Abstract
Bone homeostasis is maintained by the synergistic actions of bone-resorbing osteoclasts and bone-forming osteoblasts. Here, we show that the transcriptional coactivator/repressor interferon-related developmental regulator 1 (Ifrd1) is expressed in osteoclast lineages and represents a component of the machinery that regulates bone homeostasis. Ifrd1 expression was transcriptionally regulated in preosteoclasts by receptor activator of nuclear factor κB (NF-κB) ligand (RANKL) through activator protein 1. Global deletion of murine Ifrd1 increased bone formation and decreased bone resorption, leading to a higher bone mass. Deletion of Ifrd1 in osteoclast precursors prevented RANKL-induced bone loss, although no bone loss was observed under normal physiological conditions. RANKL-dependent osteoclastogenesis was impaired in vitro in Ifrd1-deleted bone marrow macrophages (BMMs). Ifrd1 deficiency increased the acetylation of p65 at residues K122 and K123 via the inhibition of histone deacetylase-dependent deacetylation in BMMs. This repressed the NF-κB-dependent transcription of nuclear factor of activated T cells, cytoplasmic 1 (NFATc1), an essential regulator of osteoclastogenesis. These findings suggest that an Ifrd1/NF-κB/NFATc1 axis plays a pivotal role in bone remodeling in vivo and represents a therapeutic target for bone diseases.
INTRODUCTION
Interferon-related developmental regulator 1 (Ifrd1) is a transcriptional coactivator/repressor that controls gene expression by interacting with transcription factors or histone deacetylase (HDAC) complexes. Ifrd1 expression in vivo is upregulated by acute tissue injuries, including cerebral ischemia/reperfusion and muscle trauma (1). It is also upregulated by various growth factors and cytokines in vitro (2). Ifrd1 is implicated in the pathophysiology of cystic fibrosis through regulation of the function of neutrophil effector cells (3). In addition, Ifrd1 is a candidate gene for autosomal-dominant sensory/motor neuropathy with ataxia (4). In summary, Ifrd1 has been implicated in the regulation of cell growth and differentiation, in addition to the pathogenesis of various diseases, by modulating gene expression patterns.
Bone-forming osteoblasts and bone-resorbing osteoclasts regulate the integrity of the skeleton in a sophisticated fashion (5, 6). The osteoblast lineage is derived from primitive multipotent mesenchymal stem cells with the potential to differentiate into bone marrow stromal cells, chondrocytes, muscles, and adipocytes (7). Osteoclasts are multinucleated cells derived from hematopoietic stem cells and share the same lineage as macrophages and dendritic cells (8). The modulation of bone remodeling and bone homeostasis is tightly regulated by three essential mechanisms. These are the cell-autonomous regulation of osteoblastogenesis, the cell-autonomous regulation of osteoclastogenesis, and the osteoblast-dependent regulation of osteoclastogenesis (9). Recently, we demonstrated that Ifrd1 expression in osteoblasts represses osteoblastogenesis and activates osteoclastogenesis by modulating the nuclear factor κB (NF-κB)/Smad/Osterix and β-catenin/osteoprotegerin (OPG) pathways, respectively (10). These results indicate that Ifrd1 expression in osteoblasts regulates bone homeostasis by modulating the cell-autonomous regulation of osteoblastogenesis (bone formation) and the osteoblast-dependent regulation of osteoclastogenesis (bone resorption).
To further establish the pivotal role of Ifrd1 in bone homeostasis, we generated osteoclast-specific Ifrd1-deficient mice. Here, we showed that Ifrd1 is upregulated in preosteoclasts by the receptor activator of the NF-κB ligand (RANKL), an osteoclastogenic cytokine, through the activator protein 1 (AP-1) pathway. Moreover, we demonstrated a crucial role for Ifrd1 in the cell-autonomous regulation of osteoclastogenesis and bone resorption in vitro and in vivo. Subsequent analyses identified NF-κB and the nuclear factor of activated T cells, cytoplasmic 1 (NFATc1), as essential factors in Ifrd1-mediated regulation of osteoclastogenesis.
MATERIALS AND METHODS
Mouse generation.
Ifrd1fl/fl mice (10) were crossed with either Nestin-Cre (11), cathepsin K (Ctsk)-Cre (12), or CD11b-Cre mice (13). These mutant mice were backcrossed more than five generations with C57BL/6J mice. Mice were bred under standard animal housing conditions at 23 ± 1°C with humidity of 55% and a light/dark cycle of 12 h, with free access to food and water. Genotyping was performed by PCR using tail genomic DNA. The study protocol meets the guidelines of the Japanese Pharmacological Society and was approved by the Committee for the Ethical Use of Experimental Animals at Kanazawa University, Kanazawa, Japan. The numbers of animals used per experiment are stated in the figure legends.
Bone histomorphometric analyses and μCT analyses.
Bone histomorphometric analyses were performed on vertebrae as previously described (14). Briefly, the vertebrae were fixed with 10% formalin, followed by dehydration in different concentrations of ethanol and subsequent embedding in methyl methacrylate resin according to standard protocols. The ratio of bone volume to tissue volume (BV/TV), one of the trabecular bone structural parameters, was measured by von Kossa staining, which detects calcium deposits in tissue. Bone formation rate (BFR), which is the calculated rate at which cancellous bone surface and bone volume are being replaced annually, was analyzed by the calcein double-labeling method. Calcein was injected into mice twice with an interval of 3 days, and the mice were sacrificed 2 days after the last injection. Osteoblast and osteoclast parameters were analyzed by staining with toluidine blue or tartrate-resistant acid phosphatase (TRAP), respectively. Analyses were performed using the OsteoMeasure analysis system (OsteoMetrics, Inc., Atlanta, GA) according to standard protocols (15). Two-dimensional images of the distal femurs were obtained by μCT scanning using a Scan Xmate-L090 (Comscan Tecno, Yokohama, Japan) with a high resolution of 12 μm/pixel, voltage of 75 kV, and current of 100 μA, as described previously (16, 17).
RANKL administration.
The glutathione S-transferase (GST)–RANKL vector provided by S. L. Teitelbaum (Washington University) was expressed in Escherichia coli strain BL21 cultured with 50 μM isopropyl-β-d-thiogalactopyranoside at 30°C for 24 h. The bacterial cells were centrifuged, and subsequently the bacterial pellets were subjected to freeze-thaw treatment 3 times, followed by incubation in B-PER (bacterial protein extraction reagent; Thermo Fisher Scientific, Waltham, MA) containing 0.1 mg/ml lysozyme and 5 U DNase for 30 min. GST-RANKL proteins in the supernatant were purified by a column packed with glutathione-Sepharose 4 fast flow (GE Health Care, Little Chalfont, United Kingdom) with elution buffer (50 mM Tris-HCl, 20 mM glutathione, pH 8.0) at 4°C. The eluted proteins were then dialyzed for 24 h at 4°C against repeated changes of dialysis buffer (10 mM Tris-HCl, pH 7.5).
Eight-week-old mice were intraperitoneally (i.p.) administered GST-RANKL fusion protein at a rate of 2 mg/kg of body weight daily for 2 days. Mice were sacrificed by decapitation 12 h after the final injection, followed by the dissection of femurs, vertebrae, or calvariae, and subsequent bone histomorphometric analyses or TRAP staining.
Real-time quantitative PCR.
Total RNA was extracted from cells or tissues, followed by the synthesis of cDNA using reverse transcriptase and oligo(dT) primer. The cDNA samples were then used as templates for real-time PCR analysis, which was performed on an MX3005P quantitative PCR (qPCR) system (Agilent Technologies, Inc., Santa Clara, CA) using specific primers for each gene (see Table S1 in the supplemental material). Expression levels of the genes examined were normalized using 36b4 expression as an internal control for each sample (18).
Immunoblot analysis.
Cultured cells were solubilized in lysis buffer (10 mM Tris-HCl, 150 mM NaCl, 0.5 mM EDTA, 10 mM NaF, 1% Nonidet P-40, pH 7.4) containing protease inhibitor cocktail (1 mM p-amidinophenylmethanesulfonyl fluoride [APMSF], 1 μg/ml leupeptin, 1 μg/ml pepstatin A, 1 μg/ml antipain). Samples were then subjected to SDS-PAGE, followed by transfer to polyvinylidene difluoride (PVDF) membranes and subsequent immunoblotting. The primary antibodies used were anti-Ifrd1 and anti-HDAC1 antibodies (Abcam, Cambridge, United Kingdom); anti-glyceraldehyde-3-phosphate dehydrogenase (anti-GAPDH), anti-c-Fos, and anti-β-actin antibodies (Santa Cruz Biotechnology, Santa Cruz, CA); anti-NFATc1, anti-p65, and anti-acetyl-lysine antibodies (Cell Signaling Technologies, Danvers, MA); and anti-FLAG antibody (Wako, Osaka, Japan). Quantification was performed by densitometry using ImageJ (National Institutes for Health, Bethesda, MD).
Procedures for transient transfection and the luciferase assay.
Cells of the murine preosteoclast cell line RAW 264.7 (ATCC, Manassas, VA) were transiently transfected with vectors or short interfering RNA (siRNA) using the lipofection method as previously described (19). For the luciferase assay, cells were transfected with reporter vectors, followed by the preparation of cell lysates and subsequent determination of luciferase activity using specific substrates in a luminometer (ATTO Corp., Tokyo, Japan). Ifrd1-luc (−4442/+160) was previously generated by a PCR-based cloning method in our laboratory (20), and NF-κB-luc was obtained from Stratagene (Santa Clara, CA). NFATc1-luc (−1026/+1) was obtained by PCR-based cloning with the forward primer 5′-GAATTCTTTGGCAGCAGAGTTGCACC-3′ and the reverse primer 5′-GAATTCGCAGGGCCGGGCTCTGCCT-3′ with mouse genomic DNA. Transfection efficiency was normalized by determining the activity of Renilla-luc. Predesigned c-Fos, c-Jun, TRAF6, and Ifrd1 siRNAs were purchased from Invitrogen (Carlsbad, CA).
ChIP assay.
Chromatin immunoprecipitation (ChIP) experiments were performed by following the protocol of the ChIP assay kit (Merck Millipore, Billerica, MA). Cells were treated with formaldehyde for cross-linking and subsequently subjected to sonication in lysis buffer. Immunoprecipitation was performed with the anti-c-Fos antibody or anti-p-p65 antibody, followed by PCR with specific primers (see Table S2 in the supplemental material).
Immunoprecipitation assay.
Cells were solubilized in lysis buffer (10 mM Tris-HCl, 150 mM NaCl, 0.5 mM EDTA, 10 mM NaF, 1% Nonidet P-40, pH 7.4) containing protease inhibitor cocktail. After centrifugation, the supernatant was treated with antibodies indicated in Results for 1 h at 4°C and subsequent incubation with protein G-Sepharose for an additional 1 h at 4°C, followed by elution with SDS sample buffer (10 mM Tris-HCl, 2% SDS, 10% glycerol, 5% 2-mercaptoethanol). Samples were then separated by SDS-PAGE, followed by immunoblotting as previously described (20).
Oligonucleotide pulldown assay.
A double-stranded biotinylated oligonucleotide containing NF-κB consensus sequence (5′-AAACAGGGGGCTTTCCCTCCTC-3′ and 5′-GAGGAGGGAAAGCCCCCTGTTT-3′) was incubated with streptavidin-agarose (Sigma-Aldrich, St. Louis, MO) for 1 h at 4°C. Streptavidin-oligonucleotide beads were washed three times with binding buffer (10 mM Tris-HCl, 30 mM NaCl, 1 mM EDTA, 1 mM dithiothreitol [DTT], 5% glycerol, 1 mg/ml bovine serum albumin [BSA], pH 7.4), followed by incubation with protein extracts for 1 h at 4°C. Streptavidin-oligonucleotide beads were then washed three times with binding buffer and subjected to elution with SDS sample buffer (10 mM Tris-HCl, 2% SDS, 10% glycerol, 5% 2-mercaptoethanol), followed by SDS-PAGE for immunoblotting.
Generation of retroviral vectors and infection.
The pMX-p65 and pMX-p50 vectors were generated by subcloning from pcDNA3-p65 (20012) and pcDNA3-p50 (20018) vectors obtained from Addgene (Cambridge, MA) (10). The pMX-NFATc1 vector was generated by subcloning into pMX vector from pcDNA3-NFATc1 obtained from Thermo Fisher Scientific. The pMX-Ifrd1 vector was constructed by PCR cloning with the forward primer 5′-GGTACCGATGCCGAAGAACAAGAAG-3′ and the reverse primer 5′-TCTAGACTACAAGAATTCTCCAACATC-3′ using mouse cDNA. These vectors were transfected into PLAT-E cells using the calcium carbonate method. The virus supernatant was collected 48 h after transfection, and the cells were then infected with this viral supernatant for 72 h in the presence of 4 μg/ml Polybrene. The cells were then selected using 1 μg/ml puromycin for 3 days before experimentation.
Culture of osteoclasts and TRAP staining, the actin ring assay, and the pit formation assay.
Bone marrow macrophages (BMMs) were cultured in the presence of 20 ng/ml macrophage colony-stimulating factor (M-CSF) and 20 ng/ml RANKL for 4 days consecutively. TRAP staining, the actin ring assay, and the pit formation assay were performed as previously described (21). Briefly, cells were cultured on bone slices and subsequent fixation with 4% paraformaldehyde for 10 min, followed by permeabilization with 0.5% Triton X-100–phosphate-buffered saline (PBS) for 10 min. For the actin ring assay, the bone slices were then incubated in Alexa Fluor 546-phalloidin-PBS solution (Thermo Fisher Scientific). For the pit assay, the bone slices were scrubbed with a brush to remove attached cells. The bone slices were then treated with 20 μg/ml lectin-PBS solution (Sigma-Aldrich) for 1 h and subsequent incubation with 0.5 mg/ml DAB (Dojindo, Kumamoto, Japan) in PBS, including 0.01% H2O2 for 10 min in the dark.
Data analysis.
All results are expressed as the means ± standard errors of the means, and statistical significance was determined using the two-tailed, unpaired Student's t test or one-way analysis of variance with the Bonferroni/Dunnett post hoc test.
RESULTS
Ifrd1 expression is upregulated by RANKL through AP-1 in preosteoclasts.
We first tested whether Ifrd1 expression was altered in preosteoclastic RAW 264.7 cells by osteoclastogenic stimulation. Ifrd1 expression was significantly increased by RANKL, tumor necrosis factor alpha (TNF-α), and lipopolysaccharide (LPS) in preosteoclastic RAW 264.7 cells (Fig. 1A). We also verified that Ifrd1 protein was induced in preosteoclasts by RANKL (Fig. 1B). To identify putative transcription factors responsible for the induction of Ifrd1 expression by RANKL in preosteoclasts, Ifrd1 promoter activity next was monitored in RAW 264.7 cells that were transfected with various transcription factor expression vectors. The introduction of the AP-1 complex (composed of c-Fos/c-Jun) dominantly upregulated mouse Ifrd1 promoter (−4442 to +160) activity in preosteoclasts (Fig. 1C). Moreover, the introduction of c-Fos/c-Jun significantly increased the endogenous Ifrd1 mRNA expression in preosteoclasts (Fig. 1D). In addition, RANKL-induced endogenous Ifrd1 mRNA expression was completely blocked by siRNA-mediated knockdown of c-Fos, c-Jun, or TNF receptor-associated factor 6 (TRAF6) (Fig. 1E). Computational analysis of the 5′-flanking region of the Ifrd1 gene identified at least three putative AP-1-responsive elements (ARE) in the 5′-flanking region of the highly conserved mouse Ifrd1 and human IFRD1 genes (Fig. 1F), suggesting AP-1-dependent transcription of both mouse and human Ifrd1 genes. A ChIP assay was conducted to further verify the binding of AP-1 to putative binding sites (Fig. 1F, sites 1 to 3) in the Ifrd1 promoter. Recruitment of c-Fos to the Ifrd1 promoter region encompassing site 3 was significantly enhanced in RANKL-treated preosteoclasts (Fig. 1G and H). These results demonstrate that Ifrd1 expression is transcriptionally regulated by a RANKL-TRAF6-AP-1 pathway in preosteoclasts (Fig. 1I).
FIG 1.
Ifrd1 is upregulated by RANKL in preosteoclasts. Upregulation of Ifrd1 expression in RAW 264.7 cells by RANKL stimulation. (A) RAW 264.7 cells were stimulated with osteoclastogenic stimulants, 50 ng/ml RANKL, 10 ng/ml TNF-α, and 100 ng/ml LPS for 4 h, followed by the determination of Ifrd1 mRNA expression. (B) RAW 264.7 cells were stimulated with RANKL at 50 ng/ml for the indicated times and subjected to determination of Ifrd1 protein expression. Activation of Ifrd1 promoter activity by c-Fos/c-Jun in RAW 264.7 cells. (C) RAW 264.7 cells were transiently cotransfected with Ifrd1-luc and expression vectors bearing c-Fos and/or c-Jun, followed by determination of luciferase activity. Upregulation of Ifrd1 mRNA expression by c-Fos/c-Jun in RAW 264.7 cells. (D) RAW 264.7 cells were transiently transfected with c-Fos/c-Jun expression vectors, followed by determination of endogenous Ifrd1 mRNA expression. Inhibition of RANKL-induced Ifrd1 mRNA expression by knockdown of c-Fos, c-Jun, or TRAF6 in RAW 264.7 cells is shown. (E) RAW 264.7 cells were transfected with si-c-Fos, si-c-Jun, or si-TRAF6 and then subjected to 50 ng/ml RANKL treatment, followed by determination of endogenous Ifrd1 mRNA expression. (F) Schematic representation of the alignment of mouse and human Ifrd1 promoter regions with putative AP-1 binding sites. Highly conserved regions between mouse and human were identified using VISTA tools (http://genome.lbl.gov/vista/index.shtml). Enhancement of c-Fos recruitment on Ifrd1 promoter by RANKL in RAW 264.7 cells is shown. (G and H) RAW 264.7 cells were treated with 50 ng/ml RANKL and ChIP assay was performed using anti-c-Fos antibody and specific primers to recognize sites 1 (a-b), 2 (c-d), and 3 (e-f), shown in panel F. (I) Schematic model of this part of the study. Asterisks indicate values significantly different (*, P < 0.05; **, P < 0.01) from each control value obtained in cells treated with PBS (A and E) or transfected with empty vector (E.V.) (C and D). #, P < 0.05, significantly different from the value obtained for RANKL-treated cells transfected with si-Control.
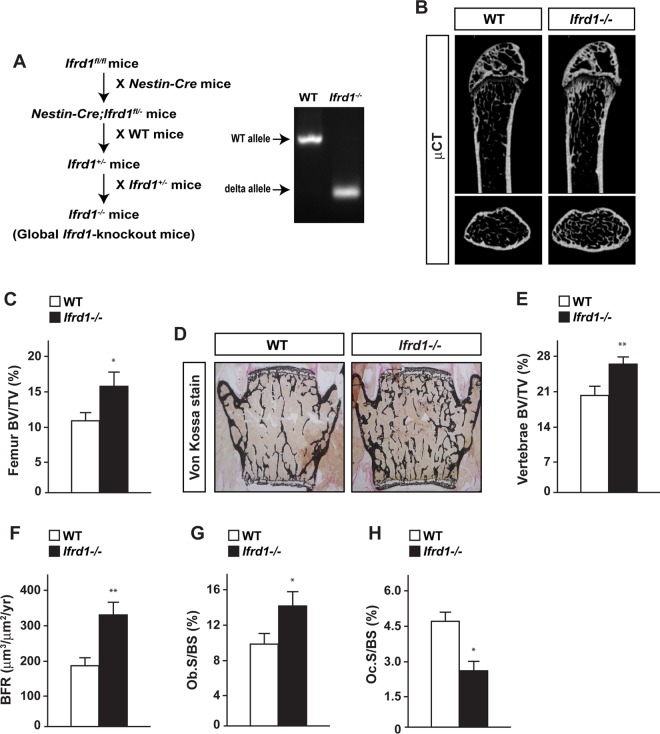
Germ line deletion of Ifrd1 increases bone formation and decreases bone resorption, leading to higher bone mass.
To evaluate the physiological importance of Ifrd1 in bone homeostasis in vivo, Ifrd1-floxed mice were crossed with Nestin-Cre transgenic mice to generate global Ifrd1-knockout mice (here referred to as Ifrd1−/− mice) (Fig. 2A) as a result of Nestin-Cre activity in the male germ line (11). Ifrd1−/− mice were indistinguishable from wild-type (WT) mice in terms of physical appearance, body weight, and nasoanal length (data not shown). However, Ifrd1−/− mice displayed a significantly higher BV/TV ratio in the femurs and vertebrae than WT mice (Fig. 2B to E). Bone histomorphometric analyses revealed that bone formation indices, such as BFR and osteoblast surface/bone surface (Ob.S/BS) values, were significantly increased in Ifrd1−/− mice compared to levels in WT mice (Fig. 2F and G). Conversely, osteoclast surface/bone surface (Oc.S/BS), an index of osteoclastic function, was significantly lower in Ifrd1−/− mice than in WT mice (Fig. 2H). These results indicated that Ifrd1 regulates both bone formation and bone resorption.
FIG 2.
Germ line deletion of Ifrd1 increases bone formation and decreases bone resorption, leading to high bone mass. (A) Strategy and confirmation for the generation of germ line Ifrd1-knockout mice. Higher bone volume in long bones and vertebrae of Ifrd1 knockout mice was found. (B and C) μCT analysis (B) and BV/TV ratios (C) of femurs of WT and Ifrd1 knockout mice at 12 weeks old. (D to H) von Kossa staining (D), BV/TV ratios (E), BFR (F), Ob.S/BS values (G), and Oc.S/BS values (H) of vertebrae of WT and Ifrd1 knockout mice at 12 weeks old (WT, n = 6 to 8; Ifrd1−/−, n = 5 to 8). Asterisks indicate values significantly different from the value obtained for WT mice: *, P < 0.05; **, P < 0.01.
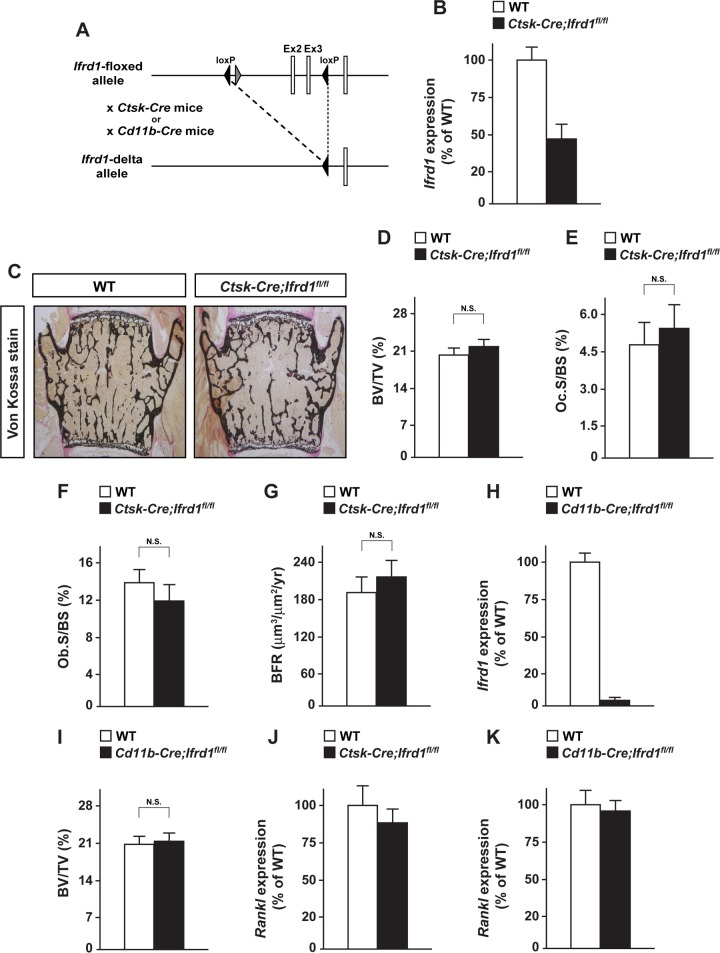
Conditional deletion of Ifrd1 in mature osteoclasts or osteoclast precursors does not affect the bone phenotype under normal physiological conditions.
To reveal the functional importance of Ifrd1 in osteoclasts, mature osteoclast-specific Ifrd1 knockout mice were generated by crossing Ifrd1-floxed mice with Ctsk-Cre knock-in mice, in which Cre recombinase is expressed in mature osteoclasts under the control of the Ctsk promoter (Fig. 3A). No changes in bone mass or osteoblastic and osteoclastic parameters were apparent in mature osteoclast-specific Ifrd1-knockout mice, termed Ctsk-Cre; Ifrd1fl/fl mice (Fig. 3C to G), irrespective of a marked reduction of Ifrd1 mRNA expression in osteoclasts (Fig. 3B). To further ascertain the effect of Ifrd1 deletion in osteoclast lineages, we used CD11b-Cre transgenic mice expressing Cre recombinase in the myeloid-osteoclast lineage under the control of the CD11b promoter to target Ifrd1 deletion from osteoclast progenitor cells (Fig. 3A), with a marked decrease in Ifrd1 expression in CD11b-positive cells in the bone marrow of CD11b-Cre; Ifrd1fl/fl mice (Fig. 3H). Similar to Ctsk-Cre; Ifrd1fl/fl mice, the bone phenotype of WT mice and CD11b-Cre; Ifrd1fl/fl mice was similar (Fig. 3I), indicating that Ifrd1 deletion in both early and mature osteoclasts does not affect bone mass under normal physiological conditions. Rankl expression was normal in the bones of Ctsk-Cre; Ifrd1fl/fl and CD11b-Cre; Ifrd1fl/fl mice (Fig. 3J and K).
FIG 3.
Osteoclast lineage-specific deletion of Ifrd1 does not show any abnormalities in bone phenotype under physiological conditions. (A) Schematic diagram of generating conditional Ifrd1 knockout. (B) RNA was isolated from osteoclasts of Ctsk-Cre; Ifrd1fl/fl mice and subjected to determination of Ifrd1 mRNA expression by qPCR. No apparent bone phenotype in Ctsk-Cre; Ifrd1fl/fl mice was seen. (C to G) von Kossa staining (C), BV/TV ratios (D), Oc.S/BS values (E), Ob.S/BS values (F), and BFR (G) of the vertebrae of WT and Ctsk-Cre; Ifrd1fl/fl mice (WT, n = 6; Ctsk-Cre; Ifrd1fl/fl, n = 5) at 12 weeks old. (H) RNA was isolated from CD11b-positive cells in bone marrow of CD11b-Cre; Ifrd1fl/fl mice and subjected to determination of Ifrd1 mRNA expression by qPCR. No apparent bone phenotype in CD11b-Cre; Ifrd1fl/fl mice was seen. (I) BV/TV ratios of the vertebrae of WT and CD11b-Cre; Ifrd1fl/fl mice (WT and CD11b-Cre; Ifrd1fl/fl; n = 5) at 12 weeks old. (J and K) RNA was isolated from femur of Ctsk-Cre; Ifrd1fl/fl mice and CD11b-Cre; Ifrd1fl/fl mice, followed by determination of Rankl expression by qPCR.
We have previously demonstrated that osteoblast-specific Ifrd1 knockout mice exhibited a higher bone mass caused by a concomitant increase in bone formation and decrease in bone resorption (10). Taken together with our previous findings, the present results suggest that under physiological conditions, Ifrd1 regulates bone homeostasis through osteoblasts rather than osteoclasts.
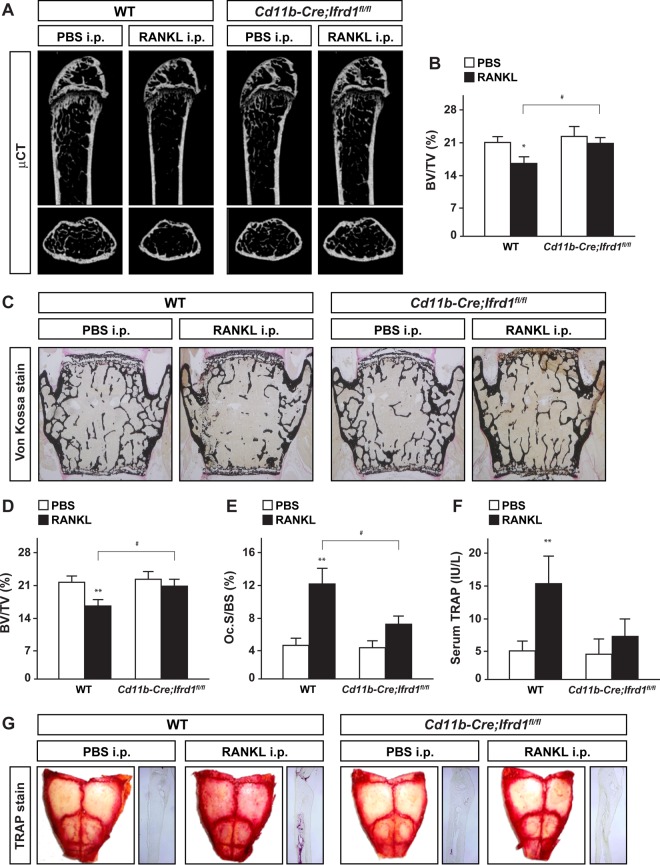
Conditional deletion of Ifrd1 in osteoclast precursors protects against RANKL-induced bone loss.
Osteoclast lineage-specific knockout of Ifrd1 did not cause any abnormalities in bone homeostasis under physiological conditions (Fig. 3). However, osteoclastogenesis was markedly impaired in coculture experiments using BMMs derived from Ifrd1−/− mice in our previous study (10). This suggested that Ifrd1 in cells of the osteoclast lineage regulate osteoclastogenesis and bone homeostasis under pathological conditions. To test this hypothesis, we investigated whether Ifrd1 expression in osteoclasts is important for the pathogenesis of osteoporosis by injecting mice with RANKL. CD11b+ osteoclast precursors differentiate into mature Ctsk-producing osteoclasts (13), indicating that the effect of Ifrd1 deficiency on bone homeostasis under pathological conditions can be investigated at all stages of osteoclast differentiation using the CD11b-Cre driver instead of the Ctsk-Cre driver. RANKL was injected into WT and CD11b-Cre; Ifrd1fl/fl mice daily for 2 days, and then the bone phenotype was analyzed. RANKL injection induced marked bone loss (indicated by a decreased BV/TV ratio) in the femurs and vertebrae of WT mice as a consequence of osteoclast activation (indicated by increased Oc.S/BS and serum TRAP levels). However, in CD11b-Cre; Ifrd1fl/fl mice, RANKL-induced osteoclast activation and bone loss were significantly impaired (Fig. 4A to F). Similarly, bone loss was not observed in Ifrd1−/− mice after RANKL treatment, despite the higher bone volumes under normal physiological conditions (see Fig. S1 in the supplemental material). Furthermore, RANKL treatment increased the number of TRAP-positive osteoclasts in the calvariae of WT mice but not CD11b-Cre; Ifrd1fl/fl mice (Fig. 4G). Therefore, Ifrd1 may modulate RANKL-stimulated osteoclastogenesis and bone resorption in vivo under pathological conditions.
FIG 4.
Ifrd1 deficiency in osteoclast precursors protects against RANKL-induced bone loss. Protection against RANKL-induced bone loss and bone resorption in CD11b-Cre; Ifrd1fl/fl mice. WT mice and CD11b-Cre; Ifrd1fl/fl mice were intraperitoneally administered GST-RANKL at 2 mg/kg daily for 2 days, and subsequently mice were sacrificed 12 h after the final injection, followed by determination of bone phenotypes. (A to F) μCT analysis (A) and BV/TV ratios (B) of femurs, von Kossa staining (C), BV/TV ratios (D), Oc.S/BS values of the vertebrae (E), and serum TRAP levels (F) of PBS- or RANKL-injected mice (WT-PBS, n = 6; WT-RANKL, n = 9; CD11b-Cre; Ifrd1fl/fl-PBS, n = 6; CD11b-Cre; Ifrd1fl/fl-RANKL, n = 8). Protection against RANKL-induced osteoclast activation in calvariae of CD11b-Cre; Ifrd1fl/fl mice was seen. (G) TRAP staining of the calvariae of PBS- or RANKL-injected mice. Asterisks indicate values significantly different from the value obtained for PBS-injected mice: *, P < 0.05; **, P < 0.01. #, P < 0.05, significantly different from the value obtained for RANKL-injected WT mice.
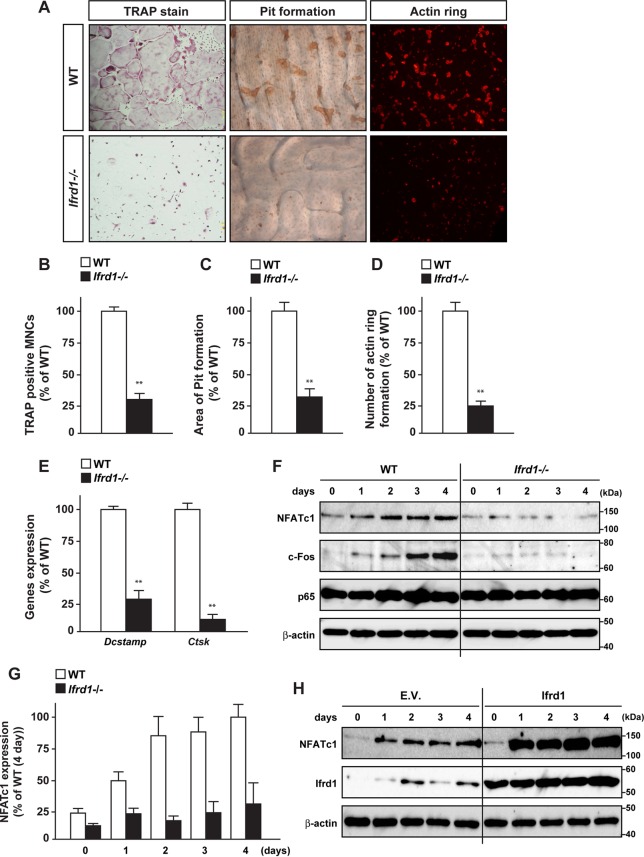
Ifrd1 deficiency abolishes RANKL-induced osteoclastogenesis.
We next evaluated whether Ifrd1 expression in osteoclasts regulates osteoclastogenesis in vitro. BMM differentiation into osteoclasts was induced by RANKL, and the differentiation was monitored by TRAP staining, actin ring formation assays (phalloidin labeling of F-actin), and pit assays (detection of resorption pits). Osteoclast differentiation and maturation were dramatically impaired in BMMs derived from Ifrd1−/− mice compared with WT mice, as determined by the number of TRAP-positive multinucleated cells, actin ring formation, and pit formation (Fig. 5A to D). Moreover, the expression of the osteoclast differentiation and fusion markers Ctsk and transmembrane 7 superfamily member 4 (DC-STAMP) was markedly decreased in osteoclasts derived from Ifrd1−/− mice (Fig. 5E). Finally, expression of NFATc1 and c-Fos protein was markedly decreased in osteoclasts derived from Ifrd1−/− mice, although p65 expression was not altered in whole-cell lysates (Fig. 5F and G). Conversely, retroviral introduction of Ifrd1 markedly upregulated the expression of NFATc1 in BMMs derived from WT mice (Fig. 5H).
FIG 5.
Ifrd1 deficiency in osteoclast precursors represses osteoclastogenesis. (A to E) Inhibition of cell differentiation and maturation by Ifrd1 deficiency in osteoclasts. BMMs from WT mice or Ifrd1−/− mice were stimulated with M-CSF (20 ng/ml) and RANKL (20 ng/ml) for 4 days, followed by TRAP staining (A and B), pit formation assay (A and C), actin ring assay (A and D), and the determination of mRNA expression of osteoclast marker genes (E). MNCs, multinucleated cells. (F and G) Downregulation of NFATc1 protein expression by Ifrd1 deficiency in osteoclasts. (F) BMMs from WT mice or Ifrd1−/− mice were stimulated with M-CSF and RANKL for the indicated days, followed by determination of protein expression of transcription factors involved in osteoclastogenesis. (G) Quantification of immunoblotting data of NFATc1 shown in panel F. (H) Upregulation of NFATc1 protein expression by Ifrd1 introduction in osteoclasts is shown. WT mouse-derived BMMs were retrovirally infected with Ifrd1 expression vector and subsequent stimulation with M-CSF and RANKL for the indicated days, followed by determination of NFATc1 protein expression by immunoblotting. **, P < 0.01, significantly different from the value obtained from WT cells.
Ifrd1 enhances osteoclastogenesis by modulating the acetylation of p65 and enhancing NF-κB/NFATc1 signaling in osteoclasts.
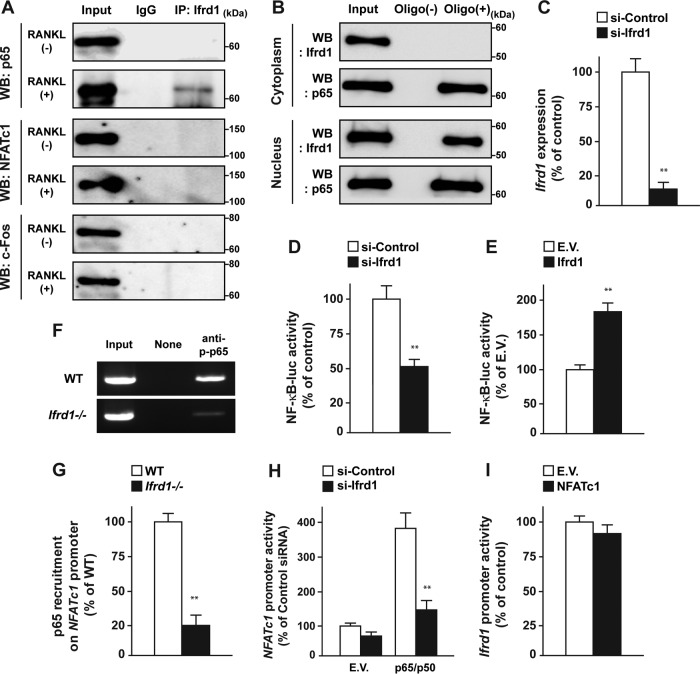
To elucidate the regulatory mechanisms of osteoclast differentiation by Ifrd1, we examined the physical interactions between Ifrd1 and NFATc1, c-Fos, and p65 in BMMs in the presence or absence of RANKL. As shown in Fig. 6A, Ifrd1 formed a complex with p65 in the presence of RANKL but did not form a complex with NFATc1 or c-Fos in BMMs in the presence or absence of RANKL (Fig. 6A). Ifrd1 and the heavy chain of IgG have similar molecular weights; therefore, it was not possible to perform reciprocal immunoprecipitation or confirm the amount of immunoprecipitated Ifrd1 in Fig. 6A. Therefore, we performed an oligonucleotide pulldown assay using the consensus NF-κB binding sequence. Ifrd1 was recruited to a consensus NF-κB binding sequence in the nucleus but not in the cytoplasm of RANKL-treated BMMs. This indicated that the physical interaction between Ifrd1 and p65 occurred selectively in the nucleus of BMMs (Fig. 6B).
FIG 6.
Ifrd1 interacts with p65 and regulates NF-κB-dependent transcription. Interaction between Ifrd1 and p65 in the nucleus of BMMs by RANKL stimulation is shown. (A) WT mouse-derived BMMs were stimulated with RANKL at 20 ng/ml and subjected to immunoprecipitation with anti-Ifrd1 antibody, followed by immunoblotting with the antibodies indicated. WB, Western blotting. (B) WT mouse-derived BMMs were stimulated with RANKL at 20 ng/ml and prepared as nucleus and cytoplasm fractions, followed by oligonucleotide pulldown assay with NF-κB consensus sequence and immunoblotting with the antibodies indicated. Regulation of NF-κB-luc activity by Ifrd1 in RAW 264.7 cells is shown. (C) RAW 264.7 cells were transfected with si-Ifrd1, and determination of Ifrd1 mRNA expression by qPCR was performed. (D and E) RAW 264.7 cells were cotransfected with NF-κB-luc and si-Ifrd1 (D) or Ifrd1 (E) expression vector and subjected to 50 ng/ml RANKL treatment, followed by determination of luciferase activity. (F and G) Inhibition of p65 recruitment on NFATc1 promoter in Ifrd1-deficient BMMs. BMMs from WT mice or Ifrd1−/− mice were stimulated with RANKL at 20 ng/ml, and ChIP assay was performed using anti-p-p65 antibody and specific primers to recognize NF-κB binding sites listed in Table S2 in the supplemental material. Inhibition of p65/p50-dependent NFATc1 promoter activity by Ifrd1 knockdown is shown. (H and I) RAW 264.7 cells were transiently cotransfected with p65/p50 expression vectors NFATc1-luc and si-Ifrd1 (H) or NFATc1 expression vector and Ifrd1-luc (I), followed by determination of luciferase activities. **, P < 0.01, significantly different from the value obtained in cells transfected with si-Control (C, D, and H), E.V. (E), or WT (G) cells.
Furthermore, the NF-κB-luc activity was significantly decreased by Ifrd1 knockdown and increased by Ifrd1 overexpression in the presence of RANKL (Fig. 6C to E). NFATc1, an established regulator of osteoclastogenesis (22), is a direct target of NF-κB (23). ChIP assays revealed that p65 recruitment by the NFATc1 promoter was also decreased in Ifrd1-deficient osteoclasts in the presence of RANKL (Fig. 6F and G). In addition, p65/p50-induced NFATc1 promoter activity was significantly repressed by Ifrd1 knockdown (Fig. 6H). Conversely, Ifrd1 promoter activity was not influenced by NFATc1 (Fig. 6I). These results suggested that Ifrd1 activates the NF-κB-dependent transcription of the NFATc1 gene in osteoclasts, consistent with the decreased NFATc1 expression in Ifrd1−/− BMMs (Fig. 5F).
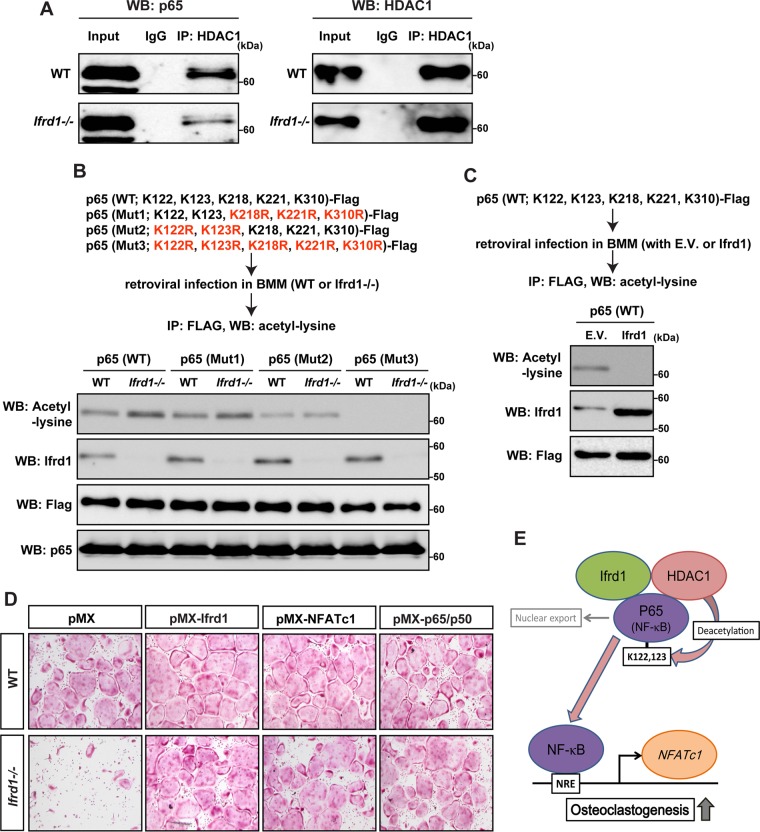
Ifrd1 modifies the acetylation status of p65 by HDACs (24). The physical interaction between HDAC1 and p65 was dramatically decreased in Ifrd1−/− BMMs (Fig. 7A). The transcriptional activity of NF-κB was enhanced by the acetylation of p65 at residues K218, K221, and K310 and repressed by the acetylation of p65 at residues K122 and K123 (25, 26). WT or Ifrd1−/− BMMs were retrovirally infected with three mutant p65 constructs: a Mut1 construct, in which K218, K221, and K310 were replaced with arginine (FLAG–p65-K218R,K221R,K310R); a Mut2 construct, in which K122 and K123 were replaced with arginine (FLAG–p65-K122R,K123R); and a Mut3 construct, in which all major lysine residues were replaced with arginine (FLAG–p65-K122R,K123R,K218R,K221R,K310R). After the mutants were introduced, we performed immunoprecipitations with a FLAG antibody, followed by immunoblotting with an anti-acetyl-lysine antibody. Compared with WT cells, acetylated p65 was markedly increased in Ifrd1−/− BMMs infected with the WT construct [p65 (WT)] and the Mut1 construct [p65 (Mut1)]. Acetylation was comparable for WT and Ifrd1−/− BMMs infected with the Mut2 construct [p65 (Mut2)] and was not detected in BMMs infected with the Mut3 construct [p65 (Mut3)] irrespective of the genotype (Fig. 7B). This indicated that Ifrd1 deficiency inhibits the deacetylation of p65 by HDAC1 at residues K122 and K123, subsequently repressing the NF-κB activity. Conversely, acetylated p65 was markedly repressed by Ifrd1 overexpression in WT mouse-derived BMMs infected with the WT construct [p65 (WT)] (Fig. 7C).
FIG 7.
Ifrd1 deficiency represses osteoclastogenesis by modulating the NF-κB/NFATc1 pathway by reducing HDAC-dependent deacetylation of p65 in osteoclasts at residues K122 and K123. Repression of interaction between HDAC1 and p65 in Ifrd1-deficient BMMs is shown. (A) BMMs from WT mice or Ifrd1−/− mice were stimulated with RANKL at 20 ng/ml and subjected to immunoprecipitation with anti-HDAC1 antibody, followed by immunoblotting with the antibodies indicated. Regulation of acetylation status of p65 at residues K122 and K123 by Ifrd1 in BMMs is shown. (B) BMMs from WT mice or Ifrd1−/− mice were infected with FLAG-tagged p65 constructs and subjected to 20 ng/ml RANKL treatment, followed by immunoprecipitation with anti-FLAG antibody and immunoblotting with the antibodies indicated. (C) WT mouse-derived BMMs were retrovirally infected with FLAG-tagged p65 (WT) vector and Ifrd1 expression vector and subjected to 20 ng/ml RANKL treatment, followed by immunoprecipitation with FLAG antibody and immunoblotting with anti-acetyl lysine antibody. Rescue of osteoclastogenesis by NFATc1 and P65/p50 in Ifrd1-deficient BMMs. (D) BMMs from WT mice and Ifrd1−/− mice were retrovirally infected with Ifrd1, NFATc1, and the p65/p50 expression vectors and subjected to stimulation with M-CSF and RANKL for 4 days, followed by TRAP staining. (E) Schematic model of this part of the study.
Finally, and importantly, impaired osteoclastogenesis was almost completely rescued by the retroviral transduction of p65/p50, NFATc1, and Ifrd1 into BMMs derived from Ifrd1−/− mice (Fig. 7D). These results suggest that Ifrd1 activates osteoclast differentiation by modulating the acetylation of p65 by HDAC1, thus enhancing NF-κB-dependent NFATc1 gene transcription (Fig. 7E).
DISCUSSION
Bone homeostasis is coordinately maintained by three known mechanisms: (i) the cell-autonomous regulation of osteoblastogenesis and bone formation; (ii) the cell-autonomous regulation of osteoclastogenesis and bone resorption; and (iii) the osteoblast-dependent regulation of osteoclastogenesis and bone resorption by the RANKL-OPG axis (7–9). Recently, we have revealed that Ifrd1 expression in osteoblasts modulates osteoblastogenesis via the NF-κB/Smad/Osterix pathway and osteoclastogenesis via the β-catenin/OPG pathway (10). In this study, we further demonstrated that Ifrd1 expression in osteoclast lineage cells modulates osteoclastogenesis via NF-κB/NFATc1 signaling in a cell-autonomous fashion under pathological conditions. These findings demonstrated that Ifrd1 regulates all three mechanisms of bone homeostasis maintenance through its coordinated expression in osteoblast- and osteoclast-lineage cells.
Osteoblast-specific Ifrd1 knockout mice have a higher bone mass because of a combination of increased bone formation and decreased bone resorption (10). This phenotype resembles that of the complete Ifrd1 knockout mouse, indicating that under physiological conditions, Ifrd1 mainly regulates bone homeostasis via its expression in osteoblasts rather than osteoclasts. Bone loss is increased in postmenopausal osteoporosis due to accelerated bone turnover, with bone resorption exceeding bone formation. This leads to the increased fragility of bone integrity and therefore an increased risk of bone fractures (27). The anti-RANKL antibody increases bone mineral density and decreases bone resorption in postmenopausal women and in ovariectomized monkeys (28, 29), suggesting that an elevated RANKL ratio plays a role in bone loss in postmenopausal osteoporosis. RANKL induces the nuclear localization and phosphorylation of p65 in BMMs (6, 30). In this study, we observed that Ifrd1 associated with p65 in the nucleus of BMMs only when cells were treated with RANKL, along with a marked upregulation of Ifrd1 expression by RANKL. These findings suggest that Ifrd1 controls bone resorption and bone homeostasis during postmenopausal osteoporosis through its expression in the osteoclast lineage. CD11b-Cre is on the Y chromosome (13); therefore, it was not possible to generate an osteoporosis model in CD11b-Cre; Ifrd1fl/fl mice by ovariectomy. However, we demonstrated that Ifrd1 deficiency in the osteoclast lineage prevented RANKL-induced bone loss and osteoclast activation in CD11b-Cre; Ifrd1fl/fl mice, revealing a crucial role of osteoclastic Ifrd1 in bone homeostasis under pathological conditions.
NF-κB is a transcription factor that plays a central role in inflammation, autoimmune responses, cell proliferation, differentiation, and apoptosis (31–33). NF-κB is a positive regulator of osteoclastogenesis and a negative regulator of osteoblastogenesis (34, 35). In addition, previous studies have demonstrated that Ifrd1 decreases and increases the activity of NF-κB-dependent transcription in myoblasts and neutrophils, respectively (3, 24). This suggests that depending on the cellular lineages, Ifrd1 may both negatively and positively regulate NF-κB signaling. In this study, we found that Ifrd1 regulates osteoclastogenesis in a cell-autonomous manner by modulating the acetylation of p65 by HDAC1, as shown in Fig. 7E. Although acetylation of the K218, K221, and K310 residues of p65 increases the transcriptional activity of NF-κB, acetylation of the K122 and K123 residues reduces the transcriptional activity of NF-κB by facilitating the nuclear export of p65 (25, 26). In addition, acetylation at residues K314 and K315 of p65 has been shown to be important for NF-κB-dependent transcription (36). In this study, we found that the acetylation of residues K314 and K315 of p65 was, at least in osteoclasts, of less importance for NF-κB-dependent transcription. In contrast, we found that Ifrd1 deficiency increased the acetylation of residues K122 and K123 of p65 but not of K218, K221, and K310 in osteoclasts. Accordingly, Ifrd1 may decrease the acetylation of residues K122 and K123 of p65 in an HDAC-dependent manner by modulating their interaction. This may lead to an increase in NF-κB-dependent transcription as a result of the nuclear accumulation of p65 and the subsequent activation of osteoclastogenesis by the augmentation of NFATc1 expression in osteoclasts (Fig. 7E). We have previously demonstrated that the acetylation of p65 at residues K122 and K123 was augmented by impaired interaction between p65 and HDAC1 in Ifrd1-deficient osteoblasts, leading to the nuclear export of p65 and the repression of NF-κB activity. This consequently resulted in the repression of Smad7, a target gene of NF-κB signaling, in osteoblasts (10). The same mechanism of NF-κB-dependent transcription by the Ifrd1 gene may operate in both osteoclast and osteoblast lineages to control bone homeostasis. Moreover, we have shown that Ifrd1 deficiency attenuates the interaction between β-catenin and HDAC1, subsequently increasing the acetylation of β-catenin at K49 and leading to accumulation in the nucleus and activation of β-catenin-dependent transcription of Opg in osteoblasts (10). β-Catenin controls osteoclastogenesis (37, 38); thus, it would be worth investigating whether Ifrd1 controls osteoclastogenesis by regulating β-catenin-dependent transcription, as observed in osteoblasts.
In conclusion, Ifrd1 modulates all three essential pathways of bone homeostasis: the cell-autonomous regulation of osteoblastogenesis, the osteoblast-dependent regulation of osteoclastogenesis, and the cell-autonomous regulation of osteoclastogenesis. Although the transcriptional programs regulating the three pathways are well defined, the mechanisms that regulate and coordinate them remain poorly understood. Ifrd1 may represent a novel target for the discovery and development of therapies and treatments for a variety of bone disorders, including osteoporosis, rheumatoid arthritis, and/or bone metastases of tumors.
Supplementary Material
ACKNOWLEDGMENTS
We thank S. L. Teitelbaum (Washington University, St. Louis, MO) and T. Kitamura (Tokyo University, Tokyo, Japan) for kindly providing GST-RANKL vector and PLAT-E cells, respectively.
This work was supported in part by Grants-in-Aid for Scientific Research to E.H. (23689004) from the Ministry of Education, Culture, Sports, Science and Technology, Japan.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.01075-15.
REFERENCES
- 1.Vadivelu SK, Kurzbauer R, Dieplinger B, Zweyer M, Schafer R, Wernig A, Vietor I, Huber LA. 2004. Muscle regeneration and myogenic differentiation defects in mice lacking TIS7. Mol Cell Biol 24:3514–3525. doi: 10.1128/MCB.24.8.3514-3525.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tirone F, Shooter EM. 1989. Early gene regulation by nerve growth factor in PC12 cells: induction of an interferon-related gene. Proc Natl Acad Sci U S A 86:2088–2092. doi: 10.1073/pnas.86.6.2088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gu Y, Harley IT, Henderson LB, Aronow BJ, Vietor I, Huber LA, Harley JB, Kilpatrick JR, Langefeld CD, Williams AH, Jegga AG, Chen J, Wills-Karp M, Arshad SH, Ewart SL, Thio CL, Flick LM, Filippi MD, Grimes HL, Drumm ML, Cutting GR, Knowles MR, Karp CL. 2009. Identification of IFRD1 as a modifier gene for cystic fibrosis lung disease. Nature 458:1039–1042. doi: 10.1038/nature07811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brkanac Z, Spencer D, Shendure J, Robertson PD, Matsushita M, Vu T, Bird TD, Olson MV, Raskind WH. 2009. IFRD1 is a candidate gene for SMNA on chromosome 7q22-q23. Am J Hum Genet 84:692–697. doi: 10.1016/j.ajhg.2009.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harada S, Rodan GA. 2003. Control of osteoblast function and regulation of bone mass. Nature 423:349–355. doi: 10.1038/nature01660. [DOI] [PubMed] [Google Scholar]
- 6.Boyle WJ, Simonet WS, Lacey DL. 2003. Osteoclast differentiation and activation. Nature 423:337–342. doi: 10.1038/nature01658. [DOI] [PubMed] [Google Scholar]
- 7.Karsenty G, Kronenberg HM, Settembre C. 2009. Genetic control of bone formation. Annu Rev Cell Dev Biol 25:629–648. doi: 10.1146/annurev.cellbio.042308.113308. [DOI] [PubMed] [Google Scholar]
- 8.Nakashima T, Hayashi M, Takayanagi H. 2012. New insights into osteoclastogenic signaling mechanisms. Trends Endocrinol Metab 23:582–590. doi: 10.1016/j.tem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 9.Silva I, Branco JC. 2011. Rank/Rankl/opg: literature review. Acta Reumatol Port 36:209–218. [PubMed] [Google Scholar]
- 10.Iezaki T, Onishi Y, Ozaki K, Fukasawa K, Takahata Y, Nakamura Y, Fujikawa K, Takarada T, Yoneda Y, Yamashita Y, Shioi G, Hinoi E. 2015. The transcriptional modulator interferon-related developmental regulator 1 in osteoblasts suppresses bone formation and promotes bone resorption. J Bone Miner Res doi: 10.1002/jbmr.2720. [DOI] [PubMed] [Google Scholar]
- 11.Chen RZ, Akbarian S, Tudor M, Jaenisch R. 2001. Deficiency of methyl-CpG binding protein-2 in CNS neurons results in a Rett-like phenotype in mice. Nat Genet 27:327–331. doi: 10.1038/85906. [DOI] [PubMed] [Google Scholar]
- 12.Nakamura T, Imai Y, Matsumoto T, Sato S, Takeuchi K, Igarashi K, Harada Y, Azuma Y, Krust A, Yamamoto Y, Nishina H, Takeda S, Takayanagi H, Metzger D, Kanno J, Takaoka K, Martin TJ, Chambon P, Kato S. 2007. Estrogen prevents bone loss via estrogen receptor alpha and induction of Fas ligand in osteoclasts. Cell 130:811–823. doi: 10.1016/j.cell.2007.07.025. [DOI] [PubMed] [Google Scholar]
- 13.Ferron M, Vacher J. 2005. Targeted expression of Cre recombinase in macrophages and osteoclasts in transgenic mice. Genesis 41:138–145. doi: 10.1002/gene.20108. [DOI] [PubMed] [Google Scholar]
- 14.Hinoi E, Nakatani E, Yamamoto T, Iezaki T, Takahata Y, Fujita H, Ishiura R, Takamori M, Yoneda Y. 2012. The transcription factor paired box-5 promotes osteoblastogenesis through direct induction of osterix and osteocalcin. J Bone Miner Res 27:2526–2534. doi: 10.1002/jbmr.1708. [DOI] [PubMed] [Google Scholar]
- 15.Dempster DW, Compston JE, Drezner MK, Glorieux FH, Kanis JA, Malluche H, Meunier PJ, Ott SM, Recker RR, Parfitt AM. 2013. Standardized nomenclature, symbols, and units for bone histomorphometry: a 2012 update of the report of the ASBMR Histomorphometry Nomenclature Committee. J Bone Miner Res 28:2–17. doi: 10.1002/jbmr.1805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bouxsein ML, Boyd SK, Christiansen BA, Guldberg RE, Jepsen KJ, Muller R. 2010. Guidelines for assessment of bone microstructure in rodents using micro-computed tomography. J Bone Miner Res 25:1468–1486. doi: 10.1002/jbmr.141. [DOI] [PubMed] [Google Scholar]
- 17.Hinoi E, Takarada T, Tsuchihashi Y, Fujimori S, Moriguchi N, Wang L, Uno K, Yoneda Y. 2006. A molecular mechanism of pyruvate protection against cytotoxicity of reactive oxygen species in osteoblasts. Mol Pharmacol 70:925–935. doi: 10.1124/mol.106.024398. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Hinoi E, Takemori A, Takarada T, Yoneda Y. 2005. Abolition of chondral mineralization by group III metabotropic glutamate receptors expressed in rodent cartilage. Br J Pharmacol 146:732–743. doi: 10.1038/sj.bjp.0706358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hinoi E, Ochi H, Takarada T, Nakatani E, Iezaki T, Nakajima H, Fujita H, Takahata Y, Hidano S, Kobayashi T, Takeda S, Yoneda Y. 2012. Positive regulation of osteoclastic differentiation by growth differentiation factor 15 upregulated in osteocytic cells under hypoxia. J Bone Miner Res 27:938–949. doi: 10.1002/jbmr.1538. [DOI] [PubMed] [Google Scholar]
- 20.Nakamura Y, Hinoi E, Iezaki T, Takada S, Hashizume S, Takahata Y, Tsuruta E, Takahashi S, Yoneda Y. 2013. Repression of adipogenesis through promotion of Wnt/beta-catenin signaling by TIS7 up-regulated in adipocytes under hypoxia. Biochim Biophys Acta 1832:1117–1128. doi: 10.1016/j.bbadis.2013.03.010. [DOI] [PubMed] [Google Scholar]
- 21.Yamamoto T, Hinoi E, Fujita H, Iezaki T, Takahata Y, Takamori M, Yoneda Y. 2012. The natural polyamines spermidine and spermine prevent bone loss through preferential disruption of osteoclastic activation in ovariectomized mice. Br J Pharmacol 166:1084–1096. doi: 10.1111/j.1476-5381.2012.01856.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Takayanagi H, Kim S, Koga T, Nishina H, Isshiki M, Yoshida H, Saiura A, Isobe M, Yokochi T, Inoue J, Wagner EF, Mak TW, Kodama T, Taniguchi T. 2002. Induction and activation of the transcription factor NFATc1 (NFAT2) integrate RANKL signaling in terminal differentiation of osteoclasts. Dev Cell 3:889–901. doi: 10.1016/S1534-5807(02)00369-6. [DOI] [PubMed] [Google Scholar]
- 23.Asagiri M, Sato K, Usami T, Ochi S, Nishina H, Yoshida H, Morita I, Wagner EF, Mak TW, Serfling E, Takayanagi H. 2005. Autoamplification of NFATc1 expression determines its essential role in bone homeostasis. J Exp Med 202:1261–1269. doi: 10.1084/jem.20051150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Micheli L, Leonardi L, Conti F, Maresca G, Colazingari S, Mattei E, Lira SA, Farioli-Vecchioli S, Caruso M, Tirone F. 2011. PC4/Tis7/IFRD1 stimulates skeletal muscle regeneration and is involved in myoblast differentiation as a regulator of MyoD and NF-kappaB. J Biol Chem 286:5691–5707. doi: 10.1074/jbc.M110.162842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chen LF, Mu Y, Greene WC. 2002. Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-kappaB. EMBO J 21:6539–6548. doi: 10.1093/emboj/cdf660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiernan R, Bres V, Ng RW, Coudart MP, El Messaoudi S, Sardet C, Jin DY, Emiliani S, Benkirane M. 2003. Post-activation turn-off of NF-kappa B-dependent transcription is regulated by acetylation of p65. J Biol Chem 278:2758–2766. doi: 10.1074/jbc.M209572200. [DOI] [PubMed] [Google Scholar]
- 27.Feng X, McDonald JM. 2011. Disorders of bone remodeling. Annu Rev Pathol 6:121–145. doi: 10.1146/annurev-pathol-011110-130203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McClung MR, Lewiecki EM, Cohen SB, Bolognese MA, Woodson GC, Moffett AH, Peacock M, Miller PD, Lederman SN, Chesnut CH, Lain D, Kivitz AJ, Holloway DL, Zhang C, Peterson MC, Bekker PJ. 2006. Denosumab in postmenopausal women with low bone mineral density. N Engl J Med 354:821–831. doi: 10.1056/NEJMoa044459. [DOI] [PubMed] [Google Scholar]
- 29.Ominsky MS, Stouch B, Schroeder J, Pyrah I, Stolina M, Smith SY, Kostenuik PJ. 2011. Denosumab, a fully human RANKL antibody, reduced bone turnover markers and increased trabecular and cortical bone mass, density, and strength in ovariectomized cynomolgus monkeys. Bone 49:162–173. doi: 10.1016/j.bone.2011.04.001. [DOI] [PubMed] [Google Scholar]
- 30.Huang H, Ryu J, Ha J, Chang EJ, Kim HJ, Kim HM, Kitamura T, Lee ZH, Kim HH. 2006. Osteoclast differentiation requires TAK1 and MKK6 for NFATc1 induction and NF-kappaB transactivation by RANKL. Cell Death Differ 13:1879–1891. doi: 10.1038/sj.cdd.4401882. [DOI] [PubMed] [Google Scholar]
- 31.Perkins ND. 2007. Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat Rev Mol Cell Biol 8:49–62. doi: 10.1038/nrm2083. [DOI] [PubMed] [Google Scholar]
- 32.Baud V, Karin M. 2009. Is NF-kappaB a good target for cancer therapy? Hopes and pitfalls. Nat Rev Drug Discov 8:33–40. doi: 10.1038/nrd2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hinoi E, Iezaki T, Ozaki K, Yoneda Y. 2014. Nuclear factor-kappaB is a common upstream signal for growth differentiation factor-5 expression in brown adipocytes exposed to proinflammatory cytokines and palmitate. Biochem Biophys Res Commun 452:974–979. doi: 10.1016/j.bbrc.2014.09.022. [DOI] [PubMed] [Google Scholar]
- 34.Jimi E, Aoki K, Saito H, D'Acquisto F, May MJ, Nakamura I, Sudo T, Kojima T, Okamoto F, Fukushima H, Okabe K, Ohya K, Ghosh S. 2004. Selective inhibition of NF-kappa B blocks osteoclastogenesis and prevents inflammatory bone destruction in vivo. Nat Med 10:617–624. doi: 10.1038/nm1054. [DOI] [PubMed] [Google Scholar]
- 35.Chang J, Wang Z, Tang E, Fan Z, McCauley L, Franceschi R, Guan K, Krebsbach PH, Wang CY. 2009. Inhibition of osteoblastic bone formation by nuclear factor-kappaB. Nat Med 15:682–689. doi: 10.1038/nm.1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buerki C, Rothgiesser KM, Valovka T, Owen HR, Rehrauer H, Fey M, Lane WS, Hottiger MO. 2008. Functional relevance of novel p300-mediated lysine 314 and 315 acetylation of RelA/p65. Nucleic Acids Res 36:1665–1680. doi: 10.1093/nar/gkn003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wei W, Zeve D, Suh JM, Wang X, Du Y, Zerwekh JE, Dechow PC, Graff JM, Wan Y. 2011. Biphasic and dosage-dependent regulation of osteoclastogenesis by beta-catenin. Mol Cell Biol 31:4706–4719. doi: 10.1128/MCB.05980-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Albers J, Keller J, Baranowsky A, Beil FT, Catala-Lehnen P, Schulze J, Amling M, Schinke T. 2013. Canonical Wnt signaling inhibits osteoclastogenesis independent of osteoprotegerin. J Cell Biol 200:537–549. doi: 10.1083/jcb.201207142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.