Abstract
Deafness in humans is a common neurosensory disorder and is genetically heterogeneous. Across diverse ethnic groups, mutations of MYO15A at the DFNB3 locus appear to be the third or fourth most common cause of autosomal recessive, nonsyndromic deafness. In 49 of the 67 exons of MYO15A, there are currently 192 recessive mutations identified, including 14 novel mutations reported here. These mutations are distributed uniformly across MYO15A with one enigmatic exception; the alternatively spliced giant exon 2, encoding 1,233 residues, has 17 truncating mutations but no convincing deafness-causing missense mutations. MYO15A encodes three distinct isoform classes, one of which is 395 kDa (3,530 residues), the largest member of the myosin superfamily of molecular motors. Studies of Myo15 mouse models that recapitulate DFNB3 revealed two different pathogenic mechanisms of hearing loss. In the inner ear, myosin 15 is necessary both for the development and the long-term maintenance of stereocilia, mechanosensory sound-transducing organelles that extend from the apical surface of hair cells. The goal of this Mutation Update is to provide a comprehensive review of mutations and functions of MYO15A.
Keywords: DFNB3, deafness, myosin 15, MYO15A, shaker 2, giant exon, micro exon
Introduction
The village of Bengkala, Bali was chartered in the twelfth century. Today, two percent of Bengkala’s population of 2,200 is profoundly deaf from childbirth. Over several generations, the deaf people of this village developed a unique sign language, which is shared with many hearing people in the community (Hinnant, 2000). In Bengkala, sensorineural deafness segregates as an autosomal recessive disorder (Friedman et al., 1995; Winata et al., 1995). A genome-wide screen for homozygosity of STR (Short Tandem Repeat) markers co-segregating with deafness in Bengkala revealed a 3.0 centiMorgan (cM) linkage interval on chromosome 17p11.2. This deafness locus was designated DFNB3 (OMIM #600316, deafness neurosensory autosomal recessive 3)(Friedman et al., 1995). Many of the genes on human chromosome 17p11.2 were not annotated in 1995, but of those that were, pathogenic variants were not detected in the genomic DNA of affected deaf individuals from Bengkala. In mouse, the map location of the deaf-circling shaker 2 (sh2) phenotype is in a region of conserved synteny with DFNB3 (Deol, 1956; Probst et al., 1998; Wang et al., 1998). Hence, we sought to first identify the mutation underlying the mouse sh2 phenotype with the expectation this would subsequently reveal the cause of human DFNB3 deafness.
In Sally Camper’s laboratory, wild type bacterial artificial chromosomes (BACs) that spanned the sh2 locus were individually introduced by pronuclear injection into homozygous sh2 fertilized oocytes. One transgene (BAC425p24, derived from chromosome 11) heritably restored hearing (Probst et al., 1998; Kanzaki et al., 2006). The entire sequence of the 140 kb insert of BAC425p24 included only two genes, Drg2 and a novel member of the myosin ATPase superfamily of molecular motors that had not previously been described. The predicted gene on BAC425p24 had all of the consensus hallmarks of a functional myosin molecular motor, yet the amino acid sequence was sufficiently dissimilar to other known members of the myosin superfamily that it was assigned its own class: myosin 15. In the sh2 mouse, a transition mutation of the Myo15 gene was identified that substituted a tyrosine for an evolutionarily conserved cysteine (p.Cys1779Tyr; NP_034992) in the motor domain of myosin 15 (Probst et al., 1998). Similarly, homozygosity for the missense mutation c.6337A>T, p.Ile2113Phe in the human ortholog MYO15A (MIM# 602666) segregated with deafness in individuals from Bengkala. This pathogenic variant was not present in genomic DNA of 390 control chromosomes (Wang et al., 1998), and is not present in the 120,000 chromosomes of the Exome Aggregation Consortium (ExAC).
In addition to myosin 15, several other members of the myosin superfamily are also required for sound transduction (Table 1). In humans, expression of wild type myosin 3a (MYO3A; MIM# 606808), myosin 6 (MYO6; MIM# 600970), myosin 7a (MYO7A; MIM# 276903), myosin 9 (MYH9; MIM# 160775), and myosin 14 (MYH14; MIM# 608568) are each necessary for normal hearing and do not compensate for a loss of myosin 15 function, at least not in the inner ear (Liu et al., 1997a; Liu et al., 1997b; Weil et al., 1997; Lalwani et al., 2000; Melchionda et al., 2001; Walsh et al., 2002; Ahmed et al., 2003; Donaudy et al., 2004). Only a few mutations of these other myosin genes have been associated with human nonsyndromic deafness. However, mutations in MYO15A are now recognized as one of the more common causes of severe to profound autosomal recessive nonsyndromic deafness. This Mutation Update focuses only on MYO15A and includes (1) a description of the 178 reported and 14 novel mutations of MYO15A, (2) data supporting a novel exon defining an additional isoform class of MYO15A in human and mouse, (3) details of an emerging genotype-phenotype relationship, (4) molecular mechanisms of inner ear pathogenesis, (5) the wild type functions of myosin 15 gleaned from biochemical studies and animal models of DFNB3 deafness, and (6) puzzling questions about the molecular genetics and biology of myosin 15.
Table 1.
Myosin superfamily members necessary for human hearing
| Gene | Myosin | Locus1 | MIM | Reference |
|---|---|---|---|---|
| MYO3A | myosin 3a | DFNB30 | 607101 | Walsh et al., 2002 |
| MYO6 | myosin 6 | DFNB37/DFNA22 | 607821/606346 | Melchionda et al., 2001; Ahmed et al., 2003 |
| MYO7A | myosin 7a | DFNB2/DFNA11/USH1B | 600060/601317/276900 | Liu et al., 1997a; Liu et al., 1997b; Weil et al., 1997 |
| MYH9 | myosin 9 | DFNA17 | 603622 | Lalwani et al., 2000 |
| MYH14 | myosin 14 | DFNA4 | 600652 | Donaudy et al., 2004 |
| MYO15A | myosin 15 | DFNB3 | 600316 | Friedman et al., 1995; Wang et al., 1998 |
DFNA locus mapped in a family segregating nonsyndromic deafness as a dominant trait. A DFNB locus mapped in a family segregating nonsyndromic deafness as a recessive trait. USH1B is a locus for Usher syndrome type 1 (OMIM# 276900). Mutations of MYO7A are associated with DFNA, DFNB or USH.
Gene Structure and Domains of MYO15A
Myosins are a superfamily of molecular motors that bind cytoskeletal actin filaments and produce force to power motility and tension generation within the cell. All myosin molecules share a core ATPase ‘motor’ domain, a conserved structural fold that binds nucleotide (ATP) and actin filaments, and is the minimal structural unit required for force production (Sweeney and Houdusse, 2010). The human genome encodes 39 different myosin proteins that are grouped into 12 functional classes based upon amino acid conservation within the motor domain (http://www.genenames.org/cgi-bin/genefamilies/set/656). The human MYO15A gene was designated with a suffix “A” to distinguish it from MYO15B, a transcribed non-processed pseudogene on human chromosome 17q25 that originated from an ancient genomic duplication that also included a few closely linked genes. MYO15B has since accumulated a large subdomain deletion as well as mutations of key conserved residues that are necessary for ATPase activity (Boger et al., 2001). Although the motor domain of myosin 15B is likely non-functional, it remains possibile that this expressed pseudogene may indeed still be functional as a decoy mRNA transcript that binds regulatory molecules, or as a dominant negative protein to regulate endogenous myosin 15 function.
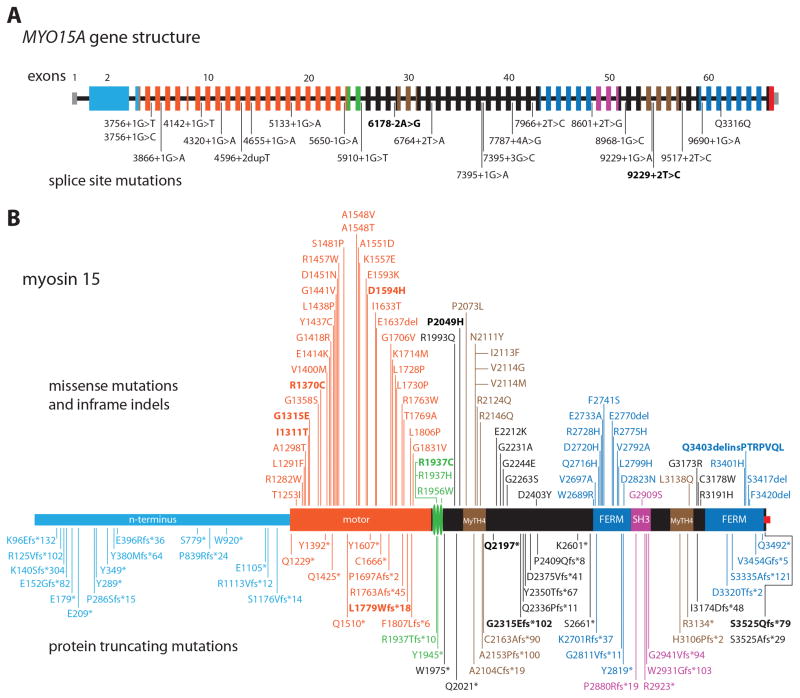
The myosin 15 polypeptide is encoded by 66 reported exons in the MYO15A gene that spans 71 kb of genomic DNA on chromosome 17p11.2 (chr17:18,012,020-18,083,116; hg19 assembly). We further provide new data in this manuscript supporting an additional protein-coding alternate first exon (see below). The longest human MYO15A transcript encodes a protein of 3,530 residues (isoform 1) that has a predicted molecular weight of 395 kDa, making it the largest of all known myosins in the mammalian proteome. Myosin 15 protein has four distinct structural regions: 1) a proline-rich 1,233 residue N-terminal domain (133-kDa), 2) the ATPase motor domain, 3) a lever arm, consisting of two consensus IQ motifs and a predicted third highly divergent IQ, acts to amplify conformational changes within the motor domain, and 4) a globular tail domain that contains myosin tail homology 4 (MyTH4), band 4.1, ezrin, radixin and moesin (FERM), Src homology 3 (SH3) domain2s in addition to a carboxy-terminus PDZ ligand (Fig. 1).
Figure 1.
Human MYO15A gene structure, the location of mutations associated with nonsyndromic deafness DFNB3 and the domains of myosin 15. Only pathogenic or likely pathogenic mutations are shown here while the variants of unknown significance are listed in Table 2. The c.6487delG (p.Ala2153fs) allele is not included in this figure (see Supp. Table S1). New mutations reported in this study are in bold font (pedigrees in Supp. Figure S2). A: Each rectangle represents one of the 66 reported exons of MYO15A while the horizontal black line represents intronic sequence. The 5′ and 3′ untranslated regions (UTRs) are denoted by light grey rectangles (exon 1 and part of exon 66). Exons and the encoded protein domains of myosin 15 in panel B have the same color. B: Missense mutations and inframe indels are shown on top of the drawing whereas protein truncating mutations are shown below. The three vertical green disks represent two consensus IQ motifs and a third IQ-like motif. Black colored rectangles denote regions of myosin 15 with no predicted domains. The small red square at the C-terminus represents a class 1 PDZ binding motif that interacts with whirlin. MyTh4, myosin tail homology 4 domain; FERM (band 4.1, ezrin, radixin, moesin) domain; SH3, Src homology 3 domain.
Several proteins have been identified that associate with these functional domains of myosin 15, although there are likely more to be discovered. The molecular chaperones UNC-45B and HSP90AA1 transiently interact with the motor domain, and are required for the polypeptide to mature into a functionally active ATPase (Bird et al., 2014). Essential (MYL6) and regulatory light chains (MYL12B) bind to the IQ domains of myosin 15 and stabilize the alpha-helical lever arm to aid force transmission from the motor domain (Bird et al., 2014). Although the IQ domains of myosin 15 can associate with calmodulin (CALM), the canonical IQ binding protein, CALM binds with a lower affinity than MYL6 and MYL12B (Bird et al., 2014). So far, only two proteins whirlin and EPS8 have been reported to interact with the tail domain of myosin 15. Whirlin (WHRN) is a scaffold protein with three PDZ domains and is essential in humans for normal hearing (DFNB31, OMIM #607084) and vision (USH2D, OMIM #611383) (Mburu et al., 2003; Belyantseva et al., 2005; Delprat et al., 2005; Ebermann et al., 2007). EPS8 is an actin-binding and capping protein and is similarly required for human hearing (DFNB102, OMIM #615974) (Manor et al., 2011; Behlouli et al., 2014). No proteins have yet been identified that associate with the N-terminal domain encoded by giant exon 2.
Alternative Splice Isoforms of Human and Mouse Myosin 15
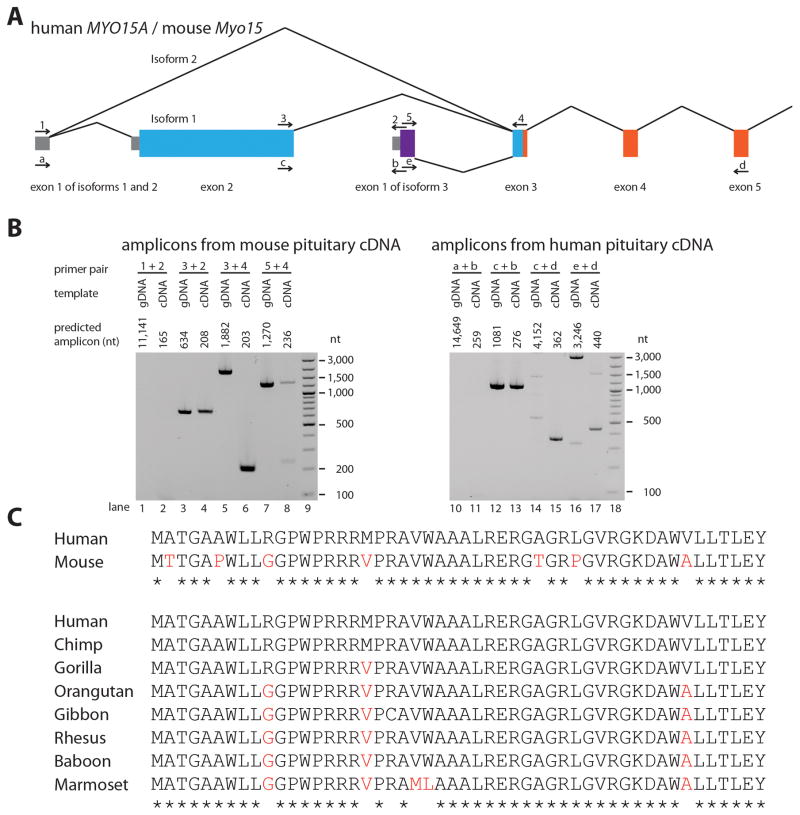
The majority of large protein-coding genes in vertebrates have multiple alternatively spliced mRNA transcripts (Ahmed et al., 2008; Scotti and Swanson, 2016). With 67 exons, MYO15A has the potential for extensive transcriptional diversity. Although the full scope of this has not been explored, two isoform classes have been experimentally demonstrated and previously reported for mouse Myo15 and human MYO15A (Liang et al., 1999; Fang et al., 2015). For the longest transcript class, exon 1 (encoding a 5′ UTR) splices to giant exon 2 (3,828 nt), which contains 1203 codons including a translation start codon and splices downstream to exon 3. This produces an 11,876 nt isoform 1 mRNA (NM_016239.3) that encodes a 395 kDa polypeptide (NP_057323.3). Alternatively, exon 1 can be spliced to exon 3 (Fig. 2). This class of transcript is 8,048 nt long and encodes a 262 kDa polypeptide denoted as isoform 2. The inclusion or exclusion of exon 2 encoding the 1,203 amino acid residues of the N-terminal domain appears to be the sole difference between isoform classes 1 and 2, respectively. In mouse, these two alternatively spliced transcripts of Myo15 encode isoforms with distinct temporal patterns of expression and functions in the mouse auditory system (Fang et al., 2015), which we presume to be also true for human MYO15A.
Figure 2.
Two alternative transcription start sites of MYO15A, three isoform classes and data supporting a novel exon 1 of isoform 3 identified in both human and mouse. A: Transcription for isoforms 1 and 2 starts from exon 1, whereas isoform 3 uses an alternate transcription start site of a second exon 1 located in intron 2. We identified this novel exon 1 of isoform 3 through 5′RACE from mouse pituitary cDNA and later confirmed it by PCR amplification and Sanger sequencing. Location of primers used to amplify and sequence novel exon 1 from mouse and human pituitary tissue cDNA are shown by arrows above and below exons, respectively. Lines indicate splicing of the primary transcript. Coloring of exons is the same as in Fig. 1A, except for the novel exon 1 (purple) of isoform 3 not illustrated in Fig 1. B: Images of 2% agarose gels used to size-separate amplicons. Each primer pair used to amplify cDNA was also used to amplify genomic DNA as a control template. Bands in lanes 4, 8 (upper band) and 13 show that the commercial cDNA libraries also contained some genomic DNA. C: Clustal Omega alignments of the 50 amino acid residues encoded by exon 1 of isoform 3. Non-identical amino acids are highlighted in red font whereas identical amino acids for all species examined are noted with a star. gDNA, genomic DNA; cDNA, complementary DNA; nt, nucleotide.
A novel alternative transcription start site newly reported here begins with an unannotated 218 nt exon 1. We previously noted a conserved sequence in the second intron of MYO15A and speculated that it might be either a cis-acting regulatory element or an unannotated alternatively spliced exon (Liang et al., 1999). We report here that this conserved sequence is a second alternative transcription start site for MYO15A defining isoform class 3 (Fig. 2). We first detected exon 1 of isoform 3 in a 5′RACE experiment using mouse cDNA from pituitary tissue and later independently confirmed it by PCR amplification and Sanger sequencing of cDNA derived from mouse and human pituitary tissues (Fig. 2). Exon 1 of isoform 3 encodes 50 residues that have 86% amino acid sequence identity between human and mouse and 94% identity among eight primates (Fig. 2). Functional studies of this new isoform class of myosin 15 with its conserved 50 residue novel N-terminal domain await a mouse model that deletes this novel exon 1 and specific antisera to examine its range of expression and intracellular localization.
Additional transcriptional complexity arises from the alternative splicing of cassette exons 8 and 26 (Liang et al., 1999). Vertebrate MYO15A genes have a conserved six nucleotide (ATAAAG) microexon 8, encoding isoleucine and lysine that are predicted to insert into loop 1; a surface exposed flexible region of the motor domain that is close to the ATP nucleotide binding pocket (Liang et al., 1999; Supp. Figure S1). Insertions into loop 1 have been described in other myosin classes and reported to modulate activity of the ATPase domain (Murphy and Spudich, 2000; Sellers, 2000). It is currently unknown if these two residues alter the biochemical properties of the myosin 15 ATPase cycle directly, or whether they might promote an interaction with a protein binding partner of the motor domain.
While the function of exon 8 sequence has yet to be determined, transcripts that include the 18 inframe codons of cassette exon 26 appear to be necessary for inner ear function of human myosin 15, as evidenced by a homozygous mutant allele p.Trp1975* (Supp. Table S1) that segregates with deafness in two Iranian families (Fattahi et al., 2012; Sloan-Heggen et al., 2015). Additionally, when an upstream alternative acceptor site of exon 26 is utilized, the additional 162 nucleotides include premature translation stop codons (PTCs) that would result in a truncation after the IQ motifs and thus a version of myosin 15 lacking a tail domain, if a protein is synthesized at all (Liang et al., 1999). PTCs can activate translation-dependent nonsense-mediated decay (NMD) of the transcript, a phenomenon referred to as regulated unproductive splicing and translation [RUST; (Lewis et al., 2003; Drummond and Friderici, 2013; Hug et al., 2016)]. Intermittent inclusion of PTCs-containing exon 26 sequence may be a post-transcriptional mechanism to fine-tune the optimal amount of myosin 15 in a cell.
Pathogenic Mutations of MYO15A
The only documented phenotype associated with mutations of human MYO15A is the combination of prelingual deafness and vestibular dysfunction (Friedman and Riazuddin, 2014). This is surprising in view of abundant MYO15A expression not only in the inner ear but also in the pituitary and other neuroendocrine tissues (Liang et al., 1999; Lloyd et al., 2001). If there are additional clinically relevant phenotypes associated with mutations of MYO15A, they may have been overlooked because of the initial and continued narrow focus on deafness (Friedman and Riazuddin, 2014). Alternatively, there may be a functional redundancy with other myosin classes that could substitute for the loss of myosin 15 in organs other than the inner ear. In the deaf-circling ci2 rat, a missense mutation in a MyTh4 domain of myosin 15 is associated with deafness (Loscher, 2010; Held et al., 2011), and some ci2 rats are also blind. However, the association of vision loss with a mutant myosin 15 is uncertain in these animals and a variant elsewhere in the ci2 genome was not conclusively ruled out. In one human subject, compound heterozygosity for two truncating mutations of MYO15A was reported to be associated with deafness and loss of vision (Neveling et al., 2013). However, recent personal communication from the senior author indicated that the deaf patient has no loss of vision. Thus, there is no evidence to date that mutations of MYO15A cause a syndromic form of deafness.
The first few mutations of MYO15A that cause DFNB3 deafness were identified 20 years ago in deaf individuals from Bengkala, Bali and in two consanguineous families from India (Wang et al., 1998). At that time, screening for variants in the reported 66 exons was a daunting and expensive task. As a result, MYO15A was rarely sequenced in families segregating deafness, unless there was significant genetic linkage data implicating the DFNB3 locus. Rather, effort was focused on screening for variants of smaller genes that were already established as significant contributors to human hereditary deafness such as GJB2, which has only a single protein coding exon (DFNB1, OMIM #121011) or SLC26A4 (DFNB4, OMIM #600791; Pendred syndrome, OMIM #274600). The extensive global contribution of MYO15A to human deafness went unrecognized until massively parallel sequencing became economical and widely adopted.
Mutations of MYO15A are now a well-known cause of recessively inherited nonsyndromic deafness globally. Such mutations are perhaps the third most common cause of deafness, and have been identified in sequence encoding all of the domains and motifs of this protein (Fig. 1). A total of 192 recessive variants of MYO15A are associated with hearing loss including 14 novel variants presented in this review (Supp. Table S1). Of the 192 reported and novel variants, we have categorized 82 as pathogenic, 73 as likely pathogenic (Fig. 1) and the remaining 37 variants as having unknown significance (Table 2). We used the following criteria for this assignment; frameshift, nonsense, and ±1 or 2 splice site variants are categorized as pathogenic if (a) they have an allele frequency <0.5% in controls (except for p.Trp1975*, discussed later in this review) and (b) deaf individuals are homozygous for the variant, or if in compound heterozygosity, both likely pathogenic alleles have been described. We did not include in Fig. 1 or in Supp. Table S1 heterozygous variants of MYO15A when a second mutation was not identified that could explain recessively inherited deafness (Woo et al., 2013; Yang et al., 2013; Besnard et al., 2014; Park et al., 2014), except for heterozygous missense mutations of giant exon 2 for reasons discussed later in this review. Inframe insertions/deletions were categorized as “likely pathogenic”. Missense and splice site variants, other than canonical ±1 or 2, were also categorized as likely pathogenic only if, in addition to satisfying the aforementioned two criteria, they were predicted as damaging by the four in silico programs for evaluating missense variants and the two in silico programs for splice site variants that we used. Missense and splice site variants that did not meet these criteria, and for which there is no other experimental data to support pathogenicity, were considered to be of “unknown significance” (Supp. Table S1).
Table 2.
MYO15A variants of unknown significance
| Exon/ intron | Location on chromosome 17 (GRCh37/hg19) | Nucleotide change (NM_016239.3) | Protein change (NP_057323.3) | References |
|---|---|---|---|---|
| 2 | 18022628C>T | c.514C>T | p.(Leu172Phe) | Miyagawa et al., 2013 |
| 2 | 18022668G>A | c.554G>A | p.(Gly185Asp) | Miyagawa et al., 2013 |
| 2 | 18022727T>C | c.613T>C | p.(Phe205Leu) | Miyagawa et al., 2013 |
| 2 | 18022785A>G | c.671A>G | p.(Tyr224Cys) | Miyagawa et al., 2013 |
| 2 | 18022856C>G | c.742C>G | p.(Arg248Gly) | this study |
| 2 | 18023337C>T | c.1223C>T | p.(Ala408Val) | Brownstein et al., 2014 |
| 2 | 18023501A>G | c.1387A>G | p.(Met463Val) | Fattahi et al., 2012 |
| 2 | 18023568T>C | c.1454T>C | p.(Val485Ala) | Sloan-Heggen et al., 2015 |
| 2 | 18023748C>T | c.1634C>T | p.(Ala545Val) | Sloan-Heggen et al., 2016 |
| 2 | 18025140C>A | c.3026C>A | p.(Pro1009His) | Yang et al., 2013 |
| 11 | 18035776G>A | c.4216G>A | p.(Glu1406Lys) | Miyagawa et al., 2013 |
| 16 | 18040946G>A | c.4828G>A | p.(Glu1610Lys) | Miyagawa et al., 2013 |
| 17 | 18041441C>G | c.4888C>G | p.(Arg1630Gly) | Miyagawa et al., 2013 |
| 17 | 18041505C>T | c.4952C>T | p.(Ser1651Leu) | Sloan-Heggen et al., 2015 |
| 19 | 18043829A>G | c.5212-2A>G | splice site | Atik et al., 2015 |
| 20 | 18043955T>C | c.5336T>C | p.(Leu1779Pro) | Ammar-Khodja et al., 2015 |
| 26 | 18046936G>A | c.5964+3G>A | splice site | Gao et al., 2013 |
| 29 | 18047850C>T | c.6217C>T | p.(Pro2073Ser) | Shearer et al., 2009 |
| 31 | 18051447C>T | c.6614C>T | p.(Thr2205Ile) | Liburd et al., 2001 |
| 32 | 18051821T>C | c.6703T>C | p.(Ser2235Pro) | Miyagawa et al., 2015 |
| 33 | 18052106G>A | c.6796G>A | p.(Val2266Met) | Nal et al., 2007 |
| 33 | 18052203G>A | c.6893G>A | p.(Arg2298Gln) | Sloan-Heggen et al., 2015 |
| 33 | 18052275C>G | c.6956+9C>G | splice site | Yang et al., 2013 |
| 37 | 18054082G>A | c.7395+3G>A | splice site | Riahi et al., 2014 |
| 39 | 18054500C>G | c.7550C>G | p.(Thr2517Ser) | Sloan-Heggen et al., 2016 |
| 40 | 18054733G>A | c.7679G>A | p.(Arg2560Gln) | Sloan-Heggen et al., 2016 |
| 42 | 18055426G>T | c.7894G>T | p.(Val2632Leu) | Bademci et al., 2015 |
| 45 | 18058072A>G | c.8224+3A>G | splice site | this study |
| 47 | 18058737G>A | c.8450G>A | p.(Arg2817His) | Gu et al., 2015 |
| 51 | 18061059G>A | c.8812G>A | p.(Gly2938Arg) | Sloan-Heggen et al., 2016 |
| 57 | 18064722C>T | c.9478C>T | p.(Leu3160Phe) | Nal et al., 2007; Miyagawa et al., 2013; Miyagawa et al., 2015 |
| 59 | 18066565G>A | c.9620G>A | p.(Arg3207His) | Bademci et al., 2015 |
| 60 | 18067146A>T | c.9781A>T | p.(Asn3261Tyr) | Miyagawa et al., 2013 |
| 61 | 18069795A>G | c.9908A>G | p.(Lys3303Arg) | Sloan-Heggen et al., 2016 |
| 63 | 18075050C>T | c.10181C>T | p.(Ala3394Val) | Sloan-Heggen et al., 2016 |
| 64 | 18075517C>G | c.10263C>G | p.(Ile3421Met) | Miyagawa et al., 2015 |
| 65 | 18077138G>A | c.10394G>A | p.(Arg3465Gln) | Sloan-Heggen et al., 2016 |
See Supp. Table S1 for more details.
Overall, the most common type of mutation of MYO15A are missense alleles (67/155, 43.2%, variants of unknown significance not included in the denominator), followed by frameshift (36/155, 23.2%,), nonsense (24/155, 15.5%), splice site (23/155,14.9), and in-frame indels (5/155, 3.2%). These mutations are located in 49 of the 66 protein coding exons of the three isoforms of MYO15A. The 17 exons of MYO15A in which no mutations have yet been found are randomly distributed and have an average size of 94 nucleotides as compared to 116 nucleotides for exons that have at least one mutation (calculated excluding giant ~ 4 kb exon 2). No pathogenic mutations have been reported in either the newly discovered exon 1 of isoform 3 or in mini-exon 8 possibly because of the small target size for mutagenic events. Given that the pathogenic mutations of MYO15A appear to occur randomly, with further study deafness-associated mutations maybe discovered in all exons of MYO15A.
The nomenclature of the mutations described in Supp. Table S1 is based on accession numbers NM_016239.3 and NP_057323.3 for MYO15A cDNA and protein, respectively. The genomic coordinates we used are from human genome build GRCh37/hg19. Mutation nomenclature in this update adheres to the recommendations of the Human Genome Variation Society (HGVS) and was double-checked for accuracy using Mutalyzer (http://lovd.nl/mutalyzer/). In order to conform to the guidelines of HGVS, we standardized the nomenclature for 37 published mutations of MYO15A that are noted in Supp. Table S1.
We report 14 novel mutations of MYO15A that co-segregate with deafness in 13 consanguineous families (Fig. 1 bold font; Supp. Figure S2; Table 2). These 14 additional variants include six missense, five protein-truncating, and three splice site mutations, none of which are found in 114 to 352 population-matched control chromosomes and are also not present in the ExAC database as of December 1, 2015. The 14 novel mutations and corresponding pedigrees are described in Supp. Table S1 and Supp. Figure S2. All novel variants identified in this study were submitted to ClinVar (http://www.ncbi.nlm.nih.gov/clinvar/). The study was approved by the Internal Review Boards, the National Institutes of Health, Bethesda (Combined Neurosciences Blue Panel), the Baylor College of Medicine, Houston, the University of Maryland, Baltimore, the Quaid-i- Azam University, Islamabad, Pakistan, the Centre of Excellence in Molecular Biology, University of the Punjab, Lahore, Pakistan and the Allama Iqbal Medical College in Lahore, Pakistan.
Table 3 lists 40 mutations of MYO15A identified more than once in unrelated deaf individuals and therefore are either founder or recurrent mutations. Five of these mutations have a conserved haplotype of closely linked markers suggesting a founder origin. The p.Ile2113Phe is responsible for deafness in Bengkala, Bali. The p.Arg1937Thrfs*10 and p.Ser3335Alafs*121 are likely to be founder alleles in the Turkish population (Cengiz et al., 2010; Bademci et al., 2015). A missense mutation p.Asp2720His occurs on a conserved haplotype segregating in four Pakistani families while the p.Tyr1392* is an apparent founder allele in the Iranian population and also reported in a family living in Pakistan (Nal et al., 2007; Sloan-Heggen et al., 2015). Another likely founder allele of MYO15A in the South Asian population is p.Trp1975*. This variant is located in the fifth of the 18 codons of cassette exon 26 and is associated with deafness in two Iranian families (Fattahi et al., 2012; Sloan-Heggen et al., 2015). The p.Trp1975* has an allele frequency of 1.7% (138/8086 chromosomes, 2 homozygotes) in the ExAC database for South Asians, suggesting that this mutation may be a common cause of DFNB3 deafness in this population but the certitude of its pathogenicity awaits direct experimental confirmation. A high allele frequency for a variant doesn’t necessarily indicate that it is benign. For example, the carrier frequency of the nonsyndromic deafness-causing 167delT allele of GJB2 (DFNB1) among Ashkenazi Jews is approximately 4% (Morell et al., 1998). Therefore, screening for c.5925G>A (p.Trp1975*) of MYO15A may be a consideration for sporadic deaf South Asians and South Asian families segregating hearing loss linked to genetic markers of DFNB3.
Table 3.
Pathogenic and likely pathogenic variants of MYO15A detected in multiple unrelated deaf individuals
Affected individuals were reported to share the same haplotype.
Same allele in three families described in Cengiz et al., 2010 who share a haplotype and two families reported in Bademci et al., 2015 who share a haplotype. Whether or not there is one or two haplotypes is unresolved.
p.(Asp2403Tyr) results from a nucleotide change of the second to last codon of exon 35 and was experimentally demonstrated to affect normal splicing of MYO15A.
N/A, not available.
Polymorphisms of MYO15A and Variants of Unknown Significance
As of January 2016, the ExAC database of ~120,000 chromosomes contains 2,087 DNA variants in exons and splice regions of MYO15A. Seventy-one of these variants have an allele frequency of ≥ 0.5% (based on at least 200 chromosomes) in one or more of the seven populations in the ExAC database. The 71 variants include 34 missense, 32 synonymous, four splice regions, and one nonsense allele (p.Trp1975* described above). Four of the 34 missense variants (p.Thr2205Ile, p.Val2266Met, p.Leu3160Phe and p.Pro1009His) have been associated with hearing loss. The p.Thr2205Ile was detected in the hemizygous state in combination with a Smith-Magenis Syndrome deletion of chromosome 17p11.2 in a North American patient who has a moderately severe high-frequency hearing loss (SMS OMIM #182290) (Liburd et al., 2001; Nal et al., 2007). The pathogenicity of p.Thr2205Ile is open to question. Its allele frequency in the ExAC database is 0.42% [(504/119760 chromosomes, five homozygotes of unknown phenotype; 2.1% in the Finnish population (141/6604 chromosomes)]. This allele frequency suggests that p.Thr2205Ile is a polymorphism but predicted to be deleterious by Polyphen-2, SIFT, MutationTaster, fathmm, and Mutationassessor. Two variants, p.Val2266Met and p.Leu3160Phe were reported as pathogenic but were re-categorized as benign based on both their elevated allele frequency in normal populations (>0.5%) and in silico predictions suggesting non-pathogenicity (Nal et al., 2007; Shearer et al., 2014). Using the same criteria, Shearer and co-authors annotated p.Met463Val and p.Ala1551Asp as benign variants of MYO15A (Shearer et al., 2014).
A p.Pro1009His mutation is located in giant exon 2, encoding the N-terminal domain of myosin 15 and was reported as pathogenic in a single individual from East Asia (Yang et al., 2013). The allele frequency for p.Pro1009His is ≥ 0.5% in the ExAC database and 4.6% in East Asians including 14 homozygotes of unknown hearing phenotype. There are four additional missense variants in giant exon 2 that co-segregate with recessive deafness and their pathogenicity is questionable under a similar rationale. The p.Ala408Val was identified in compound heterozygous state in a Palestinian Arab family with two deaf siblings (Brownstein et al., 2014), but is now predicted to be benign by MutationTaster, PolyPhen2, PROVEAN and SIFT. The p.Arg248Gly observed in family 4524 (Supp. Figure S2), is predicted to be benign by MutationTaster and PROVEAN, and the arginine 248 residue is not fully conserved among mammals. We note that affected individuals from family 4524 are also homozygous for p.Asp1594His of MYO15A, which is predicted to be deleterious by four different programs (Supp. Table S1). Additionally, c.4780G>C (p.Asp1594His) changes the first nucleotide of exon 16, affecting the consensus splice acceptor site (agG to agC), is predicted by Human Splicing Finder to affect the normal splicing of exon 15 to exon 16, hence, homozygosity for p.Asp1594His is a likely cause of deafness DFNB3. A homozygous p.Met463Val allele in giant exon 2 was reported and predicted (Table S1) to be benign notwithstanding co-segregation with deafness in an Iranian family. However, p.Met463Val is in disequilibrium with the likely causative mutation of MYO15A. Affected individuals of this family are also homozygous for a truncating mutation of MYO15A (p.Phe1807Leufs*6), which the authors conclude is the likely cause of deafness (Fattahi et al., 2012). Another missense variant (p.Val485Ala) in giant exon 2 reported in a family segregating deafness was designated as an allele of unknown significance in the supplemental information (Sloan-Heggen et al., 2015). There are another four heterozygous missense variants reported for giant exon 2 (Miyagawa et al., 2013a), each of which were only found once in 216 deaf patients from Japan, and there is no mention of a second pathogenic variant of MYO15A that would explain recessively inherited deafness. Therefore, we have categorized these as variants of unknown significance.
Forty-four percent of pathogenic DFNB3 mutations in MYO15A are missense alleles overall. However, we conclude that there are no convincing reports of pathogenic missense mutations in giant exon 2. However, there are 17 truncating mutations in this giant exon 2 that are associated with deafness segregating in DFNB3 families from 9 different ethnicities (Fig. 1). This demonstrates that myosin 15 isoform 1 is critical for normal hearing in humans (Fig. 1). Moreover, a mouse model of the human p.Glu1105* DFNB3 allele in giant exon 2 causes deafness (Nal et al., 2007; Fang et al., 2015).
Given an equal distribution of mutations across the genomic interval, we expect pathogenic missense alleles in giant exon 2, since it comprises one third of the entire myosin 15 isoform 1 protein. The absence of missense mutations in giant exon 2 is not explained by skewed codon usage. For example, four of the six codons for arginine have a CpG dinucleotide that is more prone to mutation and has the potential for nonsynonymous substitutions (Cooper and Krawczak, 1990; Vitkup et al., 2003). There is no underrepresentation of arginine in the N-terminal domain of myosin 15. Indeed, arginine is the second most frequently observed residue encoded by giant exon 2. Therefore, we considered other explanations for the absence of pathogenic missense mutations in giant exon 2. One possibility is that pathogenic missense variants are unreported because giant exon 2 encodes no consensus protein domains and consequently missense variants are predicted to be benign or tolerated by algorithms such as Mutation Taster, SIFT, or Polyphen-2. However, we also considered this to be an unlikely explanation because there are missense mutations in the tail of myosin 15 that are located outside of reported domains (Fig. 1) and predicted to be “deleterious” by in silico tools. Another possibility is that there is ascertainment bias in studies of hereditary deafness. If missense mutations in giant exon 2 are associated only with a mild hearing loss or a late onset hearing loss, these phenotypes would have been ignored when only severely to profoundly deaf probands are specifically sought, often through schools for the deaf. There are clinical and experimental data in human subjects and mouse models that support ascertainment bias explanation for the dearth of missense mutations in giant exon 2. Five homozygous protein truncating mutations in this exon (p.Tyr289*, p.Glu396Argfs*36, p.Glu1105*, p.Arg1113Valfs*12, and Ser1176Valfs*14), which entirely ablate the function of MYO15A isoform 1, are associated with less than severe to profound hearing loss in many family members (Nal et al., 2007; Cengiz et al., 2010; Bashir et al., 2012; Li et al., 2016). In addition, a recently reported Myo15a mouse model homozygous for an engineered nonsense mutation in exon 2, referred to as ΔN (equivalent to human mutation p.Glu1105* in exon 2, Fig. 1), does have some minimal hearing initially, unlike the sh2 allele that results in profound deafness at the earliest time after birth that auditory function can be quantitatively measured by auditory brain stem analyses (Fang et al., 2015). With these observations in mind, we hypothesize that amino acid substitutions of the predicted intrinsically disordered N-terminal domain are structurally less menacing, causing a more subtle hearing loss. To test this idea, subjects with mild to moderate hearing loss could be ascertained in collaboration with professionals who prescribe and fit hearing aids.
Myosin 15 is a Master Regulator of Stereocilia Development and Conservation
Humans can hear sound with extraordinary sensitivity and frequency selectivity. At the heart of this remarkable system are hair cells, the primary sensory receptors within the cochlea that detect sound using mechano-sensitive protrusions; called stereocilia. Each hair cell typically has > 50 individual stereocilia, arranged into ranks of increasing height that are cross-linked with extracellular filaments that create a cohesive bundle (Frolenkov et al., 2004). Deflection of the hair bundle by sound waves leads to opening of mechano-electric transduction (MET) channels that reside at the tips of individual stereocilia (Fig. 3). This ultimately depolarizes the hair cell receptor potential and is the beginning of afferent neurotransmission to the brainstem via the 8th cranial nerve (Schwander et al., 2010). Given the overall complexity of the auditory system, it is not surprising that human deafness is a highly genetically heterogeneous neurosensory disorder (Dror and Avraham, 2010; Drummond et al., 2012; Griffith and Friedman, 2016).
Figure 3.
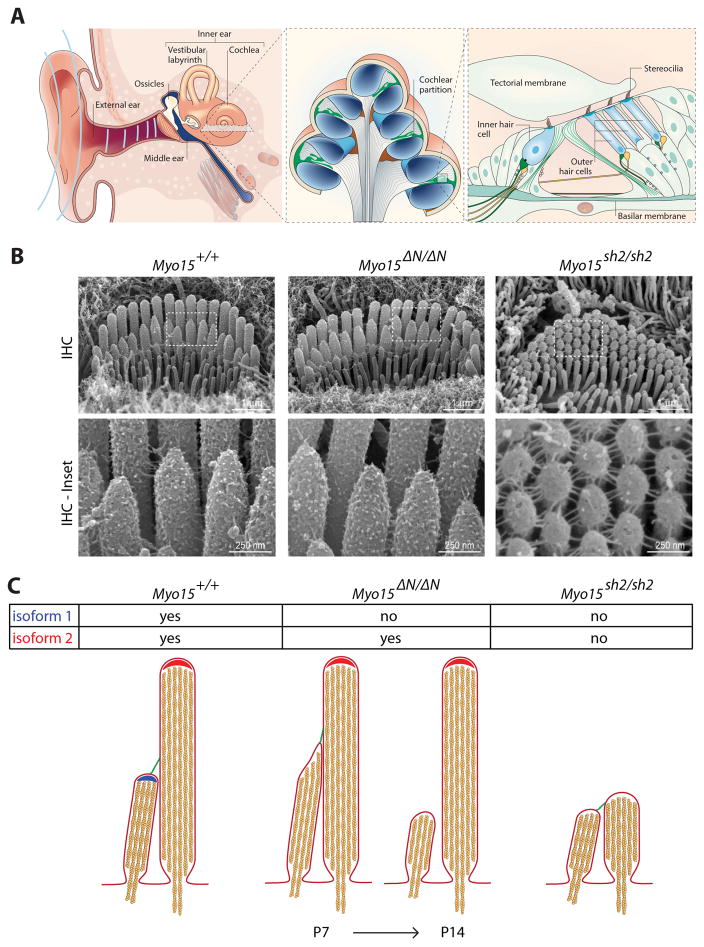
A: The cochlea is the sensory organ that detects sound within the inner ear. Sound waves enter the external auditory canal and are transmitted to the inner ear via the tympanic membrane and ossicles. Sound energy sets up oscillations within the fluid-filled ducts of the cochlea that displaces the cochlea partition and organ of Corti. Mechanosensory hair cells within the organ of Corti detect these displacements and release neurotransmitters to trigger afferent neural signaling. This panel reproduced from Frolenkov et al., (2004) with permission from Gregory Frolenkov and the publisher. B: Scanning electron microscopy (SEM) showing that mutations in mouse myosin 15 (Myo15) can have different effects upon development and maintenance of mechanosensory stereocilia. The missense (sh2) mutation in exon 20 blocks stereocilia elongation, while conversely, the nonsense (ΔN) mutation in exon 2 does not. Both mouse models exhibit profound deafness. Figure modified with permission from the publisher of Fang et al., (2015). C: Summary of myosin 15 isoform localization and their functions within stereocilia. In maturing stereocilia (P7 and older), isoforms 1 and 2 are concentrated at the tips of different stereocilia rows in inner hair cells. Isoform 2 is required to drive the developmental elongation of stereocilia, whilst isoform 1 is essential postnatally to stabilize shorter stereocilia rows that elicit mechano-electric transduction (MET) currents when the stereocilia bundle is defelected.
In general, molecular and cellular studies of the human cochlea are not feasible, since this organ is encased in bone, severely limiting non-invasive experimental access. The potential to directly differentiate hair cells in vitro from patient-derived pluripotent stem cells is an exciting new approach to this longstanding problem; though extremely promising, these systems have yet to replicate the full architectural complexity of the cochlea (Oshima et al., 2010; Ronaghi et al., 2014). Animal models recapitulating human deafness have thus provided the critical biological material to study ultrastructure, cell biology and physiology of hair cells (Frolenkov et al., 2004), and more recently to test potential therapies for auditory system pathology (Askew et al., 2015; Chien et al., 2015; Muller and Barr-Gillespie, 2015). Using animal models, the majority of genes mutated in hereditary deafness have been shown to encode proteins involved in development, maintenance and normal physiology of stereocilia (Frolenkov et al., 2004; Petit and Richardson, 2009; Schwander et al., 2010; Barr-Gillespie, 2015; Drummond et al., 2015; Griffith and Friedman, 2016). Stereocilia develop from microvilli that undergo a complex program of lengthening and thickening as they metamorphose into the mature mechanosensitive organelles (Tilney et al., 1992; Barr-Gillespie, 2015; Drummond et al., 2015). The final geometry of the hair bundle is critical for mechanotransduction and, remarkably, this is reliably reproduced with nanometer tolerances from hair cell to hair cell. The molecular mechanisms that specify, assemble and maintain this architecture are poorly understood, but are central to the acquisition and continued operation of these delicate mechanosensors.
Myosin 15 was the first protein found to control stereocilia development as they transform from microvilli and start to grow. Stereocilia bundles are short and fail to elongate in mice homozygous for the sh2 (Myo15sh2) or shaker 2J (Myo15sh2-J) allele, resulting in profound deafness and vestibular dysfunction (Probst et al., 1998; Anderson et al., 2000; Belyantseva et al., 2003; Stepanyan et al., 2006). The sh2 allele is a substitution of cysteine for tyrosine p.Cys1779Tyr within the motor domain, whilst the shaker 2J allele contains a genomic deletion that removes 6 exons and truncates the last FERM domain from the myosin 15 molecule. Exactly how these mutations alter the molecular properties of myosin 15 is unknown. However, they functionally prevent myosin 15 from trafficking to its normal location at the tips of stereocilia; the major site of actin polymerization during stereocilia elongation (Drummond et al., 2015; Narayanan et al., 2015). Myosin 15 must interact with at least two other proteins, Eps8 and whirlin, in order to drive stereocilia growth (Belyantseva et al., 2005; Manor et al., 2011; Zampini et al., 2011). Both proteins localize with myosin 15 at the tips of elongating stereocilia. Individual mouse mutants of Eps8 or whirlin have short stereocilia similar to the phenotype of the sh2 mouse, despite the fact that myosin 15 trafficks normally to stereocilia tips. In sh2 mice, Eps8 and whirlin are absent from stereocilia, demonstrating that they are dependent upon myosin 15 for their trafficking to the tip compartment. Critically, reintroduction of wild-type EGFP-myosin 15 isoform 2 into sh2 hair cells in culture can restore the normal localization of whirlin and Eps8 (Belyantseva et al., 2005; Manor et al., 2011) and rescue stereocilia elongation in vitro (Belyantseva et al., 2005). These data show that myosin 15 traffics Eps8 and whirlin within hair cells, and that the overall macromolecular complex drives the critical transformation of microvilli into mature stereocilia.
Recently, a new role for myosin 15 was discovered that is independent of its function in stereocilia development (Fang et al., 2015). This new function emerged from the finding that hair cells express both isoforms 1 and 2 of myosin 15, and that the sh2 and shaker 2J alleles were simultaneously ablating both isoforms (Fang et al., 2015). To examine how loss of one isoform might impact hearing function, a humanized mouse allele was engineered with a nonsense p.Glu1086* mutation in giant exon 2 (Myo15E1086X; abbreviated to ΔN) to model the equivalent p.Glu1105* DFNB3 allele in humans (Nal et al., 2007; Fang et al., 2015). Since giant exon 2 is only included in isoform 1 transcripts (Fig. 2), the ΔN allele specifically eliminates isoform 1 without perturbing the expression of isoform 2. Homozygous ΔN/ΔN mice are profoundly deaf at six weeks of age, similar to sh2 mutants (Fang et al., 2015). Remarkably, despite the profound deafness in isoform 1-null ΔN/ΔN mice, stereocilia still initially develop normally (Fig. 3) in striking contrast to those in sh2 hair cells (Fang et al., 2015). Isoform 2 is still produced in ΔN/ΔN hair cells and is detected at the tips of stereocilia, suggesting this is sufficient to drive stereocilia elongation in the absence of isoform 1. Consistent with this interpretation, isoform 2 is able to traffic Eps8 and whirlin to their wild-type locations in ΔN/ΔN hair cell stereocilia (Fang et al., 2015).
If isoform 1 is dispensable for stereocilia development, then what role does it have in DFNB3 pathology? Although isoform 1-null ΔN/ΔN hair cell stereocilia are initially indistinguishable from wild-type hair cells, degenerative changes are detected from postnatal day 6 (P6) onwards. These changes are specifically observed on the shorter stereocilia ranks that house active mechano-electric transduction (MET) channels (Beurg et al., 2009). The actin cytoskeleton becomes dysregulated in these shorter rows and stereocilia decrease in both length and diameter, eventually being completely resorbed into the hair cell (Fig. 3). The onset of degeneration coincides with a switch in cochlear mRNA splicing, a pivot from producing mainly isoform 2 at P1 when stereocilia are actively elongating, to predominantly isoform 1 by P7 (Fang et al., 2015). As isoform 1 protein is synthesized in auditory hair cells, it accumulates at the tips of shorter-row mechano-transducing stereocilia, where it functions to prevent disassembly of core actin cytoskeleton. In contrast, for inner hair cells, isoform 2 traffics predominantly to the tips of stereocilia in the tallest row (Fang et al., 2015). Extrapolating from these Myo15 DFNB3 mouse models, human DFNB3 deafness presumably develops from at least two fundamentally different pathologies. Mutations affecting isoform 2 block stereocilia development, while mutations in giant exon 2 (affecting solely isoform 1) interfere with stereocilia maintenance.
How does isoform 1 regulate the actin cytoskeleton in shorter-row mechanosensitive stereocilia? Unlike actin filaments in filopodia and microvilli, which continuously treadmill, the mature actin cores of stereocilia are not continuously renewed on a daily basis (Schneider et al., 2002; Rzadzinska et al., 2004). Rather, the stereocilia actin core appears to be remarkably stable with localized zones of turnover, and has an overall half-life on the order of months, if not longer (Zhang et al., 2012; Drummond et al., 2015; Narayanan et al., 2015). Identification of the isoform 1 protein interactome is now critical to understand how this molecule potentially interacts with other actin-regulatory proteins to set the rate of actin filament turnover in stereocilia. Progress towards this goal will be boosted by a recent advance in purifying recombinant myosin 15 molecules, which will allow for detailed biochemical and biophysical analyses of its properties (Bird et al., 2014). Combined with whole animal physiology and cell biology approaches, these biochemical studies will provide unprecedented insight into the mechanisms that control development and maintenance of hair cell stereocilia, both of which are central to healthy human hearing.
Conclusions
Variants of MYO15A are now documented to be one of the leading causes of human hereditary hearing loss worldwide. Of the nearly two hundred mutations in MYO15A identified, we preliminarily categorized some as being of unknown significance. These variants may yet be reassigned as likely benign or pathogenic once their effect upon myosin 15 has been experimentally determined. Several roles for myosin 15 are now emerging in the auditory system, where it is responsible for both the development, as well separately for the maintenance of sensory hair cell stereocilia. The presence of two protein splice isoforms precisely correlates with these activities, establishing a clear genotype-phenotype relationship. Isoform 2 traffics molecular cargo to the tips of differentiating hair cell stereocilia and drives elongation of the core actin cytoskeleton. Isoform 1 is dispensable for initial development, but is then critical to stabilize the actin cytoskeleton and prevent disassembly of mature mechano-transducing stereocilia. We speculate that myosin 15 may have additional functions, given the number of different, but currently unstudied alternative splice isoforms of this gene. In conclusion, human DFNB3 deafness results from a spectrum of sensory hair cell pathologies. This complex disease etiology necessitates detailed understanding of how myosin 15 normally functions, so that restorative small molecules or gene-editing therapies can be targeted to the appropriate isoform, at the appropriate time.
Supplementary Material
Acknowledgments
Grant Sponsor: This research was supported in part by the Intramural Research Program of the NIH, NIDCD through DC000048-19 to TBF. The work was also supported by funds from Higher Education Commission of Pakistan (HEC) to ShR, NIDCD grants R01 DC003594 and R01 DC011651 to SML, R01DC012564 to ZMA and R01DC011803 to SR.
We thank Benjamin Perrin, Davide Risso, and Dennis Drayna for critical review of the manuscript. We also thank the University of Washington Center for Mendelian Genomics. This study utilized the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health, Bethesda, MD (http://biowulf.nih.gov), and was supported by funds from Higher Education Commission of Pakistan (HEC) to ShR, NIDCD grants R01 DC003594 and R01 DC011651 to SML, R01DC012564 to ZMA, R01DC011803 to SR, and was supported in part by the Intramural Research Program of the NIDCD/NIH DC000048-19 to TBF.
Footnotes
Disclosure statement: All authors declare no conflicts of interest
Articles cited in the online supporting information:
(Ammar-Khodja et al., 2015), (Atik et al., 2015), (Bademci et al., 2015),(Bashir et al., 2012), (Belguith et al., 2009), (Brownstein et al., 2014), (Brownstein et al., 2011), (Cengiz et al., 2010), (Chang et al., 2015), (Chen et al., 2015), (Diaz-Horta et al., 2012), (Duman et al., 2011), (Fattahi et al., 2012), (Gao et al., 2013), (Gu et al., 2015), (Imtiaz et al., 2011), (Kalay et al., 2007), (Lezirovitz et al., 2008), (Li et al., 2016), (Liburd et al., 2001), (Miyagawa et al., 2013a; Miyagawa et al., 2013b), (Miyagawa et al., 2015), (Moteki et al., 2015), (Nal et al., 2007), (Neveling et al., 2013), (Park et al., 2014), (Rehman et al., 2015), (Riahi et al., 2014), (Schrauwen et al., 2013), (Shafique et al., 2014), (Shahin et al., 2010), (Shearer et al., 2009), (Sloan-Heggen et al., 2015), (Sloan-Heggen et al., 2016), (Vona et al., 2014), (Vozzi et al., 2014), (Wang et al., 1998), (Woo et al., 2013), (Xia et al., 2015), (Yano et al., 2013), (Yang et al., 2013)
References
- Ahmed ZM, Morell RJ, Riazuddin S, Gropman A, Shaukat S, Ahmad MM, Mohiddin SA, Fananapazir L, Caruso RC, Husnain T, Khan SN, Riazuddin S, et al. Mutations of myo6 are associated with recessive deafness, dfnb37. Am J Hum Genet. 2003;72:1315–1322. doi: 10.1086/375122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmed ZM, Riazuddin S, Aye S, Ali RA, Venselaar H, Anwar S, Belyantseva PP, Qasim M, Riazuddin S, Friedman TB. Gene structure and mutant alleles of pcdh15: Nonsyndromic deafness dfnb23 and type 1 usher syndrome. Hum Genet. 2008;124:215–223. doi: 10.1007/s00439-008-0543-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ammar-Khodja F, Bonnet C, Dahmani M, Ouhab S, Lefevre GM, Ibrahim H, Hardelin JP, Weil D, Louha M, Petit C. Diversity of the causal genes in hearing impaired algerian individuals identified by whole exome sequencing. Mol Genet Genomic Med. 2015;3:189–196. doi: 10.1002/mgg3.131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson DW, Probst FJ, Belyantseva IA, Fridell RA, Beyer L, Martin DM, Wu D, Kachar B, Friedman TB, Raphael Y, Camper SA. The motor and tail regions of myosin xv are critical for normal structure and function of auditory and vestibular hair cells. Hum Mol Genet. 2000;9:1729–1738. doi: 10.1093/hmg/9.12.1729. [DOI] [PubMed] [Google Scholar]
- Askew C, Rochat C, Pan B, Asai Y, Ahmed H, Child E, Schneider BL, Aebischer P, Holt JR. Tmc gene therapy restores auditory function in deaf mice. Sci Transl Med. 2015;7:295ra108. doi: 10.1126/scitranslmed.aab1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atik T, Onay H, Aykut A, Bademci G, Kirazli T, Tekin M, Ozkinay F. Comprehensive analysis of deafness genes in families with autosomal recessive nonsyndromic hearing loss. PLoS One. 2015;10:e0142154. doi: 10.1371/journal.pone.0142154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bademci G, Foster J, 2nd, Mahdieh N, Bonyadi M, Duman D, Cengiz FB, Menendez I, Diaz-Horta O, Shirkavand A, Zeinali S, Subasioglu A, Tokgoz-Yilmaz S, et al. Comprehensive analysis via exome sequencing uncovers genetic etiology in autosomal recessive nonsyndromic deafness in a large multiethnic cohort. Genet Med. 2015 doi: 10.1038/gim.2015.1089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barr-Gillespie PG. Assembly of hair bundles, an amazing problem for cell biology. Mol Biol Cell. 2015;26:2727–2732. doi: 10.1091/mbc.E14-04-0940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bashir R, Fatima A, Naz S. Prioritized sequencing of the second exon of myo15a reveals a new mutation segregating in a pakistani family with moderate to severe hearing loss. Eur J Med Genet. 2012;55:99–102. doi: 10.1016/j.ejmg.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behlouli A, Bonnet C, Abdi S, Bouaita A, Lelli A, Hardelin JP, Schietroma C, Rous Y, Louha M, Cheknane A, Lebdi H, Boudjelida K, et al. Eps8, encoding an actin-binding protein of cochlear hair cell stereocilia, is a new causal gene for autosomal recessive profound deafness. Orphanet J Rare Dis. 2014;9:55. doi: 10.1186/1750-1172-9-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belguith H, Aifa-Hmani M, Dhouib H, Ben Said M, Mosrati MA, Lahmar I, Moalla J, Charfeddine I, Driss N, Ben Arab S, Ghorbel A, Ayadi H, et al. Screening of the dfnb3 locus: Identification of three novel mutations of myo15a associated with hearing loss and further suggestion for two distinctive genes on this locus. Genet Test Mol Biomarkers. 2009;13:147–151. doi: 10.1089/gtmb.2008.0077. [DOI] [PubMed] [Google Scholar]
- Belyantseva IA, Boger ET, Friedman TB. Myosin xva localizes to the tips of inner ear sensory cell stereocilia and is essential for staircase formation of the hair bundle. Proc Natl Acad Sci U S A. 2003;100:13958–13963. doi: 10.1073/pnas.2334417100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belyantseva IA, Boger ET, Naz S, Frolenkov GI, Sellers JR, Ahmed ZM, Griffith AJ, Friedman TB. Myosin-xva is required for tip localization of whirlin and differential elongation of hair-cell stereocilia. Nat Cell Biol. 2005;7:148–156. doi: 10.1038/ncb1219. [DOI] [PubMed] [Google Scholar]
- Besnard T, Garcia-Garcia G, Baux D, Vache C, Faugere V, Larrieu L, Leonard S, Millan JM, Malcolm S, Claustres M, Roux AF. Experience of targeted usher exome sequencing as a clinical test. Mol Genet Genomic Med. 2014;2:30–43. doi: 10.1002/mgg3.25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beurg M, Fettiplace R, Nam JH, Ricci AJ. Localization of inner hair cell mechanotransducer channels using high-speed calcium imaging. Nat Neurosci. 2009;12:553–558. doi: 10.1038/nn.2295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bird JE, Takagi Y, Billington N, Strub MP, Sellers JR, Friedman TB. Chaperone-enhanced purification of unconventional myosin 15, a molecular motor specialized for stereocilia protein trafficking. Proc Natl Acad Sci U S A. 2014;111:12390–12395. doi: 10.1073/pnas.1409459111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boger ET, Sellers JR, Friedman TB. Human myosin xvbp is a transcribed pseudogene. J Muscle Res Cell Motil. 2001;22:477–483. doi: 10.1023/a:1014507705858. [DOI] [PubMed] [Google Scholar]
- Brownstein Z, Abu-Rayyan A, Karfunkel-Doron D, Sirigu S, Davidov B, Shohat M, Frydman M, Houdusse A, Kanaan M, Avraham KB. Novel myosin mutations for hereditary hearing loss revealed by targeted genomic capture and massively parallel sequencing. Eur J Hum Genet. 2014;22:768–775. doi: 10.1038/ejhg.2013.232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownstein Z, Friedman LM, Shahin H, Oron-Karni V, Kol N, Abu Rayyan A, Parzefall T, Lev D, Shalev S, Frydman M, Davidov B, Shohat M, et al. Targeted genomic capture and massively parallel sequencing to identify genes for hereditary hearing loss in middle eastern families. Genome Biol. 2011;12:R89. doi: 10.1186/gb-2011-12-9-r89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cengiz FB, Duman D, Sirmaci A, Tokgoz-Yilmaz S, Erbek S, Ozturkmen-Akay H, Incesulu A, Edwards YJK, Ozdag H, Liu XZ, Tekin M. Recurrent and private myo15a mutations are associated with deafness in the turkish population. Genet Test Mol Biomarkers. 2010;14:543–550. doi: 10.1089/gtmb.2010.0039. [DOI] [PubMed] [Google Scholar]
- Chang MY, Kim AR, Kim NK, Lee C, Lee KY, Jeon WS, Koo JW, Oh SH, Park WY, Kim D, Choi BY. Identification and clinical implications of novel myo15a mutations in a non-consanguineous korean family by targeted exome sequencing. Mol Cells. 2015;38:781–788. doi: 10.14348/molcells.2015.0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Wang Z, Wang Z, Chen D, Chai Y, Pang X, Sun L, Wang X, Yang T, Wu H. Targeted next-generation sequencing in uyghur families with non-syndromic sensorineural hearing loss. PLoS One. 2015;10:e0127879. doi: 10.1371/journal.pone.0127879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chien WW, Monzack EL, McDougald DS, Cunningham LL. Gene therapy for sensorineural hearing loss. Ear Hear. 2015;36:1–7. doi: 10.1097/AUD.0000000000000088. [DOI] [PubMed] [Google Scholar]
- Cooper DN, Krawczak M. The mutational spectrum of single base-pair substitutions causing human genetic disease: Patterns and predictions. Hum Genet. 1990;85:55–74. doi: 10.1007/BF00276326. [DOI] [PubMed] [Google Scholar]
- Delprat B, Michel V, Goodyear R, Yamasaki Y, Michalski N, El-Amraoui A, Perfettini I, Legrain P, Richardson G, Hardelin JP, Petit C. Myosin xva and whirlin, two deafness gene products required for hair bundle growth, are located at the stereocilia tips and interact directly. Hum Mol Genet. 2005;14:401–410. doi: 10.1093/hmg/ddi036. [DOI] [PubMed] [Google Scholar]
- Deol MS. The anatomy and development of the mutants pirouette, shaker-1 and waltzer in the mouse. Proc R Soc Lond B Biol Sci. 1956;145:206–213. doi: 10.1098/rspb.1956.0028. [DOI] [PubMed] [Google Scholar]
- Diaz-Horta O, Duman D, Foster J, Sirmaci A, Gonzalez M, Mahdieh N, Fotouhi N, Bonyadi M, Cengiz FB, Menendez I, Ulloa RH, Edwards YJK, et al. Whole-exome sequencing efficiently detects rare mutations in autosomal recessive nonsyndromic hearing loss. PLoS One. 2012;7:e50628. doi: 10.1371/journal.pone.0050628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donaudy F, Snoeckx R, Pfister M, Zenner HP, Blin N, Di Stazio M, Ferrara A, Lanzara C, Ficarella R, Declau F, Pusch CM, Nurnberg P, et al. Nonmuscle myosin heavy-chain gene myh14 is expressed in cochlea and mutated in patients affected by autosomal dominant hearing impairment (dfna4) Am J Hum Genet. 2004;74:770–776. doi: 10.1086/383285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dror AA, Avraham KB. Hearing impairment: A panoply of genes and functions. Neuron. 2010;68:293–308. doi: 10.1016/j.neuron.2010.10.011. [DOI] [PubMed] [Google Scholar]
- Drummond MC, Barzik M, Bird JE, Zhang DS, Lechene CP, Corey DP, Cunningham LL, Friedman TB. Live-cell imaging of actin dynamics reveals mechanisms of stereocilia length regulation in the inner ear. Nature Communications. 2015;6 doi: 10.1038/ncomms7873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond MC, Belyantseva IA, Friderici KH, Friedman TB. Actin in hair cells and hearing loss. Hear Res. 2012;288:89–99. doi: 10.1016/j.heares.2011.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drummond MC, Friderici KH. A novel actin mrna splice variant regulates actg1 expression. PLoS Genet. 2013;9:e1003743. doi: 10.1371/journal.pgen.1003743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duman D, Sirmaci A, Cengiz FB, Ozdag H, Tekin M. Screening of 38 genes identifies mutations in 62% of families with nonsyndromic deafness in turkey. Genet Test Mol Biomarkers. 2011;15:29–33. doi: 10.1089/gtmb.2010.0120. [DOI] [PubMed] [Google Scholar]
- Ebermann I, Scholl HP, Charbel Issa P, Becirovic E, Lamprecht J, Jurklies B, Millan JM, Aller E, Mitter D, Bolz H. A novel gene for usher syndrome type 2: Mutations in the long isoform of whirlin are associated with retinitis pigmentosa and sensorineural hearing loss. Hum Genet. 2007;121:203–211. doi: 10.1007/s00439-006-0304-0. [DOI] [PubMed] [Google Scholar]
- Fang Q, Indzhykulian AA, Mustapha M, Riordan GP, Dolan DF, Friedman TB, Belyantseva IA, Frolenkov GI, Camper SA, Bird JE. The 133-kda n-terminal domain enables myosin 15 to maintain mechanotransducing stereocilia and is essential for hearing. Elife. 2015;4:e08627. doi: 10.7554/eLife.08627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fattahi Z, Shearer AE, Babanejad M, Bazazzadegan N, Almadani SN, Nikzat N, Jalalvand K, Arzhangi S, Esteghamat F, Abtahi R, Azadeh B, Smith RJ, et al. Screening for myo15a gene mutations in autosomal recessive nonsyndromic, gjb2 negative iranian deaf population. Am J Med Genet A. 2012;158A:1857–1864. doi: 10.1002/ajmg.a.34411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman TB, Liang Y, Weber JL, Hinnant JT, Barber TD, Winata S, Arhya IN, Asher JH., Jr A gene for congenital, recessive deafness dfnb3 maps to the pericentromeric region of chromosome 17. Nat Genet. 1995;9:86–91. doi: 10.1038/ng0195-86. [DOI] [PubMed] [Google Scholar]
- Friedman TB, Riazuddin S. Nonsyndromic deafness: It ain’t necessarily so. In: Popper AN, Fay RR, editors. Perspectives on auditory research. Springer-Verlag; New York: 2014. pp. 149–161. [Google Scholar]
- Frolenkov GI, Belyantseva IA, Friedman TB, Griffith AJ. Genetic insights into the morphogenesis of inner ear hair cells. Nature Reviews Genetics. 2004;5:489–498. doi: 10.1038/nrg1377. [DOI] [PubMed] [Google Scholar]
- Gao X, Zhu QY, Song YS, Wang GJ, Yuan YY, Xin F, Huang SS, Kang DY, Han MY, Guan LP, Zhang JG, Dai P. Novel compound heterozygous mutations in the myo15a gene in autosomal recessive hearing loss identified by whole-exome sequencing. J Transl Med. 2013;11:284. doi: 10.1186/1479-5876-11-284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffith AJ, Friedman TB. In: Ballenger’s otorhinolaryngology head and neck surgery. 18. Wackym PA, Snow JB, editors. USA: People’s Medical Publishing House - USA; 2016. pp. 329–345. [Google Scholar]
- Gu X, Guo L, Ji H, Sun S, Chai R, Wang L, Li H. Genetic testing for sporadic hearing loss using targeted massively parallel sequencing identifies 10 novel mutations. Clin Genet. 2015;87:588–593. doi: 10.1111/cge.12431. [DOI] [PubMed] [Google Scholar]
- Held N, Smits BM, Gockeln R, Schubert S, Nave H, Northrup E, Cuppen E, Hedrich HJ, Wedekind D. A mutation in myo15 leads to usher-like symptoms in lew/ztm-ci2 rats. PLoS One. 2011;6:e15669. doi: 10.1371/journal.pone.0015669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinnant JT. Adaptation to deafness in a balinese community. In: Berlin CI, Keats BJB, editors. Genetics and hearing loss. San Diego, CA: Singular Publishing Group; 2000. pp. 111–123. [Google Scholar]
- Hug N, Longman D, Caceres JF. Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 2016;44:1483–1495. doi: 10.1093/nar/gkw010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imtiaz F, Taibah K, Ramzan K, Bin-Khamis G, Kennedy S, Al-Mubarak B, Trabzuni D, Allam R, Al-Mostafa A, Sogaty S, Al-Shaikh AH, Bamukhayyar SS, et al. A comprehensive introduction to the genetic basis of non-syndromic hearing loss in the saudi arabian population. BMC Med Genet. 2011;12:91. doi: 10.1186/1471-2350-12-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalay E, Uzumcu A, Krieger E, Caylan R, Uyguner O, Ulubil-Emiroglu M, Erdol H, Kayserili H, Hafiz G, Baserer N, Heister AJ, Hennies HC, et al. Myo15a (dfnb3) mutations in turkish hearing loss families and functional modeling of a novel motor domain mutation. Am J Med Genet A. 2007;143A:2382–2389. doi: 10.1002/ajmg.a.31937. [DOI] [PubMed] [Google Scholar]
- Kanzaki S, Beyer L, Karolyi IJ, Dolan DF, Fang Q, Probst FJ, Camper SA, Raphael Y. Transgene correction maintains normal cochlear structure and function in 6-month-old myo15a mutant mice. Hear Res. 2006;214:37–44. doi: 10.1016/j.heares.2006.01.017. [DOI] [PubMed] [Google Scholar]
- Lalwani AK, Goldstein JA, Kelley MJ, Luxford W, Castelein CM, Mhatre AN. Human nonsyndromic hereditary deafness dfna17 is due to a mutation in nonmuscle myosin myh9. Am J Hum Genet. 2000;67:1121–1128. doi: 10.1016/s0002-9297(07)62942-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BP, Green RE, Brenner SE. Evidence for the widespread coupling of alternative splicing and nonsense-mediated mrna decay in humans. Proc Natl Acad Sci U S A. 2003;100:189–192. doi: 10.1073/pnas.0136770100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lezirovitz K, Pardono E, de Mello Auricchio MT, de Carvalho ESFL, Lopes JJ, Abreu-Silva RS, Romanos J, Batissoco AC, Mingroni-Netto RC. Unexpected genetic heterogeneity in a large consanguineous brazilian pedigree presenting deafness. Eur J Hum Genet. 2008;16:89–96. doi: 10.1038/sj.ejhg.5201917. [DOI] [PubMed] [Google Scholar]
- Li W, Guo L, Li Y, Wu Q, Li Q, Li H, Dai C. A novel recessive truncating mutation in myo15a causing prelingual sensorineural hearing loss. Int J Pediatr Otorhinolaryngol. 2016;81:92–95. doi: 10.1016/j.ijporl.2015.12.013. [DOI] [PubMed] [Google Scholar]
- Liang Y, Wang A, Belyantseva IA, Anderson DW, Probst FJ, Barber TD, Miller W, Touchman JW, Jin L, Sullivan SL, Sellers JR, Camper SA, et al. Characterization of the human and mouse unconventional myosin xv genes responsible for hereditary deafness dfnb3 and shaker 2. Genomics. 1999;61:243–258. doi: 10.1006/geno.1999.5976. [DOI] [PubMed] [Google Scholar]
- Liburd N, Ghosh M, Riazuddin S, Naz S, Khan S, Ahmed Z, Riazuddin S, Liang Y, Menon PSN, Smith T, Smith ACM, Chen KS, et al. Novel mutations of myo15a associated with profound deafness in consanguineous families and moderately severe hearing loss in a patient with smith-magenis syndrome. Hum Genet. 2001;109:535–541. doi: 10.1007/s004390100604. [DOI] [PubMed] [Google Scholar]
- Liu XZ, Walsh J, Mburu P, KendrickJones J, Cope MJTV, Steel KP, Brown SDM. Mutations in the myosin viia gene cause non-syndromic recessive deafness. Nat Genet. 1997a;16:188–190. doi: 10.1038/ng0697-188. [DOI] [PubMed] [Google Scholar]
- Liu XZ, Walsh J, Tamagawa Y, Kitamura K, Nishizawa M, Steel KP, Brown SDM. Autosomal dominant non-syndromic deafness caused by a mutation in the myosin viia gene. Nat Genet. 1997b;17:268–269. doi: 10.1038/ng1197-268. [DOI] [PubMed] [Google Scholar]
- Lloyd RV, Vidal S, Jin L, Zhang S, Kovacs K, Horvath E, Scheithauer BW, Boger ET, Fridell RA, Friedman TB. Myosin xva expression in the pituitary and in other neuroendocrine tissues and tumors. Am J Pathol. 2001;159:1375–1382. doi: 10.1016/S0002-9440(10)62524-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loscher W. Abnormal circling behavior in rat mutants and its relevance to model specific brain dysfunctions. Neurosci Biobehav Rev. 2010;34:31–49. doi: 10.1016/j.neubiorev.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Manor U, Disanza A, Grati M, Andrade L, Lin H, Di Fiore PP, Scita G, Kachar B. Regulation of stereocilia length by myosin xva and whirlin depends on the actin-regulatory protein eps8. Curr Biol. 2011;21:167–172. doi: 10.1016/j.cub.2010.12.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mburu P, Mustapha M, Varela A, Weil D, El-Amraoui A, Holme RH, Rump A, Hardisty RE, Blanchard S, Coimbra RS, Perfettini I, Parkinson N, et al. Defects in whirlin, a pdz domain molecule involved in stereocilia elongation, cause deafness in the whirler mouse and families with dfnb31. Nat Genet. 2003;34:421–428. doi: 10.1038/ng1208. [DOI] [PubMed] [Google Scholar]
- Melchionda S, Ahituv N, Bisceglia L, Sobe T, Glaser F, Rabionet R, Arbones ML, Notarangelo A, Di Iorio E, Carella M, Zelante L, Estivill X, et al. Myo6, the human homologue of the gene responsible for deafness in snell’s waltzer mice, is mutated in autosomal dominant nonsyndromic hearing loss. Am J Hum Genet. 2001;69:635–640. doi: 10.1086/323156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa M, Naito T, Nishio SY, Kamatani N, Usami S. Targeted exon sequencing successfully discovers rare causative genes and clarifies the molecular epidemiology of japanese deafness patients. PLoS One. 2013a;8:e71381. doi: 10.1371/journal.pone.0071381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa M, Nishio S, Ikeda T, Fukushima K, Usami S. Massively parallel DNA sequencing successfully identifies new causative mutations in deafness genes in patients with cochlear implantation and eas. PLoS One. 2013b;8:e75793. doi: 10.1371/journal.pone.0075793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyagawa M, Nishio SY, Hattori M, Moteki H, Kobayashi Y, Sato H, Watanabe T, Naito Y, Oshikawa C, Usami S. Mutations in the myo15a gene are a significant cause of nonsyndromic hearing loss: Massively parallel DNA sequencing-based analysis. Ann Otol Rhinol Laryngol. 2015;124(Suppl 1):158S–168S. doi: 10.1177/0003489415575058. [DOI] [PubMed] [Google Scholar]
- Morell RJ, Kim HJ, Hood LJ, Goforth L, Friderici K, Fisher R, Van Camp G, Berlin CI, Oddoux C, Ostrer H, Keats B, Friedman TB. Mutations in the connexin 26 gene (gjb2) among ashkenazi jews with nonsyndromic recessive deafness. N Engl J Med. 1998;339:1500–1505. doi: 10.1056/NEJM199811193392103. [DOI] [PubMed] [Google Scholar]
- Moteki H, Azaiez H, Booth KT, Shearer AE, Sloan CM, Kolbe DL, Nishio S, Hattori M, Usami S, Smith RJ. Comprehensive genetic testing with ethnic-specific filtering by allele frequency in a japanese hearing-loss population. Clin Genet. 2015;89:466–472. doi: 10.1111/cge.12677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller U, Barr-Gillespie PG. New treatment options for hearing loss. Nat Rev Drug Discov. 2015;14:346–365. doi: 10.1038/nrd4533. [DOI] [PubMed] [Google Scholar]
- Murphy CT, Spudich JA. Variable surface loops and myosin activity: Accessories to a motor. J Muscle Res Cell Motil. 2000;21:139–151. doi: 10.1023/a:1005610007209. [DOI] [PubMed] [Google Scholar]
- Nal N, Ahmed ZM, Erkal E, Alper OM, Luleci G, Dinc O, Waryah AM, Ain Q, Tasneem S, Husnain T, Chattaraj P, Riazuddin S, et al. Mutational spectrum of myo15a: The large n-terminal extension of myosin xva is required for hearing. Hum Mutat. 2007;28:1014–1019. doi: 10.1002/humu.20556. [DOI] [PubMed] [Google Scholar]
- Narayanan P, Chatterton P, Ikeda A, Ikeda S, Corey DP, Ervasti JM, Perrin BJ. Length regulation of mechanosensitive stereocilia depends on very slow actin dynamics and filament-severing proteins. Nature Communications. 2015;6 doi: 10.1038/ncomms7855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neveling K, Feenstra I, Gilissen C, Hoefsloot LH, Kamsteeg EJ, Mensenkamp AR, Rodenburg RJ, Yntema HG, Spruijt L, Vermeer S, Rinne T, van Gassen KL, et al. A post-hoc comparison of the utility of sanger sequencing and exome sequencing for the diagnosis of heterogeneous diseases. Hum Mutat. 2013;34:1721–1726. doi: 10.1002/humu.22450. [DOI] [PubMed] [Google Scholar]
- Oshima K, Shin K, Diensthuber M, Peng AW, Ricci AJ, Heller S. Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell. 2010;141:704–716. doi: 10.1016/j.cell.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JH, Kim NKD, Kim AR, Rhee J, Oh SH, Koo JW, Nam JY, Park WY, Choi BY. Exploration of molecular genetic etiology for korean cochlear implantees with severe to profound hearing loss and its implication. Orphanet J Rare Dis. 2014;9:167. doi: 10.1186/s13023-014-0167-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petit C, Richardson GP. Linking genes underlying deafness to hair-bundle development and function. Nat Neurosci. 2009;12:703–710. doi: 10.1038/nn.2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Probst FJ, Fridell RA, Raphael Y, Saunders TL, Wang A, Liang Y, Morell RJ, Touchman JW, Lyons RH, Noben-Trauth K, Friedman TB, Camper SA. Correction of deafness in shaker-2 mice by an unconventional myosin in a bac transgene. Science. 1998;280:1444–1447. doi: 10.1126/science.280.5368.1444. [DOI] [PubMed] [Google Scholar]
- Rehman AU, Santos-Cortez RL, Drummond MC, Shahzad M, Lee K, Morell RJ, Ansar M, Jan A, Wang X, Aziz A, Riazuddin S, Smith JD, et al. Challenges and solutions for gene identification in the presence of familial locus heterogeneity. Eur J Hum Genet. 2015;23:1207–1215. doi: 10.1038/ejhg.2014.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riahi Z, Bonnet C, Zainine R, Louha M, Bouyacoub Y, Laroussi N, Chargui M, Kefi R, Jonard L, Dorboz I, Hardelin JP, Salah SB, et al. Whole exome sequencing identifies new causative mutations in tunisian families with non-syndromic deafness. PLoS One. 2014;9:e99797. doi: 10.1371/journal.pone.0099797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ronaghi M, Nasr M, Ealy M, Durruthy-Durruthy R, Waldhaus J, Diaz GH, Joubert LM, Oshima K, Heller S. Inner ear hair cell-like cells from human embryonic stem cells. Stem Cells Dev. 2014;23:1275–1284. doi: 10.1089/scd.2014.0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rzadzinska AK, Schneider ME, Davies C, Riordan GP, Kachar B. An actin molecular treadmill and myosins maintain stereocilia functional architecture and self-renewal. J Cell Biol. 2004;164:887–897. doi: 10.1083/jcb.200310055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider ME, Belyantseva IA, Azevedo RB, Kachar B. Rapid renewal of auditory hair bundles. Nature. 2002;418:837–838. doi: 10.1038/418837a. [DOI] [PubMed] [Google Scholar]
- Schrauwen I, Sommen M, Corneveaux JJ, Reiman RA, Hackett NJ, Claes C, Claes K, Bitner-Glindzicz M, Coucke P, Van Camp G, Huentelman MJ. A sensitive and specific diagnostic test for hearing loss using a microdroplet pcr-based approach and next generation sequencing. Am J Med Genet A. 2013;161A:145–152. doi: 10.1002/ajmg.a.35737. [DOI] [PubMed] [Google Scholar]
- Schwander M, Kachar B, Muller U. The cell biology of hearing. J Cell Biol. 2010;190:9–20. doi: 10.1083/jcb.201001138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scotti MM, Swanson MS. Rna mis-splicing in disease. Nat Rev Genet. 2016;17:19–32. doi: 10.1038/nrg.2015.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sellers JR. Myosins: A diverse superfamily. Biochim Biophys Acta. 2000;1496:3–22. doi: 10.1016/s0167-4889(00)00005-7. [DOI] [PubMed] [Google Scholar]
- Shafique S, Siddiqi S, Schraders M, Oostrik J, Ayub H, Bilal A, Ajmal M, Seco CZ, Strom TM, Mansoor A, Mazhar K, Shah STA, et al. Genetic spectrum of autosomal recessive non-syndromic hearing loss in pakistani families. PLoS One. 2014;9:e100146. doi: 10.1371/journal.pone.0100146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahin H, Walsh T, Rayyan AA, Lee MK, Higgins J, Dickel D, Lewis K, Thompson J, Baker C, Nord AS, Stray S, Gurwitz D, et al. Five novel loci for inherited hearing loss mapped by snp-based homozygosity profiles in palestinian families. Eur J Hum Genet. 2010;18:407–413. doi: 10.1038/ejhg.2009.190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, Eppsteiner RW, Booth KT, Ephraim SS, Gurrola J, 2nd, Simpson A, Black-Ziegelbein EA, Joshi S, Ravi H, Giuffre AC, Happe S, Hildebrand MS, et al. Utilizing ethnic-specific differences in minor allele frequency to recategorize reported pathogenic deafness variants. Am J Hum Genet. 2014;95:445–453. doi: 10.1016/j.ajhg.2014.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shearer AE, Hildebrand MS, Webster JA, Kahrizi K, Meyer NC, Jalalvand K, Arzhanginy S, Kimberling WJ, Stephan D, Bahlo M, Smith RJ, Najmabadi H. Mutations in the first myth4 domain of myo15a are a common cause of dfnb3 hearing loss. Laryngoscope. 2009;119:727–733. doi: 10.1002/lary.20116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan-Heggen CM, Babanejad M, Beheshtian M, Simpson AC, Booth KT, Ardalani F, Frees KL, Mohseni M, Mozafari R, Mehrjoo Z, Jamali L, Vaziri S, et al. Characterising the spectrum of autosomal recessive hereditary hearing loss in iran. J Med Genet. 2015;52:823–829. doi: 10.1136/jmedgenet-2015-103389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sloan-Heggen CM, Bierer AO, Shearer AE, Kolbe DL, Nishimura CJ, Frees KL, Ephraim SS, Shibata SB, Booth KT, Campbell CA, Ranum PT, Weaver AE, et al. Comprehensive genetic testing in the clinical evaluation of 1119 patients with hearing loss. Hum Genet. 2016;135:441–450. doi: 10.1007/s00439-016-1648-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stepanyan R, Belyantseva IA, Griffith AJ, Friedman TB, Frolenkov GI. Auditory mechanotransduction in the absence of functional myosin-xva. J Physiol. 2006;576:801–808. doi: 10.1113/jphysiol.2006.118547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney HL, Houdusse A. Structural and functional insights into the myosin motor mechanism. Annual Review of Biophysics. 2010;39:539–557. doi: 10.1146/annurev.biophys.050708.133751. [DOI] [PubMed] [Google Scholar]
- Tilney LG, Tilney MS, Derosier DJ. Actin-filaments, stereocilia, and hair-cells - how cells count and measure. Annu Rev Cell Biol. 1992;8:257–274. doi: 10.1146/annurev.cb.08.110192.001353. [DOI] [PubMed] [Google Scholar]
- Vitkup D, Sander C, Church GM. The amino-acid mutational spectrum of human genetic disease. Genome Biol. 2003;4:R72. doi: 10.1186/gb-2003-4-11-r72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vona B, Muller T, Nanda I, Neuner C, Hofrichter MA, Schroder J, Bartsch O, Lassig A, Keilmann A, Schraven S, Kraus F, Shehata-Dieler W, et al. Targeted next-generation sequencing of deafness genes in hearing-impaired individuals uncovers informative mutations. Genet Med. 2014;16:945–953. doi: 10.1038/gim.2014.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vozzi D, Morgan A, Vuckovic D, D’Eustacchio A, Abdulhadi K, Rubinato E, Badii R, Gasparini P, Girotto G. Hereditary hearing loss: A 96 gene targeted sequencing protocol reveals novel alleles in a series of italian and qatari patients. Gene. 2014;542:209–216. doi: 10.1016/j.gene.2014.03.033. [DOI] [PubMed] [Google Scholar]
- Walsh T, Walsh V, Vreugde S, Hertzano R, Shahin H, Haika S, Lee MK, Kanaan M, King MC, Avraham KB. From flies’ eyes to our ears: Mutations in a human class iii myosin cause progressive nonsyndromic hearing loss dfnb30. Proc Natl Acad Sci U S A. 2002;99:7518–7523. doi: 10.1073/pnas.102091699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang A, Liang Y, Fridell RA, Probst FJ, Wilcox ER, Touchman JW, Morton CC, Morell RJ, Noben-Trauth K, Camper SA, Friedman TB. Association of unconventional myosin myo15 mutations with human nonsyndromic deafness dfnb3. Science. 1998;280:1447–1451. doi: 10.1126/science.280.5368.1447. [DOI] [PubMed] [Google Scholar]
- Weil D, Kussel P, Blanchard S, Levy G, LeviAcobas F, Drira M, Ayadi H, Petit C. The autosomal recessive isolated deafness, dfnb2, and the usher 1b syndrome are allelic defects of the myosin-viia gene. Nat Genet. 1997;16:191–193. doi: 10.1038/ng0697-191. [DOI] [PubMed] [Google Scholar]
- Winata S, Arhya IN, Moeljopawiro S, Hinnant JT, Liang Y, Friedman TB, Asher JH., Jr Congenital non-syndromal autosomal recessive deafness in bengkala, an isolated balinese village. J Med Genet. 1995;32:336–343. doi: 10.1136/jmg.32.5.336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woo HM, Park HJ, Baek JI, Park MH, Kim UK, Sagong B, Koo SK. Whole-exome sequencing identifies myo15a mutations as a cause of autosomal recessive nonsyndromic hearing loss in korean families. BMC Med Genet. 2013;14:72. doi: 10.1186/1471-2350-14-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia H, Huang X, Guo Y, Hu P, He G, Deng X, Xu H, Yang Z, Deng H. Identification of a novel myo15a mutation in a chinese family with autosomal recessive nonsyndromic hearing loss. PLoS One. 2015;10:e0136306. doi: 10.1371/journal.pone.0136306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang T, Wei X, Chai Y, Li L, Wu H. Genetic etiology study of the non-syndromic deafness in chinese hans by targeted next-generation sequencing. Orphanet J Rare Dis. 2013;8:85. doi: 10.1186/1750-1172-8-85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yano T, Ichinose A, Nishio S, Kobayashi Y, Sato H, Usami S. A novel mutation of myo15a associated with hearing loss in a japanese family. Journal of Clinical Case Reports. 2013;3:10. 4172/2165-7920.1000319. [Google Scholar]
- Zampini V, Ruttiger L, Johnson SL, Franz C, Furness DN, Waldhaus J, Xiong H, Hackney CM, Holley MC, Offenhauser N, Di Fiore PP, Knipper M, et al. Eps8 regulates hair bundle length and functional maturation of mammalian auditory hair cells. PLoS Biol. 2011;9:e1001048. doi: 10.1371/journal.pbio.1001048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang DS, Piazza V, Perrin BJ, Rzadzinska AK, Poczatek JC, Wang M, Prosser HM, Ervasti JM, Corey DP, Lechene CP. Multi-isotope imaging mass spectrometry reveals slow protein turnover in hair-cell stereocilia. Nature. 2012;481:520–524. doi: 10.1038/nature10745. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.