ABSTRACT
ADP-ribosylation is a posttranslational protein modification in which ADP-ribose is transferred from NAD+ to specific acceptors to regulate a wide variety of cellular processes. The macro domain is an ancient and highly evolutionarily conserved protein domain widely distributed throughout all kingdoms of life, including viruses. The human TARG1/C6orf130, MacroD1, and MacroD2 proteins can reverse ADP-ribosylation by acting on ADP-ribosylated substrates through the hydrolytic activity of their macro domains. Here, we report that the macro domain from hepatitis E virus (HEV) serves as an ADP-ribose-protein hydrolase for mono-ADP-ribose (MAR) and poly(ADP-ribose) (PAR) chain removal (de-MARylation and de-PARylation, respectively) from mono- and poly(ADP)-ribosylated proteins, respectively. The presence of the HEV helicase in cis dramatically increases the binding of the macro domain to poly(ADP-ribose) and stimulates the de-PARylation activity. Abrogation of the latter dramatically decreases replication of an HEV subgenomic replicon. The de-MARylation activity is present in all three pathogenic positive-sense, single-stranded RNA [(+)ssRNA] virus families which carry a macro domain: Coronaviridae (severe acute respiratory syndrome coronavirus and human coronavirus 229E), Togaviridae (Venezuelan equine encephalitis virus), and Hepeviridae (HEV), indicating that it might be a significant tropism and/or pathogenic determinant.
IMPORTANCE Protein ADP-ribosylation is a covalent posttranslational modification regulating cellular protein activities in a dynamic fashion to modulate and coordinate a variety of cellular processes. Three viral families, Coronaviridae, Togaviridae, and Hepeviridae, possess macro domains embedded in their polyproteins. Here, we show that viral macro domains reverse cellular ADP-ribosylation, potentially cutting the signal of a viral infection in the cell. Various poly(ADP-ribose) polymerases which are notorious guardians of cellular integrity are demodified by macro domains from members of these virus families. In the case of hepatitis E virus, the adjacent viral helicase domain dramatically increases the binding of the macro domain to PAR and simulates the demodification activity.
INTRODUCTION
The ADP ribosyltransferase (ART) enzymes covalently attach the ADP-ribose moiety of NAD+ to either amino acid side chain of a target protein or a growing poly(ADP-ribose) (PAR) chain. Downstream of this signaling event, at least four families of protein modules bind ADP-ribose and its metabolites: macro domains, PAR binding zinc finger (PBZ) domains, WWE domains, and the PAR binding motif (PBM) (1, 2). They participate in a variety of cellular processes, such as DNA damage response, protein stability, assembly of stress granules, chromatin structure, and cell death (3, 4).
In these pathways, ADP-ribose acts as a posttranslational tag that must be efficiently added, recognized, or removed in a timely manner. ADP-ribosylation is thus a highly dynamic and reversible process (3). To date, three kinds of proteins have been reported to reverse protein ADP-ribosylation: poly(ADP-ribose) glycohydrolases (PARG), ADP-ribosyl hydrolases (ARH), and several cellular macro domain-containing proteins (2, 3, 5).
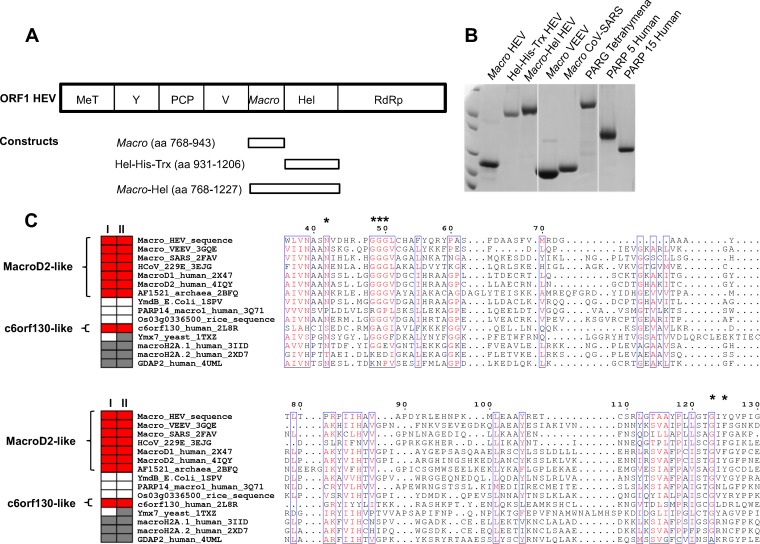
The macro domain fold is ancient and highly conserved through evolution (Fig. 1). It is widely distributed throughout all kingdoms of life, including viruses (reviewed in reference 6). Macro domains bind to ADP-ribose, PAR, poly(A), and O-acetyl-ADP-ribose and may act as enzymes (7–11). The human TARG1/C6orf130 protein removes the whole PAR chain from poly(ADP)-ribosylated proteins, including the last ADP-ribose unit covalently connected to the protein surface in reactions, here termed de-PARylation and de-MARylation for the removal of poly(ADP-ribose) and mono-ADP-ribose (MAR), respectively (5, 11).
FIG 1.
Proteins relevant to this study. Predicted macro domain (amino acids [aa] 789 to 902) and Hel domain (amino acids 974 to 1185) inside ORF1 of the HEV genome (MeT, methyltransferase; Y, Y domain; PCP, papain-like cysteine protease; V, hypervariable proline-rich hinge; macro: macro domain; Hel, helicase; RdRP, RNA-dependent RNA polymerase). Rectangles show the produced proteins, as indicated. (B) Coomassie blue-stained SDS-PAGE gel showing the purified macro domain (macro-HEV), Hel (Hel-His-Trx HEV), macro-Hel, VEEV macro domain, SARS-CoV macro domain, Tetrahymena thermophila PARG, PARP5a catalytic domain (PARP5 human), and PARP15 catalytic domain (PARP15 human). (C) Structure-based sequence alignment of selected macro domains. Red rectangles, confirmed A1″Pase (I) or de-MARylation (II) activity; gray rectangles, no activity; blank rectangle, not tested. Activity data are based on the results of this study and previously published data (2). Macro domains carrying both A1″Pase and de-MARylation activities are further divided into MacroD2- and c6orf130-like subgroups, based on sequence conservation and catalytic mechanisms. Asterisks indicate important amino acids for PAR binding and/or de-MARylation activity of the MacroD2-like subgroup. E. coli, Escherichia coli; Trx, thioredoxin.
A set of positive-strand RNA viruses, including Coronaviridae, Togaviridae, and Hepeviridae, encode a macro domain. The crystal structures of macro domains from several coronaviruses (e.g., severe acute respiratory syndrome coronavirus [SARS-CoV])and Togaviridae (e.g., chikungunya virus [CHIKV] and Venezuelan equine encephalitis virus [VEEV]) have been determined with and without bound ADP-ribose (8, 12). These studies suggested a close phylogenetic and most probably functional relationship between viral and cellular macro domains. Although several studies have shown that viral macro domains carry ADP-ribose, PAR binding, and ADR-ribose-1″-phosphate phosphatase (A1″Pase) activities, an ADP-ribose–protein hydrolase activity has not been demonstrated yet. Viral macro domains interact with poly(ADP-ribose) polymerase 1 (PARP1) in intact cells (13) and possibly modulate virus production (14, 15), virulence (16), apoptotic cell death (17), and type I interferon (IFN) induction (18).
Hepatitis E virus (HEV) is the sole member of the Hepeviridae family. It currently represents the most frequent cause of acute viral hepatitis and jaundice in the world (19). HEV infection occurs sporadically or in epidemics, causing significant morbidity and death, especially during pregnancy, with a fatality/case rate of up to 20% (20). No specific antiviral drug or vaccine is licensed globally. HEV has a single-stranded positive-sense 5′-capped and 3′-polyadenylated RNA genome of 7.2 kb. The genome contains three open reading frames (ORFs), of which ORF1 encodes the nonstructural protein, ORF2 encodes the capsid protein, and ORF3 encodes a small multifunctional protein (21). ORF1 is translated into a nonstructural polyprotein of ∼186 kDa. This ORF1 polyprotein contains several putative functional domains including a macro domain adjacent to an SF1 family RNA helicase (Hel) domain (Fig. 1A) (22).
The HEV macro domain is endowed with A1″Pase activity. It binds to poly(ADP-ribose) and poly(A) in vitro (8, 9) and is a putative IFN antagonist that modulates the host immune response (14, 18). The recombinant HEV helicase is able to unwind RNA duplexes bearing 5′ overhangs in an ATPase-dependent manner (23) and carries a 5′ RNA triphosphatase (RTPase) activity presumably involved in RNA capping (24). Interestingly, in addition to the macro domain, the Hel domain has also been shown to modulate HEV pathogenicity (25–27), supposedly through interaction with viral and/or host proteins other than helicase-associated enzymatic activities (28).
Here, we show that the HEV macro domain can reverse auto-ADP-ribosylation of several ARTs in vitro. Abrogation of this activity through mutations in the macro domain dramatically decreases replication of an HEV subgenomic replicon in Huh7 cells. The de-MARylation activity is also observed in macro domains from SARS-CoV, human coronavirus 229E (HCoV-229E), and VEEV. The HEV helicase domain greatly stimulates binding of the macro domain to free PAR and PARylated protein. The reversion of cellular ADP-ribosylation by a viral macro domain suggests a novel connection between viral enzymes and host cell signaling proteins.
MATERIALS AND METHODS
PAR binding assay.
Radiolabeled PAR was synthesized by auto-ADP-ribosylation of the catalytic fragment of PARP5a (29). A 500-μl reaction mixture containing 2 μM PARP5, 200 μM NAD+, 50 μCi of [32P]NAD+ (800 Ci/mmol; PerkinElmer), 100 mM Tris (pH 8.0), 10 mM MgCl2, and 2 mM dithiothreitol (DTT) was incubated at room temperature (RT) for 2 h. The reaction mixture was used with no additional treatment for binding assays. Alternatively, free PAR was obtained by treating the reaction product with 200 nM proteinase K at 37°C for 1 h, followed by heating at 70°C for 30 min.
PAR binding assays used either the polymer bound to PARP5a or a protein-free preparation (9). Serial dilutions of macro domain-containing proteins were blotted onto a nitrocellulose membrane using a Minifold I slot blot apparatus (Acrylic). The membrane was blocked for 1 h in TBS-T (10 mM Tris [pH 7.4], 150 mM NaCl, and 0.05% Tween) containing 5% nonfat milk powder. The membrane was incubated for 1 h with the poly(ADP-ribose) preparation (500 μl) diluted in 30 ml of TBS-T, subjected to three 10-min washes in TBS-T, dried, and autoradiographed.
Competitive binding assays between poly(ADP-ribose) and RNA to macro-Hel.
Synthetic PAR concentration (29; also data not shown) was determined using the following equation: [PAR] = (A258) cm−1/(13,500 cm−1 M−1). PAR was analyzed using denaturing PAGE and stained with SYBR Gold stain (S-11494; Thermo Fisher). Binding to a macro domain-Hel fusion (macro-Hel) used fluorescein-labeled RNA (Cy3-5′-CCAGGCGACAUCAGCG-3′) in fluorescence polarization (FP) assays and standard procedures with a microplate reader (PHERAstar FS; BMG Labtech). The Kd (dissociation constant) was calculated from standard saturating binding analysis. A competitive binding assay was carried out in 50 μl of reaction buffer [50 mM Tris, pH 7.5, 100 mM NaCl, 2 mM DTT, 100 nM Cy3-RNA, 500 μM macro-Hel, and decreasing concentrations of poly(ADP-ribose) or unlabeled RNA (5′-CCAGGCGACAUCAGCG-3′)].
De-MARylation and de-PARylation assays.
Mono- or poly(ADP)-ribosylated substrates were prepared by auto-ADP-ribosylation of the catalytic fragment of PARP1, PARP3, PARP5a, PARP10, or PARP15. PARP1, PARP3, and PARP10 PARylations and de-PARylations have been described previously (30). For a single reaction, 0.1 μM PARP1 was used in a 20-μl final volume with either a 0.5 μM or 5 μM NAD+ concentration, including [32P]NAD+. The reaction was terminated with PARP1 inhibitor PJ34 (2.5 μM) to avoid additional PARylation activity that could compete with the de-PARylation reaction. The reaction products were then incubated with macro domains at 0.5 μM final concentrations. For PARP3, the final NAD+ concentration in the reaction mixture was 0.5 μM. For PARP5a and PARP15, a 200-μl reaction mixture containing 2 μM PARP5a or PARP15, 5 μM (for mono-ADP-ribosylation) or 50 μM [for poly(ADP)-ribosylation] NAD+, 50 μCi of [32P]NAD+ (800 Ci/mmol; PerkinElmer), 100 mM Tris (pH 8.0), and 10 mM MgCl2 was incubated at RT for 2 h. The reaction mixture was loaded onto His-Mag agarose beads (Merck Millipore), subjected to three 5-min washes in 200 μl of washing buffer (50 mM Tris, pH 7.5, 1 M NaCl, 10 μM ATP, and 100 μM NAD+) (31), followed by three 5-min washes in 200 μl of reaction buffer (50 mM Tris, pH 7.5, 50 mM NaCl, 2 mM DTT, 50 mM imidazole, and 100 μM NAD+), and finally resuspended in 100 μl of the reaction buffer. Unless stated otherwise, de-MARylation and de-PARylation reactions were performed at 30°C for 30 min or 1 h. For the mutants shown in Table 1, quantification of released 32P-labeled ADP-ribose was done in triplicate, and results were normalized to the value of the negative control (wild type). After incubation, half of the reaction mixture was subjected to SDS-PAGE (10%) to analyze the remaining radioactive signal of either PARP15 or PARP5, and the second half was loaded on a 7 M urea-PAGE (20%) gel to resolve the released products, mono- and/or poly(ADP-ribose), which were autoradiographed (Fujifilm FLA-3000) and quantitated (Image Gauge, version 4.0). Cold ADP-ribose detected by UV shadowing was used as a migration standard.
TABLE 1.
Effect of amino acid substitutions on macro-Hel-mediated de-MARylation of the PARP15 catalytic domain
| Hel protein | Location of the mutation(s) | Relative activity (%)a |
|---|---|---|
| Wild-type macro-Hel | 100 | |
| N42A | Macro domain | 40.3 ± 7.5 |
| G50A | Macro domain | 44.9 ± 14.1 |
| G48S G49S G50A | Macro domain | 8.1 ± 5.6 |
| G48S G49S | Macro domain | 16.5 ± 3.0 |
| G123A | Macro domain | 28.7 ± 4.6 |
| I124A | Macro domain | 85.3 ± 2.3 |
| Y125F | Macro domain | 98.1 ± 3.6 |
| K215A | Helicase domain (Walker A motif) | 107.3 ± 15.0 |
| Q291A | Helicase domain (motif III) | 110.0 ± 10.3 |
| R321A | Helicase domain (motif IIIa) | 90.7 ± 13.6 |
| Q378A | Helicase domain (motif Va) | 90.9 ± 5.7 |
| R401A | Helicase domain (motif VI) | 100.8 ± 9.9 |
| D431A | Helicase domain | 99.2 ± 11.5 |
Compared to that of the wild-type macro-Hel, set at 100%.
Replication assay.
The genotype 1 Sar55/S17/luc replicon-encoding plasmid was a kind gift from Suzanne U. Emerson (National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD). Huh7 cells were seeded into six-well plates at 2 × 105 cells per well and transfected with capped Sar55/S17/luc RNA transcripts (1 μg per well) 24 h later using Lipofectin (Life Technologies) as previously described (32). After samples were incubated for 72 h at 35°C, Gaussia luciferase activity was measured in 20 μl of culture medium with a Renilla luciferase assay system (Promega).
RESULTS
The HEV macro domain interacts with PAR in a helicase-stimulated manner.
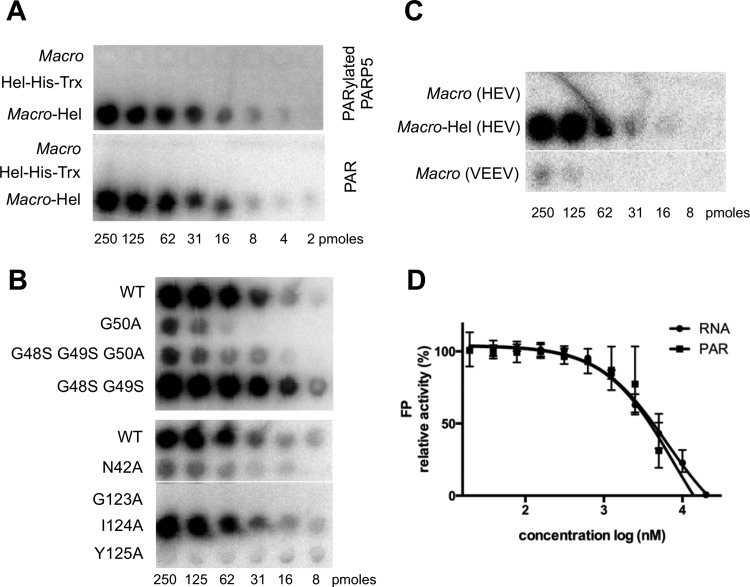
Macro domains, e.g., MacroD1 and viral macro domains of HEV, Semliki Forest virus (SFV), VEEV, and SARS-CoV, bind PAR (9, 12). Since these domains are often included in multidomain proteins, we assessed the binding ability of the macro domain, Hel, and macro-Hel from HEV to PAR and the poly(ADP)ribosylated-PARP5a catalytic domain. The macro-Hel fusion strongly binds PAR and poly(ADP)-ribosylated PARP5a, whereas either the macro domain or helicase domain alone does not (Fig. 2A).
FIG 2.
PAR binding studies. 32P-labeled PARylated PARP5a or free PAR was incubated with decreasing concentrations of proteins, dot blotted, and imaged. (A) Binding of the macro domain, Hel, and macro-Hel of HEV to PARylated PARP5a and free PAR. (B) Effects of mutations on the binding of macro-Hel to free PAR. (C) Binding of the HEV macro domain, macro-Hel (HEV), and the VEEV macro domain to free PAR. (D) Poly(ADP-ribose) competitively inhibits the binding of Cy3-labeled RNA to macro-Hel. WT, wild type.
Replacing N42, G50, G123, and Y125 with alanine (A) residues dramatically decreased the binding of macro-Hel to PAR (Fig. 2B), whereas mutations within the helicase domain did not (data not shown). The PAR binding ability of the VEEV macro domain was compared using the same assay. An approximately 10-fold-weaker PAR binding is observed for VEEV than for macro-Hel (Fig. 2C): loss of detectable binding occurs around 62 nM and 8 nM, respectively. The presence of the helicase domain confers a PAR-binding ability to the HEV macro-Hel construct that is much higher than that of the VEEV or HEV macro domain alone.
Since helicases bind RNA and DNA, the competition between PAR and a labeled RNA was measured to further understand the binding mode of macro-Hel to PAR. Exogenous PAR inhibits the binding of a labeled RNA to macro-Hel, with a potency similar to that of RNA (Fig. 2D) (apparent Ki of PAR [Ki App(PAR)] = 7.4 ± 2 μM; Ki App(unlabeled RNA) = 6.0 ± 1.1 μM), indicating that the HEV helicase domain in cis helps the binding of PAR and/or RNA, possibly by a common binding surface.
The HEV macro domain removes PAR from poly(ADP)-ribosylated ART substrates.
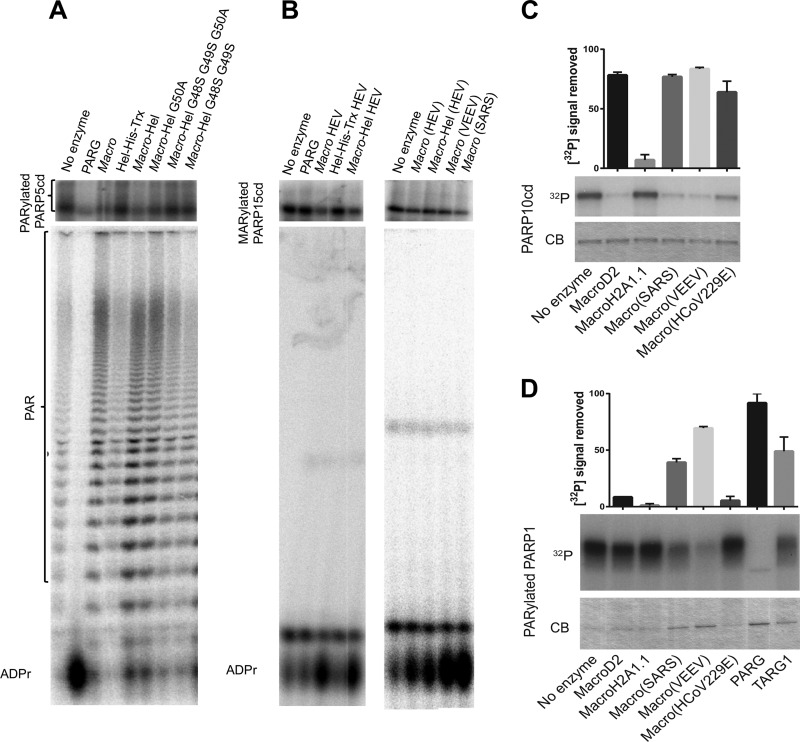
We tested the ability of the macro domain, Hel, and macro-Hel to remove PAR attached to the catalytic domain of PARP5a (Fig. 3A). Macro domain and macro-Hel proteins remove PAR chains from poly(ADP)-ribosylated PARP5a, as judged by a decrease in radiolabel from this protein (Fig. 3A, top) with a concomitant increase of free PAR (Fig. 3A, bottom). Substitutions of the three conserved glycines (positions 48, 49, and 50) of the macro domain (Fig. 1A) decrease this activity. In contrast to PARG that acts on internal bonds within the PAR chain to release free ADP-ribose, the macro and macro-Hel domains remove the whole PAR chain from PARP5a. Similar results were obtained using SARS-CoV and VEEV macro domains together with auto-PARylated PARP1 and PARP3 substrates (data not shown). The macro domain-mediated PAR removal activity resembles that of the human macro domain-containing protein TARG1/c6orf130 (5, 31). We thus measured the de-MARylation activity of the macro domain and macro-Hel.
FIG 3.
Effects of HEV macro-Hel and other macro domains on PARylated PARP5 and PARP 1 and on MARylated PARP15 and PARP10. SDS–15% PAGE (upper) and 7 M urea–20% PAGE (lower) autoradiography showing the following: de-PARylation of the PARP5a catalytic domain (PARP5cd) by T. thermophila PARG, the macro domain, Hel (Hel-His-Trx), macro-Hel wild type and mutants G50A, G48S-G49S-G50A, and G48S-G49S (A); de-MARylation of the PARP15 catalytic domain by the human PARG, the macro domain, Hel, macro-Hel as well as by the VEEV and SARS macro domains on mono-ADP-ribosylated. Identification of ADP-ribose was based on comigration with cold ADP-ribose detected using UV shadowing. (C) De-MARylation of the PARP10 catalytic domain by different macro domains as observed by SDS-PAGE with Coomassie blue staining (CB) or autoradiography (32P). (D) De-PARylation of PARP1 by different macro domains as observed by SDS-PAGE with Coomassie blue staining (CB) or autoradiography (32P). Quantifications of 32P signal removed are shown above. Error bars indicate standard deviations (n = 2). ADPr, ADP-ribose.
Viral macro domains release ADP-ribose from mono-ADP-ribosylated ART substrates.
The macro and macro-Hel domains remove the mono-ADP-ribose from a radiolabeled mono-ADP-ribosylated PARP15 catalytic domain (Fig. 3B). The Hel domain and human PARG are unable to do so under a variety of experimental conditions (Fig. 3B). In addition, macro domains of VEEV and SARS-CoV have a slightly higher ability to excise mono-ADP-ribose from PARP15 than the HEV macro domain (Fig. 3B), suggesting that the reversal of ADP-ribosylation from proteins is a common function carried by viral macro domains. To further probe the substrate spectrum of various viral macro domains, macro domains of VEEV, SARS-CoV, and human coronavirus 229E (HCoV-229E) were assayed using ADP-ribosylated PARP1 and PARP10 catalytic domains and compared to the already characterized MacroD2. All the viral macro domains exhibit activity in a range comparable to the activity of MacroD2 (Fig. 3C and D). Interestingly, the macro domain from the mildly pathogenic HCoV-229E does not show significant activity on PARP1 specifically (Fig. 3D and data not shown). Table 2 summarizes the macro domain activities addressed in the present study.
TABLE 2.
Summary of macro domain activities addressed in this studya
| Activity | MacroD2 | MacroH2A1.1 | HEV macro domain | HEV macro-Hel | SARS macro domain | VEEV macro domain | HCoV-229E macro domain |
|---|---|---|---|---|---|---|---|
| De-MARylation of PARP10cd and/or PARP15cd | + | − | + | + | + | + | + |
| De-PARylation of PARP1 and/or PARP5cd | − | − | ± | + | + | ± | − |
cd, catalytic domain; +, activity detected; −, no activity detected; ±, low-level activity detected.
Amino acid substitutions were introduced into the macro-Hel domain to determine critical amino acids for macro-Hel-mediated de-MARylation of PARP15 (Table 1). The N42A, G50A, G123A, and I124A substitutions moderately decreased the catalytic activity of macro-Hel toward mono-ADP-ribosylated PARP15. The G48S-G49S and the triple G48S-G49S-G50A substitutions dramatically decreased the activity. In light of the PAR binding results (Fig. 2), these results suggest that N42, G50, and G123 are critical for substrate binding, thereby moderating catalytic activity. The Y125A mutation yielded an inactive enzyme (data not shown), but the purified protein behaved anomalously in solution, an indication that it was probably misfolded (data not shown). The Y125 position was further studied through replacement with phenylalanine (F) since F is also frequently present at this position in various macro domains (Fig. 1C). That the catalytic activity of Y125F is comparable to that of the wild-type protein indicates that the presence of an aromatic ring in this position might be essential for this activity. Interestingly, the double G48S-G49S substitution dramatically decreased the de-MARylation activity, with almost no effect on PAR binding (Fig. 2B), suggesting that the conserved macro domain glycine loop is into or close to the catalytic site.
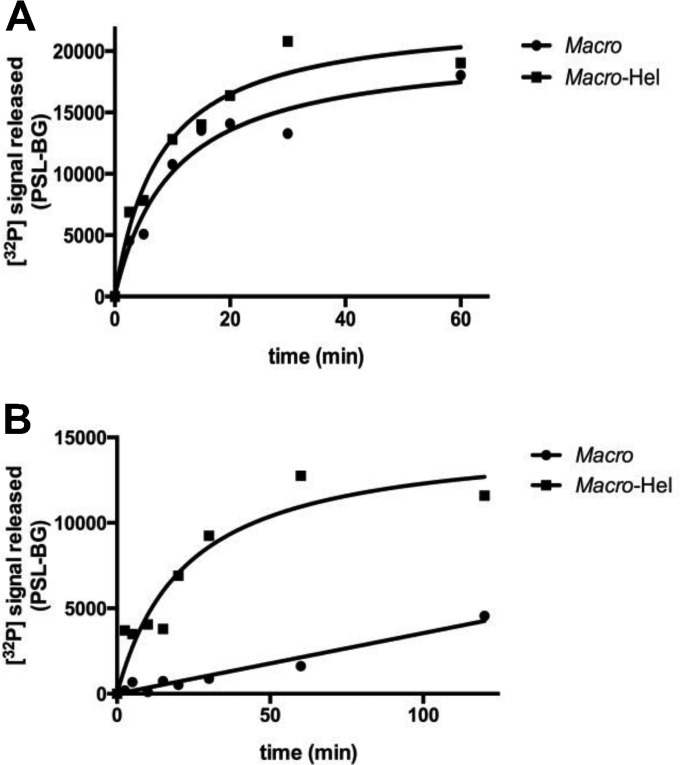
The HEV helicase domain stimulates de-PARylation but not de-MARylation.
We performed time courses of de-MARylation and de-PARylation to further understand the role of the HEV helicase domain in these reactions. The macro and macro-Hel domains show comparable kinetics for de-MARylation (Fig. 4A and data not shown), whereas macro-Hel shows de-PARylation activity ∼11-fold higher than that of the macro domain alone (Fig. 4B and data not shown). Single amino acid mutations within functionally essential helicase domains, including the RNA binding site (Table 3), have no significant effect on PAR binding (data not shown) or de-MARylation (Table 1). PAR can compete with RNA (Fig. 2D), indicating that both polymers might bind to the same surface. We could not find a mutation abolishing specifically RNA binding. Single amino acid substitutions are not sufficient to impact RNA binding to the HEV helicase (33), which generally involves numerous atomic contacts on a protein surface. We propose that the presence of the Hel domain stimulates de-PARylation through PAR binding, whereas it has no effect on de-MARylation.
FIG 4.
Time course of macro domain and macro-Hel activity on mono-ADP-ribosylated PARP15 (A) and poly(ADP)-ribosylated PARP5a (B) catalytic domains. The auto-ADP-ribosylated PARP5a or -15 catalytic domain was incubated with 100 nM macro domain or macro-Hel at 30°C for 1 h or 2 h. PSL-BG, photo-stimulated luminescence counts minus background counts.
TABLE 3.
List of macro-Hel mutants in the helicase domain
| Hel protein | Predicted functiona | Location of mutationa | Relative activity (%)b |
|
|---|---|---|---|---|
| ATPase | ssRNA binding | |||
| Wild type | 100 | 100 | ||
| K215A | NTP binding | Walker A motif | 5.6 ± 4.5 | 54.9 ± 1.6 |
| Q291A | NTP binding | Motif III | 0 | 115.7 ± 7.11 |
| R321A | NTP binding | Motif IIIa | 0.4 ± 1.4 | 54.2 ± 3.4 |
| Q378A | NTP binding | Motif Va | 39.0 ± 4.2 | 120.7 ± 2.7 |
| R401A | ssRNA binding | Motif VI | 72.6 ± 8.6 | 90.7 ± 2.6 |
| D431A | NDc | 79.0 ± 2.5 | 59 ± 3.6 | |
The function and location of mutated residues were predicted based on the sequence alignment with the SF1 helicase from tomato mosaic virus (50).
The ATPase and ssRNA binding activities were determined relative to that of the wild type, which was set as 100%. Values are means ± standard deviations (n = 3).
ND, not determined.
Amino acids involved in de-MARylation and de-PARylation are essential for HEV replication.
Amino acid substitutions were introduced into an HEV genotype 1 Sar55/S17/luc replicon. As expected, a single amino acid K215A or R321A substitution within the nucleoside triphosphate (NTP)-binding/hydrolysis site (Table 3) of the helicase domain completely abrogated HEV replication in Huh7 cells (Fig. 5). Of other mutant replicons bearing a mutation within the macro domain, N42A, G50A, and G123A resulted in a moderate decrease of replication of the HEV replicon, whereas G48S-G49S and the triple mutant G48S-G49S-G50A showed poor replication compared to that of the wild type (∼20% and 10%, respectively). In light of the macro-Hel enzymatic assays, these results suggest that the macro domain fulfills an essential role in HEV replication, possibly through expression of its de-MARylation or de-PARylation activities.
FIG 5.
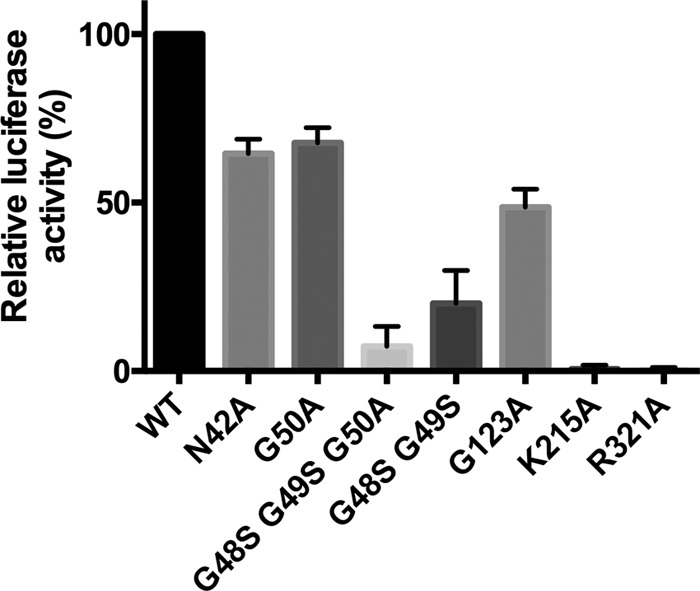
Effect on HEV replication of mutations within the macro-Hel domain. Huh7 cells were transfected with HEV genotype 1 Sar55/S17/luc wild-type (WT) or capped mutant replicon RNA. Luciferase activity was measured after 72 h of incubation at 35°C.
The putative catalytic mechanism de-MARylation is consistent with the known A1″Pase activity.
There are three documented protein families grouping hydrolases that are able to use ADP-ribose-containing substrates (34), i.e., ARH type, PARG type, and macro domain-containing proteins. All of these proteins can be classified into two main groups. The first group comprises poly(ADP-ribose) glycohydrolases like PARG and ARH3, which act on the glycosidic ribose-ribose bond to leave MARylated proteins. The second group comprises ADP-ribose protein hydrolases like ARH1 (35), MacroD2 (11), and TARG1/c6orf130 (5), which cleave an ADP-ribose–amino acid bond to release an intact protein.
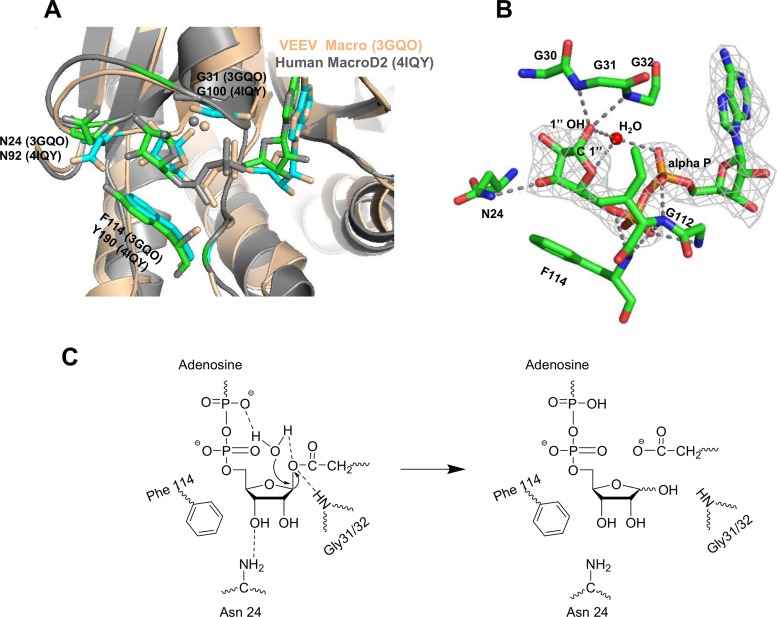
Crystal structure models of viral macro domains superimpose well with many macro domains (8, 12). The spatial arrangement of catalytic amino acid residues indicates that both SARS-CoV and VEEV macro domains belong to the MacroD2 type (Fig. 6A). Likewise, examination of the alignment of conserved amino acids within viral macro domains (Fig. 1C) suggests that they all belong to the MacroD2 family. In line with this proposition, the mono-ADP-ribose hydrolysis activity of the macro domain and macro-Hel, like human MacroD2 (11), is not markedly affected by the addition of magnesium, calcium, or manganese as a cofactor (36).
FIG 6.
The mode of ADP-ribose coordination in the VEEV macro domain pocket and proposed catalytic mechanism for de-MARylation activity. (A) Superimposition of VEEV macro domain (Protein Data Bank [PDB] accession number 3GQO) with human MacroD2 (PDB accession number 4IQY). (B) Close-up view of the 2.6Å-resolution X-ray structure of the VEEV macro domain in complex with ADP-ribose. ADP-ribose is shown with the corresponding electron density map (light gray). Hydrogen bonds between protein and ADP-ribose are indicated by dashed lines (dark gray). The H2O molecule is coordinated by hydrogen bonds to the 1″-OH, and oxygen from the α-phosphate is shown. (C) Proposed scheme for a catalytic mechanism (see the text for details). The 2mFo − DFc electron density map is contoured at 1.0 σ. The figure was generated using PyMOL.
A close-up of the VEEV macro domain active site and a proposal for a catalytic mechanism for the hydrolase activity, similar to that of the MacroD2 domain (36), are presented in Fig. 6B and C. The VEEV macro domain binds ADP-ribose in a deep cleft; the ADP-ribose is tightly coordinated by N24, G31, G32, G112, and F114, which is in agreement with the PAR-binding results of HEV macro-Hel. A water molecule is located in this ADP-ribose pocket, held in place by the hydrogen bond between the oxygen carried by the C1″ atom and the oxygen carried by the α-phosphate of ADP-ribose. The latter, rather than an elusive Asp-His dyad (11), presumably acts as the general base (36); as in the case of MacroD2, this water molecule is proposed to be activated by one oxygen of the α-phosphate of ADP-ribose, allowing the nucleophilic attack on the C1″ atom of ADP-ribose (Fig. 6C).
DISCUSSION
The macro domain is an example of a very conserved protein domain through evolution in all kingdoms of life, including the three positive-sense single-stranded RNA [(+)ssRNA] virus families Coronaviridae, Togaviridae, and Hepeviridae. All macro domains contain a typical protein fold with a deep ADP-ribose binding crevice around which several enzyme activities have been demonstrated, such as ADP-ribose deacetylation (7), de-MARylation or de-PARylation (5, 11, 36), or A1″Pase (9, 37). The last activity is generally found in the nucleus for intron processing where rapid turnover of ADP-ribose metabolites may fine-tune cellular ADP-ribose pools (37). The catalytic efficiency demonstrated for the viral A1″Pase is too low and topologically inconsistent with such a regulatory mechanism (8, 9).
Here, we propose that de-MARylation is the main enzyme activity of viral macro domains. Our data reconcile both experiments, demonstrating the essential role of the macro domain (15, 38) and an altered interferon response (16, 18) with an enzymatic activity acting on innate immunity guardians.
We had previously established that viral macro domains bind PAR efficiently (8) but failed to detect any free PAR hydrolysis by coronavirus, alphavirus, and HEV macro domains (8, 9). The recent discovery that the human MacroD2 is able to remove ADP-ribose from a mono-ADP-ribosylated protein prompted us to examine this activity for viral macro domains. Our results show that the de-MARylation activity is a common feature of at least one macro domain from all macro domain-containing viral families examined so far. Remarkably, these viral macro domains are also able to remove PAR from PARylated proteins, which represents an additional feature relative to the activity of MacroD2-like macro domains. When the C-terminal region of the HEV macro domain is fused to the helicase domain, as occurs in vivo, the de-PARylation activity carried by the HEV macro domain is greatly stimulated with a concomitant increase of affinity for PAR. As domain design might not be optimal, this stimulation could merely be the result of a better folding—and thus PAR binding—of the C-terminal region of the macro domain when it is fused to the helicase. However, extensive domain design was performed, from which the most stable macro domain was selected (39). Additionally, the macro-Hel construct binds PAR and RNA at the micromolar range (Fig. 2), whereas the active HEV macro domain exhibits a poor affinity for either PAR or RNA (9). We thus propose that the helicase domain serves as an auxiliary protein helping the macro domain to access its substrate with better efficiency, a finding that could connect viral helicases to cellular innate immunity (25, 28).
Most viral macro domains so far are homologous to the human MacroD2 type. Hence, we infer that a related catalytic mechanism occurs whereby an activated water molecule acts as a nucleophile to attack a nucleophilic center (11, 36). Since MacroD2 acts on aspartic or glutamic esters, we infer that viral macro domains exhibit the same bond specificity.
In a previous work, we showed that it is difficult to model either a 1″-phosphate or a 1″-carboxyl ester on the bound ADP-ribose. A mechanism similar to the one proposed here has been suggested for Saccharomyces cerevisiae Ymx7 (40), and the A1″Pase activity observed for most viral macro domains is consistent with such a mechanism. The absence of de-MARylation activity of Ymx7 from S. cerevisiae (36) confirms that an aromatic ring at position 125 (HEV numbering) is essential for the removal of mono- or poly(ADP-ribose) by MacroD2-like macro domains (Fig. 1C and 3E).
ARTs and ART-related proteins are thought to play significant roles in host-virus interactions (30, 41). Some of these ARTs exert antiviral effects through a wide range of responses. For example, the ZAP protein (PARP13), a presumably nonfunctional ART for protein PARylation/MARylation, affects the replication of representative viruses from several virus families, including Retroviridae, Hepadnaviridae, Filoviridae, and Togaviridae (42–45). The PARP13 antiviral effect is mediated through recruitment of viral RNA to promote its degradation by the exosome (46). PARP12 and its PAPR12L isoform repress viral replication through cellular translation shutdown (47). Thus, the roles of ADP-ribosylation in host immunity and antiviral effects might well be counteracted by viral macro domains.
The fact that all viral macro domains belong to the MacroD2 family suggests that they all act on Asp/Glu-grafted ADP-ribose molecules. Given the diversity of target amino acids (Lys, Arg, Glu, and Asp) in the cell for ADP-ribosylation and of the corresponding catalytic mechanisms, this observation suggests that viral macro domains target a narrow class of ADP-ribosylated proteins which respond to virus infection. PARP1, -3, -6, -10, -11, -12, and -16 have been demonstrated to be ADP-ribosylated on carboxylic residues (31). Among these ARTs, at least PARP1, -10, and -12 have already been shown to be involved in the antiviral response against several RNA viruses (48). Recent developments in proteomics now enable a fine-mapping of candidates directly ADP-ribosylated by ARTs (reviewed in reference 49). ART-mediated ADP-ribosylation may reveal ADP-ribosylated proteins involved in yet unknown, specific, innate antiviral response pathways in which viral macro domains might play a decisive role.
ACKNOWLEDGMENTS
We thank Hélène Malet, Hélène Dutartre, Marie-Pierre Egloff, Antoine Frangeul, Saïd Jamal, and Benjamin Morin for initial work on the project, as well as Julie Lichière for excellent technical assistance.
This work was supported by funding from EU-FP7/2011-2014 (Project EUVIRNA grant 264286 and Project SILVER grant 260644), the Agence Nationale de Recherche SIDA-Hépatites (ANRS), BELSPO (BELVIR grant), the Wellcome Trust, and the European Research Council to I.A. Y.D. was a fellow of the Fund for Scientific Research (FWO), Flanders.
The funders had no role in study design, data collection and interpretation, or the decision to submit the work for publication.
REFERENCES
- 1.Gibson BA, Kraus WL. 2012. New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13:411–424. doi: 10.1038/nrm3376. [DOI] [PubMed] [Google Scholar]
- 2.Karlberg T, Langelier MF, Pascal JM, Schuler H. 2013. Structural biology of the writers, readers, and erasers in mono- and poly(ADP-ribose) mediated signaling. Mol Aspects Med 34:1088–1108. doi: 10.1016/j.mam.2013.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barkauskaite E, Jankevicius G, Ladurner AG, Ahel I, Timinszky G. 2013. The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J 280:3491–3507. doi: 10.1111/febs.12358. [DOI] [PubMed] [Google Scholar]
- 4.Krietsch J, Rouleau M, Pic E, Ethier C, Dawson TM, Dawson VL, Masson JY, Poirier GG, Gagne JP. 2013. Reprogramming cellular events by poly(ADP-ribose)-binding proteins. Mol Aspects Med 34:1066–1087. doi: 10.1016/j.mam.2012.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sharifi R, Morra R, Appel CD, Tallis M, Chioza B, Jankevicius G, Simpson MA, Matic I, Ozkan E, Golia B, Schellenberg MJ, Weston R, Williams JG, Rossi MN, Galehdari H, Krahn J, Wan A, Trembath RC, Crosby AH, Ahel D, Hay R, Ladurner AG, Timinszky G, Williams RS, Ahel I. 2013. Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. EMBO J 32:1225–1237. doi: 10.1038/emboj.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Han W, Li X, Fu X. 2011. The macro domain protein family: structure, functions, and their potential therapeutic implications. Mutat Res 727:86–103. doi: 10.1016/j.mrrev.2011.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chen D, Vollmar M, Rossi MN, Phillips C, Kraehenbuehl R, Slade D, Mehrotra PV, von Delft F, Crosthwaite SK, Gileadi O, Denu JM, Ahel I. 2011. Identification of macrodomain proteins as novel O-acetyl-ADP-ribose deacetylases. J Biol Chem 286:13261–13271. doi: 10.1074/jbc.M110.206771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Egloff MP, Malet H, Putics A, Heinonen M, Dutartre H, Frangeul A, Gruez A, Campanacci V, Cambillau C, Ziebuhr J, Ahola T, Canard B. 2006. Structural and functional basis for ADP-ribose and poly(ADP-ribose) binding by viral macro domains. J Virol 80:8493–8502. doi: 10.1128/JVI.00713-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Neuvonen M, Ahola T. 2009. Differential activities of cellular and viral macro domain proteins in binding of ADP-ribose metabolites. J Mol Biol 385:212–225. doi: 10.1016/j.jmb.2008.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Peterson FC, Chen D, Lytle BL, Rossi MN, Ahel I, Denu JM, Volkman BF. 2011. Orphan macrodomain protein (human C6orf130) is an O-acyl-ADP-ribose deacylase: solution structure and catalytic properties. J Biol Chem 286:35955–35965. doi: 10.1074/jbc.M111.276238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rosenthal F, Feijs KL, Frugier E, Bonalli M, Forst AH, Imhof R, Winkler HC, Fischer D, Caflisch A, Hassa PO, Luscher B, Hottiger MO. 2013. Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat Struct Mol Biol 20:502–507. doi: 10.1038/nsmb.2521. [DOI] [PubMed] [Google Scholar]
- 12.Malet H, Coutard B, Jamal S, Dutartre H, Papageorgiou N, Neuvonen M, Ahola T, Forrester N, Gould EA, Lafitte D, Ferron F, Lescar J, Gorbalenya AE, de Lamballerie X, Canard B. 2009. The crystal structures of chikungunya and Venezuelan equine encephalitis virus nsP3 macro domains define a conserved adenosine binding pocket. J Virol 83:6534–6545. doi: 10.1128/JVI.00189-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Park E, Griffin DE. 2009. Interaction of Sindbis virus non-structural protein 3 with poly(ADP-ribose) polymerase 1 in neuronal cells. J Gen Virol 90:2073–2080. doi: 10.1099/vir.0.012682-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lhomme S, Garrouste C, Kamar N, Saune K, Abravanel F, Mansuy JM, Dubois M, Rostaing L, Izopet J. 2014. Influence of polyproline region and macro domain genetic heterogeneity on HEV persistence in immunocompromised patients. J Infect Dis 209:300–303. doi: 10.1093/infdis/jit438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Park E, Griffin DE. 2009. The nsP3 macro domain is important for Sindbis virus replication in neurons and neurovirulence in mice. Virology 388:305–314. doi: 10.1016/j.virol.2009.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fehr AR, Athmer J, Channappanavar R, Phillips JM, Meyerholz DK, Perlman S. 2015. The nsp3 macrodomain promotes virulence in mice with coronavirus-induced encephalitis. J Virol 89:1523–1536. doi: 10.1128/JVI.02596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Nargi-Aizenman JL, Simbulan-Rosenthal CM, Kelly TA, Smulson ME, Griffin DE. 2002. Rapid activation of poly(ADP-ribose) polymerase contributes to Sindbis virus and staurosporine-induced apoptotic cell death. Virology 293:164–171. doi: 10.1006/viro.2001.1253. [DOI] [PubMed] [Google Scholar]
- 18.Nan Y, Yu Y, Ma Z, Khattar SK, Fredericksen B, Zhang YJ. 2014. Hepatitis E virus inhibits type I interferon induction by ORF1 products. J Virol 88:11924–11932. doi: 10.1128/JVI.01935-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Perez-Gracia MT, Suay B, Mateos-Lindemann ML. 2014. Hepatitis E: an emerging disease. Infect Genet Evol 22:40–59. doi: 10.1016/j.meegid.2014.01.002. [DOI] [PubMed] [Google Scholar]
- 20.Kamar N, Dalton HR, Abravanel F, Izopet J. 2014. Hepatitis E virus infection. Clin Microbiol Rev 27:116–138. doi: 10.1128/CMR.00057-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao D, Meng XJ. 2012. Molecular biology and replication of hepatitis E virus. Emerg Microbes Infect 1:e17. doi: 10.1038/emi.2012.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ahmad I, Holla RP, Jameel S. 2011. Molecular virology of hepatitis E virus. Virus Res 161:47–58. doi: 10.1016/j.virusres.2011.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Karpe YA, Lole KS. 2010. NTPase and 5′ to 3′ RNA duplex-unwinding activities of the hepatitis E virus helicase domain. J Virol 84:3595–3602. doi: 10.1128/JVI.02130-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Karpe YA, Lole KS. 2010. RNA 5′-triphosphatase activity of the hepatitis E virus helicase domain. J Virol 84:9637–9641. doi: 10.1128/JVI.00492-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Inoue J, Takahashi M, Mizuo H, Suzuki K, Aikawa T, Shimosegawa T, Okamoto H. 2009. Nucleotide substitutions of hepatitis E virus genomes associated with fulminant hepatitis and disease severity. Tohoku J Exp Med 218:279–284. doi: 10.1620/tjem.218.279. [DOI] [PubMed] [Google Scholar]
- 26.Mishra N, Walimbe AM, Arankalle VA. 2013. Hepatitis E virus from India exhibits significant amino acid mutations in fulminant hepatic failure patients. Virus Genes 46:47–53. doi: 10.1007/s11262-012-0833-7. [DOI] [PubMed] [Google Scholar]
- 27.Takahashi K, Okamoto H, Abe N, Kawakami M, Matsuda H, Mochida S, Sakugawa H, Suginoshita Y, Watanabe S, Yamamoto K, Miyakawa Y, Mishiro S. 2009. Virulent strain of hepatitis E virus genotype 3, Japan. Emerg Infect Dis 15:704–709. doi: 10.3201/eid1505.081100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devhare P, Sharma K, Mhaindarkar V, Arankalle V, Lole K. 2014. Analysis of helicase domain mutations in the hepatitis E virus derived from patients with fulminant hepatic failure: effects on enzymatic activities and virus replication. Virus Res 184:103–110. doi: 10.1016/j.virusres.2014.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tan ES, Krukenberg KA, Mitchison TJ. 2012. Large-scale preparation and characterization of poly(ADP-ribose) and defined length polymers. Anal Biochem 428:126–136. doi: 10.1016/j.ab.2012.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Daugherty MD, Young JM, Kerns JA, Malik HS. 2014. Rapid evolution of PARP genes suggests a broad role for ADP-ribosylation in host-virus conflicts. PLoS Genet 10:e1004403. doi: 10.1371/journal.pgen.1004403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Vyas S, Matic I, Uchima L, Rood J, Zaja R, Hay RT, Ahel I, Chang P. 2014. Family-wide analysis of poly(ADP-ribose) polymerase activity. Nat Commun 5:4426. doi: 10.1038/ncomms5426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Debing Y, Gisa A, Dallmeier K, Pischke S, Bremer B, Manns M, Wedemeyer H, Suneetha PV, Neyts J. 2014. A mutation in the hepatitis E virus RNA polymerase promotes its replication and associates with ribavirin treatment failure in organ transplant recipients. Gastroenterology 147:1008–1011.e7. doi: 10.1053/j.gastro.2014.08.040. [DOI] [PubMed] [Google Scholar]
- 33.Mhaindarkar V, Sharma K, Lole KS. 2014. Mutagenesis of hepatitis E virus helicase motifs: effects on enzyme activity. Virus Res 179:26–33. doi: 10.1016/j.virusres.2013.11.022. [DOI] [PubMed] [Google Scholar]
- 34.Steffen JD, Pascal JM. 2013. New players to the field of ADP-ribosylation make the final cut. EMBO J 32:1205–1207. doi: 10.1038/emboj.2013.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mashimo M, Kato J, Moss J. 2014. Structure and function of the ARH family of ADP-ribosyl-acceptor hydrolases. DNA Repair (Amst) 23:88–94. doi: 10.1016/j.dnarep.2014.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jankevicius G, Hassler M, Golia B, Rybin V, Zacharias M, Timinszky G, Ladurner AG. 2013. A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nat Struct Mol Biol 20:508–514. doi: 10.1038/nsmb.2523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shull NP, Spinelli SL, Phizicky EM. 2005. A highly specific phosphatase that acts on ADP-ribose 1″-phosphate, a metabolite of tRNA splicing in Saccharomyces cerevisiae. Nucleic Acids Res 33:650–660. doi: 10.1093/nar/gki211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Eriksson KK, Cervantes-Barragan L, Ludewig B, Thiel V. 2008. Mouse hepatitis virus liver pathology is dependent on ADP-ribose-1″-phosphatase, a viral function conserved in the alpha-like supergroup. J Virol 82:12325–12334. doi: 10.1128/JVI.02082-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bignon C, Li C, Lichiere J, Canard B, Coutard B. 2013. Improving the soluble expression of recombinant proteins by randomly shuffling 5′ and 3′ coding-sequence ends. Acta Crystallogr D Biol Crystallogr 69:2580–2582. doi: 10.1107/S0907444913018751. [DOI] [PubMed] [Google Scholar]
- 40.Kumaran D, Eswaramoorthy S, Studier FW, Swaminathan S. 2005. Structure and mechanism of ADP-ribose-1″-monophosphatase (Appr-1″-pase), a ubiquitous cellular processing enzyme. Protein Sci 14:719–726. doi: 10.1110/ps.041132005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kerns JA, Emerman M, Malik HS. 2008. Positive selection and increased antiviral activity associated with the PARP-containing isoform of human zinc-finger antiviral protein. PLoS Genet 4:e21. doi: 10.1371/journal.pgen.0040021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bick MJ, Carroll JW, Gao G, Goff SP, Rice CM, MacDonald MR. 2003. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol 77:11555–11562. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao G, Guo X, Goff SP. 2002. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science 297:1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 44.Mao R, Nie H, Cai D, Zhang J, Liu H, Yan R, Cuconati A, Block TM, Guo JT, Guo H. 2013. Inhibition of hepatitis B virus replication by the host zinc finger antiviral protein. PLoS Pathog 9:e1003494. doi: 10.1371/journal.ppat.1003494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Muller S, Moller P, Bick MJ, Wurr S, Becker S, Gunther S, Kummerer BM. 2007. Inhibition of filovirus replication by the zinc finger antiviral protein. J Virol 81:2391–2400. doi: 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guo X, Ma J, Sun J, Gao G. 2007. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci U S A 104:151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atasheva S, Akhrymuk M, Frolova EI, Frolov I. 2012. New PARP gene with an anti-alphavirus function. J Virol 86:8147–8160. doi: 10.1128/JVI.00733-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Atasheva S, Frolova EI, Frolov I. 2014. Interferon-stimulated poly(ADP-Ribose) polymerases are potent inhibitors of cellular translation and virus replication. J Virol 88:2116–2130. doi: 10.1128/JVI.03443-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Daniels CM, Ong SE, Leung AK. 2015. The promise of proteomics for the study of ADP-ribosylation. Mol Cell 58:911–924. doi: 10.1016/j.molcel.2015.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nishikiori M, Sugiyama S, Xiang H, Niiyama M, Ishibashi K, Inoue T, Ishikawa M, Matsumura H, Katoh E. 2012. Crystal structure of the superfamily 1 helicase from Tomato Mosaic Virus. J Virol 86:7565–7576. doi: 10.1128/JVI.00118-12. [DOI] [PMC free article] [PubMed] [Google Scholar]