Abstract
Phenotypic plasticity involves a process in which cells transiently acquire phenotypic traits of another lineage. Two commonly studied types of phenotypic plasticity are epithelial-mesenchymal transition (EMT) and mesenchymal-epithelial transition (MET). In carcinomas, EMT drives invasion and metastatic dissemination, while MET is proposed to play a role in metastatic colonization. Phenotypic plasticity in sarcomas is not well studied; however, there is evidence that a subset of sarcomas undergo an MET-like phenomenon. While the exact mechanisms by which these transitions occur remain largely unknown, it is likely that some of the same master regulators that drive EMT and MET in carcinomas also act in sarcomas. In this study, we combined mathematical models with bench experiments to identify a core regulatory circuit that controls MET in sarcomas. This circuit comprises the microRNA 200 (miR-200) family, ZEB1, and GRHL2. Interestingly, combined expression of miR-200s and GRHL2 further upregulates epithelial genes to induce MET. This effect is phenocopied by downregulation of either ZEB1 or the ZEB1 cofactor, BRG1. In addition, an MET gene expression signature is prognostic for improved overall survival in sarcoma patients. Together, our results suggest that a miR-200, ZEB1, GRHL2 gene regulatory network may drive sarcoma cells to a more epithelial-like state and that this likely has prognostic relevance.
INTRODUCTION
Phenotypic plasticity is defined as the reversible conversion of cellular phenotypes from one state to another. The two most commonly studied types of plasticity are epithelial-mesenchymal transition (EMT) and the reverse, mesenchymal-epithelial transition (MET). These phenotypic transitions play important roles in normal development (reviewed in references 1, to ,4) and wound healing (reviewed in reference 5); however, similar pathways and gene expression programs can also be coopted by the cell during fibrosis (reviewed in references 6, to ,8) and carcinoma progression (reviewed in references 9, to ,12). In the context of carcinoma progression, a subset of cells within the tumor are thought to undergo an EMT, which enables those cells to break free from the tumor mass via loss of cell-cell adhesions (13, 14) and upregulate invasive programs that facilitate dissemination (13). In addition to these phenotypic changes, EMT also contributes to alterations in cancer cell metabolism (10), drug resistance (15, 16), tumor initiation ability (17, 18), and perhaps even host immune evasion (19). EMT is often accompanied by downregulation of proliferation (20, 21), and, in some cases, MET is important for reinitiation of proliferation during metastatic colonization (22). It is important to note that as the field of phenotypic plasticity has matured, particularly in the context of carcinoma progression, EMT and MET have become recognized as more of a spectrum of phenotypes, rather than discrete states of fully differentiated epithelial and mesenchymal phenotypes. These metastable, or hybrid, E/M transition states have been observed in clinical specimens, including the circulation (23, 24) and the metastatic niche (reviewed in reference 25). Yet despite these observations, the importance of the metastable E/M state in metastasis remains unknown.
The relevance of phenotypic plasticity as a critical pathway in the metastatic cascade in carcinoma progression has been studied for decades now; however, more recent observations suggest that some of the drivers of plasticity in carcinomas may also contribute to sarcoma aggressiveness. For example, the EMT transcription factor and master regulator, SNAIL (SNAI1), was shown to be associated with poorer overall survival in human sarcomas, and ectopic expression of SNAIL in fibroblasts induced sarcomas in immunodeficient mice (26). Similarly, ZEB1, another EMT transcription factor, was upregulated in osteosarcomas compared to normal bone, and osteosarcoma patients with metastases had higher ZEB1 levels than those without metastases (27). In addition, sarcoma patients with higher levels of the epithelial marker E-cadherin have improved survival compared to those with low or no E-cadherin (28). It is perhaps not surprising that just as epithelium-derived cancers are capable of awakening the embryologic pathways related to phenotypic plasticity, so, too, could mesenchymal cancers transition to a more epithelial-like phenotype.
In the present work, we used both predictive mathematical models and validation experiments to develop a conceptual framework for the control of cellular phenotype in sarcomas. Our results predict that the combined expression of the microRNA 200 (miR-200) family and upregulation of an epithelial gene activator, GRHL2, drive MET in sarcomas. Indeed, both GRHL2 overexpression and downregulation of ZEB1—by either RNA interference (RNAi)-mediated silencing or miR-200 overexpression—act synergistically to control the upregulation of epithelial genes, including the E-cadherin gene, and consequently, MET. Together, our results highlight the functional interplay between epithelial and mesenchymal regulators in driving MET in sarcomas.
MATERIALS AND METHODS
Model construction and sensitivity analysis.
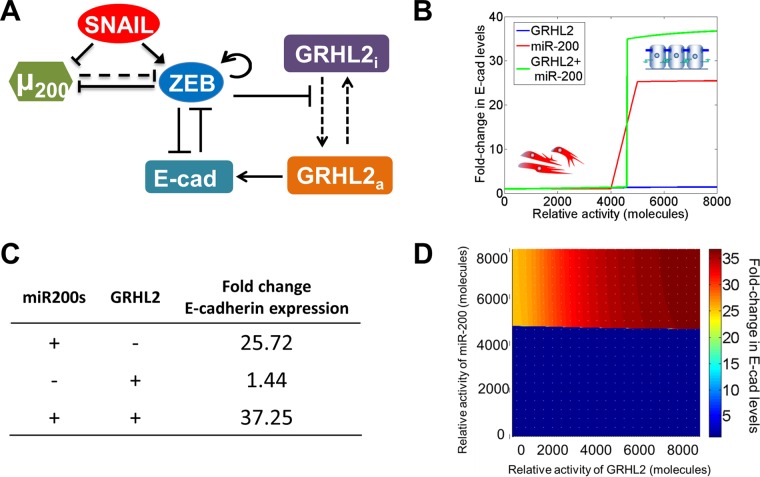
We generalized the modeling framework of miRNA-mRNA interactions developed for the miR-200/ZEB/SNAIL circuit by Lu et al. (29) to include the connections with E-cadherin and GRHL2. The initial model (see Fig. 1A) consists of eight species—SNAIL mRNA, SNAIL protein, ZEB mRNA, ZEB protein, the miR-200 family (miR-200a, -b, and -c, miR-141, and miR-429), E-cadherin mRNA, GRHL2 mRNA, and GRHL2 protein. In the revised model (see Fig. 4A), we introduced two species to consider the active and inactive forms of GRHL2 protein. The model construction and parameter estimation details and the sensitivity analysis are given in the supplemental material. The model was simulated in MATLAB (Mathworks Inc.), and bifurcation plots were constructed using MATCONT (30). The term relative activity in Fig. 1B and 4B and D represents the external activation that was applied on respective moieties.
FIG 1.
E-cadherin induction by GRHL2 is cell line dependent. (A) Construction of a regulatory system using literature-based interactions suggests that a ZEB1-GRHL2 feedback loop controls E-cadherin. (B) Based on the model proposed in panel A, activation of GRHL2 enhances E-cadherin levels to control cellular phenotype. (C) Ectopic expression of GRHL2 has no effect on ZEB1 mRNA levels. (D and E) Expression of GRHL2 activates E-cadherin mRNA (D) and protein (E) in MDA-MB-231 cells but not in RD or 143B sarcoma cells. *, P < 0.05.
FIG 4.
A proposed regulatory network of E-cadherin control. (A) ZEB1 represses GRHL2-induced activation of E-cadherin. Dashed lines indicate the transition of GRHL2 from inactive to active and miR-200 translational repression of ZEB1; solid arrows and bars show transcriptional activation and repression, respectively, of a given gene. (B) According to the proposed model, activation of GRHL2 results in almost no change in E-cadherin. Conversely, miR-200 upregulation induces E-cadherin expression, while the combined expression of miR-200s and GRHL2 has a synergistic effect on E-cadherin. (C) Relative levels of E-cadherin upon combined miR-200 and GRHL2 upregulation are synergistic. (D) Heat map of the miR-200- and GRHL2-induced E-cadherin expression model predicts that at a threshold of miR-200 expression, E-cadherin is upregulated. This effect is enhanced by GRHL2 expression (upper right, in dark red).
Cell culture and GRHL2 transduction.
All cells were cultured in Dulbecco's modified Eagle's medium (DMEM), high glucose, supplemented with 10% fetal bovine serum (FBS) and 1% penicillin-streptomycin (pen-strep) at 37°C and 5% CO2 in a humidified incubator. To generate lentiviruses, 3 × 105 HEK293T cells were cotransfected with either 2 μg of empty vector (pCMV-UBC-EGFP) or pCMV-GRHL2-UBC-EGFP, 1.8 μg of pΔ8.9, and 0.2 μg of pGAG-POL. Viral medium was collected 2 and 3 days later, filtered, and added to target cells, along with 20 μg/ml Polybrene. Cells positive for enhanced green fluorescent protein (EGFP) were sorted by flow cytometry at the Duke University Flow Cytometry Shared Resource.
siRNA-mediated knockdowns and miR-200 transfections.
A total of 20 nM Allstars nonsilencing (Qiagen) or target gene small interfering RNA (siRNA; Qiagen) was diluted in Opti-MEM and mixed with Lipofectamine RNAiMax diluted in Opti-MEM. Lipofectamine-siRNA mixtures were incubated for 20 min at room temperature, and cells were reverse transfected overnight in DMEM–10% FBS–1% pen-strep. The medium was replaced the next day. The same protocol was used for reverse transfection of miR-200s.
RNA extraction, RT, and RT-qPCR.
RNA was extracted using either the Promega RNA Mini-prep kit or the Zymo RNA extraction kit according to the manufacturers' protocols. A minimum of 100 ng of total RNA was used in each reverse transfection reaction using random hexamer primers in a 20-μl total reaction volume by following the ABI high-capacity cDNA reverse transcription (RT) kit protocol (Thermo Fisher). RT reaction mixtures were incubated by following the manufacturer's protocol in a SimpliAmp thermocycler (Life Technologies). RT reaction mixtures were diluted 1:5 in nuclease-free H2O, and RT-quantitative PCR (RT-qPCR) was performed using 2 μl of RT, 0.06 μl of each primer (10 μM stock), and 5 μl of KAPA SYBRFAST Universal 2× qPCR master mix in a 10-μl total reaction volume. Reactions were performed using a Vii7 real time-PCR detection system (Applied Biosystems).
Western blotting and immunofluorescence staining.
For Western blotting, total protein was extracted from cultured cells by washing cells twice in 1× ice-cold phosphate-buffered saline (PBS) and lysing them in 1× radioimmunoprecipitation assay (RIPA) buffer supplemented with 1× protease inhibitor cocktail (Roche). Cell lysates were incubated at 4°C for 15 min with rocking and clarified by centrifugation at high speed in a benchtop centrifuge for 5 min, and clarified lysates were incubated for 5 min at 95°C in 1× Laemmli sample loading buffer. Lysates were separated in 4 to 12% NuPAGE Novex bis-Tris gels (Thermo Fisher). Proteins were transferred onto nitrocellulose membranes (GE Healthcare Life Sciences) in 1× NuPAGE transfer buffer (Thermo Fisher) at 50 V for 1 to 2 h at room temperature. Membranes were blocked for 1 h at room temperature or overnight at 4°C in StartingBlock (Thermo Fisher). Primary antibodies were diluted in StartingBlock, incubated for 1 h at room temperature or overnight at 4°C, washed twice for 5 min in 1× PBS with 0.05% Tween 20 (PBS-T), and incubated with IRDye-coupled donkey anti-rabbit or donkey anti-mouse secondary antibodies diluted 1:20,000 in StartingBlock. Images were captured on an Odyssey Fc imager (LI-COR). For immunofluorescence imaging, cells were fixed for 15 min at room temperature in 4% paraformaldehyde, permeabilized for 30 min at room temperature in PBS with 0.2% Triton X-100, blocked for 30 min at room temperature in 5% bovine serum albumin (BSA) diluted in PBS, and incubated overnight at 4°C in primary antibodies diluted in BSA-PBS. Subsequent to washing with PBS, cells were incubated with Alexa Fluor secondary antibodies (1:2,000 dilution in BSA-PBS), along with 1 μg/ml of Hoechst dye (Sigma), for 1 h at room temperature in the dark. Images were captured on an inverted Olympus IX 71 epifluorescence microscope at a total magnification of ×200 unless otherwise specified in the figure legends. A complete list of primary antibodies and their dilutions is provided in the supplemental material.
Analysis of methylation data from The Cancer Genome Atlas (TCGA).
GRHL2 and E-cadherin methylation (DNA Methylation 450K) data were downloaded from the UCSC Cancer Genomics Browser (https://genome-cancer.soe.ucsc.edu/). Boxplots were created using RStudio (version 0.98.1091). To investigate whether GRHL2 or E-cadherin methylation was significantly different in epithelial-like or mesenchymal-like patient samples, each patient sample was assigned a value of “epithelial-like” or “mesenchymal-like” by parsing samples based on their transcriptome sequencing (RNA-Seq) data using a previously published EMT signature (48). The EMT signature is as follows: (FN1 + VIM + ZEB1 + ZEB2 + TWIST1 + TWIST2 + SNAI1 + SNAI2 + CDH2) − (CLDN4 + CLDN7 + TJP3 + MUC1 + CDH1). Kaplan-Meier curves were created using the KM function, and the value of the upper or lower curve was used to set the E-like versus M-like state. Any sample above the value for the line on the Kaplan-Meier curve corresponding to the E-like state was considered “mesenchymal-like,” while any sample equal to or below the value for the E-like state was considered “epithelial-like.”
Analysis of EMT gene expression profiles and clinical outcomes.
TCGA RNA-Seq data from 250 soft tissue sarcomas was downloaded using the UCSC Cancer Genomics Browser. Samples were designated mesenchymal-like or epithelial-like using the upper and lower values on the KM curve function, as described above, and Kaplan-Meier curves were generated in JMP Pro 12. Kaplan-Meier curves for osteosarcoma microarray data were generated using the Kaplan-Meier plot function in the R2 genomics analysis and visualization platform (http://hgserver1.amc.nl/cgi-bin/r2/main.cgi) with the “Kaplan scan a single gene” function and the “cutoff_modus” set to scan.
Statistical analyses.
All data were analyzed for statistically significant differences using the Student t test (for two comparisons) or analysis of variance (for multiple comparisons) in JMP Pro 12. Any P value of <0.05 was considered statistically significant.
RESULTS
Modeling an EMT/MET regulatory network.
E-cadherin is a key protein involved in epithelial cell-cell adhesion, and the loss of E-cadherin is a hallmark of EMT. We used experimentally derived information based on published literature to construct a gene regulatory network based on the known mediators of E-cadherin and EMT (see Materials and Methods and supplemental material for a detailed description of the model construction). The mutually inhibitory feedback loop between the miR-200 family and the transcription factor family ZEB1/2 (31, 32) has been proposed to act as a three-way decision-making switch, enabling the coexistence of three phenotypes: epithelial (high miR-200 and low ZEB), hybrid epithelial/mesenchymal (moderate miR-200 and moderate ZEB), and mesenchymal (low miR-200 and high ZEB) (29). Our model couples this miR200/ZEB loop with another mutually inhibitory loop including miR-34/SNAIL (33) by incorporating their interconnections: activation of ZEB by SNAIL (34), inhibition of miR-200 by SNAIL (32, 35), and inhibition of miR-34 by ZEB (36). We treat SNAIL as the external input signal to the miR-200/ZEB circuit (Fig. 1A), as these interconnections do not change the qualitative behavior of EMT decision-making (29).
The miR-200 family consists of two subgroups: one containing miR-141 and miR-200a and the other including miR-200b, miR-200c, and miR-429. The ZEB transcription factor family includes two orthologues, ZEB1 and ZEB2. The 3′ untranslated region (UTR) of ZEB1 has a total of eight conserved binding sites for miR-200: three for the first subgroup and five for the second subgroup. The 3′ UTR of ZEB2 has nine conserved binding sites for miR-200 (three for the first subgroup and six for the second subgroup). Experiments suggest that the expression of miR-200c can restore E-cadherin and induce MET (37). Therefore, in our miR-200/ZEB module, we considered six binding sites (number of binding sites of the second subgroup on ZEB2) on the 3′ UTR of ZEB for the binding of miR-200. Also, the members of miR-200 family are located on two different chromosomes: miR-200c and miR-141 on chromosome 12 (with three conserved ZEB-binding sites) and miR-200b, miR-200a, and miR-141 on chromosome 1 (with two conserved ZEB-binding sites). We consider three ZEB-binding sites on the promoter region of miR-200 (31, 32). ZEB can also activate its own transcription both via stabilizing SMAD complexes (38) and by activating CD44s (39); hence, we assume two binding sites for ZEB self-activation. SNAIL, a well-known EMT inducer, can transcriptionally inhibit miR-200 and activate ZEB (32, 34, 35). Both of these regulatory steps have been postulated to happen through two binding sites (29).
GRHL2, a key regulator of morphogenesis (40), forms a mutually inhibitory loop with ZEB. The promoter region of GRHL2 has three binding sites for ZEB1, and the promoter of ZEB1 has one binding site for GRHL2 (41, 42). GRHL2 can also activate E-cadherin (43), but E-cadherin is repressed by ZEB1 via binding to E-boxes in its promoter region (Fig. 1A) (44). Also, E-cadherin sequesters β-catenin at the membrane; β-catenin can translocate to the nucleus and activate ZEB via binding to TCF/LEF (45). This cascade has been represented as an inhibitory link from E-cadherin to ZEB. Thus, GRHL2 acts in two ways to upregulate E-cadherin: (i) GRHL2 represses the E-cadherin repressor ZEB1 (46), and (ii) GRHL2 directly activates E-cadherin transcription (43). This model predicted that activating GRHL2 would increase E-cadherin expression to levels commensurate with MET (Fig. 1B).
Combined expression of GRHL2 and miR-200s increases E-cadherin and induces MET in sarcoma cells.
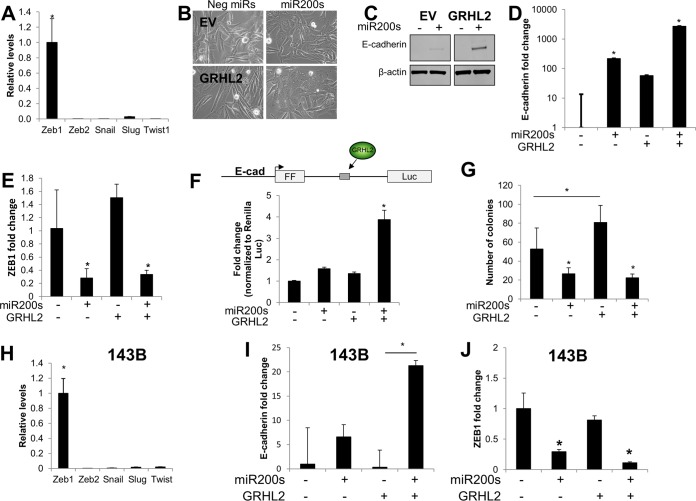
Based on our model, we sought to experimentally validate the importance of GRHL2 in driving MET and E-cadherin expression. We selected several model cell lines to better understand the potential ability for GRHL2 to influence epithelial plasticity in cells from different lineages (e.g., epithelial versus mesenchymal). The MDA-MB-231 cell line is a post-EMT breast cancer model that is frequently used in studies of epithelial plasticity. RD and 143B cells represent models of soft tissue sarcoma (specifically, rhabdomyosarcoma) and osteosarcoma, respectively. To test the capacity of GRHL2 to activate E-cadherin and induce MET, we ectopically overexpressed GRHL2 using lentiviral transduction in each of the three cell lines (see Fig. S1 in the supplemental material). GRHL2 was expressed to similar levels in all three cell lines, as quantified by RT-qPCR (see Fig. S1A), and localized to the nucleus in all three lines (see Fig. S1B). Surprisingly, GRHL2 expression had no significant effect on ZEB1 in any of the cell lines (Fig. 1C); however, GRHL2 expression led to a relatively modest, but significant, induction of E-cadherin mRNA (Fig. 1D) and protein (Fig. 1E) in MDA-MB-231 cells. Conversely, GRHL2 expression had no effect on E-cadherin in either of the sarcoma lines (Fig. 1D and E). We postulated that GRHL2 expression was insufficient to overcome the repression of E-cadherin by other factors. RT-qPCR of five major EMT-TFs in RD cells revealed that the E-cadherin transcriptional repressor and EMT master regulator, ZEB1, was expressed at least 35-fold higher than the other EMT-TFs (Fig. 2A). This prompted us to ask whether repression of ZEB1 by the miR-200 family was able to enhance the effect of GRHL2 on E-cadherin and MET in RD cells. Transfection of miR-200 family members miR-200a, -b, and -c (here referred to as miR-200s) had no obvious effect on the morphology of RD cells. However, addition of miR-200s in the context of GRHL2 expression led to a morphological change consistent with MET; cells changed from a spindle-shaped appearance with few cell-cell contacts to a rounded, cobblestone-like appearance with increased cell-cell contacts (Fig. 2B). The morphological change was also accompanied by upregulation of epithelial markers E-cadherin (Fig. 2C), EpCAM (see Fig. S2A in the supplemental material), and TJP1 (also known as zona occludens 1 [ZO-1]) at cell-cell contacts (see Fig. S2B, white arrows). Interestingly, E-cadherin was upregulated with addition of miR-200s, but the effect of miR-200s on E-cadherin was enhanced in a synergistic fashion in the presence of GRHL2 overexpression (Fig. 2D; note log scale). Although GRHL2 had no effect on ZEB1, transfection of miR-200s reduced ZEB1 mRNA (Fig. 2E). To test whether the synergistic effect of GRHL2 and miR-200s was due to sequence elements within the E-cadherin gene, we created a luciferase-based reporter in which firefly luciferase (FFLuc) is driven by the E-cadherin promoter. In addition to promoter control, the FFLuc open reading frame is interrupted by a constitutively spliced intron, modified from one described in reference 47, harboring a region of E-cadherin intron 2 in which GRHL2 has been demonstrated to bind (43) (Fig. 2F). Consistent with the results from the endogenous gene, transfection of this E-cadherin/GRHL2 reporter into RD cells in the presence of GRHL2 and miR-200s led to a synergistic increase in FFLuc expression (Fig. 2F). Soft-agar assays revealed that MET induction in RD cells significantly reduced the number of colonies capable of growing in an anchorage-independent setting (Fig. 2G). This MET-associated reduction in anchorage-independent growth was driven exclusively by the miR-200s (Fig. 2G).
FIG 2.
Combined expression of miR-200s and GRHL2 induces MET. (A) ZEB1 is the most highly expressed of five major EMT master regulators in RD cells. (B) Combined expression of miR-200s and GRHL2 induces a morphological change consistent with MET. (C and D) Expression of miR-200s induces E-cadherin expression, while combined expression of miR-200s and GRHL2 has a multiplicative effect on E-cadherin protein (C) and mRNA (D) levels (note log scale). (E) ZEB1 mRNA levels are inhibited by miR-200s but not by GRHL2. (F) Schematic of the E-cadherin/GRHL2 reporter. The firefly luciferase transcript is driven by the E-cadherin promoter. The transcript is interrupted by a portion of E-cadherin intron 2 that contains the GRHL2 binding site. The E-cadherin/GRHL2 reporter recapitulates the synergistic effect of GRHL2 and miR-200s on the endogenous E-cadherin gene. (G) Anchorage-independent growth of RD cells is inhibited by miR-200s and increased by GRHL2 in soft-agar assays. (H) ZEB1 is a highly expressed EMT master regulator in 143B osteosarcoma cells. (I) GRHL2 and miR-200s have a multiplicative effect on E-cadherin mRNA in 143B cells. (J) ZEB1 is downregulated by miR-200s but not by GRHL2 in 143B cells. *, P < 0.05.
Similar to the RD cells, the 143B osteosarcoma cell line also expressed high levels of ZEB1 compared to other EMT-TFs (Fig. 2H), and combined transfection of miR-200s with GRHL2 expression led to a multiplicative increase in E-cadherin (Fig. 2I). Also similar to the results with RD cells, GRHL2 alone had no effect on ZEB1, and miR-200s inhibited ZEB1 (Fig. 2J). Conversely, in MDA-MB-231 breast cancer cells, which have almost equal levels of expression of Zeb1 and Slug (see Fig. S3A in the supplemental material), overexpression of miR-200 was sufficient to induce upregulation of E-cadherin, EpCAM, and CGN, and downregulation of ZEB1. GRHL2, transfected either alone or in combination with miR-200s, had only a modest effect on the expression of E-cadherin, ZEB1, or other MET markers (see Fig. S3) in MDA-MB-231 cells. We also tested whether combined expression of GRHL2 and miR-200s induced MET using three additional lines, including BT-549 triple-negative breast cancer cells (see Fig. S4), U2-OS osteosarcoma cells (see Fig. S5), and SK-LMS leiomyosarcoma cells (see Fig. S6). Overall, the results were consistent with an increased effect on MET induction with combined expression of GRHL2 and miR-200s, with the miR-200s driving the majority of MET-like gene expression changes (see Fig. S4 to S6).
ZEB1 repression and GRHL2 expression induce E-cadherin expression.
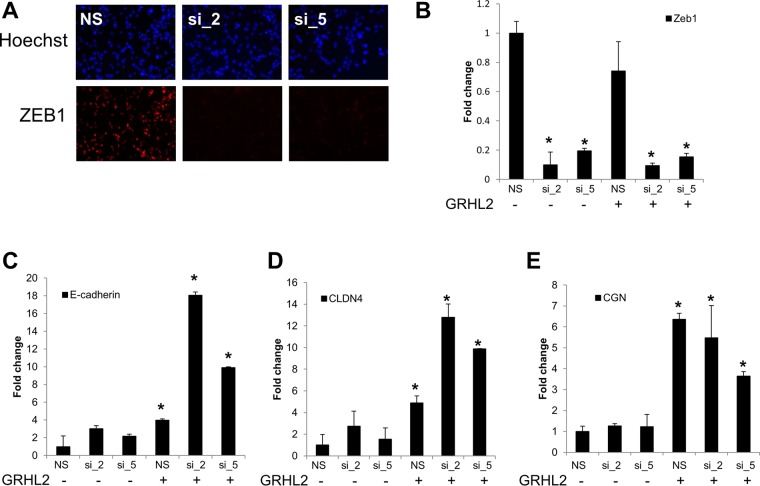
Given the relatively broad effects of miR-200s on potentially hundreds of targets, we next asked whether the combinatorial effect we observed on E-cadherin and MET in sarcoma cells was through ZEB1. To determine this, we performed siRNA-mediated knockdown of ZEB1 using two independent siRNAs to mitigate the potential for off-target effects. Both siRNAs significantly reduced ZEB1 protein (Fig. 3A) and mRNA (Fig. 3B) in RD cells. Consistent with the hypothesis that miR-200s repressed ZEB1 to facilitate GRHL2 activation of E-cadherin, we observed a larger increase in E-cadherin with ZEB1 knockdown and GRHL2 expression than with either factor alone (Fig. 3C). Similarly, we also found this combinatorial upregulation of another epithelial cell-cell adhesion molecule, claudin 4 (CLDN4), with ZEB1 knockdown and GRHL2 expression (Fig. 3D). Conversely, a third epithelial cell adhesion molecule, cingulin (CGN), appeared to be solely regulated by GRHL2; knockdown of ZEB1 had no effect (Fig. 3E). Together, our results suggest that downregulating ZEB1 via miR-200s has no effect on any of these epithelial genes without the presence of an epithelial gene activator, GRHL2.
FIG 3.
GRHL2 competes with ZEB1 to activate E-cadherin. (A and B) siRNA-mediated knockdown of ZEB1 protein (A) and mRNA (B) in RD cells. NS, nonsilencing control siRNA; si_2 and si_5, two independent siRNAs targeting ZEB1. (C) E-cadherin is upregulated upon ZEB1 knockdown only in the presence of GRHL2 expression. (D) The cell-cell adhesion molecule CLDN4 is upregulated upon ZEB1 knockdown only in the presence of GRHL2 expression. (E) Unlike E-cadherin and claudin 4, ZEB1 knockdown has no additional effect on the GRHL2-induced upregulation of the gene for the cell-cell adhesion molecule cingulin (CGN). *, P < 0.05.
A revised model predicts that GRHL2 exists in two possible states.
Based on our experimental results, we revised our predictive model. In the revised model, we parsed GRHL2 into two categories: inactive (i) and active (a) forms. Although we refer to the inactive and active forms as conceptual constructions to view the potential states of GRHL2, these states can potentially reflect one of two scenarios: GRHL2 being blocked from activating E-cadherin (inactive) and GRHL2 capable of activating E-cadherin (active). We constructed the model so that rather than inhibiting GRHL2 levels directly, ZEB1 prevents GRHL2 from transitioning to the active state (Fig. 4A). This realization is consistent with our experimental data that show upregulation of epithelial gene targets by GRHL2 only upon ZEB1 knockdown (Fig. 3C and D). Importantly, the revised model recapitulates the synergistic effect of miR-200s and GRHL2 on E-cadherin. GRHL2 has little effect on E-cadherin (Fig. 4B, blue line; quantified in Fig. 4C), while addition of miR-200s alone increases E-cadherin (Fig. 4B, red line; quantified in Fig. 4C). Overexpression of both GRHL2 and miR-200s is predicted to enhance E-cadherin expression in a synergistic fashion (Fig. 4B, green line; quantified in Fig. 4C). When we plotted various degrees of overexpression of GRHL2 and miR-200s on the x and y axes, respectively, we observed the largest effect on E-cadherin levels with the activation of miR-200s, while GRHL2 alone contributed very little to E-cadherin activation (Fig. 4D). Yet the effect of GRHL2 in increasing E-cadherin is visible only once miR-200s reach a threshold value and activate E-cadherin (Fig. 4D). While the exact numerical values of fold change of levels of E-cadherin depend on changes in model parameters, the relatively small effect of GRHL2 in activating E-cadherin in the absence of miR-200s and the synergistic induction of E-cadherin levels upon combinatorial overexpression of miR-200s and GRHL2 are robust features of the model (see the supplemental material). All of these observations are consistent with our experimental data, suggesting that the modeled regulatory network is capable of generating biologically relevant outputs.
GRHL2-mediated regulation of E-cadherin and cellular phenotype depends on BRG1 and GRHL2 methylation.
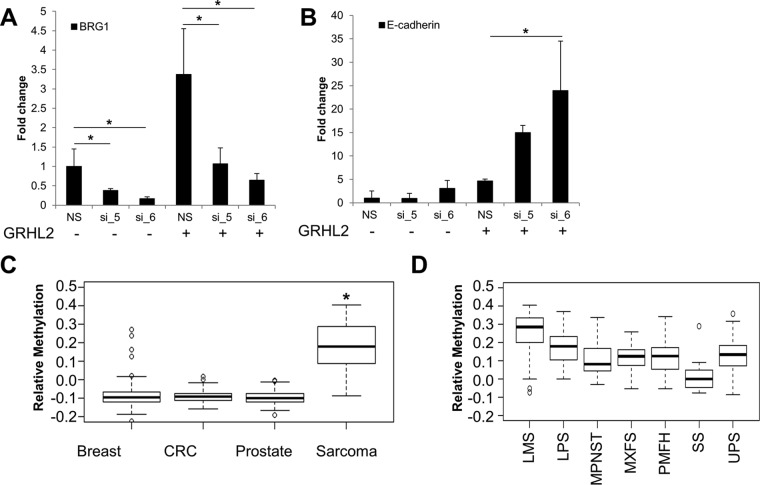
The revised predictive model prompted us to hypothesize that regulation of E-cadherin by GRHL2 could be controlled as follows: by (i) GRHL2-mediated activation of E-cadherin and/or (ii) GRHL2 expression levels. To address the first hypothesis, we knocked down the chromatin remodeling protein BRG1, which has been shown to act as a cofactor of ZEB1 in repressing E-cadherin by condensing the chromatin around the E-cadherin promoter (44). We knocked down BRG1 using two independent siRNAs in RD cells containing either an empty vector or GRHL2 (Fig. 5A). While siRNA-mediated silencing of BRG1 had no effect on E-cadherin expression in empty vector cells, knockdown of BRG1 in the context of GRHL2 expression led to a significant increase in E-cadherin mRNA (Fig. 5B). The increase in E-cadherin was also dose dependent; E-cadherin was higher in cells for which BRG1 was knocked down to a greater degree (Fig. 5B). These results suggest that GRHL2-based activation of E-cadherin may be dependent on the degree to which the chromatin surrounding the E-cadherin promoter is available to be bound by GRHL2.
FIG 5.
Regulatory steps in the GRHL2-mediated activation of E-cadherin. (A) BRG1 mRNA expression upon siRNA-mediated knockdown of BRG1. (B) E-cadherin is upregulated in a GRHL2-dependent manner upon BRG1 knockdown. (C) Analysis of GRHL2 methylation status from The Cancer Genome Atlas indicates that GRHL2 methylation is significantly increased in sarcomas compared to epithelium-derived breast, colorectal (CRC), and prostate cancers. (D) GRHL2 methylation is lower in synovial sarcoma than in other sarcoma subtypes. *, P < 0.05.
To test the second hypothesis, that GRHL2 itself could be regulated as a mechanism to control E-cadherin expression and cellular phenotype, we queried data from The Cancer Genome Atlas (TCGA). Interestingly, we found that methylation of the DNA upstream of GRHL2 was significantly higher in mesenchyme-derived sarcoma cancer specimens than in epithelium-derived breast, colorectal (CRC), and prostate cancers (Fig. 5C). When we further analyzed GRHL2 methylation by sarcoma subtype, we found that GRHL2 methylation was lowest in synovial sarcomas (Fig. 5D). Synovial sarcomas are classically characterized by a biphasic histologic appearance consisting of a distinct cellular population with mesenchymal morphology (and expressing mesenchymal biomarkers) and another distinct population with epithelial morphology (and expressing epithelial biomarkers). In addition, when we used a previously published signature of EMT (48) to separate sarcomas into more epithelial-like and mesenchymal-like samples, we found that GRHL2 methylation was significantly higher in the more mesenchymal-like samples (see Fig. S7 in the supplemental material). While this was not the case for breast and prostate cancers, GRHL2 methylation was also significantly higher in mesenchymal-like CRC samples than in epithelial-like samples (see Fig. S7). These analyses suggest that GRHL2 methylation patterns differ based on their cellular phenotype.
An epithelial-like gene expression profile has prognostic value in sarcomas.
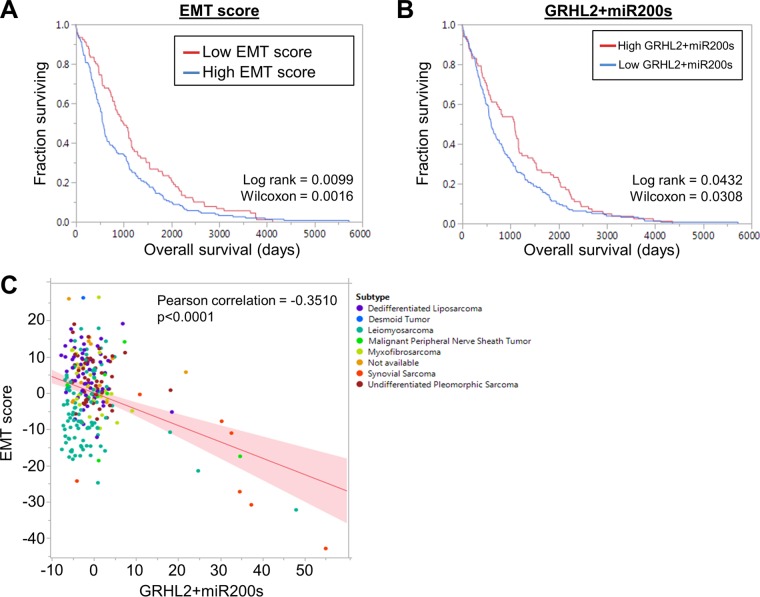
Our results suggest that some of the gene expression pathways that control EMT/MET in carcinomas may also regulate these phenotypic transitions in sarcomas. To better understand the clinical significance of these phenotypic plasticity pathways in sarcomas, we analyzed RNA-Seq data from 250 soft tissue sarcomas from patients available through The Cancer Genome Atlas. Tumors were separated into two groups based on a previously published EMT signature (48). Partitioning the tumors in this way revealed a significant difference in patient prognosis based on EMT signature; patients with a low EMT score (more epithelial-like tumors) had a more favorable prognosis than did patients with a high EMT score (more mesenchymal-like tumors) (Fig. 6A). Similarly, the sum of RNA-Seq scores from GRHL2 and miR200 family members was prognostic for overall survival, albeit with modest significance, with higher levels of GRHL2 and miR200s associated with a more favorable prognosis (Fig. 6B). As expected, the combined RNA-Seq values from GRHL2 and miR200s negatively correlated with EMT score (Fig. 6C). Analysis of GRHL2 and ZEB1 expression in an independent osteosarcoma microarray data set (n = 84; GSE28974 [49]) showed that patients with increased GRHL2 expression had significantly improved metastasis-free survival and trended toward improved overall survival (see Fig. S8A and B in the supplemental material). Conversely, patients with higher ZEB1 trended toward having worse metastasis-free survival (P = 0.056) and significantly worse overall survival (P = 0.032) (see Fig. S8C and D). Together, these data indicate that a more epithelial-like gene expression program may be prognostic for improved clinical outcomes in sarcomas. It is worth noting that although more epithelial-like soft tissue sarcoma samples were evident within the group of soft tissue sarcomas (see Fig. S9A), the EMT biomarker expression and EMT scores were strongly mesenchymal-like when normalized to all cancers (PANCAN normalized) within the TCGA data set (see Fig. S9B). These observations suggest that the more “epithelial-like” sarcomas, while being shifted more toward an epithelial phenotype, still maintain the signature of a mesenchymal-like lineage compared to epithelium-derived cancers (see Fig. S9).
FIG 6.
Epithelial biomarkers are prognostic for improved survival in soft tissue sarcomas. (A) RNA-Seq data from The Cancer Genome Atlas was used to construct Kaplan-Meier curves for patients with soft tissue sarcomas. Patients were segregated into two groups based on a previously published EMT signature (48). Patients with a more epithelial-like gene expression profile (red line) have a better prognosis than patients with a more mesenchymal-like gene expression profile (blue line). (B) Soft tissue sarcoma patients with higher GRHL2 and miR200 expression have improved survival compared to patients with lower GRHL2 and miR200s. (C) Expression of GRHL2 and miR200s negatively correlates with a mesenchymal-like gene expression profile. Individual patients are color coded by their soft tissue sarcoma subtype.
DISCUSSION
Maintenance of cellular phenotype is critical to the proper homeostasis of multicellular organisms. For proper division of labor to take place within tissues and organs, each cell must operate within the context of its lineage. The breakdown of this lineage commitment can lead to dedifferentiation, trans-differentiation, and/or other events related to phenotypic plasticity, all phenomena that are observed across multiple cancer types and that correlate with poor prognosis. In the context of sarcoma, phenotypic plasticity appears to have various associations with clinical aggression. For example, dedifferentiated liposarcomas are particularly aggressive and have a high rate of metastasis compared to their well-differentiated and intermediate-grade counterparts (50). Similarly, others have argued that sarcomas arise from reprogramming of mesenchymal stem-like cells that become fixed in a more undifferentiated state by oncogenes (51). Conversely, in the case of MET, it appears that the transition to a more epithelial-like gene expression program is associated with a favorable prognosis in a subset of soft tissue sarcomas and osteosarcomas (26–28, 52, 53). Interestingly, our computational model and laboratory experiments suggest that sarcoma cells may require multiple inputs to undergo a phenotypic transition to a more epithelial-like state. It is possible that under normal conditions, this multilevel regulation limits phenotypic transitions and provides a system to buffer noise from internal or external cues that feed into the transcriptional circuit. Only when the signals are strong enough to activate multiple factors is the phenotypic transition able to occur. More intriguing is the question of what factors would favor phenotypic transitions in sarcomas. In both carcinomas and sarcomas, presumably a more mesenchymal-like state would favor the fitness of the cancer cell, especially in the setting of competition for limited local resources, where such a state would permit metastasis and colonization of a new, less crowded niche (10–12). What, if any, selective pressures would favor MET by sarcomas has yet to be explored. It is possible that such transitions in sarcomas lack the same selective pressures in carcinomas or even that such transitions reflect innate biologic traits of particular tumors and hinder those tumors from lethal behaviors.
One of the mechanisms by which a cell can be poised toward phenotypic plasticity is through epigenetic modifications (54). Along these lines, differentiated mammary cells with high levels of E-cadherin carry an active H3K4me3 modification on the E-cadherin promoter, while the more stem-like cells harbor both the active H3K4me3 mark and a repressive H3K27me3 modification (54). As Tam and Weinberg postulate, it is logical to conclude that the bivalent modifications enable a rapid transition to a differentiated, epithelial-like state (54). Our results showing that downregulation of the chromatin remodeling protein BRG1 phenocopies the effect of ZEB1 knockdown on E-cadherin expression (Fig. 5B) fit with the idea that the chromatin conformation may dictate where on the spectrum of phenotypes a given cell may exist. Further study of the relationship between epigenetic modification, phenotype, and capacity for phenotypic plasticity in sarcomas may elucidate the context of phenotypic plasticity among mesenchymal tumors.
Our coupling of dynamic modeling with experimentally based refinements and validations has enabled the establishment of a framework by which to understand regulation of MET in sarcomas, a phenomenon that appears to have clinical relevance (28, 52, 53). A deeper understanding of how these transitions occur in sarcoma and what selective pressures influence them could offer immediate prognostic value and, one day, lead to therapeutically shifting sarcomas to a more epithelial-like state, thereby attenuating their aggressiveness and improving patient outcomes.
Supplementary Material
ACKNOWLEDGMENTS
The results published here are in part based upon data generated by the TCGA Research Network (http://cancergenome.nih.gov).
J.A.S. acknowledges support from the Duke Cancer Institute, the Duke University Genitourinary Oncology Laboratory, and the Duke University Department of Orthopaedics. H.L. was supported by the National Science Foundation (NSF) Center for Theoretical Biological Physics (NSF grant PHY-1427654) and NSF grant DMS-1361411 and as a CPRIT (Cancer Prevention and Research Institute of Texas) Scholar in Cancer Research of the State of Texas at Rice University. K.E.W. was supported by NIH grant F32 CA192630.
M.K.J. and H.L. benefited from useful discussions with Mary C. Farach-Carson, J. N. Onuchic, Samir M. Hanash, Kenneth J. Pienta, and Donald S. Coffey.
Footnotes
Supplemental material for this article may be found at http://dx.doi.org/10.1128/MCB.00373-16.
REFERENCES
- 1.Nakaya Y, Sheng G. 2013. EMT in developmental morphogenesis. Cancer Lett 341:9–15. doi: 10.1016/j.canlet.2013.02.037. [DOI] [PubMed] [Google Scholar]
- 2.Lim J, Thiery JP. 2012. Epithelial-mesenchymal transitions: insights from development. Development 139:3471–3486. doi: 10.1242/dev.071209. [DOI] [PubMed] [Google Scholar]
- 3.Thiery JP, Acloque H, Huang RY, Nieto MA. 2009. Epithelial-mesenchymal transitions in development and disease. Cell 139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 4.Thiery JP, Sleeman JP. 2006. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol 7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 5.Weber CE, Li NY, Wai PY, Kuo PC. 2012. Epithelial-mesenchymal transition, TGF-beta, and osteopontin in wound healing and tissue remodeling after injury. J Burn Care Res 33:311–318. doi: 10.1097/BCR.0b013e318240541e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galichon P, Finianos S, Hertig A. 2013. EMT-MET in renal disease: should we curb our enthusiasm? Cancer Lett 341:24–29. doi: 10.1016/j.canlet.2013.04.018. [DOI] [PubMed] [Google Scholar]
- 7.Carew RM, Wang B, Kantharidis P. 2012. The role of EMT in renal fibrosis. Cell Tissue Res 347:103–116. doi: 10.1007/s00441-011-1227-1. [DOI] [PubMed] [Google Scholar]
- 8.Willis BC, Borok Z. 2007. TGF-beta-induced EMT: mechanisms and implications for fibrotic lung disease. Am J Physiol Lung Cell Mol Physiol 293:L525–L534. doi: 10.1152/ajplung.00163.2007. [DOI] [PubMed] [Google Scholar]
- 9.Ye X, Weinberg RA. 2015. Epithelial-mesenchymal plasticity: a central regulator of cancer progression. Trends Cell Biol 25:675–686. doi: 10.1016/j.tcb.2015.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li L, Li W. 2015. Epithelial-mesenchymal transition in human cancer: comprehensive reprogramming of metabolism, epigenetics, and differentiation. Pharmacol Ther 150:33–46. doi: 10.1016/j.pharmthera.2015.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Tsai JH, Yang J. 2013. Epithelial-mesenchymal plasticity in carcinoma metastasis. Genes Dev 27:2192–2206. doi: 10.1101/gad.225334.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bitting RL, Schaeffer D, Somarelli JA, Garcia-Blanco MA, Armstrong AJ. 2014. The role of epithelial plasticity in prostate cancer dissemination and treatment resistance. Cancer Metastasis Rev 33:441–468. doi: 10.1007/s10555-013-9483-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schaeffer D, Somarelli JA, Hanna G, Palmer GM, Garcia-Blanco MA. 2014. Cellular migration and invasion uncoupled: increased migration is not an inexorable consequence of epithelial-to-mesenchymal transition. Mol Cell Biol 34:3486–3499. doi: 10.1128/MCB.00694-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Mathow D, Chessa F, Rabionet M, Kaden S, Jennemann R, Sandhoff R, Grone HJ, Feuerborn A. 2015. Zeb1 affects epithelial cell adhesion by diverting glycosphingolipid metabolism. EMBO Rep 16:321–331. doi: 10.15252/embr.201439333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ware KE, Hinz TK, Kleczko E, Singleton KR, Marek LA, Helfrich BA, Cummings CT, Graham DK, Astling D, Tan AC, Heasley LE. 2013. A mechanism of resistance to gefitinib mediated by cellular reprogramming and the acquisition of an FGF2-FGFR1 autocrine growth loop. Oncogenesis 2:e39. doi: 10.1038/oncsis.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yauch RL, Januario T, Eberhard DA, Cavet G, Zhu W, Fu L, Pham TQ, Soriano R, Stinson J, Seshagiri S, Modrusan Z, Lin CY, O'Neill V, Amler LC. 2005. Epithelial versus mesenchymal phenotype determines in vitro sensitivity and predicts clinical activity of erlotinib in lung cancer patients. Clin Cancer Res 11:8686–8698. doi: 10.1158/1078-0432.CCR-05-1492. [DOI] [PubMed] [Google Scholar]
- 17.Jolly MK, Huang B, Lu M, Mani SA, Levine H, Ben-Jacob E. 2014. Towards elucidating the connection between epithelial-mesenchymal transitions and stemness. J R Soc Interface 11:20140962. doi: 10.1098/rsif.2014.0962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, Campbell LL, Polyak K, Brisken C, Yang J, Weinberg RA. 2008. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 133:704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, Gibbons DL, Goswami S, Cortez MA, Ahn YH, Byers LA, Zhang X, Yi X, Dwyer D, Lin W, Diao L, Wang J, Roybal JD, Patel M, Ungewiss C, Peng D, Antonia S, Mediavilla-Varela M, Robertson G, Jones S, Suraokar M, Welsh JW, Erez B, Wistuba II, Chen L, Peng D, Wang S, Ullrich SE, Heymach JV, Kurie JM, Qin FX. 2014. Metastasis is regulated via microRNA-200/ZEB1 axis control of tumour cell PD-L1 expression and intratumoral immunosuppression. Nat Commun 5:5241. doi: 10.1038/ncomms6241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vega S, Morales AV, Ocana OH, Valdes F, Fabregat I, Nieto MA. 2004. Snail blocks the cell cycle and confers resistance to cell death. Genes Dev 18:1131–1143. doi: 10.1101/gad.294104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang SS, Huang JS. 2005. TGF-beta control of cell proliferation. J Cell Biochem 96:447–462. doi: 10.1002/jcb.20558. [DOI] [PubMed] [Google Scholar]
- 22.Tsai JH, Donaher JL, Murphy DA, Chau S, Yang J. 2012. Spatiotemporal regulation of epithelial-mesenchymal transition is essential for squamous cell carcinoma metastasis. Cancer Cell 22:725–736. doi: 10.1016/j.ccr.2012.09.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Armstrong AJ, Marengo MS, Oltean S, Kemeny G, Bitting RL, Turnbull JD, Herold CI, Marcom PK, George DJ, Garcia-Blanco MA. 2011. Circulating tumor cells from patients with advanced prostate and breast cancer display both epithelial and mesenchymal markers. Mol Cancer Res 9:997–1007. doi: 10.1158/1541-7786.MCR-10-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yu M, Bardia A, Wittner BS, Stott SL, Smas ME, Ting DT, Isakoff SJ, Ciciliano JC, Wells MN, Shah AM, Concannon KF, Donaldson MC, Sequist LV, Brachtel E, Sgroi D, Baselga J, Ramaswamy S, Toner M, Haber DA, Maheswaran S. 2013. Circulating breast tumor cells exhibit dynamic changes in epithelial and mesenchymal composition. Science 339:580–584. doi: 10.1126/science.1228522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jolly MK, Boareto M, Huang B, Jia D, Lu M, Ben-Jacob E, Onuchic JN, Levine H. 2015. Implications of the hybrid epithelial/mesenchymal phenotype in metastasis. Front Oncol 5:155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Alba-Castellón L, Batlle R, Franci C, Fernandez-Acenero MJ, Mazzolini R, Pena R, Loubat J, Alameda F, Rodriguez R, Curto J, Albanell J, Munoz A, Bonilla F, Ignacio Casal J, Rojo F, Garcia de Herreros A. 2014. Snail1 expression is required for sarcomagenesis. Neoplasia 16:413–421. doi: 10.1016/j.neo.2014.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Shen A, Zhang Y, Yang H, Xu R, Huang G. 2012. Overexpression of ZEB1 relates to metastasis and invasion in osteosarcoma. J Surg Oncol 105:830–834. doi: 10.1002/jso.23012. [DOI] [PubMed] [Google Scholar]
- 28.Wang N, He YL, Pang LJ, Zou H, Liu CX, Zhao J, Hu JM, Zhang WJ, Qi Y, Li F. 2015. Down-regulated E-cadherin expression is associated with poor five-year overall survival in bone and soft tissue sarcoma: results of a meta-analysis. PLoS One 10:e0121448. doi: 10.1371/journal.pone.0121448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu M, Jolly MK, Levine H, Onuchic JN, Ben-Jacob E. 2013. MicroRNA-based regulation of epithelial-hybrid-mesenchymal fate determination. Proc Natl Acad Sci U S A 110:18144–18149. doi: 10.1073/pnas.1318192110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dhooge A, Govaerts W, Kuznetsov YA. 2003. MATCONT: a MATLAB package for numerical bifurcation analysis of ODEs. ACM Trans Math Softw 29:141–164. doi: 10.1145/779359.779362. [DOI] [Google Scholar]
- 31.Bracken CP, Gregory PA, Kolesnikoff N, Bert AG, Wang J, Shannon MF, Goodall GJ. 2008. A double-negative feedback loop between ZEB1-SIP1 and the microRNA-200 family regulates epithelial-mesenchymal transition. Cancer Res 68:7846–7854. doi: 10.1158/0008-5472.CAN-08-1942. [DOI] [PubMed] [Google Scholar]
- 32.Burk U, Schubert J, Wellner U, Schmalhofer O, Vincan E, Spaderna S, Brabletz T. 2008. A reciprocal repression between ZEB1 and members of the miR-200 family promotes EMT and invasion in cancer cells. EMBO Rep 9:582–589. doi: 10.1038/embor.2008.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Siemens H, Jackstadt R, Hunten S, Kaller M, Menssen A, Gotz U, Hermeking H. 2011. miR-34 and SNAIL form a double-negative feedback loop to regulate epithelial-mesenchymal transitions. Cell Cycle 10:4256–4271. doi: 10.4161/cc.10.24.18552. [DOI] [PubMed] [Google Scholar]
- 34.Guaita S, Puig I, Franci C, Garrido M, Dominguez D, Batlle E, Sancho E, Dedhar S, De Herreros AG, Baulida J. 2002. Snail induction of epithelial to mesenchymal transition in tumor cells is accompanied by MUC1 repression and ZEB1 expression. J Biol Chem 277:39209–39216. doi: 10.1074/jbc.M206400200. [DOI] [PubMed] [Google Scholar]
- 35.Gill JG, Langer EM, Lindsley RC, Cai M, Murphy TL, Kyba M, Murphy KM. 2011. Snail and the microRNA-200 family act in opposition to regulate epithelial-to-mesenchymal transition and germ layer fate restriction in differentiating ESCs. Stem Cells 29:764–776. doi: 10.1002/stem.628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahn YH, Gibbons DL, Chakravarti D, Creighton CJ, Rizvi ZH, Adams HP, Pertsemlidis A, Gregory PA, Wright JA, Goodall GJ, Flores ER, Kurie JM. 2012. ZEB1 drives prometastatic actin cytoskeletal remodeling by downregulating miR-34a expression. J Clin Invest 122:3170–3183. doi: 10.1172/JCI63608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hurteau GJ, Carlson JA, Roos E, Brock GJ. 2009. Stable expression of miR-200c alone is sufficient to regulate TCF8 (ZEB1) and restore E-cadherin expression. Cell Cycle 8:2064–2069. doi: 10.4161/cc.8.13.8883. [DOI] [PubMed] [Google Scholar]
- 38.Hill L, Browne G, Tulchinsky E. 2013. ZEB/miR-200 feedback loop: at the crossroads of signal transduction in cancer. Int J Cancer 132:745–754. doi: 10.1002/ijc.27708. [DOI] [PubMed] [Google Scholar]
- 39.Preca BT, Bajdak K, Mock K, Sundararajan V, Pfannstiel J, Maurer J, Wellner U, Hopt UT, Brummer T, Brabletz S, Brabletz T, Stemmler MP. 2015. A self-enforcing CD44s/ZEB1 feedback loop maintains EMT and stemness properties in cancer cells. Int J Cancer 137:2566–2577. doi: 10.1002/ijc.29642. [DOI] [PubMed] [Google Scholar]
- 40.Varma S, Cao Y, Tagne JB, Lakshminarayanan M, Li J, Friedman TB, Morell RJ, Warburton D, Kotton DN, Ramirez MI. 2012. The transcription factors Grainyhead-like 2 and NK2-homeobox 1 form a regulatory loop that coordinates lung epithelial cell morphogenesis and differentiation. J Biol Chem 287:37282–37295. doi: 10.1074/jbc.M112.408401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cieply B, Riley P, Pifer PM, Widmeyer J, Addison JB, Ivanov AV, Denvir J, Frisch SM. 2012. Suppression of the epithelial-mesenchymal transition by Grainyhead-like-2. Cancer Res 72:2440–2453. doi: 10.1158/0008-5472.CAN-11-4038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cieply B, Farris J, Denvir J, Ford HL, Frisch SM. 2013. Epithelial-mesenchymal transition and tumor suppression are controlled by a reciprocal feedback loop between ZEB1 and Grainyhead-like-2. Cancer Res 73:6299–6309. doi: 10.1158/0008-5472.CAN-12-4082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Werth M, Walentin K, Aue A, Schonheit J, Wuebken A, Pode-Shakked N, Vilianovitch L, Erdmann B, Dekel B, Bader M, Barasch J, Rosenbauer F, Luft FC, Schmidt-Ott KM. 2010. The transcription factor grainyhead-like 2 regulates the molecular composition of the epithelial apical junctional complex. Development 137:3835–3845. doi: 10.1242/dev.055483. [DOI] [PubMed] [Google Scholar]
- 44.Sánchez-Tilló E, Lazaro A, Torrent R, Cuatrecasas M, Vaquero EC, Castells A, Engel P, Postigo A. 2010. ZEB1 represses E-cadherin and induces an EMT by recruiting the SWI/SNF chromatin-remodeling protein BRG1. Oncogene 29:3490–3500. doi: 10.1038/onc.2010.102. [DOI] [PubMed] [Google Scholar]
- 45.Schmalhofer O, Brabletz S, Brabletz T. 2009. E-cadherin, beta-catenin, and ZEB1 in malignant progression of cancer. Cancer Metastasis Rev 28:151–166. doi: 10.1007/s10555-008-9179-y. [DOI] [PubMed] [Google Scholar]
- 46.Werner S, Frey S, Riethdorf S, Schulze C, Alawi M, Kling L, Vafaizadeh V, Sauter G, Terracciano L, Schumacher U, Pantel K, Assmann V. 2013. Dual roles of the transcription factor grainyhead-like 2 (GRHL2) in breast cancer. J Biol Chem 288:22993–23008. doi: 10.1074/jbc.M113.456293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Oltean S, Febbo PG, Garcia-Blanco MA. 2008. Dunning rat prostate adenocarcinomas and alternative splicing reporters: powerful tools to study epithelial plasticity in prostate tumors in vivo. Clin Exp Metastasis 25:611–619. doi: 10.1007/s10585-008-9186-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Salt MB, Bandyopadhyay S, McCormick F. 2014. Epithelial-to-mesenchymal transition rewires the molecular path to PI3K-dependent proliferation. Cancer Discov 4:186–199. doi: 10.1158/2159-8290.CD-13-0520. [DOI] [PubMed] [Google Scholar]
- 49.Kuijjer ML, Rydbeck H, Kresse SH, Buddingh EP, Lid AB, Roelofs H, Burger H, Myklebost O, Hogendoorn PC, Meza-Zepeda LA, Cleton-Jansen AM. 2012. Identification of osteosarcoma driver genes by integrative analysis of copy number and gene expression data. Genes Chromosomes Cancer 51:696–706. doi: 10.1002/gcc.21956. [DOI] [PubMed] [Google Scholar]
- 50.Crago AM, Singer S. 2011. Clinical and molecular approaches to well differentiated and dedifferentiated liposarcoma. Curr Opin Oncol 23:373–378. doi: 10.1097/CCO.0b013e32834796e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Eid JE, Garcia CB. 2015. Reprogramming of mesenchymal stem cells by oncogenes. Semin Cancer Biol 32:18–31. doi: 10.1016/j.semcancer.2014.05.005. [DOI] [PubMed] [Google Scholar]
- 52.Yin K, Liao Q, He H, Zhong D. 2012. Prognostic value of Twist and E-cadherin in patients with osteosarcoma. Med Oncol 29:3449–3455. doi: 10.1007/s12032-012-0317-6. [DOI] [PubMed] [Google Scholar]
- 53.Tian W, Wang G, Yang J, Pan Y, Ma Y. 2013. Prognostic role of E-cadherin and Vimentin expression in various subtypes of soft tissue leiomyosarcomas. Med Oncol 30:401. doi: 10.1007/s12032-012-0401-y. [DOI] [PubMed] [Google Scholar]
- 54.Tam WL, Weinberg RA. 2013. The epigenetics of epithelial-mesenchymal plasticity in cancer. Nat Med 19:1438–1449. doi: 10.1038/nm.3336. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.