SUMMARY
Pax9 encodes a paired-box homeodomain transcription factor and is critical for the development of multiple organs. Using CrispR/Cas9-mediated homologous directed repair (HDR), we generated a new Pax9-CreER knock-in mouse line in which the CreERT2 fusion protein is produced after synthesis of endogenous Pax9 protein. We found that tdTomato reporter expression in Pax9-CreER;tdTomato reporter mice is detectable in a similar pattern to the endogenous Pax9 expression, faithfully recapitulating the Pax9 expression domains throughout the embryo and in the adult mouse. At early embryonic stages, the tdTomato reporter is expressed first in the pharyngeal pouch region and later in the craniofacial mesenchyme, somites, limbs, and lingual papillae in the adult tongue. These results demonstrate that this new Pax9-CreER knock-in mouse line can be used for lineage tracing and genetic targeting of Pax9-expressing cells and their progeny in a temporally and spatially controlled manner during development and organogenesis.
Keywords: Pax9, Pax9-CreER, genetic labeling, mouse development
INTRODUCTION
Pax9 belongs to a family of highly conserved genes that encode paired-box homeodomain (Pax) transcription factors and is expressed in a wide variety of mouse embryonic tissues during development. Expression of Pax9 mRNA is first detectable in the pharyngeal pouch around embryonic day E9.0 (Peters et al., 1998) and, subsequently, in the derivatives of the pharyngeal pouch, sclerotome, and limb buds (Neubüser et al., 1995; Peters et al., 1998). In addition, Pax9 is also expressed in the craniofacial mesenchyme, including the nasal processes and mandibular and maxillary arch (Peters et al., 1998). Pax9 expression also marks sites of prospective tooth development before morphological signs of odontogenesis appear, and is maintained in the developing embryonic tooth mesenchyme (Neubüser et al., 1997; Peters et al., 1998).
In Pax9 null mutant mice (Pax9LacZ/LacZ), not all the structures expressing Pax9 are affected, suggesting that Pax9 is not indispensible for development of all Pax9+ tissues. These Pax9 knock-out mice showed defects in derivatives of the third and fourth pharyngeal pouches, such as the thymus and parathyroid glands (Peters et al., 1998). Although the elbows, knees and tails of Pax9LacZ/LacZ knock-out mice appeared unaffected, they did exhibit abnormal digit formation and preaxial digit duplication of the fore- and hindlimbs (Peters et al., 1998). Pax9 is also critical for the development of craniofacial mesenchymal tissues, particularly those that are derived from the cranial neural crest. Both Pax9 null and Pax9 conditional knock-out (Wnt1-Cre;Pax9fl/fl) mice, in which Pax9 is only deleted in neural crest derivatives, exhibit cleft secondary palate with absent palatal processes of the premaxilla. In the mandible region, alveolar bone and coronoid process formation was also impaired. Furthermore, tooth development is arrested at the bud stage (Kist et al., 2007; Peters et al., 1998).
Interestingly, although Pax9 is not expressed in the skin, it is expressed in the epithelium of the tongue at late embryonic stages and in adults (Jonker et al., 2004). Moreover, this Pax9 expression is required for maintaining the morphology and permeability of tongue epithelial cells and may also prevent these cells from adopting a skin-specific epithelial fate (Jonker et al., 2004). In adults, Pax9 expression is also detectable in epithelial tissues of the digestive system, such as the esophagus (Jonker et al., 2004; Peters et al., 1997). Although the function of Pax9 in later stages remains unclear, previous studies have suggested a role in malignancy and cell survival of cancerous epithelium (Gerber et al., 2002; Lee et al., 2008).
Despite the critical requirement for Pax9 during embryonic development, the specific contribution of the Pax9+ subpopulation remains unclear. Moreover, the molecular regulation associated with maintaining distinct cellular identities in Pax9+ cells remains unknown. CrispR/Cas9-mediated gene knock-out/knock-in methods have recently emerged as useful tools for the efficient generation of transgenic mouse models (Wang et al., 2013; Yang et al., 2013). Using the CrispR/Cas9-mediated homologous directed repair (HDR) mechanism, we generated a new Pax9-CreER knock-in allele allowing the production of the tamoxifen-inducible CreERT2 recombinase (Feil et al., 1997) in cells normally expressing Pax9. Therefore, Pax9-driven Cre-mediated recombination can be used to leave indelible genetic markers that identify Pax9+ cell lineages during embryonic developmental stages as well as in adults. These mice will help identify the molecular mechanisms regulating Pax9+ cells. Here, we describe this new allele and its utility for lineage tracing and genetic targeting of Pax9 expressing cells and their progeny during embryonic and postnatal stages.
RESULTS AND DISCUSSION
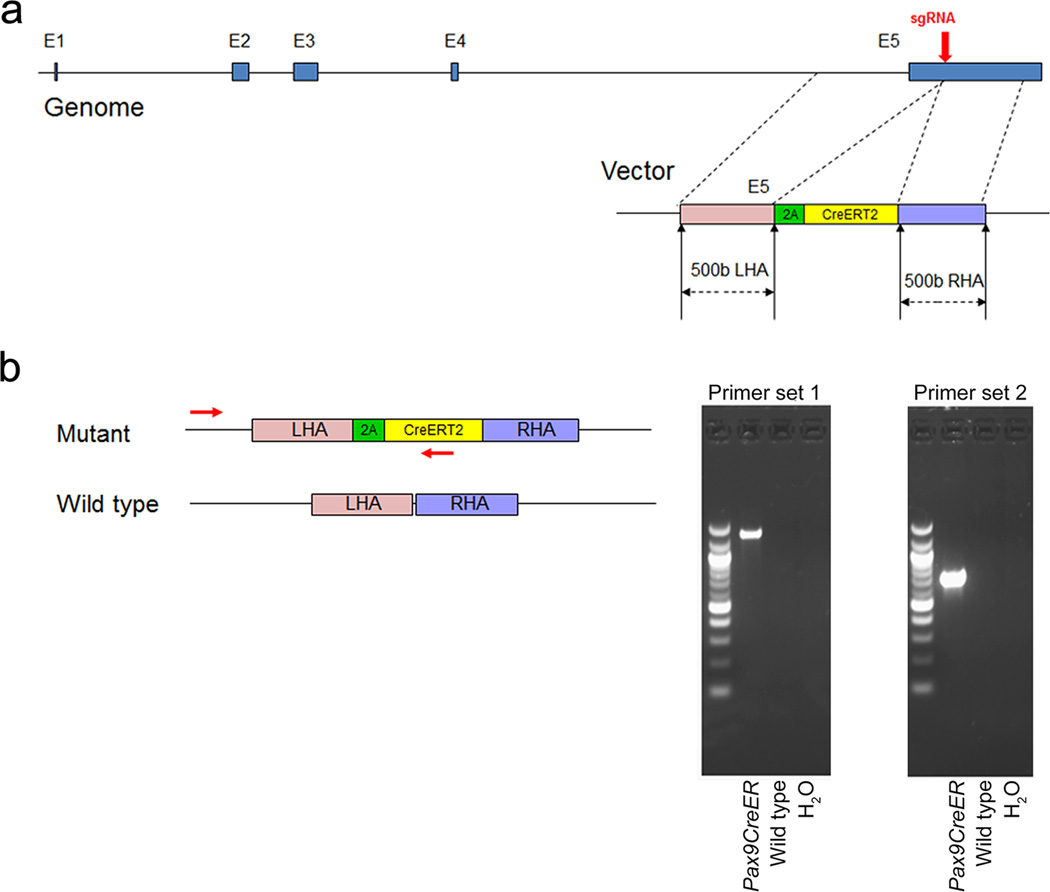
The mouse Pax9 gene is located on Chromosome 12:56,691,767-56,712,822. It consists of 5 exons interrupted by 4 introns (Ensemble, ENSMUSG00000001497). In order to create an allele in which CreERT2 expression recapitulates endogenous Pax9 expression without interfering with Pax9 function, we integrated the 2A-CreERT2 coding sequence into the last exon of Pax9, immediately before the stop codon (Fig. 1a). Following sgRNA-mediated double strand DNA breakage by Cas9 enzyme, the homology arms guide the 2A-CreER to insert in-frame with the Pax9 open-reading frame by homologous directed repair (HDR). As a result, CreER expression is regulated by the Pax9 promoter in tandem with endogenous Pax9 expression.
FIG. 1.
Generation of Pax9-CreER knock-in mice. (a) Schematic diagram of the knock-in strategy showing the insertion site of the Pax9 donor construct into the Pax9 gene. (b) Primers and PCR products for the identification of Pax9-CreER mice. Schematic diagram depicts the location of the PCR primers detecting the mutant allele.
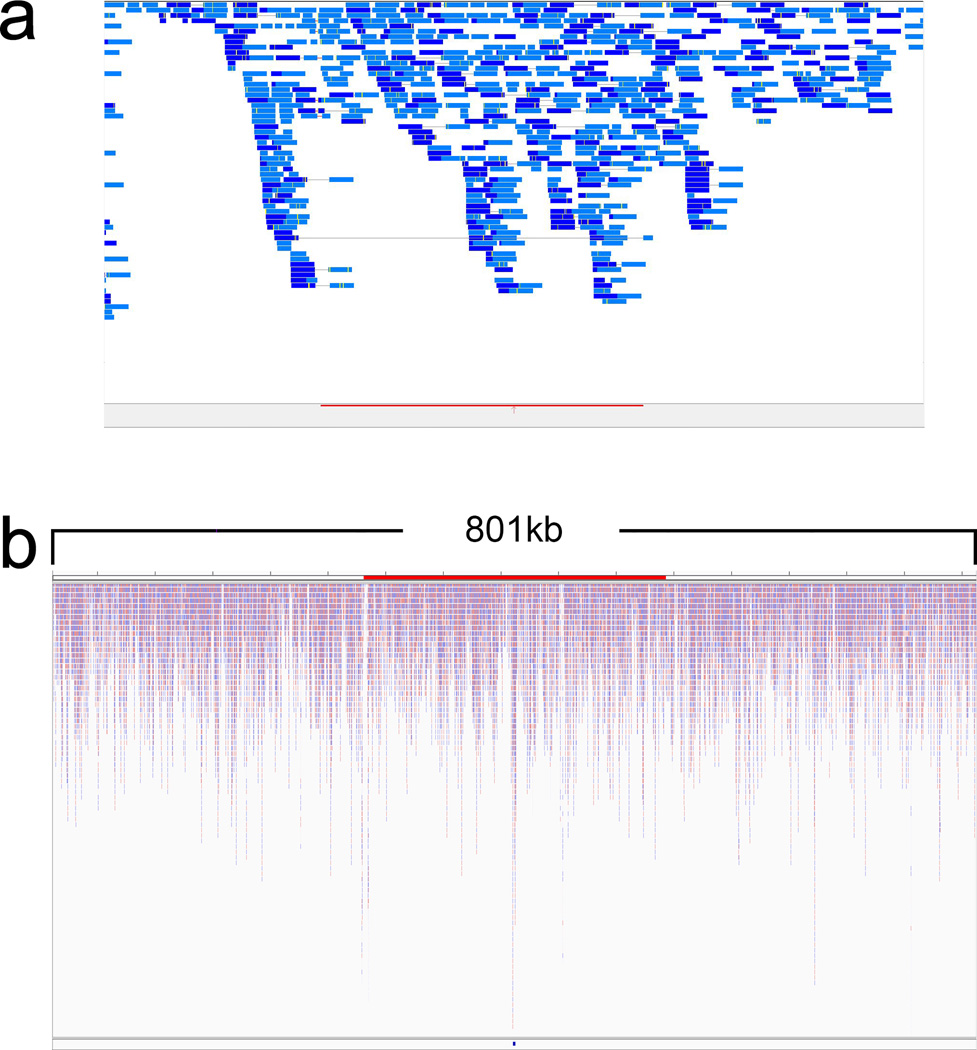
To detect the insertion of CreERT2 into the Pax9 gene, we designed polymerase chain reaction (PCR) primers to target the altered DNA fragments spanning the Pax9 gene outside the left homology arm and the CreER cassette inside the donor construct (Fig. 1b). We identified one mouse from 24 littermates that contained mutant PCR products consistent with the presence of the CreER cassette in the 3’ end of the Pax9 gene. To examine the insertion site in greater detail, we performed whole-genome sequencing and aligned the sequence reads to a mouse reference genome, in which Chromosome 12 was replaced with an artificial Chromosome 12 sequence containing the Pax9 donor construct (left homology arm+2A-CreER+right homology arm). We confirmed that the Pax9 donor construct is inserted in the 3’end of the Pax9 gene (Fig. 2a). Moreover, analysis of the genome sequencing map from a wider region of Chromosome 12 indicates that the Pax9 donor construct read is comparable to that of other regions (Fig. 2b), consistent with insertion of only one copy of the Pax9 donor construct. Taken together, our results show that the Pax9 donor construct was only inserted at the predicted site, not in an ectopic site.
FIG. 2.
Whole-genome sequencing analysis of Pax9-CreER knock-in mice. (a) Focused snapshot image of whole-genome sequencing reads that aligned to the 800kb region of the Pax9 donor construct (orange bar at the bottom of the figure) within mouse Chromosome 12. Light and dark blue horizontal bars reflect the paired-end read direction with a minimum read depth of 20× with no sequencing gaps located within the insert. (b) Zoomed out view of the Pax9 donor construct (small blue rectangle at the bottom of the figure) and surrounding genomic region demonstrating no significant deviation of the average coverage and typical read depth. The figures were produced using the Genetrix Viewer (Epicenter Software) and the Integrative Genomics Viewer (IGV) from the Broad Institute.
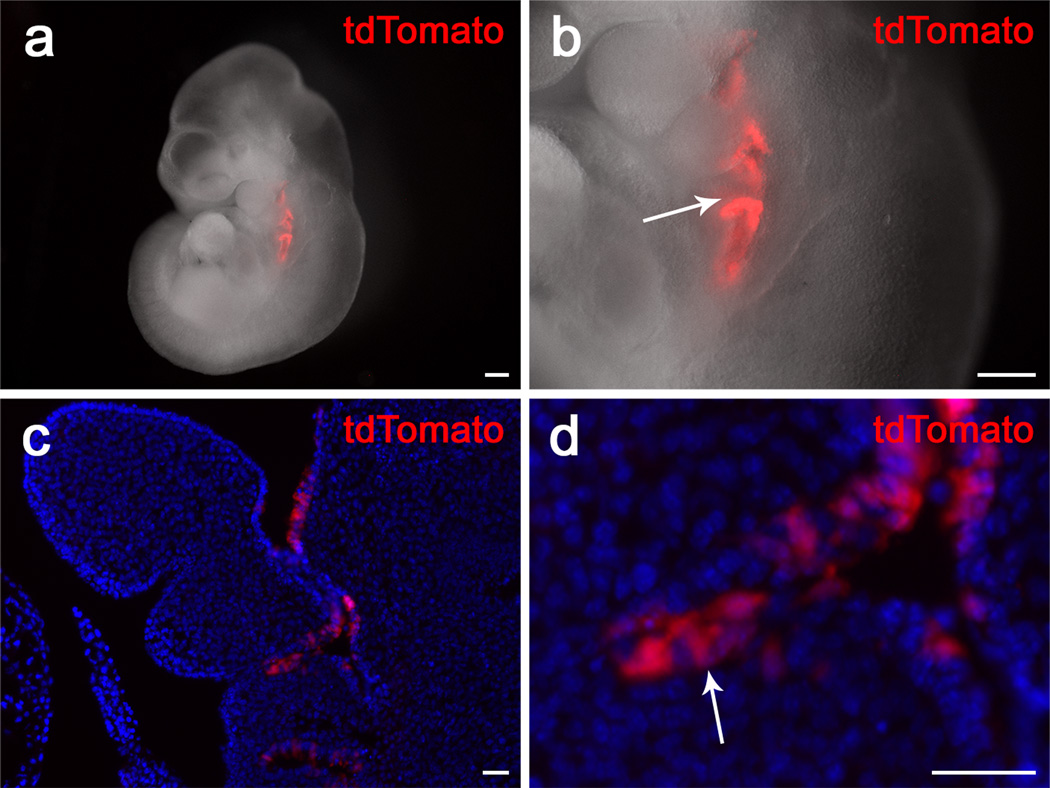
To analyze the CreER expression pattern in vivo, we generated Pax9-CreER;tdTomato reporter mice to detect Cre activity driven by dynamic Pax9 expression at various stages. Throughout this study, mice were analyzed 48 hours post-tamoxifen induction, in order to allow 12–24 hours for the tamoxifen to take effect and another 12–24 hours for the tdTomato protein to be synthesized. To confirm that the CreER recombination reflects the expression of endogenous Pax9, we compared the pattern of tdTomato expression to that previously reported for Pax9. Previously, Pax9 expression was first reported in the pharyngeal pouch region around E9.0. Consistent with this, after Cre activity was induced with tamoxifen at E8.5, tdTomoto reporter expression was detectable in a similar region 48 hours later (Fig. 3).
FIG. 3.
tdTomato expression in the pharyngeal pouch region of Pax9-CreER;tdTomato mice induced at E8.5. (a, b) Visualization of whole-mount Pax9-CreER;tdTomato mice 48 hours after tamoxifen induction at E8.5. Arrow indicates tdTomato staining in the pharyngeal pouch regions (c, d) Visualization of sections of Pax9-CreER;tdTomato mice 48 hours after tamoxifen induction at E8.5. Note that tdTomato signaling is restricted to the epithelium of the pharyngeal pouches (arrow). Scale bars, (a) and (b), 200μm; (c) and (d), 25μm.
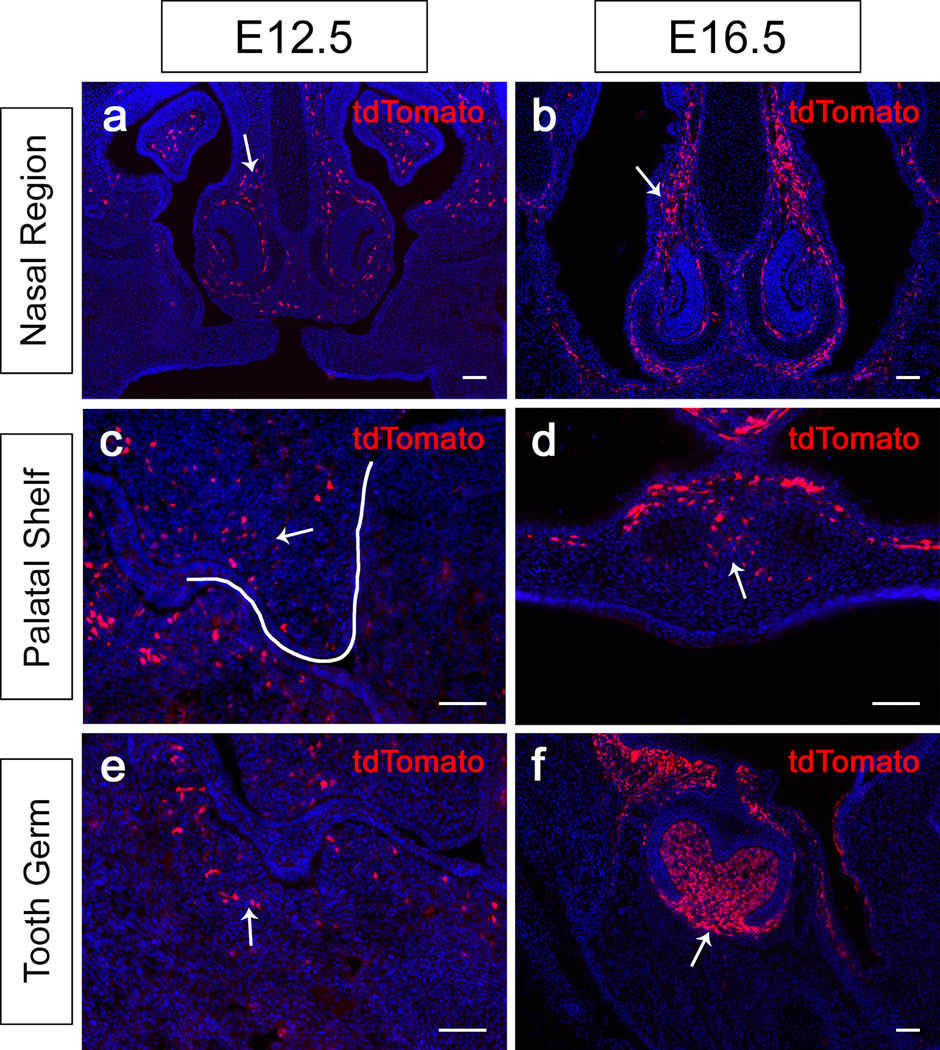
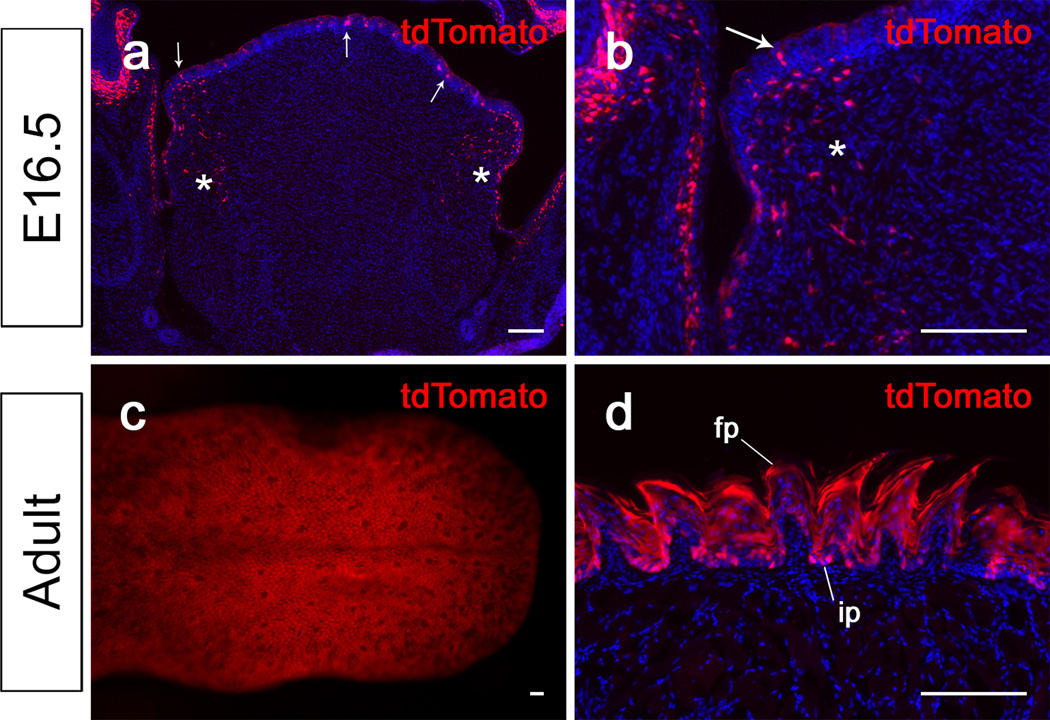
Pax9 is a critical regulator of the craniofacial region during development (Kist et al., 2007; Peters et al., 1998), so we examined Pax9 expression at various stages of craniofacial development. Two days after tamoxifen induction of E10.5 and E14.5 Pax9-CreER;tdTomato reporter mice, tdTomato expression was detectable in the craniofacial region. In sections, expression was detectable in the nasal mesenchyme (Fig. 4a,b). Furthermore, we found that tdTomato was expressed in the palatal shelf mesenchyme (Fig. 4c,d). In the mandible, we detected tdTomato-expressing cells in the mesenchymal region closely associated with the tooth bud at E12.5 (Fig. 4e). At the bell stage, tdTomato expression was more restricted to the dental mesenchyme, dental follicle and mesenchymal region immediately adjacent to the dental epithelium (Fig. 4f). Overall, this pattern of tdTomato expression is consistent with that of Pax9 in previously published studies.
FIG. 4.
tdTomato expression in the craniofacial region of Pax9-CreER;tdTomato mice induced at E10.5 and E14.5. (a–i) Visualization of coronal sections of the nasal region (a–b), the palatal shelves (c–d), and the tooth germs (e–f) of Pax9-CreER;tdTomato mice 48 hours after tamoxifen induction at E10.5 and E14.5. White line indicates the palatal shelf prior to elevation and fusion (c). Arrows indicate tdTomato staining in the mesenchyme of these regions. Scale bars, 50μm.
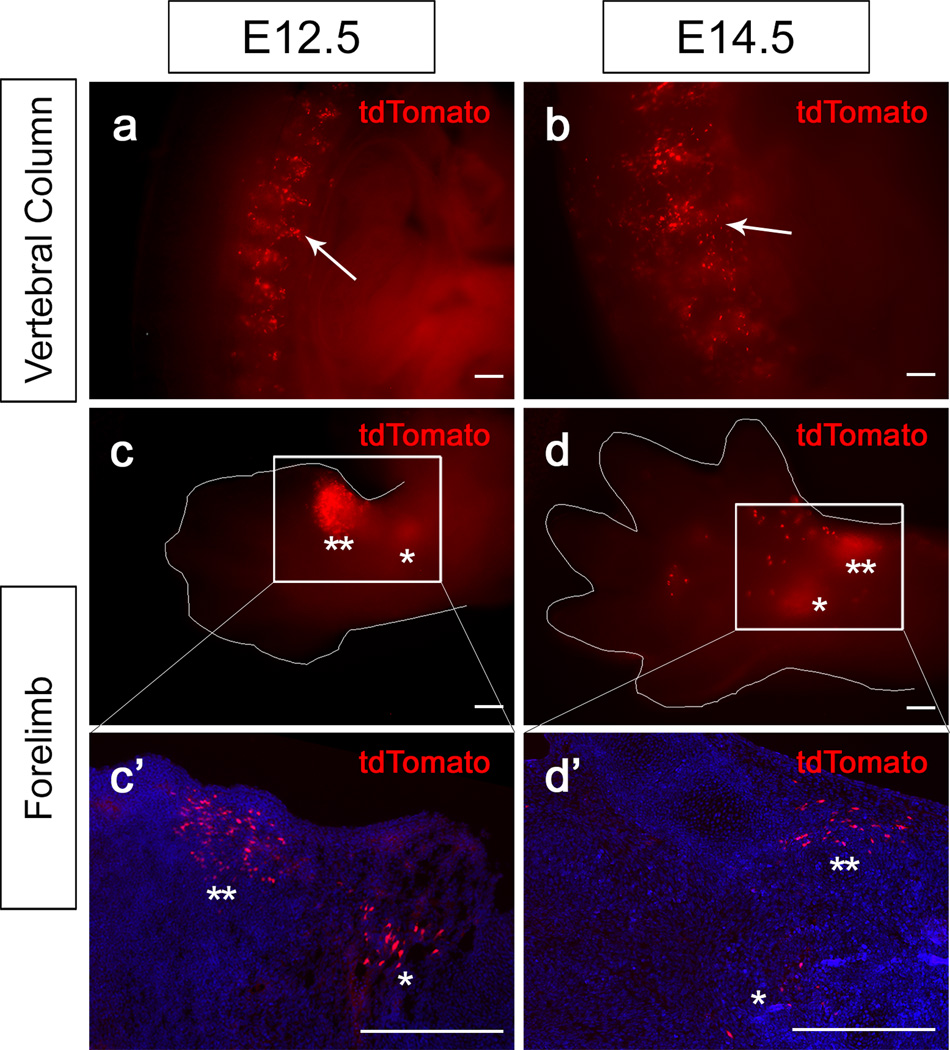
Next, we examined tdTomato expression in other tissues of the body during development. Two days after tamoxifen induction of E10.5 and E12.5 Pax9-CreER;tdTomato reporter mice, tdTomato expression was detectable in the vertebral column (Fig. 5a,b). Specifically, we detected tdTomato expression in the condensation of the intervertebral disk primordia at both stages. In addition, we detected tdTomato expression in the limb region (Fig. 5c,d). At E12.5, strong expression of tdTomato was detectable in the anterior proximal region of the forelimb mesenchyme, with weaker expression more posteriorly (Fig. 5c,c’). At E14.5, tdTomato expression was detectable ventral to the primordia of the phalangeal bones and in the pawpad (Fig. 5d,d’). At later stages, we also observed Pax9 expression in other regions where Pax9 was previously reported to be expressed, such as the tongue (Fig. 6a–d). Two days after tamoxifen induction of E14.5 Pax9-CreER;tdTomato reporter mice, tdTomato expression was detectable in the tongue epithelium and the mesenchyme of the lateral region of the tongue (Fig. 6a,b). In adults, Pax9 expression was previously reported in the filiform papillae of the tongue. Consistently, in adult Pax9-CreER;tdTomato reporter mice, we detected tdTomoto reporter expression throughout the lingual papillae, including the filiform papillae (Fig. 6c,d).
FIG. 5.
tdTomato expression in the vertebral column and forelimbs of Pax9-CreER;tdTomato mice induced at E10.5 and E12.5. (a, b) Visualization of whole mount transverse section view of the vertebral column of E12.5 and E14.5 Pax9-CreER;tdTomato mice 48 hours after tamoxifen induction. Arrows indicates tdTomato expression in the intervertebral disk primodia. (c, d) Ventral views of forelimbs from E12.5 and E14.5 Pax9-CreER;tdTomato mice 48 hours after tamoxifen induction in whole mounts (c, d) and sections (c’, d’). ** and * in (c) and (c’) indicate strong and weaker expression region of tdTomato in the limb bud. ** and * in (d) and (d’) indicate expression close to the phalangeal bone primodia and in the pawpad region. Boxed areas in (c) and (d) are shown enlarged in (c’) and (d’), respectively. Thin white line outlines the forelimb. Scale bars, 200μm.
FIG. 6.
tdTomato expression in the tongue of Pax9-CreER;tdTomato mice induced in E14.5 embryos and 4-week old adults. (a, b) Visualization of coronal sections of tongues of Pax9-CreER;tdTomato mice 48 hours after tamoxifen induction at E14.5. Arrows indicate tdTomato expression in the tongue epithelium. Asterisks indicate tdTomato expressing mesenchymal cells on the lateral side of the tongue. Panel (b) is a magnified view from (a). (c,d) Visualization of whole mount (c) and coronal sections (d) of tongues from adult Pax9-CreER;tdTomato mice 48 hours after tamoxifen induction. fp, filiform papilla; ip, interpapillary region. Scale bars,100μm.
Taken together, the expression of tdTomato in Pax9-CreER;tdTomato reporter mice recapitulated the previously reported expression pattern of endogenous Pax9. Therefore, our Pax9-CreER knock-in mice can serve as a useful model for lineage tracing and genetic targeting of the craniofacial and limb mesenchyme, tongue epithelium and pharyngeal pouch derivatives during embryonic development and in adult life, and will be available to the research community after appropriate arrangements are made.
METHODS
Single guided RNA (sgRNA) and donor vector construct
Pax9 sgRNA GTGCAGAAGCGGTCACAGAATGG was designed to target the 3' end of Pax9, optimized for T7E1 assay efficiency and against off-targets (PNABio). pCAG-CreERT2 plasmids were obtained from Addgene (Plasmid #14797) for donor vector construction (Matsuda and Cepko, 2007). An E2A peptide cleavage site was inserted in front of the CreERT2 coding sequence, which was flanked by a left homology arm containing 500bp of the last intron and exon of Pax9 and right homology arm containing 500bp of 3' UTR of Pax9 (PNABio).
One-cell stage embryo injection
All animal procedures were performed according to NIH guidelines and approved by the University of Southern California Institutional Animal Care and Use Committee (IACUC). B6D2F1 (C57BL/6×DBA2) and CD-1 mouse strains were used as embryo donors and foster mothers, respectively. Super-ovulated female B6D2F1 mice (8 weeks old) were mated to B6D2F1 stud males, and fertilized embryos were collected from oviducts. After 100–200 ng/μl Cas9 mRNA (L-6125, TriLink Biotechnologies) and 50–100 ng/μl sgRNA were mixed on ice for 10 minutes, 5–10 ng/μl purified Pax9 donor DNA was added. The mixture was then injected into the cytoplasm and pronucleus of fertilized eggs with well-recognized pronuclei in M2 medium (M2112, Cytospreen). Injected eggs were cultured in M16 medium (M6111, Cytospreen) at 37 °C under 5% CO2 overnight. Approximately 20–25 two-cell stage embryos were transferred into oviducts of the pseudopregnant CD-1 females at 0.5dpc.
Genotyping of the Pax9-CreER founder mouse
The mutant allele of the Pax9-CreER knock-in mouse was confirmed by genotyping PCR products that span from the upstream region of the left homology arm of the Pax9 gene to the CreER insert in the donor construct. We used two sets of primers to detect the mutant allele independently. Primer set 1 (forward TCTGCCTTTCTCTCCTGGAA and reverse ATTCTCCCACCGTCAGTACG) generates a 1373bp PCR product for the mutant allele, and primer set 2 (forward CAGTGCTGAGCAGAATTCCA and reverse AGGCAAATTTTGGTGTACGG) generates PCR products of 722bp. Both PCR primer pairs identified the same one founder mouse containing the mutant allele from 24 littermates.
Whole-genome sequencing of the Pax9-CreER founder mouse
We performed whole-genome sequencing using genomic DNA from the founder Pax9-CreER knock-in mouse (Fulgent Diagnostics). The sequenced reads were aligned to the mouse reference genome mm9 downloaded from the UCSC database (http://hgdownload.cse.ucsc.edu/goldenPath/mm9/chromosomes/). The reference sequence reads in Chromosome 12 of mm9 were replaced in silico with an artificial Chromosome 12 sequence containing the Pax9 donor construct. The modified mm9 genome was indexed using the Burrows-Wheeler Aligner (BWA) BWA Index command to prepare it for use with the BWA aligner. Alignment was performed using the BWA software using the BWA-Mem algorithm. The fastq file containing the sequenced reads was aligned against the modified mm9 genome and a .bam file was generated. Duplicate reads in the .bam file were marked using Samblaster software and then sorted and indexed using Samtools software. To view the aligned .bam file in the Integrative Genomics Viewer (IGV), a genome file was created using the modified mm9 reference genome fasta file with the artificial Chromosome 12 within the IGV software using the Create.genome File tool. The genome was loaded into IGV followed by the aligned .bam file and the reads aligning to the Pax9 donor construct were analyzed.
Genetic labeling of Pax9-CreER knock-in mice
The Pax9-CreER founder mouse (F0) was mated with Bl6 females to generate F1 Pax9-CreER mice. ROSA26LoxP−STOP−LoxP−tdTomato (tdTomato reporter) mice were obtained from the Jackson Laboratory (JAX no. 007905) (Madisen et al., 2010). tdTomato reporter female mice were mated with Pax9-CreER F1 male mice and received 1.5mg/10g bodyweight tamoxifen (Sigma T5648) at E8.5 through E14.5. The female mice were euthanized 48 hours after injection to collect Pax9-CreER;tdTomato embryos. Adult Pax9-CreER;tdTomato mice received 5 daily injections of 1.5 mg/10 g bodyweight tamoxifen and were euthanized 48 hours after the last injection. Whole samples were imaged using a Leica MZ10F. For cryosections, samples at various stages were fixed in 4% paraformaldehyde, passed through a sucrose series, then embedded in optimal cutting temperature (OCT) compound (Tissue-Tek, Sakura) and frozen onto a dry ice block to solidify. Embedded samples were cryosectioned at 7μm thickness using a cryostat (Leica CM1850). Sections were counterstained with DAPI (Sigma, D9542). Images were captured using a fluorescence microscope (Leica DMI 3000B) with filter settings for DAPI/FITC/TRITC.
Acknowledgments
We are grateful to Dr. Julie Mayo and Bridget Samuels for critical reading and editing of the manuscript. This work was supported by the National Institute of Dental and Craniofacial Research, National Institutes of Health (DE022503; DE012711 and DE025221 to Y.C.).
LITERATURE CITED
- Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre Recombinase Activity by Mutated Estrogen Receptor Ligand-Binding Domains. Biochemical and Biophysical Research Communications. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- Gerber J-K, Richter T, Kremmer E, Adamski J, Höfler H, Balling R, Peters H. Progressive loss of PAX9 expression correlates with increasing malignancy of dysplastic and cancerous epithelium of the human oesophagus. The Journal of Pathology. 2002;197:293–297. doi: 10.1002/path.1115. [DOI] [PubMed] [Google Scholar]
- Jonker L, Kist R, Aw A, Wappler I, Peters H. Pax9 is required for filiform papilla development and suppresses skin-specific differentiation of the mammalian tongue epithelium. Mechanisms of Development. 2004;121:1313–1322. doi: 10.1016/j.mod.2004.07.002. [DOI] [PubMed] [Google Scholar]
- Kist R, Greally E, Peters H. Derivation of a mouse model for conditional inactivation of Pax9. genesis. 2007;45:460–464. doi: 10.1002/dvg.20295. [DOI] [PubMed] [Google Scholar]
- Lee J-C, Sharma M, Lee Y-H, Lee N-H, Kim S-Y, Yun J-S, Nam S-Y, Hwang P-H, Jhee E-C, Yi H-K. Pax9 mediated cell survival in oral squamous carcinoma cell enhanced by c-myb. Cell Biochemistry and Function. 2008;26:892–899. doi: 10.1002/cbf.1522. [DOI] [PubMed] [Google Scholar]
- Madisen L, Zwingman TA, Sunkin SM, Oh SW, Zariwala HA, Gu H, Ng LL, Palmiter RD, Hawrylycz MJ, Jones AR, Lein ES, Zeng H. A robust and high-throughput Cre reporting and characterization system for the whole mouse brain. Nat Neurosci. 2010;13:133–140. doi: 10.1038/nn.2467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda T, Cepko CL. Controlled expression of transgenes introduced by in vivo electroporation. Proceedings of the National Academy of Sciences. 2007;104:1027–1032. doi: 10.1073/pnas.0610155104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubüser A, Koseki H, Balling R. Characterization and Developmental Expression of Pax9, a Paired-Box-Containing Gene Related to Pax1. Developmental Biology. 1995;170:701–716. doi: 10.1006/dbio.1995.1248. [DOI] [PubMed] [Google Scholar]
- Neubüser A, Peters H, Balling R, Martin GR. Antagonistic Interactions between FGF and BMP Signaling Pathways: A Mechanism for Positioning the Sites of Tooth Formation. Cell. 1997;90:247–255. doi: 10.1016/s0092-8674(00)80333-5. [DOI] [PubMed] [Google Scholar]
- Peters H, Neubüser A, Kratochwil K, Balling R. Pax9-deficient mice lack pharyngeal pouch derivatives and teeth and exhibit craniofacial and limb abnormalities. Genes & Development. 1998;12:2735–2747. doi: 10.1101/gad.12.17.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peters H, Schuster G, Neubüser A, Richter T, Höfler H, Balling R. Isolation of the PAX9 cDNA from adult human esophagus. Mammalian Genome. 1997;8:62–64. doi: 10.1007/s003359900351. [DOI] [PubMed] [Google Scholar]
- Wang H, Yang H, Shivalila Chikdu S, Dawlaty Meelad M, Cheng Albert W, Zhang F, Jaenisch R. One-Step Generation of Mice Carrying Mutations in Multiple Genes by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;153:910–918. doi: 10.1016/j.cell.2013.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H, Wang H, Shivalila Chikdu S, Cheng Albert W, Shi L, Jaenisch R. One-Step Generation of Mice Carrying Reporter and Conditional Alleles by CRISPR/Cas-Mediated Genome Engineering. Cell. 2013;154:1370–1379. doi: 10.1016/j.cell.2013.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]