Abstract
Primary open angle glaucoma (POAG) is a complex disease and is one of the major leading causes of blindness worldwide. Genome-wide association studies have successfully identified several common variants associated with glaucoma; however, most of these variants only explain a small proportion of the genetic risk. Apart from the standard approach to identify main effects of variants across the genome, it is believed that gene-gene interactions can help elucidate part of the missing heritability by allowing for the test of interactions between genetic variants to mimic the complex nature of biology. To explain the etiology of glaucoma, we first performed a genome-wide association study (GWAS) on glaucoma case-control samples obtained from electronic medical records (EMR) to establish the utility of EMR data in detecting non-spurious and relevant associations; this analysis was aimed at confirming already known associations with glaucoma and validating the EMR derived glaucoma phenotype. Our findings from GWAS suggest consistent evidence of several known associations in POAG. We then performed an interaction analysis for variants found to be marginally associated with glaucoma (SNPs with main effect p-value <0.01) and observed interesting findings in the electronic MEdical Records and GEnomics Network (eMERGE) network dataset. Genes from the top epistatic interactions from eMERGE data (Likelihood Ratio Test i.e. LRT p-value <1e-05) were then tested for replication in the NEIGHBOR consortium dataset. To replicate our findings, we performed a gene-based SNP-SNP interaction analysis in NEIGHBOR and observed significant gene-gene interactions (p-value <0.001) among the top 17 gene-gene models identified in the discovery phase. Variants from gene-gene interaction analysis that we found to be associated with POAG explain 3.5% of additional genetic variance in eMERGE dataset above what is explained by the SNPs in genes that are replicated from previous GWAS studies (which was only 2.1% variance explained in eMERGE dataset); in the NEIGHBOR dataset, adding replicated SNPs from gene-gene interaction analysis explain 3.4% of total variance whereas GWAS SNPs alone explain only 2.8% of variance. Exploring gene-gene interactions may provide additional insights into many complex traits when explored in properly designed and powered association studies.
Author Summary
The complex nature of primary-open angle glaucoma (POAG) has left researchers exploring the genetic architecture and searching for the missing heritability using a number of different study designs. Over the past decade, many studies have been conducted to explain the etiology of POAG; however, a high proportion of estimated heritability still remains unexplained. GWA studies for POAG have identified significant associations but these associations have only explained a small proportion of the genetic risk (odds ratios range between 1–3). In this paper, we sought to confirm the primary genome-wide significant associations that have been discovered so far for glaucoma in phenotypes developed from EMR data in an effort to show that EMR data can be a powerful resource for finding genetic variants influencing POAG susceptibility. Next, we tested for statistical interactions, which can be presented as an important tool in an attempt to explain POAG heritability. We used a reduced list of variants filtered by marginal main effect analysis to look for epistatic interactions. We present our results from replication of gene-based interaction analyses performed in eMERGE and the NEIGHBOR consortium data. Using expression data and annotations from various publicly available databases, the most significant genes that replicated in our analyses show expression in the eye and trabecular meshwork. Analysis for estimation of genetic variance explained by significant associations from previous GWAS and replicated variants from gene-based interactions suggest that these explain 5.6% of variance in eMERGE dataset and also explain 3.4% variance in NEIGHBOR dataset.
Introduction
Glaucoma, a chronic degenerative optic neuropathy that results in loss of retinal ganglion cells and axons, is one of the primary causes of irreversible visual impairment worldwide affecting approximately 70 million people[1]. Twin and relative studies have estimated the heritability of primary open angle glaucoma (POAG) to be between 16–20%[2,3]. Genetic linkage studies have only identified a common mutation found in MYOC which explains a very small fraction of total risk in different populations[4]. Genome-wide association studies (GWAS) have proven to be a successful tool in identifying several loci and genes associated with POAG which have elucidated important biological information that has enhanced some understanding of the genetic architecture of this disease[5–8]. Family based studies and GWAS taken together still only explain less than 10% of the heritability of POAG[9].
Phenotypes from electronic medical records (EMR) data are curated and linked to genetic bio-repositories in the eMERGE (electronic MEdical Records and GEnomics) network at several participating institutions nationwide[10–12]. In eMERGE, genotypes in more than 55,000 samples from 9 sites and 9 different genotyping platforms have been imputed to the 1000 Genomes reference panel[13]. We conducted a GWAS on 5,090 eMERGE samples (961 cases and 4,129 controls) that were extracted from the dataset containing ~55,000 samples for the purpose of confirming known signals from previous POAG GWAS to show that EMRs are a powerful resource that can be utilized to explore genetic associations for complex traits. In addition to disease- associated SNPs identified by GWAS, some studies have also identified interactions between variants in genes OPTN and OLFM2 to be significantly associated with open angle glaucoma (OAG)[14]. The genetic architecture of traits with complex inheritance such as POAG are expected to include complex genetic interactions, that in part, can also account for disease heritability [15–19]. Many studies in model organisms such as drosophila and mouse have shown strong evidence of epistasis, or gene-gene interactions, in complex phenotypes [20,21]. Because of the complex nature of POAG, it is possible that disease risk is not mediated only by single loci but also by a harmonious links between different genes.
Many computational techniques have emerged in recent years to explore genetic interactions, yet numerous challenges must be overcome to conduct these analyses, including extensive computational resources and time required to run analyses. Further, exhaustively searching for pairwise SNP-SNP interactions leads to issues with multiple testing corrections, which makes discovering interactions even more difficult. To limit the search space, it is important to reduce the number of variants being tested[22]. To address this issue, we limited our SNP-SNP interaction analysis in the eMERGE dataset (Discovery dataset) to only those SNPs attaining marginal to strong association (p-value < 0.01) in the eMERGE POAG GWAS.
Additional analytic challenges include biological interpretation of all SNP-SNP interactions. Replication of the exact SNP pairs is extremely difficult and still remains an exasperating task as it is complicated by linkage disequilibrium (LD) pattern differences as well as genetic and/or clinical heterogeneity [23–25]. Genetic heterogeneity possesses a challenge in replicating exact variants associated with a disease in different populations, as the variants are spread across multiple loci in a gene or are in high linkage disequilibrium (LD) with other variants. It has been previously reported from studies in model organisms such as yeast that understanding the genetic architecture of diseases precisely in heterogeneous populations such as humans is an arduous task [26–28]. Therefore, to overcome the problem of replicating exact SNP-SNP pairs and address the challenge of heterogeneity, we annotated SNPs to genes and then extracted all SNPs in the top interacting models (results with likelihood ratio test (LRT) p-value<1e-05 in the Discovery dataset) to perform a gene-based SNP-SNP interaction analyses followed by replication in the NEIGHBOR consortium (Replication dataset; previously genotyped and reported) [29]. The dataset, which includes 2,132 POAG cases and 2,290 non-POAG controls, was then imputed to the 1000Genomes reference dataset[30]. In this study, we report our top replicated findings from the first gene-based interaction study in POAG conducted on two independent datasets.
Results
Generalization results from GWAS
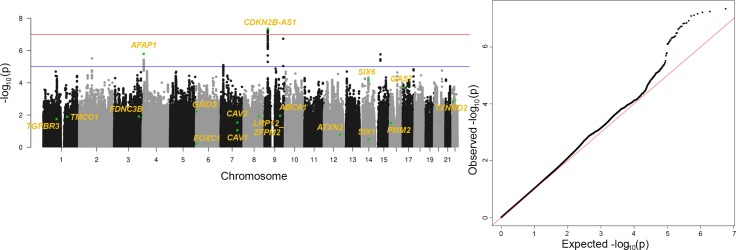
To identify previously reported associations between SNPs and POAG disease status as defined by the EMR algorithm, we performed a genome-wide scan with samples classified as POAG cases (N = 961) and controls (N = 4,129). The POAG cases were electronically phenotyped using the EMR data in eMERGE (details on the phenotypic algorithm for POAG are provided in supplementary material S1 Text). Our analysis was adjusted for the first six principal components, age, sex, genotyping platform, and eMERGE site. Fig 1A illustrates the results for the primary GWAS analysis in a Manhattan plot. SNPs in genes that have known association with glaucoma were annotated with gene names for simplicity of interpretation of results. Fig 1B shows a quantile-quantile plot of observed and expected p-values. The genomic inflation factor (λ) is 1.04, which suggests that there is minimal inflation in our results.
Fig 1. POAG GWAS Results in eMERGE dataset.
(A) Manhattan plot shows chromosome and base pair positions on x-axis and -log10 of p-value on y-axis. Each point here represents a SNP. Red line is the genome-wide significance line at 5e-08. Highlighted points indicate annotation of SNPs with previously reported GWAS associated genes. (B) Shows a QQ-plot from GWAS analysis.
From the GWAS analysis, we reviewed 17 previously reported glaucoma-associated loci that have attained genome-wide significance (i.e. p-values<5e-08) according to the NHGRI GWAS catalog in one or more studies[31]. Table 1shows all of the known loci as of March 25, 2016 and with the p-values obtained from our analyses. Although only one of our results reaches genome-wide significance level (5e-08), six of our results reach Bonferroni significance (0.05/17 tests), and 13/17 exceed a p-value <0.05; thus, we observe p-values for known associations also clearly shows that EMR data can produce non-spurious and relevant findings. Thus, we believe that the data derived from the EMR were able to replicate many of the signals and demonstrate that in our EHR-derived phenotype we can identify association signals with glaucoma. A point to note here is that based on previous GWAS’s as cited from NHGRI GWAS catalog, not all loci reach genome-wide significance in all studies. Here we have compiled lists of all loci that have reached genome-wide significance in a minimum of 1 study. For example, variants in gene PMM2 have so far only being replicated in Chinese populations. Based on power calculations, we had approximately 45% power to detect a common allele with odds ratio greater than 1.5 at genome wide significance (S1 Table shows power calculation results considering odds ratio in range on 1–3 as reported in previous GWAS studies for POAG in the NHGRI GWAS catalog and MAF range of 0.05–0.5). Power calculations were done using Quanto available at: http://biostats.usc.edu/Quanto.html. Thus, we demonstrate that existing EMR data can be extensively utilized in confirming known GWAS hits and are powerful resources for applying new methodologies to explore the genetic architecture of complex traits.
Table 1. Results from GWAS analysis, showing previously reported gene regions that are known to be associated with glaucoma; the lowest p-value of the gene region in eMERGE GWAS analysis for glaucoma is reported.
| Gene Region | Chromosome | Start Position | End Position | SNP | Lowest P-value | Odds Ratio | Reference Study |
|---|---|---|---|---|---|---|---|
| CDKN2B-AS | 9 | 21984777 | 22131096 | rs7866783 | 4.49E-08 | 1.34 | [57]Osman W (PMID: 22419738) |
| AFAP1 | 4 | 7750440 | 7951653 | rs1320074 | 1.61E-06 | 0.77 | [5]Gharahkhani P (PMID: 25173105) |
| SIX6 | 14 | 60965669 | 60989568 | rs1018533 | 6.65E-05 | 0.80 | [5]Gharahkhani P (PMID: 25173105) |
| [57]Osman W (PMID: 22419738) | |||||||
| GAS7 | 17 | 9803926 | 10111868 | rs12150284 | 1.95E-04 | 1.22 | [58]van Koolwijk LM (PMID: 22570627) |
| TXNRD2 | 22 | 19853040 | 19939359 | rs58714937 | 1.43E-03 | 1.26 | [30]Cooke Bailey J (PMID: 26752265) |
| GMDS | 6 | 1614035 | 2255868 | rs13217030 | 6.11E-03 | 1.21 | [5]Gharahkhani P (PMID: 25173105) |
| ABCA1 | 9 | 107533283 | 107790527 | rs2472495 | 1.10E-02 | 1.17 | [45]Chen Y (PMID: 25173107) |
| FNDC3B | 3 | 171747418 | 172128493 | rs12897 | 1.24E-02 | 0.87 | [32]Lu Y(PMID: 23291589) |
| 8q22 Region: LRP12_ZFPM2 | 8 | 105491459 | 106826767 | rs1545699 | 1.23E-02 | 0.86 | [29]Wiggs J (PMID: 22570617) |
| TMCO1 | 1 | 165683528 | 165806992 | rs4233408 | 1.28E-02 | 0.82 | [58]van Koolwijk LM (PMID: 22570627) |
| [5]Gharahkhani P (PMID: 25173105) | |||||||
| TGFBR3 | 1 | 92135900 | 92381559 | rs2810903 | 1.77E-02 | 1.24 | [33]Li Z (PMID: 25861811) |
| PMM2 | 16 | 8890670 | 8944194 | rs8057024 | 3.18E-02 | 1.11 | [45]Chen Y (PMID: 25173107) |
| CAV2 | 7 | 115917434 | 116158595 | rs75347112 | 2.91E-02 | 0.79 | [7]Thorleifsson G (PMID: 20835238) |
| CAV1 | 7 | 116154839 | 116211239 | rs7795510 | 8.84E-02 | 1.09 | [7]Thorleifsson G (PMID: 20835238) |
| ATXN2 | 12 | 111880018 | 112047480 | rs653178 | 1.60E-01 | 1.08 | [30]Cooke Bailey J (PMID: 26752265) |
| SIX1 | 14 | 61100133 | 61134977 | rs7152548 | 3.17E-01 | 0.90 | [57]Osman W (PMID: 22419738) |
| FOXC1 | 6 | 1600681 | 1624132 | rs2235718 | 6.15E-01 | 0.96 | [30] Cooke Bailey J (PMID: 26752265) |
Gene based SNP-SNP Interactions
Detection of epistasis is of interest in exploring the etiology of complex diseases. In the present study, markers that showed suggestive marginal association with POAG from the eMERGE GWAS (4,624 SNPs with p-value<0.01) were chosen as potential candidates to detect epistatic interactions between genes, a strategy also used by many others (reviewed by Sun et al 2014) [32]. We performed this analysis in all 5,090 eMERGE samples and also in samples only from European American (EA) ancestry (4,840 samples). In testing for epistasis, we performed a pairwise SNP-SNP interaction analysis i.e. each from the 4,624 SNPs were tested against the remaining 4,623 SNPs. To examine the distribution of observed p-values in this discovery dataset, we plotted the p-values in a quantile-quantile plot to compare against random expected p-values. This qq-plot is shown in S1 Fig. We also calculated the inflation factor for the observed values using the GenABEL package in R[34] and the inflation factor was 1.03. SNPs within or 10kb upstream or downstream of the gene location were mapped to 2,433 genes using the Library of Knowledge Integration (LOKI) database compiler in Biofilter 2.0[33]. From the eMERGE dataset, we chose a p-value threshold (1e-05) to obtain approximately the top 100 results to consider for replication. Next, to empirically estimate the null distribution of the test statistics in the discovery dataset, we conducted the Kolmogorov-Smirnov test for all p-values above the chosen threshold (1e-05) to test the null hypothesis that p-values come from a uniform distribution. The test resulted in a p-value of 0.28 which suggests that we did not have proper evidence to reject the null hypothesis. Using a LRT p-value <1e-05 as a threshold, we observed 117 SNP-SNP models that mapped to 91 genes. SNPs (151 out of 224) not located within 10Kb region of a gene were not tested in replication based on the definition of a gene as described above. A total of 3,839 SNPs were available in the NEIGHBOR dataset that mapped to the 91 genes from eMERGE and were subsequently moved forward to the replication stage. There were 1,361 results that passed p-value criteria for marginal replication i.e. LRT p-value <0.01 in the NEIGHBOR dataset (S2 Table). Considering LRT p-value <0.001 (to get the top 100 models) for the NEIGHBOR dataset, we observed 117 SNP-SNP models in 33 unique genes that replicated consistently with the eMERGE results. The flow of this process is explained in detail in Fig 2. Since we tested 91 unique genes in NEIGHBOR dataset, we calculated Bonferroni significance at alpha = 0.05 based on these 91 genes (i.e. 0.05/4095 = 1.2E-05 where 4095 is the number of tests for 91 genes), 3 models (ALX4-RBFOX1, OPCML-RYR3 and ZNF385B-ELMO1) passed Bonferroni corrected p-value (1.2E-05) at alpha = 0.05. Fig 3illustrates all unique gene-gene models (resulted in 17 unique models from a total of 117 SNP-SNP non-unique models) and their respective lowest p-values from the models in both eMERGE and NEIGHBOR datasets. Supplementary S2 Table shows all non-unique results that were replicated from both all eMERGE samples as well as only EA eMERGE samples. It is evident from the table that many tag SNPs spread across the same genes show similar interactions, supporting the approach of gene-based interaction analysis rather than finding exact SNP-SNP interaction in both discovery and replication analyses.
Fig 2. Flow chart explaining all the steps for gene based interaction analysis in discovery (eMERGE) and replication dataset (NEIGHBOR).
Fig 3. Synthesis view plot representing unique gene-gene interaction models and their p-values in discovery and replication dataset.
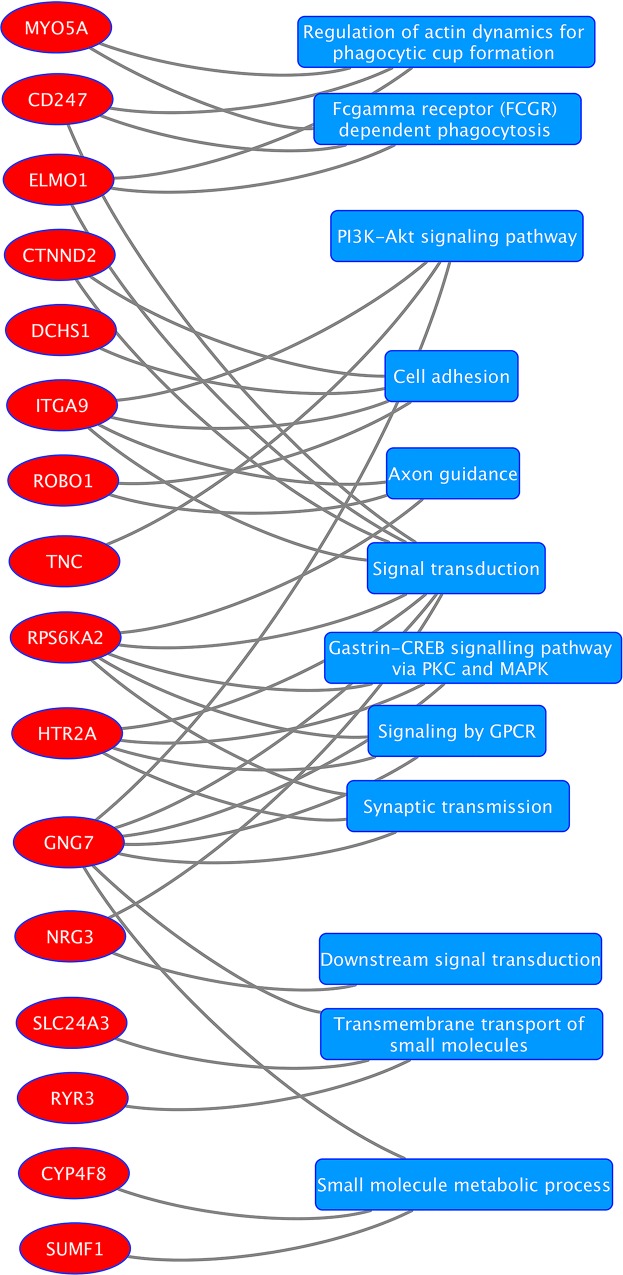
As additional follow-up, we explored which of the 33 unique genes comprising the 17 unique gene-gene models (described above) showed evidence of expression in eye tissue using two online databases: Tissue-specific Gene Expression and Regulation (TiGER) and BioGPS[35]. Fig 4shows a gene-gene interaction circular plot among the 33 unique genes color-coded by the genes that are expressed (red) and not expressed (grey) in eye based on these databases. Next, we looked for expression of these genes in different eye tissues using the ocular tissue database[36] and reported the expression of the genes found in the database in 10 different eye tissues in Fig 5. We also annotated these 33 unique replicating genes to pathways using Biofilter 2.0. To evaluate the presence of genes in major pathways, only pathways supported by at least 2 sources in Biofilter were considered. Since Biofilter reports results from all the data sources, we observed some annotations whose relevance could not be established and these annotations were discarded. We carefully reviewed pathway annotations from Biofilter and removed results that were not applicable. Fig 6represents a Cytoscape hierarchal plot[37] for 16 of the 33 genes that were annotated to 11 major pathways.
Fig 4. Circular plot representing only unique gene-gene interactions.
Genes are colored as red if they are expressed in eye and as grey if not expressed in the eye. All genes are ordered by chromosome. Links between the genes represent gene-gene interactions that are replicated.
Fig 5. Heat map representing expression of the unique genes found in the ocular tissue database from GXG interaction results.
Here, the x-axis lists all genes and y-axis lists the names of 10 tissues from the eye. Scale and color intensity represent PLIER values as reported in the ocular tissue database.
Fig 6. Hierarchal plot showing 16 genes in red circles and 11 pathways in blue rectangles.
Connection between the genes and pathways was established using Biofilter 2.0.
Lastly, to investigate genetic variance explained by these SNPs, we combined 17 SNPs from Table 1 with the 34 SNPs from Table 2 to estimate variance of POAG risk. Analysis in eMERGE data using GCTA[38] suggests that GWAS SNPs explain 2.1% of the variance. Adding top significant SNPs from the interaction analysis performed here increases the explained estimated variance to 5.6%. Analysis in NEIGHBOR dataset for GWAS as well as SNPs replicated from interaction study explain 3.4% of the total variance. Variance explained in the NEIGHBOR dataset is less than the variance explained in eMERGE mainly because the disease prevalence (1.86% as suggested by National Eye Institute (https://nei.nih.gov/eyedata/glaucoma)) used in this analysis is low as compared to the proportion of cases and controls as suggested in GCTA manual[38].
Table 2. All replicating gene-gene interactions at LRT p-value <1e-04 for replication dataset.
| Gene1_Gene2 | eMERGE Var1 | eMERGE Var2 | eMERGE LRT P-val | Neighbor Var1 | Neighbor Var2 | Neighbor LRT P-val |
|---|---|---|---|---|---|---|
| ALX4_RBFOX1 | rs10838251 | rs653127 | 7.29E-06 | rs7126447 | rs11077011 | 1.62E-06 |
| OPCML_RYR3 | rs2246352 | rs2467565 | 4.34E-06 | rs2659614 | rs8028442 | 1.01E-05 |
| ZNF385B_ELMO1 | rs2138196 | rs918978 | 6.53E-06 | rs13408749 | rs2392477 | 1.23E-05 |
| CTNND2_NRG3 | rs1302802 | rs11191761 | 9.23E-06 | rs2973515 | rs660464 | 1.60E-05 |
| TMEM97P2_SLC24A3 | rs7836543 | rs7264540 | 9.24E-06 | rs7826784 | rs11907443 | 1.12E-04 |
| CD247_CYP4F8 | rs864537 | rs2239367* | 8.88E-06 | rs704856 | rs3794989 | 1.24E-04 |
| CCDC3_GTF3C1 | rs2399892 | rs4787965 | 6.78E-06 | rs2580904 | rs7184872 | 1.91E-04 |
| ROBO1_HTR2A | rs6802127 | rs2070039 | 5.44E-06 | rs1444480 | rs9534510 | 2.18E-04 |
| ERC2_TNC | rs7637114 | rs7035322 | 4.88E-06 | rs815443 | rs7847271 | 2.42E-04 |
| CARS2_SIRPB2 | rs389656* | rs12480584 | 1.83E-06 | rs2304767 | rs2422575 | 3.83E-04 |
| FAM167A_NAV2 | rs7018324 | rs10734289 | 6.59E-06 | rs2618433 | rs2584848 | 3.90E-04 |
| SUMF1_RPS6KA2 | rs11919486 | rs12208871 | 2.39E-06 | rs13096374 | rs1883361 | 4.32E-04 |
| C8orf12_NAV2 | rs7018324 | rs10734289 | 6.59E-06 | rs10105588 | rs2243437 | 6.09E-04 |
| SETDB1_ITGA9 | rs34207591 | rs4678971 | 9.66E-06 | rs198325 | rs78196452 | 6.47E-04 |
| NLN_DCHS1 | rs10096 | rs937856 | 2.97E-06 | rs1309822 | rs2659863 | 7.03E-04 |
| MYO5A_GNG7 | rs1669859 | rs759065 | 3.01E-06 | rs2693467 | rs7258864 | 7.84E-04 |
| LOC101928114_LOC101928703 | rs12491137 | rs6840100 | 9.30E-06 | rs62248525 | rs75134552 | 7.96E-04 |
Discussion
We have successfully completed a discovery and replication study to test gene-based interactions in two large POAG datasets: eMERGE and the NEIGHBOR. We have identified several gene-gene interactions associated with glaucoma and showed that most of the genes are expressed in the eye. We annotated all replicating genes from the gene-gene interaction analysis to pathways and observed that 16 genes from the analysis were mapped to 11 major pathways based on the sources available in Biofilter.
Our results show several genes involved in cell adhesion, axonal guidance and signaling pathways, previously hypothesized to impact glaucoma-related optic nerve degeneration [39]. Expression analysis from the ocular tissue database also shows that many genes identified by this analysis show high expression in optic nerve and optic nerve head especially three genes in particular GNG7, RYR3 and CTNND2 (See Fig 5).
Zinc finger proteins are also known to be associated with glaucoma[40]. Our results indicate associations with POAG involving interactions between several zinc finger proteins and other protein coding genes such as interaction between genes ZNF385B and ELMO1 (LRT p-value 6.53–06 and 1.23E-05 for eMERGE and NEIGHBOR respectively).
Among the top associations, is the interaction between protein coding genes OPCML and RYR3 (LRT p-value 4.34E-06 and 1.01E-05 for eMERGE and NEIGHBOR respectively). The OPCML gene is an opioid binding protein/cell-adhesion molecule that is closely related to other proteins including, the NTN (neurotrimin), and genes in the immunoglobin family (IgLON family)[41]. NTN has been found to be nominally associated with glaucoma in previous studies[42].
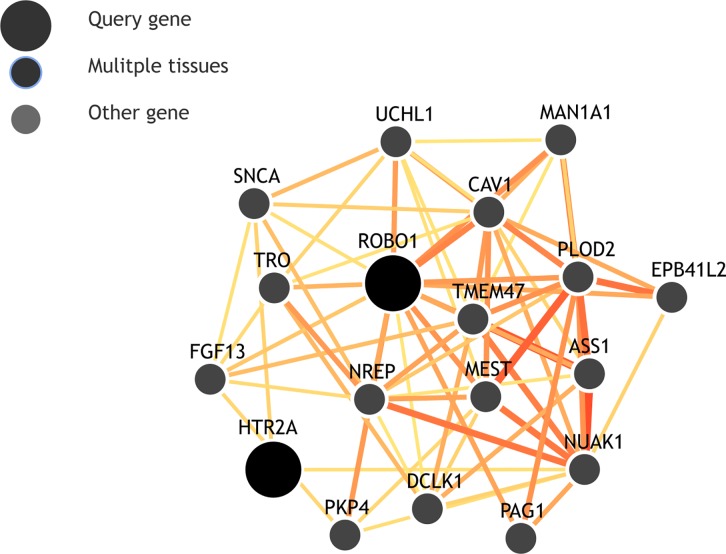
Our results also indicate interaction for ROBO1 and HTR2A (LRT p-values 5.44–06 and 2.18–04 for eMERGE and NEIGHBOR respectively as shown in Table 2). Both of these genes show co-expression with SNCA as predicted by the GIANT network [43] (Genome-scale Integrated Analysis of gene Networks in Tissues: http://giant.princeton.edu/) web based interface. Fig 7shows an interaction network generated by GIANT with genes that show functional interaction between query genes and other genes expressed only in the eye. HTR2A encodes receptors of 5-HT2A serotonin molecules. Serotonin molecules work as neurotransmitters and these molecules also function as receptors for many drugs[44].
Fig 7. GIANT network representing interaction between ROBO1 and HTR2A query genes and other genes for expression in the eye.
Scale represents the confidence in the relationship shown.
Another interesting finding is an interaction between CTNND2 and NRG3 (LRT p-value 9.23E-06 and 1.60E-05 for eMERGE and NEIGHBOR respectively as shown in Table 2). CTNND2, i.e. Catenin Delta 2 gene, encodes adhesive junction proteins that are involved in retinal morphogenesis and cell adhesion [45,46].
Cadherin molecules are known to support the migration of axons during optic nerve morphogenesis [47][50] and have been previously investigated as POAG susceptibility genes [48,49]. Our results identify a statistical interaction between NLN and glycoprotein DCHS1 also known as PCDH7 (LRT p-value 2.97E-06 and 7.035E-04 for eMERGE and the NEIGHBOR respectively) and also an interaction between NGR3 (neuregulin 3) and cadherin-associated protein CTNND2 (LRT p-value 9.23E-06 and 1.60E-05 for eMERGE and the NEIGHBOR respectively).
Pathway analysis using the gene-gene models revealed several interesting pathways including signal transduction, cell adhesion and regulation of actin dynamics for phagocytic cup formation. Myocilin, a protein involved in mendelian forms of glaucoma, may impact focal adhesion and migration of cells [50] but the interaction among other genes with similar functions is unknown. Our analysis highlights several genes with these functions and their role in glaucoma may require further investigation.
Our interaction analysis showed at least two genes (CTNND2 and RBFOX1) that have been previously associated with myopia, and are also associated with glaucoma in our datasets. CTNND2 is known to be associated with myopia[51] and myopia can influence glaucoma risk [52]. CTNND2 is significantly associated with myopia in an Asian GWAS and RBFOX1 has been associated with myopia in a multi-ethnic cohort and also in a Chinese population-based study [53–55]. In eMERGE, POAG cases and controls were not screened for myopia. Therefore, these associations will need further evaluation to understand if they are due to the co-morbidities of POAG and myopia in these patients. These results suggest that exploring interaction among genes associated with myopia may also be of interest in POAG studies. In conclusion, interactions among genes with only marginal POAG association can demonstrate interesting interactions that may contribute to the complex etiology of POAG.
This is the first replication study to identify gene-gene interactions in POAG in a large case-control dataset. We found 17 gene-gene replicating models at LRT p-value less than 0.001 and many of these interacting genes play important roles in cell adherence junction and signal transduction. Most drugs for glaucoma target synthesis of aqueous humor, which could be achieved by manipulating trabecular meshwork that maintains the outflow of aqueous humor in the eye. In most of the glaucoma patients, the outflow mechanism from the trabecular meshwork is negotiated when the progression of the disease is accompanied by an increase in intraocular pressure. For all the genes that are expressed in eye, we looked at the genes that are also expressed in trabecular meshwork via literature search and in the ocular tissue database as shown in Fig 5[53,54]. While most of the genes identified in this study show significant expression in the eye, 9 genes in particular (GNG7, ROBO1, SUMF1, RYR3, SLC24A3, CCDC3, CARS2, RPS6KA, SETDB1) also show expression in trabecular meshwork. Thus, this suggests potential genes that could be involved in ocular hypertension and elevated IOP could be linked to the progression of POAG. Many complex diseases are caused by multiple variants in the same genes or in different genes. SNP-based association tests do not account for locus based genetic heterogeneity, and when looking for replication it is highly unlikely that the same variant might be causal for a complex disease in an independent population. Our analysis highlights the importance of performing gene based interaction analysis to utilize complexity in biology and considering heterogeneity for testing statistical epistasis. Our analysis also indicates that testing for interactions in association studies can help explain additional genetic variance beyond that accounted for by SNPs associated with a disease via main effects only. Despite of the advantages to look for gene-based interactions, one limitation of this approach is that focusing only on the genes could result in missing interactions among the intragenic regulatory regions such as distal enhancers. Furthermore, we also did not test for all possible pairwise combinations but instead picked variants that show significant to marginal main effect which could also result in missing some potentially significant epistatic variants. Future studies could include testing for interactions based on prior-biological knowledge and testing for regions that are involved in similar pathways or are linked based on regulatory information available[32]. More studies or meta-analysis to replicate the interaction results can provide increased confidence in our analyses.
In conclusion, we identified many significant gene-gene models that are associated with POAG and our study clearly indicates that we can consider heterogeneity among independent populations by looking for replication in gene regions rather than exact SNPs. Thus presenting a gene based SNP-SNP interaction analysis, which has been done by others including Ma et al. 2012 and 2013[56,57], as a powerful approach in replication studies.
Materials and Methods
Discovery dataset
eMERGE-II is a consortium in the United States that consists of seven adult sites and two pediatric sites across the country. There are a total of 55,289 unique samples in eMERGE that have been genotyped on different platforms and imputed to the 1000 Genomes March 2012 reference dataset using SHAPEIT2 for phasing and IMPUTE2 for imputation [13]. Five adult sites (Marshfield Clinic, Group Health, Geisinger Clinic, Northwestern University, and Mayo Clinic) contributed samples to the glaucoma phenotype. There are a total of 5090 samples that include 961 cases and 4,129 controls, (2,247 males and 2,843 females). Table 3shows the distribution of samples across all sites and gender. Glaucoma case and control status was obtained from EMR data. All cases and controls selected were age 40 or above and had a recent eye exam within 2 years. Patients with two or more occurrences for ICD-9 codes for open angle (365.10), primary open angle (365.11) or low-tension open angle glaucoma (365.12) were diagnosed as cases and all the other samples were further evaluated for controls by following criteria: excluded any samples that had diagnosis for Ocular Hypertension; patients with Current Procedural Terminology (CPT) codes for glaucoma related surgery; patients that were given intraocular pressure lowering medications (details in supplementary S1 Text). Glaucoma samples in the eMERGE dataset were extracted from the imputed version3 dataset (dbGaP accession: phs000888.v1.p1}; all markers and samples below 99% call rate were excluded from the analysis. For the purpose of running GWAS and GXG common variant analysis, we only kept markers at MAF > = 0.05. This resulted in 2,914,185 SNPs tested for GWAS. Principal component analysis was performed using Eigensoft [58] on all non-related samples to look for individual level population variations. A plot of eigenvector 1 versus 2 from the eigen-analysis is shown in Fig 8. Adjusting for principal components helps to identify non-spurious and relevant associations that are merely due to ancestral difference.
Table 3. Demographics for glaucoma samples in eMERGE discovery dataset.
| Variable | Type | Category | Observations | % |
|---|---|---|---|---|
| Glaucoma Phenotype | Cases | 961 | 0.189 | |
| Controls | 4129 | 0.811 | ||
| Site | Cases | Group Health | 409 | 0.080 |
| Vanderbilt | 71 | 0.014 | ||
| Marshfield | 289 | 0.057 | ||
| Mayo | 86 | 0.017 | ||
| Geisinger | 85 | 0.017 | ||
| Northwestern | 21 | 0.004 | ||
| Controls | Group Health | 1441 | 0.283 | |
| Vanderbilt | 253 | 0.050 | ||
| Marshfield | 1245 | 0.245 | ||
| Mayo | 411 | 0.081 | ||
| Geisinger | 382 | 0.075 | ||
| Northwestern | 397 | 0.078 | ||
| Sex | Cases | Male | 404 | 0.079 |
| Female | 557 | 0.109 | ||
| Controls | Male | 1843 | 0.362 | |
| Female | 2286 | 0.449 | ||
| Race | Cases | Black | 14 | 0.003 |
| White | 922 | 0.181 | ||
| Other | 25 | 0.005 | ||
| Controls | Black | 82 | 0.016 | |
| White | 3918 | 0.770 | ||
| Other | 129 | 0.025 |
Fig 8. Principal component analysis (PCA) of 5090 samples (glaucoma cases and controls) from eMERGE network.
Eigenvector 1 is on x-axis and eigenvector 2 is on y-axis. Each point is a sample and they are color coded by case and control status.
Replication dataset
DNA from more than 4,900 POAG cases and controls (primarily European Americans) was collected for the NEIGHBOR study from 12 sites including Massachusetts Eye and Ear Infirmary, Brigham and Women’s Hospital, Duke University, Johns Hopkins University, Marshfield Clinic, Stanford University, University of Pittsburgh, University of West Virginia, University of Miami, University of Michigan, University of California, San Diego, and Vanderbilt University as previously reported[6,29]. POAG was defined in cases by the presence of optic nerve disease with visual field loss consistent with nerve fiber layer dropout; when this information was not available, cases were determined based on a cup-to-disc ratio greater than 0.8 in at least one eye. POAG controls had normal optic nerves, normal intraocular pressure, and presumed normal visual fields, or a cup to disc ratio less than 0.7. Samples were genotyped on the Illumina 660W Quad platform at the Center for Inherited Diseases Research (CIDR). Samples with call rate <97% were excluded, as were samples determined to be related with a kinship coefficient >0.0312. This resulted in 4,422 individuals (2,132 POAG cases, 2,290 controls). Principal components analysis was run using Eigenstrat and eigenvectors were tested for association with disease status using a logistic regression model in Stata; eigenvectors 1,2,3, and 10 were significant. The GWAS data were phased and imputed to the March 2012 version of the 1000Genomes reference data using ShapeIt2 and IMPUTE2. Markers with call rate <97% and MAF<0.05 were removed, as were markers with imputation quality <0.7; this resulted in 6,287,101 variants to be examined for further analysis. Out of ~6M variants, 3839 SNPs were used for the purpose of replication as described earlier in methods section.
SNP-SNP interaction analysis
Exploring pairwise SNP-SNP interaction is a two-step process. Steps for filtering of SNPs at different stages are explained in detail in Fig 2. From GWAS results, we extracted all SNPs with main effect p-value <0.01 and also to test only independent models, we removed any SNPs in high linkage disequilibrium (LD r2 threshold >0.6). We removed these SNPs in LD using PLINK. Secondly, we investigated all pairwise SNP-SNP interaction models using logistic regression in PLATO (https://ritchielab.psu.edu/software/plato-download) assuming an additive encoding model. All models were adjusted for age, sex, site, platform and first 6 principal components. Likelihood ratio tests were performed to determine significance of each pairwise interaction model above and beyond its main effects. This resulted in 117 SNP-SNP models with LRT p-value <1e-05. To perform replication, these SNPs from the top 117 SNP-SNP models were annotated to genes using Biofilter 2.0[59] which resulted in 91 unique genes that were considered for replication. All SNPs in these genes were extracted from NEIGHBOR data and then SNPs passing an info score threshold of 0.7, marker call rate of 99%, MAF 0.05 and SNPs passing LD pruned r2 threshold 0.6 were considered for replication (i.e. 3,839 SNPs). All genes based on expanded SNP-SNP models were exhaustively tested in the replication dataset. A total of 7,367,042 pairwise SNP-SNP models (from 3,839 SNPs) in 91 genes were evaluated in the replication dataset. Unique models with replicating LRT p-value< 0.001 are reported in Table 2.
Ethics statement
All eMERGE and NEIGHBOR consortium sites have appropriate IRB approvals for this de-identified research. All relevant data are within the paper and its Supporting Information files. Raw genotype files for eMERGE (phs000888.v1.p1) and NEIGHBOR (phs000238.v1.p1) are at dbGaP.
Supporting Information
Flowchart as well as pesudocode for phenotypic algorithm to extract samples from Electronic Health Record data to classify as cases and controls for POAG.
(DOC)
Power calculation as performed by Quanto considering odds ratio in range of 1–3. Two separate sheets for disease risk of 0.0001 and 0.018
(XLSX)
All SNP-SNP interaction results replicated in eMERGE and NEIGHBOR analysis. This table lists p-values for all eMERGE samples analysis and also analysis in European American samples only. P-values for each variable in the model as well as likelihood ratio test p-value for interactive effect is also provided.
(XLSX)
QQ-plot showing distribution of p-values from interaction analyses in discovery dataset.
(TIFF)
Acknowledgments
eMERGE II contributors
Boston Children’s Hospital: Jonathan Bickel MD, Ariel Chandler, Ingrid Holm MD, MPH, Isaac Kohane MD, PhD, Shannon Manzi PharmD, Kyriacos Markianos PhD, Nathan Palmer PhD, Cassandra Perry MS, Guergana Savova PhD, Wendy Wolf PhD, Sek Wong Kong MD; Cincinnati Children’s Hospital Medical Center: Armand Antommaria MD, PhD, FAAP, Beth Cobb MBA, John Harley MD, PhD, John Hutton MD, Todd Lingren MS, John Lynch PhD, Keith Marsolo PhD, Melanie Myers PhD, Bahram Namjou MD, Cindy Prows MSN, RN, Imre Solti MA, Sander Vinks PharmD, PhD, FCP, Michael Wagner PhD; Children’s Hospital of Philadelphia: Debbie Abrams MGC, Berta Almoguera PhD, Rosetta Chiavacci BSN, CCRC, John Connolly PhD, Marcella Devoto PhD, Hakon Hakonarson MD, PhD, Brendan Keating PhD, Cecilia Kim, Frank Mentch PhD, Haijun Qui, Patrick Sleiman PhD, Lifeng Tian PhD, Lyam Vazquez MD; Geisinger Health System: Brynn Bartko, Jonathan Bock, Ken Borthwick, David Carey PhD, Ryan Colonie, Robert Elston PhD, Andy Faucett MS, Samantha Fetterolf, Glenn Gerhard MD, Helena Kuivaniemi MD, PhD, FAHA, David Ledbetter PhD, Eric Lipsky, Sarah Pendergrass PhD, MS, Agnes Sundaresan MD, Diane Smelser PhD, Gerard Tromp PhD, FAHA, Janet Williams MS, Marc Williams MD, Craig Wood; Group Health/University of Washington: Jen Albrecht, Chris Carlson PhD, David Carrell PhD, Paul Crane MD, MPH, David Cronkite MS, David Crosslin PhD, Colleen Davis, Kelly Ehrlich MS, Malia Fullterton DPhil, Carlos Gallego MD, Adam Gordon, Jane Grafton, Kelly Hansen, Andrea Hartzler PhD, Gail Jarvik MD, PhD, Eric Larson MD, MPH, MACP, Kathy Leppig MD, Shubhabrata Mukherjee PhD, James Ralston MD, MPH, Aaron Scrol MA, Peter Tarczy-Hornoch MD, Sue Trinidad MA; Mayo Clinic: Sue Bielinski PhD, Kevin Bruce, Chris Chute MD, DrPH, FACMI, Mariza De Andrade PhD, Hayan Jouni MD, John Heit MD, Iyad Isseh, Barbara Koenig PhD, Iftikhar Kullo MD, Dingcheng Li, Jen McCormick PhD, Jyoti Pathak PhD, Janey Russell, Cloann Schultz MS, Carolyn Vitek MS, Luanne Wussow, Richard Sharp PhD, Adelaide Olson MD, Hutchens Chandra; Marshfield Clinic/Essentia Rural Health/The Pennsylvania State University: Gretta Armstrong MA, Richard Berg MS, Ariel Brautbar MD, Murray Brilliant PhD, Joe Finamore, Norm Frost MD, MPH, Molly Hall PhD, MS, Max He PhD, Terrie Kitchner, Simon Lin MD, Jim Linneman MS, Cathy McCarty PhD, MPH, RD, Peggy Peissig PhD, Marylyn D. Ritchie PhD, MS, Setia Shefali MS, Robert Strenn, Bryan Weichelt MBA; Mt. Sinai: Emilia Bagiella PhD, Erwin Bottinger MD, Douglas Dieterich PhD, Steve Ellis, Kadija Ferryman MS, Carol Friedman PhD, Scott Friedman MD, Genevieve Galarneau, Alanna Gomez, Omri Gottesman MD, Jean Sebastien Hulot MD, Carol Horowitz MD, MPH, George Hripcsak MD, Joseph Kannry MD, Eimear Kenny PhD, Yolanda Keppel, Jenny Lin MD, Vaneet Lotay MS, Ana Mejia MS, Rajiv Nadukuru, Inga Peter PhD, Rosamond Rhodes PhD, Saskia Sanderson PhD, Stuart Scott PhD, Thomas Ullman MD, Zhang Weijia PhD, Chunhua Weng PhD, Randi Zinberg MS, CGC; Northwestern University: Loren Armstrong, Sharon Aufox MS, CGC, David Baker MD, MPH, Kenzie Cameron PhD, MPH, Rex Chisholm PhD, Carl Christensen MS, Phil Greenland MD, Geoff Hayes PhD, Michael Heathcote, Abel Kho MD, Bill Lowe MD, Kathryn Jackson, Jen Pacheco, Vivian Pan CGC, Steve Persell MD, MPH, Luke Rasmussen, Laura Rasmussen-Torvik PhD, MPH, Maureen Smith MS, CGC, Justin Starren MD, PhD, Will Thompson PhD, Cathy Wicklund MS, CGC; Vanderbilt University: Kyle Brothers MD, Lixin Chen MS, Ellen Clayton MD, JD, James Cowan MS, Dana Crawford PhD, Josh Denny MD, Todd Edwards PhD, Paul Harris PhD, Nanibaa' Garrison PhD, Brad Malin PhD, Tracy McGregor MD, Josh Peterson MD, MPH, Dan Roden MD, Peter Speltz, Sarah Stallings PhD, Mary Stroud RN, Digna Velez, Sara VanDriest MD, PhD, Russ Wilke MD, PhD; Coordinating Center: Melissa Basford MBA, Mollie Bodin Claar MS, Adam Hardebeck BS, Paul Harris PhD, Jacqueline Kirby MS, Sarah Stallings PhD, Brad Malin PhD, Marylyn Ritchie PhD, Gretta Armstrong MA, Shefali Setia MS; NHGRI: Dave Kaufman, Rongling Li MD, PhD, MPH, Teri Manolio MD, PhD
NEIGHBOR contributors
Janey L.Wiggs, MD, PhD, Michael A. Hauser, PhD, Robert Rand Allingham, MD, Donald L. Budenz, MD, Jae H. Kang, DSc, Douglas Gaasterland, MD, Terry Gaasterland, PhD, Richard K. Lee, MD, Paul R. Lichter, MD, Yutao Liu, PhD, Sayoko E. Moroi, MD, Anthony Realini, MD, Robert Ritch, MD, Julia E. Richards, PhD, Joel S. Schuman, MD, Bill Scott, Kuldev Singh, MD, Douglas Vollrath, MD, PhD, Robert N. Weinreb, MD, Gadi Wollstein, MD, Brian L. Yaspan, PhD, Sachiko Yoneyama, PhD, Don Zack, MD, PhD, Kang Zhang, MD, PhD, Margaret Pericak-Vance, PhD, Louis R. Pasquale, MD, and Jonathan L. Haines, PhD
Data Availability
All relevant data are within the paper and its Supporting Information files. Raw genotype files for eMERGE (phs000888.v1.p1) and NEIGHBOR (phs000238.v1.p1) are at dbGaP.
Funding Statement
The eMERGE Network was initiated and funded by NHGRI through the following grants: U01HG006828 (Cincinnati Children’s Hospital Medical Center/Boston Children’s Hospital); U01HG006830 (Children’s Hospital of Philadelphia); U01HG006389 (Essentia Institute of Rural Health, Marshfield Clinic Research Foundation and Pennsylvania State University); U01HG006382 (Geisinger Clinic); U01HG006375 (Group Health Cooperative/University of Washington); U01HG006379 (Mayo Clinic); U01HG006380 (Icahn School of Medicine at Mount Sinai); U01HG006388 (Northwestern University); U01HG006378 (Vanderbilt University Medical Center); and U01HG006385 (Vanderbilt University Medical Center serving as the Coordinating Center); U01HG006389 and NCATS/NIH grant UL1TR000427 (Marshfield);U01HG004438 (CIDR) and U01HG004424 (the Broad Institute). NEIGHBORHOOD data collection and analysis are supported by US National Institutes of Health/National Eye Institute (NIH/NEI) grant R01EY022305 (JLW). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Resnikoff S, Pascolini D, Etya’ale D, Kocur I, Pararajasegaram R, Pokharel GP, et al. Global data on visual impairment in the year 2002. Bull World Health Organ. 2004;82: 844–851. /S0042-96862004001100009 [PMC free article] [PubMed] [Google Scholar]
- 2.Teikari JM, Airaksinen PJ, Kaprio J, Koskenvuo M. Primary open-angle glaucoma in 2 monozygotic twin pairs. Acta Ophthalmologica. 1987;65: 607–611. 10.1111/j.1755-3768.1987.tb07050.x [DOI] [PubMed] [Google Scholar]
- 3.Wang X, Harmon J, Zabrieskie N, Chen Y, Grob S, Williams B, et al. Using the Utah Population Database to assess familial risk of primary open angle glaucoma. Vision Res. 2010;50: 2391–2395. 10.1016/j.visres.2010.09.018 [DOI] [PubMed] [Google Scholar]
- 4.Fan BJ, Wiggs JL. Glaucoma: genes, phenotypes, and new directions for therapy. Journal of Clinical Investigation. 2010;120: 3064–3072. 10.1172/JCI43085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gharahkhani P, Burdon KP, Fogarty R, Sharma S, Hewitt AW, Martin S, et al. Common variants near ABCA1, AFAP1 and GMDS confer risk of primary open-angle glaucoma. Nat Genet. 2014;46: 1120–1125. 10.1038/ng.3079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiggs JL, Hauser MA, Abdrabou W, Allingham RR, Budenz DL, Delbono E, et al. The NEIGHBOR consortium primary open-angle glaucoma genome-wide association study: rationale, study design, and clinical variables. J Glaucoma. 2013;22: 517–525. 10.1097/IJG.0b013e31824d4fd8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Thorleifsson G, Walters GB, Hewitt AW, Masson G, Helgason A, DeWan A, et al. Common variants near CAV1 and CAV2 are associated with primary open-angle glaucoma. Nat Genet. 2010;42: 906–909. 10.1038/ng.661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stone EM, Fingert JH, Alward WLM, Nguyen TD, Polansky JR, Sunden SLF, et al. Identification of a Gene That Causes Primary Open Angle Glaucoma. Science. 1997;275: 668–670. 10.1126/science.275.5300.668 [DOI] [PubMed] [Google Scholar]
- 9.Burdon KP. Genome-wide association studies in the hunt for genes causing primary open-angle glaucoma: a review. Clin Experiment Ophthalmol. 2012;40: 358–363. 10.1111/j.1442-9071.2011.02744.x [DOI] [PubMed] [Google Scholar]
- 10.Crawford DC, Crosslin DR, Tromp G, Kullo IJ, Kuivaniemi H, Hayes MG, et al. eMERGEing progress in genomics-the first seven years. Front Genet. 2014;5: 184 10.3389/fgene.2014.00184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gottesman O, Kuivaniemi H, Tromp G, Faucett WA, Li R, Manolio TA, et al. The Electronic Medical Records and Genomics (eMERGE) Network: past, present, and future. Genet Med. 2013;15: 761–771. 10.1038/gim.2013.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McCarty CA, Chisholm RL, Chute CG, Kullo IJ, Jarvik GP, Larson EB, et al. The eMERGE Network: a consortium of biorepositories linked to electronic medical records data for conducting genomic studies. BMC Med Genomics. 2011;4: 13 10.1186/1755-8794-4-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Verma SS, de Andrade M, Tromp G, Kuivaniemi H, Pugh E, Namjou-Khales B, et al. Imputation and quality control steps for combining multiple genome-wide datasets. Front Genet. 2014;5: 370 10.3389/fgene.2014.00370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Funayama T, Mashima Y, Ohtake Y, Ishikawa K, Fuse N, Yasuda N, et al. SNPs and interaction analyses of noelin 2, myocilin, and optineurin genes in Japanese patients with open-angle glaucoma. Invest Ophthalmol Vis Sci. 2006;47: 5368–5375. 10.1167/iovs.06-0196 [DOI] [PubMed] [Google Scholar]
- 15.Maher B. Personal genomes: The case of the missing heritability. Nature. 2008;456: 18–21. 10.1038/456018a [DOI] [PubMed] [Google Scholar]
- 16.Cordell HJ. Detecting gene-gene interactions that underlie human diseases. Nat Rev Genet. 2009;10: 392–404. 10.1038/nrg2579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, et al. Missing heritability and strategies for finding the underlying causes of complex disease. Nat Rev Genet. 2010;11: 446–450. 10.1038/nrg2809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Frazer KA, Murray SS, Schork NJ, Topol EJ. Human genetic variation and its contribution to complex traits. Nat Rev Genet. 2009;10: 241–251. 10.1038/nrg2554 [DOI] [PubMed] [Google Scholar]
- 19.Zuk O, Hechter E, Sunyaev SR, Lander ES. The mystery of missing heritability: Genetic interactions create phantom heritability. PNAS. 2012;109: 1193–1198. 10.1073/pnas.1119675109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shao H, Burrage LC, Sinasac DS, Hill AE, Ernest SR, O’Brien W, et al. Genetic architecture of complex traits: Large phenotypic effects and pervasive epistasis. PNAS. 2008;105: 19910–19914. 10.1073/pnas.0810388105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mackay TFC. Epistasis and quantitative traits: using model organisms to study gene-gene interactions. Nat Rev Genet. 2014;15: 22–33. 10.1038/nrg3627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sun X, Lu Q, Mukherjee S, Mukheerjee S, Crane PK, Elston R, et al. Analysis pipeline for the epistasis search—statistical versus biological filtering. Front Genet. 2014;5: 106 10.3389/fgene.2014.00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Greene CS, Penrod NM, Williams SM, Moore JH. Failure to replicate a genetic association may provide important clues about genetic architecture. PLoS ONE. 2009;4: e5639 10.1371/journal.pone.0005639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wade MJ. Epistasis, complex traits, and mapping genes. Genetica. 2001;112–113: 59–69. [PubMed] [Google Scholar]
- 25.Roberts SB, MacLean CJ, Neale MC, Eaves LJ, Kendler KS. Replication of linkage studies of complex traits: an examination of variation in location estimates. Am J Hum Genet. 1999;65: 876–884. 10.1086/302528 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foss EJ, Radulovic D, Shaffer SA, Ruderfer DM, Bedalov A, Goodlett DR, et al. Genetic basis of proteome variation in yeast. Nat Genet. 2007;39: 1369–1375. 10.1038/ng.2007.22 [DOI] [PubMed] [Google Scholar]
- 27.Brem RB, Kruglyak L. The landscape of genetic complexity across 5,700 gene expression traits in yeast. PNAS. 2005;102: 1572–1577. 10.1073/pnas.0408709102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Steinmetz LM, Sinha H, Richards DR, Spiegelman JI, Oefner PJ, McCusker JH, et al. Dissecting the architecture of a quantitative trait locus in yeast. Nature. 2002;416: 326–330. 10.1038/416326a [DOI] [PubMed] [Google Scholar]
- 29.Wiggs JL, Yaspan BL, Hauser MA, Kang JH, Allingham RR, Olson LM, et al. Common variants at 9p21 and 8q22 are associated with increased susceptibility to optic nerve degeneration in glaucoma. PLoS Genet. 2012;8: e1002654 10.1371/journal.pgen.1002654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey JNC, Loomis SJ, Kang JH, Allingham RR, Gharahkhani P, Khor CC, et al. Genome-wide association analysis identifies TXNRD2, ATXN2 and FOXC1 as susceptibility loci for primary open-angle glaucoma. Nat Genet. 2016; 10.1038/ng.3482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Welter D, MacArthur J, Morales J, Burdett T, Hall P, Junkins H, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42: D1001–1006. 10.1093/nar/gkt1229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lu Y, Vitart V, Burdon KP, Khor CC, Bykhovskaya Y, Mirshahi A, et al. Genome-wide association analyses identify multiple loci associated with central corneal thickness and keratoconus. Nat Genet. 2013;45: 155–163. 10.1038/ng.2506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Li Z, Allingham RR, Nakano M, Jia L, Chen Y, Ikeda Y, et al. A common variant near TGFBR3 is associated with primary open angle glaucoma. Hum Mol Genet. 2015;24: 3880–3892. 10.1093/hmg/ddv128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Aulchenko YS, Ripke S, Isaacs A, van Duijn CM. GenABEL: an R library for genome-wide association analysis. Bioinformatics. 2007;23: 1294–1296. 10.1093/bioinformatics/btm108 [DOI] [PubMed] [Google Scholar]
- 35.Wu C, Orozco C, Boyer J, Leglise M, Goodale J, Batalov S, et al. BioGPS: an extensible and customizable portal for querying and organizing gene annotation resources. Genome Biology. 2009;10: R130 10.1186/gb-2009-10-11-r130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wagner AH, Anand VN, Wang W-H, Chatterton JE, Sun D, Shepard AR, et al. Exon-level expression profiling of ocular tissues. Exp Eye Res. 2013;111: 105–111. 10.1016/j.exer.2013.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smoot ME, Ono K, Ruscheinski J, Wang P-L, Ideker T. Cytoscape 2.8: new features for data integration and network visualization. Bioinformatics. 2011;27: 431–432. 10.1093/bioinformatics/btq675 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang J, Lee SH, Goddard ME, Visscher PM. GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet. 2011;88: 76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swarup G, Vaibhava V, Nagabhush A. Functional Defects Caused by Glaucoma–Associated Mutations in Optineurin In: Rumelt S, editor. Glaucoma—Basic and Clinical Aspects. InTech; 2013. Available: http://www.intechopen.com/books/glaucoma-basic-and-clinical-aspects/functional-defects-caused-by-glaucoma-associated-mutations-in-optineurin [Google Scholar]
- 40.Hoehn R, Zeller T, Verhoeven VJM, Grus F, Adler M, Wolfs RC, et al. Population-based meta-analysis in Caucasians confirms association with COL5A1 and ZNF469 but not COL8A2 with central corneal thickness. Hum Genet. 2012;131: 1783–1793. 10.1007/s00439-012-1201-3 [DOI] [PubMed] [Google Scholar]
- 41.Sellar GC, Watt KP, Rabiasz GJ, Stronach EA, Li L, Miller EP, et al. OPCML at 11q25 is epigenetically inactivated and has tumor-suppressor function in epithelial ovarian cancer. Nat Genet. 2003;34: 337–343. 10.1038/ng1183 [DOI] [PubMed] [Google Scholar]
- 42.Ulmer M, Li J, Yaspan BL, Ozel AB, Richards JE, Moroi SE, et al. Genome-Wide Analysis of Central Corneal Thickness in Primary Open-Angle Glaucoma Cases in the NEIGHBOR and GLAUGEN Consortia. Invest Ophthalmol Vis Sci. 2012;53: 4468–4474. 10.1167/iovs.12-9784 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Greene CS, Krishnan A, Wong AK, Ricciotti E, Zelaya RA, Himmelstein DS, et al. Understanding multicellular function and disease with human tissue-specific networks. Nat Genet. 2015;47: 569–576. 10.1038/ng.3259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knauer CS, Campbell JE, Chio CL, Fitzgerald LW. Pharmacological characterization of mitogen-activated protein kinase activation by recombinant human 5-HT2C, 5-HT2A, and 5-HT2B receptors. Naunyn Schmiedebergs Arch Pharmacol. 2009;379: 461–471. 10.1007/s00210-008-0378-4 [DOI] [PubMed] [Google Scholar]
- 45.Duparc R-H, Boutemmine D, Champagne M-P, Tétreault N, Bernier G. Pax6 is required for delta-catenin/neurojugin expression during retinal, cerebellar and cortical development in mice. Dev Biol. 2006;300: 647–655. 10.1016/j.ydbio.2006.07.045 [DOI] [PubMed] [Google Scholar]
- 46.Paffenholz R, Kuhn C, Grund C, Stehr S, Franke WW. The arm-repeat protein NPRAP (neurojungin) is a constituent of the plaques of the outer limiting zone in the retina, defining a novel type of adhering junction. Exp Cell Res. 1999;250: 452–464. 10.1006/excr.1999.4534 [DOI] [PubMed] [Google Scholar]
- 47.Matsunaga M, Hatta K, Nagafuchi A, Takeichi M. Guidance of optic nerve fibres by N-cadherin adhesion molecules. Nature. 1988;334: 62–64. 10.1038/334062a0 [DOI] [PubMed] [Google Scholar]
- 48.Lin H-J, Tsai F-J, Hung P, Chen W-C, Chen H-Y, Fan S-S, et al. Association of E-cadherin gene 3’-UTR C/T polymorphism with primary open angle glaucoma. Ophthalmic Res. 2006;38: 44–48. 10.1159/000089523 [DOI] [PubMed] [Google Scholar]
- 49.Wecker T, Han H, Börner J, Grehn F, Schlunck G. Effects of TGF-β2 on cadherins and β-catenin in human trabecular meshwork cells. Invest Ophthalmol Vis Sci. 2013;54: 6456–6462. 10.1167/iovs.13-12669 [DOI] [PubMed] [Google Scholar]
- 50.Kwon HS, Tomarev SI. Myocilin, a glaucoma-associated protein, promotes cell migration through activation of integrin-focal adhesion kinase-serine/threonine kinase signaling pathway. J Cell Physiol. 2011;226: 3392–3402. 10.1002/jcp.22701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Verhoeven VJM, Hysi PG, Saw S-M, Vitart V, Mirshahi A, Guggenheim JA, et al. Large scale international replication and meta-analysis study confirms association of the 15q14 locus with myopia. The CREAM consortium. Hum Genet. 2012;131: 1467–1480. 10.1007/s00439-012-1176-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chen S-J, Lu P, Zhang W-F, Lu J-H. High myopia as a risk factor in primary open angle glaucoma. Int J Ophthalmol. 2012;5: 750–753. 10.3980/j.issn.2222-3959.2012.06.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, Allingham RR, Qin X, Layfield D, Dellinger AE, Gibson J, et al. Gene expression profile in human trabecular meshwork from patients with primary open-angle glaucoma. Invest Ophthalmol Vis Sci. 2013;54: 6382–6389. 10.1167/iovs.13-12128 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Paylakhi SH, Yazdani S, April C, Fan J-B, Moazzeni H, Ronaghi M, et al. Non-housekeeping genes expressed in human trabecular meshwork cell cultures [Internet]. 28 January 2012. [cited 9 Feb 2015]. Available: http://www.molvis.org/molvis/v18/a28/ [PMC free article] [PubMed] [Google Scholar]
- 55.Sun X, Lu Q, Mukherjee S, Mukheerjee S, Crane PK, Elston R, et al. Analysis pipeline for the epistasis search—statistical versus biological filtering. Front Genet. 2014;5: 106 10.3389/fgene.2014.00106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ma L, Brautbar A, Boerwinkle E, Sing CF, Clark AG, Keinan A. Knowledge-driven analysis identifies a gene-gene interaction affecting high-density lipoprotein cholesterol levels in multi-ethnic populations. PLoS Genet. 2012;8: e1002714 10.1371/journal.pgen.1002714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ma L, Clark AG, Keinan A. Gene-based testing of interactions in association studies of quantitative traits. PLoS Genet. 2013;9: e1003321 10.1371/journal.pgen.1003321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Price AL, Patterson NJ, Plenge RM, Weinblatt ME, Shadick NA, Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat Genet. 2006;38: 904–909. 10.1038/ng1847 [DOI] [PubMed] [Google Scholar]
- 59.Ritchie MD. Using biological knowledge to uncover the mystery in the search for epistasis in genome-wide association studies. Ann Hum Genet. 2011;75: 172–182. 10.1111/j.1469-1809.2010.00630.x [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Flowchart as well as pesudocode for phenotypic algorithm to extract samples from Electronic Health Record data to classify as cases and controls for POAG.
(DOC)
Power calculation as performed by Quanto considering odds ratio in range of 1–3. Two separate sheets for disease risk of 0.0001 and 0.018
(XLSX)
All SNP-SNP interaction results replicated in eMERGE and NEIGHBOR analysis. This table lists p-values for all eMERGE samples analysis and also analysis in European American samples only. P-values for each variable in the model as well as likelihood ratio test p-value for interactive effect is also provided.
(XLSX)
QQ-plot showing distribution of p-values from interaction analyses in discovery dataset.
(TIFF)
Data Availability Statement
All relevant data are within the paper and its Supporting Information files. Raw genotype files for eMERGE (phs000888.v1.p1) and NEIGHBOR (phs000238.v1.p1) are at dbGaP.