); ZAK (1.6); THUMPD1 (1.6); CELF2 (1.8) |
| mmu-miR-671-5p | -2.6 | MPEG1 (1.0); ANKS1A (1.2); RBMS3 (1.2); NFYA (1.3); MYOM3 (1.4); KIAA0430 (1.5); ATXN7L1 (1.6); SEPP1 (1.7); SAMD12 (1.8); SPTBN1 (2.9) |
| mmu-miR-216b-5p | -2.6 | E2F4 (1.1); NRK (1.2); MCM4 (1.2); NFYA (1.3); FBXO8 (1.3); CHMP1B (1.5); GLTSCR1L (1.5); SDHC (1.6); SAR1B (1.6); CELF2 (1.8); TPM3 (1.9); SPTBN1 (2.9); GFRA1 (3.8) |
| mmu-miR-223-3p | -2.7 | NUCKS1 (1.1); EBNA1BP2 (1.2); PARP1 (1.2); CNOT2 (1.2); ATG7 (1.2); AEBP2 (1.2); SYNCRIP (1.3); DDIT4 (1.3); FBXO8 (1.3); NUP210 (1.4); ARFIP1 (1.6); ATXN7L1 (1.6); WDR77 (1.6); PRR14L (1.8); PRKCE (1.8); ERC1 (4.8) |
| mmu-miR-199a-5p | -3.2 | TOX3 (1.3); NFYA (1.3); MCFD2 (1.3); CCNJ (1.3); NUP210 (1.4); ARHGEF12 (1.5); ADD3 (1.5); RALGAPA1 (1.5); ATXN7L1 (1.6); CELF2 (1.8) |
| mmu-miR-146a-5p | -3.2 | CAMSAP1 (1.1); NUCKS1 (1.1); CCNA2 (1.2); LFNG (1.2); ATG7 (1.2); CCNJ (1.3); NAIF1 (1.3); EDNRB (1.5); NLGN1 (1.6); CELF2 (1.8); PRKCE (1.8); NOTCH2 (2.0); SBSPON (2.0); STAT1 (2.7) |
| mmu-miR-142-3p | -3.3 | NUCKS1 (1.1); ANKS1A (1.2); HGS (1.2); MORF4L2 (1.2); ZCCHC24 (1.3); CCNJ (1.3); ARHGEF12 (1.5); MYLK (1.7); SAMD12 (1.8); ERC1 (4.8) |
| mmu-miR-125a-3p | -3.4 | MAPK1IP1L (1.2); AEBP2 (1.2); SWAP70 (1.4); FYCO1 (1.8); RAB22A (2.2) |
| mmu-miR-145-5p | -10.5 | CCNA2 (1.2); SMC1A (1.2); DENND4B (1.2); KIAA0930 (1.2); KATNBL1 (1.2); PPP2R3A (1.3); GABARAPL1 (1.3); HABP4 (1.3); PTGR2 (1.3); GCLM (1.3); AKAP12 (1.3); GXYLT1 (1.3); SWAP70 (1.4); VSTM4 (1.4); ARHGEF12 (1.5); ADD3 (1.5); PHACTR2 (1.6); DOK6 (1.6); ATXN7L1 (1.6); QSER1 (1.8); COMMD5 (1.9); TPM3 (1.9); ANKRD28 (1.9); ANGPT2 (2.7); GFRA1 (3.8) |
Transcription factors are shown in bold; genes involved in synapse formation or function are underlined.
Fig 4. Gene ontology categories affected in myopic retina.
Graph shows top 18 biological processes which were modified in the myopic retina.
Analysis of miRNA-regulated signaling pathways in the mouse form-deprivation myopia
Our initial analysis revealed that each differentially expressed miRNA regulates an extended network of protein-coding genes within a limited number of gene ontology categories, therefore we then explored interactions between 21 differentially expressed miRNAs and 135 mRNAs which were targeted by these miRNAs. This analysis revealed that these miRNAs and mRNAs are organized into 9 overlapping miRNA-mRNA signaling pathways (Fig 5; S2–S10 Figs). Myopia pathways (MP) 1 and 2 (MP1 and MP2) were the largest pathways, which comprised 16 miRNAs, followed by MP3 (15 miRNAs), MP4 (14 miRNAs), MP5 (14 miRNAs), MP6 (13 miRNAs), MP7 (12 miRNAs), MP8 (4 miRNAs) and MP9 (1 miRNA). MP1, MP2, MP3, MP4, MP5, MP6 and MP7 had a common core comprised of mmu-miR-1-3p, mmu-miR-145-5p, mmu-miR-18a-5p, mmu-miR-199a-5p, mmu-miR-200b-3p, mmu-miR-223-3p, mmu-miR-291a-3p, mmu-miR-34a-5p and their target mRNAs. Two of these miRNAs, i.e., mmu-miR-145-5p and mmu-miR-200b-3p, served as a common core for all pathways except for MP9. MP9 comprised only 1 miRNA (mmu-miR-1903) and 3 mRNAs (Ascl1, Dcx and Notch2), but was linked to 6 larger pathways, i.e., MP1, MP2, MP3, MP4, MP5 and MP7, via Notch2 (indirect target of mmu-miR-1903). In all pathways, miRNAs played a role of major regulatory hubs targeting transcription factors and/or regulatory proteins. The absolute majority of miRNAs targeted at least one transcription factor while the average number of transcription factors targeted by each differentially expressed miRNA was 1.7 ± 1.2 (Table 2). At the same time, several transcription factors were targeted by multiple often overlapping miRNAs. For example, Hlf was targeted by mmu-miR-18b-5p, mmu-miR-429-3p and mmu-miR-291a-3p; Cnot6 and Zfp91 were targeted by mmu-miR-206-3p and mmu-miR-291a-3p; Rbfox2 was targeted by mmu-miR-429-3p and mmu-miR-449c-5p; Aebp2 was targeted by mmu-miR-125a-3p, mmu-miR-223-3p and mmu-miR-496-3p; Nfya was targeted by mmu-miR-199a-5p, mmu-miR-216b-5p and mmu-miR-671-5p; Nucks1 was targeted by mmu-miR-142-3p, mmu-miR-146a-5p and mmu-miR-223-3p; Notch2 was targeted by mmu-miR-146a-5p and indirectly via Ascl1 by mmu-miR-1903. Interestingly, mmu-miR-145-5p and mmu-miR-429-3p (mmu-miR-200b-3p cluster), which formed a common core of all pathways, were among the most differential miRNAs and targeted the largest number of mRNAs. Mmu-miR-145-5p was strongly down-regulated in myopic retina (FC = -10.5, p = 8.87 × 10−09) and targeted 25 mRNAs, while mmu-miR-429-3p was strongly up-regulated in myopic retina (FC = 7.8, p = 2.05 × 10−03) and targeted 17 mRNAs (Table 2).
Fig 5. Overlap between miRNA-regulated signaling pathways affected in myopic retina.
Diagram depicts miRNA contributions to the 9 miRNA-mRNA signaling cascades associated with form-deprivation myopia in mice.
Among the mRNAs targeted by differentially expressed miRNAs, there were several (Prkar1a, Rims2, Map2, Gabarapl1, Htr7, Add3, Erc1, Nlgn1) which were involved in synapse formation and function suggesting that synaptic function was one of the biological processes affected in form-deprivation myopia. However, IPA gene ontology analysis revealed that the most prominent biological processes associated with the 21 differentially expressed miRNAs and their target mRNAs were quantity of neurons (p = 1.08 × 10−4, activation z-score (z) = 2.04), migration of neurons (p = 2.24 × 10−6, z = 1.33), growth of axons (p = 7.21 × 10−4, z = 1.10), outgrowth of neurites (p = 1.83 × 10−5, z = 0.74), growth of neurites (p = 8.51 × 10−7, z = 0.54), neuritogenesis (p = 1.03 × 10−6, z = 0.41), proliferation of neuronal cells (p = 9.93 × 10−7, z = 0.33) and differentiation of neurons (p = 1.45 × 10−3, z = 0.17) (S4 Table). Therefore, we have analyzed the overlap between all 9 pathways and GO categories linked to neurogenesis. We found that each pathway was indeed involved in regulation of neurogenesis and had a net positive effect on neurogenesis-associated processes (S2–S10 Figs). Thus, pathway analysis suggests that miRNAs serve as key regulators of several signaling cascades underlying development of form-deprivation myopia.
Discussion
Development of myopia is associated with remodeling of several ocular tissues, including the retina, RPE, choroid and sclera [14, 69–72]. Several studies analyzed gene expression in various animal models of myopia using oligonucleotide-based microarrays and each identified between 15 and 280 differentially expressed genes [10–14, 61, 62], including a total of 611 mRNAs in the retina [10, 13, 14, 61, 62]. Although involvement of coding genes in myopia is a well-established fact, to date only 1 miRNA was shown to be associated with myopia development. It was reported that a SNP located within the miR-328 binding site in the 3’-UTR of PAX6 reduced PAX6 protein levels and was significantly associated with extreme myopia in a Chinese cohort [54, 55]. Considering that miRNAs often serve as regulatory hubs in many signaling pathways [21–24] and provide higher-order coordination of signaling pathways underlying the same biological process [68, 73], understanding the role of miRNAs in refractive eye development is critical for the understanding of myopia development.
In this work, we sought to identify miRNAs differentially expressed during the development of form-deprivation myopia in the mouse model and to explore miRNA-mRNA genetic networks underlying myopia. Considering that mice are most susceptible to visually-guided myopia during active phase of ocular growth from P21 through P40 [60, 74], we analyzed early changes in miRNA expression in the retina and sclera in P34 mice 10 days after induction of form-deprivation myopia. Ten days of visual form deprivation induced -6.93 ± 2.44 diopters of myopia and large-scale changes in miRNA expression in the retina indicating that early retinal response in visually-guided myopia is associated with substantial changes in retinal signaling. Surprisingly, we did not find differences in miRNA expression in the sclera, which suggests that early myopia-associated scleral remodeling in response to visual form deprivation may not involve miRNAs, or that changes in miRNA expression are very subtle at this early stage of myopia development. In the retina, we identified 53 differentially expressed miRNAs, including 19 miRNAs with more than 5-fold change in expression. Importantly, 18 of these highly differential miRNAs were upregulated in the myopic retina and only 1 was down-regulated suggesting that the “net effect” may be the down-regulation of the corresponding target genes in myopia. Seven of the up-regulated miRNAs (miR-465b, miR-466f, miR-669o, miR-18b, miR-291a, miR-696, miR-101a) were also much more (≥ 5 fold) expressed in the retina compared to the sclera suggesting that they might be involved in the regulation of retina-specific processes. The down-regulated miR-145 was 25.4 times less abundant in the retina versus sclera suggesting that it is likely to be expressed in a very small population of retinal cells. Several of the highly differential miRNAs were previously shown to be key regulators of various developmental processes. For example, miR-200a, miR-429 and miR-141 were shown to play important roles in neurogenesis, epithelial-to-mesenchymal transition and Notch signaling [73, 75–84], miR-214 was found to be overexpressed in fetal sclera versus adult sclera and shown to play important role in brain and retina development and function [36, 85–89], miR-18b, miR-21, miR-101a, miR-200a and miR-429 were found to be involved in stem cell function and differentiation [90–100], miR-1306 negatively regulated Alzheimer’s disease gene ADAM10 [101]. MicroRNA miR-145 regulates stem cell, smooth muscle cell, corneal epithelium and mesenchymal stem cell differentiation [102–107], as well as intestine and neural crest development [108, 109]. MiR-145 was also shown to regulate L-DOPA decarboxylase [110], which is one of the key enzymes synthesizing dopamine in the dopaminergic amacrine cells in the retina [111]. Thus, differential expression of these microRNAs suggests that form-deprivation myopia is associated with changes in neurogenesis, as well as in neuronal and synaptic functions.
Analysis of the mRNAs targeted by the differentially expressed miRNAs revealed that putative mRNA targets could be found for 21 out of 53 differential miRNAs (i.e., for 40% of all differential miRNAs), suggesting that the currently available list of mRNAs differentially expressed in the myopic retina is not complete and many retinal mRNAs underlying myopia are still unknown. However, the 21 differential miRNAs and their associated targets provided a sufficient foundation for the analysis of main biological processes and pathways associated with myopia development. Gene ontology analysis revealed that such biological processes as generation of new neurons, migration of neurons, growth of axons, outgrowth of neurites, cellular growth and proliferation, nervous and visual systems development, which were suggested to be affected based on the analysis of the most differential miRNAs, were significantly enhanced in myopic retina. Interestingly, this finding is consistent with the recent report that form-deprivation myopia in primates is associated with increased proliferation of retinal progenitor cells and retinal growth [14], thus, providing an outline of the putative molecular network underlying retinal growth associated with myopia development. Several target mRNAs were also linked to synaptic structure and function, which is consistent with the observations in animal models that synaptic signaling at the level of amacrine cells is involved in myopia development [112–123]. Remarkably, analysis of the miRNA-mRNA networks formed by the differential miRNAs and their targets revealed that myopia development is, in fact, regulated by a small number of highly integrated signaling pathways. MicroRNAs played a role of master regulators, which targeted large number of mRNAs often involved in the same biological process. Furthermore, miRNAs often had common transcription factors among the targets, which provides an additional level of integration. Interestingly, two miRNAs miR-145 and miR-200b seemed to play a role of the integrative core for all pathways.
Taken together, our findings suggest that the miRNAs differentially expressed in the retina of myopic eyes play important regulatory roles in the development of myopia by regulating a highly integrated genetic network. We analyzed expression of 56% of mouse miRNAs deposited in the miRBase database and identified 53 miRNAs differentially expressed in the retina during development of form-deprivation myopia. We also identified 135 target genes for 21 of these miRNAs and reconstructed putative miRNA-mRNA pathways underlying key biological processes associated with development of form-deprivation myopia. These results expand our understanding of the molecular mechanisms of myopia and demonstrate that the development of myopia is associated with large-scale changes in expression of both coding and none-coding RNAs. The power of our analysis was limited by the cellular complexity of the retina as well as some differences in cell composition of the retina in different species. Although it was shown that processing of defocus and refractive eye development are regulated by a relatively small subset of retinal cells (i.e., amacrine cells) [112–123], detecting small changes in gene expression might be challenging in heterogeneous tissues such as retina, resulting in an underestimate of miRNA influences. It is also a challenge to place differential miRNAs and their corresponding target mRNAs in the proper cellular context when analyzing gene expression in complex tissues. Future studies of the signaling pathways underlying myopia development at the single-cell level should provide more accurate information. Our data also suggest that more comprehensive genome-wide approaches should be applied to reconstruct signaling pathways underlying myopia in their entirety. This study lays a strong foundation for such future studies and provides a framework for the development of potential novel microRNA-based therapies for myopia.
Supporting Information
Logarithmic values (base 2) of Agilent total gene signal for differentially expressed miRNAs (cutoff: FC > 2, FDR-adjusted p-value < 0.05) were quantile normalized, shifted to mean zero, scaled to standard deviation of 1.0 and subjected to hierarchical clustering using Euclidean dissimilarity and average linkage. The color scale indicates transcript abundance: red identifies an increase in relative miRNA abundance; blue identifies a decrease in relative miRNA abundance. Columns show individual samples, whereas rows show individual miRNAs.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #1 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #2 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #3 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #4 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #5 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #6 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #7 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #8 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #9 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(XLSX)
(XLSX)
(DOCX)
(XLSX)
Acknowledgments
This project was supported by NIH Grants R01EY023839 to AVT, R01EY023287 and R01EY016228 to PG, R01EY014685 to TLY, P30EY019007 (Core Support for Vision Research), Duke Eye Center Exploring Grant and unrestricted funds from Research to Prevent Blindness (New York, NY) to the Department of Ophthalmology, Columbia University and the Department of Ophthalmology and Visual Sciences, University of Wisconsin-Madison. These research was funded with support from the Acquavella Family Foundation, Joseph C. Connors, and the University of Wisconsin Centennial Scholar Fund.
Data Availability
All relevant data are within the paper and its Supporting Information files except for the gene expression data, which were deposited in the Gene Expression Omnibus (Accession: GSE84220).
Funding Statement
This project was supported by National Institutes of Health/National Eye Institute grant R01EY023839, (www.nih.gov) (to AVT), National Institutes of Health/National Eye Institute grants R01EY023287 and R01EY016228, (www.nih.gov) (to PG), National Institutes of Health/National Eye Institute grant R01EY014685, (www.nih.gov) (to TLY), National Institutes of Health/National Eye Institute grant P30EY019007 (Core Support for Vision Research) (to AVT), Duke Eye Center Exploring Grant and unrestricted funds from Research to Prevent Blindness (New York, NY) to the Department of Ophthalmology, Columbia University and the Department of Ophthalmology and Visual Sciences, University of Wisconsin-Madison. These research was funded with support from the Acquavella Family Foundation, Joseph C. Connors, and the University of Wisconsin Centennial Scholar Fund.
References
- 1.Pararajasegaram R. VISION 2020-the right to sight: from strategies to action. Am J Ophthalmol. 1999;128(3):359–60. [DOI] [PubMed] [Google Scholar]
- 2.Vitale S, Ellwein L, Cotch MF, Ferris FL III, Sperduto R. Prevalence of refractive error in the United States, 1999–2004. Arch Ophthalmol. 2008;126(8):1111–9. 10.1001/archopht.126.8.1111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lin LL, Shih YF, Hsiao CK, Chen CJ. Prevalence of myopia in Taiwanese schoolchildren: 1983 to 2000. Ann Acad Med Singapore. 2004;33(1):27–33. [PubMed] [Google Scholar]
- 4.Lam CS, Goldschmidt E, Edwards MH. Prevalence of myopia in local and international schools in Hong Kong. Optom Vis Sci. 2004;81(5):317–22. [DOI] [PubMed] [Google Scholar]
- 5.Alexander LJ. Primary care of the posterior segment 2 ed. Connecticut: Appleton & Lange; 1994. [Google Scholar]
- 6.Saw SM, Gazzard G, Shih-Yen EC, Chua WH. Myopia and associated pathological complications. Ophthalmic Physiol Opt. 2005;25(5):381–91. [DOI] [PubMed] [Google Scholar]
- 7.Flitcroft DI. The complex interactions of retinal, optical and environmental factors in myopia aetiology. Prog Retin Eye Res. 2012;31(6):622–60. 10.1016/j.preteyeres.2012.06.004 [DOI] [PubMed] [Google Scholar]
- 8.Curtin BJ. The myopias: basic science and clinical management Philadelphia: Harper & Row; 1985. [Google Scholar]
- 9.Oyster CW. The human eye: structure and function Sunderland, MA: Sinauer Associates; 1999. [Google Scholar]
- 10.McGlinn AM, Baldwin DA, Tobias JW, Budak MT, Khurana TS, Stone RA. Form-deprivation myopia in chick induces limited changes in retinal gene expression. Invest Ophthalmol Vis Sci. 2007;48(8):3430–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brand C, Schaeffel F, Feldkaemper MP. A microarray analysis of retinal transcripts that are controlled by image contrast in mice. Mol Vis. 2007;13:920–32. [PMC free article] [PubMed] [Google Scholar]
- 12.Shelton L, Troilo D, Lerner MR, Gusev Y, Brackett DJ, Rada JS. Microarray analysis of choroid/RPE gene expression in marmoset eyes undergoing changes in ocular growth and refraction. Mol Vis. 2008;14:1465–79. [PMC free article] [PubMed] [Google Scholar]
- 13.Schippert R, Schaeffel F, Feldkaemper MP. Microarray analysis of retinal gene expression in chicks during imposed myopic defocus. Mol Vis. 2008;14:1589–99. [PMC free article] [PubMed] [Google Scholar]
- 14.Tkatchenko AV, Walsh PA, Tkatchenko TV, Gustincich S, Raviola E. Form deprivation modulates retinal neurogenesis in primate experimental myopia. Proc Natl Acad Sci U S A. 2006;103(12):4681–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Frost MR, Norton TT. Alterations in protein expression in tree shrew sclera during development of lens-induced myopia and recovery. Invest Ophthalmol Vis Sci. 2012;53(1):322–36. 10.1167/iovs.11-8354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gao H, Frost MR, Siegwart JT Jr, Norton TT. Patterns of mRNA and protein expression during minus-lens compensation and recovery in tree shrew sclera. Mol Vis. 2011;17:903–19. [PMC free article] [PubMed] [Google Scholar]
- 17.Siegwart JT Jr, Norton TT. The time course of changes in mRNA levels in tree shrew sclera during induced myopia and recovery. Invest Ophthalmol Vis Sci. 2002;43(7):2067–75. [PMC free article] [PubMed] [Google Scholar]
- 18.Gregory RI, Chendrimada TP, Cooch N, Shiekhattar R. Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell. 2005;123(4):631–40. [DOI] [PubMed] [Google Scholar]
- 19.Hutvagner G. Small RNA asymmetry in RNAi: function in RISC assembly and gene regulation. FEBS Lett. 2005;579(26):5850–7. [DOI] [PubMed] [Google Scholar]
- 20.Pattanayak D, Agarwal S, Sumathi S, Chakrabarti SK, Naik PS, Khurana SM. Small but mighty RNA-mediated interference in plants. Indian journal of experimental biology. 2005;43(1):7–24. [PubMed] [Google Scholar]
- 21.Qureshi IA, Mehler MF. Emerging roles of non-coding RNAs in brain evolution, development, plasticity and disease. Nature reviews Neuroscience. 2012;13(8):528–41. 10.1038/nrn3234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Miska EA. How microRNAs control cell division, differentiation and death. Curr Opin Genet Dev. 2005;15(5):563–8. [DOI] [PubMed] [Google Scholar]
- 23.Shivdasani RA. MicroRNAs: regulators of gene expression and cell differentiation. Blood. 2006;108(12):3646–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paschou M, Paraskevopoulou MD, Vlachos IS, Koukouraki P, Hatzigeorgiou AG, Doxakis E. miRNA regulons associated with synaptic function. PLoS One. 2012;7(10):e46189 10.1371/journal.pone.0046189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–97. [DOI] [PubMed] [Google Scholar]
- 26.Gurtan AM, Sharp PA. The role of miRNAs in regulating gene expression networks. J Mol Biol. 2013;425(19):3582–600. 10.1016/j.jmb.2013.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sun K, Lai EC. Adult-specific functions of animal microRNAs. Nat Rev Genet. 2013;14(8):535–48. 10.1038/nrg3471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xu S. microRNA expression in the eyes and their significance in relation to functions. Prog Retin Eye Res. 2009;28(2):87–116. 10.1016/j.preteyeres.2008.11.003 [DOI] [PubMed] [Google Scholar]
- 29.Arora A, Guduric-Fuchs J, Harwood L, Dellett M, Cogliati T, Simpson DA. Prediction of microRNAs affecting mRNA expression during retinal development. BMC Dev Biol. 2010;10:1 10.1186/1471-213X-10-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bak M, Silahtaroglu A, Moller M, Christensen M, Rath MF, Skryabin B, et al. MicroRNA expression in the adult mouse central nervous system. RNA. 2008;14(3):432–44. 10.1261/rna.783108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Deo M, Yu JY, Chung KH, Tippens M, Turner DL. Detection of mammalian microRNA expression by in situ hybridization with RNA oligonucleotides. Dev Dyn. 2006;235(9):2538–48. [DOI] [PubMed] [Google Scholar]
- 32.Hackler L Jr, Wan J, Swaroop A, Qian J, Zack DJ. MicroRNA profile of the developing mouse retina. Invest Ophthalmol Vis Sci. 2010;51(4):1823–31. 10.1167/iovs.09-4657 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jin ZB, Hirokawa G, Gui L, Takahashi R, Osakada F, Hiura Y, et al. Targeted deletion of miR-182, an abundant retinal microRNA. Mol Vis. 2009;15:523–33. [PMC free article] [PubMed] [Google Scholar]
- 34.Karali M, Peluso I, Gennarino VA, Bilio M, Verde R, Lago G, et al. miRNeye: a microRNA expression atlas of the mouse eye. BMC Genomics. 2010;11:715 10.1186/1471-2164-11-715 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ryan DG, Oliveira-Fernandes M, Lavker RM. MicroRNAs of the mammalian eye display distinct and overlapping tissue specificity. Mol Vis. 2006;12:1175–84. [PubMed] [Google Scholar]
- 36.Metlapally R, Gonzalez P, Hawthorne FA, Tran-Viet KN, Wildsoet CF, Young TL. Scleral Micro-RNA Signatures in Adult and Fetal Eyes. PLoS One. 2013;8(10):e78984 10.1371/journal.pone.0078984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Genini S, Guziewicz KE, Beltran WA, Aguirre GD. Altered miRNA expression in canine retinas during normal development and in models of retinal degeneration. BMC Genomics. 2014;15:172 10.1186/1471-2164-15-172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soundara Pandi SP, Chen M, Guduric-Fuchs J, Xu H, Simpson DA. Extremely complex populations of small RNAs in the mouse retina and RPE/choroid. Invest Ophthalmol Vis Sci. 2013;54(13):8140–51. 10.1167/iovs.13-12631 [DOI] [PubMed] [Google Scholar]
- 39.La Torre A, Georgi S, Reh TA. Conserved microRNA pathway regulates developmental timing of retinal neurogenesis. Proc Natl Acad Sci U S A. 2013;110(26):E2362–70. 10.1073/pnas.1301837110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Sousa E, Walter LT, Higa GS, Casado OA, Kihara AH. Developmental and functional expression of miRNA-stability related genes in the nervous system. PLoS One. 2013;8(5):e56908 10.1371/journal.pone.0056908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Conte I, Banfi S, Bovolenta P. Non-coding RNAs in the development of sensory organs and related diseases. Cell Mol Life Sci. 2013;70(21):4141–55. 10.1007/s00018-013-1335-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maiorano NA, Hindges R. Non-coding RNAs in retinal development. International journal of molecular sciences. 2012;13(1):558–78. 10.3390/ijms13010558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wei L, Zhou Q, Hou S, Bai L, Liu Y, Qi J, et al. MicroRNA-146a and Ets-1 gene polymorphisms are associated with pediatric uveitis. PLoS One. 2014;9(3):e91199 10.1371/journal.pone.0091199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gonzalez P, Li G, Qiu J, Wu J, Luna C. Role of microRNAs in the trabecular meshwork. Journal of ocular pharmacology and therapeutics: the official journal of the Association for Ocular Pharmacology and Therapeutics. 2014;30(2–3):128–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Qi J, Hou S, Zhang Q, Liao D, Wei L, Fang J, et al. A functional variant of pre-miRNA-196a2 confers risk for Behcet's disease but not for Vogt-Koyanagi-Harada syndrome or AAU in ankylosing spondylitis. Hum Genet. 2013;132(12):1395–404. 10.1007/s00439-013-1346-8 [DOI] [PubMed] [Google Scholar]
- 46.Lumayag S, Haldin CE, Corbett NJ, Wahlin KJ, Cowan C, Turturro S, et al. Inactivation of the microRNA-183/96/182 cluster results in syndromic retinal degeneration. Proc Natl Acad Sci U S A. 2013;110(6):E507–16. 10.1073/pnas.1212655110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hwang W, Hackler L Jr, Wu G, Ji H, Zack DJ, Qian J. Dynamics of regulatory networks in the developing mouse retina. PLoS One. 2012;7(10):e46521 10.1371/journal.pone.0046521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ishida W, Fukuda K, Higuchi T, Kajisako M, Sakamoto S, Fukushima A. Dynamic changes of microRNAs in the eye during the development of experimental autoimmune uveoretinitis. Invest Ophthalmol Vis Sci. 2011;52(1):611–7. 10.1167/iovs.10-6115 [DOI] [PubMed] [Google Scholar]
- 49.Loscher CJ, Hokamp K, Kenna PF, Ivens AC, Humphries P, Palfi A, et al. Altered retinal microRNA expression profile in a mouse model of retinitis pigmentosa. Genome Biol. 2007;8(11):R248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Shen J, Yang X, Xie B, Chen Y, Swaim M, Hackett SF, et al. MicroRNAs regulate ocular neovascularization. Mol Ther. 2008;16(7):1208–16. 10.1038/mt.2008.104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kovacs B, Lumayag S, Cowan C, Xu S. MicroRNAs in early diabetic retinopathy in streptozotocin-induced diabetic rats. Invest Ophthalmol Vis Sci. 2011;52(7):4402–9. 10.1167/iovs.10-6879 [DOI] [PubMed] [Google Scholar]
- 52.Iliff BW, Riazuddin SA, Gottsch JD. A single-base substitution in the seed region of miR-184 causes EDICT syndrome. Invest Ophthalmol Vis Sci. 2012;53(1):348–53. 10.1167/iovs.11-8783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hughes AE, Bradley DT, Campbell M, Lechner J, Dash DP, Simpson DA, et al. Mutation altering the miR-184 seed region causes familial keratoconus with cataract. Am J Hum Genet. 2011;89(5):628–33. 10.1016/j.ajhg.2011.09.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen KC, Hsi E, Hu CY, Chou WW, Liang CL, Juo SH. MicroRNA-328 may influence myopia development by mediating the PAX6 gene. Invest Ophthalmol Vis Sci. 2012;53(6):2732–9. 10.1167/iovs.11-9272 [DOI] [PubMed] [Google Scholar]
- 55.Liang CL, Hsi E, Chen KC, Pan YR, Wang YS, Juo SH. A functional polymorphism at 3'UTR of the PAX6 gene may confer risk for extreme myopia in the Chinese. Invest Ophthalmol Vis Sci. 2011;52(6):3500–5. 10.1167/iovs.10-5859 [DOI] [PubMed] [Google Scholar]
- 56.Mattapallil MJ, Wawrousek EF, Chan CC, Zhao H, Roychoudhury J, Ferguson TA, et al. The Rd8 Mutation of the Crb1 Gene Is Present in Vendor Lines of C57BL/6N Mice and Embryonic Stem Cells, and Confounds Ocular Induced Mutant Phenotypes. Invest Ophthalmol Vis Sci. 2012;53(6):2921–7. 10.1167/iovs.12-9662 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Chase HB. Studies on an Anophthalmic Strain of Mice. III. Results of Crosses with Other Strains. Genetics. 1942;27(3):339–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kalter H. Sporadic congenital malformations of newborn inbred mice. Teratology. 1968;1(2):193–9. [DOI] [PubMed] [Google Scholar]
- 59.Koch FLP, Gowen JW. Spontaneous ophthalmic mutation in a laboratory mouse. Arch Pathol Lab Med. 1939;28:171–6. [Google Scholar]
- 60.Tkatchenko TV, Shen Y, Tkatchenko AV. Mouse experimental myopia has features of primate myopia. Invest Ophthalmol Vis Sci. 2010;51(3):1297–303. 10.1167/iovs.09-4153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Rada JA, Wiechmann AF. Ocular expression of avian thymic hormone: changes during the recovery from induced myopia. Mol Vis. 2009;15:778–92. [PMC free article] [PubMed] [Google Scholar]
- 62.Ashby RS, Feldkaemper MP. Gene expression within the amacrine cell layer of chicks after myopic and hyperopic defocus. Invest Ophthalmol Vis Sci. 2010;51(7):3726–35. 10.1167/iovs.09-4615 [DOI] [PubMed] [Google Scholar]
- 63.Altenhoff AM, Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput Biol. 2009;5(1):e1000262 10.1371/journal.pcbi.1000262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Agarwal V, Bell GW, Nam JW, Bartel DP. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kozomara A, Griffiths-Jones S. miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 2014;42(Database issue):D68–73. 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, et al. Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 2016;44(D1):D733–45. 10.1093/nar/gkv1189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.McGarvey KM, Goldfarb T, Cox E, Farrell CM, Gupta T, Joardar VS, et al. Mouse genome annotation by the RefSeq project. Mamm Genome. 2015;26(9–10):379–90. 10.1007/s00335-015-9585-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Xu J, Wong C. A computational screen for mouse signaling pathways targeted by microRNA clusters. RNA. 2008;14(7):1276–83. 10.1261/rna.997708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rada JA, Shelton S, Norton TT. The sclera and myopia. Exp Eye Res. 2006;82(2):185–200. [DOI] [PubMed] [Google Scholar]
- 70.Rymer J, Wildsoet CF. The role of the retinal pigment epithelium in eye growth regulation and myopia: a review. Vis Neurosci. 2005;22(3):251–61. [DOI] [PubMed] [Google Scholar]
- 71.Morgan IG. The biological basis of myopic refractive error. Clin Exp Optom. 2003;86(5):276–88. [DOI] [PubMed] [Google Scholar]
- 72.Nickla DL, Wallman J. The multifunctional choroid. Prog Retin Eye Res. 2010;29(2):144–68. 10.1016/j.preteyeres.2009.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sass S, Dietmann S, Burk UC, Brabletz S, Lutter D, Kowarsch A, et al. MicroRNAs coordinately regulate protein complexes. BMC systems biology. 2011;5:136 10.1186/1752-0509-5-136 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Tkatchenko TV, Shen Y, Tkatchenko AV. Analysis of postnatal eye development in the mouse with high-resolution small animal magnetic resonance imaging. Invest Ophthalmol Vis Sci. 2010;51(1):21–7. 10.1167/iovs.08-2767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Trumbach D, Prakash N. The conserved miR-8/miR-200 microRNA family and their role in invertebrate and vertebrate neurogenesis. Cell Tissue Res. 2015;359(1):161–77. 10.1007/s00441-014-1911-z [DOI] [PubMed] [Google Scholar]
- 76.Bracken CP, Li X, Wright JA, Lawrence DM, Pillman KA, Salmanidis M, et al. Genome-wide identification of miR-200 targets reveals a regulatory network controlling cell invasion. EMBO J. 2014;33(18):2040–56. 10.15252/embj.201488641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Engelsvold DH, Utheim TP, Olstad OK, Gonzalez P, Eidet JR, Lyberg T, et al. miRNA and mRNA expression profiling identifies members of the miR-200 family as potential regulators of epithelial-mesenchymal transition in pterygium. Exp Eye Res. 2013;115:189–98. 10.1016/j.exer.2013.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wang G, Guo X, Hong W, Liu Q, Wei T, Lu C, et al. Critical regulation of miR-200/ZEB2 pathway in Oct4/Sox2-induced mesenchymal-to-epithelial transition and induced pluripotent stem cell generation. Proc Natl Acad Sci U S A. 2013;110(8):2858–63. 10.1073/pnas.1212769110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Xia H, Cheung WK, Sze J, Lu G, Jiang S, Yao H, et al. miR-200a regulates epithelial-mesenchymal to stem-like transition via ZEB2 and beta-catenin signaling. J Biol Chem. 2010;285(47):36995–7004. 10.1074/jbc.M110.133744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gregory PA, Bert AG, Paterson EL, Barry SC, Tsykin A, Farshid G, et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nature cell biology. 2008;10(5):593–601. 10.1038/ncb1722 [DOI] [PubMed] [Google Scholar]
- 81.Diaz-Martin J, Diaz-Lopez A, Moreno-Bueno G, Castilla MA, Rosa-Rosa JM, Cano A, et al. A core microRNA signature associated with inducers of the epithelial-to-mesenchymal transition. J Pathol. 2014;232(3):319–29. 10.1002/path.4289 [DOI] [PubMed] [Google Scholar]
- 82.Vallejo DM, Caparros E, Dominguez M. Targeting Notch signalling by the conserved miR-8/200 microRNA family in development and cancer cells. EMBO J. 2011;30(4):756–69. 10.1038/emboj.2010.358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Lin CH, Jackson AL, Guo J, Linsley PS, Eisenman RN. Myc-regulated microRNAs attenuate embryonic stem cell differentiation. EMBO J. 2009;28(20):3157–70. 10.1038/emboj.2009.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Li Z, Gou J, Jia J, Zhao X. MicroRNA-429 functions as a regulator of epithelial-mesenchymal transition by targeting Pcdh8 during murine embryo implantation. Human reproduction. 2015;30(3):507–18. 10.1093/humrep/dev001 [DOI] [PubMed] [Google Scholar]
- 85.Sinha M, Ghose J, Bhattarcharyya NP. Micro RNA -214,-150,-146a and-125b target Huntingtin gene. RNA Biol. 2011;8(6):1005–21. 10.4161/rna.8.6.16035 [DOI] [PubMed] [Google Scholar]
- 86.Zhang HY, Zheng SJ, Zhao JH, Zhao W, Zheng LF, Zhao D, et al. MicroRNAs 144, 145, and 214 are down-regulated in primary neurons responding to sciatic nerve transection. Brain Res. 2011;1383:62–70. 10.1016/j.brainres.2011.01.067 [DOI] [PubMed] [Google Scholar]
- 87.Calissano M, Latchman DS. Cell-specific regulation of the pro-survival Brn-3b transcription factor by microRNAs. Mol Cell Neurosci. 2010;45(4):317–23. 10.1016/j.mcn.2010.06.015 [DOI] [PubMed] [Google Scholar]
- 88.Chen H, Shalom-Feuerstein R, Riley J, Zhang SD, Tucci P, Agostini M, et al. miR-7 and miR-214 are specifically expressed during neuroblastoma differentiation, cortical development and embryonic stem cells differentiation, and control neurite outgrowth in vitro. Biochem Biophys Res Commun. 2010;394(4):921–7. 10.1016/j.bbrc.2010.03.076 [DOI] [PubMed] [Google Scholar]
- 89.Decembrini S, Bressan D, Vignali R, Pitto L, Mariotti S, Rainaldi G, et al. MicroRNAs couple cell fate and developmental timing in retina. Proc Natl Acad Sci U S A. 2009;106(50):21179–84. 10.1073/pnas.0909167106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Wang HC, Greene WA, Kaini RR, Shen-Gunther J, Chen HI, Cai H, et al. Profiling the microRNA Expression in Human iPS and iPS-derived Retinal Pigment Epithelium. Cancer informatics. 2014;13(Suppl 5):25–35. 10.4137/CIN.S14074 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Kushwaha R, Thodima V, Tomishima MJ, Bosl GJ, Chaganti RS. miR-18b and miR-518b Target FOXN1 during epithelial lineage differentiation in pluripotent cells. Stem Cells Dev. 2014;23(10):1149–56. 10.1089/scd.2013.0262 [DOI] [PubMed] [Google Scholar]
- 92.Singh SK, Marisetty A, Sathyan P, Kagalwala M, Zhao Z, Majumder S. REST-miR-21-SOX2 axis maintains pluripotency in E14Tg2a.4 embryonic stem cells. Stem cell research. 2015;15(2):305–11. 10.1016/j.scr.2015.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Di Bernardini E, Campagnolo P, Margariti A, Zampetaki A, Karamariti E, Hu Y, et al. Endothelial lineage differentiation from induced pluripotent stem cells is regulated by microRNA-21 and transforming growth factor beta2 (TGF-beta2) pathways. J Biol Chem. 2014;289(6):3383–93. 10.1074/jbc.M113.495531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Lu TX, Lim EJ, Itskovich S, Besse JA, Plassard AJ, Mingler MK, et al. Targeted ablation of miR-21 decreases murine eosinophil progenitor cell growth. PLoS One. 2013;8(3):e59397 10.1371/journal.pone.0059397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Mei Y, Bian C, Li J, Du Z, Zhou H, Yang Z, et al. miR-21 modulates the ERK-MAPK signaling pathway by regulating SPRY2 expression during human mesenchymal stem cell differentiation. J Cell Biochem. 2013;114(6):1374–84. 10.1002/jcb.24479 [DOI] [PubMed] [Google Scholar]
- 96.Niu Z, Goodyear SM, Rao S, Wu X, Tobias JW, Avarbock MR, et al. MicroRNA-21 regulates the self-renewal of mouse spermatogonial stem cells. Proc Natl Acad Sci U S A. 2011;108(31):12740–5. 10.1073/pnas.1109987108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Kim YJ, Hwang SJ, Bae YC, Jung JS. MiR-21 regulates adipogenic differentiation through the modulation of TGF-beta signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells. 2009;27(12):3093–102. 10.1002/stem.235 [DOI] [PubMed] [Google Scholar]
- 98.Singh SK, Kagalwala MN, Parker-Thornburg J, Adams H, Majumder S. REST maintains self-renewal and pluripotency of embryonic stem cells. Nature. 2008;453(7192):223–7. 10.1038/nature06863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen J, Wang G, Lu C, Guo X, Hong W, Kang J, et al. Synergetic cooperation of microRNAs with transcription factors in iPS cell generation. PLoS One. 2012;7(7):e40849 10.1371/journal.pone.0040849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Li D, Zhan S, Wang Y, Wang L, Zhong T, Li L, et al. Role of microRNA-101a in the regulation of goat skeletal muscle satellite cell proliferation and differentiation. Gene. 2015;572(2):198–204. 10.1016/j.gene.2015.07.010 [DOI] [PubMed] [Google Scholar]
- 101.Augustin R, Endres K, Reinhardt S, Kuhn PH, Lichtenthaler SF, Hansen J, et al. Computational identification and experimental validation of microRNAs binding to the Alzheimer-related gene ADAM10. BMC Med Genet. 2012;13:35 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Turczynska KM, Sadegh MK, Hellstrand P, Sward K, Albinsson S. MicroRNAs are essential for stretch-induced vascular smooth muscle contractile differentiation via microRNA (miR)-145-dependent expression of L-type calcium channels. J Biol Chem. 2012;287(23):19199–206. 10.1074/jbc.M112.341073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang B, Guo H, Zhang Y, Chen L, Ying D, Dong S. MicroRNA-145 regulates chondrogenic differentiation of mesenchymal stem cells by targeting Sox9. PLoS One. 2011;6(7):e21679 10.1371/journal.pone.0021679 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Lee SK, Teng Y, Wong HK, Ng TK, Huang L, Lei P, et al. MicroRNA-145 regulates human corneal epithelial differentiation. PLoS One. 2011;6(6):e21249 10.1371/journal.pone.0021249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Cordes KR, Sheehy NT, White MP, Berry EC, Morton SU, Muth AN, et al. miR-145 and miR-143 regulate smooth muscle cell fate and plasticity. Nature. 2009;460(7256):705–10. 10.1038/nature08195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chivukula RR, Mendell JT. Abate and switch: miR-145 in stem cell differentiation. Cell. 2009;137(4):606–8. 10.1016/j.cell.2009.04.059 [DOI] [PubMed] [Google Scholar]
- 107.Xu N, Papagiannakopoulos T, Pan G, Thomson JA, Kosik KS. MicroRNA-145 regulates OCT4, SOX2, and KLF4 and represses pluripotency in human embryonic stem cells. Cell. 2009;137(4):647–58. 10.1016/j.cell.2009.02.038 [DOI] [PubMed] [Google Scholar]
- 108.Banerjee P, Dutta S, Pal R. Dysregulation of Wnt-signaling and a candidate set of miRNAs underlie the effect of metformin on neural crest cell development. Stem Cells. 2015. [DOI] [PubMed] [Google Scholar]
- 109.Zeng L, Carter AD, Childs SJ. miR-145 directs intestinal maturation in zebrafish. Proc Natl Acad Sci U S A. 2009;106(42):17793–8. 10.1073/pnas.0903693106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Papadopoulos EI, Fragoulis EG, Scorilas A. Human L-DOPA decarboxylase mRNA is a target of miR-145: A prediction to validation workflow. Gene. 2015;554(2):174–80. 10.1016/j.gene.2014.10.043 [DOI] [PubMed] [Google Scholar]
- 111.Nguyen-Legros J, Krieger M, Simon A. Immunohistochemical localization of L-dopa and aromatic L-amino acid-decarboxylase in the rat retina. Invest Ophthalmol Vis Sci. 1994;35(7):2906–15. [PubMed] [Google Scholar]
- 112.Stone RA, Laties AM, Raviola E, Wiesel TN. Increase in retinal vasoactive intestinal polypeptide after eyelid fusion in primates. Proc Natl Acad Sci U S A. 1988;85(1):257–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Seltner RL, Stell WK. The effect of vasoactive intestinal peptide on development of form deprivation myopia in the chick: a pharmacological and immunocytochemical study. Vision Res. 1995;35(9):1265–70. [DOI] [PubMed] [Google Scholar]
- 114.Fischer AJ, Seltner RL, Stell WK. N-methyl-D-aspartate-induced excitotoxicity causes myopia in hatched chicks. Can J Ophthalmol. 1997;32(6):373–7. [PubMed] [Google Scholar]
- 115.Fischer AJ, McGuire JJ, Schaeffel F, Stell WK. Light- and focus-dependent expression of the transcription factor ZENK in the chick retina. Nat Neurosci. 1999;2(8):706–12. [DOI] [PubMed] [Google Scholar]
- 116.Feldkaemper MP, Schaeffel F. Evidence for a potential role of glucagon during eye growth regulation in chicks. Vis Neurosci. 2002;19(6):755–66. [DOI] [PubMed] [Google Scholar]
- 117.Zhong X, Ge J, Smith EL III, Stell WK. Image defocus modulates activity of bipolar and amacrine cells in macaque retina. Invest Ophthalmol Vis Sci. 2004;45(7):2065–74. [DOI] [PubMed] [Google Scholar]
- 118.Vessey KA, Lencses KA, Rushforth DA, Hruby VJ, Stell WK. Glucagon receptor agonists and antagonists affect the growth of the chick eye: a role for glucagonergic regulation of emmetropization? Invest Ophthalmol Vis Sci. 2005;46(11):3922–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen JC, Brown B, Schmid KL. Evaluation of inner retinal function in myopia using oscillatory potentials of the multifocal electroretinogram. Vision Res. 2006;46(24):4096–103. [DOI] [PubMed] [Google Scholar]
- 120.Mathis U, Schaeffel F. Glucagon-related peptides in the mouse retina and the effects of deprivation of form vision. Graefes Arch Clin Exp Ophthalmol. 2007;245(2):267–75. [DOI] [PubMed] [Google Scholar]
- 121.Feldkaemper MP, Neacsu I, Schaeffel F. Insulin acts as a powerful stimulator of axial myopia in chicks. Invest Ophthalmol Vis Sci. 2009;50(1):13–23. 10.1167/iovs.08-1702 [DOI] [PubMed] [Google Scholar]
- 122.Ashby R, Kozulin P, Megaw PL, Morgan IG. Alterations in ZENK and glucagon RNA transcript expression during increased ocular growth in chickens. Mol Vis. 2010;16:639–49. [PMC free article] [PubMed] [Google Scholar]
- 123.Tkatchenko AV, Tkatchenko TV, Guggenheim JA, Verhoeven VJ, Hysi PG, Wojciechowski R, et al. APLP2 Regulates Refractive Error and Myopia Development in Mice and Humans. PLoS Genet. 2015;11(8):e1005432 10.1371/journal.pgen.1005432 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Logarithmic values (base 2) of Agilent total gene signal for differentially expressed miRNAs (cutoff: FC > 2, FDR-adjusted p-value < 0.05) were quantile normalized, shifted to mean zero, scaled to standard deviation of 1.0 and subjected to hierarchical clustering using Euclidean dissimilarity and average linkage. The color scale indicates transcript abundance: red identifies an increase in relative miRNA abundance; blue identifies a decrease in relative miRNA abundance. Columns show individual samples, whereas rows show individual miRNAs.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #1 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #2 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #3 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #4 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #5 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #6 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #7 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #8 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(A) Diagram showing interactions between miRNAs differentially expressed in myopic retina and their target mRNAs. Arrows show relationships between different miRNAs and mRNAs. (B) Overlap between signaling pathway #9 and top gene ontology categories. Red identifies genes/categories which are up-regulated in myopic retina, whereas green identifies genes/categories which are down-regulated in myopic retina.
(TIF)
(XLSX)
(XLSX)
(DOCX)
(XLSX)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files except for the gene expression data, which were deposited in the Gene Expression Omnibus (Accession: GSE84220).