ABSTRACT
Previous studies have shown that highly conserved residues in the inner domain of gp120 are required for HIV-1 envelope glycoprotein (Env) transitions to the CD4-bound conformation (A. Finzi, S. H. Xiang, B. Pacheco, L. Wang, J. Haight, et al., Mol Cell 37:656–667, 2010, http://dx.doi.org/10.1016/j.molcel.2010.02.012; A. Desormeaux, M. Coutu, H. Medjahed, B. Pacheco, A. Herschhorn, et al., J Virol 87:2549–2562, 2013, http://dx.doi.org/10.1128/JVI.03104-12). Moreover, W69, a highly conserved residue located at the interface between layer 1 and layer 2 of the inner domain, was recently shown to be important for efficient Env recognition by CD4-induced (CD4i) antibodies capable of potent antibody-dependent cellular cytotoxicity (W. D. Tolbert, N. Gohain, M. Veillette, J. P. Chapleau, C. Orlandi, et al., 2016, Structure 24:697–709, http://dx.doi.org/10.1016/j.str.2016.03.005; S. Ding, M. Veillette, M. Coutu, J. Prevost, L. Scharf, et al., 2016, J Virol 90:2127–2134, http://dx.doi.org/10.1128/JVI.02779-15). We evaluated the contribution of the hydrophobicity of W69 to conformational changes of Env by replacing it with a series of residues with aliphatic or aromatic side chains of decreasing chain length. We have found that the hydrophobicity of residue 69 is important for Env processing, CD4 binding, and its transition to the CD4-bound conformation. The most deleterious effect was observed when W69 was replaced with alanine or glycine residues. However, the functions lost due to W69 mutations could be progressively restored with amino acids of increasing aliphatic chain length and fully recovered with residues bearing an aromatic ring. Interestingly, poor CD4 binding of W69A could be fully restored by introducing a compensatory mutation within layer 2 (S115W). Structural studies of HIV-1 gp120 coree W69A/S115W mutant bound to the CD4 peptide mimetic M48U1 and Fab of anti-cluster A antibody N60-i3 revealed no perturbations to the overall structure of the double mutant compared to the wild-type protein but identified higher mobility within the interface between layer 1 and layer 2, the bridging sheet region, and the CD4 binding site.
IMPORTANCE HIV-1 Env transitions to the CD4-bound conformation are required for viral entry. Previous studies identified a highly conserved residue of the inner domain, W69, as being involved in these conformational transitions (A. Finzi, S. H. Xiang, B. Pacheco, L. Wang, J. Haight, et al., Mol Cell 37:656–667, 2010, http://dx.doi.org/10.1016/j.molcel.2010.02.012). Here, we show that W69, located at the interface between gp120 and gp41 in the PGT151-bound trimer, plays a critical role in the interprotomer signaling induced by CD4 binding. This new information might be useful in immunogen design.
INTRODUCTION
HIV-1 envelope glycoproteins (Env) play a key role during the first step of viral infection. Mature HIV-1 Env trimers are the products of a proteolytic cleavage of gp160 precursor polypeptides into gp120 (SU) and gp41 (TM) subunits. Mature metastable Env is a consolidation of three gp120 with three gp41 transmembrane subunits related in a labile noncovalent manner (1). Binding of gp120 exterior subunit to its cellular receptor, CD4, initiates viral entry (2, 3). CD4 binding allows gp120 conformational rearrangement, which is required for its attachment to CCR5 or CXCR4 chemokine coreceptors (4–11), upon which further conformational changes expose the gp41 helical heptad repeat segment (HR1), resulting in the formation of the prehairpin intermediates followed by a transition to a six-helix bundle composed of HR1 and HR2. These conformational changes lead to viral and cellular membrane fusion (12–14). CD4-induced changes allow the trimer to switch from a metastable high-energy unliganded form to a low-energy stable state.
Recent studies have shown that highly conserved residues in the inner domain of gp120 are required for HIV-1 Env transitions to the CD4-bound conformation and efficient CD4 binding (15, 16). Moreover, among these residues, W69 was recently shown to be important for efficient Env recognition by antibody-dependent cellular cytotoxicity (ADCC)-mediating anti-cluster A antibodies and also by sera from HIV-1-infected individuals (17–20). Since W69 is located at the interface between layer 1 and layer 2 of the inner domain, we evaluated the contribution of hydrophobicity at this position to Env conformational changes. A set of Env variants with substitutions at position 69 with residues of aliphatic or aromatic chains were generated and evaluated for binding to CD4, CD4-induced (CD4i), and CD4-binding site (CD4bs) antibodies. We found that the hydrophobicity of residue 69, which is normally imparted by a tryptophan in HIV-1 Env, is essential for the ability of Env to undergo functional transitions required to assume the CD4-bound state.
MATERIALS AND METHODS
Cell lines.
293T human embryonic kidney cells, Cf2Th canine thymocytes (American Type Culture Collection; previously used to measure infectivity and neutralization of luciferase coding viruses due to their low luciferase background [15, 16, 21–25]), and TZM-bl cell lines (NIH AIDS Research and Reference Reagent Program) were grown at 37°C and 5% CO2 in Dulbecco's modified Eagle's medium (Invitrogen) containing 10% fetal bovine serum (Sigma) and 100 μg/ml of penicillin-streptomycin (Mediatech). Cf2Th cells stably expressing human CD4 and CCR5 (26) were grown in medium supplemented with 0.4 mg/ml of G418 (Invitrogen) and 0.15 mg/ml of hygromycin B (Roche Diagnostics). The TZM-bl cell line is a HeLa cell line stably expressing high levels of CD4 and CCR5 and possessing an integrated copy of the luciferase gene under the control of the HIV-1 long terminal repeat (27).
Site-directed mutagenesis.
Mutations were introduced individually or in combination into the previously described pSVIIIenv vector expressing the HIV-1YU2 envelope glycoproteins or in the pCDNA3.1 codon-optimized YU2 (for SPR analysis, see below) (15). Site-directed mutagenesis was performed using the QuikChange II XL site-directed mutagenesis protocol (Stratagene). The presence of the desired mutations was determined by automated DNA sequencing. The numbering of the HIV-1 envelope glycoprotein amino acid residues is based on that of the prototypic HXBc2 strain of HIV-1, where position 1 is the initial methionine (28).
Immunoprecipitation of envelope glycoproteins.
For pulse-labeling experiments, 3 × 105 293T cells were transfected by the calcium phosphate method with envelope expressors. One day after transfection, cells were metabolically labeled for 16 h with 100 μCi/ml [35S]methionine-cysteine ([35S] protein labeling mix; Perkin-Elmer) in Dulbecco's modified Eagle's medium lacking methionine and cysteine and supplemented with 5% dialyzed fetal bovine serum. Cells were subsequently lysed in radioimmunoprecipitation assay (RIPA) buffer (140 mM NaCl, 8 mM Na2HPO4, 2 mM NaH2PO4, 1% NP-40, 0.05% sodium dodecyl sulfate [SDS]). Precipitation of radiolabeled HIV-1YU2 envelope glycoproteins from cell lysates or medium was performed with a mixture of sera from HIV-1-infected individuals. Alternatively, radiolabeled gp120 envelope glycoprotein in medium was precipitated with various amounts of anti-gp120 monoclonal antibodies or the recombinant CD4-Ig protein for 1 h at 37°C in the presence of 50 μl of 10% protein A-Sepharose (American BioSciences).
Processing and association indices were determined by precipitation of radiolabeled cell lysates and supernatants with mixtures of sera from HIV-1-infected individuals. The association index is a measure of the ability of the mutant gp120 molecule to remain associated with the Env trimer complex on the expressing cell relative to that of the wild-type (WT) Env trimers. The association index was calculated with the following formula: association index = ([mutant gp120]cell × [WT gp120]supernatant)/([mutant gp120]supernatant × [WT gp120]cell). The processing index is a measure of the conversion of the mutant gp160 Env precursor to mature gp120 relative to that of the wild-type Env trimers. The processing index was calculated with the following formula: processing index = ([total gp120]mutant × [gp160]WT)/([gp160]mutant × [total gp120]WT).
Recombinant luciferase viruses.
Recombinant viruses containing the firefly luciferase gene were produced by calcium phosphate transfection of 293T cells with the HIV-1 proviral vector pNL4.3 Env− Luc and the pSVIIIenv plasmid expressing the wild-type or mutant HIV-1YU2 envelope glycoproteins at a ratio of 2:1. Two days after transfection, the cell supernatants were harvested. The reverse transcriptase (RT) activities of all viruses were measured as described previously (29). The virus-containing supernatants were stored in aliquots at −80°C.
Infection by single-round luciferase viruses.
Cf2Th-CD4-CCR5 target cells were seeded at a density of 5 × 103 cells/well in 96-well luminometer-compatible tissue culture plates (PerkinElmer) 24 h before infection. Recombinant viruses (10,000 reverse transcriptase units) in a final volume of 100 μl were then added to the target cells, followed by incubation for 48 h at 37°C; the medium was then removed from each well, and the cells were lysed by the addition of 30 μl of passive lysis buffer (Promega) and three freeze-thaw cycles. An LB 941 TriStar luminometer (Berthold Technologies) was used to measure the luciferase activity of each well after the addition of 100 μl of luciferin buffer (15 mM MgSO4, 15 mM KPO4 [pH 7.8], 1 mM ATP, and 1 mM dithiothreitol) and 50 μl of 1 mM d-luciferin potassium salt (Prolume).
Protein purification.
For protein expression for surface plasmon resonance (SPR) analysis, FreeStyle 293F cells (Invitrogen) were grown in FreeStyle 293F medium (Invitrogen) to a density of 1 × 106 cells/ml at 37°C with 8% CO2 with regular agitation (125 rpm). Cells were transfected with a codon-optimized plasmid expressing His6-tagged wild-type or mutant HIV-1YU2 gp120 using the 293Fectin reagent, as directed by the manufacturer (Invitrogen). One week later, the cells were pelleted and discarded. The supernatants were filtered (0.22-μm filter) (Corning) and the gp120 glycoproteins were purified by nickel affinity columns, as directed by the manufacturer (Invitrogen). Monomeric gp120 was subsequently purified by fast protein liquid chromatography (FPLC), as reported previously (30). The gp120 preparations were dialyzed against phosphate-buffered saline (PBS) and stored in aliquots at −80°C. To assess purity, recombinant proteins were loaded on SDS-PAGE polyacrylamide gels in the absence of beta-mercaptoethanol and stained with Coomassie blue, as reported previously (30, 31).
SPR biosensor analysis.
SPR biosensor data were collected on a BIACORE 3000 optical biosensor (General Electric), as reported previously (15, 16, 30). Briefly, CD4-Ig and anti-gp120 monoclonal antibodies were immobilized onto separate flow cells within the same sensor chip (CM5; GE) to a surface density of ∼500 response units (RU) using standard amine coupling chemistry (32). The binding capacities of CD4 and anti-gp120 antibody surfaces were kept low to avoid mass transport effects and steric hindrance. Flow cell 1 or 3 was left blank as a control for nonspecific binding and refractive index changes. With the instrument operating in a parallel sensing mode, soluble gp120 was injected over flow cells 1 and 2 or 3 and 4 at different concentrations, ranging from 100 to 750 nM, at a flow rate of 30 μl/min for 3 min. This was followed by a 10-min dissociation phase to allow an estimation of off rates and binding affinities. Sensor data were prepared for kinetic analysis by subtracting binding responses collected from the blank reference surface. To examine the binding of the 17b antibody to gp120-CD4 complexes, a mixture of soluble gp120 and sCD4 (molar ratio, 1:1) at concentrations ranging from 100 to 750 nM was passed over the 17b chip. The association and dissociation phase data were fitted simultaneously with BIAevalution 3.2 RC1 software using a 1:1 Langmuir model of binding.
Molecular modeling and simulation.
All molecular modeling and simulations were performed in Discovery Studio Client 4.0. (Biovia, San Diego, CA). The full-length gp120 model was built using Modeler version 9.1 (33). The simulations for the interaction energies were estimated using CHARMm implicit solvation models.
Structural studies of HIV-1 gp120 coree W69A/S115W mutant.
gp120 core W69A/S115W mutant was crystalized in a ternary complex with CD4 peptide mimetic M48U1 (34) and Fab of anti-cluster A monoclonal antibody (MAb) N60-i3. MAb N60-i3 for crystallization studies was purified by HiTrap protein A column (GE Healthcare) chromatography from 293T supernatants prepared by transfecting plasmids encoding the heavy- and light-chain genes of N60-i3 MAb. The Fab was prepared from the purified IgG (10 mg/ml) by proteolytic digestion with immobilized papain (Pierce) and purified using a protein A column to remove Fc (GE Healthcare), followed by gel filtration chromatography on a Superdex 200 16/60 column (GE Healthcare).
The W69A and S115W mutations (Hxbc numbering) were added to the sequence of gp120 coree (lacking the N and C termini and variable loops 1, 2, and 3) (35) of clade A/E strain 93TH057 using the QuikChange mutagenesis kit per the manufacturer's protocol (Stratagene). W69A/S115W gp12093TH057 coree was expressed, purified, and deglycosylated as previously described (19). gp120 coree was combined with a 20% molar excess of the CD4 mimetic M48U1 and purified by size exclusion chromatography (Hiload 26/60 Superdex S200 preparation grade [GE Healthcare] in 0.15 M NaCl, 5 mM Tris, pH 7.2). After concentration, the gp120-M48U1 complex was mixed with a 20% molar excess of Fab, passed again through a size exclusion column, and concentrated to ∼10 mg/ml in 0.15 M NaCl, 5 mM Tris, pH 7.2, for crystallization experiments.
Crystals of N60-i3 Fab W69A/S115W gp12093TH057-M48U1 were grown by the hanging-drop method. Protein and well solution were mixed in equal volumes on a coverslip, but before sealing the well, 1.0 M NaCl was added to the well solution to a final concentration of 0.065 M. Crystallization trials were incubated at 22°C. N60-i3 complex crystals were grown from well solutions containing 10 to 16% polyethylene glycol (PEG) 8000 or PEG 10000 and 0.1 M Tris-HCl, pH 8.5. Complex crystals were frozen in liquid nitrogen after being soaked briefly in a cryoprotectant solution of 18% 2-methyl-2,4-pentanediol (MPD), 16% PEG 8000 or 10000, 0.1 M Tris-HCl, pH 8.5, and 0.065 M sodium chloride. Diffraction data were collected at the Stanford Synchrotron Radiation Light Source (SSRL), beam line BL14-1, equipped with an ADSC Quantum 315 area detector. All data were processed and reduced with HKL2000 (36). The complex of W69A/S115W gp12093TH057 crystallized in the same space group as the complex of the wild-type gp12093TH057 determined by us previously at 3.2-Å resolution (19), P21212, with slightly different cell dimensions, a = 95.2 Å, b = 101.2 Å, c = 108.7 Å, an approximately 3-Å (or 3%) change in a, and a 1-Å (<1%) change in the other dimensions. The structure was solved by molecular replacement with Phaser (37) from the CCP4 suite (38) based on the coordinates of the wild-type complex, N60-i3 Fab-gp12093TH057-M48U1 (Protein Data Bank [PDB] entry 4RFO). Refinement was carried out with Refmac (39) and Phenix (40) and was coupled with manual rebuilding in COOT (41). The structure was refined to an R/Rfree of 0.249/0.306 (see Table 4).
TABLE 4.
Crystallographic data collection and refinement statistics
| Parameter | Value(s)a for Fab N60-i3-gp120 (W69A/S115W)93TH057-M48U1 |
|---|---|
| Data collection | |
| Wavelength (Å) | 0.9795 |
| Space group | P21212 |
| Cell parameters | |
| a, b, c (Å) | 95.2, 101.2, 108.7 |
| α, β, γ (°) | 90, 90, 90 |
| Molecules (a.u.f) | 4 |
| Resolution (Å) | 50–3.05 (3.05–3.00) |
| No. of reflections | |
| Total | 78,160 |
| Unique | 19,540 |
| Rmergeb (%) | 22.0 (69.1) |
| I/σ | 5.1 (1.2) |
| Completeness (%) | 87.6 (67.6) |
| Redundancy | 4.0 (3.7) |
| Refinement statistics | |
| Resolution (Å) | 108.8–3.0 |
| Rc (%) | 24.9 |
| Rfreed (%) | 30.6 |
| No. of atoms | |
| Protein | 6,014 |
| Water | 9 |
| Ligand | 180 |
| Overall B value (Å2) | |
| Protein | 31.2 |
| Water | 21.7 |
| Ligand | 60.8 |
| Root mean square deviation | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.52 |
| Ramachandrane (%) | |
| Favored | 78.4 |
| Allowed | 12.9 |
| Outliers | 8.7 |
Values in parentheses are for the highest-resolution shell.
Rmerge = ∑|I − <I>|/∑I, where I is the observed intensity and <I> is the average intensity obtained from multiple observations of symmetry-related reflections after rejections.
R = ∑‖Fo|− |Fc‖/∑|Fo|, where Fo and Fc are the observed and calculated structure factors, respectively.
Rfree was defined by Brünger (50).
Calculated with MolProbity.
a.u., arbitrary units.
RESULTS
The HIV-1 gp120 layer 1 inner domain W69 mutants.
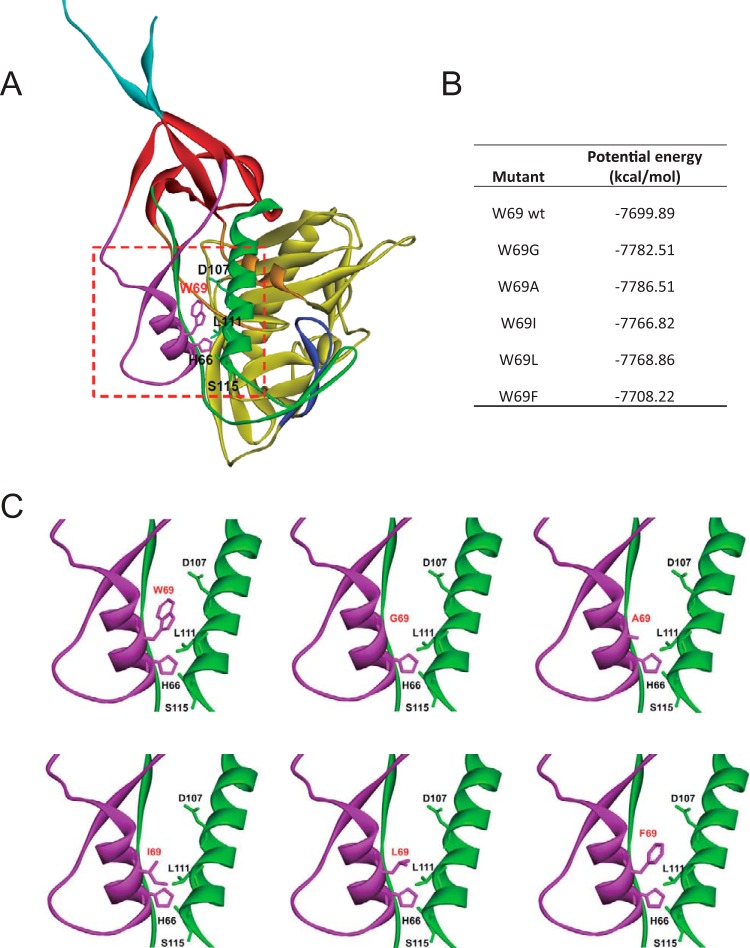
We recently reported that the interaction between layer 1 and layer 2 of the gp120 inner domain are important for the transition of Env to the CD4-bound conformation (15). Moreover, we found that a bulky, hydrophobic residue located at the layer 1-layer 2 interface, tryptophan 69 (W69), was critical for ADCC responses mediated by anti-cluster A antibodies and some HIV+ sera (20). To get a better understanding of how hydrophobicity at position 69 affects Env conformation, we generated a series of mutants containing aliphatic/aromatic amino acids of decreasing hydrophobicity at this position with the goal of progressively impacting the layer 1-layer 2 interface. We first modeled these variants on monomeric gp120, and interaction energies were estimated using CHARMm (33) (Fig. 1). As shown in Fig. 1B, by replacing W69 with phenylalanine, leucine, isoleucine, alanine, and glycine, residues with decreasing hydrophobicity index as described by the Wimley-White whole-residue hydrophobicity scale (42), we progressively impacted the hydrophobicity energy potential of gp120, with the biggest impact for the residue with the shortest aliphatic or no side chain (Gly and Ala, respectively) and almost no difference for the residue with the bulky aromatic chain (Phe). We also generated a double mutant in which W69 was exchanged for alanine and S115 for tryptophan. The S115 mutation was modeled on available structural information on CD4-triggered gp120 cores (19, 43) with the assumption that the hydrophobicity lost due to the W69A mutation at the layer 1-layer 2 interface could be compensated for by a tryptophan at an alternative interlayer position. All mutant variants were evaluated with respect to the following properties: proteolytic processing of the gp160 precursor, association of the gp120 and gp41 subunits, CD4 binding, functional ability to mediate cell-cell fusion, and virus infectivity (Table 1). The conformation of gp120 in cell supernatants was assessed by precipitation and SPR with a panel of monoclonal antibodies that recognize conformation-dependent epitopes (44) (Table 1).
FIG 1.
Structure of HIV-1 gp120 inner domain layers 1 and 2 in the CD4-bound conformation. (A) The structure of HIV-1HXBc2 gp120 (ribbon) complexed with two-domain CD4 (omitted for clarity purposes) is shown from the approximate perspective of the Env gp trimer axis. The outer domain of gp120 is colored yellow. The N and C termini are colored cyan. The components of the gp120 inner domain are the β-sandwich (red) and three loop-like extensions: layer 1 (magenta), layer 2 (green), and layer 3 (orange). The b20-b21 strands of gp120 (blue) project from the outer domain and, in the CD4-bound conformation, are composed of two of the strands of the four-stranded bridging sheet. The other two strands of the bridging sheet are derived from the distal portion of layer 2. (B) The interaction energies of W69 variants were simulated and are listed using CHARMm implicit solvation models. (C) A closeup of the interface between layer 1 and layer 2 (red square presented in panel A) for each modeled W69 variant. The most relevant residues for layer 1-layer 2 interaction (W69, H66, D107, L111, and S115) are shown and labeled.
TABLE 1.
Characterization of HIV-1YU2 gp120 W69 variantsg
| Env | Association indexa | Processing indexb | Infectivityc | IC50 (μg/ml) of CD4bs ligandd |
Cell-cell fusione | Immunoprecipitation resultf (gp120 monomer) |
|||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| sCD4 | VRC01 | VRC13 | VRC16 | CoRBS antibody |
Anti-cluster A antibody |
CD4bs antibody |
|||||||||||||
| 17b | 48d | A32 | N5-i5 | N60-i3 | CD4-Ig | VRC01 | VRC03 | VRC13 | VRC16 | b12 | |||||||||
| WT | 1.00 | 1.00 | 1.00 | 2.25 | 0.59 | 2.90 | 0.56 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 | 1.00 |
| W69G | 0.58 | 0.20 | 0.01 | NA | NA | NA | NA | 0.10 | 0.47 | 0.18 | 0.09 | 0.05 | 0.03 | 0.73 | 1.12 | 1.67 | 1.72 | 4.57 | 1.56 |
| W69A | 0.57 | 0.27 | 0.05 | 2.77 | 0.23 | 1.07 | 0.13 | 0.54 | 0.38 | 0.18 | 0.08 | 0.15 | 0.01 | 0.69 | 1.05 | 1.50 | 1.41 | 3.94 | 1.72 |
| W69I | 0.81 | 0.51 | 0.19 | 5.43 | 0.24 | 2.07 | 0.24 | 0.51 | 0.34 | 0.12 | 0.07 | 0.10 | 0.01 | 0.57 | 0.95 | 1.60 | 1.24 | 2.33 | 1.81 |
| W69L | 1.40 | 0.55 | 0.51 | >10 | 0.21 | 1.65 | 0.18 | 0.75 | 0.36 | 0.14 | 0.15 | 0.10 | 0.05 | 0.52 | 1.02 | 1.25 | 1.29 | 2.87 | 1.98 |
| W69F | 1.31 | 0.91 | 1.16 | 2.51 | 0.66 | 3.30 | 0.66 | 0.88 | 0.90 | 0.83 | 1.09 | 1.17 | 0.94 | 0.69 | 0.93 | 0.59 | 0.89 | 0.66 | 0.92 |
| W69A/S115W | 0.44 | 0.28 | 0.01 | NA | NA | NA | NA | 0.03 | 0.51 | 0.21 | 0.15 | 0.10 | 0.00 | 1.03 | 1.16 | 1.65 | 1.63 | 3.39 | 1.38 |
The association index is a measure of the ability of the mutant gp120 molecule to remain associated with the envelope glycoprotein complex on the expressing cell relative to that of the wild-type envelope glycoproteins. The calculation method is described in Materials and Methods.
The processing index is a measure of the conversion of the mutant gp160 envelope glycoprotein precursor to mature gp120 relative to that of the wild-type envelope glycoproteins. The calculation method is described in Materials and Methods.
The infectivity was assessed on Cf2Th-CD4/CCR5 cells using RT-normalized amounts of pseudoviruses of the WT and W69 variants. Data shown here are the ratios of mutant to wild-type virus infectivity.
The ligand (sCD4, VRC01, VRC13, and VRC16) concentrations that inhibited the infection of pseudoviruses of the WT and W69 variants by 50% (IC50) are reported. NA, not applicable, since infectivity was too low to perform the neutralization experiment.
Cell-cell fusion activity was assessed by coincubation between 293T cells expressing Env variants and TZM-bl cells for 6 h at 37°C. Luciferase activity in the mixture of cell lysate was measured to determine the cell-to-cell fusion efficiency.
Comparable amounts of radiolabeled monomeric gp120 from the WT and W69 variants were incubated with 5 μg/ml of coreceptor binding antibodies (17b and 48d), anti-cluster A antibodies (A32, N5-i5, and N60-i3), CD4-Ig, and CD4-binding site antibodies (VRC01, VRC03, VRC13, VRC16, and b12) for 1 h at 37°C. The precipitates were washed, run on SDS-polyacrylamide gels, and analyzed by densitometry.
Values presented in this table represent the means of data from at least three independent experiments, with experimental variation typically not more than 20% of the value reported. When WT values were normalized to 1, signals of ≤0.5 are in boldface, and values of ≥1.5 are in italics.
Ability of W69 mutants to bind CD4.
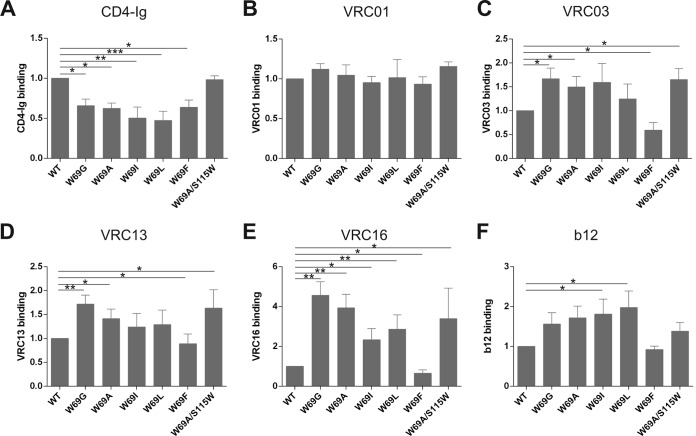
The effects of the changes introduced into W69 on the affinity of monomeric full-length gp120 for CD4 were examined first. The amounts of radiolabeled WT and mutant gp120 in supernatants of Env-expressing 293T cells were normalized, and equivalent amounts of gp120 were used for precipitation by CD4-Ig. As shown in Fig. 2A, all five W69 changes significantly decreased the efficiency of mutant gp120's interaction with CD4. A detailed analysis of this interaction by SPR indicated that the decreased CD4 recognition by W69 variants was due to enhanced off rates compared to that of WT gp120 (Table 2). This is in agreement with previous observations for layer 1 and layer 2 variants, including W69L (15). Interestingly, we observed that the accelerated off-rate appeared to inversely correlate with the hydrophobicity of the residue used to replace W69. Indeed, replacing tryptophan 69 with a glycine (W69G) resulted in a pronounced acceleration of the off rate, whereas replacing it with bulky residues, particularly a phenylalanine, restored the WT phenotype (Table 2). Thus, gp120 mutants with alterations at W69 fail to engage CD4 efficiently. Interestingly, the double W69A/S115W mutant showed binding properties comparable to those of its WT counterpart, indicating that the hydrophobicity lost due to the W69A mutation could be compensated for by CD4 binding by introducing a new tryptophan at residue 115 at the layer 1-layer 2 interface (Fig. 2A and Table 2).
FIG 2.
Ability of W69 variants to interact with CD4-binding site ligands. Comparable amounts of radiolabeled monomeric gp120 from WT and W69 variants were incubated with 5 μg/ml of CD4-Ig (A), VRC01 (B), VRC03 (C), VRC13 (D), VRC16 (E), and b12 (F) for 1 h at 37°C. The precipitates were washed, run on SDS-polyacrylamide gels, and analyzed by densitometry. All values shown here were evaluated by the ligand binding capacity of WT gp120 (set as 1). Data represent the averages ± standard deviations (SD) from at least three independent experiments. Statistical significance was tested using an unpaired two-tailed t test. Shown are P values with significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
TABLE 2.
Characterization of CD4, CD4i, and CD4-binding site antibody binding to HIV-1YU2 gp120 W69 variants by surface plasmon resonancea
| gp120 variant | CD4-Ig |
17b |
17b plus sCD4 |
A32 |
N60-i3 |
VRC16 |
b12 |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| On rate (M−1 s−1) | Off rate (s−1) | KD (M) (fold change) | On rate (M−1 s−1) | Off rate (s−1) | KD (M) (fold change) | On rate (M−1 s−1) | Off rate s−1) | KD (M) (fold change) | On rate (M−1 s−1) | Off rate (s−1) | KD (M) (fold change) | On rate (M−1 s−1) | Off rate (s−1) | KD (M) (fold change) | On rate (M−1 s−1) | Off rate (s−1) | KD (M) (fold change) | On rate (M−1 s−1) | Off rate (s−1) | KD (M) (fold change) | |
| WT | 9.71 × 103 | 2.23 × 10−4 | 2.29 × 10−8 (1.0) | 1.78 × 104 | 3.41 × 10−5 | 1.92 × 10−9 (1.0) | 3.14 × 104 | 2.15 × 10−5 | 1.92 × 10−10 (1.0) | 3.14 × 104 | 3.01 × 10−5 | 9.57 × 10−10 (1.0) | 2.68 × 104 | 5.88 × 10−5 | 2.19 × 10−9 (1.0) | 5.52 × 103 | 9.16 × 10−3 | 1.66 × 10−6 (1.00) | 2.74 × 104 | 4.82 × 10−3 | 1.76 × 10−7 (1.00) |
| W69G | 9.65 × 103 | 9.31 × 10−4 | 9.65 × 10−8 (4.2) | 2.61 × 102 | 2.40 × 10−4 | 9.20 × 10−7 (479) | 9.44 × 103 | 6.76 × 10−5 | 9.20 × 10−9 (10.5) | ND | ND | ND | ND | ND | ND | 4.91 × 103 | 3.12 × 10−3 | 6.35 × 10−7 (0.38) | 2.61 × 104 | 2.91 × 10−3 | 1.11 × 10−7 (0.63) |
| W69A | 9.76 × 103 | 8.35 × 10−4 | 8.56 × 10−8 (3.7) | 3.01 × 102 | 2.07 × 10−4 | 6.90 × 10−7 (359) | 1.01 × 104 | 6.47 × 10−5 | 6.90 × 10−9 (9.3) | ND | ND | ND | ND | ND | ND | 5.55 × 103 | 4.05 × 10−3 | 7.30 × 10−7 (0.44) | 2.72 × 104 | 3.81 × 10−3 | 1.40 × 10−7 (0.80) |
| W69I | 9.31 × 103 | 7.24 × 10−4 | 7.77 × 10−8 (3.4) | 3.79 × 102 | 2.02 × 10−4 | 5.33 × 10−7 (277) | 1.18 × 104 | 7.00 × 10−5 | 5.33 × 10− (8.7) | ND | ND | ND | ND | ND | ND | 5.22 × 103 | 5.36 × 10−3 | 1.03 × 10−6 (0.62) | 2.59 × 104 | 4.85 × 10−3 | 1.87 × 10−7 (1.07) |
| W69L | 9.47 × 103 | 7.08 × 10−4 | 7.47 × 10−8 (3.3) | 3.09 × 102 | 1.89 × 10−4 | 6.11 × 10−7 (318) | 1.15 × 104 | 6.07 × 10−5 | 6.11 × 10−9 (7.7) | ND | ND | ND | ND | ND | ND | 6.05 × 103 | 4.15 × 10−3 | 6.86 × 10−7 (0.41) | 2.67 × 104 | 3.87 × 10−3 | 1.45 × 10−7 (0.83) |
| W69F | 9.77 × 103 | 3.00 × 10−4 | 3.07 × 10−8 (1.3) | 1.57 × 104 | 4.36 × 10−5 | 2.78 × 10−9 (1.5) | 1.81 × 104 | 3.35 × 10−5 | 2.78 × 10−9 (2.7) | 2.04 × 104 | 7.78 × 10−5 | 3.82 × 10−9 (4.0) | 2.07 × 104 | 5.56 × 10−5 | 2.68 × 10−9 (1.2) | 5.14 × 103 | 8.34 × 10−3 | 1.62 × 10−6 (0.98) | 2.84 × 104 | 5.40 × 10−3 | 1.90 × 10−7 (1.08) |
| W69A/S115W | 9.64 × 103 | 3.59 × 10−4 | 3.73 × 10−8 (1.6) | 3.22 × 102 | 2.14 × 10−4 | 6.65 × 10−7 (346) | 1.85 × 104 | 6.19 × 10−5 | 6.65 × 10−9 (4.9) | ND | ND | ND | ND | ND | ND | 5.07 × 103 | 3.00 × 10−3 | 5.92 × 10−7 (0.36) | 2.81 × 104 | 4.03 × 10−3 | 1.44 × 10−7 (0.82) |
CD4-Ig,17b, A32, N60-i3, VRC16 or b12 was immobilized directly onto a CM5 sensor chip, and the binding of the indicated gp120 protein was evaluated as described in Materials and Methods. KD, equilibrium dissociation constant. ND, signal too low to be detected.
Recognition of W69 variants by CD4i antibodies.
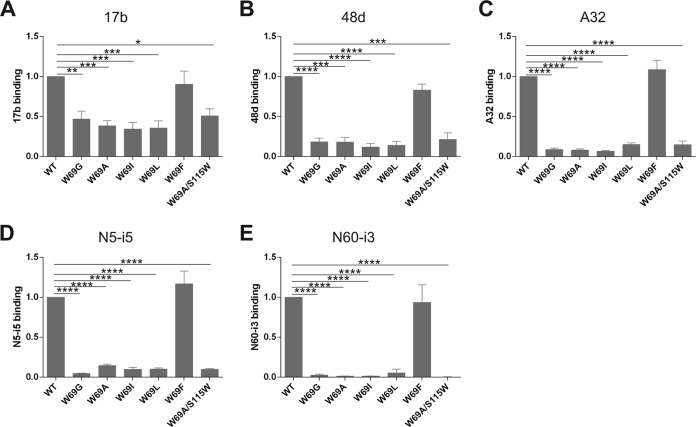
Since it has been previously reported that the inner domain layers modulate recognition of gp120 by CD4i antibodies (15, 16, 30), we next examined the binding capacity of our panel of W69 gp120 variants to CD4i antibodies recognizing the coreceptor binding site (CoRBS; 17b and 48d) and anti-cluster A antibodies, known to be potent mediators of ADCC responses (A32, N5-i5, and N60-i3) (15, 19, 43, 45, 46). It was previously shown that CD4i antibodies preferentially recognize the CD4-triggered monomeric gp120, but they are still capable of binding to unliganded gp120 with roughly 10- to 20-fold lower affinities (15, 18). Figure 3 and Table 2 show the binding of CoRBS and anti-cluster A antibodies to W69 mutant variants as quantified by immunoprecipitation and SPR, respectively. With the exception of W69F, the W69 variants were significantly less recognized by these antibodies (Fig. 3), which were mainly due to on-rate effects, as evaluated by SPR. In the case of anti-cluster A antibodies, this is consistent with previous mutagenesis and structural data indicating that tryptophan 69 is localized within the cluster A epitope, contributing to the conformation of this region recognized by this family of antibodies (15, 18–20, 43). However, in the case of CoRBS antibodies, these results are consistent with a model in which mutants W69G, W69A, W69I, and W69L do not spontaneously sample the CD4-bound conformation. Accordingly, incubation with soluble CD4 (sCD4) partially restored 17b binding (Table 2), indicating that these inner domain alterations do not directly disrupt the 17b epitope. Interestingly, while the compensatory layer 2 mutation S115W was sufficient to restore CD4 binding for the W69A variant (Fig. 2A and Table 2), it was unable to restore recognition by CoRBS antibodies (Fig. 3 and Table 2). These data indicate that the S115W mutation does not compensate for the conformational changes induced by the W69A mutation within the bridging sheet required for CoRBS antibody binding.
FIG 3.
Ability of W69 variants to interact with ligands that prefer the CD4-bound conformation. Comparable amounts of radiolabeled monomeric gp120 from WT and W69 variants were incubated with 5 μg/ml of 17b (A), 48d (B), A32 (C), N5-i5 (D), and N60-i3 (E) for 1 h at 37°C. The precipitates were washed, run on SDS-polyacrylamide gels, and analyzed by densitometry. All values shown here were evaluated by the ligand binding capacity of WT gp120 (set as 1). Data represent the averages ± SD from at least three independent experiments. Statistical significance was tested using an unpaired two-tailed t test. Shown are P values with significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Capacity of W69 variants to recognize CD4-binding site antibodies.
Since changes in W69 affected to different degrees the ability of monomeric gp120 to interact with CD4-Ig, we decided to evaluate their effect on recognition by other CD4-binding site ligands. Therefore, we performed immunoprecipitation of normalized amounts of gp120 with potent CD4bs neutralizing antibodies with large (VRC01, VRC03, and VRC13)- and reduced (VRC16 and b12)-breadth neutralization profiles. Interestingly, while no major differences were observed for the broadly neutralizing VRC01 antibody, we found that W69 variants had a significant positive effect in the recognition of monomeric gp120 by the other CD4bs antibodies tested (Fig. 2). More pronounced phenotypes were observed for VRC16 and b12 antibodies, with an enhanced recognition of more than 4-fold for the W69G variant (VRC16). Of note, the double W69A/S115W mutant was recognized by CD4bs antibodies with affinities comparable to or elevated compared to that of the wild type. Altogether, these data indicate that CD4bs antibodies, in contrast to CD4 and CD4i antibodies, most effectively recognize monomeric gp120 variants that poorly sample the CD4-bound conformation. As such, they represent a unique group of Env ligands which bind to Env independently of the presence of a hydrophobic residue at position 69.
Subunit association and processing of the Env glycoprotein mutants.
Previous work identified residues located in alpha helix α1 (layer 2) but not α0 (layer 1) as important for trimer stability (15). However, when the contribution of tryptophan 69 to trimer stability was evaluated in α0, only the W69L variant was tested and no effects on subunit association or Env processing were observed (15). Here, we extended these observations by characterizing the proteolytic processing of gp160 and association of the gp120 and gp41 subunits of each W69 variant. While all of the mutants were expressed efficiently, we observed a marked decrease in proteolytic processing of the gp160 Env precursor that inversely correlated with the size of the residue used to replace W69 (Fig. 4 and Table 1). A similar but less pronounced effect was observed when trimer stability was assessed (Fig. 4 and Table 1); replacing W69 with relatively small residues, such as Gly, Ala, and Ile, enhanced gp120 shedding, but as the size of the replacing residue increased (W69L, W69F) and the gap within the layer 1-layer 2 interface reduced, trimer stability was restored. Of note, mutating S115 to Trp did not restore the processing and association defects of W69A (Fig. 4 and Table 1).
FIG 4.
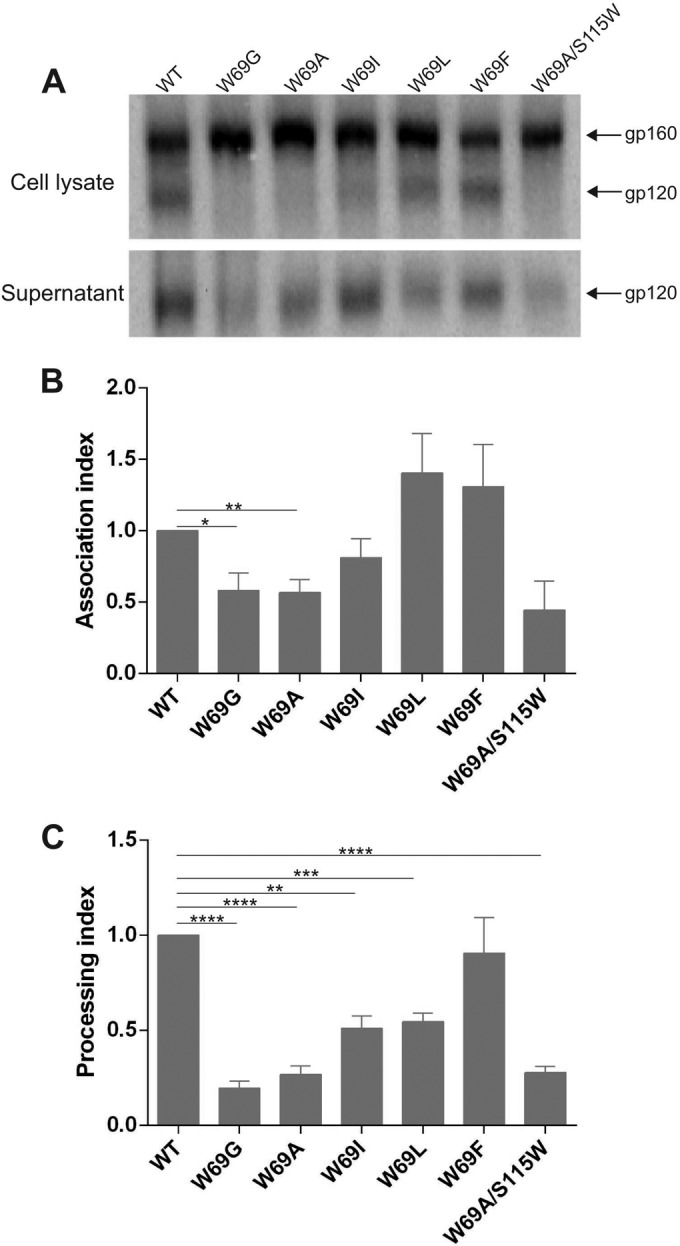
Trimer stability and processing of W69 variants. (A) Cell lysates and supernatants from 35S-labeled cells transiently expressing the HIV-1YU2 WT and W69 variants were precipitated with a mixture of sera from HIV-1-infected patients. The precipitated proteins were loaded onto SDS-PAGE polyacrylamide gels and analyzed by autoradiography and densitometry to calculate their association (B) and processing (C) indexes. The processing index is a measure of the conversion of the mutant gp160 Env precursor to mature gp120 relative to that of the wild-type Env trimers, and the association index is a measure of the ability of the mutant gp120 molecule to remain associated with the envelope glycoprotein complex on the expressing cell relative to that of the wild-type envelope glycoproteins. Both indexes were calculated as described in Materials and Methods. Data shown in panel A are representative of 3 experiments. Data shown in panels B and C represent averages ± SD from at least three independent experiments. Statistical significance was tested using an unpaired two-tailed t test. Shown are P values with significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001).
Function of HIV-1 Env W69 mutants.
The mutant HIV-1 Env trimers were assessed for their ability to mediate cell-cell fusion and their ability to support virus entry into cells expressing CD4 and CCR5. In accordance with their processing and shedding phenotypes, replacing W69 with an alanine dramatically impacted viral infectivity, and the W69G change totally abrogated it. However, as the size of the replacing residue increased, we observed a progressive recovery of viral infectivity, with W69F presenting no infectivity defects (Fig. 5A and Table 1). The phenotype of the W69 mutants with regard to their ability to mediate cell-to-cell fusion was less pronounced but correlated with their infectivity phenotype. Indeed, W69G presented the lowest levels of cell-to-cell fusion, but increasing the size of the residue at position W69 progressively restored this activity to wild-type levels (Fig. 5B). Mutating S115 to Trp did not restore the infectivity defect of W69A (Fig. 5 and Table 1).
FIG 5.
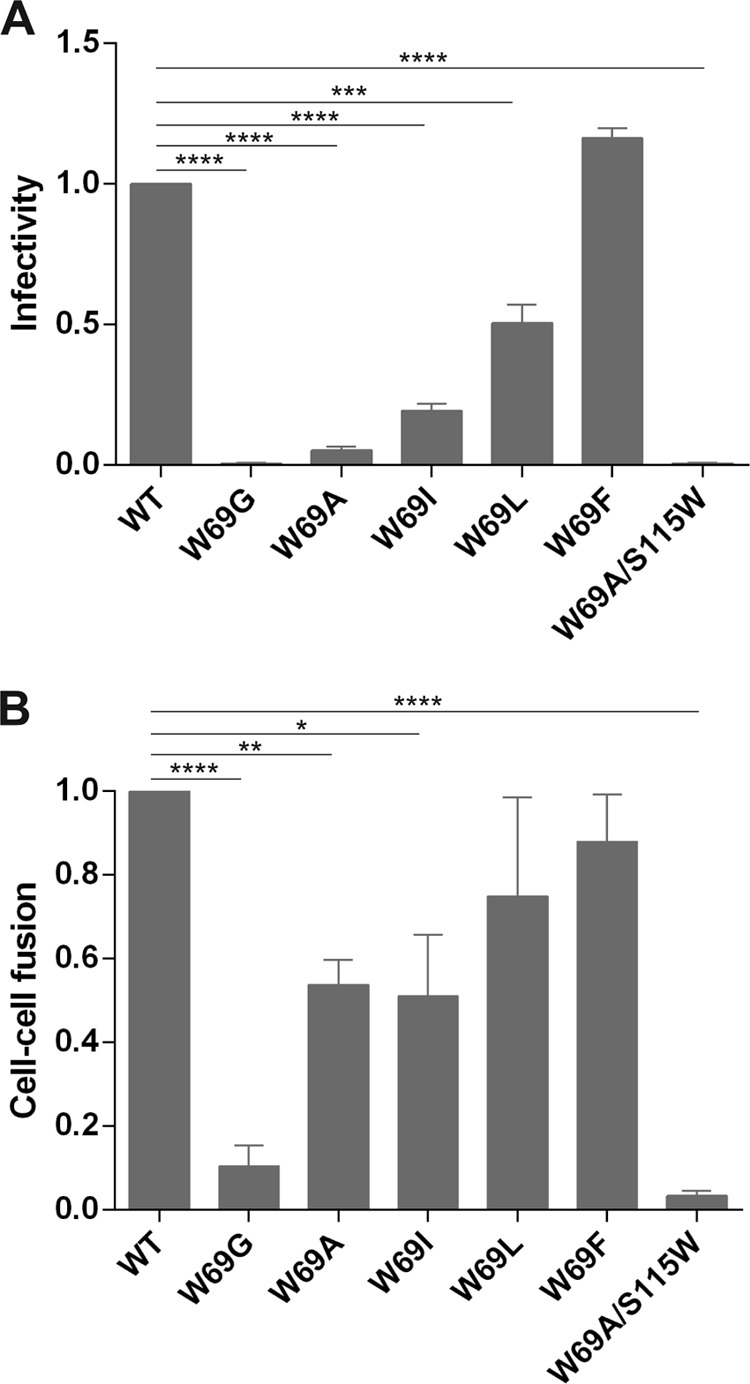
Capacity of W69 variants to mediate viral infectivity and cell-to-cell fusion. (A) To measure the infectivity mediated by WT and W69 Env variants, Cf2Th-CD4-CCR5 cells were infected with RT-normalized amounts of HIV-1 pseudoviruses and maintained at 37°C for 48 h. Luciferase activity in the cell lysate was then measured to determine the infectivity of each virus. Data shown are the ratios of mutant to wild-type virus infectivity. (B) 293T cells expressing Env from WT and W69 variants were incubated with TZM-bl cells for 6 h at 37°C. Luciferase activity in the mixture of cell lysate was measured to determine the cell-to-cell fusion efficiency. Data shown are representative of results from at least three independent experiments performed in quadruplicate. Statistical significance was tested using unpaired two-tailed t test. Shown are P values with significant differences (*, P < 0.05; **, P < 0.01; ***, P < 0.001; ****, P < 0.0001).
Differences in the cell-to-cell fusion and viral infectivity assays can be explained by the fact that in cell-to-cell fusion, Env is constantly transported to the cell membrane; therefore, even if a mutant has a shedding phenotype, there are newly synthesized Envs able to engage with CD4 on the target cell. On the contrary, in the virus infectivity assay, once gp120 is shed from the viral particle, it is impossible to replace. Therefore, viral infectivity assays are more sensitive to decreases in the stability of gp120-trimer association, as has been previously reported (15, 16, 24).
Sensitivity of the mutant viruses to neutralization by sCD4 and CD4bs antibodies.
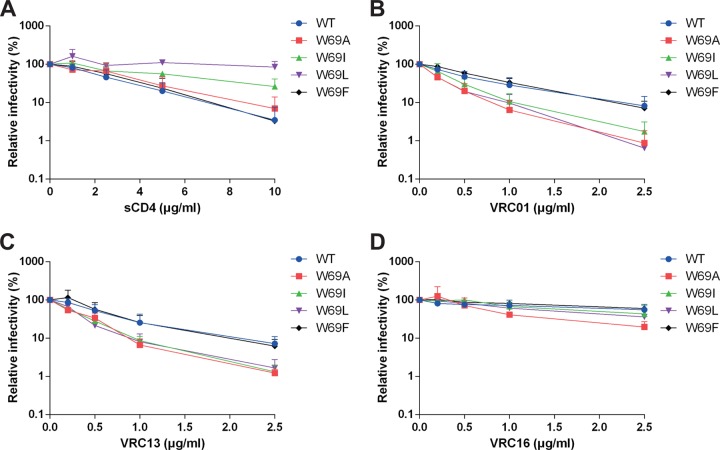
Since W69G and W69A/S115W were not infectious (Fig. 5A and Table 1), we could not evaluate their sensitivity to sCD4; however, W69A, W69I, W69L, and W69F variants retained enough infectivity potential for neutralization experiments. As previously reported, W69L was resistant to sCD4 neutralization (Fig. 6A) (15) and W69I presented the same level of resistance, both of which are consistent with their decreased CD4 binding. W69F bound CD4 better than these mutants, and accordingly it was sensitive to sCD4 neutralization in a manner similar to that of the wild-type envelope. A similar inverse correlation between ligand interaction and neutralization was observed when assessing neutralization by VRC01, VRC13, and VRC16 (not shown). Indeed, VRC01 recognized the wild-type and W69F variants equivalently and thus neutralized viral particles bearing these variants equivalently, but it appeared to neutralize W69A, W69I, and W69L slightly more efficiently. However, VRC13 and VRC16 bound more efficiently to the W69A, W69I, and W69L variants (Fig. 2) and therefore were able to neutralize viral particles presenting these Env mutants more readily than their wild-type or W69F counterpart (Fig. 6).
FIG 6.
Viral neutralization by CD4-binding site ligands. Recombinant HIV-1 expressing luciferase and bearing WT or W69 Env variants were normalized by reverse transcriptase activity. Equal amounts of viruses were incubated with serial dilutions of sCD4 (A), VRC01 (B), VRC13 (C), and VRC16 (D) for 1 h at 37°C before infecting Cf2Th-CD4-CCR5 cells. Luciferase activity in cell lysates was measured to determine the infectivity. Infectivity was normalized to 100% in the absence of the ligand. Data shown are representative of results from at least three independent experiments performed in quadruplicate.
Structural analysis of the W69A/S115W mutant.
To evaluate if and to what extent the double W69A/S115W mutant restored structural properties of the wild-type protein, we obtained a 3.0-Å-resolution crystal structure of the complex formed by W69A/S115W gp120 coree, CD4 peptide mimetic M48U1, and Fab of anti-cluster A antibody N60-i3 (PDB entry 5KJR). Of note, while the double mutant abrogated N60-i3 interaction within the gp120 full-length context (Fig. 3E), we observed that when introduced within coree, the mutations permitted sufficient interaction with N60-i3 (not shown), allowing cocrystallization and further supporting a role of the variable regions in gp120 conformation (35). The complex crystallized in the same space group with unit parameters (Table 3) very similar to those of the assembly of the wild-type gp120 coree complexed with the same CD4 mimetic and N60-i3 Fab, as determined by us previously (43). Since both complexes were obtained in the same space group and with the same crystal packing (Table 4 and Fig. 7A), we could analyze changes to the structure solely attributed to the mutations (i.e., not induced by crystal packing).
TABLE 3.
Details of the N60-i3-gp12093TH057 and -gp12093TH057(W69A/S115W) coree interfacesa
| Component | BSA (Å2) of: |
|
|---|---|---|
| N60-i3 Fab-gp120(W69A/S115W)93TH057 coree-M48U1 |
N60-i3 Fab-gp12093TH057 coree-M48U1 |
|
| gp12093TH057 coree total | 695 | 736 |
| 7-Stranded β-sandwich | 2 | 1 |
| Layer 1 | 546 | 575 |
| Layer 2 | 147 | 160 |
| Heavy chain total | 657 | 673 |
| FWR | 17 | 6 |
| CDR H1 | 109 | 117 |
| CDR H2 | 154 | 159 |
| CDR H3 | 377 | 391 |
| Light chain total | 114 | 133 |
| CDR L1 | 75 | 94 |
| CDR L2 | 0 | 0 |
| CDR L3 | 39 | 39 |
| Heavy and light chain total | 771 | 806 |
| Fab-gp12093TH057 coree total | 1,466 | 1,542 |
Details were calculated by the EBI PISA server (http://www.ebi.ac.uk/msd-srv/prot_int/cgi-bin/piserver).
FIG 7.
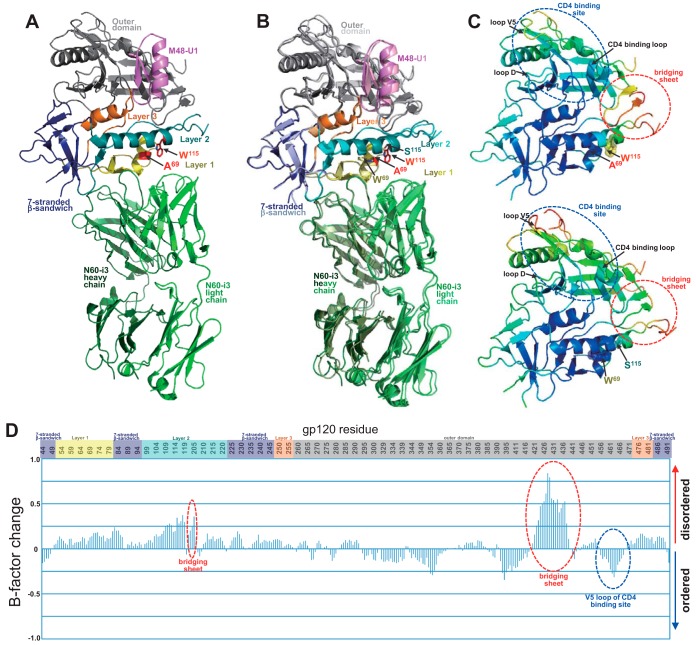
Structure of monomeric gp120 (W69A/S115W) in complex with N60-i3. (A) N60-i3 Fab-g12093TH057(W69A/S115W)-M48-U1 complex ribbon diagram. N60-i3 Fab heavy and light chains are colored dark/light green, respectively. The gp120 outer domain is in gray, the 7-stranded β-sandwich in blue, mobile layer 1 in yellow, layer 2 in cyan, and layer 3 in orange. M48-U1 is in purple. Mutated residues are shown with sticks in red. (B) Superposition of the mutant N60-i3 Fab-g12093TH057(W69A/S115W)-M48-U1 complex on the wild-type N60-i3 Fab-g12093TH057-M48-U1 complex. The mutant and wild-type gp120 complexes are colored with dark/light colors, respectively, as described for panel A. Mutated residues are shown with sticks in red if mutated and with gp120 colors if wild type. (C) gp120 mutant (top) and wild-type (bottom) colors are based on the normalized B factors, with blue representing lower B factor and red higher B factor. B factors were scaled to range from 2 to 120 and the average shifted to 60 to allow comparison between the two structures. (D) Relative B factor difference for the mutant compared to wild-type gp120 scaled so that the maximum difference is 1.
At first glance, both complexes show very close similarity, as indicated by a root mean square deviation (RMSD) of 0.972 Å for 3,132 main-chain atoms of the complex (Table 5) and fully preserved interfaces between the N60-i3 Fab and gp120 coree variants (Table 6). Despite variable intramolecular contacts mediated by A69 and W115 in the mutant compared to W69 and Ser115 (Table 6) in the wild-type protein, respectively, the layer 1-layer 2 interface is well maintained in both structures.
TABLE 5.
Root mean square deviations for gp120(W69A/S115W)93TH057 and wild-type gp12093TH057 corees
| Component | Value (RMSD; Å) fora: |
||
|---|---|---|---|
| 1 | 2 | 3 | |
| Complex | 0.972 | ||
| Fab | 0.924 | ||
| gp12093TH057 coree | 0.719 | 0.926 | 0.868 |
| 7-Stranded β-sandwich | 0.511 | 0.643 | 0.804 |
| Layer 1 | 0.448 | 0.581 | 0.683 |
| Layer 2 | 0.725 | 1.18 | 0.985 |
| Layer 3 | 0.376 | 0.269 | 0.372 |
| Outer domain | 0.786 | 0.838 | 0.843 |
Root mean square deviations (RMSD) for gp120 coree atoms of N60-i3 Fab-gp120(W69A/S115W)93TH057 coree-M48U1 and N60-i3 Fab-gp12093TH057-M48U1 (PDB entry 4RFO) complexes (column 1), N60-i3 Fab-gp120(W69A/S115W)93TH057 coree-M48U1 and N5-i5- gp12093TH057-d1d2CD4 (PDB entry 4H8W) complexes (column 2), and N60-i3 Fab-gp12093TH057-M48U1 (PDB entry 4RFO) and N5-i5- gp12093TH057-d1d2CD4 (PDB entry 4H8W) complexes (column 3) are shown.
TABLE 6.
Interactions at the N60-i3-gp120(W69A/S115W) coree and N60-i3-gp120 coree interfacea
| Component | Mutant |
WT |
||
|---|---|---|---|---|
| ASA (Å2) | BSA (Å2) | ASA (Å2) | BSA (Å2) | |
| N60-i3 heavy chain | ||||
| FWR H1 | ||||
| H:ALA 27 | 45.17 | 4.66 || | ||
| H:SER 28 | 48.90 | 1.31 | | ||
| H:ILE 29 | 72.85 | 0.34 | | ||
| H:SER 30 | 66.54 | 16.09 ||| | ||
| CDR H1 | ||||
| H:SER 31 | 88.82 | 38.65 ||||| | 57.48 | 40.16 ||||||| |
| H:GLY 32 | 26.76 | 22.11 ||||||||| | 21.24 | 18.92 ||||||||| |
| H:GLY 33 | 26.92 | 26.92 |||||||||| | 24.18 | 24.18 ||||||||||| |
| H:TYR 34 | 58.03 | 17.42 |||| | 36.53 | 10.69 ||| |
| H:PHE 35 | 15.83 | 11.75 |||||||| | 20.18 | 15.32 |||||||| |
| CDR H2 | ||||
| H:TYR 52 | 48.97 | 44.26 |||||||||| | 43.42 | 40.61 |||||||||| |
| H:TYR 53H | 67.42 | 28.88 ||||| | 68.62 | 36.74 |||||| |
| H:ILE 54 | 101.52 | 56.12 |||||| | 98.99 | 52.24 |||||| |
| H:ASN 56 | 83.65 | 17.72 ||| | 64.79 | 17.84 ||| |
| H:TYR 58 | 88.46 | 12.06 || | 88.33 | 6.31 | |
| CDR H3 | ||||
| H:ARG 97H | 121.87 | 87.91 |||||||| | 113.20 | 76.08 ||||||| |
| H:LEU 98H | 126.83 | 87.51 ||||||| | 120.56 | 84.50 |||||||| |
| H:ARG 99HS | 238.01 | 191.35 ||||||||| | 239.92 | 194.91 ||||||||| |
| H:GLY 100 | 35.50 | 14.38 ||||| | 37.93 | 15.68 ||||| |
| H:ASN 100B | 93.19 | 10.29 || | 79.51 | 5.97 | |
| gp12093TH057(W69A/S115W) coree/gp12093TH057 coree | ||||
| Layer 1 | ||||
| G:THR 50 | 35.52 | 1.23 | | ||
| G:THR 51 | 109.67 | 46.63 ||||| | 112.44 | 47.77 ||||| |
| G:LEU 52H | 16.59 | 16.26 |||||||||| | 16.68 | 16.01 |||||||||| |
| G:PHE 53 | 83.81 | 72.25 ||||||||| | 80.16 | 70.31 ||||||||| |
| G:CYS 54H | 7.43 | 7.09 |||||||||| | 7.28 | 7.28 ||||||||||| |
| G:ALA 69/TRP 69 | 17.88 | 1.88 || | ||
| G:THR 71H | 60.43 | 10.53 || | 77.17 | 9.56 || |
| G:HIS 72 | 110.56 | 19.66 || | 101.61 | 21.96 ||| |
| G:ALA 73 | 86.34 | 66.14 |||||||| | 60.28 | 50.80 ||||||||| |
| G:CYS 74H | 22.83 | 9.29 ||||| | 12.93 | 10.32 |||||||| |
| G:VAL 75 | 72.69 | 69.69 |||||||||| | 65.92 | 61.92 |||||||||| |
| G:PRO 76 | 115.14 | 83.42 |||||||| | 118.17 | 84.65 |||||||| |
| G:THR 77 | 45.72 | 7.35 || | 41.33 | 7.37 || |
| G:ASP 78H | 74.63 | 37.45 |||||| | 72.18 | 20.65 ||| |
| G:PRO 79 | 120.58 | 17.10 || | 127.06 | 28.45 ||| |
| Layer 2 | ||||
| G:GLN 103H | 21.50 | 10.73 ||||| | 27.08 | 17.37 ||||||| |
| G:GLU 106HS | 104.31 | 15.47 || | 116.19 | 12.91 || |
| G:ASP 107S | 57.72 | 23.58 ||||| | 32.53 | 25.32 |||||||| |
| G:TYR 217 | 9.77 | 5.44 |||||| | 8.67 | 4.62 |||||| |
| G:THR 219 | 18.62 | 5.65 |||| | 10.55 | 5.89 |||||| |
| G:PRO 220 | 40.49 | 30.69 |||||||| | 45.37 | 34.28 |||||||| |
| G:ALA 221 | 106.84 | 48.67 ||||| | 92.55 | 43.36 ||||| |
| 7-Stranded β-sandwich | ||||
| G:GLN 246 | 90.39 | 0.61 | | 74.22 | 1.65 | |
| N60-i3 light chain | ||||
| CDR L1 | mutant BSA | wt ASA | wt BSA | |
| L:GLY 29 | 44.03 | 13.32 |||| | 41.52 | 0.70 | |
| L:TYR 30 | 65.84 | 33.72 |||||| | 68.43 | 28.71 ||||| |
| L:TYR 32H | 108.15 | 47.52 ||||| | 109.87 | 45.39 ||||| |
| CDR L3 | ||||
| L:TYR 91 | 50.16 | 28.11 |||||| | 48.19 | 31.11 ||||||| |
| L:SER 94 | 101.39 | 0.17 | | ||
| L:SER 95 | 75.49 | 10.69 || | 73.08 | 7.80 || |
| gp12093TH057(W69A/S115W) coree/gp12093TH057 coree | ||||
| Layer 1 | ||||
| G:ALA 60 | 67.49 | 3.35 | | 86.42 | 0.25 | |
| G:VAL 68 | 51.15 | 2.00 | | 73.47 | 2.51 | |
| G:THR 71 | 60.43 | 13.90 ||| | 77.17 | 17.32 ||| |
| G:HIS 72H | 110.56 | 80.78 |||||||| | 101.61 | 77.40 |||||||| |
| G:ALA 73 | 86.34 | 11.26 || | 60.28 | 9.48 || |
| Layer 2 | ||||
| G:GLN 114 | 114.52 | 13.12 || | 90.05 | 3.08 | |
| G:TRP 115/SER 115 | 60.07 | 6.86 || | ||
Accessible surface area (ASA) and buried surface area (BSA) were calculated using the EBI PISA server (http://www.ebi.ac.uk/msd-srv/prot_int/cgi-bin/piserver). Bars represent the buried surface/accessible surface percentage (10% per bar). Residues contributing to the interface through H bonds or salt bridges are indicated by a superscript H or S, respectively.
Despite close similarities of overall structures, there are several noticeable differences between these two structures, revealed clearly when B factors for gp120 molecules within both complexes are analyzed (Fig. 7C and D). First of all, the bridging sheet within the W69A/S115W mutant is largely destabilized, as indicated by several residues in this region that are disordered and missing from the refined model (Fig. 7A and B, residues 121 to 123), supporting the observed decreased binding of the double mutant to CoRBS antibodies (Fig. 3A and B). High B factors for remaining bridging sheet residues also indicate high mobility of this region compared to the wild-type protein (Fig. 7C, top, and D, shift from blue to yellow). Destabilization, but to a lower extend, is also observed for components of the CD4-binding site, as indicated by higher B factors for V5 and CD4 binding loops. Also, although compensatory, the S115W mutation is unable to fully restore the rigidity of the wild-type protein at the interface between the α0 (layer 1) and α1 (layer 2) helices. In the wild-type structure the side chain of W69 is highly ordered, with lower B factors (Fig. 7C, bottom, and D, blue), whereas A69 is less ordered, with the C terminus of the α1-helix partially destabilized (Fig. 7C, top, and D, shift from blue to green). S115W compensates for destabilization of α1-helix but is unable to completely restore order within layer 1 (Fig. 7C, bottom, and D, yellow). Together these data, although obtained with the mutant bearing a compensating S115W mutation, identify the bridging sheet region (i.e., the region involved in CoRBS antibody binding), the CD4 binding site region (i.e., the region involved in CD4 and CD4bs antibody binding), and the interface between the α0 (layer 1) and α1 (layer 2) helices (i.e., the region involved in anti-cluster A antibody binding) as the most affected by the W69A mutation.
DISCUSSION
The inner domain of HIV-1 gp120 can be subdivided into two parts: (i) a β-sandwich, which mutagenesis studies suggest contributes to gp41 association (1, 47), and (ii) three layers (or loop-like extensions) that emanate from the strands of the β-sandwich and project toward the target cell. These layers form topologically separate and structurally plastic components that are capable of acting as shape-changing spacers stabilizing the unliganded Env trimer and required to facilitate the transition of Env to the CD4-bound conformation (15, 16, 24).
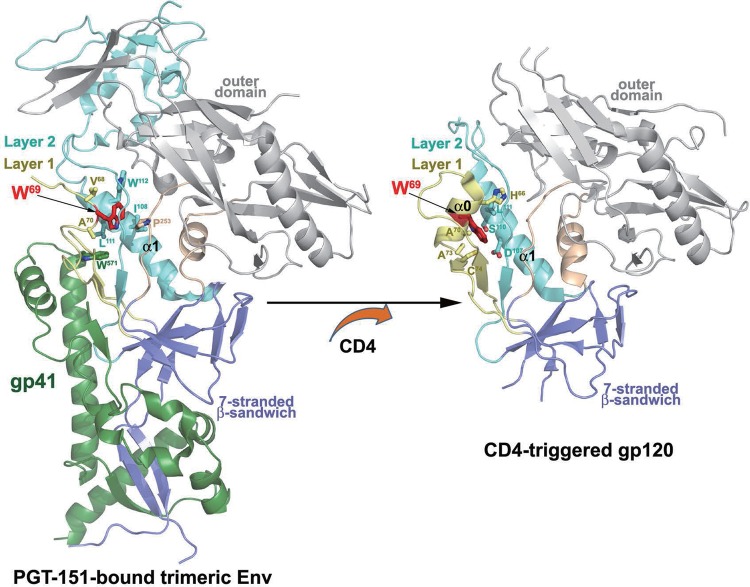
Residues located at the interface between these layers, such as His 66 or Leu 111, were previously shown to modulate this transition (15, 16, 21, 22). The recently solved cryoelectron microscopy (cryo-EM) structure of an antibody-bound, fully glycosylated, cleaved HIV-1 envelope trimer (48) and crystal structures of several complexes of monomeric gp120 in its CD4-bound conformation (18, 19, 43, 49) provided us with atomic-resolution details about the assembly of the inner domain layers and localization of tryptophan 69 (W69) in the context of the PGT151-bound trimer and monomeric gp120 core in its CD4-bound state. As shown in Fig. 8, W69 sits between layer 1 and layer 2 of the inner domain in the PGT151-stabilized trimer and the CD4-triggered gp120 but is involved with different contact residues. In the PGT151-stabilized trimer, the α0 helix is not formed and W69 reaches deeper inside the gp120 molecule, making intraprotomer contacts with V68 and A70 (layer 1); I108, L111, and W112 (layer 2); P253 (layer 3); and W571 of gp41 HR1. In contrast, upon CD4 binding, W69 is localized at the center of the newly formed α0 helix and sandwiched between layers 1 and 2, contacting H66, A70, A73, and C74 (layer 1) and D107, S110, and L111 (layer 2). Overall, the buried surface area (BSA) of W69 in the PGT151-stabilized trimer is 2.3 Å, whereas it is 7.9 Å in the CD4-triggered gp120 core (calculated based on PDB entry 4H8W). These data indicate that W69 is localized in the region that undergoes the most significant structural changes during the CD4-induced rearrangements of Env. In addition, the bulky, hydrophobic side chain of W69, which is completely buried at the layer 1-layer 2 interface of the CD4-triggered gp120 (Fig. 8), contributes to the stability of the CD4-bound conformation of gp120. Accordingly, by evaluating the role of hydrophobicity at position 69 on Env transition to the CD4-bound conformation, we observed that residues with decreasing hydrophobicity indexes progressively impacted the spontaneous sampling of the CD4-bound state. Indeed, we observed the biggest impact on CD4 and CD4i antibody binding for the residues with the shortest aliphatic chain and almost no difference for residues with bulky aromatic chains. Interestingly, adding a bulky residue on layer 2 at position 115 (S115W) rescued CD4 binding for the W69A mutant, indicating that the hydrophobicity lost due to the W69A mutation at the layer 1-layer 2 interface can be compensated for by adding a hydrophobic residue at a different interlayer position. Similarly, it was previously reported that replacing Leu 111 with a Trp (L111W) could compensate for some W69L phenotypes (15). However, while S115W was able to rescue CD4 binding of W69A, it did not recover CD4i antibody binding, indicating that this double mutant is unable to fully restore the capacity of gp120 to transition to the CD4-bound conformation. This could be explained by small but noticeable structural differences between the inner domain double mutant and its WT counterpart. Indeed, the W69A/S115W mutant presented a largely destabilized bridging sheet, explaining the lack of recognition by CoRBS antibodies. Additionally, we observed increased B factors and, thus, higher mobility for residues forming the CD4 binding site and the V5 and the CD4 binding loops in particular (Fig. 8). Interestingly, destabilization of the CD4-binding site region seems to have variable effects on the capacity of different CD4-binding site ligands to recognize Env. Whereas CD4 shows decreased recognition of Env mutants bearing less hydrophobic (than Trp) residues at position 69, CD4bs neutralizing antibodies with large- and reduced-breadth neutralization preferentially recognized variants with residues other than Trp. These findings indicate that CD4bs antibodies are capable of tolerating a wide range of Env conformations, including one with altered or destroyed interlayer contacts. This is in sharp contrast to CD4, which is very sensitive to perturbations at the layer1-layer2 interface.
FIG 8.
Localization of W69 within the PGT151-bound HIV-1 envelope trimer and CD4-triggered gp120. The gp120-gp41 protomer from the cryo-EM structure of a fully glycosylated, PGT151-bound, cleaved HIV-1 envelope trimer (48) and gp120 coree from the Fab N5-i5-gp120 coree-d1d2CD4 structure (19) is shown as a ribbon diagram with W69 highlighted in red. Residues that contact W69 (as determined by 4-Å distance cutoff) are shown as a ball and sticks.
The role played by W69 on gp120-gp41 interaction and the transition to the CD4-bound conformation suggest that this highly conserved residue, located at the interface between gp120 and gp41 in the PGT151-bound trimer, plays a critical role in the interprotomer signaling induced by CD4 binding.
ACKNOWLEDGMENTS
We thank John Mascola (VRC, NIH) for kindly providing VRC01, VRC03, VRC13, and VRC16. We thank Joseph Sodroski for critical discussions.
This work was supported by a Canada Foundation for Innovation Program Leader grant, by CIHR operating grants 119334 and 134117, by FRQS Establishment of Young Scientist grant 26702 to A.F., by the FRQS AIDS and Infectious Diseases Network, and by NIH AI100645, Center for HIV/AIDS Vaccine Immunology and Immunogen Design (CHAVI-ID), and NIAID, NIH, grant R01AI116274 to M.P. A.F. is the recipient of a Canada Research Chair on Retroviral Entry.
The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. We have no conflicts of interest to report.
REFERENCES
- 1.Helseth E, Olshevsky U, Furman C, Sodroski J. 1991. Human immunodeficiency virus type 1 gp120 envelope glycoprotein regions important for association with the gp41 transmembrane glycoprotein. J Virol 65:2119–2123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dalgleish AG, Beverley PC, Clapham PR, Crawford DH, Greaves MF, Weiss RA. 1984. The CD4 (T4) antigen is an essential component of the receptor for the AIDS retrovirus. Nature 312:763–767. doi: 10.1038/312763a0. [DOI] [PubMed] [Google Scholar]
- 3.Klatzmann D, Champagne E, Chamaret S, Gruest J, Guetard D, Hercend T, Gluckman JC, Montagnier L. 1984. T-lymphocyte T4 molecule behaves as the receptor for human retrovirus LAV. Nature 312:767–768. doi: 10.1038/312767a0. [DOI] [PubMed] [Google Scholar]
- 4.Alkhatib G, Combadiere C, Broder CC, Feng Y, Kennedy PE, Murphy PM, Berger EA. 1996. CC CKR5: a RANTES, MIP-1alpha, MIP-1beta receptor as a fusion cofactor for macrophage-tropic HIV-1. Science 272:1955–1958. doi: 10.1126/science.272.5270.1955. [DOI] [PubMed] [Google Scholar]
- 5.Choe H, Farzan M, Sun Y, Sullivan N, Rollins B, Ponath PD, Wu L, Mackay CR, LaRosa G, Newman W, Gerard N, Gerard C, Sodroski J. 1996. The beta-chemokine receptors CCR3 and CCR5 facilitate infection by primary HIV-1 isolates. Cell 85:1135–1148. doi: 10.1016/S0092-8674(00)81313-6. [DOI] [PubMed] [Google Scholar]
- 6.Deng H, Liu R, Ellmeier W, Choe S, Unutmaz D, Burkhart M, Di Marzio P, Marmon S, Sutton RE, Hill CM, Davis CB, Peiper SC, Schall TJ, Littman DR, Landau NR. 1996. Identification of a major co-receptor for primary isolates of HIV-1. Nature 381:661–666. doi: 10.1038/381661a0. [DOI] [PubMed] [Google Scholar]
- 7.Doranz BJ, Rucker J, Yi Y, Smyth RJ, Samson M, Peiper SC, Parmentier M, Collman RG, Doms RW. 1996. A dual-tropic primary HIV-1 isolate that uses fusin and the beta-chemokine receptors CKR-5, CKR-3, and CKR-2b as fusion cofactors. Cell 85:1149–1158. doi: 10.1016/S0092-8674(00)81314-8. [DOI] [PubMed] [Google Scholar]
- 8.Dragic T, Litwin V, Allaway GP, Martin SR, Huang Y, Nagashima KA, Cayanan C, Maddon PJ, Koup RA, Moore JP, Paxton WA. 1996. HIV-1 entry into CD4+ cells is mediated by the chemokine receptor CC-CKR-5. Nature 381:667–673. doi: 10.1038/381667a0. [DOI] [PubMed] [Google Scholar]
- 9.Feng Y, Broder CC, Kennedy PE, Berger EA. 1996. HIV-1 entry cofactor: functional cDNA cloning of a seven-transmembrane, G protein-coupled receptor. Science 272:872–877. doi: 10.1126/science.272.5263.872. [DOI] [PubMed] [Google Scholar]
- 10.Trkola A, Dragic T, Arthos J, Binley JM, Olson WC, Allaway GP, Cheng-Mayer C, Robinson J, Maddon PJ, Moore JP. 1996. CD4-dependent, antibody-sensitive interactions between HIV-1 and its co-receptor CCR-5. Nature 384:184–187. doi: 10.1038/384184a0. [DOI] [PubMed] [Google Scholar]
- 11.Wu L, Gerard NP, Wyatt R, Choe H, Parolin C, Ruffing N, Borsetti A, Cardoso AA, Desjardin E, Newman W, Gerard C, Sodroski J. 1996. CD4-induced interaction of primary HIV-1 gp120 glycoproteins with the chemokine receptor CCR-5. Nature 384:179–183. doi: 10.1038/384179a0. [DOI] [PubMed] [Google Scholar]
- 12.Lu M, Blacklow SC, Kim PS. 1995. A trimeric structural domain of the HIV-1 transmembrane glycoprotein. Nat Struct Biol 2:1075–1082. doi: 10.1038/nsb1295-1075. [DOI] [PubMed] [Google Scholar]
- 13.Chan DC, Fass D, Berger JM, Kim PS. 1997. Core structure of gp41 from the HIV envelope glycoprotein. Cell 89:263–273. doi: 10.1016/S0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 14.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. 1997. Atomic structure of the ectodomain from HIV-1 gp41. Nature 387:426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 15.Finzi A, Xiang SH, Pacheco B, Wang L, Haight J, Kassa A, Danek B, Pancera M, Kwong PD, Sodroski J. 2010. Topological layers in the HIV-1 gp120 inner domain regulate gp41 interaction and CD4-triggered conformational transitions. Mol Cell 37:656–667. doi: 10.1016/j.molcel.2010.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Desormeaux A, Coutu M, Medjahed H, Pacheco B, Herschhorn A, Gu C, Xiang SH, Mao Y, Sodroski J, Finzi A. 2013. The highly conserved layer-3 component of the HIV-1 gp120 inner domain is critical for CD4-required conformational transitions. J Virol 87:2549–2562. doi: 10.1128/JVI.03104-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Veillette M, Coutu M, Richard J, Batraville LA, Dagher O, Bernard N, Tremblay C, Kaufmann DE, Roger M, Finzi A. 2014. The HIV-1 gp120 CD4-bound conformation is preferentially targeted by ADCC-mediating antibodies in sera from HIV-1-infected individuals. J Virol 89:545–551. doi: 10.1128/JVI.02868-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tolbert WD, Gohain N, Veillette M, Chapleau JP, Orlandi C, Visciano ML, Ebadi M, DeVico AL, Fouts TR, Finzi A, Lewis GK, Pazgier M. 2016. Paring down HIV Env: design and crystal structure of a stabilized inner domain of HIV-1 gp120 displaying a major ADCC target of the A32 region. Structure 24:697–709. doi: 10.1016/j.str.2016.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Acharya P, Tolbert WD, Gohain N, Wu X, Yu L, Liu T, Huang W, Huang CC, Kwon YD, Louder RK, Luongo TS, McLellan JS, Pancera M, Yang Y, Zhang B, Flinko R, Foulke JS Jr, Sajadi MM, Kamin-Lewis R, Robinson JE, Martin L, Kwong PD, Guan Y, DeVico AL, Lewis GK, Pazgier M. 2014. Structural definition of an antibody-dependent cellular cytotoxicity response implicated in reduced risk for HIV-1 infection. J Virol 88:12895–12906. doi: 10.1128/JVI.02194-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ding S, Veillette M, Coutu M, Prevost J, Scharf L, Bjorkman PJ, Ferrari G, Robinson JE, Sturzel C, Hahn BH, Sauter D, Kirchhoff F, Lewis GK, Pazgier M, Finzi A. 2016. A highly conserved residue of the HIV-1 gp120 inner domain is important for antibody-dependent cellular cytotoxicity responses mediated by anti-cluster A antibodies. J Virol 90:2127–2134. doi: 10.1128/JVI.02779-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kassa A, Finzi A, Pancera M, Courter JR, Smith AB III, Sodroski J. 2009. Identification of a human immunodeficiency virus (HIV-1) envelope glycoprotein variant resistant to cold inactivation. J Virol 83:4476–4488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kassa A, Madani N, Schon A, Haim H, Finzi A, Xiang SH, Wang L, Princiotto A, Pancera M, Courter J, Smith AB III, Freire E, Kwong PD, Sodroski J. 2009. Transitions to and from the CD4-bound conformation are modulated by a single-residue change in the human immunodeficiency virus type 1 gp120 inner domain. J Virol 83:8364–8378. doi: 10.1128/JVI.00594-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Xiang SH, Finzi A, Pacheco B, Alexander K, Yuan W, Rizzuto C, Huang CC, Kwong PD, Sodroski J. 2010. A V3 loop-dependent gp120 element disrupted by CD4 binding stabilizes the human immunodeficiency virus envelope glycoprotein trimer. J Virol 84:3147–3161. doi: 10.1128/JVI.02587-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Finzi A, Pacheco B, Xiang SH, Pancera M, Herschhorn A, Wang L, Zeng X, Desormeaux A, Kwong PD, Sodroski J. 2012. Lineage-specific differences between human and simian immunodeficiency virus regulation of gp120 trimer association and CD4 binding. J Virol 86:8974–8986. doi: 10.1128/JVI.01076-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Medjahed H, Pacheco B, Desormeaux A, Sodroski J, Finzi A. 2013. The HIV-1 gp120 major variable regions modulate cold inactivation. J Virol 87:4103–4111. doi: 10.1128/JVI.03124-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.LaBonte JA, Patel T, Hofmann W, Sodroski J. 2000. Importance of membrane fusion mediated by human immunodeficiency virus envelope glycoproteins for lysis of primary CD4-positive T cells. J Virol 74:10690–10698. doi: 10.1128/JVI.74.22.10690-10698.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Platt EJ, Wehrly K, Kuhmann SE, Chesebro B, Kabat D. 1998. Effects of CCR5 and CD4 cell surface concentrations on infections by macrophagetropic isolates of human immunodeficiency virus type 1. J Virol 72:2855–2864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Korber B, Foley BT, Kuiken C, Pillai SK, Sodroski JG. 1998. Numbering positions in HIV relative to HXB2CG, p 102–111, vol III In Human retroviruses and AIDS, vol III Los Alamos National Laboratory, Los Alamos, NM. [Google Scholar]
- 29.Rho HM, Poiesz B, Ruscetti FW, Gallo RC. 1981. Characterization of the reverse transcriptase from a new retrovirus (HTLV) produced by a human cutaneous T-cell lymphoma cell line. Virology 112:355–360. doi: 10.1016/0042-6822(81)90642-5. [DOI] [PubMed] [Google Scholar]
- 30.Coutu M, Finzi A. 2015. HIV-1 gp120 dimers decrease the overall affinity of gp120 preparations for CD4-induced ligands. J Virol Methods 215-216:37–44. [DOI] [PubMed] [Google Scholar]
- 31.Finzi A, Pacheco B, Zeng X, Kwon YD, Kwong PD, Sodroski J. 2010. Conformational characterization of aberrant disulfide-linked HIV-1 gp120 dimers secreted from overexpressing cells. J Virol Methods 168:155–161. doi: 10.1016/j.jviromet.2010.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Johnsson B, Lofas S, Lindquist G. 1991. Immobilization of proteins to a carboxymethyldextran-modified gold surface for biospecific interaction analysis in surface plasmon resonance sensors. Anal Biochem 198:268–277. doi: 10.1016/0003-2697(91)90424-R. [DOI] [PubMed] [Google Scholar]
- 33.Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen MY, Pieper U, Sali A. 2006. Comparative protein structure modeling using Modeller. Curr Protoc Bioinformatics Chapter 5:Unit 5.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Acharya P, Luongo TS, Louder MK, McKee K, Yang Y, Kwon YD, Mascola JR, Kessler P, Martin L, Kwong PD. 2013. Structural basis for highly effective HIV-1 neutralization by CD4-mimetic miniproteins revealed by 1.5 A cocrystal structure of gp120 and M48U1. Structure 21:1018–1029. doi: 10.1016/j.str.2013.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kwon YD, Finzi A, Wu X, Dogo-Isonagie C, Lee LK, Moore LR, Schmidt SD, Stuckey J, Yang Y, Zhou T, Zhu J, Vicic DA, Debnath AK, Shapiro L, Bewley CA, Mascola JR, Sodroski JG, Kwong PD. 2012. Unliganded HIV-1 gp120 core structures assume the CD4-bound conformation with regulation by quaternary interactions and variable loops. Proc Natl Acad Sci U S A 109:5663–5668. doi: 10.1073/pnas.1112391109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Otwinowski Z, Minor W, Carter CW Jr. 1997. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol 276:307–326. [DOI] [PubMed] [Google Scholar]
- 37.McCoy AJ. 2007. Solving structures of protein complexes by molecular replacement with Phaser. Acta Crystallogr D Biol Crystallogr 63:32–41. doi: 10.1107/S0907444906045975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Collaborative Computational Project Number 4. 1994. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr 50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 39.Murshudov GN, Vagin AA, Dodson EJ. 1997. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr 53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 40.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. 2002. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr 58:1948–1954. doi: 10.1107/S0907444902016657. [DOI] [PubMed] [Google Scholar]
- 41.Emsley P, Cowtan K. 2004. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr 60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 42.Wimley WC, White SH. 1996. Experimentally determined hydrophobicity scale for proteins at membrane interfaces. Nat Struct Biol 3:842–848. doi: 10.1038/nsb1096-842. [DOI] [PubMed] [Google Scholar]
- 43.Gohain N, Tolbert WD, Acharya P, Yu L, Liu T, Zhao P, Orlandi C, Visciano ML, Kamin-Lewis R, Sajadi MM, Martin L, Robinson JE, Kwong PD, DeVico AL, Ray K, Lewis GK, Pazgier M. 2015. Cocrystal structures of antibody N60-i3 and antibody JR4 in complex with gp120 define more cluster A epitopes involved in effective antibody-dependent effector function against HIV-1. J Virol 89:8840–8854. doi: 10.1128/JVI.01232-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moore JP, Sodroski J. 1996. Antibody cross-competition analysis of the human immunodeficiency virus type 1 gp120 exterior envelope glycoprotein. J Virol 70:1863–1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Guan Y, Pazgier M, Sajadi MM, Kamin-Lewis R, Al-Darmarki S, Flinko R, Lovo E, Wu X, Robinson JE, Seaman MS, Fouts TR, Gallo RC, DeVico AL, Lewis GK. 2013. Diverse specificity and effector function among human antibodies to HIV-1 envelope glycoprotein epitopes exposed by CD4 binding. Proc Natl Acad Sci U S A 110:E69–E78. doi: 10.1073/pnas.1217609110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Veillette M, Desormeaux A, Medjahed H, Gharsallah NE, Coutu M, Baalwa J, Guan Y, Lewis G, Ferrari G, Hahn BH, Haynes BF, Robinson JE, Kaufmann DE, Bonsignori M, Sodroski J, Finzi A. 2014. Interaction with cellular CD4 exposes HIV-1 envelope epitopes targeted by antibody-dependent cell-mediated cytotoxicity. J Virol 88:2633–2644. doi: 10.1128/JVI.03230-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang X, Mahony E, Holm GH, Kassa A, Sodroski J. 2003. Role of the gp120 inner domain beta-sandwich in the interaction between the human immunodeficiency virus envelope glycoprotein subunits. Virology 313:117–125. doi: 10.1016/S0042-6822(03)00273-3. [DOI] [PubMed] [Google Scholar]
- 48.Lee JH, Ozorowski G, Ward AB. 2016. Cryo-EM structure of a native, fully glycosylated, cleaved HIV-1 envelope trimer. Science 351:1043–1048. doi: 10.1126/science.aad2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pancera M, Majeed S, Ban YE, Chen L, Huang CC, Kong L, Kwon YD, Stuckey J, Zhou T, Robinson JE, Schief WR, Sodroski J, Wyatt R, Kwong PD. 2010. Structure of HIV-1 gp120 with gp41-interactive region reveals layered envelope architecture and basis of conformational mobility. Proc Natl Acad Sci U S A 107:1166–1171. doi: 10.1073/pnas.0911004107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Brünger AT. 1992. Free R value: a novel statistical quantity for assessing the accuracy of crystal structures. Nature 355:472–475. [DOI] [PubMed] [Google Scholar]