Abstract
Some genes involved in complex human diseases are particularly vulnerable to genetic variations such as single nucleotide polymorphism, copy number variations, and mutations. For example, Ras mutations account for over 30% of all human cancers. Additionally, there are some genes that can display different variations with functional impact in different diseases that are unrelated. One such gene stands out: δ-catenin/NPRAP/Neurojungin with gene designation as CTNND2 on chromosome 5p15.2. Recent advances in genome wide association as well as molecular biology approaches have uncovered striking involvement of δ-catenin gene variations linked to complex human disorders. These disorders include cancer, bipolar disorder, schizophrenia, autism, Cri-du-chat syndrome, myopia, cortical cataract-linked Alzheimer’s disease, and infectious diseases. This list has rapidly grown longer in recent years, underscoring the pivotal roles of δ-catenin in critical human diseases. δ-Catenin is an adhesive junction-associated protein in the delta subfamily of the β-catenin superfamily. δ-Catenin functions in Wnt signaling to regulate gene expression and modulate Rho GTPases of the Ras superfamily in cytoskeletal reorganization. δ-Catenin likely lies where Wnt signaling meets Rho GTPases and is a unique and vulnerable common target for mutagenesis in different human diseases.
Keywords: Genetic variations, mutations, cancer, neurological disorders, Rho GTPases, Wnt signaling
Introduction
The advancement in sequencing technology in the last decade has allowed the identification of a large amount of gene variations in the genomes of the animal kingdom. These efforts have produced the whole genome data from yeast to C. elegans, Drosophila, zebrafish, mouse, and human. These efforts are also slated to provide holistic information about genomic landscapes of critical human diseases such as cancer. Next generation sequencing (NGS) not only confirmed many gene variations in human diseases, such as Ras mutations in cancer, but also helped identify many disease-related gene variants that were not known before (Vogelstein et al. 2013). Many such discovered gene variations, like copy number variations (CNV) and single nucleotide polymorphism (SNP), may or may not be so obviously associated with the pathological mechanisms of diseases. However, a growing number of genes have turned out to be critically involved in human disorders. For example, δ-Catenin/NPRAP/Neurojungin with gene designation as CTNND2 on chromosome 5p15.2, is increasingly recognized as one such important gene. Especially, as it will become clear from this review, δ-catenin alterations are unlike many other genes, such as Ras mutations that are recurring and enriched in cancer. Functional δ-catenin gene alterations that do not necessarily recur at the same nucleotide sides are now reported in a number of high impact studies. They can be linked to different diseases that are unrelated, underscoring the unique and pivotal roles of δ-catenin in human physiology.
Discovery of δ-catenin
1. Nomenclature of catenin of the delta subfamily
δ-Catenin sequences were initially discovered as neural specific. It was identified in Germany as NPRAP for neural protein related to armadillo protein in a screening of brain cDNA library based on its homology to armadillo domain containing proteins (Paffenholz and Franke 1997). At about the same time, its partial sequence was cloned in the USA in a two-hybrid system of searching for protein interactions with the Alzheimer’s disease (AD) gene presenilin 1 and was named as δ-catenin since it shares armadillo repeating sequence homology to β-catenin (Zhou et al. 1997). It was later found that the gene designation for catenin of the delta subfamily already existed for p120ctn (Reynolds et al. 1992), which has been previously reviewed (Lu 2010). Therefore, this subfamily now has two members: CTNND1 for p120ctn and CTNND2 for δ-catenin/NPRAP. In 2000, NPRAP was renamed to neurojungin in a study that showed δ-catenin expression and localization in the outer-limiting membrane of the retina (Paffenholz et al. 1999). Nevertheless, the field has widely adopted the name δ-catenin. While p120ctn is ubiquitously expressed in mammalian tissues, δ-catenin is neurally enriched in the central nervous system and the retina (Lu et al. 1999; Ho et al. 2000).
The p120ctn protein was identified in a screen for substrates of the Src tyrosine kinase whose tyrosine phosphorylation correlated with cellular transformation (Reynolds et al. 1989). The various functions of p120ctn and its role in cell adhesion and cancer have been reviewed (Reynolds and Roczniak-Ferguson 2004; Reynolds 2007). We have also reviewed the interactions of p120ctn and δ-Catenin with cadherins (Lu 2010).
δ-Catenin expression begins during early embryogenesis. In mouse brain development, δ-catenin is localized to the ventricular zone of the cortex at embryonic day 9~10 (Ho et al. 2000). δ-Catenin expression decreases as neurons migrate to the cortical plate during embryogenesis and then its expression increases again and reaches the highest level at postnatal day 7. δ-Catenin expression becomes highly enriched at postsynaptic density (PSD) when neurons fully mature in the adult brain. Besides its enrichment in the dendritic spines in association with the synaptic junction, δ-catenin maintains expression at the ependymal apical junction of the stem cell layer in the adult brain (Lu et al. 1999). However, it is unclear whether and how δ-catenin may be involved in stem cell activity.
2. The functions of δ-catenin
We now know that δ-catenin interacts with classical cadherins, the calcium regulated cell-cell junction proteins (Lu et al. 1999; Ho et al. 2000). In neurons, δ-catenin interacts with N-cadherin, glutamate receptors at PSD, and PSD95, as well as a whole array of other synaptic junction associated proteins (Lu et al. 1999; Ide et al. 1999; Jones et al. 2002; Silverman et al. 2007; Brigidi et al. 2014; Yuan et al. 2015). Based on all the known protein binding partners for δ-catenin, it strongly suggests that δ-catenin plays important roles in brain functions that involve synaptic regulation, especially learning and emotions according to a gene deletion study (Israely et al. 2004). Indeed, δ-catenin knockdown regulates spine architecture related to cadherin and PDZ-dependent interactions (Yuan et al. 2015). A gene knockout (KO) study revealed that δ-catenin is required for the maintenance of neural structure and function in mature cortex in vivo (Matter et al. 2009). Another study that employed the same KO strain showed that δ-catenin regulates spine and synapse morphogenesis and function in hippocampal neurons during development (Arikkath et al. 2009).
When overexpressed in the epithelial cells, δ-catenin is localized to the cell-cell junction and interacts with E-cadherin. Expression of δ-catenin caused the cell monolayer to rise and cells to extend processes in a morphogenetic movement. When treated with hepatocyte growth factor (scattering factor), MDCK cells overexpressing δ-catenin promoted the cell scattering and the disintegration of the adherens junction (Lu et al. 1999).
When ectopically expressed in the otherwise δ-catenin-free fibroblast, δ-catenin induced the exuberant formation of branched cellular processes reminiscent of inhibition of small GTPase RhoA (Kim et al. 2002). In fact, fibroblastic cells expressing δ-catenin are morphologically indistinguishable from neurons. However, these cells do not express neural specific genes such as tau and microtubule associated protein 2 (MAP2). Therefore, the effect of δ-catenin-induced neuron-like phenotype is perhaps limited to the morphological properties and the architectural framework of neurons. Additional genes are required in order to allow stem cells to become fully differentiated into neurons.
Genetic Alterations of δ-Catenin: Functional Implications in Human Diseases
1. δ-Catenin gene alterations in neurocognitive disorders
a. Alzheimer’s disease
Smoking, estrogen supplements, hypertension, depression, stroke, heart disease, arthritis, and diabetes are risk factors for AD but some may also be early signs of the disease (Ridge et al. 2013). Although aging is the most evident risk (Herrup 2010), several genetic risk factors have been identified to contribute to AD (Ridge et al. 2013; Guerreiro and Hardy 2014). Interestingly, cortical cataract and δ-catenin may be linked to AD. A recent study that monitored brain damage and lens opacity within the same individual and between siblings found that cortical cataract may predict the development of several AD-related brain changes and that siblings of patients are at greater risk (Jun et al. 2012). They further reported that the δ-catenin SNP rs17183619 displayed significant association with amyloid precursor protein (APP) and a rare missense δ-catenin mutation (G810R) alters δ-catenin localization and increases secreted Aβ42 in HEK293 cells (Jun et al. 2012). Lastly, lens tissue from AD subjects showed increased δ-catenin expression compared to control, which authors proposed formed light scattering deposits that contribute to decreased lens opacity (Jun et al. 2012).
b. Cri-du-chat syndrome
Recent studies reported that the partial deletion of δ-catenin in 5p15.2 and partial duplication of its promoter region led to a mild case of Cri-du-chat syndrome (Sardina et al. 2014). On the other hand, the hemizygous loss of δ-catenin is associated with severe mental retardation in Cri-du-chat children (Medina et al. 2000). In this study, it was found that there are two critical regions for Cri-du-chat. Semaphorin F is located in the critical region 1 whereas δ-catenin is located in the critical region 2. When the deletion included semaphorin F, mental retardation was not manifested. However, when the deletion penetrated δ-catenin to remove the carboxyl terminal PDZ binding domain, the mental retardation association became more evident (Medina et al. 2000). Since Cri-du-chat is a genetic disorder that results from the variable deletion of chromosome 5p, other genes in addition to δ-catenin may also contribute to the disease when they are deleted.
There has been an attempt to elucidate the physiological changes of δ-catenin in Cri-du-chat. A mouse model showed that the deletion of δ-catenin impeded cognitive functions and significantly reduced N-cadherin and PSD95 (Israely et al. 2004). This is in line with a previous report that showed that δ-catenin regulated the maintenance of dendrites and dendritic spines in mature cortex of the mouse but did not appear to be necessary for the initial establishment of these structures during development (Matter et al. 2009).
2. δ-Catenin gene variations in other neurological disorders
a. Autism spectrum disorder
Autism spectrum disorder (ASD) is a multifactorial neurodevelopmental disorder that affects social and behavioral traits (Schieve et al. 2012). The genetics of ASD has been reviewed elsewhere (Yoo 2015). The hunt for ASD candidate genes has led to a pair of Nature publications that identified chromosome 5p as a significant factor in a genome wide association study (GWAS) (Weiss et al. 2009) and the association of δ-catenin with potential critical roles in severe ASD (Turner et al. 2015). In female-enriched multiplex families, the authors found that δ-catenin harbored significantly more deleterious missense and CNV in autistic patients (Turner et al. 2015). Five (G34S, G275C, Q507P, R713C, T862M) out of seven mutations were validated in ASD patients and conserved in zebrafish. δ-Catenin CNV was enriched in ASD patients when compared to control (odds ratio = 5.9, P = 4.10 × 10−4 (Turner et al. 2015). What is significant is that these variants, by functional testing, are loss-of-function and affect Wnt signaling. The expression of δ-catenin is highly correlated with other autism genes further implicating its potential roles in autistic etiology. Besides the well-established roles of δ-catenin in dendritic morphogenesis and maintenance, a novel and interesting finding of this study is that δ-catenin correlated genes are enriched for chromatin and histone modification (Turner et al. 2015).
b. Schizophrenia
Rare CNVs may play a significant role in schizophrenia (Sklar et al. 2008; Stefansson et al. 2008). δ-Catenin was one of thirteen genes affected by rare CNVs found in patients. The δ-catenin CNV in a female patient resulted in a duplication of several genes on 5p15.2 with a breakpoint in δ-catenin (Vrijenhoek et al. 2008). SNPs also play a role in schizophrenia (Sklar et al. 2008; Stefansson et al. 2008). Analysis of GWAS from the Psychiatric Genomics Consortium revealed several SNPs in δ-catenin correlated to major depressive disorder and schizophrenia. The δ-catenin SNP rs1012176 (P= 0.0009997) showed the strongest association with anxiety disorders, rs10059890 (P= 0.0002496) for major depressive disorder, and s4524507 (P= 0.0005537) showed the strongest for schizophrenia (Nivard et al. 2014).
c. Intellectual disturbance
Abnormalities in δ-catenin also contribute to intellectual disability that is frequently associated with the aforementioned neurologic disorders. A breakpoint to intron 9 of δ-catenin on chromosome 5 was identified in a mother and daughter with borderline intelligence and dyslexia-related learning problems (Hofmeister et al. 2015). Further, a microdeletion of δ-catenin observed in a boy presented apparent learning disabilities and dyslexia (Hofmeister et al. 2015). Additionally, CNVs leading to deletion of δ-catenin was detected in two patients with mild intellectual disability (Belcaro et al. 2015). Thus, these studies appear to further support that the decrease or loss of δ-catenin impairs cognitive functions.
d. Myopia
Interestingly, δ-catenin is located in a myopia susceptible locus highlighting its potential role in myopia (Lam et al. 2008). Together with the 11q21.1 genomic region, the δ-catenin SNP rs1479617 significantly differed between the pathological myopia and control groups of Chinese patients (Yu et al. 2012). A meta-analysis revealed that SNPs of δ-catenin (rs12716080 and rs6885224) showed a strong correlation with high myopia in both Chinese and Japanese patients (Liu and Zhang 2014). Alternatively, an independent case-control series study identified that the δ-catenin SNP rs6885224 displayed protective properties when compared in Chinese patients with high or moderate myopia with controls (Lu et al. 2011). Another case-control study with Jiangsu Chinese did not associate SNPs of δ-catenin with high myopia (Wang et al. 2014). Therefore, it remains to be seen whether genetic variations in δ-catenin plays a functional role in myopia. A summary of reported genetic modifications in various neurodegenerative and neurodevelopmental disorders is shown in Table 1.
Table 1.
Summary of reported genetic modifications in various neurodegenerative and neurodevelopmental disorders.
| Modification | Description | Ref |
|---|---|---|
| Alzheimer’s Disease | ||
| rs17183619 | GWAS-Statistical interaction with APP SNP rs2096488 | (Jun et al. 2012) |
| rs61754599 | GWAS-Missense mutation (G810R) altered δ-catenin localization and increased secreted AB1-42 in HEK293 cells | (Jun et al. 2012) |
| Cri-du-chat | ||
| CNV | Case study: eight-year-old African-American female- Promoter region and alternative exons 1–14 are duplicated and exons 15–68 of δ-catenin are deleted. | (Sardina et al. 2014) |
| Hemizygous deletion | Case studies: correlation analysis from three previous reports- Hemizygous deletion within 5p15.2pter leads to severe mental retardation | (Medina et al. 2000) |
| Deletion (knockout) | δ-cat−/− mice were viable but showed decline in cognitive functions and significantly reduced N-cadherin and PSD95 protein level. | (Israely et al. 2004) |
| Autism | ||
| G34S, P189L, P224L, R454H, Q507P | Case study: female-enriched multiplex families- Hypomorphic mutations found in patients and conserved in zebrafish embryos model | (Turner et al. 2015) |
| CNV | Case studies: reexamination of previously reported cases and literature review- Ten deletions and two duplications, seven overlapping one or more exons. Deletions leading to loss-of-function suggest happloinsufficiency. | (Turner et al. 2015) |
| Schizophrenia | ||
| CNV | Case studies: patients from the Netherlands- CNV of seven genes in 5p15.2 with a breakpoint disrupting the terminal portion of CTNND2 gene in a female patient. | (Vrijenhoek et al. 2008) |
| rs6873547 | Case study: patients from the Netherlands- CTNND2 SNP with the strongest association with Schizophrenia. Top ten SNPs for anxiety disorder, major depressive disorder, and bipolar disorder were also reported. | (Nivard et al. 2014) |
| Intellectual Disability | ||
| Translocation breakpoint | Case study: mother and daughter- balanced translocation of t(1;8)(p22;q24) and t(5;18)(p15;q11) resulted in problems with learning (dyslexia, attention deficit, speech delay). The breakpoint was located in intron 9 of the δ-catenin resulting in a loss-of-function. | (Hofmeister et al. 2015) |
| Deletion of exons 12–18 | Case study: fourteen-year-old male- Presented similar phenotype with more pronounced learning disabilities and dyslexia. | (Hofmeister et al. 2015) |
| CNV | Case study: two males with delayed developmental milestones- intragenic deletion leads to mild intellectual disability. | (Belcaro et al. 2015) |
| Myopia | ||
| rs1479617 | Case study: patients from China- This SNP and 11q21.1 genomic region significantly differed between the pathological myopia and control groups of Chinese patients. | (Yu et al. 2012) |
| rs12716080 rs6885224 |
Meta-analysis - Chinese and Japanese patients revealed a strong correlation with high myopia. | (Liu and Zhang 2014) |
| rs6885224 | Case-control series study- displayed protective properties when compared in Chinese patients with high or moderate myopia with controls. | (Lu et al. 2011) |
3. δ-Catenin mutations in cancer
a. Sanger database
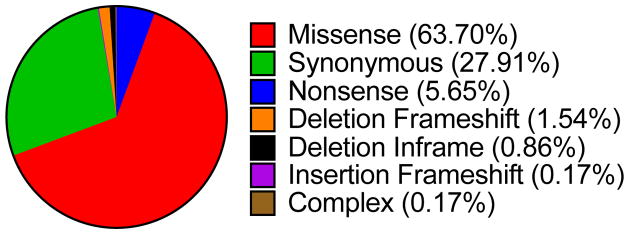
At the time of this review, the Sanger COSMIC (Catalogue of Somatic Mutations in Cancer) database reported 541 unique mutations for δ-catenin from 25124 samples in 38 different tissues (Table 2). We have previously reviewed the role of δ-catenin in cancer and indicated that overexpression is often observed in cancers (Lu 2010). To date, only cancers of the central nervous system (1.29%) are reported to display reduced expression of δ-catenin. Point mutations are more commonly observed in skin (9.22%), large intestine (7.31%), stomach (6.93%), and lung (6.47%) cancers. Missense mutations are the most common (66.55%) followed by synonymous substitutions (29.16%, Fig. 1). CNV for δ-catenin was reported for several cancers such as cancers of the cervix (Huang et al. 2006) and bladder (Zheng et al. 2004). CNV leading to a gain is more common in cancers. These cancers include esophagus (9.26%), urinary tract (8%), soft tissue (7.75%), cervix (7.47%), and upper aero-digestive tract (5.77%). Lastly, δ-catenin is overexpressed in several cancers such as prostate (10.84%), liver (9.65%), breast (6.52%), ovary (6.02%), endometrium (5.56%), and lung (5.50%). Thus, δ-catenin may presents itself as a potential target for cancers, especially for lung cancer and prostate cancer that showed an increased incidence of point mutations and gene expression.
Table 2.
Summary of reported point mutations and copy number variations in a variety of cancers. Percentages and totals are taken from COSMIC (accessed March 2016).
| Tissue | Point Mutations | Copy Number Variations | Gene Expression | Methylation | |||||
|---|---|---|---|---|---|---|---|---|---|
| mutated | total | gain | loss | total | +over -under |
total | +hyper -hypo |
total | |
| Adrenal gland | 0.42 | 240 | 11.11 | - | 72 | +3.8 | 79 | - | - |
| Autonomic ganglia | 0.27 | 748 | - | - | - | - | - | - | - |
| Biliary tract | 1.82 | 329 | - | - | - | - | - | - | - |
| Bone | 0.20 | 509 | - | - | - | - | - | - | - |
| Breast | 0.71 | 1406 | 2.51 | 0.20 | 997 | +6.52 | 1104 | - | - |
| Central nervous system | 0.46 | 2165 | 0.49 | - | 811 | +4.32 −1.29 |
695 | - | - |
| Cervix | 0.62 | 322 | 7.47 | - | 174 | +4.23 | 307 | - | - |
| Endometrium | 3.13 | 640 | - | - | 424 | +5.65 | 602 | +0.75 | 398 |
| Eye | 2.56 | 39 | - | - | - | - | - | - | - |
| Fallopian tube | - | 2 | - | - | - | - | - | - | - |
| Genital tract | - | 28 | - | - | - | - | - | - | - |
| Hematopoietic/lymphoid | 0.35 | 2820 | - | - | - | +1.81 | 221 | - | - |
| Kidney | 0.45 | 1571 | 0.72 | 0.24 | 418 | +2.33 | 600 | −4.37 | 206 |
| Large intestine | 7.31 | 1519 | 0.43 | 1.14 | 699 | +2.47 | 607 | +0.36 −10.32 |
281 |
| Liver | 1.78 | 1627 | 0.16 | - | 644 | +9.65 | 373 | −7.38 | 244 |
| Lung | 6.47 | 1838 | 13.13 | 0.09 | 1112 | +5.50 | 1018 | −18.37 | 294 |
| Meninges | - | 65 | - | - | - | - | - | - | - |
| NS | 5.56 | 54 | - | - | - | - | - | - | - |
| Esophagus | 3.23 | 929 | 9.26 | 0.93 | 108 | +2.40 | 125 | - | - |
| Ovary | 1.20 | 831 | 3.61 | - | 721 | +6.02 | 266 | - | - |
| Pancreas | 0.78 | 1539 | 0.28 | 0.14 | 706 | +4.47 | 179 | - | - |
| Parathyroid | - | 25 | - | - | - | - | - | - | - |
| Peritoneum | - | 10 | - | - | - | - | - | - | - |
| Pituitary | - | 15 | - | - | - | - | - | - | - |
| Placenta | - | 2 | - | - | - | - | - | - | - |
| Pleura | - | 77 | - | - | - | - | - | - | - |
| Prostate | 0.17 | 1145 | - | - | - | +10.84 | 498 | - | - |
| Salivary gland | 1.59 | 63 | - | - | - | - | - | - | - |
| Skin | 9.22 | 1074 | 1.86 | 0.41 | 484 | +2.12 | 472 | - | - |
| Small intestine | - | 40 | - | - | - | - | - | - | - |
| Soft tissue | 0.66 | 456 | 7.75 | 0.70 | 142 | +0.76 | 263 | - | - |
| Stomach | 7.09 | 592 | 3.68 | - | 353 | +4.56 | 285 | - | - |
| Testis | - | 20 | - | - | - | - | - | - | - |
| Thymus | - | 28 | - | - | - | - | - | - | - |
| Thyroid | 0.88 | 568 | - | - | - | +4.48 | 513 | - | 510 |
| Upper aero-digestive tract | 1.25 | 1117 | 5.77 | 0.21 | 468 | +4.41 | 522 | +0.20 −9.27 |
496 |
| Urinary tract | 1.50 | 667 | 8.00 | 0.89 | 225 | +4.41 | 408 | −19.41 | 273 |
| Vulva | - | 3 | - | - | - | - | - | - | - |
Figure 1. Distribution of CTNND2 mutations in cancer.
Taken from COSMIC (accessed March 2016).
b. Non-coding region
δ-Catenin is overexpressed in roughly 11% of prostate cancers (Table 2) and has been described as a potential diagnostic biomarker (Burger et al. 2002; Lu et al., 2005; Lu et al., 2009). Thus, amplified δ-catenin translation resulting in increased δ-catenin protein expression may play an important role in prostate cancer, highlighting the significance of processes that control translation. Although hypomethylation is more common than hypermethlation (Table 2) in some cancers (endometrium, kidney, large intestine, liver, lung, upper aerodigestive tract, and urinary tract), δ-catenin methylation did not appear to regulate protein synthesis in prostate cancer. Methylation was not observed in normal prostate (PZ-HPV-7) with low δ-catenin expression or prostate cancer (CWR22Rv-1) with high δ-catenin expression (Wang et al. 2009). Further, demethylation of δ-catenin following 5′Aza-DC treatment had no effect on δ-catenin mRNA levels. Hence, other regulatory factors may be playing a more prominent role. In fact, a point mutation (-9 G>A) was identified in the 5′-untranslated region of δ-catenin that promoted translation (Wang et al. 2009). This mutation was found only in prostate tumor tissues and not in benign adjacent prostate tissue, and is associated with a high Gleason’s score.
c. Coding region
The Sanger COSMIC database recorded several mutations in the coding region of δ-catenin. More recently, gene variations were observed after sequencing and analyzing the entire coding region of δ-catenin from a total of 24 out of 40 human prostatic adenocarcinomas (Nopparat et al. 2015). Most functional mutations occurred after exon 9 while missense or frameshifting mutations occurred after exon 13. Control tissue showed little or no missense or frameshifting mutations. In this study, δ-catenin was ectopically overexpressed into two prostate cancer cell lines. Surprisingly, the stable cell lines produced truncated δ-catenin resulting from a frameshift mutation (p.fs, c. 2640insA) that led to premature termination. CWR22-Rv1 prostate cancer cells expressing truncated δ-catenin increased cell number beyond full confluency, highlighting a potential role in tumor promotion. Indeed, mice with δ-catenin gene disrupted in exon 9 and Myc oncogene conditionally expressed in the prostate showed increased tumor size and progression from prostatic intraepithelial neoplasia (PIN) to adenocarcinomas when compared to Myc expression alone. In addition, CWR22-Rv1 cells with truncated δ-catenin tolerated glucose starvation. Altogether, these findings highlight the ability of cancer cells to alter genes resulting in increased cancer cell survival and/or transformation along with modifying cellular metabolism (Nopparat et al. 2015).
4. δ-Catenin in malaria resistance
While the above literature showed that δ-catenin mostly contributed to diseases, in an environmental correlation analysis that examined genotype data from 12,425 newborns in coastal Kenya, δ-catenin (P=0.003) was one of the top 10% of genes identified to contribute to malaria resistance (Mackinnon et al. 2016). Furthermore, other genes (MAGI2, FN1, NRP2, NRXN3, RGS6, ADGRL2) associated with cadherin-mediated adhesion and signaling at cell-cell junctions in the brain and/or vascular endothelium were among the top 1% of loci. These results suggest the activation of these pathways and/or previously reported pathways, CDH13 (Band et al. 2013) and HS3ST3B1 (Atkinson et al. 2012) may protect against death from malaria.
δ-Catenin at crossroad: Wnt signaling and Rho GTPase modulation
With the wealth of genetic information, we can begin to speculate on the mechanisms of δ-catenin functions in human diseases. For example, δ-catenin is amplified in esophageal carcinoma cell lines that show increased Wnt signaling (Brown et al. 2011). δ-Catenin mutations alter protein expression and distribution in the Wnt/β-catenin and hypoxia pathways (Nopparat et al. 2015). Radiation hybrid mapping of genes in the lithium-sensitive Wnt signaling pathway included Wnt, frizzled, disheveled, GSK-3α/β, axin, α-catenin, δ-catenin and ARVCF, and a frizzled-like protein (frpHE) on human chromosomes (Rhoads et al. 1999).
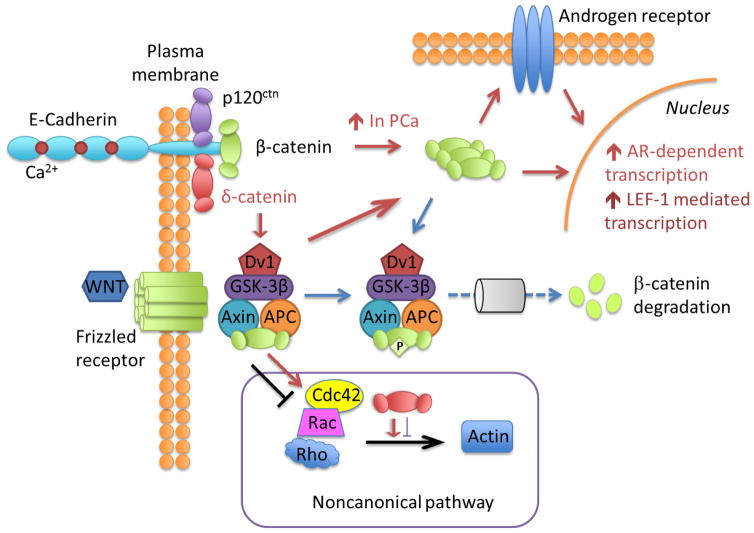
Recently, the finding that δ-catenin interacts with cadherin suggests its mechanistic context in Wnt/β-catenin signaling (Fig. 2). β-Catenin binds to E-cadherin at cell-cell junction but Wnt signaling leads to its translocation to the nucleus. GSK-3α/β kinase phosphorylates δ-catenin and negatively regulates its stability via ubiquitination/proteosome-mediated proteolysis (Oh et al. 2009). This finding is confirmed in the neurons because δ-catenin is in the GSK-3α/β signaling complex that promotes β-catenin turnover in neurons (Bareiss et al. 2010). In prostate cancer, δ-catenin promotes E-cadherin processing and activates β-catenin-mediated signaling. Overexpressing δ-catenin resulted in an elevated total β-catenin level and increased its nuclear distribution, leading to the activation of β-catenin/LEF-1-mediated transcription and their downstream target genes as well as androgen receptor-mediated transcription (Kim et al. 2012). Further study showed that C-Src-mediated phosphorylation of δ-catenin increases its protein stability and the ability of inducing nuclear distribution of β-catenin (He et al. 2014).
Figure 2. Summary of Wnt signaling involving δ-catenin and Rho GTPases.
The signaling of δ-catenin is highlighted in brown arrows. Overexpression of δ-catenin in prostate cancer (PCa) leads to increased β-catenin levels leading to its translocation to the nucleus to promote LEF-1-mediated and androgen receptor (AR) dependent transcription. Wnt-signaling also leads to β-catenin translocation. Wnt-GSK-dependent proteolysis of β-catenin is highlighted in blue. GSK phosphorylates β-catenin that signals ubiquitination/proteasome degradation. The box highlights the noncanonical/Rho GTPase pathway of δ-catenin involvement in Wnt signaling. Rho GTPases are mediators of Wnt signaling (highlighted in black). δ-Catenin can interrupt Rho GTPase signaling thus affecting actin-mediated dendritic and spine morphogenesis.
During the past several years, Rho GTPases have emerged as key mediators of Wnt signals, most notably in the noncanonical pathways that involve polarized cell shape changes and migrations, but also more recently in the canonical pathway leading to β-catenin/LEF-1-dependent transcription (Schlessinger et al. 2009). Rho GTPases integrate Wnt-induced signals spatially and temporally to promote morphological and transcriptional changes affecting cell behavior (Fig. 2). It is known that the morphogenetic effects of δ-catenin is caused by its ability to interfere with Rho family small GTPases. Specifically, δ-catenin interacts with p190RhoGEF, and Akt1-mediated phosphorylation at Thr-454 residue of δ-catenin is important in this interaction. δ-Catenin overexpression decreased the binding between p190RhoGEF and RhoA, and significantly lowered the levels of GTP-RhoA. δ-Catenin T454A significantly reduced the length and number of mature mushroom shaped spines in primary hippocampal neurons. These studies highlight Rho GTPase signaling in the regulation of δ-catenin-induced dendritic and spine morphogenesis in neurons (Kim et al. 2008a).
When HEK293 cells were grown in low cell densities, only 15% of cells overexpressing δ-catenin showed dendrite-like processes with no changes in RhoA activity. δ-Catenin was localized mainly in the cytoplasm and was associated with p190RhoGEF. However, at high cell densities, δ-catenin localization was shifted to the plasma membrane. The association of δ-catenin with E-cadherin was strengthened, whereas its interaction with p190RhoGEF was weakened. These studies support that δ-catenin induces cellular processes through interaction with p190RhoGEF, which is diminished when δ-catenin binds to E-cadherin at the cell-cell junction (Kim et al. 2008b).
Summary
Recent GWAS and NGS technologies have enabled big data to link certain genes to complex human diseases. Many gene alterations are in the introns and non-coding regions. However, δ-catenin/CTNND2 often displays a variety of functional mutations that are linked to unrelated diseases and reported in a number of high profile publications in recent years. As an armadillo repeat containing protein in the β-catenin superfamily, δ-catenin contains multiple domains for extensive protein-protein interactions. It is predicted to be capable of nuclear and cytoplasmic localization as well as lipid modification to interact with the plasma membrane. As important as these domain structures, it is possible that mutations that occur at the different sites can have different consequences in nervous system as compared to cancers in the peripheral tissues. At the molecular level, δ-catenin is at the crossroad with interactions to such signaling pathways of Wnt and Rho family small GTPases in the Ras superfamily, which are known to drive human disease pathogenesis. Understanding the genetic variations in δ-catenin in cancer, cortical cataract-linked Alzheimer’s disease, autism, schizophrenia, mental retardation, myopia, and infectious diseases would provide a wealth of information to guide future functional genomic studies for treating human diseases.
Acknowledgments
Authors wish to acknowledge the funding from US National Institutes of Health CA111891 (Q. Lu), CA165202 (Q. Lu), HL085752 (Y. H. C), ES016888 (Y. H. C), Department of Defense PC040569 (Q. Lu), Alzheimer’s North Carolina (Q. Lu) and The Wooten Foundation.
Footnotes
Conflict of interest statement:
On behalf of all authors, the corresponding author states that there is no conflict of interest.
References
- Arikkath J, Peng I-FF, Ng YG, et al. Delta-catenin regulates spine and synapse morphogenesis and function in hippocampal neurons during development. J Neurosci. 2009;29:5435–42. doi: 10.1523/JNEUROSCI.0835-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson A, Garnier S, Afridi S, et al. Genetic variations in genes involved in heparan sulphate biosynthesis are associated with Plasmodium falciparum parasitaemia: a familial study in Burkina Faso. Malar J. 2012;11:108. doi: 10.1186/1475-2875-11-108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Band G, Le QS, Jostins L, et al. Imputation-based meta-analysis of severe malaria in three African populations. PLoS Genet. 2013;9:e1003509. doi: 10.1371/journal.pgen.1003509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bareiss S, Kim K, Lu Q. Delta-catenin/NPRAP: A new member of the glycogen synthase kinase-3beta signaling complex that promotes beta-catenin turnover in neurons. J Neurosci Res. 2010;88:2350–63. doi: 10.1002/jnr.22414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belcaro C, Dipresa S, Morini G, et al. CTNND2 deletion and intellectual disability. Gene. 2015;565:146–9. doi: 10.1016/j.gene.2015.03.054. [DOI] [PubMed] [Google Scholar]
- Brigidi GS, Sun Y, Beccano-Kelly D, et al. Palmitoylation of δ-catenin by DHHC5 mediates activity-induced synapse plasticity. Nat Neurosci. 2014;17:522–32. doi: 10.1038/nn.3657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown J, Bothma H, Veale R, Willem P. Genomic imbalances in esophageal carcinoma cell lines involve Wnt pathway genes. World J Gastroenterol. 2011;17:2909–23. doi: 10.3748/wjg.v17.i24.2909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burger MJ, Tebay MA, Keith PA, et al. Expression analysis of delta-catenin and prostate-specific membrane antigen: their potential as diagnostic markers for prostate cancer. Int J Cancer. 2002;100:228–37. doi: 10.1002/ijc.10468. [DOI] [PubMed] [Google Scholar]
- Guerreiro R, Hardy J. Genetics of Alzheimer’s disease. Neurotherapeutics. 2014;11:732–7. doi: 10.1007/s13311-014-0295-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y, Kim H, Ryu T, et al. C-Src-mediated phosphorylation of δ-catenin increases its protein stability and the ability of inducing nuclear distribution of β-catenin. Biochim Biophys Acta. 2014;1843:758–68. doi: 10.1016/j.bbamcr.2013.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrup K. Reimagining Alzheimer’s disease--an age-based hypothesis. J Neurosci. 2010;30:16755–62. doi: 10.1523/JNEUROSCI.4521-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ho C, Zhou J, Medina M, et al. delta-catenin is a nervous system-specific adherens junction protein which undergoes dynamic relocalization during development. J Comp Neurol. 2000;420:261–76. [PubMed] [Google Scholar]
- Hofmeister W, Nilsson D, Topa A, et al. CTNND2-a candidate gene for reading problems and mild intellectual disability. J Med Genet. 2015;52:111–22. doi: 10.1136/jmedgenet-2014-102757. [DOI] [PubMed] [Google Scholar]
- Huang FY, Chiu PM, Tam KF, et al. Semi-quantitative fluorescent PCR analysis identifies PRKAA1 on chromosome 5 as a potential candidate cancer gene of cervical cancer. Gynecol Oncol. 2006;103:219–25. doi: 10.1016/j.ygyno.2006.02.028. [DOI] [PubMed] [Google Scholar]
- Ide N, Hata Y, Deguchi M, et al. Interaction of S-SCAM with neural plakophilin-related Armadillo-repeat protein/delta-catenin. Biochem Biophys Res Commun. 1999;256:456–61. doi: 10.1006/bbrc.1999.0364. [DOI] [PubMed] [Google Scholar]
- Israely I, Costa RM, Xie CW, et al. Deletion of the neuron-specific protein delta-catenin leads to severe cognitive and synaptic dysfunction. Curr Biol. 2004;14:1657–63. doi: 10.1016/j.cub.2004.08.065. [DOI] [PubMed] [Google Scholar]
- Jones SB, Lanford GW, Chen Y-HH, et al. Glutamate-induced delta-catenin redistribution and dissociation from postsynaptic receptor complexes. Neuroscience. 2002;115:1009–21. doi: 10.1016/s0306-4522(02)00532-8. [DOI] [PubMed] [Google Scholar]
- Jun G, Moncaster JA, Koutras C, et al. δ-Catenin is genetically and biologically associated with cortical cataract and future Alzheimer-related structural and functional brain changes. PLoS ONE. 2012;7:e43728. doi: 10.1371/journal.pone.0043728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Han J-RR, Park J, et al. Delta-catenin-induced dendritic morphogenesis. An essential role of p190RhoGEF interaction through Akt1-mediated phosphorylation. J Biol Chem. 2008a;283:977–87. doi: 10.1074/jbc.M707158200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, He Y, Yang I, et al. δ-Catenin promotes E-cadherin processing and activates β-catenin-mediated signaling: implications on human prostate cancer progression. Biochim Biophys Acta. 2012;1822:509–21. doi: 10.1016/j.bbadis.2011.12.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Oh M, Lu Q, Kim K. E-Cadherin negatively modulates delta-catenin-induced morphological changes and RhoA activity reduction by competing with p190RhoGEF for delta-catenin. Biochem Biophys Res Commun. 2008b;377:636–41. doi: 10.1016/j.bbrc.2008.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim K, Sirota A, Chen YH, et al. Dendrite-like process formation and cytoskeletal remodeling regulated by delta-catenin expression. Exp Cell Res. 2002;275:171–84. doi: 10.1006/excr.2002.5503. [DOI] [PubMed] [Google Scholar]
- Lam CY, Tam PO, Fan DS, et al. A genome-wide scan maps a novel high myopia locus to 5p15. Invest Ophthalmol Vis Sci. 2008;49:3768–78. doi: 10.1167/iovs.07-1126. [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang HX. Polymorphism in the 11q24.1 genomic region is associated with myopia: a comprehensive genetic study in Chinese and Japanese populations. Mol Vis. 2014;20:352–8. [PMC free article] [PubMed] [Google Scholar]
- Lu B, Jiang D, Wang P, et al. Replication study supports CTNND2 as a susceptibility gene for high myopia. Invest Ophthalmol Vis Sci. 2011;52:8258–61. doi: 10.1167/iovs.11-7914. [DOI] [PubMed] [Google Scholar]
- Lu Q. δ-Catenin dysregulation in cancer: interactions with E-cadherin and beyond. J Pathol. 2010;222:119–23. doi: 10.1002/path.2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Paredes M, Medina M, et al. delta-catenin, an adhesive junction-associated protein which promotes cell scattering. J Cell Biol. 1999;144:519–32. doi: 10.1083/jcb.144.3.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu Q, Dobbs LJ, Gregory CW, Lanford GW, Revelo MP, Shappell S, Chen YH. Increased expression of delta-catenin/neural plakophilin-related armadillo protein is associated with the down-regulation and redistribution of E-cadherin and p120ctn in human prostate cancer. Hum Pathol. 2005;36(10):1037–1048. doi: 10.1016/j.humpath.2005.07.012. [DOI] [PubMed] [Google Scholar]
- Lu Q, Zhang J, Allison R, Gay H, Yang WX, Bhowmick NA, Frelix G, Shappell S, Chen YH. Identification of extracellular delta-catenin accumulation for prostate cancer detection. Prostate. 2009;69(4):411–418. doi: 10.1002/pros.20902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackinnon MJ, Ndila C, Uyoga S, et al. Environmental Correlation Analysis for Genes Associated with Protection against Malaria. Mol Biol Evol. 2016 doi: 10.1093/molbev/msw004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matter C, Pribadi M, Liu X, Trachtenberg JT. Delta-catenin is required for the maintenance of neural structure and function in mature cortex in vivo. Neuron. 2009;64:320–7. doi: 10.1016/j.neuron.2009.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medina M, Marinescu RC, Overhauser J, Kosik KS. Hemizygosity of delta-catenin (CTNND2) is associated with severe mental retardation in cri-du-chat syndrome. Genomics. 2000;63:157–64. doi: 10.1006/geno.1999.6090. [DOI] [PubMed] [Google Scholar]
- Nivard MG, Mbarek H, Hottenga JJ, et al. Further confirmation of the association between anxiety and CTNND2: replication in humans. Genes Brain Behav. 2014;13:195–201. doi: 10.1111/gbb.12095. [DOI] [PubMed] [Google Scholar]
- Nopparat J, Zhang J, Lu J-PP, et al. δ-Catenin, a Wnt/β-catenin modulator, reveals inducible mutagenesis promoting cancer cell survival adaptation and metabolic reprogramming. Oncogene. 2015;34:1542–52. doi: 10.1038/onc.2014.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M, Kim H, Yang I, et al. GSK-3 phosphorylates delta-catenin and negatively regulates its stability via ubiquitination/proteosome-mediated proteolysis. J Biol Chem. 2009;284:28579–89. doi: 10.1074/jbc.M109.002659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paffenholz R, Franke WW. Identification and localization of a neurally expressed member of the plakoglobin/armadillo multigene family. Differentiation. 1997;61:293–304. doi: 10.1046/j.1432-0436.1997.6150293.x. [DOI] [PubMed] [Google Scholar]
- Paffenholz R, Kuhn C, Grund C, et al. The arm-repeat protein NPRAP (neurojungin) is a constituent of the plaques of the outer limiting zone in the retina, defining a novel type of adhering junction. Exp Cell Res. 1999;250:452–64. doi: 10.1006/excr.1999.4534. [DOI] [PubMed] [Google Scholar]
- Reynolds AB. p120-catenin: Past and present. Biochim Biophys Acta. 2007;1773:2–7. doi: 10.1016/j.bbamcr.2006.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reynolds AB, Herbert L, Cleveland JL, et al. p120, a novel substrate of protein tyrosine kinase receptors and of p60v-src, is related to cadherin-binding factors beta-catenin, plakoglobin and armadillo. Oncogene. 1992;7:2439–45. [PubMed] [Google Scholar]
- Reynolds AB, Roczniak-Ferguson A. Emerging roles for p120-catenin in cell adhesion and cancer. Oncogene. 2004;23:7947–56. doi: 10.1038/sj.onc.1208161. [DOI] [PubMed] [Google Scholar]
- Reynolds AB, Roesel DJ, Kanner SB, Parsons JT. Transformation-specific tyrosine phosphorylation of a novel cellular protein in chicken cells expressing oncogenic variants of the avian cellular src gene. Mol Cell Biol. 1989;9:629–38. doi: 10.1128/mcb.9.2.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoads AR, Karkera JD, Detera-Wadleigh SD. Radiation hybrid mapping of genes in the lithium-sensitive wnt signaling pathway. Mol Psychiatry. 1999;4:437–42. doi: 10.1038/sj.mp.4000538. [DOI] [PubMed] [Google Scholar]
- Ridge PG, Ebbert MT, Kauwe JS. Genetics of Alzheimer’s disease. Biomed Res Int. 2013;2013:254954. doi: 10.1155/2013/254954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sardina JM, Walters AR, Singh KE, et al. Amelioration of the typical cognitive phenotype in a patient with the 5pter deletion associated with Cri-du-chat syndrome in addition to a partial duplication of CTNND2. Am J Med Genet A. 2014;164A:1761–4. doi: 10.1002/ajmg.a.36494. [DOI] [PubMed] [Google Scholar]
- Schieve LA, Rice C, Yeargin-Allsopp M, et al. Parent-reported prevalence of autism spectrum disorders in US-born children: an assessment of changes within birth cohorts from the 2003 to the 2007 National Survey of Children’s Health. Matern Child Health J. 2012;16(Suppl 1):S151–7. doi: 10.1007/s10995-012-1004-0. [DOI] [PubMed] [Google Scholar]
- Schlessinger K, Hall A, Tolwinski N. Wnt signaling pathways meet Rho GTPases. Genes Dev. 2009;23:265–77. doi: 10.1101/gad.1760809. [DOI] [PubMed] [Google Scholar]
- Silverman JB, Restituito S, Lu W, et al. Synaptic anchorage of AMPA receptors by cadherins through neural plakophilin-related arm protein AMPA receptor-binding protein complexes. J Neurosci. 2007;27:8505–16. doi: 10.1523/JNEUROSCI.1395-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sklar P, Stone JL, O’Donovan MC, et al. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–41. doi: 10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefansson H, Rujescu D, Cichon S, et al. Large recurrent microdeletions associated with schizophrenia. Nature. 2008;455:232–6. doi: 10.1038/nature07229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner TN, Sharma K, Oh EC, et al. Loss of δ-catenin function in severe autism. Nature. 2015;520:51–6. doi: 10.1038/nature14186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogelstein B, Papadopoulos N, Velculescu VE, et al. Cancer genome landscapes. Science. 2013;339:1546–58. doi: 10.1126/science.1235122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vrijenhoek T, Buizer-Voskamp JE, van der Stelt I, et al. Recurrent CNVs disrupt three candidate genes in schizophrenia patients. Am J Hum Genet. 2008;83:504–10. doi: 10.1016/j.ajhg.2008.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yang M, Su S, et al. Association of ZNF644, GRM6 and CTNND2 genes polymorphisms with high myopia. Zhonghua Yi Xue Za Zhi. 2014;94:1289–93. [PubMed] [Google Scholar]
- Wang T, Chen Y-HH, Hong H, et al. Increased nucleotide polymorphic changes in the 5′-untranslated region of delta-catenin (CTNND2) gene in prostate cancer. Oncogene. 2009;28:555–64. doi: 10.1038/onc.2008.399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiss LA, Arking DE, Daly MJ, Chakravarti A. A genome-wide linkage and association scan reveals novel loci for autism. Nature. 2009;461:802–8. doi: 10.1038/nature08490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoo H. Genetics of Autism Spectrum Disorder: Current Status and Possible Clinical Applications. Exp Neurobiol. 2015;24:257–72. doi: 10.5607/en.2015.24.4.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu Z, Zhou J, Chen X, et al. Polymorphisms in the CTNND2 gene and 11q24.1 genomic region are associated with pathological myopia in a Chinese population. Ophthalmologica. 2012;228:123–9. doi: 10.1159/000338188. [DOI] [PubMed] [Google Scholar]
- Yuan L, Seong E, Beuscher JL, Arikkath J. δ-Catenin Regulates Spine Architecture via Cadherin and PDZ-dependent Interactions. J Biol Chem. 2015;290:10947–57. doi: 10.1074/jbc.M114.632679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng M, Simon R, Mirlacher M, et al. TRIO amplification and abundant mRNA expression is associated with invasive tumor growth and rapid tumor cell proliferation in urinary bladder cancer. Am J Pathol. 2004;165:63–9. doi: 10.1016/S0002-9440(10)63275-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou J, Liyanage U, Medina M, et al. Presenilin 1 interaction in the brain with a novel member of the Armadillo family. Neuroreport. 1997;8:2085–90. doi: 10.1097/00001756-199705260-00054. [DOI] [PubMed] [Google Scholar]