ABSTRACT
The subcellular sites of HIV-1 assembly, determined by the localization of the structural protein Gag, vary in a cell-type-dependent manner. In T cells and transformed cell lines used as model systems, HIV-1 assembles at the plasma membrane (PM). The binding and localization of HIV-1 Gag to the PM are mediated by the interaction between the matrix (MA) domain, specifically the highly basic region, and a PM-specific acidic phospholipid, phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2]. In primary macrophages, prominent accumulation of assembling or assembled particles is found in the virus-containing compartments (VCCs), which largely consist of convoluted invaginations of the PM. To elucidate the molecular mechanism of HIV-1 Gag targeting to the VCCs, we examined the impact of overexpression of polyphosphoinositide 5-phosphatase IV (5ptaseIV), which depletes cellular PI(4,5)P2, in primary macrophages. We found that the VCC localization and virus release of HIV-1 are severely impaired upon 5ptaseIV overexpression, suggesting an important role for the MA-PI(4,5)P2 interaction in HIV-1 assembly in primary macrophages. However, our analysis of HIV-1 Gag derivatives with MA changes showed that this interaction contributes to Gag membrane binding but is dispensable for specific targeting of Gag to the VCCs per se. We further determined that deletion of the NC domain abolishes VCC-specific localization of HIV-1 Gag. Notably, HIV-1 Gag localized efficiently to the VCCs when the NC domain was replaced with a leucine zipper dimerization motif that promotes Gag multimerization. Altogether, our data revealed that targeting of HIV-1 Gag to the VCCs requires NC-dependent multimerization.
IMPORTANCE In T cells and model cell lines, HIV-1 Gag localizes to the PM in a manner dependent on the MA-PI(4,5)P2 interaction. On the other hand, in primary macrophages, HIV-1 Gag localizes to convoluted intracellular membrane structures termed virus-containing compartments (VCCs). Although these compartments have been known for decades, and despite the implication of viruses in VCCs being involved in virus reservoir maintenance and spread, the viral determinant(s) that promotes Gag targeting to VCCs is unknown. In this study, we found that the MA-PI(4,5)P2 interaction facilitates efficient Gag membrane binding in macrophages but is not essential for Gag targeting to VCCs. Rather, our results revealed that NC-dependent multimerization promotes VCC targeting. Our findings highlight the differential roles played by MA and NC in HIV-1 Gag membrane binding and targeting and suggest a multimerization-dependent mechanism for Gag trafficking in primary macrophages similar to that for Gag localization to uropods in polarized T cells.
INTRODUCTION
Macrophages represent one of the primary cell targets of HIV-1 and play important roles in HIV-1 pathogenesis (1–3). They are terminally differentiated phagocytic cells of myeloid lineage and are found in various tissues in the body. Macrophages are long-lived and are more resistant than CD4+ T cells to cytopathic effects of virus replication (1–5). Some antiretroviral drugs are less potent against macrophage infection in vitro, and infected macrophages are found in infected individuals on successful antiretroviral therapy (ART) (1, 3). Moreover, macrophage infection in tissues, in particular the central nervous system, may create a condition where drug penetration is insufficient (3). Thus, HIV-1-infected macrophages are likely to represent a long-term reservoir for viruses that are not eradicated during ART (1–3). In addition, HIV-1 dissemination and establishment of infection in the brain, to which macrophages contribute, lead to HIV-associated neurocognitive disorders, which are prevalent even in the era of successful combination ART (1, 3, 6).
The subcellular site of HIV-1 assembly can be identified by the presence of immature particles as well as virus structures budding from the limiting cellular membrane by electron microscopy (EM). In T cells and other cell lines, such as 293T, COS, and HeLa cells, HIV-1 assembly structures are formed at the plasma membrane (PM) (7–9). In contrast, early ultrastructural studies of HIV-1-infected primary macrophages showed that both mature and immature HIV-1 particles and budding structures accumulate in intracellular compartments (10, 11) which share some similarity with endosomes (5, 11, 12). Subsequent EM studies over the past decade revealed that at least some of these intracellular compartments, now termed virus-containing compartments (VCCs), are deep and convoluted invaginations of the PM and appear to form connections to the cell surface and extracellular space via tubular structures in a dynamic and transient manner (5, 13–19). Notably, mature particles stored in macrophages remain infectious for weeks in the presence of a protease inhibitor (20). Moreover, viruses in VCCs are inaccessible to neutralizing antibodies (14, 17) yet can be transferred to CD4+ T cells that come into contact with the infected macrophages (21, 22). These findings collectively support the hypothesis that VCC-associated viruses are likely to play important roles in HIV-1 pathogenesis.
Recent studies focused on characterizing the membrane protein composition of VCCs. VCCs have been shown to contain tetraspanins, such as CD9, CD53, CD81, and CD82 (12, 15, 23, 24). In addition, CD18 (a leukocyte-specific β2 integrin), CD44 (a cell adhesion protein/hyaluronic acid receptor), and CD36 (a low-density lipoprotein [LDL] receptor) are also present (24–26). Cellular compartments with identical markers also exist in the absence of HIV-1. These compartments are termed intracellular PM-connected compartments (IPMCs) (24). Previous studies have also suggested the involvement of host cellular components, such as kinesins, in formation and/or dynamics of VCCs or IPMCs (27, 28).
Even though much effort has been carried out to characterize the structure and composition of VCCs, very little is known about the viral determinants that target HIV-1 assembly to the VCCs. HIV-1 assembly is driven by its structural protein, Gag. Gag is synthesized as a polyprotein (Pr55Gag) containing multiple structural domains, each of which plays a critical role during the assembly process (29–34). The matrix (MA) domain makes up the N-terminal part of the Gag protein and is responsible for Gag localization to and membrane binding at the PM. Capsid (CA) and nucleocapsid (NC) domains mediate Gag-Gag and Gag-RNA interactions, respectively, both of which promote formation of Gag multimers. The p6 domain contains the late domain motifs that recruit ESCRT proteins, which in turn aid in the release of virus particles from the cell surface.
The molecular determinants for Gag localization in cells where HIV-1 assembly takes place at the PM are relatively well understood. HIV-1 MA contains two key signals for membrane binding: the N-terminal myristoyl moiety, which mediates hydrophobic interactions with the membrane, and the highly basic region (HBR) (35–39). The HBR binds to the membrane via interactions with acidic phospholipids, in particular phosphatidylinositol-4,5-bisphosphate [PI(4,5)P2], a PM-specific phosphoinositide (39–43). Mutations in the HBR cause membrane binding defects and/or mislocalization of HIV-1 Gag to perinuclear compartments in HeLa and T cells (40, 44–49). Likewise, depletion of cellular PI(4,5)P2 by overexpression of polyphosphoinositide 5-phosphatase IV (5ptaseIV) leads to defects in both total membrane binding and specific Gag localization to the PM, which in turn result in a severe reduction in virus release from the cell (40, 50). Altogether, these results strongly suggest that the HBR-PI(4,5)P2 interaction is required for both specific Gag localization to the PM and efficient membrane binding.
In addition to PI(4,5)P2, HIV-1 MA, in particular the HBR, binds RNA (51–57). Removal of MA-bound RNA by RNase treatment reduces HIV-1 Gag specificity for PI(4,5)P2 in vitro (46). These results and other in vitro and cell-based studies (58–63) collectively support a model in which MA-bound RNA prevents HIV-1 Gag from binding to cellular membranes that do not contain PI(4,5)P2. This way, the MA-RNA interaction ensures specific binding of HIV-1 Gag to the PM in regions where PI(4,5)P2 is found. Notably, PH-GFP, a PI(4,5)P2-specific marker, was shown to overlap CD81 and Gag puncta at intracellular sites in macrophages, suggesting that PI(4,5)P2 is present at the VCCs (23, 64). Therefore, it is possible that MA promotes specific localization of Gag to the VCCs via a mechanism regulated by PI(4,5)P2 and RNA.
In polarized T cells, HIV-1 Gag is specifically targeted to the uropod part of the PM. The uropod is a protrusion at the rear end of a polarized leukocyte (65). This region is enriched in membrane proteins that are also found in the VCCs, such as CD9, CD44, CD81, and CD82 (65–67). The uropod serves as a contact site between infected and uninfected T cells and participates in the formation of virological synapses (VS) (67), where cell-to-cell transmission of HIV-1 occurs (68–73). Interestingly, Gag targeting to the uropod is not dependent on MA as long as a heterologous membrane binding domain is provided; rather, NC-mediated multimerization is required (67, 74). Gag multimerization is required for uropod localization of murine leukemia virus Gag as well (75).
As described above, various factors are involved in proper Gag localization to virus assembly sites. It is not currently known which of these factors are important for HIV-1 Gag targeting to the VCCs in macrophages. Since PI(4,5)P2 is present at the VCCs (23, 64), it is quite conceivable that HIV-1 Gag localization to VCCs is PI(4,5)P2 dependent. Alternatively or additionally, Gag multimerization may play a key role in VCC targeting as is the case with uropod targeting. To test these hypotheses, we examined (i) the effects of PI(4,5)P2 depletion on VCC targeting of HIV-1 Gag and HIV-1 assembly, (ii) the effects of mutations and substitutions of the MA domain on the localization of HIV-1 Gag in infected macrophages, and (iii) the localization phenotypes of HIV-1 Gag derivatives with alterations that affect post-membrane-binding steps in virus assembly. Our results indicate that HIV-1 Gag localization to the VCCs is dependent on NC and that while PI(4,5)P2 serves as an important membrane anchor for Gag in macrophages, the MA interaction with this lipid is dispensable for targeting of Gag to the VCCs.
MATERIALS AND METHODS
Cells and plasmids.
Monocytes were isolated by plate adhesion from peripheral blood mononuclear cells, which were obtained from buffy coats derived from healthy donors (New York Blood Center, NY). Monocytes were cultured in RPMI 1640 medium supplemented with 10% fetal bovine serum (FBS) for 7 days before being used as monocyte-derived macrophages (MDMs) for experiments. 293T cells was cultured and maintained in Dulbecco's modified Eagle's medium (DMEM; Lonza) supplemented with 10% FBS (HyClone) as described previously (67).
An HIV-1 molecular clone encoding Gag-yellow fluorescent protein (Gag-YFP) (pNL4-3/Gag-YFP) was described previously (40). pNL4-3/KFS/398/IRES-Myc-5ptaseIV and pNL4-3/KFS/398/IRES-Myc-5ptaseIVΔ1 encode Myc-5ptaseIV and an inactive deletion mutant, Myc-5ptaseIV Δ1, respectively, following an internal ribosome entry site (IRES) sequence in place of the nef gene, as previously described (76). These plasmids were derived from a parental plasmid, pNL4-3/KFS/398, which has the nef gene sequence replaced with a sequence containing multiple restriction sites derived from plasmid p398-6 (a kind gift from K. T. Jeang) and contains a frameshift mutation (KFS) that disrupts Env expression (77). pNL4-3/Gag-YFP/KFS/398/IRES-Myc-5ptaseIV and pNL4-3/Gag-YFP/398/IRES-Myc-5ptaseIVΔ1 were constructed from pNL4-3/Gag-YFP by using standard molecular cloning techniques (76). pNL4-3/1GA Gag-YFP (78), pNL4-3/29KT/31KT Gag-YFP (46), pNL4-3/Fyn(10)/ΔMA Gag-YFP (40), pNL4-3/PH/ΔMA Gag-YFP (79), pNL4-3/delNC Gag-YFP (78), pNL4-3/GagLZ-YFP (67), and pNL4-3/Gag-LZ4-YFP (66) were described previously. pNL4-3/Kmyr/ΔMA Gag-YFP, pNL4-3/HTMA Gag-YFP, and pNL4-3/EE75,76AA Gag-YFP were also constructed using standard molecular cloning techniques, using pNL4-3/Gag-YFP and previously described plasmids (61, 80, 81).
Virus stocks and infection.
Vesicular stomatitis virus G glycoprotein (VSV-G)-pseudotyped HIV-1 stocks were prepared as previously described, with modifications (67). Briefly, 3.4 × 106 293T cells were transfected with 9 μg of pNL4-3-derived molecular clones, 9 μg of pCMVNLGagPol-RRE (82), and 3 μg of pHCMV-G (83). At 2 days posttransfection, virus-containing supernatants were filtered through a 0.45-μm filter, and virus particles were pelleted by ultracentrifugation (35,000 rpm, 4°C, 45 min). Virus pellets were resuspended in 900 μl of RPMI-10 medium. VSV-G-pseudotyped simian immunodeficiency virus (SIV) virus-like particles containing Vpx (SIV-Vpx) were prepared by transfecting 9 μg of SIV3+ (a kind gift from Andrea Cimarelli [84–86]) and 3 μg of pHCMV-G into 3.4 × 106 293T cells. Virus particles were then pelleted as described above.
MDMs were first transduced with Vpx-containing virus-like particles (SIV-Vpx) for either 2 h or overnight and then incubated with pseudotyped HIV-1 particles for 6 more hours. Subsequently, cells were washed and cultured for an additional 48 h.
Virus release assay.
The virus release assay was performed as previously described (50), with modifications. Briefly, MDMs were treated with SIV-Vpx followed by infection with pseudotyped HIV-1 molecular clones as described above. At 48 h postinfection, the culture medium was changed to RPMI 1640 lacking both methionine (Met) and cysteine (Cys) and supplemented with 2% FBS [RPMI-2 (−Met/−Cys)], and the cells were incubated for 30 min. Subsequently, these cells were metabolically labeled with [35S]Met/Cys (Perkin-Elmer) in fresh RPMI-2 (−Met/−Cys) for 4 h. Cell and virion lysates were prepared and subjected to immunoprecipitation with HIV Ig (NIH AIDS Research and Reference Reagent Program). The virus release efficiency was calculated as the amount of virion-associated Gag, expressed as a fraction of the total amount of Gag synthesized during the labeling period.
Immunostaining and confocal fluorescence microscopy.
For visualization of the PM, MDMs infected with VSV-G-pseudotyped HIV-1 particles were incubated with Alexa Fluor 594-conjugated concanavalin A (ConA) (Invitrogen) for 5 min at room temperature. Cells were then washed with 1× phosphate-buffered saline (PBS), fixed with 4% paraformaldehyde (Electron Microscopy Sciences) in PBS for 30 min at room temperature, permeabilized in PBS containing 0.1% Triton X-100 for 2 min, and washed thoroughly with PBS. The cells were immunostained with anti-CD81 antibody (BD Biosciences Pharmingen, San Diego, CA) for 1 h at room temperature, washed twice with PBS, and stained with goat anti-mouse IgG conjugated to Alexa Fluor 647 (Invitrogen). The immunostained cells were subsequently washed with PBS and mounted. Cells were visualized using a Leica SP5X inverted confocal microscope system with a 60× objective. Pearson's correlation coefficients (PCCs) for ConA and Gag-YFP as well as CD81 and Gag-YFP were calculated using the Coloc2 plug-in in the ImageJ software program. At least 10 cells per donor were analyzed for each condition. MDMs from at least 2 donors were used in the experiments.
RESULTS
PI(4,5)P2 depletion reduces the efficiency of HIV-1 release from MDMs.
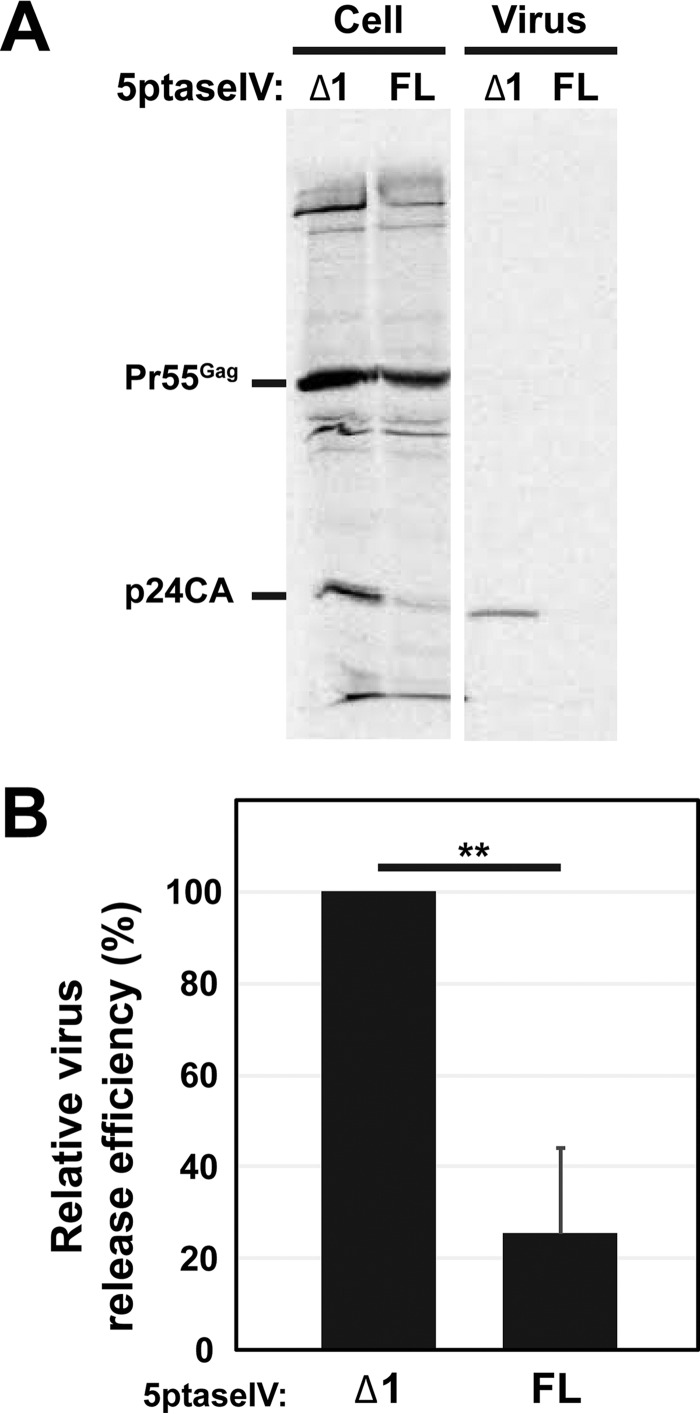
Overexpression of 5ptaseIV, which depletes cellular PI(4,5)P2, significantly reduced the HIV-1 release efficiency in HeLa and other model cell lines as well as in T cells (40, 50, 61, 76, 87–89). To assess the role of PI(4,5)P2 in HIV-1 release from MDMs, we sought to coexpress 5ptaseIV with HIV-1 in a large number of MDMs sufficient for biochemical virus release assays. Since MDMs are refractory to plasmid transfection, to achieve coexpression of 5ptaseIV with HIV-1, we chose to infect MDMs with VSV-G-pseudotyped HIV-1 carrying the 5ptaseIV gene in place of the nef open reading frame (ORF). As a control, we replaced the full-length 5ptaseIV sequence with the sequence of the catalytically inactive 5ptaseIV Δ1 mutant (76). Myeloid cells, including macrophages, express high levels of a restriction factor known as SAM domain- and HD domain-containing protein 1 (SAMHD1), which suppresses HIV-1 replication (90, 91). Notably, however, exogenous introduction of the SIV Vpx protein, for example, in the form of pseudotyped virus-like particles (SIV-Vpx), to target myeloid cells prior to or during HIV-1 infection has been shown to improve HIV-1 infection efficiency (84, 90–92). Thus, to obtain a large number of MDMs expressing HIV-1 derivatives that are incapable of spreading infection (e.g., molecular clones encoding full-length 5ptaseIV or catalytically inactive 5ptaseIV Δ1) so as to facilitate quantitative analyses, in this and subsequent experiments we pretreated MDMs with SIV-Vpx prior to infection with a VSV-G-pseudotyped HIV-1 molecular clone. We confirmed that SIV-Vpx transduction did not alter colocalization of Gag and CD81 in apparently intracellular compartments (shown below), which has been observed in previous studies (see the introduction). At 48 h postinfection, cells were metabolically labeled with [35S]Met/Cys for 4 h, and 35S-labeled HIV-1 Gag in cell and virus lysates was immunoprecipitated using HIV Ig. We found that the HIV-1 release efficiency was reduced >3-fold in 5ptaseIV-expressing cells compared to 5ptaseIV Δ1-expressing cells (Fig. 1A and B). This indicates that PI(4,5)P2 plays an important role in efficient release of HIV-1 from MDMs.
FIG 1.
HIV-1 release from MDMs is sensitive to 5ptaseIV overexpression. (A) MDMs expressing HIV-1 along with full-length 5ptaseIV (FL) or its Δ1 mutant were metabolically labeled for 4 h. Cell- and virus-associated Gag proteins were recovered by immunoprecipitation and analyzed by SDS-PAGE. (B) Relative virus release efficiencies of HIV-1 were calculated. Data for 7 donors are shown as means and standard deviations. The average virus release efficiency of MDMs that expressed HIV-1 along with 5ptaseIV Δ1 was 18.4%. P values were determined using Student's t test, using raw data. **, P < 0.001.
PI(4,5)P2 is required for localization of wild-type (WT) Gag to VCCs in MDMs.
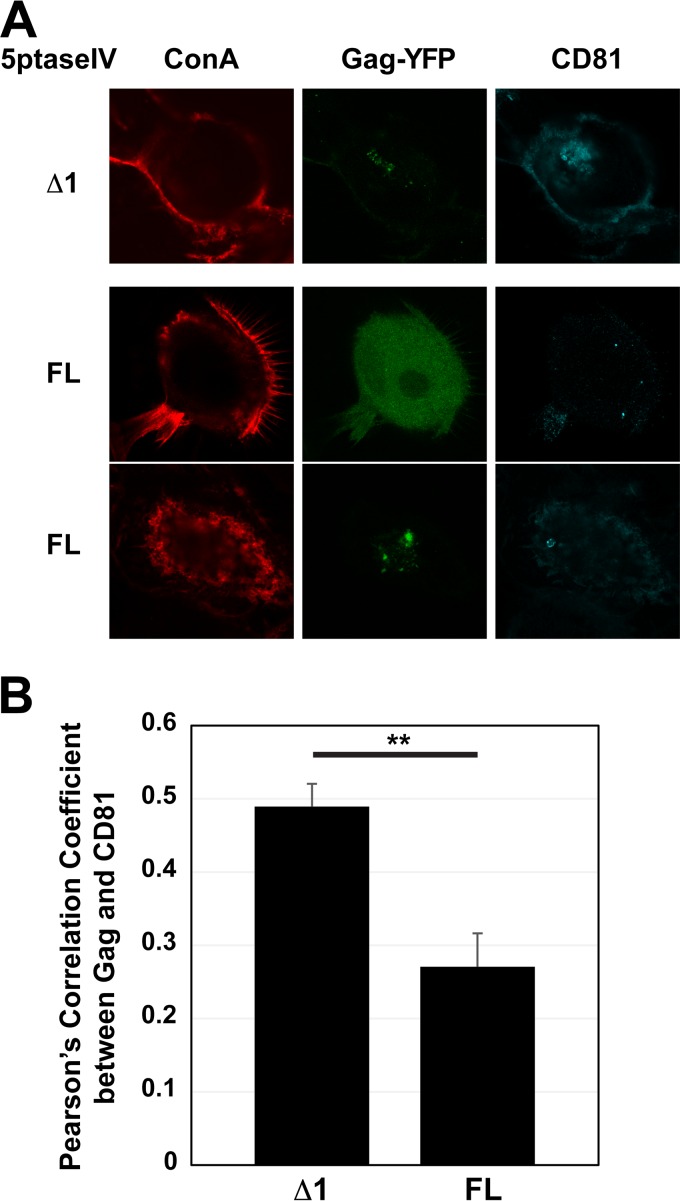
In addition to virus release, PI(4,5)P2 is required for HIV-1 Gag localization to the PM, the site of assembly in HeLa and T cells (40, 50, 61, 76). To assess whether PI(4,5)P2 depletion influences HIV-1 Gag localization to the VCCs in MDMs, we expressed HIV-1 molecular clones encoding Gag-YFP along with either full-length 5ptaseIV or 5ptaseIV Δ1. Cells were stained with fluorescently labeled concanavalin A (ConA) prior to fixation and permeabilization and subsequently immunostained for the tetraspanin CD81. Unlike another tetraspanin, CD63, which localizes predominantly to late endosomes/lysosomes in MDMs, CD81 was shown to localize at the cell surface PM and VCCs, with minimal overlap with endosomal pathways, in MDMs (15). Therefore, in MDMs, we can use CD81 to distinguish VCCs from other intracellular compartments to which Gag can possibly be targeted, such as late endosomes. Thus, in the current study, we defined VCCs as intracellular CD81-positive vesicular compartments. In 5ptaseIV Δ1-expressing cells, we found that Gag-YFP localized to VCCs (Fig. 2A). In contrast, in full-length 5ptaseIV-expressing cells, HIV-1 Gag-YFP failed to localize to intracellular CD81-positive compartments. Instead, Gag-YFP displayed a hazy cytosolic signal (Fig. 2A, middle row) or localized to CD81-negative compartments (Fig. 2A, bottom row). At this point, it is not possible to distinguish whether the latter pattern of Gag localization is due to mistargeting of Gag or if PI(4,5)P2 depletion inhibits recruitment of CD81 to VCCs.
FIG 2.
HIV-1 Gag failed to localize to VCCs upon 5ptaseIV overexpression. (A) MDMs were infected with pseudotyped HIV-1 encoding Gag-YFP along with 5ptaseIV (FL) or 5ptaseIV Δ1. At 48 h postinfection, cells were stained with ConA labeled with Alexa Fluor 594 (a PM marker), fixed, immunostained with a mouse monoclonal anti-CD81 antibody and anti-mouse IgG conjugated with Alexa Fluor 647, and analyzed using a confocal microscope. (B) Pearson's correlation coefficients for colocalization of Gag-YFP with CD81 are shown as means and standard errors of the means (SEM). Twenty to 30 cells were analyzed per condition. **, P < 0.001.
To quantitatively assess the effect of PI(4,5)P2 depletion on Gag-YFP localization to the VCCs on a single-cell basis, we measured Pearson's correlation coefficient (PCC) for Gag-YFP and CD81 signals from confocal images (Fig. 2B). In this analysis, we found that Gag-YFP and CD81 showed a higher PCC for 5ptaseIV Δ1-expressing cells than for full-length 5ptaseIV-expressing cells. Note that because CD81 is also present at the cell surface, the low PCC value for the latter cells likely reflects the absence of the Gag-YFP signal at both the cell surface PM and intracellular CD81-positive compartments. Altogether, these data support the hypothesis that PI(4,5)P2 is required for HIV-1 Gag localization to the VCCs in MDMs.
Gag targeting to VCCs requires the N-terminal myristate moiety but not an intact HBR.
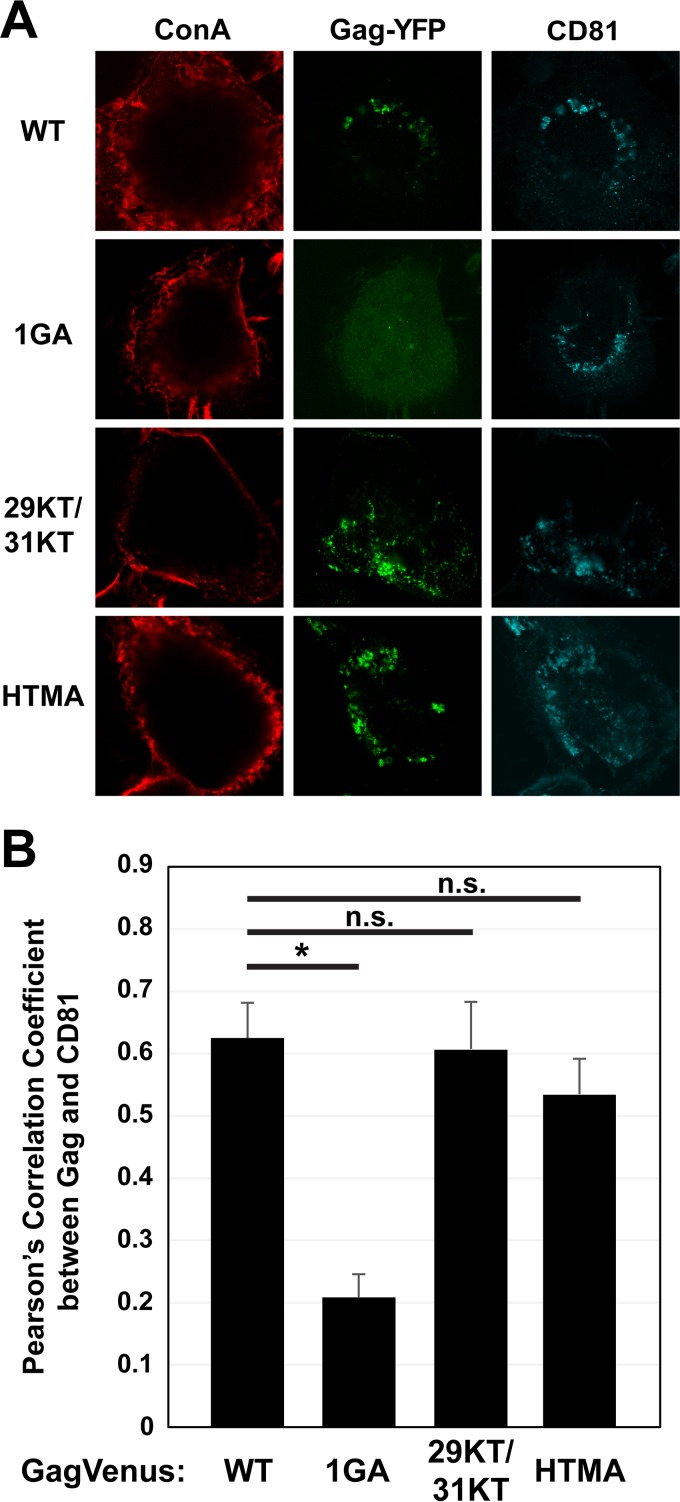
The results described above did not allow us to determine whether cellular PI(4,5)P2 is necessary primarily for Gag membrane binding, which is likely a prerequisite for Gag localization to VCCs, or whether PI(4,5)P2 also determines specific Gag localization via directly recruiting Gag to VCCs. To distinguish between these possibilities, we examined the localization of Gag derivatives with changes in MA, the interface for PI(4,5)P2 interaction. To test whether membrane binding is indeed a prerequisite for HIV-1 Gag localization to the VCCs, we expressed an HIV-1 molecular clone containing a Gag-YFP construct defective in myristoylation (1GA Gag-YFP) in MDMs (Fig. 3A). As expected, we found that 1GA Gag-YFP displayed a hazy cytosolic signal, indicative of a membrane binding defect, and failed to colocalize with CD81 (Fig. 3A). Consistent with this, the PCC for 1GA Gag-YFP and CD81 was low (∼0.2), in contrast to the PCC for WT Gag-YFP and CD81 (∼0.6) (Fig. 3B). These results indicate that the myristate moiety of HIV-1 Gag is required for membrane binding in MDMs and that membrane binding is required for VCC localization.
FIG 3.
The myristate moiety but not the intact HIV-1 MA sequence is required for VCC localization. (A) MDMs were infected with pseudotyped HIV-1 encoding WT Gag-YFP or Gag-YFP derivatives containing the indicated MA substitutions. At 48 h postinfection, cells were stained with ConA labeled with Alexa Fluor 594, fixed, immunostained with a mouse monoclonal anti-CD81 antibody and anti-mouse IgG conjugated with Alexa Fluor 647, and analyzed using a confocal microscope. (B) Pearson's correlation coefficients for colocalization of Gag-YFP with CD81 are shown as means and SEM. Twenty to 30 cells were analyzed per condition. *, P < 0.005; n.s., not significant.
Previous studies showed that mutations in HBR residues 29 and 31 result in HIV-1 Gag mislocalization to the perinuclear region in HeLa cells (45). Both basic-to-acidic (29KE/31KE) and basic-to-neutral (29KT/31KT) amino acid substitutions at these residues impair Gag interactions with PI(4,5)P2 in in vitro liposome binding assays (40). To test whether the HBR-mediated PI(4,5)P2 interaction is important for Gag targeting to the VCCs, we expressed HIV-1 encoding a Gag-YFP construct that contains 29KT/31KT mutations. Gag-YFP with the 29KT/31KT substitutions colocalized with CD81 at intracellular compartments, with a high PCC value similar to that with WT Gag-YFP (Fig. 3A and B). This observation is consistent with previous studies showing that HIV-1 Gag with 29KE/31KE changes, which is less efficient in total membrane binding than the 29KT/31KT mutant, shows colocalization with CD81 (21). Thus, these results suggest that HBR residues which are required for PI(4,5)P2 binding in vitro are not required for Gag targeting to the VCCs.
The MA-RNA interaction, which suppresses MA binding to non-PI(4,5)P2 acidic lipids, is not required for Gag localization to VCCs.
While the results described above suggest that the MA-PI(4,5)P2 interaction does not directly promote localization of HIV-1 Gag to VCCs, it is possible that RNA-mediated suppression of MA-acidic lipid interactions may enhance the specificity of this localization. In contrast to HIV-1 Gag membrane binding, membrane binding of human T-cell leukemia virus 1 (HTLV-1) Gag or chimeric HIV-1 Gag proteins containing HTLV-1 MA in place of HIV-1 MA is neither dependent on PI(4,5)P2 nor susceptible to RNA-mediated suppression (61, 93). Consistent with the lack of lipid specificity, the HTLV-MA-containing Gag constructs localize to both the PM and intracellular compartments in HeLa cells (61, 93). To test whether Gag localization to VCCs requires the MA-RNA interaction, which suppresses MA interactions with ubiquitous acidic lipids, we expressed in MDMs an HIV-1 molecular clone encoding a Gag-YFP chimera in which HIV-1 MA is replaced with HTLV-1 MA (HTMA Gag) (61). We found that HTMA Gag-YFP colocalized with CD81 in intracellular compartments, with an efficiency similar to that of WT Gag-YFP (Fig. 3A and B). These results suggest that RNA-mediated inhibition of membrane binding via non-PI(4,5)P2 acidic lipids is not required for HIV-1 Gag localization to the VCCs.
HIV-1 MA is not required for VCC localization in MDMs.
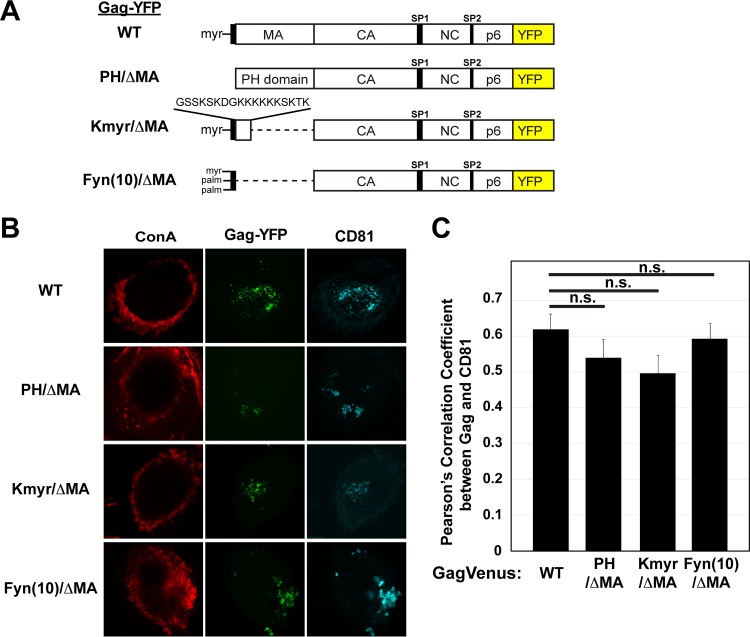
To further study the role of MA-mediated membrane binding in Gag localization to the VCCs, we replaced HIV-1 MA with the following membrane binding motifs: (i) the PH domain of phospholipase C δ1, which binds to the PI(4,5)P2 head group (79, 94–96); (ii) Kmyr, which contains a myristoylation signal and the K-Ras4B polybasic sequence (81, 97, 98); and (iii) Fyn(10), which corresponds to the N-terminal 10 residues of the Fyn kinase, containing myristoylation and dual-palmitoylation signals and hence binding to the membrane via hydrophobic interactions (40) (Fig. 4A). These MA replacement changes were introduced into HIV-1 molecular clones encoding Gag-YFP and expressed in MDMs. Interestingly, we found that, similarly to WT Gag-YFP, PH/ΔMA Gag-YFP, Kmyr/ΔMA Gag-YFP, and Fyn(10)/ΔMA Gag-YFP colocalized with CD81 at intracellular compartments, with high PCC values (0.5 or higher) (Fig. 4B and C), indicating that various modes of membrane binding [i.e., mediated by a structure binding to PI(4,5)P2 head group, myristoylation and polybasic sequence, or triple acylation] can replace MA functions with regard to HIV-1 Gag localization to VCCs. Notably, the PCC for ConA, the cell surface marker, and WT Gag-YFP was <0.1 (see below). Likewise, the PCC values for ConA and the 29KE/31KE, HTMA, and ΔMA Gag-YFP derivatives were all <0.1 (data not shown). Therefore, high PCC values for these Gag-YFP proteins and CD81 reflect Gag-YFP accumulation in intracellular CD81-positive compartments but not colocalization with the surface PM population of CD81. Altogether, our data indicate that membrane binding mediated by MA is an essential step in Gag localization to the VCCs but that the MA-PI(4,5)P2 interaction and any other MA functions, except for general membrane binding, are dispensable for VCC localization. Thus, PI(4,5)P2 serves mainly as a membrane anchor that facilitates HIV-1 Gag membrane binding, not as a molecule recruiting Gag specifically to VCCs.
FIG 4.
Heterologous membrane binding sequences can replace HIV-1 MA without affecting VCC localization. (A) Schematic illustrations of WT, PH/ΔMA, Kmyr/ΔMA, and Fyn(10)/ΔMA Gag-YFP. (B) MDMs were infected with pseudotyped HIV-1 encoding WT Gag-YFP or Gag-YFP derivatives in which MA was replaced by a heterologous membrane binding sequence. At 48 h postinfection, cells were stained with ConA labeled with Alexa Fluor 594, fixed, immunostained with a mouse monoclonal anti-CD81 antibody and anti-mouse IgG conjugated with Alexa Fluor 647, and analyzed using a confocal microscope. (C) Pearson's correlation coefficients for colocalization of Gag-YFP with CD81 are shown as means and SEM. Twenty to 30 cells were analyzed per condition. n.s., not significant.
Higher-order multimerization is required for HIV-1 Gag localization to VCCs in MDMs.
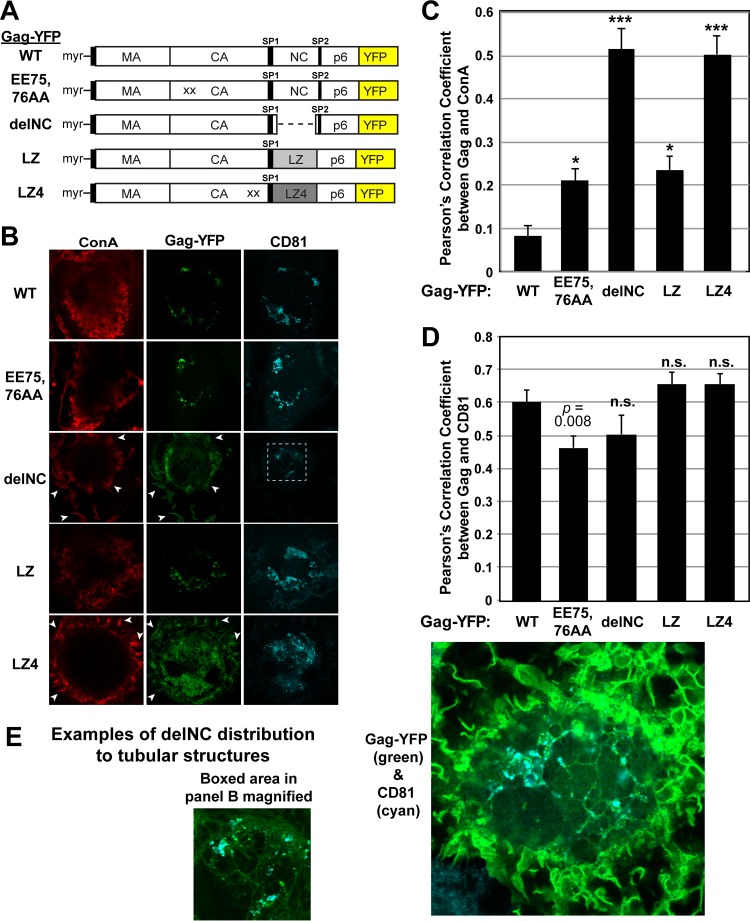
Since HIV-1 MA is dispensable for VCC localization of Gag, a downstream region of HIV-1 Gag is likely to contain the molecular determinant(s) for Gag localization to the VCCs. Previous studies demonstrated that NC-dependent multimerization is necessary for the specific localization of HIV-1 Gag to the uropod in polarized T cells (66, 67, 74). To test whether multimerization is similarly important for specific targeting to the VCCs in MDMs, we tested a panel of HIV-1 Gag constructs containing various multimerization mutations (Fig. 5A).
FIG 5.
HIV-1 Gag localization to VCCs requires higher-order multimerization. (A) Schematic illustrations of WT, EE75,76AA, delNC, LZ, and LZ4 Gag-YFP. LZ4 Gag-YFP contains the WM184,185AA CA mutation, which disrupts Gag dimerization, in addition to replacement of NC with a tetramer-forming LZ sequence. (B) MDMs were infected with pseudotyped HIV-1 encoding WT Gag-YFP or Gag-YFP derivatives containing the substitutions shown in panel A. At 48 h postinfection, cells were stained with ConA labeled with Alexa Fluor 594, fixed, immunostained with a mouse monoclonal anti-CD81 antibody and anti-mouse IgG conjugated with Alexa Fluor 647, and analyzed using a confocal microscope. Note that for cells expressing delNC Gag-YFP or LZ4 Gag-YFP, prominent YFP signals were found at the cell surface as detected by ConA (white arrowheads), whereas such surface YFP signals were nearly undetectable in WT Gag-YFP-expressing cells. (C) Pearson's correlation coefficients for colocalization of Gag-YFP with ConA are shown as means and SEM. Twenty to 40 cells were analyzed per condition. *, P < 0.005; ***, P < 0.0001. Note that while the EE75,76AA and LZ constructs showed higher PCC values than that of the WT, the values are less than 0.3, consistent with the lack of obvious PM localization of these Gag derivatives. (D) Pearson's correlation coefficients for colocalization of Gag-YFP with CD81 are shown as means and SEM. Twenty to 40 cells were analyzed per condition. n.s., not significant. (E) Examples of delNC Gag-YFP distribution to internal tubular structures. In the left panel, the boxed area from panel B is magnified. The YFP intensity was enhanced in this panel relative to that in panel B.
We first tested EE75,76AA Gag, an HIV-1 Gag mutant with mutations in the CA domain. This mutant was previously shown to form electron-dense patches at the PM, suggesting that it multimerizes but fails to cause membrane curvature or to form virus particles (66, 80, 99). We found that Gag-YFP containing the EE75,76AA mutation was able to localize to intracellular CD81-positive compartments (Fig. 5B). These results are analogous to our previous findings in that uropod localization of HIV-1 Gag does not require formation of membrane curvature or full particles (66).
We next examined whether NC is required for VCC targeting. We first compared the ability of HIV-1 delNC Gag, an NC-deleted HIV-1 Gag mutant, to localize to the VCCs. delNC Gag was previously shown to bind to the membrane but displayed lower levels of Gag-Gag interaction than those with WT Gag in HeLa and T cells (67, 78). We found that deletion of NC resulted in Gag localization not only to the intracellular CD81-positive compartments but also to other locations (Fig. 5B and C). In particular, delNC Gag was also found at the cell surface as well as at tubular compartments that appeared to interconnect the intracellular CD81-positive compartments (Fig. 5B and E). The PCC for delNC Gag-YFP and ConA, used as a PM marker, shows that they colocalized with a high efficiency (PCC, ∼0.5) (Fig. 5C). This is in stark contrast to the situation with WT Gag and EE75,76AA Gag, which showed less colocalization with ConA and thus had lower PCCs (0.1 to 0.2) (Fig. 5B and C). We note that despite the difference in PM localization, PCCs for CD81 and both delNC Gag and EE75,76AA Gag, as well as those for the NC substitution mutants described below, were around 0.5 or higher (Fig. 5D), presumably due to the presence of CD81 at the cell surface PM and the VCCs. Altogether, these results suggest that NC enhances the specificity of HIV-1 Gag localization to the VCCs.
To examine whether NC contributes to VCC targeting by promoting Gag multimerization or whether NC-RNA interactions play a role, we examined the subcellular localization of a Gag-YFP construct with NC replaced with a dimerizing leucine zipper (LZ) motif (GagLZ) (Fig. 5A). GagLZ was previously shown to multimerize efficiently and to be able to form virus-like particles that are indistinguishable from those with WT HIV-1 Gag (100). We found that the LZ sequence restored specific localization of Gag to the intracellular CD81-positive compartments and reduced the distribution of Gag to the ConA-positive PM (PCC, ∼0.2) (Fig. 5B and C). These results indicate that Gag multimerization is an essential step in VCC localization and that NC contributes to specific Gag targeting to the VCCs by functioning as a multimerization domain.
Finally, we examined the minimal level of multimerization that is needed for Gag targeting to the VCCs. Our previous studies demonstrated that an LZ derivative that forms a tetramer (LZ4) (101) is able to restore Gag localization to the uropod in T cells even when both CA- and NC-mediated multimerization is disrupted (66) (Fig. 5A). Thus, it is conceivable that the level of multimerization mediated by LZ4 is also sufficient to target Gag to VCCs in MDMs. Interestingly, however, while we observed Gag-LZ4 localization to intracellular CD81 compartments, a large portion of this Gag derivative remained at the PM and colocalized with ConA (PCC, ∼0.5) (Fig. 5B and C). These results suggest that specific Gag targeting to the VCCs in MDMs requires more extensive Gag multimerization than Gag localization to uropods in polarized T cells.
DISCUSSION
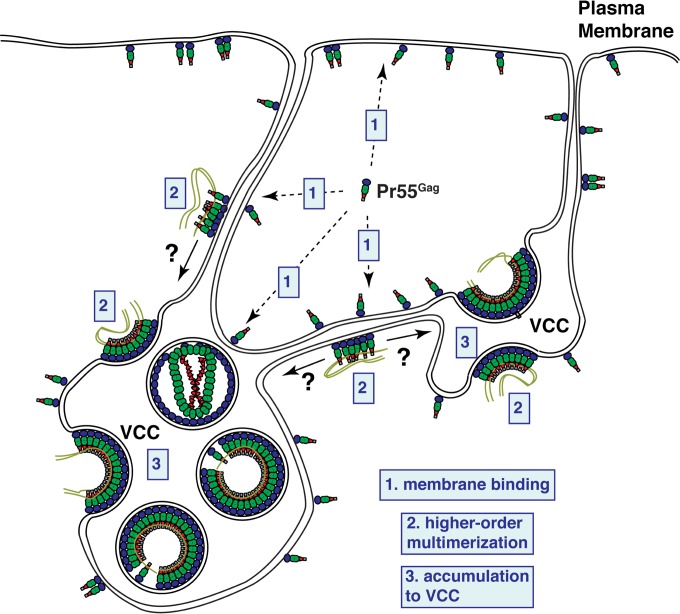
Despite the potential importance of macrophages and VCCs in maintenance of virus reservoirs and virus spread to T cells (20–22, 102–104), how HIV-1 Gag is targeted to this site of virus assembly in macrophages is not well understood. In this study, we found that the mechanism of HIV-1 Gag targeting to the VCCs is dependent on the NC domain, an aspect shared by the mechanism of Gag targeting to the uropod in polarized T cells. We observed that deletion of NC results in a failure of Gag to specifically accumulate to the CD81-enriched intracellular compartments and causes substantial Gag distribution to the cell surface PM (Fig. 5). In contrast, WT Gag or Gag mutants that are capable of higher-order multimerization (such as EE75,76AA Gag and GagLZ) are found mostly in the intracellular CD81-positive compartments. Based on these results, we propose a working model in which NC-mediated multimerization facilitates Gag movement from the cell surface PM to the VCCs, which are deep and complex invaginations of the PM (Fig. 6). In this model, we propose that HIV-1 Gag (i) initially binds to the cell surface PM in a manner dependent on myristoylation and the MA-PI(4,5)P2 interaction, (ii) begins to multimerize, and (iii) subsequently accumulates in the VCCs. Gag multimerization induces reorganization of PM microdomains in model cell types (79, 105). Therefore, it is conceivable that NC-mediated multimerization at the PM in MDMs may promote reorganization of and/or association with microdomains, which may in turn facilitate Gag trafficking to VCCs. Interestingly, delNC Gag localized not only to the PM but also to apparently tubular compartments forming a membranous network. This is consistent with an earlier electron tomography study which showed that VCCs not only are connected to the cell surface but are interconnected intracellularly as well (18). Notably, PH-GFP, a reporter construct that binds PI(4,5)P2, was observed to localize to a similar web-like intracellular structure that highly overlaps VCCs in MDMs (23). Therefore, initial membrane binding of Gag may occur not only at the cell surface PM but also anywhere in the intracellular web enriched in PI(4,5)P2. Thus, although not all VCCs are connected to the cell surface PM (16, 27), Gag may be recruited to the VCCs via these web-like structures. While our study demonstrated that multimerization of HIV-1 Gag drives its VCC localization, how HIV-1 Gag multimers distinguish VCCs from any other part of the intracellular membranous network is not known.
FIG 6.
Working model for HIV-1 Gag targeting to VCCs. HIV-1 Gag proteins are synthesized in the cytosol and bind to membranes containing PI(4,5)P2, which include the PM and VCCs as well as the tubular network connecting these locations. These membranes may also receive Gag from vesicles of endosomal pathways in cases where Gag lacks specificity for PI(4,5)P2 (e.g., HTMA Gag or some ΔMA Gag derivatives). Higher-order multimers of Gag accumulate in VCCs, potentially via lateral movement along membranous connections to the VCCs, where assembly continues to progress.
BST-2/tetherin has been shown to promote expansion of VCCs (106, 107). However, EE75,76AA Gag, which is unable to recruit BST-2/tetherin to the site of assembly in HeLa cells (80), still showed only minimal localization at the cell surface and instead localized to VCCs. Therefore, it appears that Gag targeting to VCCs and reduction of Gag distribution to the cell surface PM can occur independently of tetherin recruitment to Gag multimers (Fig. 5). Nevertheless, cell-type-specific differences may influence the phenotypes of the EE75,76AA Gag mutant (i.e., the ability to recruit tetherin or the lack thereof), and therefore more in-depth studies of the Gag multimer-tetherin association in macrophages need to be carried out.
Our study showed that HIV-1 MA can be replaced with any membrane binding motif, including the PH domain, the Kmyr sequence, the Fyn (10) sequence, and the HTLV-1 MA domain, without affecting VCC localization of Gag (Fig. 3 and 4). While these results indicate that the Gag-PI(4,5)P2 interaction is dispensable for specific localization to the VCCs, we also found that membrane binding of WT Gag and virus release are disrupted in macrophages expressing 5ptaseIV, which depletes cellular PI(4,5)P2 (Fig. 1 and 2). Thus, these data suggest that the MA-PI(4,5)P2 interaction is required for Gag binding to the membrane, which in turn allows Gag to multimerize and be targeted to VCCs. Notably, both Fyn(10)/ΔMA Gag and HTMA Gag are promiscuously distributed to both the PM and intracellular vesicles in HeLa cells (40, 61), presumably due to their lack of PI(4,5)P2- and RNA-mediated regulation of membrane binding via the MA HBR. Despite this lack of specificity, both Gag derivatives showed a WT-like localization to intracellular CD81-positive vesicular compartments, the definition of VCCs in this study, in MDMs. Therefore, we do not rule out the possibility that some population of these Gag derivatives may initially localize at non-PM, non-VCC compartments, which may subsequently fuse with either the PM or the expanding VCCs during infection.
In conclusion, the current study identified a viral determinant for Gag targeting during HIV-1 assembly in macrophages and thereby revealed that HIV-1 Gag targeting to the site of assembly in macrophages is a two-step process, i.e., successful membrane binding via the PI(4,5)P2-MA interaction followed by accumulation in VCCs, the latter being driven by NC-dependent higher-order multimerization. This multimerization-driven targeting mechanism is reminiscent of Gag targeting to the uropod of polarized T cells, another subcellular site implicated in cell-to-cell transmission of HIV-1, although the extent of Gag multimerization required for VCC localization appears to be different from that for uropod localization. Investigation into the mechanisms that drive Gag multimers to these subcellular locations is under way.
ACKNOWLEDGMENTS
We thank the members of our laboratory for helpful discussions and critical reviews of the manuscript and Leslie Goo for technical contributions during the early phase of the study. We thank A. Cimarelli and K. T. Jeang for reagents. The following reagent was obtained through the AIDS Research and Reference Reagent Program, Division of AIDS, NIAID, NIH: HIV Ig (from NABI and NHLBI).
This work was supported by an American Heart Association Midwest Predoctoral Fellowship (grant 13PRE17060006 to J.I.), University of Michigan Rackham Predoctoral Fellowships (to J.I. and V.C.), and NIH grant R01 AI071727 (to A.O.).
REFERENCES
- 1.Koppensteiner H, Brack-Werner R, Schindler M. 2012. Macrophages and their relevance in human immunodeficiency virus type I infection. Retrovirology 9:82. doi: 10.1186/1742-4690-9-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kumar A, Abbas W, Herbein G. 2014. HIV-1 latency in monocytes/macrophages. Viruses 6:1837–1860. doi: 10.3390/v6041837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campbell JH, Hearps AC, Martin GE, Williams KC, Crowe SM. 2014. The importance of monocytes and macrophages in HIV pathogenesis, treatment, and cure. AIDS 28:2175–2187. doi: 10.1097/QAD.0000000000000408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Swingler S, Mann AM, Zhou J, Swingler C, Stevenson M. 2007. Apoptotic killing of HIV-1-infected macrophages is subverted by the viral envelope glycoprotein. PLoS Pathog 3:1281–1290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carter CA, Ehrlich LS. 2008. Cell biology of HIV-1 infection of macrophages. Annu Rev Microbiol 62:425–443. doi: 10.1146/annurev.micro.62.081307.162758. [DOI] [PubMed] [Google Scholar]
- 6.Joseph SB, Arrildt KT, Sturdevant CB, Swanstrom R. 2015. HIV-1 target cells in the CNS. J Neurovirol 21:276–289. doi: 10.1007/s13365-014-0287-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jouvenet N, Neil SJ, Bess C, Johnson MC, Virgen CA, Simon SM, Bieniasz PD. 2006. Plasma membrane is the site of productive HIV-1 particle assembly. PLoS Biol 4:e435. doi: 10.1371/journal.pbio.0040435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Finzi A, Orthwein A, Mercier J, Cohen EA. 2007. Productive human immunodeficiency virus type 1 assembly takes place at the plasma membrane. J Virol 81:7476–7490. doi: 10.1128/JVI.00308-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gelderblom HR. 1991. Assembly and morphology of HIV: potential effect of structure on viral function. AIDS 5:617–637. doi: 10.1097/00002030-199106000-00001. [DOI] [PubMed] [Google Scholar]
- 10.Orenstein JM, Meltzer MS, Phipps T, Gendelman HE. 1988. Cytoplasmic assembly and accumulation of human immunodeficiency virus types 1 and 2 in recombinant human colony-stimulating factor-1-treated human monocytes: an ultrastructural study. J Virol 62:2578–2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Raposo G, Moore M, Innes D, Leijendekker R, Leigh-Brown A, Benaroch P, Geuze H. 2002. Human macrophages accumulate HIV-1 particles in MHC II compartments. Traffic 3:718–729. doi: 10.1034/j.1600-0854.2002.31004.x. [DOI] [PubMed] [Google Scholar]
- 12.Pelchen-Matthews A, Kramer B, Marsh M. 2003. Infectious HIV-1 assembles in late endosomes in primary macrophages. J Cell Biol 162:443–455. doi: 10.1083/jcb.200304008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bennett AE, Narayan K, Shi D, Hartnell LM, Gousset K, He H, Lowekamp BC, Yoo TS, Bliss D, Freed EO, Subramaniam S. 2009. Ion-abrasion scanning electron microscopy reveals surface-connected tubular conduits in HIV-infected macrophages. PLoS Pathog 5:e1000591. doi: 10.1371/journal.ppat.1000591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chu H, Wang JJ, Qi M, Yoon JJ, Wen X, Chen X, Ding L, Spearman P. 2012. The intracellular virus-containing compartments in primary human macrophages are largely inaccessible to antibodies and small molecules. PLoS One 7:e35297. doi: 10.1371/journal.pone.0035297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deneka M, Pelchen-Matthews A, Byland R, Ruiz-Mateos E, Marsh M. 2007. In macrophages, HIV-1 assembles into an intracellular plasma membrane domain containing the tetraspanins CD81, CD9, and CD53. J Cell Biol 177:329–341. doi: 10.1083/jcb.200609050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jouve M, Sol-Foulon N, Watson S, Schwartz O, Benaroch P. 2007. HIV-1 buds and accumulates in “nonacidic” endosomes of macrophages. Cell Host Microbe 2:85–95. doi: 10.1016/j.chom.2007.06.011. [DOI] [PubMed] [Google Scholar]
- 17.Koppensteiner H, Banning C, Schneider C, Hohenberg H, Schindler M. 2012. Macrophage internal HIV-1 is protected from neutralizing antibodies. J Virol 86:2826–2836. doi: 10.1128/JVI.05915-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Welsch S, Groot F, Krausslich HG, Keppler OT, Sattentau QJ. 2011. Architecture and regulation of the HIV-1 assembly and holding compartment in macrophages. J Virol 85:7922–7927. doi: 10.1128/JVI.00834-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Welsch S, Keppler OT, Habermann A, Allespach I, Krijnse-Locker J, Krausslich HG. 2007. HIV-1 buds predominantly at the plasma membrane of primary human macrophages. PLoS Pathog 3:e36. doi: 10.1371/journal.ppat.0030036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sharova N, Swingler C, Sharkey M, Stevenson M. 2005. Macrophages archive HIV-1 virions for dissemination in trans. EMBO J 24:2481–2489. doi: 10.1038/sj.emboj.7600707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gousset K, Ablan SD, Coren LV, Ono A, Soheilian F, Nagashima K, Ott DE, Freed EO. 2008. Real-time visualization of HIV-1 Gag trafficking in infected macrophages. PLoS Pathog 4:e1000015. doi: 10.1371/journal.ppat.1000015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Groot F, Welsch S, Sattentau QJ. 2008. Efficient HIV-1 transmission from macrophages to T cells across transient virological synapses. Blood 111:4660–4663. doi: 10.1182/blood-2007-12-130070. [DOI] [PubMed] [Google Scholar]
- 23.Mlcochova P, Pelchen-Matthews A, Marsh M. 2013. Organization and regulation of intracellular plasma membrane-connected HIV-1 assembly compartments in macrophages. BMC Biol 11:89. doi: 10.1186/1741-7007-11-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pelchen-Matthews A, Giese S, Mlcochova P, Turner J, Marsh M. 2012. beta2 integrin adhesion complexes maintain the integrity of HIV-1 assembly compartments in primary macrophages. Traffic 13:273–291. doi: 10.1111/j.1600-0854.2011.01306.x. [DOI] [PubMed] [Google Scholar]
- 25.Marsh M, Theusner K, Pelchen-Matthews A. 2009. HIV assembly and budding in macrophages. Biochem Soc Trans 37:185–189. doi: 10.1042/BST0370185. [DOI] [PubMed] [Google Scholar]
- 26.Berre S, Gaudin R, Cunha de Alencar B, Desdouits M, Chabaud M, Naffakh N, Rabaza-Gairi M, Gobert FX, Jouve M, Benaroch P. 2013. CD36-specific antibodies block release of HIV-1 from infected primary macrophages and its transmission to T cells. J Exp Med 210:2523–2538. doi: 10.1084/jem.20130566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Gaudin R, Berre S, Cunha de Alencar B, Decalf J, Schindler M, Gobert FX, Jouve M, Benaroch P. 2013. Dynamics of HIV-containing compartments in macrophages reveal sequestration of virions and transient surface connections. PLoS One 8:e69450. doi: 10.1371/journal.pone.0069450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gaudin R, de Alencar BC, Jouve M, Berre S, Le Bouder E, Schindler M, Varthaman A, Gobert FX, Benaroch P. 2012. Critical role for the kinesin KIF3A in the HIV life cycle in primary human macrophages. J Cell Biol 199:467–479. doi: 10.1083/jcb.201201144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bieniasz PD. 2009. The cell biology of HIV-1 virion genesis. Cell Host Microbe 5:550–558. doi: 10.1016/j.chom.2009.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sundquist WI, Krausslich HG. 2012. HIV-1 assembly, budding, and maturation. Cold Spring Harb Perspect Med 2:a006924. doi: 10.1101/cshperspect.a006924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Balasubramaniam M, Freed EO. 2011. New insights into HIV assembly and trafficking. Physiology (Bethesda) 26:236–251. doi: 10.1152/physiol.00051.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Adamson CS, Freed EO. 2007. Human immunodeficiency virus type 1 assembly, release, and maturation. Adv Pharmacol 55:347–387. doi: 10.1016/S1054-3589(07)55010-6. [DOI] [PubMed] [Google Scholar]
- 33.Freed EO. 1998. HIV-1 gag proteins: diverse functions in the virus life cycle. Virology 251:1–15. doi: 10.1006/viro.1998.9398. [DOI] [PubMed] [Google Scholar]
- 34.Swanstrom R, Wills JW. 1997. Synthesis, assembly, and processing of viral proteins, p 263–334. In Coffin JM, Hughes SH, Varmus HE (ed), Retroviruses. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY. [PubMed] [Google Scholar]
- 35.Bryant M, Ratner L. 1990. Myristoylation-dependent replication and assembly of human immunodeficiency virus 1. Proc Natl Acad Sci U S A 87:523–527. doi: 10.1073/pnas.87.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chukkapalli V, Ono A. 2011. Molecular determinants that regulate plasma membrane association of HIV-1 Gag. J Mol Biol 410:512–524. doi: 10.1016/j.jmb.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gottlinger HG, Sodroski JG, Haseltine WA. 1989. Role of capsid precursor processing and myristoylation in morphogenesis and infectivity of human immunodeficiency virus type 1. Proc Natl Acad Sci U S A 86:5781–5785. doi: 10.1073/pnas.86.15.5781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Spearman P, Wang JJ, Vander Heyden N, Ratner L. 1994. Identification of human immunodeficiency virus type 1 Gag protein domains essential to membrane binding and particle assembly. J Virol 68:3232–3242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhou W, Parent LJ, Wills JW, Resh MD. 1994. Identification of a membrane-binding domain within the amino-terminal region of human immunodeficiency virus type 1 Gag protein which interacts with acidic phospholipids. J Virol 68:2556–2569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chukkapalli V, Hogue IB, Boyko V, Hu WS, Ono A. 2008. Interaction between the human immunodeficiency virus type 1 Gag matrix domain and phosphatidylinositol-(4,5)-bisphosphate is essential for efficient Gag membrane binding. J Virol 82:2405–2417. doi: 10.1128/JVI.01614-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hill CP, Worthylake D, Bancroft DP, Christensen AM, Sundquist WI. 1996. Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly. Proc Natl Acad Sci U S A 93:3099–3104. doi: 10.1073/pnas.93.7.3099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Saad JS, Miller J, Tai J, Kim A, Ghanam RH, Summers MF. 2006. Structural basis for targeting HIV-1 Gag proteins to the plasma membrane for virus assembly. Proc Natl Acad Sci U S A 103:11364–11369. doi: 10.1073/pnas.0602818103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shkriabai N, Datta SA, Zhao Z, Hess S, Rein A, Kvaratskhelia M. 2006. Interactions of HIV-1 Gag with assembly cofactors. Biochemistry 45:4077–4083. doi: 10.1021/bi052308e. [DOI] [PubMed] [Google Scholar]
- 44.Ono A, Freed EO. 2004. Cell-type-dependent targeting of human immunodeficiency virus type 1 assembly to the plasma membrane and the multivesicular body. J Virol 78:1552–1563. doi: 10.1128/JVI.78.3.1552-1563.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ono A, Orenstein JM, Freed EO. 2000. Role of the Gag matrix domain in targeting human immunodeficiency virus type 1 assembly. J Virol 74:2855–2866. doi: 10.1128/JVI.74.6.2855-2866.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chukkapalli V, Oh SJ, Ono A. 2010. Opposing mechanisms involving RNA and lipids regulate HIV-1 Gag membrane binding through the highly basic region of the matrix domain. Proc Natl Acad Sci U S A 107:1600–1605. doi: 10.1073/pnas.0908661107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Freed EO, Orenstein JM, Buckler-White AJ, Martin MA. 1994. Single amino acid changes in the human immunodeficiency virus type 1 matrix protein block virus particle production. J Virol 68:5311–5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hermida-Matsumoto L, Resh MD. 2000. Localization of human immunodeficiency virus type 1 Gag and Env at the plasma membrane by confocal imaging. J Virol 74:8670–8679. doi: 10.1128/JVI.74.18.8670-8679.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Yuan X, Yu X, Lee TH, Essex M. 1993. Mutations in the N-terminal region of human immunodeficiency virus type 1 matrix protein block intracellular transport of the Gag precursor. J Virol 67:6387–6394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ono A, Ablan SD, Lockett SJ, Nagashima K, Freed EO. 2004. Phosphatidylinositol (4,5) bisphosphate regulates HIV-1 Gag targeting to the plasma membrane. Proc Natl Acad Sci U S A 101:14889–14894. doi: 10.1073/pnas.0405596101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Burniston MT, Cimarelli A, Colgan J, Curtis SP, Luban J. 1999. Human immunodeficiency virus type 1 Gag polyprotein multimerization requires the nucleocapsid domain and RNA and is promoted by the capsid-dimer interface and the basic region of matrix protein. J Virol 73:8527–8540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Chang CY, Chang YF, Wang SM, Tseng YT, Huang KJ, Wang CT. 2008. HIV-1 matrix protein repositioning in nucleocapsid region fails to confer virus-like particle assembly. Virology 378:97–104. doi: 10.1016/j.virol.2008.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Cimarelli A, Luban J. 1999. Translation elongation factor 1-alpha interacts specifically with the human immunodeficiency virus type 1 Gag polyprotein. J Virol 73:5388–5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hearps AC, Wagstaff KM, Piller SC, Jans DA. 2008. The N-terminal basic domain of the HIV-1 matrix protein does not contain a conventional nuclear localization sequence but is required for DNA binding and protein self-association. Biochemistry 47:2199–2210. doi: 10.1021/bi701360j. [DOI] [PubMed] [Google Scholar]
- 55.Lochrie MA, Waugh S, Pratt DG Jr, Clever J, Parslow TG, Polisky B. 1997. In vitro selection of RNAs that bind to the human immunodeficiency virus type-1 gag polyprotein. Nucleic Acids Res 25:2902–2910. doi: 10.1093/nar/25.14.2902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ott DE, Coren LV, Gagliardi TD. 2005. Redundant roles for nucleocapsid and matrix RNA-binding sequences in human immunodeficiency virus type 1 assembly. J Virol 79:13839–13847. doi: 10.1128/JVI.79.22.13839-13847.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Purohit P, Dupont S, Stevenson M, Green MR. 2001. Sequence-specific interaction between HIV-1 matrix protein and viral genomic RNA revealed by in vitro genetic selection. RNA 7:576–584. doi: 10.1017/S1355838201002023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Alfadhli A, McNett H, Tsagli S, Bachinger HP, Peyton DH, Barklis E. 2011. HIV-1 matrix protein binding to RNA. J Mol Biol 410:653–666. doi: 10.1016/j.jmb.2011.04.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Alfadhli A, Still A, Barklis E. 2009. Analysis of human immunodeficiency virus type 1 matrix binding to membranes and nucleic acids. J Virol 83:12196–12203. doi: 10.1128/JVI.01197-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chukkapalli V, Inlora J, Todd GC, Ono A. 2013. Evidence in support of RNA-mediated inhibition of phosphatidylserine-dependent HIV-1 Gag membrane binding in cells. J Virol 87:7155–7159. doi: 10.1128/JVI.00075-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Inlora J, Chukkapalli V, Derse D, Ono A. 2011. Gag localization and virus-like particle release mediated by the matrix domain of human T-lymphotropic virus type 1 Gag are less dependent on phosphatidylinositol-(4,5)-bisphosphate than those mediated by the matrix domain of HIV-1 Gag. J Virol 85:3802–3810. doi: 10.1128/JVI.02383-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jones CP, Datta SA, Rein A, Rouzina I, Musier-Forsyth K. 2011. Matrix domain modulates HIV-1 Gag's nucleic acid chaperone activity via inositol phosphate binding. J Virol 85:1594–1603. doi: 10.1128/JVI.01809-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Kutluay SB, Zang T, Blanco-Melo D, Powell C, Jannain D, Errando M, Bieniasz PD. 2014. Global changes in the RNA binding specificity of HIV-1 gag regulate virion genesis. Cell 159:1096–1109. doi: 10.1016/j.cell.2014.09.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gerber PP, Cabrini M, Jancic C, Paoletti L, Banchio C, von Bilderling C, Sigaut L, Pietrasanta LI, Duette G, Freed EO, Basile Gde S, Moita CF, Moita LF, Amigorena S, Benaroch P, Geffner J, Ostrowski M. 2015. Rab27a controls HIV-1 assembly by regulating plasma membrane levels of phosphatidylinositol 4,5-bisphosphate. J Cell Biol 209:435–452. doi: 10.1083/jcb.201409082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Sanchez-Madrid F, Serrador JM. 2009. Bringing up the rear: defining the roles of the uropod. Nat Rev Mol Cell Biol 10:353–359. doi: 10.1038/nrm2680. [DOI] [PubMed] [Google Scholar]
- 66.Llewellyn GN, Grover JR, Olety B, Ono A. 2013. HIV-1 Gag associates with specific uropod-directed microdomains in a manner dependent on its MA highly basic region. J Virol 87:6441–6454. doi: 10.1128/JVI.00040-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Llewellyn GN, Hogue IB, Grover JR, Ono A. 2010. Nucleocapsid promotes localization of HIV-1 gag to uropods that participate in virological synapses between T cells. PLoS Pathog 6:e1001167. doi: 10.1371/journal.ppat.1001167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Agosto LM, Uchil PD, Mothes W. 2015. HIV cell-to-cell transmission: effects on pathogenesis and antiretroviral therapy. Trends Microbiol 23:289–295. doi: 10.1016/j.tim.2015.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chen BK. 2012. T cell virological synapses and HIV-1 pathogenesis. Immunol Res 54:133–139. doi: 10.1007/s12026-012-8320-8. [DOI] [PubMed] [Google Scholar]
- 70.Ono A. 2010. Relationships between plasma membrane microdomains and HIV-1 assembly. Biol Cell 102:335–350. doi: 10.1042/BC20090165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Schiffner T, Sattentau QJ, Duncan CJ. 2013. Cell-to-cell spread of HIV-1 and evasion of neutralizing antibodies. Vaccine 31:5789–5797. doi: 10.1016/j.vaccine.2013.10.020. [DOI] [PubMed] [Google Scholar]
- 72.Mothes W, Sherer NM, Jin J, Zhong P. 2010. Virus cell-to-cell transmission. J Virol 84:8360–8368. doi: 10.1128/JVI.00443-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sattentau Q. 2008. Avoiding the void: cell-to-cell spread of human viruses. Nat Rev Microbiol 6:815–826. doi: 10.1038/nrmicro1972. [DOI] [PubMed] [Google Scholar]
- 74.Shen B, Fang Y, Wu N, Gould SJ. 2011. Biogenesis of the posterior pole is mediated by the exosome/microvesicle protein-sorting pathway. J Biol Chem 286:44162–44176. doi: 10.1074/jbc.M111.274803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li F, Sewald X, Jin J, Sherer NM, Mothes W. 2014. Murine leukemia virus Gag localizes to the uropod of migrating primary lymphocytes. J Virol 88:10541–10555. doi: 10.1128/JVI.01104-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Monde K, Chukkapalli V, Ono A. 2011. Assembly and replication of HIV-1 in T cells with low levels of phosphatidylinositol-(4,5)-bisphosphate. J Virol 85:3584–3595. doi: 10.1128/JVI.02266-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Freed EO, Englund G, Martin MA. 1995. Role of the basic domain of human immunodeficiency virus type 1 matrix in macrophage infection. J Virol 69:3949–3954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Hogue IB, Hoppe A, Ono A. 2009. Quantitative fluorescence resonance energy transfer microscopy analysis of the human immunodeficiency virus type 1 Gag-Gag interaction: relative contributions of the CA and NC domains and membrane binding. J Virol 83:7322–7336. doi: 10.1128/JVI.02545-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Hogue IB, Grover JR, Soheilian F, Nagashima K, Ono A. 2011. Gag induces the coalescence of clustered lipid rafts and tetraspanin-enriched microdomains at HIV-1 assembly sites on the plasma membrane. J Virol 85:9749–9766. doi: 10.1128/JVI.00743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Grover JR, Llewellyn GN, Soheilian F, Nagashima K, Veatch SL, Ono A. 2013. Roles played by capsid-dependent induction of membrane curvature and Gag-ESCRT interactions in tetherin recruitment to HIV-1 assembly sites. J Virol 87:4650–4664. doi: 10.1128/JVI.03526-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Grover JR, Veatch SL, Ono A. 2015. Basic motifs target PSGL-1, CD43, and CD44 to plasma membrane sites where HIV-1 assembles. J Virol 89:454–467. doi: 10.1128/JVI.02178-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ono A, Freed EO. 2001. Plasma membrane rafts play a critical role in HIV-1 assembly and release. Proc Natl Acad Sci U S A 98:13925–13930. doi: 10.1073/pnas.241320298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Yee JK, Friedmann T, Burns JC. 1994. Generation of high-titer pseudotyped retroviral vectors with very broad host range. Methods Cell Biol 43:99–112. doi: 10.1016/S0091-679X(08)60600-7. [DOI] [PubMed] [Google Scholar]
- 84.Goujon C, Jarrosson-Wuilleme L, Bernaud J, Rigal D, Darlix JL, Cimarelli A. 2006. With a little help from a friend: increasing HIV transduction of monocyte-derived dendritic cells with virion-like particles of SIV(MAC). Gene Ther 13:991–994. doi: 10.1038/sj.gt.3302753. [DOI] [PubMed] [Google Scholar]
- 85.Berger G, Durand S, Goujon C, Nguyen XN, Cordeil S, Darlix JL, Cimarelli A. 2011. A simple, versatile and efficient method to genetically modify human monocyte-derived dendritic cells with HIV-1-derived lentiviral vectors. Nat Protoc 6:806–816. doi: 10.1038/nprot.2011.327. [DOI] [PubMed] [Google Scholar]
- 86.Negre D, Mangeot PE, Duisit G, Blanchard S, Vidalain PO, Leissner P, Winter AJ, Rabourdin-Combe C, Mehtali M, Moullier P, Darlix JL, Cosset FL. 2000. Characterization of novel safe lentiviral vectors derived from simian immunodeficiency virus (SIVmac251) that efficiently transduce mature human dendritic cells. Gene Ther 7:1613–1623. doi: 10.1038/sj.gt.3301292. [DOI] [PubMed] [Google Scholar]
- 87.Chan R, Uchil PD, Jin J, Shui G, Ott DE, Mothes W, Wenk MR. 2008. Retroviruses human immunodeficiency virus and murine leukemia virus are enriched in phosphoinositides. J Virol 82:11228–11238. doi: 10.1128/JVI.00981-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fernandes F, Chen K, Ehrlich LS, Jin J, Chen MH, Medina GN, Symons M, Montelaro R, Donaldson J, Tjandra N, Carter CA. 2011. Phosphoinositides direct equine infectious anemia virus Gag trafficking and release. Traffic 12:438–451. doi: 10.1111/j.1600-0854.2010.01153.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Hamard-Peron E, Juillard F, Saad JS, Roy C, Roingeard P, Summers MF, Darlix JL, Picart C, Muriaux D. 2010. Targeting of murine leukemia virus Gag to the plasma membrane is mediated by PI(4,5)P2/PS and a polybasic region in the matrix. J Virol 84:503–515. doi: 10.1128/JVI.01134-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Hrecka K, Hao C, Gierszewska M, Swanson SK, Kesik-Brodacka M, Srivastava S, Florens L, Washburn MP, Skowronski J. 2011. Vpx relieves inhibition of HIV-1 infection of macrophages mediated by the SAMHD1 protein. Nature 474:658–661. doi: 10.1038/nature10195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Laguette N, Sobhian B, Casartelli N, Ringeard M, Chable-Bessia C, Segeral E, Yatim A, Emiliani S, Schwartz O, Benkirane M. 2011. SAMHD1 is the dendritic- and myeloid-cell-specific HIV-1 restriction factor counteracted by Vpx. Nature 474:654–657. doi: 10.1038/nature10117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Goujon C, Riviere L, Jarrosson-Wuilleme L, Bernaud J, Rigal D, Darlix JL, Cimarelli A. 2007. SIVSM/HIV-2 Vpx proteins promote retroviral escape from a proteasome-dependent restriction pathway present in human dendritic cells. Retrovirology 4:2. doi: 10.1186/1742-4690-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Inlora J, Collins DR, Trubin ME, Chung JY, Ono A. 2014. Membrane binding and subcellular localization of retroviral Gag proteins are differentially regulated by MA interactions with phosphatidylinositol-(4,5)-bisphosphate and RNA. mBio 5:e02202. doi: 10.1128/mBio.02202-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Falkenburger BH, Jensen JB, Dickson EJ, Suh BC, Hille B. 2010. Phosphoinositides: lipid regulators of membrane proteins. J Physiol 588:3179–3185. doi: 10.1113/jphysiol.2010.192153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Lemmon MA. 2008. Membrane recognition by phospholipid-binding domains. Nat Rev Mol Cell Biol 9:99–111. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 96.Varnai P, Balla T. 1998. Visualization of phosphoinositides that bind pleckstrin homology domains: calcium- and agonist-induced dynamic changes and relationship to myo-[3H]inositol-labeled phosphoinositide pools. J Cell Biol 143:501–510. doi: 10.1083/jcb.143.2.501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Roy MO, Leventis R, Silvius JR. 2000. Mutational and biochemical analysis of plasma membrane targeting mediated by the farnesylated, polybasic carboxy terminus of K-ras4B. Biochemistry 39:8298–8307. doi: 10.1021/bi000512q. [DOI] [PubMed] [Google Scholar]
- 98.Yeung T, Terebiznik M, Yu L, Silvius J, Abidi WM, Philips M, Levine T, Kapus A, Grinstein S. 2006. Receptor activation alters inner surface potential during phagocytosis. Science 313:347–351. doi: 10.1126/science.1129551. [DOI] [PubMed] [Google Scholar]
- 99.von Schwedler UK, Stray KM, Garrus JE, Sundquist WI. 2003. Functional surfaces of the human immunodeficiency virus type 1 capsid protein. J Virol 77:5439–5450. doi: 10.1128/JVI.77.9.5439-5450.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Accola MA, Strack B, Gottlinger HG. 2000. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J Virol 74:5395–5402. doi: 10.1128/JVI.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Liu J, Deng Y, Zheng Q, Cheng CS, Kallenbach NR, Lu M. 2006. A parallel coiled-coil tetramer with offset helices. Biochemistry 45:15224–15231. doi: 10.1021/bi061914m. [DOI] [PubMed] [Google Scholar]
- 102.Collins DR, Lubow J, Lukic Z, Mashiba M, Collins KL. 2015. Vpr promotes macrophage-dependent HIV-1 infection of CD4+ T lymphocytes. PLoS Pathog 11:e1005054. doi: 10.1371/journal.ppat.1005054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Duncan CJ, Williams JP, Schiffner T, Gartner K, Ochsenbauer C, Kappes J, Russell RA, Frater J, Sattentau QJ. 2014. High-multiplicity HIV-1 infection and neutralizing antibody evasion mediated by the macrophage-T cell virological synapse. J Virol 88:2025–2034. doi: 10.1128/JVI.03245-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Graziano F, Desdouits M, Garzetti L, Podini P, Alfano M, Rubartelli A, Furlan R, Benaroch P, Poli G. 2015. Extracellular ATP induces the rapid release of HIV-1 from virus containing compartments of human macrophages. Proc Natl Acad Sci U S A 112:E3265–E3273. doi: 10.1073/pnas.1500656112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Krementsov DN, Rassam P, Margeat E, Roy NH, Schneider-Schaulies J, Milhiet PE, Thali M. 2010. HIV-1 assembly differentially alters dynamics and partitioning of tetraspanins and raft components. Traffic 11:1401–1414. doi: 10.1111/j.1600-0854.2010.01111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chu H, Wang JJ, Qi M, Yoon JJ, Chen X, Wen X, Hammonds J, Ding L, Spearman P. 2012. Tetherin/BST-2 is essential for the formation of the intracellular virus-containing compartment in HIV-infected macrophages. Cell Host Microbe 12:360–372. doi: 10.1016/j.chom.2012.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Giese S, Marsh M. 2014. Tetherin can restrict cell-free and cell-cell transmission of HIV from primary macrophages to T cells. PLoS Pathog 10:e1004189. doi: 10.1371/journal.ppat.1004189. [DOI] [PMC free article] [PubMed] [Google Scholar]