ABSTRACT
The anterograde pathway, from the endoplasmic reticulum through the trans-Golgi network to the cell surface, is utilized by trans-membrane and secretory proteins. The retrograde pathway, which directs traffic in the opposite direction, is used following endocytosis of exogenous molecules and recycling of membrane proteins. Microbes exploit both routes: viruses typically use the anterograde pathway for envelope formation prior to exiting the cell, whereas ricin and Shiga-like toxins and some nonenveloped viruses use the retrograde pathway for cell entry. Mining a human genome-wide RNA interference (RNAi) screen revealed a need for multiple retrograde pathway components for cell-to-cell spread of vaccinia virus. We confirmed and extended these results while discovering that retrograde trafficking was required for virus egress rather than entry. Retro-2, a specific retrograde trafficking inhibitor of protein toxins, potently prevented spread of vaccinia virus as well as monkeypox virus, a human pathogen. Electron and confocal microscopy studies revealed that Retro-2 prevented wrapping of virions with an additional double-membrane envelope that enables microtubular transport, exocytosis, and actin polymerization. The viral B5 and F13 protein components of this membrane, which are required for wrapping, normally colocalize in the trans-Golgi network. However, only B5 traffics through the secretory pathway, suggesting that F13 uses another route to the trans-Golgi network. The retrograde route was demonstrated by finding that F13 was largely confined to early endosomes and failed to colocalize with B5 in the presence of Retro-2. Thus, vaccinia virus makes novel use of the retrograde transport system for formation of the viral wrapping membrane.
IMPORTANCE Efficient cell-to-cell spread of vaccinia virus and other orthopoxviruses depends on the wrapping of infectious particles with a double membrane that enables microtubular transport, exocytosis, and actin polymerization. Interference with wrapping or subsequent steps results in severe attenuation of the virus. Some previous studies had suggested that the wrapping membrane arises from the trans-Golgi network, whereas others suggested an origin from early endosomes. Some nonenveloped viruses use retrograde trafficking for entry into the cell. In contrast, we provided evidence that retrograde transport from early endosomes to the trans-Golgi network is required for the membrane-wrapping step in morphogenesis of vaccinia virus and egress from the cell. The potent in vitro inhibition of this step by the drug Retro-2 suggests that derivatives with enhanced pharmacological properties might serve as useful antipoxviral agents.
INTRODUCTION
Vaccinia virus (VACV) is the prototype member of the poxvirus family of cytoplasmic double-stranded DNA viruses (1). The first infectious form of VACV, the intracellular mature virion (MV), is a brick-shaped particle consisting of a nucleoprotein core, lateral bodies, and a surrounding lipoprotein membrane (2). A subset of cytoplasmic MVs are wrapped in an additional double membrane and transported on microtubules to the periphery of the cell where the outermost membrane fuses with the plasma membrane, resulting in exocytosis and induction of actin tail formation, which enhances virus spread (3). Thus, MVs have one outer membrane, intracellular wrapped virions (WVs) have three, and extracellular virions (EVs) have two. Interference with wrapping or subsequent steps results in decreased virus cell-to-cell spread and severe attenuation in an animal model (4, 5).
The MV membrane appears to be derived from the endoplasmic reticulum (ER) and is essential for the earliest steps in morphogenesis (6). The presence of viral proteins in the trans-Golgi network (TGN) led to the suggestion that this organelle serves as the precursor of the wrapping membrane (7, 8), whereas the presence of fluid-phase markers in the lumen of the double WV membrane suggested an origin from tubular endosomes (9). However, enhanced transport of fluid-phase markers to the TGN during VACV infection complicates these interpretations (8, 9).
Five integral membrane glycoproteins (A33, A34, A36, A56, and B5) and the palmitylated F13 protein are viral protein components of the wrapping membrane (10, 11). Of these, F13 and B5 are critical for wrapping to occur (12–14). B5 is a glycosylated type 1 integral membrane protein (15), whereas F13 is an unglycosylated protein that is dependent on palmitoylation for membrane association (7, 16, 17). Nevertheless, both B5 and F13 localize to the TGN (7, 8, 18). However, the finding that a dominant negative form of the Sar1 GTPase, which inhibits protein transport from the ER, prevents B5 but not F13 from reaching the Golgi apparatus suggests that the two proteins use different transport mechanisms (19).
There are two recognized major transport pathways to the TGN. Transmembrane and secretory proteins are transported through the biosynthetic anterograde pathway from the ER through the Golgi apparatus to the cell surface. Viral transmembrane proteins destined to form the envelope typically follow the anterograde path. Recycling transmembrane proteins and extracellular molecules that enter cells by endocytosis move in the opposite or retrograde direction from endosomes to the trans-Golgi network. Bacterial (Shiga and cholera) and plant (ricin and abrin) toxins (20) as well as some nonenveloped viruses, including simian virus 40 (SV40), polyomavirus, adeno-associated virus, and papillomavirus (21–24), exploit the retrograde path to enter cells. Retrograde transport is highly selective and depends on numerous tethering factors, small GTPases, and SNARES (25, 26). The small molecule Retro-2 interferes with retrograde transport from early endosome to the TGN specifically and protects against ricin and Shiga-like toxins (27) and virus entry (22–24), while having little or no apparent cellular toxicity.
Our particular interest in the retrograde pathway arose from a genome-wide small interfering RNA (siRNA) screen in which depletion of some retrograde transport factors significantly decreased spread of VACV (28). In principal, a block at any step from virus entry to release could affect the spread assay. Further analysis, described here, confirmed the role of retrograde transport proteins in VACV spread and revealed that this pathway is not required for VACV entry or assembly of infectious virions but instead for membrane wrapping and dissemination of virus particles. More specifically, retrograde transport from early endosomes to the TGN was required for the proper localization of the F13 protein in the same compartment as B5.
MATERIALS AND METHODS
Cells, viruses, and reagents.
BS-C-1 (ATCC CCL-26) and HeLa (ATCC CCL-2) cells were grown in minimum essential medium with Earle's salt and Dulbecco minimum essential medium (DMEM), respectively, supplemented with 10% fetal bovine serum, 100 U of penicillin, and 100 μg of streptomycin per ml (Quality Biologicals, Gaithersburg, MD). Experiments with monkeypox virus were carried out in a biosafety level 3 (BSL3) facility approved for select agents. Recombinant VACVs expressing either firefly luciferase, F13-green fluorescent protein (F13-GFP), or GFP-A4 were previously described (29–31). Plaque assays were performed in BS-C-1 cell monolayers in 12-well culture plates with 10-fold serial dilutions of virus. The virus was adsorbed to cells for 1 h at room temperature, unbound virus was removed, and the cells were washed and incubated with medium containing 0.5% methylcellulose. Retro-2 (Calbiochem, Billerica, MA) was prepared as a 60 mM solution in dimethyl sulfoxide (DMSO) and diluted in warm DMEM. Plasmids expressing the fluorescent fusion proteins mApple-TGNP, mApple-Rab5a, and mCherry-Rab7a were gifts from Michael Davidson and obtained from Addgene (catalog no. 54954, 54944, and 55127). Rabbit antibody to VACV strain WR and rat 192C monoclonal antibody (MAb) to B5 were described previously (8, 15, 32). The mouse MAb to the hemagglutinin (HA) epitope tag (catalog no. MMS 101P) was purchased from Covance, now part of BioLegend (San Diego, CA).
siRNA transfections, early gene expression, and spread assay.
Individual siRNAs (Silencer Select; ThermoFisher, Waltham, MA) were transfected with Lipofectamine 2000 (ThermoFisher) according to manufacturer protocols into triplicate cultures of HeLa cells at 48 or 72 h prior to infection. For the entry assay, cells were infected with VACV IHDJvFire at a multiplicity of 5 PFU/cell for 1 h at 4°C. The medium was replaced, and the cells were incubated at 37°C for 90 min; firefly luciferase activity was measured using luciferase assay substrate (Promega, Madison, WI). To measure virus spread, HeLa cells in 24-well plates were infected with recombinant virus expressing GFP (29) at a multiplicity of 0.01 PFU/cell for 1 h at 37°C. The cells were washed, incubated with medium for 18 h at 37°C, trypsin treated, and fixed with 2% paraformaldehyde. Cells expressing GFP were detected by flow cytometry and analyzed with FlowJo software (FlowJo, Ashland, OR).
Confocal microscopy.
HeLa cells grown on coverslips were infected for specified times, fixed with 4% paraformaldehyde, and permeabilized with 0.1% Triton X-100 in phosphate-buffered saline (PBS). The samples were blocked with 5% bovine serum albumin and 10% fetal bovine serum, followed by incubation with primary antibodies in serum for 2 h. Cells were washed with PBS and incubated with appropriate secondary antibodies conjugated to fluorescent dyes (Molecular Probes, Eugene, OR) for an additional 1 h with DAPI (4′,6′-diamidino-2-phenylindole). Coverslips were washed and mounted on a glass slide using Prolong Gold (Invitrogen). The slides were examined with a Leica SP5 inverted four-channel confocal microscope. Images were prepared using Imaris version 8.2 software (Bitplane Scientific Software, St. Paul, MN). Colocalization was determined using IMARIS on individual focal planes prior to z-stack superimposition. Pearson's coefficients were determined using the IMARIS colocalization module, in which the threshold was determined manually for each channel and the software calculated the linear relationship between the two signals. Statistical significance was determined by analyzing cells in several random fields.
Egress assay.
HeLa cells were infected for 18 h with VACV/GFP-A4, washed, and incubated with antibody to B5 for 1 h at 37°C. The cells were washed 3 times in cold PBS, fixed with 4% paraformaldehyde, and prepared for confocal microscopy. Colocalization calculations were done using IMARIS on several random fields of cells.
Western blot analysis.
Proteins of whole-cell lysates were separated in 4 to 12% Novex NuPAGE acrylamide gels with 2-(N-morpholino)ethanesulfonic acid buffer and transferred to nitrocellulose membranes using the iBlot system (ThermoFisher). The membrane was blocked with 5% nonfat milk in PBS plus 0.05% Tween 20 and then incubated for 1 h at room temperature or overnight at 4°C in the same solution with primary antibodies at appropriate dilutions. Excess antibodies were removed by washing with PBS plus Tween 20. IRDye 800-conjugated secondary antibodies against mouse and rabbit IgG were added, and the mixture was incubated for 1 h at room temperature, washed, and developed using an Odyssey infrared imager (Li-Cor Biosciences, Lincoln, NE).
Palmitoylation assay.
HeLa cells were infected with 3 PFU/cell of VACV expressing F13-GFP for 1 h at 37°C, and 100 μM Click-It palmitic acid-azide (ThermoFisher) was added. At 8 h after infection, the cells were washed, lysed with radioimmunoprecipitation assay (RIPA) buffer, and immunoprecipitated using antibody to GFP (Sigma-Aldrich, St. Louis, MO) or IgG control attached to protein G-Dynabeads (ThermoFisher). After washings, the Click-It reaction was carried out directly on the beads using the Click-It protein reaction buffer kit (ThermoFisher) to attach biotin-alkyne. In the control reaction, copper was omitted. Proteins were eluted in sample buffer, heated at 80°C, resolved by SDS-PAGE, transferred to nitrocellulose blots, and probed with IRDye 800-conjugated streptavidin.
For confocal microscopy, cells grown on coverslips were infected, incubated with palmitic acid-azide, washed and fixed with 4% paraformaldehyde for 20 min, and permeabilized with 0.1% Triton X-100 for 10 min. Alkyne-Alexa Fluor 594 was attached to palmitylated proteins in Click-It cell reaction buffer (ThermoFisher), and samples were processed for imaging.
Transmission electron microscopy.
Infected HeLa cells in 60-mm plates were fixed with 2% glutaraldehyde in 0.2 M sodium cacodylate buffer, postfixed in reduced osmium tetroxide, dehydrated in a series of ethanol incubations with final dehydrations in propylene oxide, and embedded in EmBed-812 resin (Electron Microscopy Sciences, Hatfield, PA) (33).
Procedures for cryosectioning and immunogold labeling were described previously (34). Cryosections were picked up on grids, thawed, washed free of sucrose, and stained with a mouse MAb to the HA epitope fused to F13 and/or rat MAb 19C2 to B5, followed by rabbit anti-mouse IgG and rabbit anti-rat IgG+IgM, respectively, and then protein A conjugated to 10-nm gold spheres (University Medical Center, Utrecht, Netherlands). Specimens were viewed with an FEI Tecnai Spirit transmission electron microscope (FEI, Hillsboro, OR). Double labeling was carried out using 5- and 10-nm gold as described previously (35).
Statistical analysis.
P values were calculated using the Student t test or one-way analysis of variance (ANOVA) followed by the Bonferroni posttest to correct for multiple assays using Prism from GraphPad.
RESULTS
Human genome-wide siRNA screen reveals retrograde transport proteins as host factors that increase VACV spread.
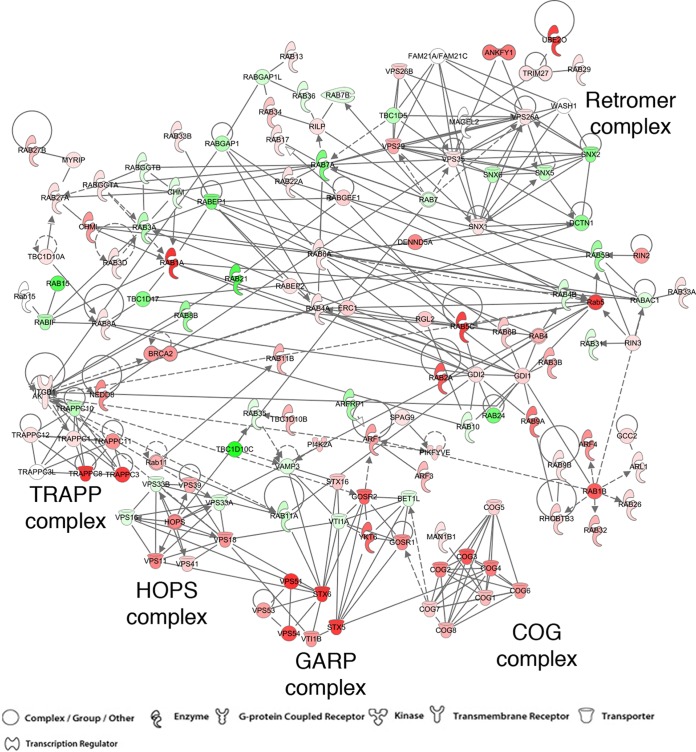
A previous comprehensive screen identified siRNAs to over 500 candidate human genes of more than 20,000 interrogated that inhibited the spread of VACV (28). In order to prioritize the hits, we looked for their enrichment in functional groups. The largest number of hits was for genes involved in mRNA translation, consistent with the absence of such genes in VACV. There was also enrichment of genes for the proteasome/ubiquitin and the ER-to-Golgi transport pathways, which were known to be important for VACV uncoating and genome replication (36–38) and for formation of EVs (19), respectively. Upon further mining of the hits, we noted many candidate genes encoding factors for retrograde transport from endosomes to the TGN; a sample of those are listed in Table 1. We prepared an interaction network by retrieving 158 genes annotated in the endosome-to-Golgi retrograde traffic category (GO:0042147) and adding human Rab membrane GTPase genes and additional literature-curated genes designated to function in the retrograde pathway. Using the Qiagen Ingenuity Pathway Analysis protein-protein interaction database (IPA; QIAGEN, Redwood City, CA), we made an interaction map from this list and overlaid the hits from our siRNA screen results (Fig. 1). VACV spread was strongly inhibited by depletion of representative components of the GARP complex, comprised of vacuolar sorting protein 52 (VPS52), VPS53, and VPS54, GARP's main binding partner syntaxin 6 (STX6), and vesicle-associated membrane protein 4 (VAMP4) as well as the small G protein ADP ribosylation factor 5A (Arl5A), which aids in the recruitment of GARP to the Golgi network (39–41). Depletion of retrograde complex protein components of the COG complex (42, 43), the HOPS complex (44), and the TRAPP complex (44) also reduced virus spread. In contrast, depletion of protein components of the Retromer complex (45) inhibited VACV spread to a lesser extent, suggesting that a specific set of transport factors are needed. Recently, the EARP complex was described, in which the VPS54 component of GARP is replaced by the coil-coil domain-containing 132 protein (CCDC132, also called syndetin) (26). Functionally, EARP is required for endosome recycling back to the membrane, while GARP is needed for endosome-to-Golgi trafficking. Depletion of syndetin had no effect on VACV spread, further suggesting that a specific retrograde endosome-to-Golgi trafficking system is needed for VACV.
TABLE 1.
Retrograde transport factor hits from a human genome-wide RNAi screen
| Gene name | Entrez GeneID | Z-scorea |
|||||
|---|---|---|---|---|---|---|---|
| SS.1 | SS.2 | SS.3 | SG | OTP | Median for all five reagents | ||
| STX6 | 10228 | −2.25 | −2.16 | −2.61 | −0.77 | −0.26 | −2.16 |
| RAB39B | 116442 | −2.15 | 0.45 | −1.93 | −0.68 | −2.77 | −1.93 |
| RAB1A | 5861 | −1.42 | −1.72 | −1.32 | −1.97 | −1.63 | −1.63 |
| VPS51 | 738 | −0.54 | −1.74 | 0.04 | −1.70 | −1.54 | −1.54 |
| STX5 | 6811 | −1.85 | −2.26 | −0.67 | −1.47 | −0.83 | −1.47 |
| Rab5C | 5878 | −0.27 | −2.33 | −1.55 | −1.42 | 0.43 | −1.42 |
| RAB5C | 5878 | −0.27 | −2.33 | −1.55 | −1.42 | 0.43 | −1.42 |
| UBE2O | 63893 | −1.42 | 1.74 | −1.70 | −1.50 | 0.11 | −1.42 |
| VPS54 | 51542 | −1.10 | −1.34 | −1.68 | −1.72 | 0.51 | −1.34 |
| VAMP4 | 8674 | −1.30 | 0.27 | −1.11 | −1.30 | −1.37 | −1.30 |
| COG3 | 83548 | −0.40 | −1.74 | −0.98 | −1.26 | −1.44 | −1.26 |
| RAB2A | 5862 | −2.01 | −1.24 | −1.09 | −1.62 | −0.49 | −1.24 |
| RAB1B | 81876 | −1.25 | −0.52 | −1.22 | −1.27 | −1.08 | −1.22 |
| GOSR2 | 9570 | −1.17 | −0.12 | −0.14 | −1.96 | −2.29 | −1.17 |
| YKT6 | 10652 | 0.06 | −1.14 | −1.47 | −1.18 | 1.81 | −1.14 |
| SNAP29 | 9342 | 0.14 | −2.21 | −1.34 | −1.13 | 1.40 | −1.13 |
| ANKFY1 | 51479 | −0.12 | 1.08 | −2.28 | −1.02 | −1.50 | −1.02 |
| RAB9A | 9367 | −1.02 | −1.47 | −0.01 | −1.99 | 0.10 | −1.02 |
| RAB38 | 23682 | −1.34 | −0.33 | −2.47 | 0.50 | −1.02 | −1.02 |
SS.1, SS.2, and SS.3 are individual siRNAs from the Ambion Silencer Select version 4 human genome library. SG is four pooled human siGenome siRNAs from Dharmacon. OTP is four pooled siRNAs from the Dharmacon On-Targetplus human genome library.
FIG 1.
Interaction network of retrograde transport factors overlaid with siRNA hits. The 158 genes annotated in the endosome-to-Golgi retrograde traffic category (GO:0042147) plus human Rab membrane GTPase genes and additional literature-curated genes designated to function in the retrograde pathway were compiled. Using the Ingenuity Pathway Analysis protein-protein interaction database, we made an interaction map from this list and overlaid the median Z-score of 5 siRNAs for each gene from our siRNA screen results (28). Decrease of virus spread is depicted in shades of red from dark to light, while increase is shown in green colors. Uncolored circles indicate spread that was unchanged or not evaluated. Continuous arrows connect proteins that have been shown to interact, dashed arrows connect proteins for which interactions have not been shown to be direct, and circular lines indicate self-associations. The symbols are defined at the bottom of the figure.
Depletion of retrograde factors decreases virus spread but not entry or formation of infectious virus.
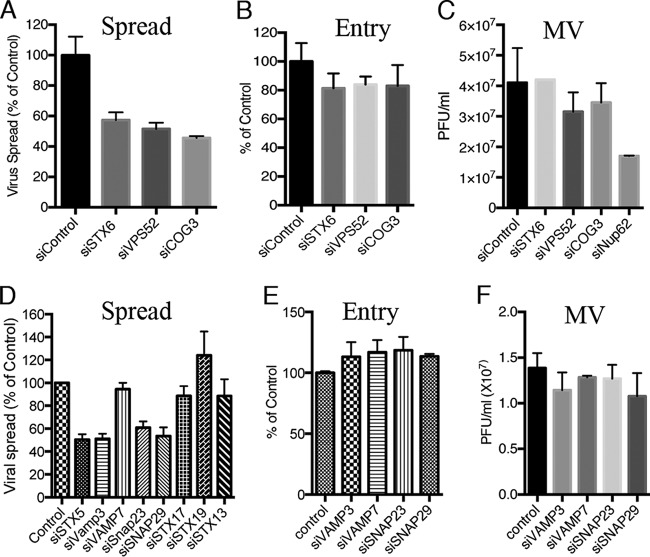
The enrichment of multiple components of the retrograde trafficking pathway in the high-throughput genome-wide screen strongly suggested that they were biologically significant. For confirmation, the effects of individual siRNAs to several high-priority genes from the screen and others involved in retrograde transfer that were not tested or did not make the list for various reasons were analyzed using flow cytometry to measure spread of a recombinant VACV expressing GFP. The siRNAs targeting STX6, the GARP component VPS52, and COG3 each reduced VACV spread by about 50% compared to a control siRNA (Fig. 2A), thereby confirming and extending results of the original screen. Incomplete depletion by siRNA, due to the stability of the proteins, can account for a consistent inhibition of approximately 50%. For example, Western blotting showed that about 20% of STX5 remained after knockdown (data not shown). Partial redundancy of transport functions could also contribute to incomplete inhibition of spread.
FIG 2.
Depletion of retrograde transport factors reduces virus spread but not entry or formation of infectious virus. (A) HeLa cells were transfected with the indicated siRNA for 48 h, followed by infection with 0.01 PFU/cell of VACV strain IHDJ expressing the A4 core protein with GFP fused to its N terminus for 18 h. GFP-positive cells were scored by flow cytometry, and results are presented as percentage of control siRNA values. (B) HeLa cells were transfected with the indicated siRNA for 48 h. Cells were infected with 5 PFU/cell of VACV IHDJ expressing firefly luciferase for 1 h at 4°C. After attachment, cells were incubated at 37°C for 90 min and luciferase activity determined. The latter was plotted as percentage of that obtained with control siRNA. (C) HeLa cells were transfected for 48 h with the indicated siRNA, followed by infection with 3 PFU/cell of VACV. After 24 h, the cells were lysed and virus titers determined by plaque assay on BS-C-1 cells. (D) The indicated siRNAs were transfected into HeLa cells for 72 h, and virus spread was determined as for panel A. (E) The indicated siRNAs were transfected into HeLa cells for 72 h, and entry and early gene expression were determined as for panel B. (F) HeLa cells were transfected for 72 h with the indicated siRNA, and virus titers were determined as for panel C. Infections were carried out in triplicate, and error bars indicate standard deviations.
Based on studies with nonenveloped viruses (21–24), we considered that VACV might use the retrograde pathway for cell entry. A convenient VACV entry assay is based on the fact that infectious poxvirus particles contain RNA polymerase and transcription factors that enable early mRNAs to be synthesized and then translated immediately after the core enters the cytoplasm. The expression of firefly luciferase regulated by an early promoter provides a sensitive and quantitative measure of entry of a recombinant VACV (46). Following synchronous infection, the levels of luciferase detected in cells treated with siRNAs that targeted STX6, VPS52, or COG3 were only modestly reduced relative to the control (Fig. 2B), in comparison to the greater inhibition of virus spread (Fig. 2A). Furthermore, siRNAs to these retrograde transport factors had much less effect on the production of cell-associated infectious virus during a 24-h single-step replication cycle than did siRNA to Nup62 (Fig. 2C), which was previously shown to inhibit virion formation (28).
Further studies were carried out to extend the above results by depleting mRNAs from additional genes, particularly those encoding SNARE proteins that mediate vesicle fusion and which may require longer times for depletion (47) than the 48 h used in the high-throughput genome-wide screen. After a 72-h preincubation, the siRNAs to vesicle-associated membrane protein 7 (VAMP7), STX13, STX17, and STX19 had no effect on virus spread, whereas those to STX5, VAMP3, synaptosome-associated protein 23 (SNAP23), and SNAP29 reduced spread by about 50% (Fig. 2D). The latter siRNAs did not reduce virus entry (Fig. 2E) or infectious virus formation (Fig. 2F). Whether the failure of siRNAs to VAMP7, STX13, STX17, and STX19 to inhibit spread was due to the redundancy of factors or inadequate depletion of the proteins was not determined. The compelling result was that inhibition of virus entry or replication could not adequately explain the reduced virus spread resulting from depletion of STX5, STX6, VPS52, COG3, VAMP3, SNAP23, and SNAP29.
Depletion of retrograde transport factors inhibits formation of extracellular virus particles.
Since the siRNAs that reduced VACV spread had only a small effect on virus entry and formation of infectious VACV, we considered an effect on virus dissemination. MVs are assembled in juxta-nuclear cytoplasmic factories, and a subset is wrapped in additional membranes and transported along microtubules to the plasma membrane where exocytosis occurs (48–52). These EVs are essentially mature virions with an additional outer membrane that are released from the infected cells (10, 53, 54, 55). However, the majority of the EVs remain attached to the outside of the plasma membrane, enabling direct cell-to-cell spread that is facilitated by actin polymerization (4, 55, 56). To ascertain whether inhibition of retrograde transport interfered with EV formation, cells were treated with a control siRNA or siRNA to STX6 or VPS52 and then infected for 18 h with a recombinant VACV in which the A4 core protein was fused to GFP, allowing detection of unwrapped and wrapped virions as previously described (57). Live unpermeabilized cells were incubated with antibody that recognizes the extracellular domain of the B5 protein component of the wrapping membrane. After removing excess antibody and fixing, confocal microcopy was used to identify extracellular virus particles by colocalization of A4-GFP and B5. In the control, numerous EVs were detected at the plasma membrane (Fig. 3A). When STX6 was depleted, transport of B5 to the plasma membrane still occurred, but almost all of the A4-GFP virions remained in a juxta-nuclear location (Fig. 3A). A similar result was obtained with siRNA to VPS52 (not shown). The extent of colocalization of A4-GFP and B5 was determined by calculation of Pearson coefficients, which confirmed the significance of the reduced colocalization resulting from depletion of STX6 or VPS52 (Fig. 3B).
FIG 3.
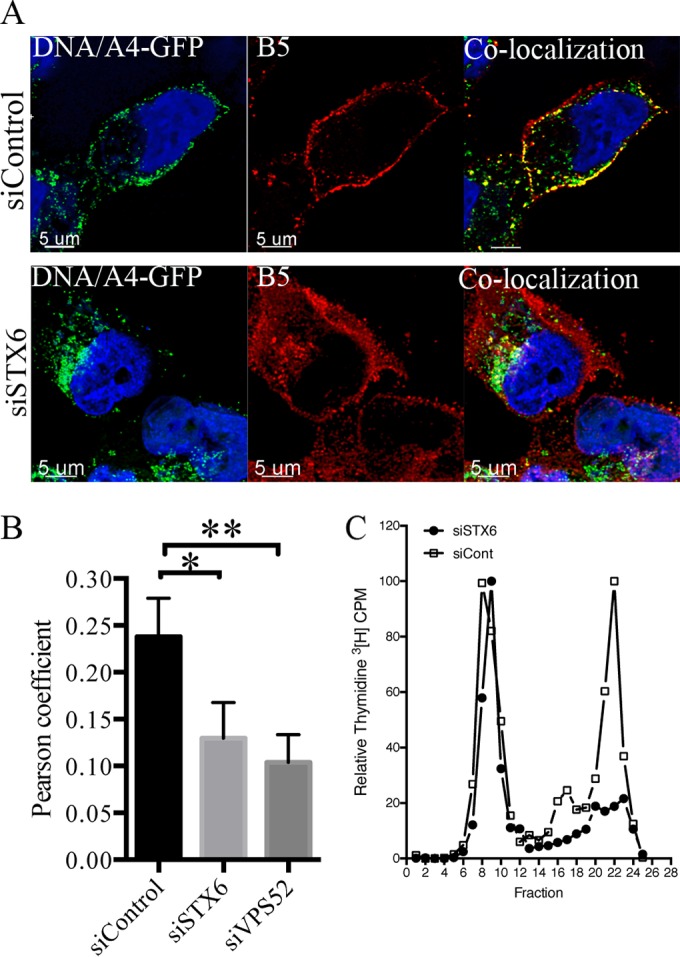
Depletion of retrograde transport factors inhibits formation of extracellular virus particles. (A) HeLa cells on coverslips were transfected with control siRNA (upper panel) or siSTX6 (lower panel) for 48 h and then infected for 18 h with 3 PFU/cell of VACV expressing the fluorescent GFP-A4 core protein. Live staining of surface B5 was performed at 37°C for 1 h by adding rat MAb specific for the external domain of B5 to the medium. Cells were washed, fixed, and processed for confocal imaging. Colocalization (yellow) of the EV marker B5 (red) and the core protein GFP-A4 (green) was calculated using IMARIS software on individual confocal planes before making the z-stack shown. (B) Pearson coefficients determined from analysis of several hundred cells in several fields. Error bars indicate standard deviations. P values of less than 0.01 and 0.05 are indicated by two or one asterisk, respectively. (C) HeLa cells were transfected with control siRNA or siRNA to STX6 for 48 h. Cells were then infected with 3 PFU/cell of VACV, labeled with [3H]thymidine, and harvested after 24 h. Cell lysates were clarified by low-speed centrifugation and analyzed by CsCl2 sedimentation as previously described (4). Fractions were collected from the bottom, and the radioactivity was determined by scintillation counting.
The absence of EVs could result from either a failure of wrapping, movement of wrapped virions to the periphery of the cell, or exocytosis. In the following experiment, we provide evidence for a defect in wrapping. The additional membranes of WVs and EVs decrease the buoyant density of virus particles so that they are separable from MVs by CsCl2 gradient sedimentation of infected-cell lysates (4). In the experiment depicted in Fig. 3C, the DNA of virus particles was labeled with [3H]thymidine during infection in cells that had been treated with a control siRNA or siRNA to STX6. The high-density peak of MVs appeared to be similar regardless of siRNA treatment, whereas the low-density peak was greatly reduced when STX6 was depleted. The confocal microscopy and sedimentation studies pointed to a role of retrograde transport factors in the transition of MVs to WVs, which would account for the reduction in virus spread.
Inhibition of VACV spread by the small molecule Retro-2.
A deeper understanding of the role of retrograde trafficking in the spread of VACV came from studies with the potent and specific inhibitor Retro-2. Retro-2 is a cell-permeative tricyclic compound that was discovered in a high-throughput screen of small molecules that could protect cells from Shiga-like toxins and ricin, which depend on retrograde transport for release into the cytoplasm (27). The activity of Retro-2 involves relocalization of STX5 and STX6 from the TGN, although the direct cellular target of Retro-2 remains to be discovered. Our finding that depletion of STX5 or STX6, as well as other retrograde trafficking factors, inhibited VACV spread suggested to us that Retro-2 might have a similar effect. Indeed, Retro-2 almost completely inhibited spread of VACV as well as the closely related monkeypox virus between concentrations of 25 and 50 μM (Fig. 4A), similar to that found for toxins (27). Like the siRNAs to retrograde transport factors, Retro-2 had little effect on entry and early gene expression as measured by luciferase activity whether administered before or shortly after infection (Fig. 4B). Moreover, Retro-2 had no discernible effect on synthesis of the major viral proteins as determined by Western blotting using VACV antiserum (Fig. 4C). During infection with VACV, progeny virions remained mostly cell associated, with a minority dissociated into the medium as shown for the DMSO control in Fig. 4D. Retro-2 diminished the amount of released virus without affecting the yield of cell-associated virus, consistent with specific inhibition of virus dissemination (Fig. 4D).
FIG 4.
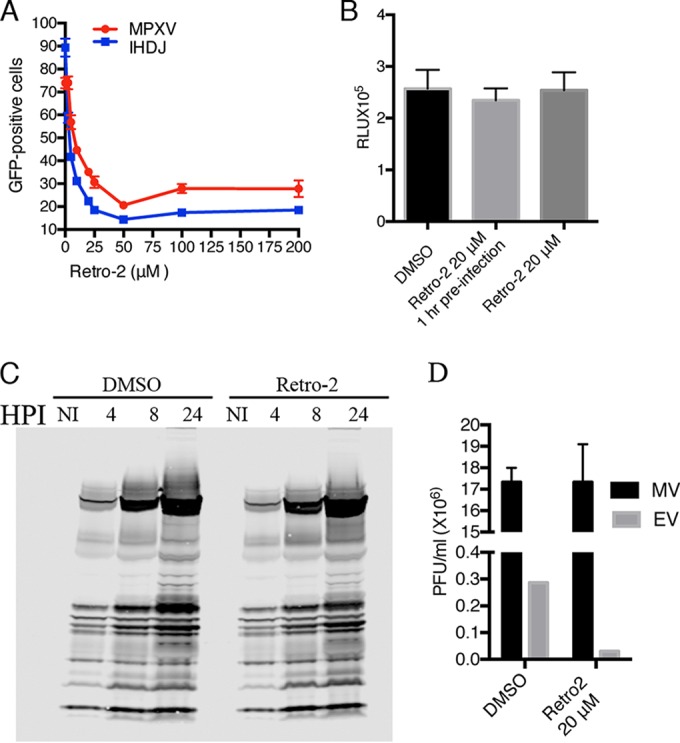
Inhibition of VACV spread by Retro-2. (A) HeLa cells were infected with 0.01 PFU/cell of VACV strain IHDJ or monkeypox virus (MPXV) expressing GFP-A4 for 1 h at 4°C. Cells were washed and incubated at 37°C with serial dilutions of Retro-2 or DMSO. After 18 h, GFP-positive cells were determined by flow cytometry and analyzed with FlowJo software. The percentage of GFP-positive cells relative to a no-Retro-2 control from four separate experiments is shown. Error bars indicate standard deviations. (B) HeLa cells were infected in triplicate with 5 PFU/cell of VACV IHDJ expressing firefly luciferase at 4°C for 1 h and transferred to a 37°C incubator for 90 min. Cells were lysed, and firefly luciferase activity was determined. Relative light units (RLU) are plotted. Error bars indicate standard deviations. (C) HeLa cells were infected with 3 PFU/cell VACV IHDJ expressing firefly luciferase for 1 h at 4°C, and the medium was replaced with fresh medium at 37°C with DMSO or 20 μM Retro-2. Cells were harvested at the indicated times and lysed, and proteins were resolved by electrophoresis in a 4 to 12% SDS-polyacrylamide gel, transferred to a nitrocellulose membrane, and probed with antibody specific for VACV proteins. HPI, hours postinfection; NI, not infected. (D) Cells were infected with 3 PFU/cell of VACV IHDJ expressing firefly luciferase in the presence of Retro-2 or DMSO carrier. After 24 h, cells and medium were collected separately and the virus titers determined by plaque assay. Infections were carried out in triplicate and standard deviations calculated.
Retro-2 prevents wrapping of MVs.
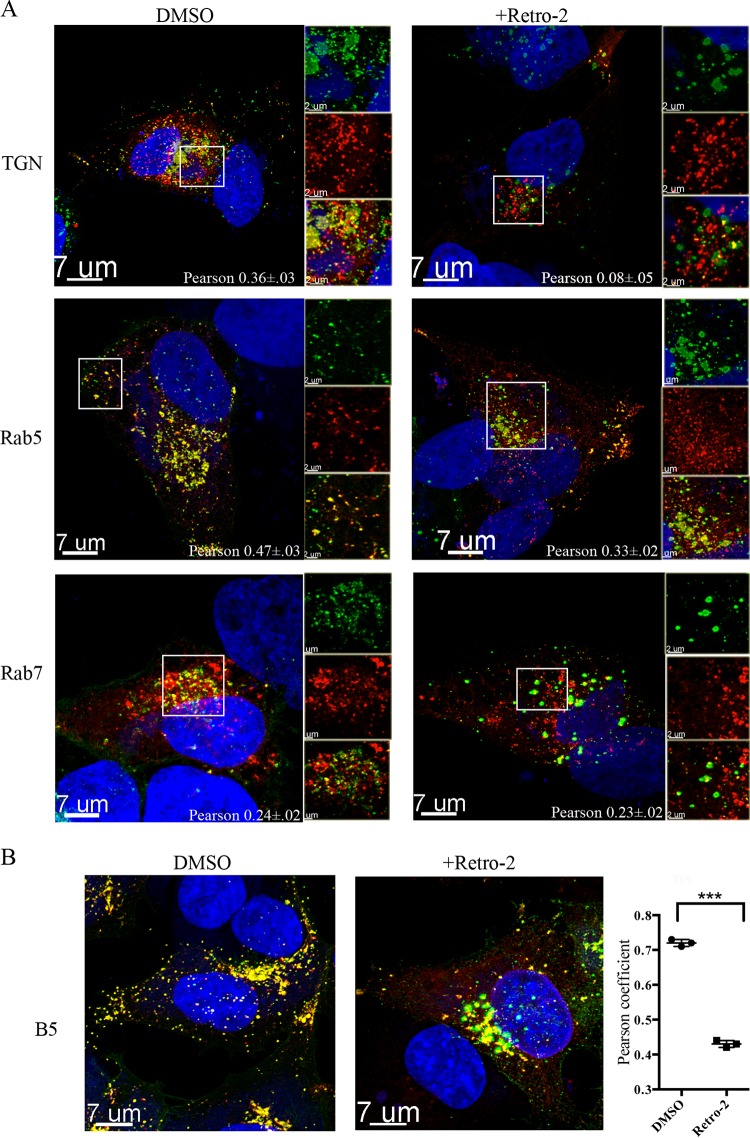
Since we had shown that depletion of STX6 led to the retention of virions in a juxta-nuclear location, we investigated whether Retro-2 had a similar effect. As a control, cells were infected in the absence of drug with recombinant VACV encoding the A4 core protein fused to GFP; at 4 and 8 h, the cells were fixed, permeabilized, and stained with MAb to the VACV B5 membrane protein. Because the cells were permeabilized, the B5 MAb interacted with both intra- and extracellular B5. During a normal VACV infection, the virus structural proteins begin to be synthesized in virus factories at 4 h, and virions are formed by 8 h. At 4 h, the GFP-A4 core protein was mainly in DNA factories adjacent to the nucleus, whereas B5 was associated with the Golgi apparatus, with little colocalization of the two viral proteins (Fig. 5). By 8 h after infection, virions had formed, and the detection of puncti that contained GFP-A4 and B5 suggested that wrapping had occurred (Fig. 5). The difference between the colocalization of virus cores with B5 at 4 and 8 h was highly significant (Fig. 5). When Retro-2 was added, the virus particles remained in the interior of the cell, whereas the B5 protein was present near the periphery (Fig. 5). ST-246, which prevents the wrapping of mature virions by targeting the F13 protein (58), also blocked the colocalization of virions with B5 (Fig. 5). The extent of colocalization of the virus cores with B5 at 8 h in the absence of drug was significantly different from that occurring in the presence of either Retro-2 or ST246 (Fig. 5).
FIG 5.
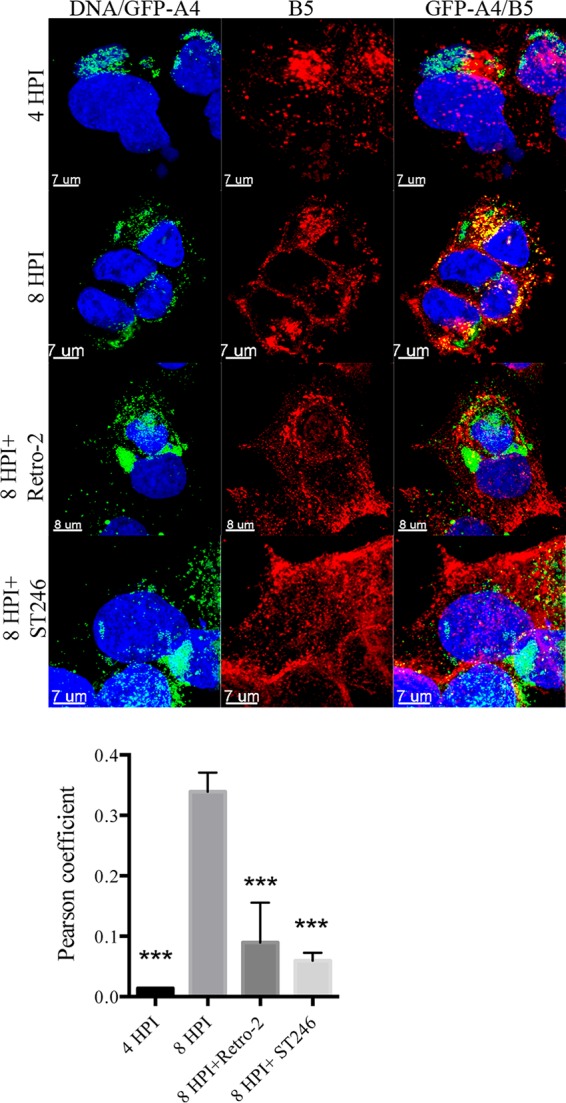
Retro-2 prevents colocalization of virions with B5 membrane protein. VACV/GFP-A4 (3 PFU/cell) was allowed to attach to HeLa cells on coverslips for 1 h, after which incubations were carried out for the indicated hours postinfection (HPI) in the presence of DMSO, 20 μM Retro-2, or 10 μM ST-246. Cells were fixed, stained with antibody specific for B5 and with DAPI to visualize DNA, and prepared for confocal microscopy. Images are superimpositions of z-stacks. The middle column shows localization of B5, the left column shows a merge of DAPI and GFP, and the right column shows a merge of DAPI, B5, and GFP. Colocalization (yellow) of B5 (red) and the core protein GFP-A4 (green) was calculated using IMARIS software on individual confocal planes before making the z-stack overlay shown. Colocalization of the B5 and GFP signals from cells in several randomly selected fields was evaluated by determining Pearson coefficients, as plotted below the images. The error bars indicate standard deviations. ***, P < 0.001.
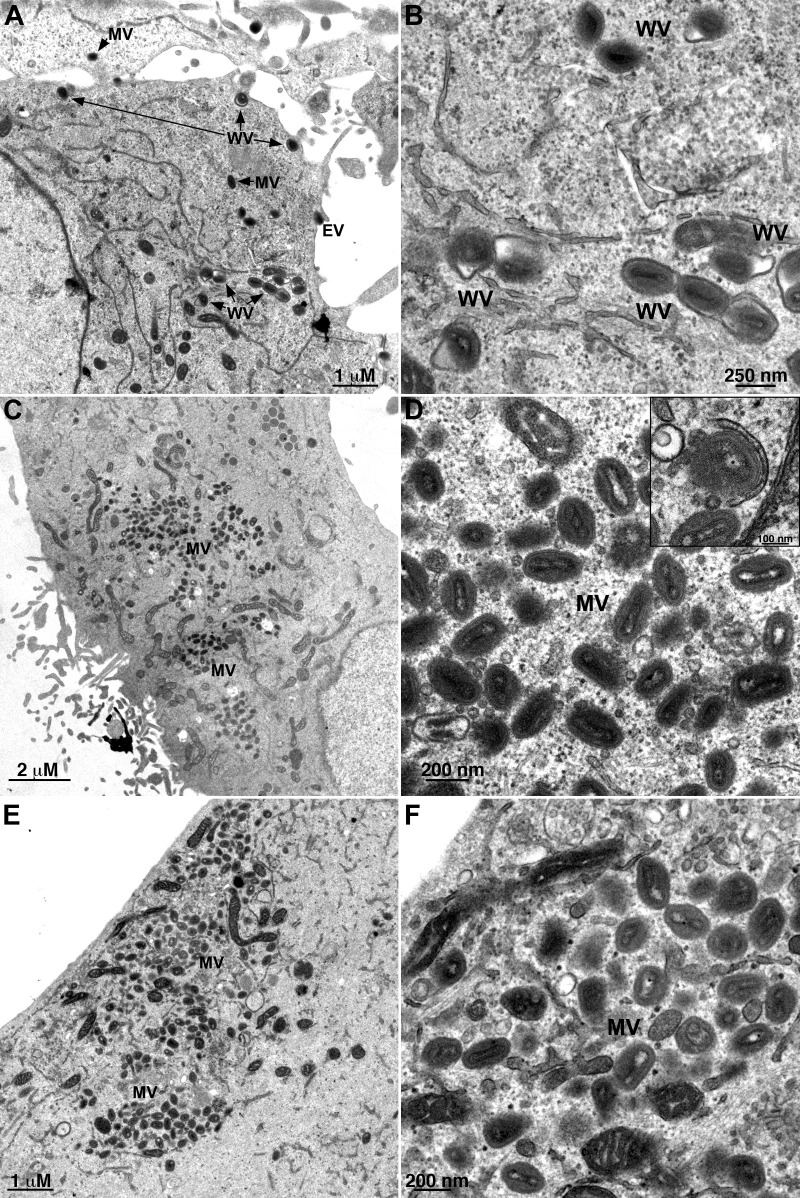
The retention of virus particles in the interior of the cell and their failure to colocalize with B5 suggested that Retro-2 blocked wrapping. However, additional defects were still possible. Therefore, transmission electron microscopy was used to investigate the effects of Retro-2 more directly. Cells were infected with VACV and treated with DMSO carrier with or without Retro-2. In the absence of Retro-2, virions surrounded by wrapping membranes were present in the cytoplasm and EVs were visualized at the cell surface (Fig. 6A and B). In the presence of Retro-2, normal-looking MVs were distributed throughout the cytoplasm, but WVs were rare and no EVs were detected (Fig. 6C and D). Some partial wrapping of MVs occurred, as shown in the inset of Fig. 6D. As anticipated, WVs and EVs were absent in cells that were infected in the presence of the inhibitor ST-246 (Fig. 6E and F). To quantify the electron microscopic data, we selected thin sections of 25 cells with MVs and determined the numbers of different types of mature virus particles formed in the absence and presence of drugs. In the cells treated with DMSO carrier, there were 299 MVs, 37 partially wrapped MVs, 125 WVs, and 37 EVs. In cells treated with Retro-2, there were 888 MVs, 28 partially wrapped MVs, 1 WV, and no EVs. In the ST246-treated cells, there were 1,094 MVs and no WVs or EVs. Thus, the block in virus spread caused by Retro-2 was due to inhibition of the wrapping step.
FIG 6.
Inhibition of WV and EV formation by Retro-2 determined by transmission electron microscopy. HeLa cells were infected with 5 PFU/cell of VACV in the presence of DMSO carrier (A and B), 20 μM Retro-2 (C and D), or 10 μM ST-246 (E and F). After 24 h, the cells were fixed and prepared for transmission electron microscopy. Left and right panels are low and high magnifications, respectively. The inset in the upper right corner of panel D shows a partially wrapped MV. MVs, mature virions; WVs, wrapped virions; EVs, extracellular virions.
Retro-2 does not prevent palmitoylation of F13.
F13, a nonglycosylated protein with two palmitoylated cysteines that provide hydrophobicity and membrane localization (7, 16, 17), is required for wrapping of virions (12). To determine whether Retro-2 interfered with palmitoylation of F13, cells were infected with a virus in which the original F13 open reading frame was replaced with a functional F13-GFP (30) and incubated with 100 μM palmitic acid-azide in the presence of Retro-2 or DMSO carrier. At 8 h after infection, the cells were washed, fixed, and labeled by Click-It chemistry with alkyne-Alexa Fluor 594. Colocalization of palmitic acid and F13 was observed with and without Retro-2 (Fig. 7A). To confirm these results, F13 was captured with antibody directed to GFP, and biotin was attached using Click-It chemistry. After SDS-PAGE and transfer to a blot, palmitoylated proteins were probed with IRDye 800-streptavidin. The intensities of the F13-GFP bands were similar in the presence and absence of Retro-2, suggesting that the failure of wrapping was not caused by inhibition of F13 palmitoylation (Fig. 7B).
FIG 7.
Palmitoylation of F13 occurs in the presence of Retro-2. (A) HeLa cells on coverslips were infected with 3 PFU/cell of VACV F13-GFP and after 1 h incubated with 100 μM palmitic acid-azide and 20 μM Retro-2 or DMSO carrier. At 8 h after infection, the cells were washed, fixed, and labeled by Click-It chemistry with alkyne-Alexa Fluor 594. Cells were examined by confocal microscopy, and superimposed z-stack projections are shown. Colocalizations were determined on individual focal planes using IMARIS software prior to stacking. Green, GFP; red, palmitate; yellow, colocalization. (B) HeLa cells were infected and labeled as for panel A. The cells were lysed, and F13 was captured with antibody to GFP attached to protein G on magnetic beads (lanes 5, 6, and 8) or control IgG (lane 7). After washing, biotin was attached with Click-It chemistry. Proteins were eluted, resolved by SDS-PAGE, blotted onto a membrane, and probed with streptavidin conjugated to IRDye 800. In lane 8, copper was omitted from the Click-It reaction mixture to provide a control.
Retro-2 alters the location of the VACV F13 membrane protein.
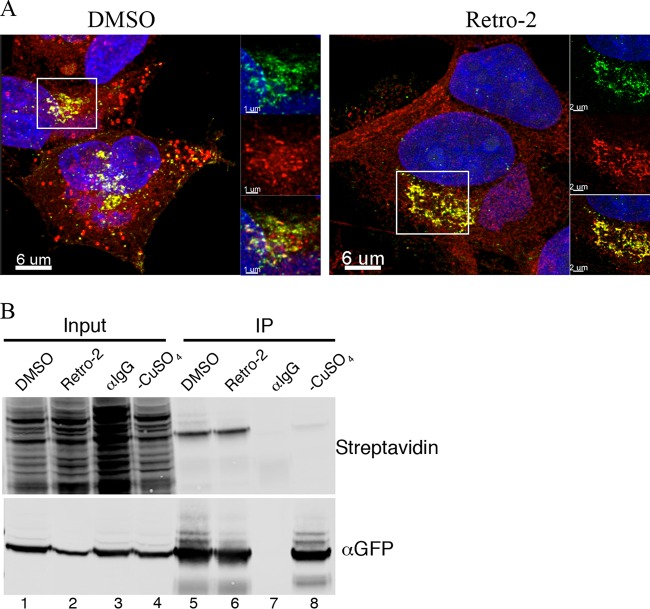
Although Retro-2 did not interfere with palmitoylation, it was of interest to determine the effect of the drug on F13 localization. Previous studies had shown that F13 associates with early and late endosomes in addition to the TGN (59, 60). Cells were transfected with cytomegalovirus (CMV) vectors expressing TGNP, Rab7, or Rab5 fused to fluorescent tags in order to localize the TGN, late endosomes, and early endosomes, respectively. After 24 h, the transfected cells were infected with a VACV expressing F13-GFP and treated with Retro-2 or the carrier DMSO alone. Cell organelle markers, F13-GFP, and DAPI-stained nuclear and cytoplasmic viral DNA were visualized by fluorescence confocal microscopy and correlation coefficients determined. In the absence of Retro-2, F13-GFP colocalized with markers for the TGN and early and late endosomes (Fig. 8A). In contrast, in the presence of Retro-2, there was little association of F13-GFP with TGN markers, but association with the endosomal markers was relatively unchanged (Fig. 8A). The difference in colocalization of F13 and the TGN marker in the absence and presence of Retro-2 was significant (P < 0.007).
FIG 8.
Retro-2 prevents TGN localization of VACV F13 membrane protein. (A) HeLa cells grown on coverslips were transfected with plasmids expressing TGNP, Rab5, and Rab7 fluorescent fusion proteins and 24 h later were infected with 3 PFU/cell of VACV expressing F13-GFP in the presence of DMSO or Retro-2. At 8 h postinfection, cells were fixed and prepared for confocal microscopy. Colocalization of B5 and GFP was calculated with IMARIS prior to superimposition of z-stacks. The area within the box is shown at higher magnification on the right as separate and colocalization images. DAPI, blue; F13-GFP, green; Rab5 and Rab7, red; colocalization, yellow. Scale bars are shown at bottom left. Pearson coefficients were determined by analyzing cells in several random fields. (B) Untransfected cells were infected with VACV F13-GFP for 8 h and then stained with rat MAb specific for the B5 protein and a secondary fluorescent antibody. DAPI, blue; B5, red; F13-GFP, green; colocalization, yellow. A Pierson coefficient plot of B5 and F13-GFP is on the right. ***, P < 0.001.
Next, we compared the localization of F13 with B5 in the absence and presence of Retro-2. In the absence of the drug, B5 and F13 colocalized near the nucleus as well as in peripheral puncti that likely represent wrapped virus particles (Fig. 8B). In contrast, there were fewer puncti and less colocalization of the two proteins in cells treated with Retro-2 (Fig. 8). The significance of the latter was confirmed by analysis of Pierson coefficients (Fig. 8).
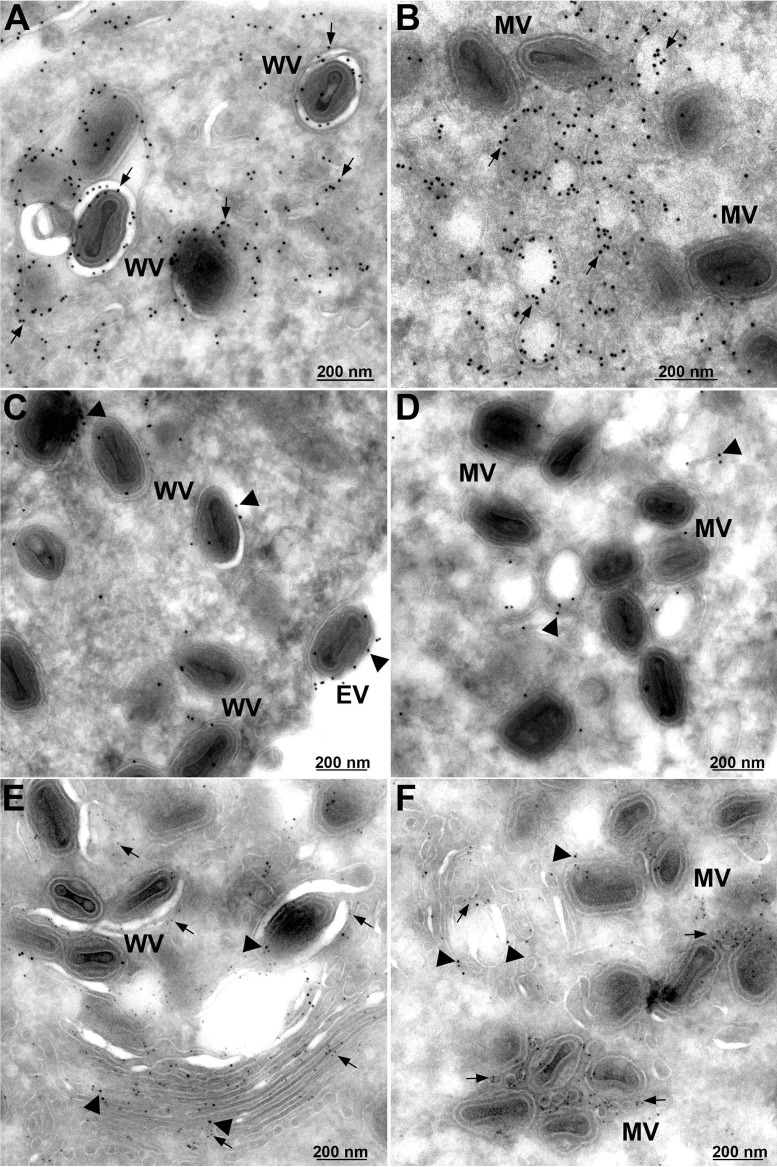
In a previous section we showed by transmission electron microscopy that Retro-2 interfered with the formation of WVs. Immunogold labeling was performed in order to localize F13 and B5 under these conditions. Cells were infected with the recombinant VACV expressing F13 with a hemagglutinin (HA) tag in the absence (Fig. 9A, C, and E) and presence (Fig. 9B, D, and F) of Retro-2 and then stained with either a mouse MAb to HA or a rat MAb to B5 followed by protein A conjugated to gold spheres. In the absence of drug, gold spheres representing F13 (Fig. 9A) and B5 (Fig. 9C) decorated WVs as well as nearby membranes. In the presence of Retro-2 (Fig. 9B), F13 was abundant on membranes that resembled cross sections of tubular early endosomes described by Tooze and Hollinshead (61) at sites of wrapping MVs. This interpretation agrees with our confocal microscopy data in Fig. 8 showing colocalization of F13-GFP with an early endosomal marker. Some B5 was also found on similar membranous structures (Fig. 9D). We also carried out double labeling in which B5 and F13 were tagged with 10- and 5-nm gold spheres, respectively. The image in Fig. 9E shows F13 and B5 in stacked membranes that appear to be in the process of enclosing virus particles in the absence of drug. In the presence of Retro-2, both F13 and B5 were associated with vesicles near MVs.
FIG 9.
Immunoelectron microscopy showing vesicular location of F13 in the presence of Retro-2. HeLa cells were infected with 5 PFU/cell of VACV expressing F13-HA for 16 h in the presence of DMSO (A, C, and E) or 20 μM Retro-2 (B, D, and F). Cells were fixed, and frozen sections were stained with a mouse MAb to the HA tag (A and B) or a rat MAb to B5 (C and D) followed by rabbit anti-mouse or rabbit anti-rat IgG and protein A conjugated to 10-nm gold spheres. Panels E and F show double label in which B5 and F13 are decorated with 10-nm and 5-nm gold spheres, respectively. WV, wrapped virions; EV, extracellular virions; MV, mature virions. Arrows point to F13 and arrowheads to B5.
DISCUSSION
The goal of the present study was to evaluate the biological significance and role of retrograde transport proteins, which had been discovered as host factors for spread of VACV in a human genome-wide RNA interference (RNAi) screen (28). As a first step, we overlaid the siRNA results on an interaction network of proteins involved in retrograde transport and found many strong hits, particularly for components of the GARP and COG complexes. Next, we tested siRNAs to several retrograde transport factors from our gene enrichment list as well as others suggested by the interaction network. In this way, we corroborated some previous hits and found new ones that had been missed in the high-throughput screen. Since the screen was based on a virus spread assay, the retrograde transport factors could be needed for any step from virus entry to egress. However, our studies showed that neither entry nor formation of infectious virus was impaired. In comparison, several families of nonenveloped viruses use retrograde transport for entry (21–24). The key results, shown by confocal microscopy and CsCl2 sedimentation, were that depletion of retrograde transport factors inhibited wrapping of MVs and egress.
The strong inhibition of VACV spread by the retrograde transport factors, and the t-SNAREs STX5 and STX6 in particular, led us to study Retro-2. Retro-2 was discovered in a high-throughput screen of small molecules that could protect cells from Shiga-like toxins and ricin, which depend on retrograde transport for release into the cytoplasm (27). Extensive studies showed that Retro-2 has no effect on binding or endosomal entry of the toxins but specifically blocks retrograde transport at the early endosome-TGN interface (27). Retro-2 does not affect targeting to the TGN through the late endosome pathway and has no discernible toxicity or effect on compartment morphology, endogenous retrograde cargoes, or other trafficking steps. The activity of Retro-2 for protein toxins involves dispersal of STX5 and STX6 from the TGN, although the cellular target that interacts with Retro-2 remains to be discovered. Whereas individual siRNAs to STX5, STX6, and other retrograde transport factors reduced VACV spread by about 50%, Retro-2 reduced spread by nearly 100% when given before or after infection. This difference is likely due to incomplete depletion of proteins by siRNAs but may also reflect use of multiple alternative factors, each of which is blocked by Retro-2. The specificity of Retro-2 on retrograde trafficking and its perturbation of the same step in the VACV reproductive cycle as depletion of retrograde transport factors strongly suggested a common mechanism of action at the endosome-TGN nexus.
Further studies with Retro-2 demonstrated that the membrane wrapping of VACV particles was abrogated as shown by confocal microscopy and transmission electron microscopy. This result focused our attention on the two proteins required for wrapping: B5 and F13 (12–14). B5 is a type 1 integral membrane glycoprotein, with a long extracellular domain and a short cytoplasmic tail, that traffics through the secretory pathway even in the absence of other viral proteins (15, 18, 59, 62). Although B5 is acylated (15), nonacylated mutants are only slightly impaired in function (63). In contrast to B5, F13 has no transmembrane domain and is dependent on palmitoylation of two cysteines for membrane localization (7, 17). F13 is found associated with the TGN as well as early and late endosomes even when COPII-mediated cargo transport from the ER is inhibited, indicating that the anterograde pathway is not utilized (59). We found that the association of F13 with the TGN and colocalization with B5 were inhibited by Retro-2, implying that F13 normally traffics from early endosomes through the retrograde pathway. F13 has another element in addition to palmitoylation that is required for membrane association and might also be responsible for localization to early endosomes. This element is a variation of a motif found in the superfamily of phospholipases and phospholipid synthases (64, 65). The canonical motif includes two copies of an HxKxxxxD sequence, whereas F13 has a single copy of an NxKxxxxD sequence. Conservative substitutions of K or D result in loss of F13 membrane localization and wrapping (66, 67). Although there are similarities in the induction of post-Golgi vesicles by phospholipase D and F13 (68), phospholipase D activity is yet to be demonstrated for recombinant F13. Moreover, an H-to-N mutation in the HxKxxxxD motif, making it more like that of VACV, abrogated the activity of phospholipase D, suggesting that F13 does not possess this activity (66). There is, however, one report of broad-specificity lipase activity of purified F13 (69), and it is also possible that F13 has an affinity for specific phospholipids that mediates early endosomal localization. Although further mechanistic studies are needed, we suggest that F13 initially localizes in early endosomes and that retrograde transport brings F13 and B5 together in wrapping membrane precursors. The use of the anterograde pathway for B5 and the retrograde pathway for F13, although seemingly inefficient, would ensure that wrapping of MVs does not occur upstream of the TGN.
There are two drugs in addition to Retro-2 and its derivatives that inhibit the wrapping and release of VACV particles: IMCBH (70–73) and ST-246 (58, 74). With both drugs, resistance mutations were mapped to the gene encoding F13 (73, 74). ST-246 is highly active in animal models (75) and is being tested clinically (76). In principle, Retro-2 could have an advantage over IMCBH and ST-246 because it targets host factors so that resistant virus mutants would be unlikely to arise. Although Retro-2 has been shown to protect mice against bacterial and plant toxins (27) and infection with enterohemorrhagic Escherichia coli (77), we were unable to demonstrate protective effects of Retro-2 in a mouse model of VACV infection, at least partly due to poor drug solubility. However, Retro-2 derivatives with greatly increased potency are in development (78–80) and will be worth testing in the future.
ACKNOWLEDGMENTS
We thank Catherine Cotter for maintaining cell lines. ST-246 was a kind gift from Dennis Hruby and SIGA Technologies, Inc.
This study was supported by the Division of Intramural Research, NIAID, NIH.
REFERENCES
- 1.Moss B. 2013. Poxviridae, p 2129–2159. In Knipe DM, Howley PM (ed), Fields virology, 6th ed, vol 2 Lippincott Williams & Wilkins, Hagerstown, MD. [Google Scholar]
- 2.Condit RC, Moussatche N, Traktman P. 2006. In a nutshell: structure and assembly of the vaccinia virion. Adv Virus Res 66:31–124. doi: 10.1016/S0065-3527(06)66002-8. [DOI] [PubMed] [Google Scholar]
- 3.Smith GL, Law M. 2004. The exit of vaccinia virus from infected cells. Virus Res 106:189–197. doi: 10.1016/j.virusres.2004.08.015. [DOI] [PubMed] [Google Scholar]
- 4.Blasco R, Moss B. 1992. Role of cell-associated enveloped vaccinia virus in cell-to-cell spread. J Virol 66:4170–4179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vliegen I, Yang G, Hruby D, Jordan R, Neyts J. 2012. Deletion of the vaccinia virus F13L gene results in a highly attenuated virus that mounts a protective immune response against subsequent vaccinia virus challenge. Antiviral Res 93:160–166. doi: 10.1016/j.antiviral.2011.11.010. [DOI] [PubMed] [Google Scholar]
- 6.Moss B. 2015. Poxvirus membrane biogenesis. Virology 479-480:619–626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hiller G, Weber K. 1985. Golgi-derived membranes that contain an acylated viral polypeptide are used for vaccinia virus envelopment. J Virol 55:651–659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schmelz M, Sodeik B, Ericsson M, Wolffe EJ, Shida H, Hiller G, Griffiths G. 1994. Assembly of vaccinia virus: the second wrapping cisterna is derived from the trans Golgi network. J Virol 68:130–147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tooze J, Hollinshead M, Reis B, Radsak K, Kern H. 1993. Progeny vaccinia and human cytomegalovirus particles utilize early endosomal cisternae for their envelopes. Eur J Cell Biol 60:163–178. [PubMed] [Google Scholar]
- 10.Payne L. 1978. Polypeptide composition of extracellular enveloped vaccinia virus. J Virol 27:28–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smith GL, Vanderplasschen A, Law M. 2002. The formation and function of extracellular enveloped vaccinia virus. J Gen Virol 83:2915–2931. doi: 10.1099/0022-1317-83-12-2915. [DOI] [PubMed] [Google Scholar]
- 12.Blasco R, Moss B. 1991. Extracellular vaccinia virus formation and cell-to-cell virus transmission are prevented by deletion of the gene encoding the 37,000 Dalton outer envelope protein. J Virol 65:5910–5920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Engelstad M, Smith GL. 1993. The vaccinia virus 42-kDa envelope protein is required for the envelopment and egress of extracellular virus and for virus virulence. Virology 194:627–637. doi: 10.1006/viro.1993.1302. [DOI] [PubMed] [Google Scholar]
- 14.Wolffe EJ, Isaacs SN, Moss B. 1993. Deletion of the vaccinia virus B5R gene encoding a 42-kilodalton membrane glycoprotein inhibits extracellular virus envelope formation and dissemination. J Virol 67:4732–4741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Isaacs SN, Wolffe EJ, Payne LG, Moss B. 1992. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J Virol 66:7217–7224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schmutz C, Rindisbacher L, Galmiche MC, Wittek R. 1995. Biochemical analysis of the major vaccinia virus envelope antigen. Virology 213:19–27. doi: 10.1006/viro.1995.1542. [DOI] [PubMed] [Google Scholar]
- 17.Grosenbach DW, Ulaeto DO, Hruby DE. 1997. Palmitylation of the vaccinia virus 37-kDa major envelope antigen. Identification of a conserved acceptor motif and biological relevance. J Biol Chem 272:1956–1964. [DOI] [PubMed] [Google Scholar]
- 18.Lorenzo MM, Galindo I, Griffiths G, Blasco R. 2000. Intracellular localization of vaccinia virus extracellular enveloped virus envelope proteins individually expressed using a Semliki Forest virus replicon. J Virol 74:10535–10550. doi: 10.1128/JVI.74.22.10535-10550.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Husain M, Moss B. 2003. Evidence against an essential role of COPII-mediated cargo transport to the endoplasmic reticulum-Golgi intermediate compartment in the formation of the primary membrane of vaccinia virus. J Virol 77:11754–11766. doi: 10.1128/JVI.77.21.11754-11766.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sandvig K, van Deurs B. 2005. Delivery into cells: lessons learned from plant and bacterial toxins. Gene Ther 12:865–872. doi: 10.1038/sj.gt.3302525. [DOI] [PubMed] [Google Scholar]
- 21.Bantel-Schaal U, Hub B, Kartenbeck J. 2002. Endocytosis of adeno-associated virus type 5 leads to accumulation of virus particles in the Golgi compartment. J Virol 76:2340–2349. doi: 10.1128/jvi.76.5.2340-2349.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lipovsky A, Popa A, Pimienta G, Wyler M, Bhan A, Kuruvilla L, Guie MA, Poffenberger AC, Nelson CD, Atwood WJ, DiMaio D. 2013. Genome-wide siRNA screen identifies the retromer as a cellular entry factor for human papillomavirus. Proc Natl Acad Sci U S A 110:7452–7457. doi: 10.1073/pnas.1302164110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nelson CD, Carney DW, Derdowski A, Lipovsky A, Gee GV, O'Hara B, Williard P, DiMaio D, Sello JK, Atwood WJ. 2013. A retrograde trafficking inhibitor of ricin and Shiga-like toxins inhibits infection of cells by human and monkey polyomaviruses. mBio 4:e00729-13. doi: 10.1128/mBio.00729-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nonnenmacher ME, Cintrat JC, Gillet D, Weber T. 2015. Syntaxin 5-dependent retrograde transport to the trans-Golgi network is required for adeno-associated virus transduction. J Virol 89:1673–1687. doi: 10.1128/JVI.02520-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bonifacino JS, Rojas R. 2006. Retrograde transport from endosomes to the trans-Golgi network. Nat Rev Mol Cell Biol 7:568–579. doi: 10.1038/nrm1985. [DOI] [PubMed] [Google Scholar]
- 26.Schindler C, Chen Y, Pu J, Guo X, Bonifacino JS. 2015. EARP is a multisubunit tethering complex involved in endocytic recycling. Nat Cell Biol 17:639–650. doi: 10.1038/ncb3129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stechmann B, Bai SK, Gobbo E, Lopez R, Merer G, Pinchard S, Panigai L, Tenza D, Raposo G, Beaumelle B, Sauvaire D, Gillet D, Johannes L, Barbier J. 2010. Inhibition of retrograde transport protects mice from lethal ricin challenge. Cell 141:231–242. doi: 10.1016/j.cell.2010.01.043. [DOI] [PubMed] [Google Scholar]
- 28.Sivan G, Martin SE, Myers TG, Buehler E, Szymczyk KH, Ormanoglu P, Moss B. 2013. Human genome-wide RNAi screen reveals a role for nuclear pore proteins in poxvirus morphogenesis. Proc Natl Acad Sci U S A 110:3519–3524. doi: 10.1073/pnas.1300708110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bengali Z, Townsley AC, Moss B. 2009. Vaccinia virus strain differences in cell attachment and entry. Virology 389:132–140. doi: 10.1016/j.virol.2009.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Husain M, Moss B. 2001. Vaccinia virus F13L protein with a conserved phospholipase catalytic motif induces colocalization of the B5R envelope glycoprotein in post-Golgi vesicles. J Virol 75:7528–7542. doi: 10.1128/JVI.75.16.7528-7542.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laliberte JP, Weisberg AS, Moss B. 2011. The membrane fusion step of vaccinia virus entry is cooperatively mediated by multiple viral proteins and host cell components. PLoS Pathog 7:e1002446. doi: 10.1371/journal.ppat.1002446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies DH, Wyatt LS, Newman FK, Earl PL, Chun S, Hernandez JE, Molina DM, Hirst S, Moss B, Frey SE, Felgner PL. 2008. Antibody profiling by proteome microarray reveals the immunogenicity of the attenuated smallpox vaccine modified vaccinia virus ankara is comparable to that of Dryvax. J Virol 82:652–663. doi: 10.1128/JVI.01706-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maruri-Avidal L, Domi A, Weisberg AS, Moss B. 2011. Participation of vaccinia virus L2 protein in the formation of crescent membranes and immature virions. J Virol 85:2504–2511. doi: 10.1128/JVI.02505-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Senkevich TG, Wyatt LS, Weisberg AS, Koonin EV, Moss B. 2008. A conserved poxvirus NlpC/P60 superfamily protein contributes to vaccinia virus virulence in mice but not to replication in cell culture. Virology 374:506–514. doi: 10.1016/j.virol.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bisht H, Weisberg AS, Szajner P, Moss B. 2009. Assembly and disassembly of the capsid-like external scaffold of immature virions during vaccinia virus morphogenesis. J Virol 83:9140–9150. doi: 10.1128/JVI.00875-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Satheshkumar PS, Anton LC, Sanz P, Moss B. 2009. Inhibition of the ubiquitin-proteasome system prevents vaccinia virus DNA replication and expression of intermediate and late genes. J Virol 83:2469–2479. doi: 10.1128/JVI.01986-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Teale A, Campbell S, Van Buuren N, Magee WC, Watmough K, Couturier B, Shipclark R, Barry M. 2009. Orthopoxviruses require a functional ubiquitin-proteasome system for productive replication. J Virol 83:2099–2108. doi: 10.1128/JVI.01753-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mercer J, Snijder B, Sacher R, Burkard C, Bleck CK, Stahlberg H, Pelkmans L, Helenius A. 2012. RNAi screening reveals proteasome- and cullin3-dependent stages in vaccinia virus infection. Cell Rep 2:1036–1047. doi: 10.1016/j.celrep.2012.09.003. [DOI] [PubMed] [Google Scholar]
- 39.Bonifacino JS, Hierro A. 2011. Transport according to GARP: receiving retrograde cargo at the trans-Golgi network. Trends Cell Biol 21:159–167. doi: 10.1016/j.tcb.2010.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Perez-Victoria FJ, Bonifacino JS. 2009. Dual roles of the mammalian GARP complex in tethering and SNARE complex assembly at the trans-Golgi network. Mol Cell Biol 29:5251–5563. doi: 10.1128/MCB.00495-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rosa-Ferreira C, Christis C, Torres IL, Munro S. 2015. The small G protein Arl5 contributes to endosome-to-Golgi traffic by aiding the recruitment of the GARP complex to the Golgi. Biol Open 4:474–481. doi: 10.1242/bio.201410975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laufman O, Hong W, Lev S. 2013. The COG complex interacts with multiple Golgi SNAREs and enhances fusogenic assembly of SNARE complexes. J Cell Sci 126:1506–1516. doi: 10.1242/jcs.122101. [DOI] [PubMed] [Google Scholar]
- 43.Laufman O, Hong W, Lev S. 2011. The COG complex interacts directly with Syntaxin 6 and positively regulates endosome-to-TGN retrograde transport. J Cell Biol 194:459–472. doi: 10.1083/jcb.201102045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sacher M, Kim YG, Lavie A, Oh BH, Segev N. 2008. The TRAPP complex: insights into its architecture and function. Traffic 9:2032–2042. doi: 10.1111/j.1600-0854.2008.00833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McGough IJ, Cullen PJ. 2011. Recent advances in retromer biology. Traffic 12:963–971. doi: 10.1111/j.1600-0854.2011.01201.x. [DOI] [PubMed] [Google Scholar]
- 46.Townsley AC, Weisberg AS, Wagenaar TR, Moss B. 2006. Vaccinia virus entry into cells via a low pH-dependent-endosomal pathway. J Virol 80:8899–8908. doi: 10.1128/JVI.01053-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gordon DE, Bond LM, Sahlender DA, Peden AA. 2010. A Targeted siRNA screen to identify SNAREs required for constitutive secretion in mammalian cells. Traffic 11:1191–1204. doi: 10.1111/j.1600-0854.2010.01087.x. [DOI] [PubMed] [Google Scholar]
- 48.Geada MM, Galindo I, Lorenzo MM, Perdiguero B, Blasco R. 2001. Movements of vaccinia virus intracellular enveloped virions with GFP tagged to the F13L envelope protein. J Gen Virol 82:2747–2760. doi: 10.1099/0022-1317-82-11-2747. [DOI] [PubMed] [Google Scholar]
- 49.Hollinshead M, Rodger G, Van Eijl H, Law M, Hollinshead R, Vaux DJ, Smith GL. 2001. Vaccinia virus utilizes microtubules for movement to the cell surface. J Cell Biol 154:389–402. doi: 10.1083/jcb.200104124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rietdorf J, Ploubidou A, Reckmann I, Holmström A, Frischknecht F, Zettl M, Zimmerman T, Way M. 2001. Kinesin dependent movement on microtubules precedes actin based motility of vaccinia virus. Nat Cell Biol 3:992–1000. doi: 10.1038/ncb1101-992. [DOI] [PubMed] [Google Scholar]
- 51.Ward BM, Moss B. 2001. Vaccinia virus intracellular movement is associated with microtubules and independent of actin tails. J Virol 75:11651–11663. doi: 10.1128/JVI.75.23.11651-11663.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Ward BM, Moss B. 2001. Visualization of intracellular movement of vaccinia virus virions containing a green fluorescent protein-B5R membrane protein chimera. J Virol 75:4802–4813. doi: 10.1128/JVI.75.10.4802-4813.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boulter EA, Appleyard G. 1973. Differences between extracellular and intracellular forms of poxvirus and their implications. Prog Med Virol 16:86–108. [PubMed] [Google Scholar]
- 54.Payne LG. 1980. Significance of extracellular virus in the in vitro and in vivo dissemination of vaccinia virus. J Gen Virol 50:89–100. doi: 10.1099/0022-1317-50-1-89. [DOI] [PubMed] [Google Scholar]
- 55.Cudmore S, Cossart P, Griffiths G, Way M. 1995. Actin-based motility of vaccinia virus. Nature 378:636–638. doi: 10.1038/378636a0. [DOI] [PubMed] [Google Scholar]
- 56.Stokes GV. 1976. High-voltage electron microscope study of the release of vaccinia virus from whole cells. J Virol 18:636–643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Herrero-Martinez E, Roberts KL, Hollinshead M, Smith GL. 2005. Vaccinia virus intracellular enveloped virions move to the cell periphery on microtubules in the absence of the A36R protein. J Gen Virol 86:2961–2968. doi: 10.1099/vir.0.81260-0. [DOI] [PubMed] [Google Scholar]
- 58.Yang G, Pevear DC, Davies MH, Collett MS, Bailey T, Rippen S, Barone L, Burns C, Rhodes G, Tohan S, Huggins JW, Baker RO, Buller RLM, Touchette E, Waller K, Schriewer J, Neyts J, DeClercq E, Jones K, Hruby D, Jordan R. 2005. An orally bioavailable antipoxvirus compound (ST-246) inhibits extracellular virus formation and protects mice from lethal orthopoxvirus challenge. J Virol 79:13139–13149. doi: 10.1128/JVI.79.20.13139-13149.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Husain M, Moss B. 2003. Intracellular trafficking of a palmitoylated membrane-associated protein component of enveloped vaccinia virus. J Virol 77:9008–9019. doi: 10.1128/JVI.77.16.9008-9019.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Chen YL, Honeychurch KM, Yang G, Byrd CM, Harver C, Hruby DE, Jordan R. 2009. Vaccinia virus p37 interacts with host proteins associated with LE-derived transport vesicle biogenesis. Virology J 6:44. doi: 10.1186/1743-422X-6-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tooze J, Hollinshead M. 1991. Tubular early endosomal networks in AtT20 and other cells. J Cell Biol 115:635–653. doi: 10.1083/jcb.115.3.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ward BM, Moss B. 2000. Golgi network targeting and plasma membrane internalization signals in vaccinia virus B5R envelope protein. J Virol 74:3771–3780. doi: 10.1128/JVI.74.8.3771-3780.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lorenzo MM, Sanchez-Puig JM, Blasco R. 2012. Mutagenesis of the palmitoylation site in vaccinia virus envelope glycoprotein B5. J Gen Virol 93:733–743. doi: 10.1099/vir.0.039016-0. [DOI] [PubMed] [Google Scholar]
- 64.Ponting CP, Kerr ID. 1996. A novel family of phospholipase D homologues that includes phospholipid synthases and putative endonucleases: identification of duplicated repeats and potential active site residues. Protein Sci 5:914–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Koonin EV. 1997. Evidence for a family of archaeal ATPases. Science 275:1489–1490. doi: 10.1126/science.275.5305.1489. [DOI] [PubMed] [Google Scholar]
- 66.Sung T-C, Roper RL, Zhang Y, Rudge SA, Temel R, Hammond SM, Morris AJ, Moss B, Engebrecht J, Frohman MA. 1997. Mutagenesis of phospholipase D defines a superfamily including a trans-Golgi viral protein required for poxvirus pathogenicity. EMBO J 16:4519–4530. doi: 10.1093/emboj/16.15.4519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Roper RL, Moss B. 1999. Envelope formation is blocked by mutation of a sequence related to the HKD phospholipid metabolism motif in the vaccinia virus F13L protein. J Virol 73:1108–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Husain M, Moss B. 2002. Similarities in the induction of post-Golgi vesicles by vaccinia virus F13L protein and phospholipase D. J Virol 76:7777–7789. doi: 10.1128/JVI.76.15.7777-7789.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Baek S-H, Kwak J-Y, Lee SH, Lee T, Ryu SH, Uhlinger DJ, Lambeth JD. 1997. Lipase activities of p37, the major envelope protein of vaccinia virus. J Biol Chem 272:32042–32049. doi: 10.1074/jbc.272.51.32042. [DOI] [PubMed] [Google Scholar]
- 70.Kato N, Eggers HJ, Rolly H. 1969. Inhibition of release of vaccinia virus by N1-isonicotinoyl-N2-3-methyl-4-chlorobenzoylhydrazine. J Exp Med 129:795–808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Payne LG, Kristenson K. 1979. Mechanism of vaccinia virus release and its specific inhibition by N1-isonicatinoyl-N2-3-methyl-4-chlorobenzoylhydrazine. J Virol 32:614–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hiller G, Eibl H, Weber K. 1981. Characterization of intracellular and extracellular vaccinia virus variants: N1-isonicotinoyl-N2-3-methyl-4-chlorobenzoylhydrazine interferes with cytoplasmic virus dissemination and release. J Virol 39:903–913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Schmutz C, Payne LG, Gubser J, Wittek R. 1991. A mutation in the gene encoding the vaccinia virus 37,000-Mr protein confers resistance to an inhibitor of virus envelopment and release. J Virol 65:3435–3442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Duraffour S, Lorenzo MM, Zoller G, Topalis D, Grosenbach D, Hruby DE, Andrei G, Blasco R, Meyer H, Snoeck R. 2015. ST-246 is a key antiviral to inhibit the viral F13L phospholipase, one of the essential proteins for orthopoxvirus wrapping. J Antimicrob Chemother 70:1367–1380. doi: 10.1093/jac/dku545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Huggins J, Goff A, Hensley L, Mucker E, Shamblin J, Wlazlowski C, Johnson W, Chapman J, Larsen T, Twenhafel N, Karem K, Damon IK, Byrd CM, Bolken TC, Jordan R, Hruby D. 2009. Nonhuman primates are protected from smallpox virus or monkeypox virus challenges by the antiviral drug ST-246. Antimicrob Agents Chemother 53:2620–2625. doi: 10.1128/AAC.00021-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chinsangaram J, Honeychurch KM, Tyavanagimatt SR, Leeds JM, Bolken TC, Jones KF, Jordan R, Marbury T, Ruckle J, Mee-Lee D, Ross E, Lichtenstein I, Pickens M, Corrado M, Clarke JM, Frimm AM, Hruby DE. 2012. Safety and pharmacokinetics of the anti-orthopoxvirus compound ST-246 following a single daily oral dose for 14 days in human volunteers. Antimicrob Agents Chemother 56:4900–4905. doi: 10.1128/AAC.00904-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Secher T, Shima A, Hinsinger K, Cintrat JC, Johannes L, Barbier J, Gillet D, Oswald E. 2015. Retrograde trafficking inhibitor of Shiga toxins reduces morbidity and mortality of mice infected with enterohemorrhagic Escherichia coli. Antimicrob Agents Chemother 59:5010–5013. doi: 10.1128/AAC.00455-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gupta N, Pons V, Noel R, Buisson DA, Michau A, Johannes L, Gillet D, Barbier J, Cintrat JC. 2014. (S)-N-methyldihydroquinazolinones are the active enantiomers of Retro-2 derived compounds against toxins. ACS Med Chem Lett 5:94–97. doi: 10.1021/ml400457j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Carney DW, Nelson CD, Ferris BD, Stevens JP, Lipovsky A, Kazakov T, DiMaio D, Atwood WJ, Sello JK. 2014. Structural optimization of a retrograde trafficking inhibitor that protects cells from infections by human polyoma- and papillomaviruses. Bioorg Med Chem 22:4836–4847. doi: 10.1016/j.bmc.2014.06.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Noel R, Gupta N, Pons V, Goudet A, Garcia-Castillo MD, Michau A, Martinez J, Buisson DA, Johannes L, Gillet D, Barbier J, Cintrat JC. 2013. N-methyldihydroquinazolinone derivatives of Retro-2 with enhanced efficacy against Shiga toxin. J Med Chem 56:3404–3413. doi: 10.1021/jm4002346. [DOI] [PubMed] [Google Scholar]