Abstract
The human papillomavirus (HPV) life cycle is tightly linked to differentiation of the infected epithelium. This means that viral proteins must exert control over epithelial gene expression in order to optimize viral production. The HPV E2 protein controls replication, transcription, and viral genome partitioning during the viral infectious life cycle. It consists of a nucleic acid-binding domain and a protein-protein interaction domain separated by a flexible serine and arginine-rich hinge region. Over the last few years, mounting evidence has uncovered an important new role for E2 in viral and cellular RNA processing. This Gem discusses the role of E2 in controlling the epithelial cellular environment and how E2 might act to coordinate late events in the viral replication cycle.
HUMAN PAPILLOMAVIRUS LIFE CYCLE
Human papillomaviruses (HPVs) are small (∼8.0-kbp) circular double-stranded DNA viruses that infect epithelial cells (keratinocytes) (1). There are more than 200 HPV genotypes. For the most part, HPVs of the α-genotype group infect mucosal epithelia, while the β-genotypes infect predominantly cutaneous epithelia (2). Most HPV infections are asymptomatic, or they manifest as benign lesions or warts. They are usually transient and are eventually cleared by the immune system (3). Despite this, HPVs cause a high burden of clinically significant disease worldwide, because the anogenital mucosa-infective high-risk group of HPVs (HR-HPVs) can cause persistent infection that leads to epithelial dysplasia and neoplasia. HR-HPVs are commonly associated with cervical cancer and tumors of other anogenital sites (2). Per annum, worldwide, there are around 500,000 new cases of cervical disease, and around 270,000 women die from cervical cancer. The two most prevalent HR-HPVs are HPV16 and -18, which are targeted by the current HPV vaccines (4). Over the last few decades, epidemic HR-HPV infection, especially in men, has been associated with a significant increase in oropharyngeal cancers (5). Certain cutaneous HPVs can also cause tumors (squamous cell carcinomas) in immunocompromised or immunosuppressed individuals (6). Thus, the causative association of HPV infection with a number of significant cancers indicates that increased understanding of the viral life cycle is essential.
The HPV life cycle is intimately linked to epithelial differentiation (Fig. 1A) (2). HPV normally infects dividing cells at the base of the epithelium (basal layer) but completes its life cycle by amplifying progeny DNA genomes in the midepithelial layers (spinous layer) and carrying out viral encapsidation in the uppermost epithelial layer (granular layer) (Fig. 1A). Viral production is achieved through a highly orchestrated and complex viral gene expression program (Fig. 1A) (2). Mature virions are released upon shedding and disintegration of dead superficial epithelial cells into the environment. As a necessity, HPV infection alters the normal differentiation pattern of the epithelium and controls cellular gene expression to support viral replication (2). In short, differentiating cells in the midepithelial layers are induced into cell division, and epithelial differentiation is somewhat delayed.
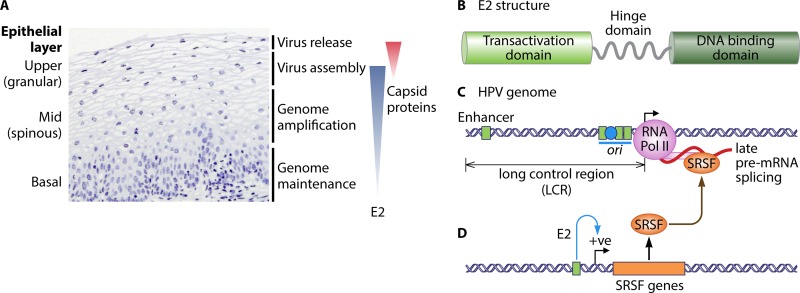
FIG 1.
Activity of E2 during the human papillomavirus (HPV) replication cycle. (A) Hematoxylin and eosin-stained (blue stain) high-risk human papillomavirus (HR-HPV)-positive low-grade cervical lesion. The various epithelial layers are indicated on the left. Note the enlarged nuclei (a hallmark of HPV infection) of cells in the midepithelial layers. The events in the HPV replication cycle and an approximate indication of where they occur within the infected epithelium are indicated on the right. The blue-shaded triangle indicates the extent of E2 expression in the epithelium, and the red-shaded triangle indicates the restricted expression of the viral capsid proteins L1 and L2 in the upper epithelial layers. (B) Diagrammatic representation of the domain structure of E2. Light green, the transactivation domain; dark green, the DNA binding domain; gray wavy line, the hinge domain. (C) A portion of an α-genotype HPV genome is shown from the enhancer in the long control region (LCR) to the late gene region. In the LCR, the four E2 homodimer binding sites are represented by green squares and the E1 binding site is indicated with a blue circle in the origin of DNA replication (ori, blue horizontal line). In mid- to upper epithelial layers, the viral late promoter (black arrow) is active, and the viral late RNAs (shown as a red curved line) are expressed. RNA polymerase II (Pol II) is shown as a pink circle, with an extension indicating the carboxy-terminal domain. SRSF proteins are indicated by an orange oval binding the late pre-mRNA. (D) An E2 binding site (green box) is shown in the promoter of SRSF genes (orange rectangle); E2 (not shown) binds to it to trans-activate (blue curved arrow, positive [+ve]) expression of SRSF genes. A brown curved arrow indicates E2-transactivated SRSF (orange oval)-regulated activity in HPV late mRNA processing (31). However, E2 may also upregulate late mRNA production directly by recruiting other cellular RNA processing factors (39) or by acting as a viral SRSF-like regulator of late mRNA splicing (21).
RNA PROCESSING
Cellular and viral gene expression is controlled at both the transcriptional and posttranscriptional levels. Posttranscriptional events include RNA processing (the expression of mature mRNA from the primary transcript [pre-mRNA], by capping, polyadenylation, and splicing), mRNA export from the nucleus to the cytoplasm, mRNA stability, and mRNA translation on the ribosomes. Tight control of all of these events is essential to ensure the appropriate profile of protein production in the cell.
Splicing is the process by which noncoding introns are removed from pre-mRNAs and exons are spliced together to form a protein-coding mRNA. However, pre-mRNAs transcribed from multiexon/intron genes can undergo a process termed alternative splicing (7). Alternative splicing is a regulated process in which a primary transcript can be spliced in more than one pattern (exon removal or retention) to generate multiple, distinct “splice isoform” mRNAs. For example, the average human gene can give rise to eight different mRNA isoforms, each encoding a different protein isoform, meaning that alternative splicing is a key player in determining the final cellular protein profile. Alternative splicing is crucial for the life cycle of some viruses, such as HIV, which expresses more than 40 different alternatively spliced mRNAs (8). Splicing is controlled by specific cellular activators (serine-arginine-rich splicing factors [SRSFs]) and repressors (heterogeneous nuclear ribonucleoproteins [hnRNPs]) (9). There are nine classical SRSFs (SRSF1 to -9) (10) that consist of at least one RNA recognition motif (RRM) that binds the pre-mRNA and one serine-arginine-rich domain that interacts with partner proteins, such as other splicing factors (11). The serine-arginine domain can undergo cycles of phosphorylation/dephosphorylation to control splicing, protein-protein interactions, and subcellular location (11). The relative levels of SRSF proteins are pivotal in maintaining the correct balance of cellular mRNA isoforms in normal cells, while overexpression of some SRSF proteins has been shown to lead to cancer progression (12).
Splicing occurs cotranscriptionally, linked through the carboxy-terminal domain of RNA polymerase II, which acts as a landing pad to recruit mRNA processing factors (13). Moreover, SRSFs have been shown to regulate elongation of transcription by RNA polymerase II (13). Apart from regulating transcription and splicing, SRSFs have multiple other roles in the cell, including regulating polyadenylation, nuclear export, mRNA stability, translation, genome stability maintenance, nucleolar stress, cell cycle progression, control of apoptosis, and protein sumoylation (12). This means that SRSFs can coordinately control transcription and splicing in the nucleus while also providing a link to other posttranscriptional processes and the cellular environment.
HPV E2
E2 plays a crucial role in the HPV life cycle (14). It possesses a DNA-binding domain and a transactivation domain that are linked by a serine-arginine-rich hinge region (Fig. 1B) (15). E2 is normally found as a homodimer that binds cognate sequences (E2-binding sites [E2BSs]) in the viral long control region (LCR) (Fig. 1C), which contains the viral early promoter, origin of replication, enhancer, and, upstream, the late 3′ untranslated region (UTR) and polyadenylation sites (not shown in Fig. 1C). For the mucosa-infective HPVs, two E2BSs are proximal to the viral early promoter, the third is located at the origin of DNA replication, and the fourth is located in the enhancer region (Fig. 1C) (14). E2 has been shown to be capable of either activating or repressing transcription in different experimental systems. However, the evidence points to a repressive function of E2 in controlling the viral early promoter (14). On the other hand, the most important role of E2 may be as an auxiliary replication factor. At the origin of DNA replication, E2 interacts with and loads the HPV E1 replication helicase, which in turn recruits the cellular DNA replication machinery (14). E2 also has an important role in partitioning viral episomal genomes during the division of infected cells by interacting with chromatin adapter proteins that tether them to host mitotic chromosomes (14).
E2 AS AN SRSF
There is evidence that E2, in addition to having DNA-related roles, can bind RNA and has properties and functions similar to those of SRSF proteins. Like the serine-arginine domain in which phosphorylation controls the function of SRSF proteins, the long hinge region (130 to 209 amino acids) of some β-type E2 proteins is rich in serine and arginine residues and can be phosphorylated (16, 17). Like all SR proteins, E2 is normally located in the nucleus (18) and, at least for HPV1 and HPV5, can be found in nuclear speckles (16, 19), the storage sites of splicing factors in the nucleus. HR-HPV E2 proteins can also shuttle between the nucleus and the cytoplasm (20). An early report indicated that the β-genotype HPV5 E2, which carries a long hinge region, could regulate both transcription and splicing of reporter pre-mRNAs (19). However, a later study could not verify this finding and instead found that the α-genotype HPV16 E2, which possesses a much shorter hinge region (14), could inhibit splicing in vitro (21). Although E2 does not possess an RNA recognition motif required by the SRSFs for RNA binding (11), one study showed that HPV16 E2 could bind RNA directly via its carboxy-terminal domain in a UV cross-linking assay (21). However, improved approaches, such as cross-linking immunoprecipitation (CLIP) in vivo, are required before E2 can be considered a true RNA-binding protein.
The cellular interactomes of E2 proteins from a range of different HPV genotypes have been dissected by two studies. Although not yet critically evaluated in most cases, as expected, many E2-interacting proteins are transcription, replication, or chromatin remodelling factors or histone-modifying enzymes (22, 23). E2 also interacts with proteins involved in apoptosis regulation, cell cycle, keratinocyte migration and differentiation, and intracellular transport. Although the majority of E2 interactors are involved in processes related to transcription, replication, or chromatin remodelling, the next most frequently identified set of proteins are involved in RNA processing. These proteins included components of the basic splicing machinery, such as SMN, SF1, and U170K (Table 1). In addition, serine-arginine protein kinases (SRPKs), which control SRSFs through phosphorylation (24), were also detected (22, 23, 25). Of particular note, E2 from most of the genotypes tested interacted with the splicing activators SRSF1, -2, and -7 and -10 (Tra2β) (22, 23). The involvement of E2 in posttranscriptional as well as transcriptional processes is perhaps unsurprising, given the close association between transcription and RNA processing. These studies suggest that E2 may integrate viral transcription and RNA processing, leading to the efficient production of processed viral mRNAs. Although further work is required to support this hypothesis, it is clear that E2 has properties suggestive of a role in controlling pre-mRNA splicing by recruiting cellular splicing factors.
TABLE 1.
Known E2 interactions with RNA splicing factors
| Protein | Alias(es) | Function(s)a | HPV type(s) |
|---|---|---|---|
| SRSF1 | ASF, SF2, SRp30a | Control of pre-mRNA splicing; paradigm SRSF protein | 1, 3, 5, 6, 8, 9, 11, 16, 18 |
| SRSF2 | SC35, PR264, SRp30b | Control of pre-mRNA splicing | 1, 5, 8, 9 |
| SRSF4 | SRp75 | Control of pre-mRNA splicing | 16 |
| SRSF5 | SRp40, HRS | Control of pre-mRNA splicing | 16 |
| SRSF7 | 9G8 | Control of pre-mRNA splicing | 1, 3, 5, 6, 8, 9, 11, 16, 32, 39 |
| Tra2β | SRSF10, PPP1R156, TRANS2B | Control of pre-mRNA splicing | 5 |
| SRPK1 | SFRSK1 | SRSF protein kinase | 1, 8, 11, 16, 18, 31 |
| SRPK2 | SFRSK1 | SRSF protein kinase | 1, 8 |
| SMN | Gemin1, BCD541, SMA | Role in assembly of snRNPs that form the splicing machinery | 16, 18, 11 |
| SF1 | ZNF162, ZFM1, BBP, D11S636, ZCCHC25 | Necessary for spliceosome assembly: branch point binding protein | 16 |
| snRNP70 | U170K, RPU1, U1AP1, Snp1, RNPU1Z | Component protein of U1 snRNP, the first RNA-protein complex to interact with pre-mRNA in a splicing reaction | 5 |
| PRPF31 | PRP31, NYBR99 | Required for the assembly of the U4.U5.U6 tri-snRNP in a splicing reaction | 1, 5, 6, 8, 9, 18 |
| EFTUD2 | SNRP116, U5116KD, HSNU114, MFDGA, MFDM | Spliceosome component | 1, 8, 11, 16, 18, 31 |
| PCBP1 | hnRNPE1, hnRNPX, HELS85 | Regulates alternative splicing | 1, 3, 5, 8, 9, 16, 18 |
| TNPO3 | Importin12, TRN-SR | Nuclear import of SRSF proteins | 5 |
snRNPs, small nuclear ribonucleoproteins.
E2 AS A REGULATOR OF SRSFs
In normal keratinocytes, E2 is detected at highest levels in the mid- to upper epithelial layers (Fig. 1A) (26), where viral vegetative replication takes place. Interestingly, expression of the late mRNAs that encode the viral capsid proteins also begins in these layers (Fig. 1A) (27, 28). In vivo, SRSF expression is highest in uninfected, undifferentiated basal epithelial cells. However, SRSF levels are high in differentiated HPV-infected keratinocytes (Fig. 1A) (29, 30). Specifically, HPV16 and HPV31 E2 can transcriptionally upregulate the expression of at least SRSF1, -2, and -3 (Fig. 1D) (31). This exciting finding implies that because the relative levels of SRSFs in a cell determine alternative splicing outcomes and thus the overall protein profile, E2 may control the cellular environment via control of SRSF expression to optimize viral replication. In support of this, a recent study has revealed that overexpression of E2 in U2OS osteosarcoma cells resulted in very significant changes in cellular alternative splicing, with 522 mRNAs affected (32). Cancer genes and genes encoding proteins involved in cell motility were the most affected. U2OS cells are tumor cells that have altered gene expression compared to that of normal cells, and this may explain the cancer-related pattern of gene expression changes. It will be of interest to see how E2 can alter splicing in the normal keratinocytes that are the target of HPV infection and whether the process of epithelial differentiation required for a productive HPV life cycle alters cellular splicing. These studies reveal that E2 has a pivotal role to play in controlling cellular and viral splicing and may regulate splicing by stimulating SRSF expression, acting as an SRSF to regulate splicing directly or by modulating SRSF activity through direct binding.
E2 CONTROLS LATE EVENTS IN VIRAL REPLICATION
Apart from E1, there is evidence that E2 can bind other viral proteins, such as E4 (33), E6 (25), E7 (34), L1 (35), and L2 (36). In particular, E2 binding L1 and L2 might affect virus capsid formation and virus release. However, E2 may have a more fundamental role to play in controlling late events in the viral replication cycle as a transcription activator. E2-mediated control of SRSF expression may have implications for the viral replication cycle, because viral proteins are translated from multiple mRNAs that are the products of SRSF-regulated alternative splicing (http://pave.niaid.nih.gov/#explore/transcriptmaps) (37). Previous studies on keratinocytes stably transfected with a genome of HPV31 (the genotype most closely related to HPV16) containing an E2 point mutation (I73L) that inactivated the E2 trans-activation function showed no effect on viral replication but a reduction of 80% in viral late mRNA expression (38). Recently, we extended this observation to show that in keratinocytes maintaining E2::I73L mutant genomes, there was a 75% decrease in capsid protein production compared with that of wild-type HPV31 genomes (31). SRSF1, -2, and -3 levels in the E2::I73L genome-containing cells were all significantly reduced, in agreement with wild-type E2 trans-activation of the promoters of the genes encoding these proteins (Fig. 1D). In order to discover which SRSF protein was responsible for controlling capsid protein expression, SRSF1, -2, -3, -5, and -7 were depleted by using small interfering RNA (siRNA) in differentiated HPV16-positive keratinocytes. Depletion only of SRSF1 and -3 caused a change in L1 capsid protein expression, with SRSF3 causing the greater reduction (50 to 55%). Conversely, SRSF3 overexpression in undifferentiated keratinocytes resulted in some induction of L1 protein expression (31). SRSF3 was required for the production of the spliced E4^L1 mRNA that encodes the L1 major capsid protein. SRSF3 likely controls viral RNA alternative splicing because, upon depletion, concomitant with a decrease in levels of the E4^L1 mRNA, there was an increase in the unspliced L2L1 mRNA that encodes the L2 minor capsid protein. Although the observations of E2-mediated transcriptional control of SRSF and capsid protein expression and SRSF3-mediated control of capsid protein expression are correlative, it seems reasonable to propose that E2 induces SRSF levels in differentiated HPV-infected keratinocytes in order to facilitate viral capsid protein expression and completion of the viral replication cycle. Interestingly, another study showed that E2 could control induction of HPV late mRNA expression posttranscriptionally, and this phenomenon was observed for HPVs of different phylogenetic types, namely, HPV1, -5, and -16 (39). In this case, both the E2 transactivation domain and the hinge region were required for control of capsid mRNA levels. E2 interacted with the 30-kDa component of the cleavage and polyadenylation specificity factor (CPSF) complex and controlled use of the viral early polyadenylation signal (39). Similar to the level of change that we observed, E2 overexpression induced a 3- to 5-fold change in late mRNA expression, although late protein expression was not examined. Therefore, the data suggest that there may be two E2-mediated posttranscriptional mechanisms for regulating viral late gene expression: SRSF-mediated control of splicing and CPSF-mediated control of polyadenylation.
SUMMARY AND PERSPECTIVES
E2 can bind viral capsid proteins directly, and, at least for L1, this interaction can enhance E2-dependent replication and transcription activation (35). However, E2-mediated posttranscriptional control of L1 expression indicates that the physical E2-L1 interaction can be ensured in differentiating keratinocytes, in which viral genome replication takes place. It is clear from the new data described above that E2 may integrate and regulate all the main late events in the viral life cycle. Aside from RNA processing, SRSF proteins may control other events in the life cycle of RNA molecules, including nuclear export, mRNA stability, and translation (11). If E2 controls SRSF expression or if E2 has SRSF-like activities, it might affect viral mRNA export to the cytoplasm, stability, and translation. A full examination of how E2 integrates the various late events in viral replication with the cellular environment via E2-mediated control of SRSF is now warranted.
ACKNOWLEDGMENTS
Thanks go to Arwa Ali A Faizo for figure development.
This work was supported by Wellcome Trust grant Wtd004098. I acknowledge funding from the Medical Research Council as core funding for the MRC-University of Glasgow Centre for Virus Research.
Funding Statement
The funders had no role in the study design, data collection and interpretation, or the decision to submit this review for publication.
REFERENCES
- 1.zur Hausen H. 2009. Papillomaviruses in the causation of human cancers—a brief historical account. Virology 384:260–265. doi: 10.1016/j.virol.2008.11.046. [DOI] [PubMed] [Google Scholar]
- 2.Egawa N, Egawa K, Griffin H, Doorbar J. 2015. Human papillomaviruses; epithelial tropisms, and the development of neoplasia. Viruses 7:2802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stanley MA. 2012. Epithelial cell responses to infection with human papillomavirus. Clin Microbiol Rev 25:215–222. doi: 10.1128/CMR.05028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stanley MA. 2006. Human papillomavirus vaccines. Rev Med Virol 16:139–149. doi: 10.1002/rmv.498. [DOI] [PubMed] [Google Scholar]
- 5.Gillison ML, Chaturvedi AK, Anderson WF, Fakhry C. 2015. Epidemiology of human papillomavirus-positive head and neck squamous cell carcinoma. J Clin Oncol 33:3235–3242. doi: 10.1200/JCO.2015.61.6995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reusser N, Downing C, Guidry J, Tyring S. 2015. HPV carcinomas in immunocompromised patients. J Clin Med 4:260. doi: 10.3390/jcm4020260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Black DL. 2003. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem 72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 8.Tazi J, Bakkour N, Marchand V, Ayadi L, Aboufirassi A, Branlant C. 2010. Alternative splicing: regulation of HIV-1 multiplication as a target for therapeutic action. FEBS J 277:867–876. doi: 10.1111/j.1742-4658.2009.07522.x. [DOI] [PubMed] [Google Scholar]
- 9.Busch A, Hertel KJ. 2012. Evolution of SR protein and hnRNP splicing regulatory factors. Wiley Interdiscip Rev RNA 3:1–12. doi: 10.1002/wrna.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Manley JL, Krainer AR. 2010. A rational nomenclature for serine/arginine-rich protein splicing factors (SR proteins). Genes Dev 24:1073–1074. doi: 10.1101/gad.1934910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Long JC, Caceres JF. 2009. The SR protein family of splicing factors: master regulators of gene expression. Biochem J 417:15–27. doi: 10.1042/BJ20081501. [DOI] [PubMed] [Google Scholar]
- 12.Das S, Krainer AR. 2014. Emerging functions of SRSF1, splicing factor and oncoprotein, in RNA metabolism and cancer. Mol Cancer Res 12:1195–1204. doi: 10.1158/1541-7786.MCR-14-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shukla S, Oberdoerffer S. 2012. Co-transcriptional regulation of alternative pre-mRNA splicing. Biochim Biophys Acta 1819:673–683. doi: 10.1016/j.bbagrm.2012.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McBride AA. 2013. The papillomavirus E2 proteins. Virology 445:57–79. doi: 10.1016/j.virol.2013.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hegde RS. 2002. The papillomavirus E2 proteins: structure, function, and biology. Annu Rev Biophys Biomol Struct 31:343–360. doi: 10.1146/annurev.biophys.31.100901.142129. [DOI] [PubMed] [Google Scholar]
- 16.Sekhar V, Reed SC, McBride AA. 2010. Interaction of the beta-papillomavirus E2 tethering protein with mitotic chromosomes. J Virol 84:543–557. doi: 10.1128/JVI.01908-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sekhar V, McBride AA. 2012. Phosphorylation regulates binding of the human papillomavirus type 8 E2 protein to host chromosomes. J Virol 86:10047–10058. doi: 10.1128/JVI.01140-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hubbert NL, Schiller JT, Lowy DR, Androphy EJ. 1988. Bovine papilloma virus-transformed cells contain multiple E2 proteins. Proc Natl Acad Sci U S A 85:5864–5868. doi: 10.1073/pnas.85.16.5864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lai M-C, Teh BH, Tarn W-Y. 1999. A human papillomavirus E2 transcriptional activator. J Biol Chem 274:11832–11841. doi: 10.1074/jbc.274.17.11832. [DOI] [PubMed] [Google Scholar]
- 20.Blachon S, Bellanger S, Demeret C, Thierry F. 2005. Nucleo-cytoplasmic shuttling of high risk human papillomavirus E2 proteins induces apoptosis. J Biol Chem 280:36088–36098. doi: 10.1074/jbc.M505138200. [DOI] [PubMed] [Google Scholar]
- 21.Bodaghi S, Jia R, Zheng Z-M. 2009. Human papillomavirus type 16 E2 and E6 are RNA-binding proteins and inhibit in vitro splicing of pre-mRNAs with suboptimal splice sites. Virology 386:32–43. doi: 10.1016/j.virol.2008.12.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muller M, Jacob Y, Jones L, Weiss A, Brino L, Chantier T, Lotteau V, Favre M, Demeret C. 2012. Large scale genotype comparison of human papillomavirus E2-host interaction networks provides new insights for E2 molecular functions. PLoS Pathog 8:e1002761. doi: 10.1371/journal.ppat.1002761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jang MK, Anderson DE, van Doorslaer K, McBride AA. 2015. A proteomic approach to discover and compare interacting partners of papillomavirus E2 proteins from diverse phylogenetic groups. Proteomics 15:13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Giannakouros T, Nikolakaki E, Mylonis I, Georgatsou E. 2011. Serine-arginine protein kinases: a small protein kinase family with a large cellular presence. FEBS J 278:570–586. doi: 10.1111/j.1742-4658.2010.07987.x. [DOI] [PubMed] [Google Scholar]
- 25.Grm HS, Massimi P, Gammoh N, Banks L. 2005. Crosstalk between the human papillomavirus E2 transcriptional activator and the E6 oncoprotein. Oncogene 24:5149–5164. doi: 10.1038/sj.onc.1208701. [DOI] [PubMed] [Google Scholar]
- 26.Xue Y, Bellanger S, Zhang W, Lim D, Low J, Lunny D, Thierry F. 2010. HPV16 E2 is an immediate early marker of viral infection, preceding E7 expression in precursor structures of cervical carcinoma. Cancer Res 70:5316–5325. doi: 10.1158/0008-5472.CAN-09-3789. [DOI] [PubMed] [Google Scholar]
- 27.Stoler MH, Wolinsky SM, Whitbeck A, Broker TR, Chow LT. 1989. Differentiation-linked human papillomavirus types 6 and 11 transcription in genital condylomata revealed by in situ hybridization with message-specific RNA probes. Virology 172:331–340. doi: 10.1016/0042-6822(89)90135-9. [DOI] [PubMed] [Google Scholar]
- 28.Stoler MH, Rhodes CR, Whitbeck A, Wolinsky SM, Chow LT, Broker TR. 1992. Human papillomavirus type 16 and 18 gene expression in cervical neoplasia. Hum Pathol 23:117–128. doi: 10.1016/0046-8177(92)90232-R. [DOI] [PubMed] [Google Scholar]
- 29.Mole S, Milligan SG, Graham SV. 2009. Human papillomavirus type 16 E2 protein transcriptionally activates the promoter of a key cellular splicing factor, SF2/ASF. J Virol 83:357–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mole S, McFarlane M, Chuen-Im T, Milligan SG, Millan D, Graham SV. 2009. RNA splicing factors regulated by HPV16 during cervical tumour progression. J Pathol 219:383–391. doi: 10.1002/path.2608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Klymenko T, Hernandez-Lopez H, MacDonald AI, Bodily JM, Graham SV. 2016. Human papillomavirus E2 regulates SRSF3 (SRp20) to promote capsid protein expression in infected differentiated keratinocytes. J Virol 90:5047–5058. doi: 10.1128/JVI.03073-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gauson EJ, Windle B, Donaldson MM, Caffarel MM, Dornan ES, Coleman N, Herzyk P, Henderson SC, Wang X, Morgan IM. 2014. Regulation of human genome expression and RNA splicing by human papillomavirus 16 E2 protein. Virology 468–470:10–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Davy C, McIntosh P, Jackson DJ, Sorathia R, Miell M, Wang Q, Khan J, Soneji Y, Doorbar J. 2009. A novel interaction between the human papillomavirus type 16 E2 and E1^E4 proteins leads to stabilization of E2. Virology 394:266–275. doi: 10.1016/j.virol.2009.08.035. [DOI] [PubMed] [Google Scholar]
- 34.Gammoh N, Grm HS, Massimi P, Banks L. 2006. Regulation of human papillomavirus type 16 E7 activity through direct protein interaction with the E2 transcriptional activator. J Virol 80:1787–1797. doi: 10.1128/JVI.80.4.1787-1797.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Siddiqa A, Léon KC, James CD, Bhatti MF, Roberts S, Parish JL. 2015. The human papillomavirus type 16 L1 protein directly interacts with E2 and enhances E2-dependent replication and transcription activation. J Gen Virol 96:2274–2285. doi: 10.1099/vir.0.000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Day PM, Roden RBS, Lowy DR, Schiller JT. 1998. The papillomavirus minor capsid protein, L2, induces localisation of the major capsid protein, L1, and the viral transcription/replication protein, E2, to PML oncogenic domains. J Virol 72:142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johansson C, Schwartz S. 2013. Regulation of human papillomavirus gene expression by splicing and polyadenylation. Nat Rev Microbiol 11:239–251. doi: 10.1038/nrmicro2984. [DOI] [PubMed] [Google Scholar]
- 38.Stubenrauch F, Colbert AME, Laimins LA. 1998. Transactivation by the E2 protein of oncogenic human papillomavirus type 31 is not essential for early and late viral functions. J Virol 72:8115–8123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Johannson C, Somberg M, Li X, Winquist EB, Fay J, Ryan F, Pim D, Banks L, Schwartz S. 2012. HPV-16 E2 contributes to induction of HPV-16 late gene expression by inhibiting early polyadenylation. EMBO J 31:3212–3227. doi: 10.1038/emboj.2012.147. [DOI] [PMC free article] [PubMed] [Google Scholar]