ABSTRACT
Antibodies are known to enhance in vitro infection by human immunodeficiency virus (HIV) and simian immunodeficiency virus (SIV). We measured the ability of antibodies induced by ALVAC-SIV/gp120 vaccination, given with alum or MF59 adjuvant, to capture infectious SIVmac251 and determined the association between capture and infection outcomes following low-dose, repeated rectal challenge of rhesus macaques. We found that capture correlated with the number of transmitted/founder (T/F) variants that established infection, such that animals whose plasma captured more virus were infected with a higher number of T/F strains. Capture also correlated with results of Env binding assays, indicating that greater immunogenicity resulted in greater capture. Although vaccination elicited negligible neutralizing activity against the challenge strain (50% inhibitory dilutions of >1/80 in all cases), animals with low capture and whose plasma, at a fixed dilution, inhibited a higher fraction of virus were infected at a lower rate than animals with high capture and low neutralization (P = 0.039); only animals with the low capture/high neutralization response profile were protected compared with unvaccinated control animals (P = 0.026). In a sieve analysis, high capture and low capture were distinguishable on the basis of polymorphisms in the V1 loop of Env at amino acids 144 and 145. Our results indicate that vaccine-induced antibody that binds to and captures infectious virus but does not inhibit its infectivity may enhance the likelihood of infection following rectal challenge with SIVmac251. Higher immunogenicity resulting in better antibody capture but similar anti-infectivity may not improve vaccine efficacy.
IMPORTANCE Vaccines generally prevent viral infections by eliciting antibodies that inhibit virus infectivity. However, antibodies, including those induced by vaccination, have the potential to enhance, rather than prevent infection. We measured the ability of vaccine-induced antibodies to capture infectious simian immunodeficiency virus (SIV) and explored the relationship between virus capture and infection outcomes. We found that capture correlated with the number of SIV variants that established infection, such that animals whose plasma captured more virus were infected with a higher number of unique strains. In addition, animals whose sera had high capture but weak anti-infectivity activity were infected at a higher rate than were animals with low capture and stronger anti-infectivity activity. These results suggest that vaccines that induce antibodies that bind to and capture infectious virus but do not inhibit virus infectivity will not be effective in preventing infection.
INTRODUCTION
Human and nonhuman primate studies have provided evidence that vaccines designed to prevent human immunodeficiency virus (HIV) or simian immunodeficiency virus (SIV) infections might, at times, increase the risk of infection (1). Such a deleterious effect can be apparent in the overall analysis of vaccine efficacy or from analyses of subject subgroups or of transmitted/founder variant number (2–4).
The mechanisms accounting for enhanced infection vary and likely include those that increase the number of CD4+ target cells and those in which vaccine-induced antibody plays a direct role. Antibodies that bind to envelope glycoproteins (Env) of lentiviruses are thought to enhance infection via complement receptors or receptors for the Fc portion of IgG antibodies (FcγRs) (5–9). Alternatively, antibodies that bind to gp120 may alter the configuration of Env and thereby allow more efficient Env-coreceptor interactions (10, 11).
Although antibodies that neutralize relevant strains of HIV-1 have been difficult to elicit by vaccination, some nonneutralizing antibodies, such as those that inhibit virus by engaging effector cells through Fc-FcγR interactions, may play a role in preventing infection (12). However, nonneutralizing antibodies and, at certain concentrations, antibodies that otherwise neutralize virus, may enhance infection (13–15). Whether an antibody enhances or inhibits virus, it must bind to infectious virions or to infected cells. Thus, the exact nature of Fab-virion binding or the manner in which effector cells or effector molecules are engaged determines whether an antibody will prevent infection, enhance infection, or do neither.
In this research, we investigated the relationship between the ability of vaccine-elicited antibodies to capture infectious virions and the role of those antibodies in enhancing infection.
MATERIALS AND METHODS
Animal trials.
All animals used in this study were colony-bred rhesus macaques (Macaca mulatta), obtained from Covance Research Products (Alice, TX). The animals were housed and handled in accordance with the standards of the Association for the Assessment and Accreditation of Laboratory Animal Care International. The care and use of the animals were in compliance with all relevant institutional (NIH) guidelines. The protocol (AUP 491) was approved by Advanced BioScience Laboratories' Institutional Animal Care and Use Committee. The study design has been described previously (16). Briefly, 54 juvenile rhesus macaques were divided into two groups and immunized intramuscularly with ALVAC-SIV (which expressed SIV Gag, Pol, and Env from a mucosally transmitted/founder [T/F] variant of SIVmac251) at weeks 0, 4, 12, and 24. Additionally, at weeks 12 and 24, both study arms (27 animals each) received a bivalent monomeric gp120 booster immunization formulated in either alum (200 μg/gp120 variant) or MF59 (100 μg/gp120 variant). The control groups consisted of 24 concurrent controls that received either alum, MF59, or nothing at weeks 12 and 24 and 23 historical controls (total, 47) (4, 17). Starting at week 28, all 54 vaccinated and 24 control macaques were challenged weekly with SIVmac251 via the rectal route at a dose of 120 50% tissue culture infective doses (TCID50). Blood and other specimens were collected at intervals for SIV RNA and DNA determinations and for various immunological assays.
Neutralization assay.
SIVmac251 was added at a 1:1 ratio by volume to serially diluted and heat-inactivated (i.e., 56°C for 30 min) preimmune and postimmune plasma samples of vaccinated rhesus macaques, followed by a 1-h incubation at 37°C. TZM-bl reporter cells were then added (1:1 by volume) at 1 × 104 cells per well in a final concentration of 10 μg/ml of DEAE-dextran. After 48 hours of incubation at 37°C, cells were washed with phosphate-buffered saline (PBS) and lysed for 10 min at room temperature with 1× cell lysis reagent (Promega). Supernatants were finally transferred into Costar 96-well white plates, and luminescence in relative light units (RLUs) was measured using a microplate luminometer (BioTek) after adding luciferase assay reagent (Promega). All plasma samples were tested at four different final dilutions (1:80, 1:240, 1:720, and 1:2,160). Assays were repeated four times, each time in duplicate. The extent of virus inhibition in the presence of SIVmac251-specific antibody is reported as the 50% inhibitory concentration (IC50) or as the fraction of neutralization at each dilution (relative to the absence of antibody).
Virus capture assay.
Ninety-six well plates (Corning) were coated with 250 ng (5 μg/ml) of goat anti-monkey IgG gamma chain-specific antibody (Rockland) per well and incubated overnight at 4°C. The next day, plates were washed 3 times with PBS and blocked with 4% nonfat dry milk in PBS at 37°C. After 1 h, plates were washed again 3 times with PBS and incubated with 50 μl of 1:20 diluted (in Dulbecco modified Eagle medium [DMEM] growth medium) preimmune and postimmune plasma samples of vaccinated (or control) rhesus macaques. Plasma samples were incubated for 1 h at 37°C, followed by three washes with PBS. SIVmac251 was diluted in DMEM growth medium and transferred to wells (1 ng p27/well). After 4 hours of incubation at 37°C, plates were washed 5 times with PBS. In a final step, the infectivity of captured virus was determined using TZM-bl reporter cells as described above for neutralization.
SGA and sieve analysis.
All sequences were obtained through single-genome amplification (SGA) and directly sequenced as described previously (16). Individual sequence fragments were assembled and edited using Sequencher 5.0 (Gene Codes). Inspection of individual chromatograms allowed for the confirmation that amplicons were derived from a single viral template. The absence of mixed bases at each nucleotide position throughout the env gene was taken as evidence of a single viral RNA (vRNA)/cDNA template. This quality control measure excluded from the analysis amplicons that resulted from PCR-generated mutations, in vitro recombination, or Taq polymerase errors. All remaining sequences proportionately represent those virions circulating in vivo. All sequence alignments and phylogenetic trees were obtained in ClustalW with manual editing in MacClade. Each low-diversity lineage was compared to a mathematical model of viral diversification over time to identify all T/F lineages. A sieve analysis was performed using the amino acid translation of gp120. Informative sites were identified as polymorphic sites found in more than one individual T/F sequence.
Statistical analyses.
Differences between two sets of continuous values were assessed using the exact Wilcoxon rank sum test. Trends and correlations between paired values were analyzed with Spearman's rank correlation or with the Jonckheere-Terpstra test when the number of discrete values was small, e.g., for the number of T/F variants. Immune outcomes with substantial fractions of zero values were summarized as two groups (= 0 versus >0) or three ordered groups (<0, =0, and >0) for analysis using the Wilcoxon rank sum, Cochran-Armitage, or Jonckheere-Terpstra tests. Rates of infection were compared using the exact log rank test. Sieve analyses were evaluated with the 2-tailed Fisher's exact test.
RESULTS
Capture of infectious virus by antibody from vaccinated animals correlates with T/F variant number.
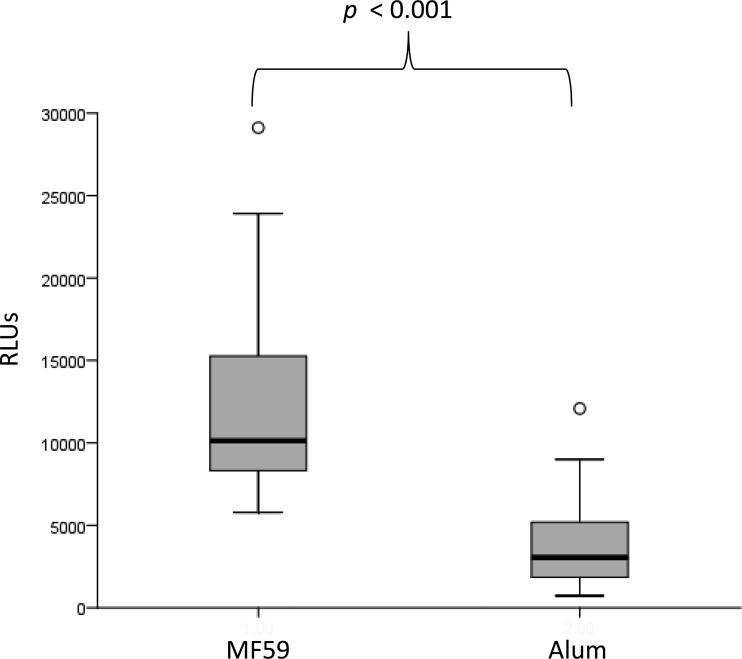
Using plasma obtained 2 weeks after vaccination, we found that capture of infectious SIVmac251 was greater among animals vaccinated with ALVAC-SIV/gp120 in MF59 than among those vaccinated with ALVAC-SIV/gp120 in alum (P < 0.001) (Fig. 1). This finding is consistent with data reported from the parent study indicating that MF59 results in higher antibody responses measured in a number of different assays (16). Although neutralizing activity against a neutralization-sensitive strain of SIV (SIVmac251.6) was higher in plasma of MF59-vaccinated animals, levels of antibody capable of neutralizing the challenge virus, SIVmac251, were very low in both groups (see below). Further analyses were done combining animals from both adjuvant groups.
FIG 1.
Plasma from animals vaccinated with ALVAC-SIV/gp120 in MF59 adjuvant (n = 27) captures more infectious SIVmac251 than that from animals receiving the same immunogen in alum (n = 27). Capture is reported as relative light units (RLUs).
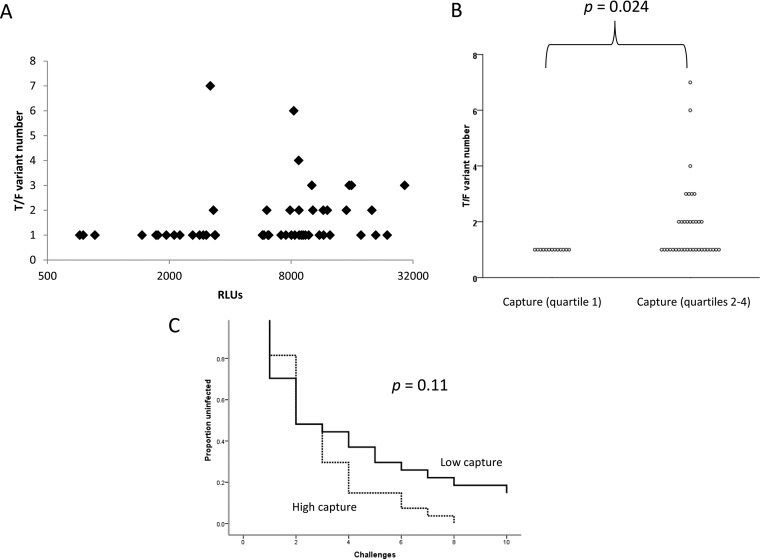
Among animals that became infected (n = 50), capture directly correlated with the number of distinct T/F variants with which the animals were infected following low-dose repeated rectal challenges (r = 0.38, P = 0.0074) (Fig. 2A). It was clear from the scatter plot, however, that the relationship between capture and T/F variant number was not linear. Thus, data were divided into quartiles for further analyses, and the number of T/F variants was found to increase significantly over the quartiles (P = 0.0042; Jonckheere-Terpstra test). Moreover, animals in the lowest quartile of capture activity were universally infected with a single T/F variant, which was significantly fewer than the number of variants infecting the vaccinated animals whose antibody captured more virus (P = 0.024; Mann-Whitney U test corrected for multiple comparisons) (Fig. 2B).
FIG 2.
Antibody capture of infectious SIVmac251 is associated with infection outcomes among animals vaccinated with ALVAC-SIV/gp120 vaccines. (A and B) Antibody capture, reported as a continuous variable in relative light units (RLUs) (A) or as quartilized data (B), is plotted against the number of T/F variants that established infection. (C) There was a trend toward higher infection rates among animals with high capture (greater than the median) than among those with low capture (less than or equal to the median).
Overall, unvaccinated control animals had numbers of T/F variants similar to those in vaccinated animals (P = 0.70), and no significant differences between control and vaccinated animals emerged when animals with high or low capture were compared with the controls. Thus, the level of antibody capable of capturing infectious SIVmac251 influenced the number of T/F variants among vaccinated animals, but no level of capture antibody predicted differences between vaccinated and control animals.
We also determined if antibody capture of infectious SIVmac251 predicted the rate of infection after challenge. When analyzed as a continuous variable, bifurcated at the median, or quartilized, capture was not significantly associated with infection rate. However, animals with more capture trended toward higher rates of infection than those with lower capture, when analyzed as binary data (P = 0.11) (Fig. 2C).
We next explored the relationship between capture antibody and other SIV-specific antibody measurements that had been previously measured in the parent animal study. Among the 23 separate antibody tests evaluated, capture correlated significantly with 13 (Table 1). When adjusted for multiple comparisons, nine of these correlations, which included those between capture and serum or rectal IgG or IgA binding antibodies to Env or V2 and those between capture and neutralization of SIVmac251.6 (a neutralization-sensitive variant of SIVmac251), antibody-dependent cell-mediated cytotoxicity (ADCC), or phagocytosis, maintained a P value of <0.05 (Table 1). Notably, the correlation between capture and neutralization of SIVmac251.6 was the strongest (r = 0.71, P < 5 × 10−6).
TABLE 1.
Correlations between capture of infectious SIVmac251 and other antibody measurementsa
| Parameter | Value for correlation with: |
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Avidity | gp130 IgG | V1V2 IgG | E660 gp140 IgG | E660 V1V2 IgG | 239 V1V2 IgG | Cyclic V2 IgG | E543 cyclic V2 IgG | Cyclic V2 IgA | Cyclic V2 E543 IgA | E660 gp140 IgA | gp140 IgA | Neutralization |
gp130 rectal IgG | Cyclic V2 rectal IgGc | Cyclic V2 sp act | E543 V2 rectal IgGc | E543 V2 rectal IgAc | V2 rectal IgAc | ADCC |
Phagocytosis | |||
| SIVmac251-6 | SIVmac251 (challenge) | Maximum kill | Titer | ||||||||||||||||||||
| Correlation coefficient | 0.002 | 0.50 | 0.31 | 0.54 | 0.17 | 0.32 | 0.49 | 0.59 | 0.16 | 0.27 | 0.53 | 0.35 | 0.71 | −0.066 | 0.64 | −0.16 | −0.33 | −0.20 | −0.27 | −0.22 | −0.16 | 0.52 | 0.45 |
| P valueb | 0.99 | 0.0004 | 0.031 | 0.0001 | 0.25 | 0.026 | 0.0004 | <0.0001 | 0.27 (MW) | 0.061 (JT) | <0.0001 | 0.014 | <0.0001 | 0.65 | <0.0001 | 0.21 (JT) | 0.049 (MW) | 0.21 | 0.085 (JT) | 0.19 (JT) | 0.26 | 0.0001 | 0.0013 |
| No. of samples | 50 | 49 | 49 | 49 | 49 | 49 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 39 | 42 | 48 | 42 | 41 | 41 | 50 | 50 | 50 |
E543 refers to SIVsmE543. Correlation coefficients and P values are Spearman's unless otherwise indicated.
MW, Mann-Whitney test for antibody values of =0 versus >0; JT, Jonckheere-Terpstra test for postvaccination minus prevaccination values of <0 versus =0 versus >0. Significant values (P < 0.05) are in bold; P < 0.004 is significant after correction for multiple comparisons.
Values obtained by subtracting prevaccination from postvaccination results.
Given the associations between capture and T/F variant number and between capture and other antibody measurements, we analyzed the relationship between the other antibody measurements and T/F strain number. Here, we found that T/F strain number correlated with two antibody measurements (Table 2); after correction for multiple comparisons, however, all P values were >0.05. When added to a binary logistic regression model with T/F strain number (bifurcated at the median) as the dependent variable, capture remained the strongest predictor of T/F strain number (not shown).
TABLE 2.
Correlations between number of transmitted/founder variants and antibody measurementsa
| Parameter | Value for correlation with: |
||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Avidity | gp130 IgG | V1V2 IgG | E660 gp140 IgG | E660 V1V2 IgG | 239 V1V2 IgG | Cyclic V2 IgG | E543 cyclic V2 IgG | Cyclic V2 IgA | Cyclic V2 E543 IgA | E660 gp140 IgA | gp140 IgA | Neutralization |
gp130 rectal IgG | Cyclic V2 rectal IgGc | Cyclic V2 sp act | E543 V2 rectal IgGc | E543 V2 rectal IgAc | V2 rectal IgAc | ADCC |
Phagocytosis | |||
| SIVmac251-6 | SIVmac251 (challenge) | Maximum kill | Titer | ||||||||||||||||||||
| Correlation coefficient | −0.073 | 0.20 | −0.028 | 0.18 | −0.056 | −0.011 | 0.12 | 0.11 | 0.017 | 0.000 | 0.25 | 0.23 | 0.30c | −0.086 | 0.22 | 0.00 | −0.28 | 0.27 | −0.056 | 0.046 | −0.27 | 0.18 | 0.32c |
| P valueb | 0.59 | 0.15 | 0.85 | 0.20 | 0.72 | 0.98 | 0.41 | 0.47 | 1.00 | 1.00 | 0.088 | 0.098 | 0.040 | 0.53 | 0.17 | 0.80 (JT) | 0.094 (CA) | 0.065 (JT) | 0.65 (JT) | 0.98 (JT) | 0.060 | 0.21 | 0.034 |
| No. of samples | 50 | 49 | 49 | 49 | 49 | 49 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 50 | 39 | 42 | 48 | 42 | 41 | 41 | 50 | 50 | 50 |
Env proteins and peptides are derived from SIVmac251 unless otherwise indicated; E543 refers to SIVsmE543. Correlation coefficients are Spearman's and P values are Jonckheere-Terpstra unless otherwise indicated.
MW, Mann-Whitney test; JT, Jonckheere-Terpstra test for postvaccination minus prevaccination values of <0 versus =0 versus >0; CA, Cochran-Armitage test. Significant values (P < 0.05) are in bold.
Values obtained by subtracting prevaccination from postvaccination results.
Influence of capture and neutralization on SIV infection after low-dose rectal challenge.
The strong correlation between capture and neutralization of a sensitive SIV strain led us to further evaluate interactions between these two antibody functions. It seemed plausible that antibodies that capture virus but have little or no neutralization activity might enhance infection as measured by the number of T/F variants or the rate at which animals became infected. Plasma neutralizing activity was measured using the neutralization-resistant tier 2 SIVmac251, the strain employed in the capture assay and in challenging the animals. As described in Materials and Methods, four dilutions of plasma were used to measure tier 2 SIVmac251 neutralization in a TZM-bl assay. Even at the lowest plasma dilution (1:80), 50% inhibition (IC50) was not achieved, consistent with the neutralization resistance of SIVmac251. However, there was considerable variability in the fraction of virus inhibited at each dilution, with a range of 0 to 43%. For further analyses, we used the fraction of virus inhibited at a plasma dilution of 1:240 (“neutralization fraction”); similar results were obtained using the fraction of virus inhibited at 1:80, which correlated well with the 1:240 data (r = 0.78, P < 0.0001) (not shown). Neutralization fractions at 1:712 and 1:2,160 were not used in further analyses.
Although there was a strong correlation between capture and neutralization of the sensitive tier 1 SIVmac251.6 (analyzed as IC50), there was no correlation between capture and neutralization fraction using the tier 2 SIVmac251 (r = 0.12, P = 0.40). Moreover, neutralization fraction did not correlate with T/F strain number (r = −0.002, P = 1; Jonckheere-Terpstra test).
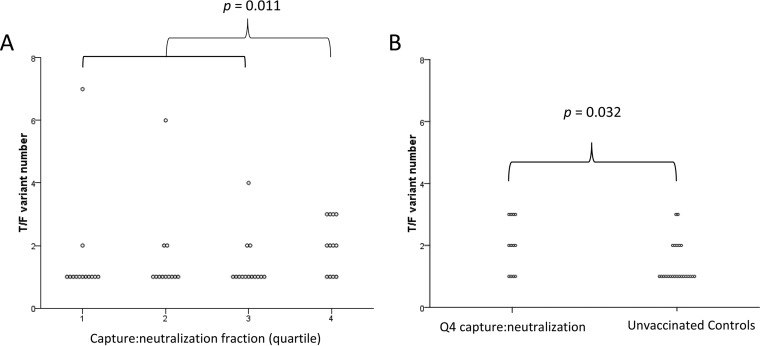
To further explore the notion that antibodies that capture infectious virus but have poor neutralizing activity might enhance infection, we calculated the ratio of capture to neutralization fraction. To a similar extent as capture alone, the ratio of capture to neutralization fraction correlated with the number of T/F variants (r = 0.35, P = 0.017; Jonckheere-Terpstra test) among infected animals. Further analysis revealed a significant association between the capture to neutralization fraction quartile and T/F number (P = 0.018; Jonckheere-Terpstra trend test). Additionally, significantly higher T/F numbers were found in animals in the highest quartile of capture neutralization fraction than in animals in the lower quartiles (P = 0.011 after correction for multiple comparisons) (Fig. 3A). Finally, animals in the highest quartile of capture to neutralization fraction were infected with more T/F strains than were unvaccinated control animals (median of 2 versus 1, respectively; P = 0.032) (Fig. 3B), providing evidence of vaccine-induced antibody-dependent enhancement of infection.
FIG 3.
Ratio of capture to neutralization of SIVmac251 predicts T/F variant number. (A) Animals in the 4th quartile of capture/neutralization ratio were infected with more T/F strains than those in quartiles 1 to 3 (P = 0.011 after adjustment for multiple comparisons). (B) Animals in the 4th quartile of capture/neutralization ratio were infected with a higher number of T/F strains than control animals.
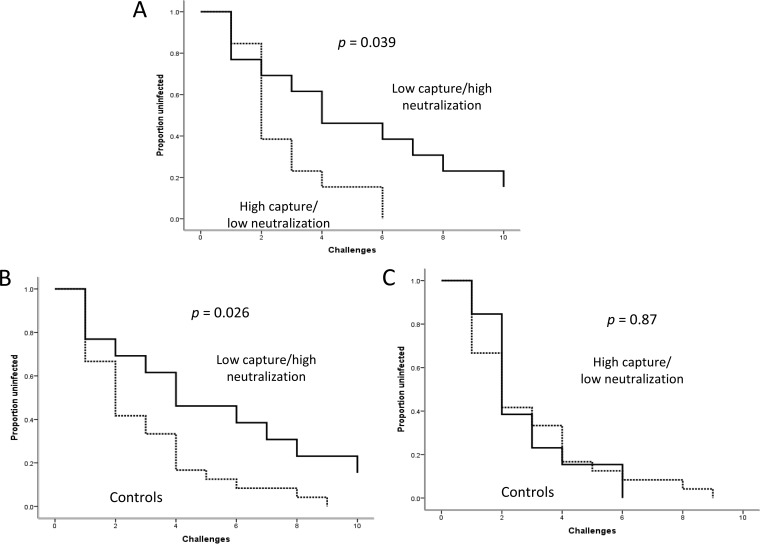
We next determined the influence of capture and neutralization fraction on the rate of infection after challenge. For this analysis, we compared what we surmised would be animals having the most beneficial responses (i.e., those with both the lowest capture and the highest neutralization fraction) with animals having the most deleterious responses (highest capture and lowest neutralization fraction). This comparison demonstrated a significantly lower rate of infection among animals with the beneficial response profile (P = 0.039) (Fig. 4A). In addition, only those animals with the beneficial response profile were protected compared with unvaccinated control animals (P = 0.026) (Fig. 4B and C). Vaccinated animals with mixed responses (i.e., low capture/low neutralization or high capture/high neutralization) were infected at a rate similar to that of those with deleterious responses (not shown). Finally, animals with the deleterious response profile were not infected at a higher rate than controls (Fig. 4C).
FIG 4.
The combination of high capture and low neutralization by vaccine-induced antibody is associated with a higher rate of infection after low-dose repeated rectal challenge with SIVmac251. (A) Survival plots comparing vaccinated animals with high capture (greater than the median) and low neutralization (less than the median) to animals with low capture and high neutralization. (B and C) Animals with low capture and high neutralization have a lower rate of infection than unvaccinated controls (B), whereas animals with high capture and low neutralization have a rate of infection similar to that in controls (C).
Sieve analysis of T/F strain Env sequences.
We analyzed gp120 sequences from T/F strains to determine if any amino acid polymorphisms in Env were associated with the level of infectious virus capture by plasma from vaccinated, infected animals. We thus sought Env polymorphisms that distinguished the highest quartile from the lowest quartile of capture and that also distinguished one of the extreme quartiles from unvaccinated controls. Note that quartiles were assigned based on the entire data set (which included animals that were not infected and thus did not contribute information to the sieve analysis). We found an association between capture and an alanine (A) at amino acid 144 and a proline (P) at amino acid 145, both in variable loop 1 (V1). 144A/145P, (found in 60% of the unique strains in the challenge stock) was observed in 11 of 14 T/F strains (78.6%) from animals with low capture and in 7 of 23 strains (30.4%) from animals with high capture (P = 0.0069) (Table 3). Six of 33 (18.2%) control animals were infected with the 144A/145P variant (P = 0.0002 compared with the low-capture group; P = 0.010 after adjustment for multiple comparisons). These results can be interpreted to indicate that an antibody response resulting in low capture exerted selective pressure on the T/F strain genotype. It is also possible that the high capture response selected non-144A/145P strains; however, this interpretation is made less likely by the fact that the high-capture animals did not differ from the controls with respect to the amino acids at positions 144 and 145. Alternatively, and perhaps more likely, the low capture responses may have uncovered another immune response that ultimately provided the selective pressure.
TABLE 3.
Association of gp120 amino acid polymorphisms with antibody functions
| Amino acid(s) | Phenotype | Fraction (%) |
|---|---|---|
| 144A/145P | High capture | 7/23 (30.4) |
| Low capture | 11/14 (78.6)a | |
| Controls | 6/33 (18.2) | |
| 141P | Low ADCC titer | 2/45 (4.4) |
| High ADCC titer | 9/37 (24.3)a | |
| Controls | 1/33 (3.0) |
Significantly different from value for other phenotype and/or controls (see text).
To determine if plasma neutralizing activity affected the relationship between capture and T/F strain selection, we used quartiles of the capture/neutralization fraction ratio. In this analysis, 13 of 20 low-quartile animals (65%) and 7 of 24 high-quartile animals (29%) were infected with the 144A/145P T/F variant (P = 0.032). Compared with controls, the low-quartile animals were again significantly more likely to be infected with a 144A/145P variant (P = 0.001; P = 0.052 after adjusting for multiple comparisons). These results suggest that the ratio was not a better predictor of the 144A/145P variant than was capture alone. The neutralization fraction itself was not significantly associated with any Env amino acid polymorphisms (not shown).
When the sieve analysis was done using quartiles of neutralization titers against the sensitive SIVmac251.6 strain, we found results nearly identical to those demonstrated for the capture analysis. That is, animals with low neutralization titers were more likely to be infected with the 144A/145P variant than were animals that mounted a high-titer neutralizing response (81.3% versus 30.0%, respectively; P = 0.003); the low-titer animals differed significantly from the control animals as well (P = 0.0001; P = 0.0052 after correction for multiple comparisons) (not shown). Given the close correlation between capture and neutralization of SIVmac251.6 (r = 0.71), it is likely that these results reflect the relationship between capture and selection at positions 144 and 145. If the SIVmac251.6 neutralization titer were itself providing selective pressure, we would likely have observed a difference between animals in the highest quartile and controls, rather than the observed difference between the lowest-quartile animals and the controls.
ADCC expressed as a titer was marginally associated with proline at amino acid 141: 2 of 45 animals with low-titer plasma (4.4%) and 9 of 37 with high-titer plasma (24.3%) were infected with 141P (P = 0.019) (Table 3). One of 33 control animals (3.0%) was infected with 141P (P = 0.015 comparing high-titer and control animals; P = 0.0017 comparing high-titer and both low-titer and control animals combined). With the adjustment for multiple comparisons, the latter P value equals 0.088. Note that because of limited variability in ADCC titers, these analyses compared the upper and lower halves of ADCC titers, rather than the upper and lower quartiles as in other analyses. When ADCC was expressed as the maximal percent target cell lysis, animals in the lowest quartile were less likely to have 141P than were those in the highest quartile (0 of 26 versus 4 of 15; P = 0.014). The highest-quartile animals were more likely to have 141P than controls (P = 0.028) or than the lowest-quartile animals combined with controls (P = 0.0052; P = 0.27 after multiple-comparison correction). Thus, it is possible, though not clear, that high ADCC activity exerted selective pressure on T/F selection during this vaccine trial.
DISCUSSION
In this study, we explored the relationship between ALVAC-SIV/gp120-vaccine-induced antibody capture of infectious SIVmac251 and outcomes of infection following low-dose, repeated rectal SIVmac251 challenge of rhesus macaques. Our principal finding is that capture by plasma antibody is positively associated with the number of transmitted/founder strains that established infection. In addition, although the vaccine elicited little neutralizing antibody activity against the challenge strain, the fraction of virus neutralized at a given concentration of plasma varied considerably from animal to animal, and animals with both low capture and high neutralization were infected at a lower rate than other animals.
Our findings suggest that antibody that captures infectious virus but does not inhibit its infectivity may enhance the likelihood of infection, measured as an increase in the number of T/F strains establishing infection or as the rate of infection (i.e., number of challenges) following exposure. Antibody-dependent enhancement of infection (ADE) with lentiviruses is a well-established in vitro phenomenon and likely plays a role in the pathogenesis of severe dengue virus infections (1, 9, 18–21). With respect to HIV and SIV, whether or not ADE occurs in vivo remains unclear. However, our results support other studies suggesting that vaccine-induced immune responses might have a deleterious effect on outcomes in both human and monkey vaccine trials (2–4, 22).
We noted a very wide range in the ability of antibody to capture virus. As might be expected, capture correlated with the ability of antibody to bind to Env or the V2 loop or to inhibit the neutralization-sensitive SIVmac251.6. Presumably, antibodies capable of binding to any antigen on infectious virions could capture virus; these antigens could be in the form of defective spikes or other nontrimeric components. Antigens on the trimer may also have been targeted by capture antibodies, although if this is true, such antibodies had little, if any, neutralizing activity.
Capture of infectious virus by itself correlated with the number of T/F strains that infected the animals. When analyzing the ratio of capture to neutralization fraction, we found a similar correlation with T/F number; thus, it appeared that neutralization sensitivity provided little additional information to explain the variability in T/F number. Nonetheless, animals in the highest quartile of capture/neutralization ratio were infected with a higher number of T/F variants than were controls, a relationship that was not seen when using quartilized capture data only. Furthermore, incorporating the neutralization data with the capture data described a phenotype that significantly predicted the rate of infection: animals with low capture and high neutralization were infected at a lower rate than animals with high capture and low neutralization. Finally, only animals with the low capture/high neutralization phenotype were afforded protection relative to the unvaccinated control animals.
Overall, our findings point toward a deleterious effect of antibodies when they capture infectious virus but do not inhibit it. Thus, based on the number of T/F variants, animals that respond to ALVAC-SIV/gp120 vaccines with antibodies that capture well but neutralize poorly have worse outcomes than animals that are not vaccinated. This implies that the vaccine-induced antibody response may have increased the likelihood of infection in some animals. Interestingly, in the parent study, we also found a direct correlation between the number of vaccine-induced CD4+ KI67+ cells that expressed either α4β7+ or CD38+ and the number of T/F variants in the MF59 group, suggesting that not only the antibodies that capture well but neutralize poorly but also an increase in virus-susceptible target cells likely affect vaccine efficacy (16).
On the other hand, we could not find direct evidence of enhanced infection relative to that in unvaccinated controls when analyzing the rate at which the animals became infected. However, the parent study was not designed to detect enhancement, and it is possible that a larger number of animals would have been required to observe differences in infection rates between unvaccinated and subgroups of vaccinated animals (16).
Our sieve analysis indicated that animals with low capture were more likely infected with a 144A/145P T/F variant than were animals with high capture or controls. Since polymorphisms in amino acids 144 and 145, which are in V1, distinguish the low-capture animals from both the high-capture and control animals, it is tempting to suggest that low capture itself provides pressure on T/F variant selection. However, another explanation, which seems more likely, is that high capture negates some beneficial vaccine-induced immune response; only in the presence of low capture does that beneficial response provide pressure. Neither neutralizing nor ADCC antibody responses were associated with the 144/145 polymorphisms. Thus, the potential beneficial functional response, if it exists, is not evident from our analysis, either because it was not measured in the parent study or because its effect was too weak to be ascertained in a study of this size (16). Of note, data from the parent study did not reveal Env polymorphisms that overall distinguished the strains infecting vaccinated animals from those infecting control animals.
Animals that received the ALVAC-SIV/gp120 in MF59 adjuvant had higher levels of antibodies that captured infectious virus than those that received the vaccine in alum. In fact, immunogenicity was generally much higher for the MF59 vaccine than for the alum vaccine, as measured by a number of assays (16). Neutralizing activity against the challenge SIVmac251 strain, however, was not detectable when measured as an IC50 in either adjuvant group; moreover, the two groups did not differ when neutralization was measured as the fraction of virus neutralized (not shown). These findings indicate that broadly increasing immunogenicity does not necessarily increase protection and underscore the critical need to evaluate the biological activity of vaccine-induced antibodies, including their antiviral and proviral effects. Finally, we emphasize that our results may apply only to the particular immunogen, challenge virus, and animal model used in the parent vaccine study. Whether or not similar immunogens and adjuvants in humans will have a similar effect is unknown.
ACKNOWLEDGMENTS
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
Funding Statement
Research reported in this publication was supported by the National Institute of Allergy and Infectious Diseases of the National Institutes of Health under award numbers R01AI102715 and R01AI118581 and in part by federal funds from the National Cancer Institute under contract HHSN261200800001E. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
REFERENCES
- 1.Gorlani A, Forthal DN. 2013. Antibody-dependent enhancement and the risk of HIV infection. Curr HIV Res 11:421–426. doi: 10.2174/1570162X113116660062. [DOI] [PubMed] [Google Scholar]
- 2.Buchbinder SP, Mehrotra DV, Duerr A, Fitzgerald DW, Mogg R, Li D, Gilbert PB, Lama JR, Marmor M, Del Rio C, McElrath MJ, Casimiro DR, Gottesdiener KM, Chodakewitz JA, Corey L, Robertson MN, Step Study Protocol Team. 2008. Efficacy assessment of a cell-mediated immunity HIV-1 vaccine (the Step Study): a double-blind, randomised, placebo-controlled, test-of-concept trial. Lancet 372:1881–1893. doi: 10.1016/S0140-6736(08)61591-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Forthal DN, Gabriel EE, Wang A, Landucci G, Phan TB. 2012. Association of Fcgamma receptor IIIa genotype with the rate of HIV infection after gp120 vaccination. Blood 120:2836–2842. doi: 10.1182/blood-2012-05-431361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pegu P, Vaccari M, Gordon S, Keele BF, Doster M, Guan Y, Ferrari G, Pal R, Ferrari MG, Whitney S, Hudacik L, Billings E, Rao M, Montefiori D, Tomaras G, Alam SM, Fenizia C, Lifson JD, Stablein D, Tartaglia J, Michael N, Kim J, Venzon D, Franchini G. 2013. Antibodies with high avidity to the gp120 envelope protein in protection from simian immunodeficiency virus SIV(mac251) acquisition in an immunization regimen that mimics the RV-144 Thai trial. J Virol 87:1708–1719. doi: 10.1128/JVI.02544-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Robinson WE Jr, Montefiori DC, Gillespie DH, Mitchell WM. 1989. Complement-mediated, antibody-dependent enhancement of HIV-1 infection in vitro is characterized by increased protein and RNA syntheses and infectious virus release. J Acquir Immune Defic Syndr 2:33–42. [PubMed] [Google Scholar]
- 6.Robinson WE Jr, Montefiori DC, Mitchell WM. 1990. Complement-mediated antibody-dependent enhancement of HIV-1 infection requires CD4 and complement receptors. Virology 175:600–604. doi: 10.1016/0042-6822(90)90449-2. [DOI] [PubMed] [Google Scholar]
- 7.Homsy J, Meyer M, Tateno M, Clarkson S, Levy JA. 1989. The Fc and not CD4 receptor mediates antibody enhancement of HIV infection in human cells. Science 244:1357–1360. doi: 10.1126/science.2786647. [DOI] [PubMed] [Google Scholar]
- 8.Takeda A, Ennis FA. 1990. FcR-mediated enhancement of HIV-1 infection by antibody. AIDS Res Hum Retroviruses 6:999–1004. [DOI] [PubMed] [Google Scholar]
- 9.Takeda A, Tuazon CU, Ennis FA. 1988. Antibody-enhanced infection by HIV-1 via Fc receptor-mediated entry. Science 242:580–583. doi: 10.1126/science.2972065. [DOI] [PubMed] [Google Scholar]
- 10.Sullivan N, Sun Y, Binley J, Lee J, Barbas CF 3rd, Parren PW, Burton DR, Sodroski J. 1998. Determinants of human immunodeficiency virus type 1 envelope glycoprotein activation by soluble CD4 and monoclonal antibodies. J Virol 72:6332–6338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Guillon C, Schutten M, Boers PH, Gruters RA, Osterhaus AD. 2002. Antibody-mediated enhancement of human immunodeficiency virus type 1 infectivity is determined by the structure of gp120 and depends on modulation of the gp120-CCR5 interaction. J Virol 76:2827–2834. doi: 10.1128/JVI.76.6.2827-2834.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haynes BF, Gilbert PB, McElrath MJ, Zolla-Pazner S, Tomaras GD, Alam SM, Evans DT, Montefiori DC, Karnasuta C, Sutthent R, Liao HX, DeVico AL, Lewis GK, Williams C, Pinter A, Fong Y, Janes H, DeCamp A, Huang Y, Rao M, Billings E, Karasavvas N, Robb ML, Ngauy V, de Souza MS, Paris R, Ferrari G, Bailer RT, Soderberg KA, Andrews C, Berman PW, Frahm N, De Rosa SC, Alpert MD, Yates NL, Shen X, Koup RA, Pitisuttithum P, Kaewkungwal J, Nitayaphan S, Rerks-Ngarm S, Michael NL, Kim JH. 2012. Immune-correlates analysis of an HIV-1 vaccine efficacy trial. N Engl J Med 366:1275–1286. doi: 10.1056/NEJMoa1113425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Burton DR, Hessell AJ, Keele BF, Klasse PJ, Ketas TA, Moldt B, Dunlop DC, Poignard P, Doyle LA, Cavacini L, Veazey RS, Moore JP. 2011. Limited or no protection by weakly or nonneutralizing antibodies against vaginal SHIV challenge of macaques compared with a strongly neutralizing antibody. Proc Natl Acad Sci U S A 108:11181–11186. doi: 10.1073/pnas.1103012108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kostrikis LG, Cao Y, Ngai H, Moore JP, Ho DD. 1996. Quantitative analysis of serum neutralization of human immunodeficiency virus type 1 from subtypes A, B, C, D, E, F, and I: lack of direct correlation between neutralization serotypes and genetic subtypes and evidence for prevalent serum-dependent infectivity enhancement. J Virol 70:445–458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lund O, Hansen J, Soorensen AM, Mosekilde E, Nielsen JO, Hansen JE. 1995. Increased adhesion as a mechanism of antibody-dependent and antibody-independent complement-mediated enhancement of human immunodeficiency virus infection. J Virol 69:2393–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Vaccari M, Gordon SN, Fourati S, Schifanella L, Liyanage NPM, Cameron M, Keele BF, Shen X, Tomaras GD, Billings E, Rao M, Chung AW, Dowell KG, Bailey-Kellogg C, Brown EP, Ackerman ME, Vargas-Inchaustequi DA, Whitney S, Doster MN, Binello N, Pegu P, Montefiori DC, Foulds K, Quinn DS, Donaldson M, Liang F, Loré K, Roederer M, Koup RA, McDermott A, Ma Z-M, Miller CJ, Phan TB, Forthal DN, Blackburn M, Caccuri F, Bissa M, Ferrari G, Kalyanaraman V, Ferrari MG, Thompson D, Robert-Guroff M, Ratto-Kim S, Kim JH, Michael NL, Phogat S, Barnett SW, Tartaglia J, Venzon D, Stablein DM, Alter G, Sekaly R-P, Franchini G. 2016. Adjuvant-dependent innate and adaptive immune signatures of risk of SIVmac251 acquisition. Nat Med 22:762–770. doi: 10.1038/nm.4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Strbo N, Vaccari M, Pahwa S, Kolber MA, Doster MN, Fisher E, Gonzalez L, Stablein D, Franchini G, Podack ER. 2013. Novel vaccination modality provides significant protection against mucosal infection by highly pathogenic simian immunodeficiency virus. J Immunol 190:2495–2499. doi: 10.4049/jimmunol.1202655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Robinson WE Jr, Montefiori DC, Mitchell WM. 1987. A human immunodeficiency virus type 1 (HIV-1) infection-enhancing factor in seropositive sera. Biochem Biophys Res Commun 149:693–699. doi: 10.1016/0006-291X(87)90423-2. [DOI] [PubMed] [Google Scholar]
- 19.Schutten M, Andeweg AC, Bosch ML, Osterhaus AD. 1995. Enhancement of infectivity of a non-syncytium inducing HIV-1 by sCD4 and by human antibodies that neutralize syncytium inducing HIV-1. Scand J Immunol 41:18–22. doi: 10.1111/j.1365-3083.1995.tb03528.x. [DOI] [PubMed] [Google Scholar]
- 20.Halstead SB, Chow JS, Marchette NJ. 1973. Immunological enhancement of dengue virus replication. Nat New Biol 243:24–26. [PubMed] [Google Scholar]
- 21.Halstead SB. 1979. In vivo enhancement of dengue virus infection in rhesus monkeys by passively transferred antibody. J Infect Dis 140:527–533. doi: 10.1093/infdis/140.4.527. [DOI] [PubMed] [Google Scholar]
- 22.Gilbert PB, Peterson ML, Follmann D, Hudgens MG, Francis DP, Gurwith M, Heyward WL, Jobes DV, Popovic V, Self SG, Sinangil F, Burke D, Berman PW. 2005. Correlation between immunologic responses to a recombinant glycoprotein 120 vaccine and incidence of HIV-1 infection in a phase 3 HIV-1 preventive vaccine trial. J Infect Dis 191:666–677. doi: 10.1086/428405. [DOI] [PubMed] [Google Scholar]