Abstract
IMPORTANCE
Sarcoidosis is a major cause of ocular or periocular inflammation. The pathogenesis of sarcoidosis is incompletely understood and diagnosis often requires a biopsy.
OBJECTIVE
To determine how gene expression in either orbital adipose tissue or the lacrimal gland affected by sarcoidosis compares with gene expression in other causes of orbital disease and how gene expression in tissue affected by sarcoidosis compares with gene expression in peripheral blood samples obtained from patients with sarcoidosis.
DESIGN, SETTING, AND PARTICIPANTS
In a multicenter, international, observational study, gene expression profiling of formalin-fixed biopsy specimens, using GeneChipp U133 Plus 2 microarrays (Affymetrix), was conducted between October 2012 and January 2014 on tissues biopsied from January 2000 through June 2013. Participants included 12 patients with orbital sarcoidosis (7 in adipose tissue; 5 affecting the lacrimal gland) as well as comparable tissue from 6 healthy individuals serving as controls or patients with thyroid eye disease, nonspecific orbital inflammation, or granulomatosis with polyangiitis. In addition, results were compared with gene expression in peripheral blood samples obtained from 12 historical individuals with sarcoidosis.
MAIN OUTCOMES AND MEASURES
Significantly differentially expressed transcripts defined as a minimum of a 1.5-fold increase or a comparable decrease and a false discovery rate of P < .05.
RESULTS
Signals from 2449 probe sets (transcripts from approximately 1522 genes) were significantly increased in the orbital adipose tissue from patients with sarcoidosis. Signals from 4050 probe sets (approximately 2619 genes) were significantly decreased. Signals from 3069 probe sets (approximately 2001 genes) were significantly higher and 3320 (approximately 2283 genes) were significantly lower in the lacrimal gland for patients with sarcoidosis. Ninety-two probe sets (approximately 69 genes) had significantly elevated signals and 67 probe sets (approximately 56 genes) had significantly lower signals in both orbital tissues and in peripheral blood from patients with sarcoidosis. The transcription factors, interferon-response factor 1, interferon-response factor 2, and nuclear factor κB, were strongly implicated in the expression of messenger RNA upregulated in common in the 3 tissues.
CONCLUSIONS AND RELEVANCE
Gene expression in sarcoidosis involving the orbit or lacrimal gland can be distinguished from gene expression patterns in control tissue and overlaps with many transcripts upregulated or downregulated in the peripheral blood of patients with sarcoidosis. These observations suggest that common pathogenic mechanisms contribute to sarcoidosis in different sites. The observations support the hypothesis that a pattern of gene expression profiles could provide diagnostic information in patients with sarcoidosis.
Sarcoidosis is a granulomatous inflammatory disease; the pathogenesis is incompletely understood. Several studies have suggested an infectious or environmental trigger. For example, catalase-peroxidase derived from Mycobacterium tuberculosis has been detected in tissue from approximately 50% of patients with sarcoidosis but not in healthy individuals serving as controls.1
Sarcoidosis is an indiscriminate disease. Although it most commonly involves the lung and hilar or mediastinal lymph nodes, sarcoidosis can affect many other sites, including the brain, skin, additional lymph nodes, liver, kidney, muscle, heart, and joints. The eye is arguably the most frequent extra-pulmonary site of involvement.2–4 Several structures within or around the eye can be affected, including the uveal tract, lacrimal gland, optic nerve, conjunctiva, and orbit.4–6
Gene expression microarray is a valuable technique to characterize molecular events within a tissue. Several groups, including ours,7 have used this type of analysis of messenger RNA transcripts to study sarcoidosis in sites, including the lungs,8–10 lymph nodes,11 peripheral blood,12–15 heart,16 and skin.17
We have organized an international consortium of ocular pathologists and orbital surgeons to advance the understanding of orbital inflammatory diseases including sarcoidosis. We collected formalin-fixed, paraffin-embedded biopsy specimens from the lacrimal gland and orbital fat from centers throughout the world. The RNA extracted from these tissues was examined by microarray analysis. Our observations relating to sarcoidosis are the subject of this report. We sought evidence to implicate specific genes that might be characteristic of ocular tissue affected by sarcoidosis. In addition, we wished to determine whether transcripts implicated in the eye were identical to transcripts implicated in other tissues affected by sarcoidosis.
Methods
This report is part of a series characterizing gene expression in orbital inflammatory diseases. Gene expression profiling was conducted between October 2012 and January 2014 on tissues biopsied from January 2000 through June 2013; analysis is ongoing. The research adhered to the tenets of the Declaration of Helsinki and was approved by the institutional review boards at Oregon Health & Science University; Columbia University; University of California, San Diego; Wake Forest University; Medical College of Wisconsin; Mount Carmel Health System; and University of Miami; as well as by the University of British Columbia Clinical Research Ethics Board, the Royal Adelaide Hospital Research Ethics Committee, and the King Khaled Eye Specialist Hospital Human Ethics Committee/Institutional Review Board. Written informed consent was obtained when required by the local review boards.
The methods for this study, including tissue collection; RNA extraction; RNA quality assessment; microarray (GeneChip U133 Plus 2; Affymetrix);Database for Annotation, Visualization, and Integrated Discovery (DAVID)18; and statistical analysis have been previously described.19 (Unpublished data; last microarray assays were run in January 2014 and last biopsy specimens were obtained from surgeries performed in June 2013; data analysis continues. The investigators were the same as the authors of the present report.) For the purpose of the present report, gene transcripts were identified as significantly differentially expressed if 1 or more of the array probe sets mapped to a gene were measured at more than 1.5-fold higher or lower levels compared with healthy tissue with a false discovery rate20 value of P < .05. DAVID18 was used to convert the lists of probe sets into gene lists.
The diagnosis of sarcoidosis was made by the referring center. All patients had noncaseating granuloma observed in either the lacrimal gland or orbital biopsy specimen. The diagnosis was independently confirmed by 2 ocular pathologists (D.J.W. and H.E.G.). Additional clinical information was unavailable for 2 of the 7 patients with sarcoidosis involving orbital adipose tissue. Among the other 5 patients, 3 had hilar and/or mediastinal adenopathy demonstrated on either chest radiography or chest tomographic scan. One patient had granulomas observed on a lip biopsy specimen. The fifth patient had an elevated serum angiotensin-converting enzyme (ACE) level as well as evidence for granuloma on the orbital biopsy specimen. Among the 5 patients with sarcoidosis affecting the lacrimal gland, 1 patient had unavailable clinical information. Three of the other 4 individuals had adenopathy demonstrated on chest radiography or chest tomographic scan. One patient with a normal chest radiograph had an elevated serum ACE level as well as evidence for disease in the lacrimal gland. Among the patients tested, the ACE level was elevated in 2 of 4 and the serum lysozyme level was elevated in 1 of 3 individuals.
Results
Demographics
Table 1 reports the characteristics of patients whose tissue was analyzed in this study. Important factors included age, sex, corticosteroid treatment, disease duration prior to biopsy, and the site of biopsy (lacrimal gland or orbital fat). Data are also provided on a control group with no known orbital inflammatory disease.
Table 1.
Participant Demographics
| Characteristic | Blood
|
Orbit
|
Lacrimal Gland
|
|||
|---|---|---|---|---|---|---|
| Normal | Sarcoidosis | Normal | Sarcoidosis | Normal | Sarcoidosis | |
| No. of samples | 12 | 12 | 6 | 7 | 6 | 5 |
|
| ||||||
| Demographics | ||||||
|
| ||||||
| Age, mean (SD), y | 53.9 (12.2) | 48.8 (21.4) | 69.7 (9.8) | 48.8 (14.7) | 68.2 (9.7) | 31.4 (15.4) |
|
| ||||||
| Female sex, No. (%) | 9 (75) | 8 (67) | 5 (83) | 5 (71) | 5 (83) | 4 (80) |
|
| ||||||
| Disease duration prior to biopsy, mean (SD), mo | NA | NA | NA | 8.8 (7.0) | NA | 2.1 (1.4) |
|
| ||||||
| Race, No. (%) | ||||||
|
| ||||||
| White | 11 (92) | 10 (83) | 3 (50) | 3 (43) | 6 (100) | 1 (20) |
|
| ||||||
| African American | 0 | 1 (8) | 1 (17) | 2 (29) | 0 | 2 (40) |
|
| ||||||
| American Indian | 0 | 0 | 1 (17) | 0 | 0 | 0 |
|
| ||||||
| Asian | 1 (8) | 1 (8) | 1 (17) | 0 | 0 | 0 |
|
| ||||||
| Unknown | 0 | 0 | 0 | 2 (29) | 0 | 2 (40) |
|
| ||||||
| Prednisone therapy at time of biopsy, No. (%) | 0 | 0 | 0 | 1 (20)a | 0 | 1 (25)b |
Abbreviation: NA, not available.
There were no data on prednisone use for 2 patients.
There were no data on prednisone use for 1 patient.
Comparisons With Healthy Orbital Tissue
The microarray data indicated substantial differences between tissues from patients with sarcoidosis and healthy controls. For orbital adipose tissue, signals from 2449 probe sets (transcripts from approximately 1522 genes) were significantly higher (>1.5-fold difference with a false discovery rate [FDR] value of P < .05) in the sarcoidosis tissues and 4050 (approximately 2619 genes) probe sets were significantly lower (eTables 1 and 2 in the Supplement). Table 2 lists genes identified by probe sets with the highest relative signals in orbital adipose tissue from patients with sarcoidosis compared with controls without inflammation. For the lacrimal gland, signals from 3069 probe sets (approximately 2001 genes)were significantly elevated and 3320 probe sets (approximately 2283 genes) were significantly decreased in sarcoidosis tissues (eTables 3 and 4 in the Supplement). Table 3 lists genes identified by probe sets with the highest relative signals in the lacrimal gland. Of the probe sets indicating higher gene expression, 1102 (approximately 739 genes) were identified for both the adipose and lacrimal gland tissue. Similarly, 1008 probe sets (approximately 744 genes) were represented in both lists, indicating significantly lower transcript levels.
Table 2.
Probe Sets With Some of the Highest Signals in Orbital Adipose Tissue Compared With Uninflamed Controlsa
| Probe Set | Gene Title | Fold Difference | P Value for FDR |
|---|---|---|---|
| 209396_s_at | Chitinase 3–like 1 (cartilage glycoprotein-39) | 93.5 | 1.5 × 10−6 |
| 1555745_a_at | Lysozyme | 93.3 | 8.98 × 10−7 |
| 214677_x_at | Immunoglobulin λ constant 1 (Mcg marker) | 53.5 | 1.84 × 10−4 |
| 224795_x_at | Immunoglobulin κ locus; immunoglobulin kappa constant | 45.0 | 2.00 × 10−4 |
| 217022_s_at | Immunoglobulin heavy locus; immunoglobulin heavy constant α 1; immunoglobulin heavy constant α 2 (A2m marker) | 42.8 | 1.22 × 10−4 |
| 203915_at | Chemokine (C-X-C motif) ligand 9 | 35.0 | 6.22 × 10−5 |
| 215121_x_at | Immunoglobulin λ light chain-like; immunoglobulin λ constant 1 (Mcg marker); immunoglobulin λ variable 1–44 | 34.8 | 3.47 × 10−4 |
| 216491_x_at | Immunoglobulin heavy constant μ | 34.2 | 7.80 × 10−5 |
| 212671_s_at | Major histocompatibility complex, class II, DQ α 1, DQ α 2, DQ α 1 chain–like | 30.9 | 1.05 × 10−5 |
| 209875_s_at | Secreted phosphoprotein 1 | 29.5 | 2.39 × 10−3 |
| 1555756_a_at | C-type lectin domain family 7, member A | 28.9 | 7.07 × 10−5 |
| 202834_at | Angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | 28.3 | 9.29 × 10−5 |
| 219386_s_at | SLAM family member 8 | 25.5 | 7.40 × 10−6 |
| 219159_s_at | SLAM family member 7 | 24.7 | 1.16 × 10−4 |
| 222838_at | SLAM family member 7 | 23.3 | 2.12 × 10−5 |
| 211429_s_at | Serpin peptidase inhibitor, clade A (α-1 antiproteinase, antitrypsin), member 1 | 21.1 | 4.50 × 10−5 |
| 209696_at | Fructose-1,6-bisphosphatase 1 | 18.4 | 1.63 × 10−7 |
| 205890_s_at | GABA B receptor, 1; ubiquitin D | 18.3 | 7.05 × 10−5 |
| 203936_s_at | Matrix metallopeptidase 9 (gelatinase B, 92-kDa gelatinase, 92-kDa type IV collagenase) | 17.7 | 1.17 × 10−6 |
| 209201_x_at | Chemokine (C-X-C motif) receptor 4 | 17.4 | 3.32 × 10−5 |
Abbreviations: FDR, false discovery rate; GABA, γ-aminobutyric acid; SLAM, signaling lymphocyte activation molecule.
Only the probe with the highest fold difference for a given gene is reported. Only probe sets with adequate annotation are included. The complete list of probe sets is reported in eTable 1 in the Supplement.
Table 3.
Probe Sets With Some of the Most Increased Signals in Lacrimal Gland Compared With Uninflamed Controlsa
| Probe Set | Gene Title | Fold Difference | P Value for FDR |
|---|---|---|---|
| 210809_s_at | Periostin, osteoblast specific factor | 54.1 | 2.52 × 10−9 |
| 209395_at | Chitinase 3–like 1 (cartilage glycoprotein-39) | 52.1 | 5.69 × 10−6 |
| 204580_at | Matrix metallopeptidase 12 (macrophage elastase) | 38.7 | 2.10 × 10−4 |
| 205890_s_at | γ-Aminobutyric acid B receptor, 1; ubiquitin D | 35.3 | 9.31 × 10−9 |
| 208168_s_at | Chitinase 1 (chitotriosidase) | 34.8 | 3.69 × 10−7 |
| 219386_s_at | SLAM family member 8 | 31.6 | 5.23 × 10−7 |
| 206134_at | ADAM-like, decysin 1 | 29.7 | 2.36 × 10−5 |
| 202834_at | Angiotensinogen (serpin peptidase inhibitor, clade A, member 8) | 29.1 | 1.76 × 10−6 |
| 209875_s_at | Secreted phosphoprotein 1 | 28.8 | 3.95 × 10−5 |
| 204416_x_at | Apolipoprotein C-I | 27.0 | 3.69 × 10−7 |
| 203936_s_at | Matrix metallopeptidase 9 (gelatinase B, 92-kDa gelatinase, 92-kDa type IV collagenase) | 26.8 | 6.87 × 10−8 |
| 211429_s_at | Serpin peptidase inhibitor, clade A (α-1 antiproteinase, antitrypsin), member 1 | 26.6 | 2.24 × 10−6 |
| 220423_at | Phospholipase A2, group IID | 24.5 | 2.00 × 10−8 |
| 218404_at | Sorting nexin 10 | 23.6 | 3.47 × 10−5 |
| 213831_at | Major histocompatibility complex, class II, DQ α 1, DQ α 1 chain–like | 22.0 | 3.17 × 10−2 |
| 1553706_at | Htra serine peptidase 4 | 21.7 | 2.18 × 10−5 |
| 206214_at | Phospholipase A2, group VII (platelet-activating factor acetylhydrolase, plasma) | 21.5 | 1.34 × 10−6 |
| 203915_at | Chemokine (C-X-C motif) ligand 9 | 20.7 | 3.75 × 10−5 |
| 206420_at | Immunoglobulin superfamily, member 6 | 20.7 | 3.09 × 10−5 |
| 32128_at | Chemokine (C-C motif) ligand 18 (pulmonary and activation-regulated); c-C motif chemokine 18–like | 19.8 | 7.65 × 10−5 |
Abbreviations: FDR, false discovery rate; SLAM, signaling lymphocyte activation molecule.
Only the probe with the highest fold difference for a given gene is reported. Only probe sets with adequate annotation are included. The complete list of probe sets is reported in eTable 3 in the Supplement.
We applied a widely used functional annotation software tool, DAVID,18 to probe the Gene Ontology (GO) Biological Process, Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway, and Biocarta pathway databases to identify relationships among the transcripts with altered expression. Both the inflamed orbit and lacrimal gland tissues were significantly enriched in transcripts characteristic of T cells, B cells, and innate immune cells. Immunoglobulin transcripts were highly enriched in the inflamed orbit but not in the lacrimal gland, perhaps owing to the abundance of plasma cells in healthy lacrimal gland tissue. Orbit adipose tissue from patients with sarcoidosis was notably deficient in transcripts related to metabolism (eg, the insulin-signaling pathway) and in the transforming growth factor β signaling pathway. The inflamed lacrimal tissues had decreased levels of genes related to protein synthesis, which is consistent with impaired lacrimal function known to be frequent in sarcoidosis.21
Comparisons With Blood From Patients With Sarcoidosis
Rosenbaum et al7 have previously reported on transcripts that are upregulated in the blood of patients with sarcoidosis. We compared the list of probe sets identified in the blood study using the same selection criteria as described above with the list of probe sets with increased signals in both the lacrimal gland (eTable 3 in the Supplement) and orbital adipose (eTable 1 in the Supplement) tissue and found 92 probe sets (OMIM 147796) in common (Table 4 and eTable 5 in the Supplement). Several of these genes are associated with interferon responses (eg, CXCL10 [OMIM 14731],22 GBP5 [OMIM 611467],23 STAT1 [OMIM 600555],24 TGM2 [OMIM 190196],25 AIM2 [OMIM 604578],26 WARS [OMIM 191050],27 ICAM1 [OMIM 147840],28 TNFAIP2 [OMIM 603300],29 SLAMF8 [OMIM 606620],30 and JAK2 [OMIM 147796]31). Similarly, there were 67 probe sets with decreased signals in peripheral blood in the prior report7 and in both orbital adipose tissue and lacrimal gland in the present study (eTable 6 in the Supplement).
Table 4.
Probe Sets With Some of the Most Increased Signals in Sarcoidosis vs Uninflamed Controls for All Tissue Types Studieda
| Probe Set | Gene Symbol | Blood
|
Orbit
|
Lacrimal Gland
|
|||
|---|---|---|---|---|---|---|---|
| Fold Difference | P Value for FDR | Fold Difference | P Value for FDR | Fold Difference | P Value for FDR | ||
| 214511_x_at | FCGR1B | 4.69 | 0.00 × 100 | 13.44 | 2.27 × 10−4 | 10.75 | 3.75 × 10−5 |
|
| |||||||
| 204533_at | CXCL10 | 2.83 | 5.03 × 10−1 | 16.53 | 6.03 × 10−4 | 7.87 | 5.15 × 10−3 |
|
| |||||||
| 229390_at | RP1-93H18.5 | 2.38 | 0.00 × 100 | 10.62 | 2.27 × 10−3 | 12.91 | 2.06 × 10−3 |
|
| |||||||
| 239196_at | ANKRD22 | 5.57 | 0.00 × 100 | 9.89 | 5.88 × 10−4 | 9.04 | 7.41 × 10−5 |
|
| |||||||
| 210029_at | INDO | 3.69 | 0.00 × 100 | 8.11 | 4.72 × 10−3 | 12.11 | 1.44 × 10−4 |
|
| |||||||
| 231577_s_at | GBP1 | 2.72 | 0.00 × 100 | 6.53 | 1.80 × 10−2 | 14.19 | 5.92 × 10−5 |
|
| |||||||
| 209969_s_at | STAT1 | 2.25 | 0.00 × 100 | 9.46 | 3.28 × 10−4 | 11.71 | 5.74 × 10−5 |
|
| |||||||
| 238581_at | GBP5 | 3.69 | 0.00 × 100 | 6.10 | 1.91 × 10−3 | 10.62 | 5.47 × 10−4 |
|
| |||||||
| 227458_at | CD274 | 4.63 | 0.00 × 100 | 6.14 | 1.27 × 10−3 | 7.42 | 3.15 × 10−4 |
|
| |||||||
| 211003_x_at | TGM2 | 2.68 | 1.19 × 100 | 4.24 | 1.29 × 10−3 | 10.13 | 2.77 × 10−6 |
|
| |||||||
| 206513_at | AIM2 | 2.00 | 2.50 × 10−1 | 6.87 | 2.12 × 10−4 | 6.57 | 5.72 × 10−5 |
|
| |||||||
| 209734_at | NCKAP1L | 2.14 | 1.19 × 100 | 3.64 | 1.97 × 10−4 | 5.88 | 6.81 × 10−5 |
|
| |||||||
| 204858_s_at | ECGF1 | 2.05 | 0.00 × 100 | 3.28 | 9.43 × 10−7 | 5.32 | 3.50 × 10−4 |
|
| |||||||
| 235574_at | GBP4 | 2.43 | 1.83 × 10−1 | 2.57 | 7.98 × 10−3 | 4.87 | 8.51 × 10−4 |
|
| |||||||
| 1561615_s_at | SLC8A1 | 2.08 | 9.25 × 10−1 | 3.00 | 5.90 × 10−3 | 4.42 | 1.24 × 10−4 |
|
| |||||||
| 238327_at | GCLMLOC440836 | 2.31 | 0.00 × 100 | 2.56 | 2.55 × 10−3 | 4.54 | 5.28 × 10−5 |
|
| |||||||
| 235643_at | SAMD9L | 2.41 | 0.00 × 100 | 2.19 | 3.08 × 10−2 | 4.41 | 1.96 × 10−4 |
|
| |||||||
| 227609_at | EPSTI1 | 3.58 | 0.00 × 100 | 2.28 | 1.99 × 10−2 | 3.11 | 4.04 × 10−3 |
|
| |||||||
| 205819_at | MARCO | 2.11 | 1.19 × 100 | 2.08 | 3.98 × 10−4 | 4.76 | 3.50 × 10−4 |
|
| |||||||
| 209773_s_at | RRM2 | 2.07 | 2.86 × 100 | 3.30 | 7.17 × 10−5 | 2.22 | 2.51 × 10−2 |
|
| |||||||
| 215761_at | DMXL2 | 2.19 | 1.16 × 10−1 | 2.99 | 4.66 × 10−5 | 2.21 | 3.93 × 10−2 |
|
| |||||||
| 232610_at | PARP14 | 2.38 | 1.83 × 10−1 | 2.14 | 2.63 × 10−3 | 2.42 | 3.76 × 10−2 |
Abbreviation: FDR, false discovery rate.
Only the probe set with the greatest fold difference for a given gene is reported. Only probe sets with adequate annotation are included. The complete list of probe sets is reported in eTable 5 in the Supplement.
Angiotensin-converting enzyme and lysozyme levels are frequently measured in serum in the investigation of possible sarcoidosis. Although transcripts for neither ACE nor lysozyme levels were elevated in the blood samples we studied, we detected a marked elevation of ACE levels in the lacrimal gland and orbital adipose tissue (P ≤ .001 for both tissues). The transcript for lysozyme levels was markedly elevated in the orbital adipose tissue (P < .001) but reduced in the lacrimal gland. Presumably, this difference results from lysozyme being normally synthesized in the lacrimal gland. Synthesis should be reduced by replacement of the healthy tissue with granulomata.
Comparisons With Genetic Studies
Polymorphisms in annexin A11 (ANXA11 [OMIM 602572]) have been implicated in predisposition to sarcoidosis.32 We found that the expression of messenger RNA for ANXA11 was substantially reduced within the orbit affected by sarcoidosis (P ≤ .01). This might imply that the genetic polymorphism reduces the function of ANXA11. We did not find alterations of ANXA11 expression in the lacrimal gland or the blood.
An “in silico” study combined genomics, proteomics, and transcriptomics to suggest 9 candidate genes in sarcoidosis: SERPIN B1 (OMIM 130135), FABP4 (OMIM 600434), S 100A8 (OMIM 123885), HBEGF (OMIM 126150), IL-7R(OMIM 146661), LRIG1(OMIM 608868), PTPN23(OMIM 606584), DPM2(OMIM 603564), and NUP214 (OMIM 114350).33 Our study found that 7 of these 9 genes were differentially regulated in either the orbit or the lacrimal gland affected by sarcoidosis (eTables 1 and 3 in the Supplement). Some of the transcripts were identified by more than 1 probe set, and not all probe sets indicated differential regulation. This is consistent with the probe set identifying a specific splice variant.
Comparisons With Other Orbital Inflammatory Diseases
Sarcoidosis is one of several systemic diseases that are known to cause inflammation in the orbit.34 In parallel studies, we investigated gene expression profiles of orbital adipose tissues from 6 individuals with granulomatosis with polyangiitis (GPA) (previously termed Wegener granulomatosis), from 25 individuals with Graves ophthalmopathy (also termed thyroid eye disease) and from 25 participants with nonspecific orbital inflammation (NSOI). (Unpublished data; last microarray assays were run in January 2014 and last biopsy specimens were obtained from surgeries performed in June 2013; data analysis continues. The investigators were the same as the authors of the present report.)To evaluate the uniqueness of the genes with increased expression in tissues from individuals with sarcoidosis, we performed 2 sets of comparisons.
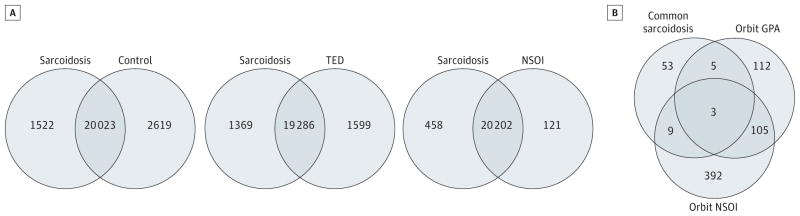
First, we compared the expression data from orbital adipose tissues from patients with sarcoidosis with those from patients with GPA, thyroid eye disease, or NSOI and from those without orbital inflammation to determine the probe sets with significantly higher or lower signals (>1.5-fold difference, FDR P < .05). In comparing sarcoidosis with GPA tissues, we identified 200 genes with probe sets that had significantly higher signals and 42 with lower signals in the sarcoidosis tissues. (Unpublished data; last microarray assays were run in January 2014 and last biopsy specimens were obtained from surgeries performed in June 2013; data analysis continues. The investigators were the same as the authors of the present report.) The counts for the other comparisons are shown in Venn diagrams (Figure, A).
Figure. Venn Diagrams Illustrate the Extent of Gene Expression Differences Between Disease Groups.
A, Estimated counts of genes with significantly different expression in comparisons of orbital adipose tissue. B, Estimated counts of genes with increased expression in blood, orbital adipose tissue, and lacrimal gland tissue from patients with sarcoidosis compared with orbital adipose tissue from other disease groups. GPA indicates granulomatosis with polyangiitis; NSOI, nonspecific orbital inflammation; and TED, thyroid eye disease.
Second, we compared the list of probe sets with higher signals in blood, orbital adipose tissue, and lacrimal gland tissue of patients with sarcoidosis vs healthy controls (eTable 5 in the Supplement) with a list of probe sets indicating increased expression in orbital adipose tissue of patients with GPA or NSOI based on the same significance criteria and the same set of un-inflamed orbit control tissues. The counts of genes in each category are displayed in Figure, B. Most of the genes with at least 1 probe set having a significantly increased signal in all 3 sarcoidosis tissues did not overlap the lists of genes with significantly increased signals in orbital tissue from patients with either GPA or NSOI. The sarcoidosis-only portion of the diagram includes 53 genes. Further analysis by querying the University of California, Santa Cruz, Transcription Factor Binding Site database with DAVID (Expression Analysis Systematic Explorer threshold, P ≤ .05) revealed that the group of 53 genes was significantly enriched in binding sites for 3 transcription factors: 17 have transcription factor–binding sequences for interferon-response factor 1, 27 have transcription factor binding sequences for interferon-response factor 2, and 23 have binding sequences for nuclear factor κB(eTable 7 in the Supplement). Neither of the interferon-response factors was enriched in the 112 genes in the GPA-only category and interferon-response factor 1 was not enriched in the NSOI-only group of 392 genes.
Discussion
The extra pulmonary involvement in sarcoidosis is a mystery. For example, it is unknown why one patient develops uveitis while another develops cutaneous disease and central nervous system manifestations. Genetic factors might contribute to the site specificity of the disease. There could also be a stochastic effect in the way that a putative inciting antigen might be distributed within the body. Our study indicates that, regardless of the site involved, the inflammation tends to activate similar genes. For example, previous studies6,7 implicated the intracellular signaling molecule, STAT1, in sarcoidosis on the basis of findings in peripheral blood and conjunctival granuloma.
Our data in the present study also point to signal transducer and activator of transcription (STAT1) as a potential contributor to sarcoidosis in the orbit or lacrimal gland since we detected elevated STAT1 transcript levels in all 3 tissues (Table 4). The validity of this finding is supported by other groups who have detected upregulation of STAT1 in peripheral blood,12 skin,17 or lung15 from patients with sarcoidosis. Interferons signal via STAT1. Interferon γ has been implicated in granuloma formation35 and the pathogenesis of sarcoidosis.22 Interferon alfa or beta therapy is known to trigger sarcoidosis.36 Both GBP5 (guanylate binding protein 5) and AIM-2 (absent in melanoma) transcripts are elevated in all 3 tissues studied and are induced by interferon γ23,26 as are other elevated transcripts that we identified, including CXCL10.22 SLAMF8 (Slam family member 8) is a macrophage activation marker also induced by interferon γ.30 Thus, upregulation of SLAMF8 fits well with the current understanding of the pathogenesis of sarcoidosis. Several other transcripts that we found to be upregulated in the orbit in tissue from patients with sarcoidosis have been implicated in studies on sarcoidosis in other tissues, including Fcγ receptor 1 (CD64 [OMIM 147840]),37 ICAM-1 (OMIM 146760),38 and IL-1β (OMIM 147720).39,40
One of the most elevated transcripts observed across all 3 tissues, ankyrin repeat domain 22 (ANKRD22), was confirmed by 2 independent probe sets on the array. It codes for a protein with no known function. The ANKRD22 transcripts are increased in the blood of patients with pancreatic cancer41 or during ovulation.42 Two transcripts, GBP5 and AIM-2, which were upregulated in all 3 of the tissues we studied, play a role in regulating inflammasomes, which are intracellular proteins that control the activation of IL-1β. Another elevated transcript that codes for the receptor, P2X7, plays an important role in the formation of multinucleated giant cells,43 which are a hallmark of sarcoidosis.
Transcripts that are increased in expression are potential targets for therapy of sarcoidosis. Transcripts for JAK2, which is activated by STAT1, were elevated in the blood, lacrimal gland, and anterior orbit. A JAK2 inhibitor is now commercially available to treat rheumatoid arthritis.44 Our work suggests that the JAK2 inhibitor may have therapeutic benefit in treating sarcoidosis as well.
A potential weakness of our study is the relatively small number of samples tested. We believe that this weakness is rebutted by finding the same transcript upregulated in 3 different tissues, by finding some transcripts detectable by more than 1 probe set, and by the independent confirmation in other studies as noted above. A previous study45 used quantitative reverse transcriptase polymerase chain reaction to validate the accuracy of our microarray technique. Our present sample size is too small to analyze additional variables, such as the effect of corticosteroid therapy, the correlation with prognosis, the effect of ethnicity, or the effect of disease duration. We chose to focus our discussion on transcripts with increased levels, but transcripts that are relatively reduced in expression could be equally important in understanding pathogenesis.
Currently available blood tests for sarcoidosis, such as an ACE or serum lysozyme level, have limited sensitivity and poor specificity.46 Biopsy of a lymph node by mediastinoscopy has morbidity and substantial expense. A chest computed tomographic scan used to search for symmetric hilar adenopathy exposes a patient to a large amount of irradiation and fails to definitively exclude entities such as lymphoma.
Based on our observations and those of others,15 we believe that detection of a relatively small number of transcripts in peripheral blood could be used to diagnose sarcoidosis. Peripheral blood gene expression can apparently distinguish between sarcoidosis and another granulomatous disease, tuberculosis.15 Although sarcoidosis usually involves hilar lymph nodes, adenopathy can resolve while residual disease is left in an organ such as the eye.5 Sarcoidosis is a potential cause of idiopathic uveitis,47 granulomatous hepatitis,48 pachymeningitis,49 orbital inflammation, and multifocal central nervous system disease.50 Biopsy of these tissues may be impossible or associated with considerable morbidity.
Conclusions
We believe that this is the first report to use microarray to analyze gene expression in tissue samples from the orbit or lacrimal gland from patients with sarcoidosis. Although additional work is indicated to devise an algorithm to enable the accurate diagnosis of sarcoidosis on the basis of gene expression in peripheral blood, we believe that these algorithms will prove more accurate than measurement of a single protein. In a recent study,51 an algorithm using gene expression in peripheral blood accurately identified active tuberculosis in African children. Furthermore, we predict that these algorithms will spare many patients the potential morbidity and expense of invasive procedures.
Supplementary Material
Acknowledgments
Funding/Support: This study was supported in part by grants EY020249, EY010572, and RR024140 from the National Health Institute and by Research to Prevent Blindness, the William and Mary Bauman Foundation, the Mas Family Foundation, and the Stan and Madelle Rosenfeld Family Trust.
Footnotes
Supplemental content at jamaophthalmology.com
Previous Presentations: Some of these data have been presented at the annual meetings of the Association for Research in Vision and Ophthalmology; May 6, 2013; Seattle, Washington, and May 7, 2014; Orlando, Florida, and also at the 2014 annual meeting of the American College of Rheumatology; November 17, 2014; Boston, Massachusetts. The raw and normalized gene expression microarray data have been uploaded to the GEO database.
Author Contributions: Drs Rosenbaum and Planck had full access to all the data in the study and take responsibility for the integrity of the data and the accuracy of the data analysis.
Study concept and design: Rosenbaum, Choi, Harrington, Tse, Selva, Planck.
Acquisition, analysis, or interpretation of data: Rosenbaum, Choi, Wilson, Grossniklaus, Harrington, Sibley, Ng, Steele, Czyz, Foster, Tse, Alabiad, Dubovy, Parekh, Harris, Kazim, Patel, White, Dolman, Korn, Kikkawa, Edward, Alkatan, al-Hussain, Yeatts, Selva, Stauffer, Planck.
Drafting of the manuscript: Rosenbaum, Choi, Wilson, Harrington, Sibley, Tse, Stauffer, Planck.
Critical revision of the manuscript for important intellectual content: Rosenbaum, Choi, Grossniklaus, Harrington, Dailey, Ng, Steele, Czyz, Foster, Tse, Alabiad, Dubovy, Parekh, Harris, Kazim, Patel, White, Dolman, Korn, Kikkawa, Edward, Alkatan, al-Hussain, Yeatts, Selva, Planck.
Statistical analysis: Choi, Sibley, Tse, Parekh.
Obtained funding: Rosenbaum, Choi, Planck.
Administrative, technical, or material support: Rosenbaum, Wilson, Grossniklaus, Harrington, Sibley, Dailey, Steele, Alabiad, Dubovy, Kazim, White, Dolman, Korn, Kikkawa, Alkatan, al-Hussain, Stauffer, Planck.
Study supervision: Rosenbaum, Harrington, Korn, Selva, Planck.
Conflict of Interest Disclosures: All authors have completed and submitted the ICMJE Form for Disclosure of Potential Conflicts of Interest. Dr Rosenbaum has consulted for Genentech and was a coinvestigator on a study funded by Genentech to evaluate the use of rituximab for orbital inflammatory diseases. No other disclosures were reported.
Role of the Funder/Sponsor: The funding organizations had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Additional Contributions: RNA extraction and microarray assays were performed in the Oregon Health & Science University Gene Profiling Shared Resource. Kristina Vartanian, BS, Integrated Genomics Laboratory, Oregon Health & Science University, provided excellent technical support for the microarray work. There was no financial compensation.
References
- 1.Song Z, Marzilli L, Greenlee BM, et al. Mycobacterial catalase-peroxidase is a tissue antigen and target of the adaptive immune response in systemic sarcoidosis. J Exp Med. 2005;201(5):755–767. doi: 10.1084/jem.20040429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Obenauf CD, Shaw HE, Sydnor CF, Klintworth GK. Sarcoidosis and its ophthalmic manifestations. Am J Ophthalmol. 1978;86(5):648–655. doi: 10.1016/0002-9394(78)90184-8. [DOI] [PubMed] [Google Scholar]
- 3.James DG. Ocular sarcoidosis. Ann N Y Acad Sci. 1986;465:551–563. doi: 10.1111/j.1749-6632.1986.tb18532.x. [DOI] [PubMed] [Google Scholar]
- 4.Jabs DA, Johns CJ. Ocular involvement in chronic sarcoidosis. Am J Ophthalmol. 1986;102(3):297–301. doi: 10.1016/0002-9394(86)90001-2. [DOI] [PubMed] [Google Scholar]
- 5.Karma A, Huhti E, Poukkula A. Course and outcome of ocular sarcoidosis. Am J Ophthalmol. 1988;106(4):467–472. doi: 10.1016/0002-9394(88)90885-9. [DOI] [PubMed] [Google Scholar]
- 6.Rosenbaum JT, Hessellund A, Phan I, Planck SR, Wilson DJ. The expression of STAT-1 and phosphorylated STAT-1 in conjunctival granulomas. Ocul Immunol Inflamm. 2010;18(4):261–264. doi: 10.3109/09273941003797934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rosenbaum JT, Pasadhika S, Crouser ED, et al. Hypothesis: sarcoidosis is a STAT1-mediated disease. Clin Immunol. 2009;132(2):174–183. doi: 10.1016/j.clim.2009.04.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lockstone HE, Sanderson S, Kulakova N, et al. Gene set analysis of lung samples provides insight into pathogenesis of progressive, fibrotic pulmonary sarcoidosis. Am J Respir Crit Care Med. 2010;181(12):1367–1375. doi: 10.1164/rccm.200912-1855OC. [DOI] [PubMed] [Google Scholar]
- 9.Christophi GP, Caza T, Curtiss C, Gumber D, Massa PT, Landas SK. Gene expression profiles in granuloma tissue reveal novel diagnostic markers in sarcoidosis. Exp Mol Pathol. 2014;96(3):393–399. doi: 10.1016/j.yexmp.2014.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crouser ED, Culver DA, Knox KS, et al. Gene expression profiling identifies MMP-12 and ADAMDEC1 as potential pathogenic mediators of pulmonary sarcoidosis. Am J Respir Crit Care Med. 2009;179(10):929–938. doi: 10.1164/rccm.200803-490OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Crouser ED, Julian MW, Crawford M, et al. Differential expression of microRNA and predicted targets in pulmonary sarcoidosis. Biochem Biophys Res Commun. 2012;417(2):886–891. doi: 10.1016/j.bbrc.2011.12.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bloom CI, Graham CM, Berry MP, et al. Transcriptional blood signatures distinguish pulmonary tuberculosis, pulmonary sarcoidosis, pneumonias and lung cancers [published correction appears in PLoS One. 2013;8(8). doi:10.1371/annotation/7d9ec449-aee0-48fe-8111-0c110850c0c1] PLoS One. 2013;8(8):e70630. doi: 10.1371/journal.pone.0070630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou T, Zhang W, Sweiss NJ, et al. Peripheral blood gene expression as a novel genomic biomarker in complicated sarcoidosis. PLoS One. 2012;7(9):e44818. doi: 10.1371/journal.pone.0044818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maertzdorf J, Weiner J, III, Mollenkopf HJ, et al. TBornot TB Network. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc Natl Acad Sci U S A. 2012;109(20):7853–7858. doi: 10.1073/pnas.1121072109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koth LL, Solberg OD, Peng JC, Bhakta NR, Nguyen CP, Woodruff PG. Sarcoidosis blood transcriptome reflects lung inflammation and overlaps with tuberculosis. Am J Respir Crit Care Med. 2011;184(10):1153–1163. doi: 10.1164/rccm.201106-1143OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lassner D, Kühl U, Siegismund CS, et al. Improved diagnosis of idiopathic giant cell myocarditis and cardiac sarcoidosis bymyocardial gene expression profiling. Eur Heart J. 2014;35(32):2186–2195. doi: 10.1093/eurheartj/ehu101. [DOI] [PubMed] [Google Scholar]
- 17.Judson MA, Marchell RM, Mascelli M, et al. Molecular profiling and gene expression analysis in cutaneous sarcoidosis: the role of interleukin-12, interleukin-23, and the T-helper 17 pathway. J Am Acad Dermatol. 2012;66(6):901–910. 910.e1–910.e2. doi: 10.1016/j.jaad.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 18.Dennis G, Jr, Sherman BT, Hosack DA, et al. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4(5):3. [PubMed] [Google Scholar]
- 19.Wong AJ, Planck SR, Choi D, et al. IgG4 immunostaining and its implications in orbital inflammatory disease. PLoS One. 2014;9(10):e109847. doi: 10.1371/journal.pone.00109847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Benjamini Y, Hochbert Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc B. 1995;57:289–300. [Google Scholar]
- 21.Drosos AA, Constantopoulos SH, Psychos D, Stefanou D, Papadimitriou CS, Moutsopoulos HM. The forgotten cause of sicca complex; sarcoidosis. J Rheumatol. 1989;16(12):1548–1551. [PubMed] [Google Scholar]
- 22.Geyer AI, Kraus T, Roberts M, et al. Plasma level of interferon γ induced protein 10 is a marker of sarcoidosis disease activity. Cytokine. 2013;64(1):152–157. doi: 10.1016/j.cyto.2013.07.010. [DOI] [PubMed] [Google Scholar]
- 23.Rupper AC, Cardelli JA. Induction of guanylate binding protein 5 by gamma interferon increases susceptibility to Salmonella enterica serovar typhimurium–induced pyroptosis in RAW 264.7 cells. Infect Immun. 2008;76(6):2304–2315. doi: 10.1128/IAI.01437-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hu X, Herrero C, Li WP, et al. Sensitization of IFN-gamma Jak-STAT signaling during macrophage activation. Nat Immunol. 2002;3(9):859–866. doi: 10.1038/ni828. [DOI] [PubMed] [Google Scholar]
- 25.Kim SY, Jeong EJ, Steinert PM. IFN-gamma induces transglutaminase 2 expression in rat small intestinal cells. J Interferon Cytokine Res. 2002;22(6):677–682. doi: 10.1089/10799900260100169. [DOI] [PubMed] [Google Scholar]
- 26.Asefa B, Klarmann KD, Copeland NG, Gilbert DJ, Jenkins NA, Keller JR. The interferon-inducible p200 family of proteins: a perspective on their roles in cell cycle regulation and differentiation. Blood Cells Mol Dis. 2004;32(1):155–167. doi: 10.1016/j.bcmd.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 27.Reano A, Richard MH, Denoroy L, Viac J, Benedetto JP, Schmitt D. Gamma interferon potently induces tryptophanyl-tRNA synthetase expression in human keratinocytes. J Invest Dermatol. 1993;100(6):775–779. doi: 10.1111/1523-1747.ep12476463. [DOI] [PubMed] [Google Scholar]
- 28.Dustin ML, Rothlein R, Bhan AK, Dinarello CA, Springer TA. Induction by IL 1 and interferon-gamma: tissue distribution, biochemistry, and function of a natural adherence molecule (ICAM-1) J Immunol. 1986;137(1):245–254. [PubMed] [Google Scholar]
- 29.Kota RS, Rutledge JC, Gohil K, Kumar A, Enelow RI, Ramana CV. Regulation of gene expression in RAW 264.7 macrophage cell line by interferon-γ. Biochem Biophys Res Commun. 2006;342(4):1137–1146. doi: 10.1016/j.bbrc.2006.02.087. [DOI] [PubMed] [Google Scholar]
- 30.Wang G, Abadía-Molina AC, Berger SB, et al. Cutting edge: Slamf8 is a negative regulator of Nox2 activity in macrophages. J Immunol. 2012;188(12):5829–5832. doi: 10.4049/jimmunol.1102620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stark GR, Kerr IM, Williams BR, Silverman RH, Schreiber RD. How cells respond to interferons. Annu Rev Biochem. 1998;67:227–264. doi: 10.1146/annurev.biochem.67.1.227. [DOI] [PubMed] [Google Scholar]
- 32.Hofmann S, Franke A, Fischer A, et al. Genome-wide association study identifies ANXA11 as a new susceptibility locus for sarcoidosis. Nat Genet. 2008;40(9):1103–1106. doi: 10.1038/ng.198. [DOI] [PubMed] [Google Scholar]
- 33.Maver A, Medica I, Peterlin B. Search for sarcoidosis candidate genes by integration of data from genomic, transcriptomic and proteomic studies. Med Sci Monit. 2009;15(12):SR22–SR28. [PubMed] [Google Scholar]
- 34.Lutt JR, Lim LL, Phal PM, Rosenbaum JT. Orbital inflammatory disease. Semin Arthritis Rheum. 2008;37(4):207–222. doi: 10.1016/j.semarthrit.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 35.Nance S, Cross R, Yi AK, Fitzpatrick EA. IFN-gamma production by innate immune cells is sufficient for development of hypersensitivity pneumonitis. Eur J Immunol. 2005;35(6):1928–1938. doi: 10.1002/eji.200425762. [DOI] [PubMed] [Google Scholar]
- 36.Buss G, Cattin V, Spring P, Malinverni R, Gilliet M. Two cases of interferon-alpha–induced sarcoidosis Koebnerized along venous drainage lines: new pathogenic insights and review of the literature of interferon-induced sarcoidosis. Dermatology. 2013;226(4):289–297. doi: 10.1159/000346244. [DOI] [PubMed] [Google Scholar]
- 37.Dubaniewicz A, Typiak M, Wybieralska M, et al. Changed phagocytic activity and pattern of Fcγ and complement receptors on blood monocytes in sarcoidosis. Hum Immunol. 2012;73(8):788–794. doi: 10.1016/j.humimm.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 38.Kim DS, Paik SH, Lim CM, et al. Value of ICAM-1 expression and soluble ICAM-1 level as a marker of activity in sarcoidosis. Chest. 1999;115(4):1059–1065. doi: 10.1378/chest.115.4.1059. [DOI] [PubMed] [Google Scholar]
- 39.Bélec L, Authier FJ, Chazaud B, Piédouillet C, Barlovatz-Meimon G, Gherardi RK. Interleukin (IL)-1 beta and IL-1 beta mRNA expression in normal and diseased skeletal muscle assessed by immunocytochemistry, immunoblotting and reverse transcriptase-nested polymerase chain reaction. J Neuropathol Exp Neurol. 1997;56(6):651–663. [PubMed] [Google Scholar]
- 40.Prior C, Knight RA, Herold M, Ott G, Spiteri MA. Pulmonary sarcoidosis: patterns of cytokine release in vitro. Eur Respir J. 1996;9(1):47–53. doi: 10.1183/09031936.96.09010047. [DOI] [PubMed] [Google Scholar]
- 41.Caba O, Prados J, Ortiz R, et al. Transcriptional profiling of peripheral blood in pancreatic adenocarcinoma patients identifies diagnostic biomarkers. Dig Dis Sci. 2014;59(11):2714–2720. doi: 10.1007/s10620-014-3291-3. [DOI] [PubMed] [Google Scholar]
- 42.Wissing ML, Kristensen SG, Andersen CY, et al. Identification of new ovulation-related genes in humans by comparing the transcriptome of granulosa cells before and after ovulation triggering in the same controlled ovarian stimulation cycle. Hum Reprod. 2014;29(5):997–1010. doi: 10.1093/humrep/deu008. [DOI] [PubMed] [Google Scholar]
- 43.Okamoto H, Mizuno K, Horio T. Human monocyte-derived multinucleated giant cells [in Japanese] Nihon Hansenbyo Gakkai Zasshi. 2004;73(3):245–251. doi: 10.5025/hansen.73.245. [DOI] [PubMed] [Google Scholar]
- 44.Maeshima K, Yamaoka K, Kubo S, et al. The JAK inhibitor to facitinib regulates synovitis through inhibition of interferon-γ and interleukin-17 production by human CD4+ T cells. Arthritis Rheum. 2012;64(6):1790–1798. doi: 10.1002/art.34329. [DOI] [PubMed] [Google Scholar]
- 45.Vartanian K, Slottke R, Johnstone T, et al. Gene expression profiling of whole blood: comparison of target preparation methods for accurate and reproducible microarray analysis. BMC Genomics. 2009;10(1):2. doi: 10.1186/1471-2164-10-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Birnbaum AD, Oh FS, Chakrabarti A, Tessler HH, Goldstein DA. Clinical features and diagnostic evaluation of biopsy-proven ocular sarcoidosis. Arch Ophthalmol. 2011;129(4):409–413. doi: 10.1001/archophthalmol.2011.52. [DOI] [PubMed] [Google Scholar]
- 47.Kosmorsky GS, Meisler DM, Rice TW, Meziane MA, Lowder CY. Chest computed tomography and media stinoscopy in the diagnosis of sarcoidosis-associated uveitis. Am J Ophthalmol. 1998;126(1):132–134. doi: 10.1016/s0002-9394(98)00081-6. [DOI] [PubMed] [Google Scholar]
- 48.Flamm SL. Granulomatous liver disease. Clin Liver Dis. 2012;16(2):387–396. doi: 10.1016/j.cld.2012.03.013. [DOI] [PubMed] [Google Scholar]
- 49.Wallace ZS, Carruthers MN, Khosroshahi A, et al. IgG4-related disease and hypertrophic pachymeningitis. Medicine (Baltimore) 2013;92(4):206–216. doi: 10.1097/MD.0b013e31829cce35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Stern BJ, Krumholz A, Johns C, Scott P, Nissim J. Sarcoidosis and its neurological manifestations. Arch Neurol. 1985;42(9):909–917. doi: 10.1001/archneur.1985.04060080095022. [DOI] [PubMed] [Google Scholar]
- 51.Anderson ST, Kaforou M, Brent AJ, et al. ILULU Consortium; KIDS TB Study Group. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N Engl J Med. 2014;370(18):1712–1723. doi: 10.1056/NEJMoa1303657. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.