Abstract
Genes of the major histocompatibility complex (MHC) exhibit heterozygote advantage in immune defense, which in turn can select for MHC-disassortative mate choice. However, many species lack this expected pattern of MHC-disassortative mating. A possible explanation lies in evolutionary processes following gene duplication: if two duplicated MHC genes become functionally diverged from each other, offspring will inherit diverse multilocus genotypes even under random mating. We used locus-specific primers for high throughput sequencing of two expressed MHC Class II B genes in Leach's storm-petrels, Oceanodroma leucorhoa, and found that exon 2 alleles fall into two gene-specific monophyletic clades. We tested for disassortative versus random mating at these two functionally diverged Class II B genes, using multiple metrics and different subsets of exon 2 sequence data. With good statistical power, we consistently found random assortment of mates at MHC. Despite random mating, birds had MHC genotypes with functionally diverged alleles, averaging 13 amino acid differences in pairwise comparisons of exon 2 alleles within individuals. To test whether this high MHC diversity in individuals is driven by evolutionary divergence of the two duplicated genes, we built a phylogenetic permutation model. The model showed that genotypic diversity was strongly impacted by sequence divergence between the most common allele of each gene, with a smaller additional impact of monophyly of the two genes. Divergence of allele sequences between genes may have reduced the benefits of actively seeking MHC-dissimilar mates, in which case the evolutionary history of duplicated genes is shaping the adaptive landscape of sexual selection.
Keywords: complementarity, high throughput sequencing, mate choice, monophyly, Oceanodroma leucorhoa, seabird
Introduction
Over the past two decades, the major histocompatibility complex (MHC) has emerged as a system for studying both molecular evolution and mate choice for genetic benefits (Balakrishnan et al. 2010; Bernatchez & Landry 2003; Kamiya et al. 2014; Milinski 2006). MHC proteins help trigger the vertebrate immune response, with Class I and Class II MHC proteins binding and presenting antigens from intracellular and extracellular pathogens, respectively. Because MHC alleles are somewhat specific in the antigens they bind, MHC genotypes can differ in their effectiveness against important pathogens in a population (Sepil et al. 2013; Wegner et al. 2003) and thereby become a target for mate choice to increase offspring health and survival.
MHC-dependent mate choice could occur via two processes which are not mutually exclusive. One is a good genes mechanism, in which individuals prefer mates with particular alleles that are effective against pathogens (Eizaguirre et al. 2009). But MHC-based mate choice could also be driven by heterozygote advantage, in which heterozygotes are better at defense against a variable pathogen landscape (Penn et al. 2002; Spurgin & Richardson 2010). The most effective way to ensure heterozygous offspring is to actively choose a mate whose MHC is different from one's own, and evidence from multiple model systems has shown that MHC alleles of potential mates can be assessed by olfaction (Boehm & Zufall 2006). Consequently, MHC-disassortative mating has been seen in a variety of taxa including mammals, fish, reptiles, and birds (Kamiya et al. 2014). In seabirds, MHC-disassortative mating has been found in two of three studies (Juola & Dearborn 2012; Knafler et al. 2012; Strandh et al. 2012) and was predicted to be common because of a combination of monogamy, long lifespan, and olfactory capability (Zelano & Edwards 2002). However, many studies do not find MHC-disassortative mating (Bichet et al. 2014; Ekblom et al. 2004; Gasparini et al. 2015; Knafler et al. 2012; Lenz et al. 2013; Sepil et al. 2015; Westerdahl 2004), and the average effect size across all studies is quite small (Kamiya et al. 2014). Thus, there is no consensus from empirical studies on the presence or strength of MHC-disassortative mating, reinforcing the idea that populations may differ in attributes that would favor disassortative versus random mate choice with respect to MHC.
In this context, little attention has been paid to another potentially important factor: the impact of gene duplication and subsequent evolution of the duplicated genes' alleles. Although the wide occurrence of duplicated MHC genes is well established (Bollmer et al. 2010; Burri et al. 2010; Nei & Rooney 2005), there is little discussion of how the relationships among the alleles of those duplicated genes might determine whether disassortative mating is needed for producing offspring whose genotypes consist of functionally divergent alleles (Lenz et al. 2013). This lack of theoretical development might be because most empirical studies of MHC-disassortative mate choice use lab methods that assay multiple MHC loci simultaneously and thus cannot assign alleles to a particular locus (see literature review in Supporting Information). If evolution after duplication of MHC genes leads to a pattern of allelic variation that is not monophyletic by locus within a species (Edwards & Hedrick 1998; Strand et al. 2013; Worley et al. 2008), selection might continue to favor MHC-disassortative mate choice as a means to create offspring with MHC-divergent alleles. In contrast, if gene duplication is followed by the evolutionary divergence of alleles into gene-specific clades within a species (Dearborn et al. 2015; Fawcett & Innan 2011; Gu & Nei 1999), random mating would always produce offspring with at least two diverged MHC alleles (i.e. at least one from each gene-specific clade), even in the absence of a mating preference for MHC-disassortative partners. Thus, the extent of divergence between duplicated MHC genes might impact the benefits of disassortative mating, though this can be robustly tested only with locus-specific primers (see literature review in Supporting Information).
In birds, MHC diversity is partly created by an ancient duplication of a Class II B gene, followed by additional birth and death of genes in different lineages (Burri et al. 2010). We investigated MHC-dependent mate choice in Leach's storm-petrel, Oceanodroma leucorhoa, a procellariiform seabird with two diverged MHC Class II B genes. In our storm-petrel population, little is known about specific pathogens and their role in shaping MHC diversity, but three other factors yield predictions about random vs. MHC-disassortative mating. First, our population has the traits hypothesized to favor MHC-disassortative mating in seabirds (Zelano & Edwards 2002): long lifespan (Haussmann & Mauck 2008), monogamy (Mauck et al. 1995), and olfactory capability (Grubb Jr 1974; Nevitt & Haberman 2003). Second, demography of our North Atlantic study population likely fosters a high level of genetic diversity in general: a large population (15,000 breeding pairs; unpubl. data) and extensive gene flow in the Atlantic basin (Bicknell et al. 2012). These factors could maintain MHC diversity to an extent that might not require disassortative mating. Third and perhaps most importantly, though, our initial data show two Class II B genes with exon 2 sequences that cluster in locus-specific clades (Dearborn et al. 2015), with the possible consequence that MHC-divergent offspring might be produced even without disassortative mating.
In this study, we first tested whether mate choice is random or disassortative with respect to MHC in this population of Leach's storm-petrel, complementing our MHC Class II B data with microsatellite-based measures of parentage and of inbreeding/outbreeding. Second, we used a modeling approach to test whether MHC diversity in individuals is influenced by monophyly in general, and in particular by divergence between the most common allele of each gene.
Materials and Methods
Study Population and Sampling
We studied Leach's storm-petrels at the Bowdoin Scientific Station on Kent Island, an 80-ha island in the Bay of Fundy, New Brunswick, Canada (44.588N, 66.818W). Field work was carried out following the Ornithological Council's Guidelines for the Use of Wild Birds in Research (Fair et al. 2010) under permits from Bowdoin College and the Canadian Wildlife Service.
We extracted DNA from blood samples collected by brachial venipuncture of 188 adults comprising 94 unique mated pairs: 52 pairs in 2010 and a non-overlapping set of 42 pairs in 2013. Birds were sampled from breeding burrows within a 1-ha area, ensuring that birds could reasonably be thought of as potential mates. In 2013, we also sampled the 3- to 7-week old nestling at the 34 of 42 nests where the nestling survived to that stage. Because storm-petrels are sexually monomorphic, we assessed the sex of all 222 birds with PCR of a sex-linked gene (Fridolfsson & Ellegren 1999) (see Supporting Information).
We have previously characterized exon 2 of these two MHC Class II B genes in this system. Repeat genotyping produced identical results in 98.7% of cases, and cDNA sequencing confirmed that both genes are expressed (Dearborn et al. 2015). The development of locus-specific nested PCR and Illumina sequencing of exon 2 was made possible by the extensive fixed differences between the two genes in intron 1 and exon 3 (Dearborn et al. 2015). In a 229 bp section of intron 1, there are fixed differences between the genes at over half of the sites, and BLAST searches of intron 1 produce two mutually exclusive sets of hits: all high-ranking matches are MHC introns from seabirds and allies, but with different sets of sequences matching the introns of the two genes. Thus, the two genes are distinctly differentiated outside of the region coding for the peptide binding groove, which is consistent with existing evidence for an ancient duplication of avian MHC genes (Burri et al. 2010). Further evidence for the distinct identity of two genes in our system comes from our parent-offspring data: we have MHC genotypes of 22 trios of mother-father-offspring whose parentage relationships were confirmed by microsatellite data (see Results), and genotypes of both genes are fully consistent with Mendelian inheritance.
MHC High Throughput Sequencing, Copy Number Variation, and Phylogenetic Analysis
Nested PCR spanning exon 2 of Ocle-DAB1 and Ocle-DAB2 was performed with locus-specific primers that were barcoded in unique combinations for each bird (Dearborn et al. 2015). As expected based on our initial characterization of these two genes, the PCR products that were subjected to Illumina sequencing were different lengths for the two genes because of primer placement – 353 bp and 395 bp for the amplicons of Ocle-DAB1 and Ocle-DAB2, respectively, including the barcoded primers. Amplicons were sequenced as part of two different Illumina MiSeq runs using a PCR-free library preparation and 2x250 bp paired end reads, with forward and reverse reads processed with the FASTX package (Dearborn et al. 2015). Details of data processing and genotyping are given in the Supporting Information and summarized here.
Copy number variation (CNV) at MHC has been reported in multiple studies and is common in some species (de Groot et al. 2015; Lighten et al. 2014). Our primary genotyping algorithm assumed an absence of CNV and instead used read-depth ratios to define genotypes based on the most abundant one or two amplicon sequences at each gene, discarding low abundance amplicons as errors. However, we also examined data with a highly permissive genotyping algorithm that liberally counts lower frequency sequences as CNV alleles (see Supporting Information). By that approach, putative CNV may occur in 1% of individuals at Ocle-DAB1 and 14% of individuals at Ocle-DAB2. We think that this is likely an overestimate of CNV in our system, for three reasons. First, preliminary Sanger sequencing of uncloned PCR products produced either clean sequences with single peaks (i.e. homozygotes) or sequences with some double peaks that could be created by a combination of two sequences from known homozygotes. Second, in the 22 parent-offspring trios, offspring genotypes determined with our primary algorithm were consistent with inheritance from their parents and showed no unattributable alleles at either MHC gene. Third, in duplicate PCR and sequencing of both genes in 39 birds using our primary genotyping algorithm, 77 of 78 (98.7%) genotypes yielded identical results (Dearborn et al. 2015). Because CNV appears to be rare, downstream analyses are based on our primary genotyping algorithm unless otherwise specified. However, to be conservative we also re-analyzed our data using the CNV-permissive genotyping algorithm. This yielded small changes in some of the descriptive statistics but had no impact on the mate choice conclusions, as detailed in the Supporting Information.
Allele sequences were aligned in Sequencher 5.2.2 (Gene Codes Corporation, Ann Arbor, MI), and putative peptide binding site (PBS) codons were determined by comparison with human HLA (Brown et al. 1993) and seabird sequences (Strandh et al. 2011). To characterize the similarity of exon 2 sequences of the 24 alleles and to ask how they cluster with respect to locus identity, we built a parsimony-based haplotype network with Haploviewer (Salzburger et al. 2011) based on the dnapars module in Phylip 3.695 (Felsenstein 2005), and we constructed maximum likelihood (ML) phylogenies from DNA sequences using a GTR + I + G model in MEGA 6.06 (Tamura et al. 2013). We also conducted a Neighbor Joining phylogenetic analysis based on a matrix of functional distances between alleles. To generate functional distances, first we mapped the 20 amino acids into a 5-dimensional space according to five physicochemical z-descriptors that measure lipophilicity (z1), steric bulk (z2), polarity (z3), and electronic effects (z4 and z5) (Sandberg et al. 1998). The five-dimensional Euclidean distance was measured between each pair of amino acids, and functional divergence between MHC alleles was calculated as the average of these Euclidean distances across all codons of the two alleles (Agbali et al. 2010; Sin et al. 2015). This measure of functional divergence is thus analogous to p-distance of amino acid sequences but with a continuous scale of physicochemical divergence for each codon position rather than a simple binary score of whether or not the two amino acids are identical. For all phylogenies, we used midpoint rooting to avoid assumptions about whether a putative outgroup gene is homologous to one, both, or neither of the duplicated genes in storm-petrels.
Paternity Analysis and Tests of Inbreeding/Outbreeding with Microsatellite Markers
Because inferences about MHC-based mate choice require data on actual genetic mates, we used microsatellite genotypes to conduct paternity exclusion analyses of social mates (Lemons et al. 2015) in Cervus 3.0.7 (Kalinowski et al. 2007). This exclusion test uses genotypes of putative mother-father-offspring trios to calculate the log of the odds (LOD scores) that an offspring's actual father is the putative father rather than some unspecified male from the population at large; critical values for LOD scores are calculated based on microsatellite data from the population. We also used microsatellite genotypes to test for genome-wide inbreeding or outbreeding, because such a pattern could bias our interpretation of MHC-based mate choice. We computed Queller and Goodnight's relatedness coefficient r (Queller & Goodnight 1989) for true mates and compared that value to 10,000 replicates of randomly pairing the 94 females and 94 males without replacement. For tests of relatedness between mates and of parentage, birds were genotyped at 15 microsatellite loci (Table S1) by Ecogenics (Balgach, Switzerland). We confirmed the absence of stutter, large-allele dropout, and null alleles with MICRO-CHECKER 2.2.3 (Van Oosterhout et al. 2004) and estimated the genotyping error rate with mother-offspring pairs in Cervus 3.0.7 (Kalinowski et al. 2007). Measures of polymorphism, observed versus expected heterozygosity, and genotypic disequilibrium were made with FSTAT 2.9.3.2 (Goudet 2002).
MHC Data Analysis
We tested for random versus MHC-disassortative mating with randomization tests in a custom Excel macro, comparing the observed MHC similarity of true mates against 10,000 replicates of randomly pairing the 94 females and 94 males without replacement. Randomization tests were two-tailed for means, to distinguish between assortative mating, random mating, and disassortative mating. Mate choice also might target partners who are intermediately dissimilar at MHC, which would manifest as random mating in a test of means but smaller-than-expected variance among pairs in a one-tailed randomization test of variances (Forsberg et al. 2007; Lenz et al. 2013). Our mate choice analyses were based on all 188 birds combined: because birds in this population breed annually, have very high breeding site fidelity, and can live more than 35 years (Haussmann & Mauck 2008; Huntington et al. 1996; Mauck et al. 2012) (unpubl. data), individuals sampled in 2010 are highly likely to be breeding in 2013, and vice versa. Similar results (not shown) were obtained when randomization tests were conducted separately for the two years.
Tests were based on two aspects of MHC similarity, both derived from amino acid sequences: allele sharing and allele sequence divergence. Allele sharing was calculated as the proportion of alleles that two birds have in common, considering both genes simultaneously. Allele sequence divergence was calculated as average p-distance for all pairwise comparisons of two birds' MHC amino acid sequences, using MEGA 6.06 (Tamura et al. 2013). Analyses were conducted with (a) all 89 codons of exon 2, (b) only the 33 putative peptide binding sites (Table S2), and (c) other subsets hypothesized to be functionally important (see Supporting Information).
We also tested for MHC-disassortative mating based on functional divergence between alleles. Mate choice randomizations were conducted with these physicochemical measures of functional divergence using (a) all polymorphic codons of exon 2, (b) only the putative PBS codons, and (c) only codons showing signatures of positive selection.
To assess the robustness of our finding of random mating (see Results), we conducted power analyses on our mate choice randomization tests following Lenz et al. (Lenz et al. 2013), as detailed in the Supporting Information.
Lastly, we tested the rarely addressed assumption that MHC differences between mates would translate to MHC diversity in offspring. For this, we assayed MHC genotypes of 22 families for which we confirmed parentage with microsatellite data.
Phylogenetic Permutation Model of MHC
We developed a phylogenetic permutation model (see Supporting Information for details) to test two evolutionary hypotheses that could explain why MHC genotypes exhibit pronounced amino acid differences between an individual's alleles, despite random mating. The first hypothesis is that MHC-divergent genotypes are generated by monophyly broadly speaking – that is, because the alleles of the two genes are diverged into locus-specific clades. The second hypothesis is that MHC-divergent genotypes are generated more specifically by divergence between the two common alleles in the population, Ocle-DAB1*004 and Ocle-DAB2*0050 (Figure 1a). To test these two hypotheses in the context of our study system, our permutation model held constant the total number of alleles per gene, the distribution of allele frequencies of each gene (from our large sample of 188 breeding adults), and the structure of the phylogeny. Within that framework, we permuted the alleles and their associated frequencies across the phylogeny, which changed the distances between particular alleles according to their new positions in the phylogeny (Figure S1). For each of 12,000 permutations of the model, we recorded the proportion of each gene's alleles that fell together into a single clade (i.e. the extent of monophyly), the distance between the most common allele of the two genes, and the resulting average within-individual diversity of MHC alleles in our set of 188 individuals. We conducted regression analyses with the model's output to test the extent to which MHC diversity of individuals was predicted by degree of monophyly and by divergence between the two most common alleles.
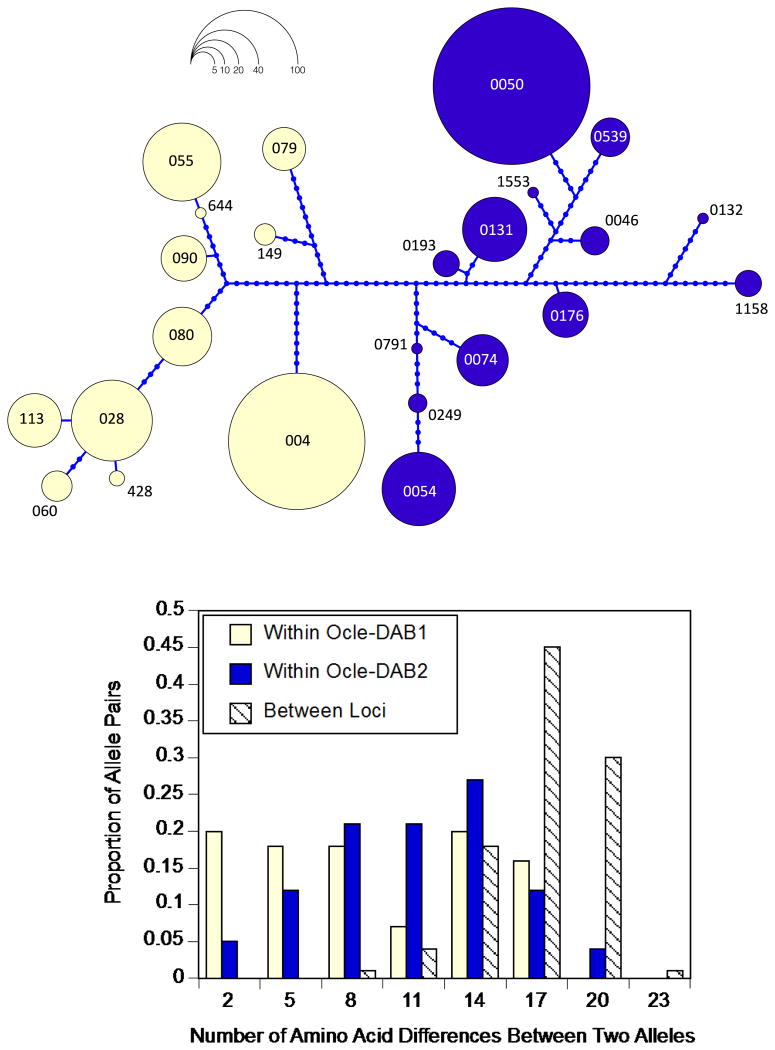
Figure 1.
Exon 2 variability at two MHC Class II B loci in 188 adult storm-petrels. (a) Haplotype network of alleles from Ocle-DAB1 (yellow) and Ocle-DAB2 (dark blue), created with Haploviewer (Salzburger et al. 2011) based on the dnapars module in Phylip 3.695 (Felsenstein 2005). Each segment corresponds to one nucleotide change. (b) Histogram of the number of amino acid differences (of 89 codons total) between alleles in the same locus or different loci. Within a locus, comparisons of an allele to itself (zero amino acid differences) are excluded.
Results
MHC Descriptives
Our sequencing efforts revealed 11 alleles from Ocle-DAB1 and 13 from Ocle-Dab2 (Figure 1a, Table S2). Nine and 12 alleles were previously described alleles from Ocle-DAB1 and Ocle-DAB2, respectively (Dearborn et al. 2015) (GenBank accession numbers for new alleles: KU232398-KU232400). Each gene was dominated by a single common allele: Ocle-DAB1*004 had a frequency of 0.431 and Ocle-DAB2*0050 a frequency of 0.569, with the remaining alleles ranging in frequency from 0.003 to 0.152 (Figure 1a, Table S2).
As parts of two different Illumina MiSeq runs of MHC Class II B, we obtained 58,864,658 sequencing reads. Valid, barcoded sequences corresponded to 19,275,393 amplicons (i.e. forward + reverse sequence with a diagnostic barcode combination) with an average FASTQ quality above Q30. For sequences identified as true alleles, there was a median read depth per bird per gene of 3,312 and 2,084 valid reads for Ocle-DAB1 and OcleDAB2. Minimum read depth was 101 and 26 for the two genes, and the number of birds with fewer than 100 reads was 0 and 4, respectively. Heterozygotes did not have greater read depth than homozygotes (Ocle-DAB1: t = 1.28, df = 208, p = 0.200; Ocle-DAB2: t = 0.12, df = 208, p = 0.902), suggesting that our ability to detect additional alleles in individuals was not limited by read depth.
Genotypes of the 188 adults met Hardy-Weinberg expectations at Ocle-DAB1 (p = 0.685), whereas Ocle-DAB2 showed an excess of homozygotes (77 observed, 67 expected; p = 0.025); at Ocle-DAB2, multiple homozygous genotypes made small contributions to this pattern, with no particular genotype showing obvious excess (e.g., no observed counts deviated from expected by more than 3 individuals). As is common in the MHC of mammals and birds (Jeffreys et al. 2001; Strand et al. 2013), these two MHC Class II B genes exhibited composite linkage disequilibrium with each other (p=0.00007), based on a test for association between diploid genotypes at both loci (Goudet 2002). Analysis of phased haplotypes in the 99 adults with 2 or 3 alleles revealed a non-significant tendency towards over-representation of the haplotype comprising the two most common alleles, Ocle-DAB1*0050 and Ocle-DAB2*004 (observed 83, expected 78.0; Fisher Exact test, p = 0.107); there also appears to be a slight overrepresentation of haplotype Ocle-DAB1*004 / Ocle-DAB2*0131 (observed 17, expected 10.1) and Ocle-DAB1*055/ Ocle-DAB2*0054 (observed 9, expected 1.6), and a slight underrepresentation of Ocle-DAB1*055/ Ocle-DAB2*0050 (observed 5, expected 13.4), but a larger sample would be required to assess whether these are robust patterns.
As with the 21 alleles described previously (Dearborn et al. 2015), the now-expanded set of 24 alleles showed signatures of positive selection at the putative PBS codons (Ocle-DAB1 dN-dS = 3.64, p = 0.0002; Ocle-DAB2 dN-dS = 2.33, p = 0.0106) and not at non-PBS codons (Ocle-DAB1 dN-dS = 0.40, p = 0.345; Ocle-DAB2 dN-dS = -0.42, p = 1.00), based on the modified Nei-Gojobori z-test in MEGA 6.06.
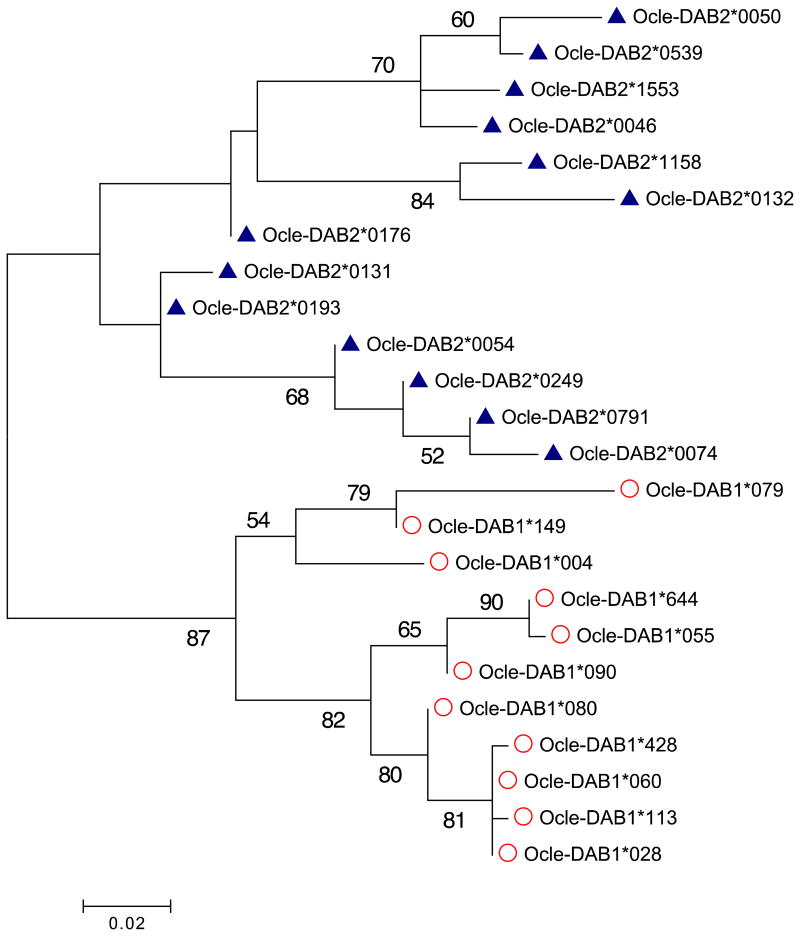
Phylogenetic analyses of alleles from Ocle-DAB1 and Ocle-DAB2 consistently gave rise to two distinct monophyletic clades corresponding to the two genes. This pattern can be visualized in a maximum likelihood tree (Figure 2), as with the previously described 21 alleles (Dearborn et al. 2015). Our parsimony-based haplotype network of DNA sequences (Figure 1a) concurred with ML trees in the clustering of alleles by locus. Furthermore, the alleles still fell into gene-specific clades in a Neighbor Joining analysis of functional divergence between alleles based on physicochemical properties of amino acids (Figure S2). Bootstrap support was not high in the maximum likelihood tree however, this is common for intraspecific phylogenies of MHC exon 2 sequences subject to diversifying selection (Anmarkrud et al. 2010; Bollmer et al. 2010). It is also important to bear in mind that, because of strong diversifying selection on the peptide binding groove encoded by exon 2, phylogenetic analyses of exon 2 sequences are expected to reveal current similarity rather than deep evolutionary history.
Figure 2.
Maximum likelihood tree of complete exon 2 sequences, with midpoint rooting to avoid assumptions about outgroup homology with duplicated genes. Bootstrap support values are shown where greater than 50%. Alleles are marked with open circles for Ocle-DAB1 and solid triangles for Ocle-DAB2. See Figure S2 for a phylogeny based on functional distance between alleles.
In line with the reciprocal monophyly of exon 2 alleles of the two genes, the average number of amino acid differences between alleles was larger between genes than within genes 17.1 ± 2.6 versus 10.1 ± 4.9 SD; t = 14.7, df = 198.5, p < 0.0001; Figure 1b); the same was true when examining functional distance (3.0 ± 0.50 versus 1.77 ± 0.89 ; t = 14.1, df = 204.6, p < 0.0001). Even the most similar Ocle-DAB1 and Ocle-DAB2 alleles differed by 8 amino acids. Furthermore, the most common allele of each gene (Ocle-DAB1*004, Ocle-DAB2*0050) differed from each other by 15 amino acids (compared to an average difference of 13.7 ± 5.2 amino acids across all pairwise comparisons of the 24 alleles, irrespective of locus); 11 of those differences occurred among the 33 putative PBS codons. Overall, the two genes are diverged, rather than homogenized, and any pair of alleles drawn from different loci will have 8 to 22 amino acid differences, with 4 to 15 of those differences being at putative peptide binding sites.
Birds in our population had high levels of MHC Class II B diversity. Considering both genes in our sample of 188 mated adults and 22 nestlings, most individuals had either three (37.6%) or four different MHC alleles (47.1%), with an average of 3.3. Pairwise comparisons of the alleles within an individual (Schwensow et al. 2008) found a mean difference of 13.5 amino acids in the 89-codon sequence (range across birds: 10.0 to 18.0), and an average difference of 9.9 amino acids in the 33 PBS codons (range across birds: 7.3 to 12.5). Importantly, 61.0% of individuals possessed at least one copy of the most common allele of each gene (Ocle-DAB1*004 and Ocle-DAB2*0050), which differ from each other by 15 amino acids.
MHC and Mate Choice
Randomization tests supported the hypothesis of random mating rather than disassortative mating at MHC Class II B: average MHC similarity between mates
showed no evidence of preference for maximally or intermediately divergent mates, whether using all 89 codons to examine allele sharing (p = 0.716 for means, p = 0.681 for variances; Figure S3) or amino acid sequence divergence (p = 0.958, p = 0.947; Figure S4) or using the 33 PBS codons to examine allele sharing (p = 0.888, p = 0.794) or amino acid sequence divergence (p = 0.910, p = 0.957). Similar results for means and variances were obtained when amino acid sequence divergence was calculated using other subsets of the exon 2 sequence that we hypothesized might be the most important (e.g., positively selected codons) or when looking at the two genes individually (Supporting Information).
Mate choice was also random with respect to functional divergence measured by physicochemical properties of alleles, whether tests of means and variances of mate similarity used all exon 2 codons (p = 0.796, p = 0.858), the putative PBS sites (p = 0.700, p = 0.847), or positively selected sites (p = 0.690, p = 0.928). We still found evidence of random mating when assuming that birds can detect only an allele's presence, and not its number of copies in potential mates (Supporting Information). Lastly, when we modified our genotype calls based on possible cases of CNV, there was still no evidence of disassortative mating (Supporting Information). Overall, then, mated storm-petrels show consistent evidence of randomly assorting by MHC genotypes.
Power analyses showed high statistical power to detect mate choice for amino acid sequence divergence between mates. The randomization test of means had an 80% likelihood of detecting a preference for mates that were 0.25 amino acids more disassortative than random matings (Figure S5a), which corresponds to a 2% increase in the mean MHC divergence between pairs compared to strictly random mating. In our test for intermediately disassortative mating, we had an 80% likelihood of detecting a smaller-than-expected variance in which the MHC divergence of randomly assigned pairs was shifted by just 0.29 amino acids (in 89 codons) inward towards the overall mean of random pairings (Figure S5b), corresponding to a 26% reduction in variance among pairs in MHC similarity. Because alternative measures of sequence divergence are highly correlated with each other (Pearson's r = 0.912 to 0.985 for measures that include both genes), power should be similarly high regardless of choice of how to measure MHC divergence.
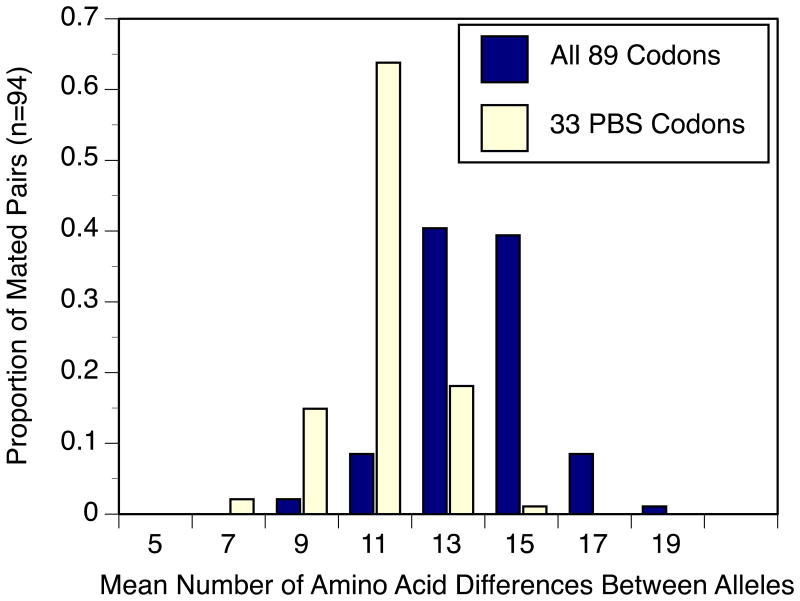
Despite random mating, mates were dissimilar at MHC. Mated pairs shared, on average, only 41.3% of their MHC Class II B alleles, and their alleles were diverged: the average pairwise divergence between alleles of mates was 12.1 amino acids out of the entire 89-codon exon 2 sequence (range 7.5 to 17.1; Figure 3), and 9.0 amino acids out of the 33 PBS codons (range 5.5 to 12.1).
Figure 3.
Distribution of exon 2 MHC amino acid differences between mates, measured as average number of amino acid differences between all pairwise combinations of a female's alleles and her mate's alleles, considering all 89 codons (dark bars) or only the 33 PBS codons (light bars).
Phylogenetic Permutation Model of MHC
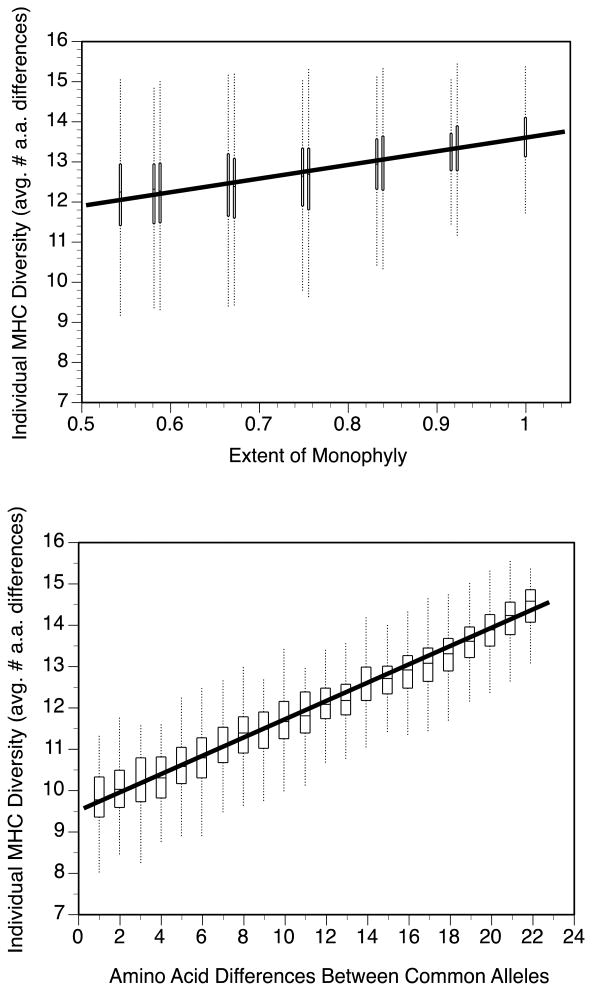
In our model, the average divergence between MHC alleles within individuals decreased in response to decreases in the extent of monophyly of the two genes' alleles (F1,10 = 190.0, p < 0.0001, r2 = 0.950; Figure 4a). However, the biological magnitude of the effect was small even in univariate regression, with average MHC diversity decreasing by only 1.4 amino acids (from 13.6 to 12.2) when moving from perfect monophyly of the two genes to a completely intermingled arrangement of their alleles. In contrast, the model showed a large effect for the hypothesized role of divergence between the most common allele at each gene. As the distance between these two alleles decreased, average divergence between an individual's alleles decreased sharply, dropping by 4.6 amino acids (from 14.4 to 9.8) across the examined range of possible distances between these two alleles in a univariate regression (F1,20 = 5,276.1, p < 0.0001, r2 = 0.996; Figure 4b). In a multiple regression model testing the two predictor variables simultaneously (F2,247 = 4,030.4, p < 0.0001, adjusted R2 = 0.970), there were significant effects of both factors: monophyly (F1,247 = 173.6, p < 0.0001) and distance between common alleles (F1,247 = 7,592.5, p < 0.0001). However, a comparison of standardized slopes in the multiple regression reveals that within-individual MHC divergence is much more influenced by the distance between common alleles (beta ± SE = 0.960 ± 0.011) than by the extent of monophyly (0.145 ± 0.011). Thus, MHC diversity in individuals is high in part because of two different aspects of the evolutionary history of MHC alleles in this population: a small influence of monophyly in general, and a larger influence of the sequence divergence between the most common allele of each gene.
Figure 4.
MHC diversity within individuals, calculated in a model that permutes the locations of alleles within the distance matrix. MHC diversity is average number of amino acid differences in pairwise comparisons of an individual's 4 alleles. Box plots show distributions of model results at each level of the predictor variable.
(a) MHC diversity as a function of the extent of monophyly of the two loci.
(b) MHC diversity as a function of sequence divergence between the most common allele at each locus.
In a multiple regression testing both factors as predictors of individual MHC diversity, there was a steeper standardized slope for the effect of distance between common alleles (beta ± SE = 0.960 ± 0.011) than for the effect of the extent of monophyly (0.145 ± 0.011).
Inheritance of MHC Diversity
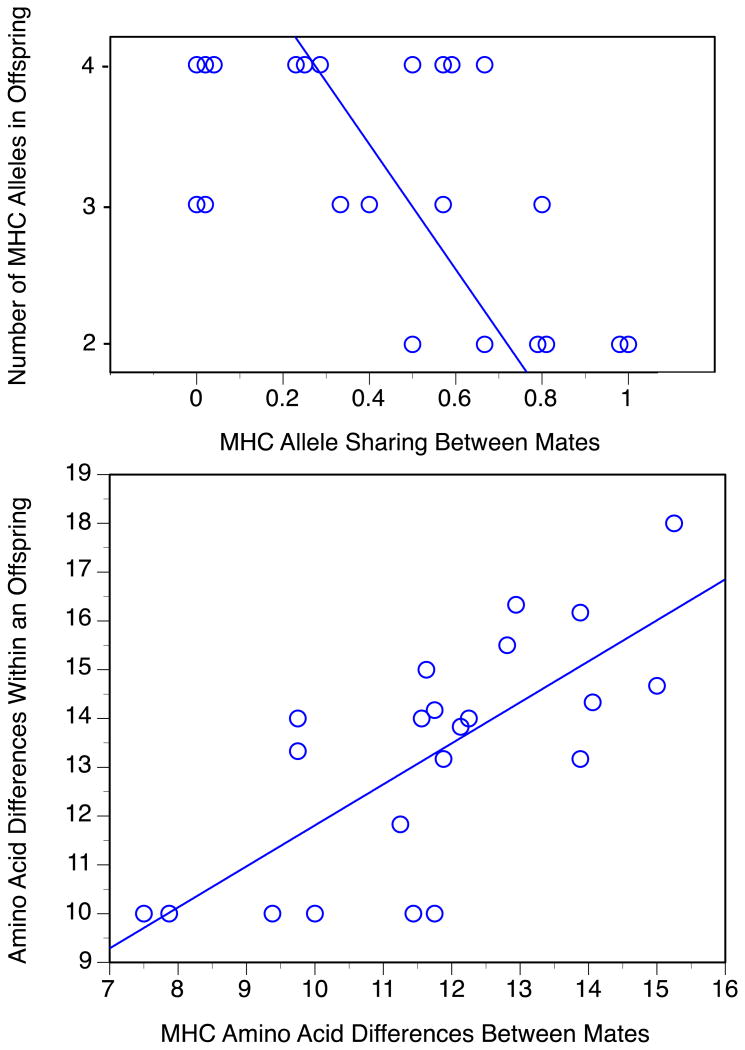
We found support for the crucial assumption that MHC divergent alleles between mates translate to MHC diversity in offspring. Parents with low MHC allele-sharing scores produced offspring with more MHC alleles (linear regression of 22 families: F1,20 = 10.9, p = 0.0035, r2 = 0.353; Figure 5a). Likewise, mates with large average divergence between their MHC amino acid sequences produced offspring whose own alleles had sequences that differed more from one another, whether considering all 89 codons (linear regression of 22 families: F1,20 = 22.7, p = 0.0001, r2 = 0.531; Figure 5b) or only the 33 PBS codons (F1,20 = 22.8, p = 0.0001, r2 = 0.533).
Figure 5.
Class II B MHC diversity in 22 nestlings and their 44 parents.
(a) Number of MHC alleles in offspring is predicted by allele-sharing between parents.
(b) Average number of amino acid differences between MHC alleles within an offspring is predicted by average divergence between its parents, based on all 89 codons of exon 2.
Paternity Analysis and Tests of Inbreeding/Outbreeding with Microsatellite Markers
Microsatellite data showed no evidence of extra-pair paternity in our sample of 34 single-chick families (Supporting Information). There were zero genotypic mismatches between chicks and putative fathers, and LOD scores for all putative fathers ranged from 3.4 to 14.3, exceeding the 95% cutoff of 0.73. Combined with previous data showing 0 extra-pair offspring in 48 families (Mauck et al. 1995), the upper 95% confidence limit on an estimate of 0% extra-pair paternity in this population is 4.5%. Thus, extra-pair paternity is unlikely to be common enough to distort patterns of MHC-based mate choice in this population. We also found no evidence of inbreeding or outbreeding, based on randomization tests of means (p = 0.566, Figure S6a) and variances (p = 0.934, Figure S6b) in microsatellite-based relatedness coefficients between mates.
Discussion
Despite random assortment of mates with respect to exon 2 of Class II B MHC, individual Leach's storm-petrels have MHC-divergent genotypes, and our model suggests that this finding is a consequence of divergence between the alleles of the duplicated genes. Random assortment of mates at MHC was consistent across the wide array of metrics employed and across different subsets of the data. Also, we found no evidence of heterozygote excess, contrary to the predictions of a multigenerational history of MHC-disassortative mating. Finally, our microsatellite data confirmed the absence of inbreeding or extra-pair parentage, which might have obscured evidence of MHC-disassortative mate choice. Thus, the data consistently show that mated Leach's storm-petrels truly are assorting randomly with respect to MHC. Despite random mating, individual genotypes typically consisted of multiple alleles that differed markedly in amino acid sequences. The results of our model support the view that the broad occurrence of MHC divergent genotypes is a consequence of two aspects of the evolutionary history of MHC in this system – a small effect of the monophyly of the alleles of each gene, and a larger effect of sequence divergence between the most common allele of each gene.
The alleles of these duplicated genes have exon 2 sequences that sort into two clades that correspond to the two genes (Figures 1a, 2, and S2). Although the two genes are distinguishable by the enormous number of fixed differences in areas upstream and downstream from the exon that encodes the peptide binding groove, it still would have been possible for mutation, gene conversion, and selection to create a set of exon 2 sequences that did not sort into two gene-specific clades (Strand et al. 2013). Such phylogenetic intermingling of alleles from different loci was not observed in our data and, consequently, the number of amino acid differences between any two alleles drawn from different genes is significantly greater than for alleles drawn from within a gene (Figure 1b); the same pattern was found when measuring functional distance between alleles. Importantly, our model showed that MHC divergence within individuals decreased when we reduced or eliminated monophyly and when we forced the two common alleles to be more similar in their amino acid sequences (Figure 4, Figure S1). Surprisingly, the effect of monophyly on within-individual MHC divergence was biologically rather small; varying the divergence between the most common allele of each gene had a stronger impact on MHC diversity (i.e. a steeper standardized slope in the multiple regression) and was also a more reliable predictor of MHC diversity (i.e., the small ranges of the boxplots in Figure 4b versus 4a). These two alleles, Ocle-DAB1*004 and Ocle-DAB2*0050, differ from each other by 15 amino acids in the 89-codon exon 2 sequence, with 11 of those differences occurring among the 33 putative PBS codons, and more than 60% of the individuals in our sample have at least one copy of both alleles (Figure 1a).
The development of locus-specific primers has allowed us to conclude that the presence of monophyly and of diverged common alleles contribute to genotypic divergence of MHC in our population. We hypothesize that this, in turn, obviates the need for MHC-disassortative mating, but the current state of the field makes it difficult to assess the generality of this idea. In most studies that test for MHC-disassortative mate choice, genotyping methods simultaneously assay multiple MHC genes (see literature review in Supporting Information), making it difficult or impossible to assess the extent of monophyly, amino acid sequence differences within versus between loci, or even simple allele frequencies. The only exception is a study of grey mouse lemurs (Huchard et al. 2013), which assayed single-locus genotypes at two MHC Class II loci that had exon 2 sequence diverged into locus-specific clades (Huchard et al. 2012). Disassortative mate choice was found at DRB but not DQB, and DRB also showed greater evidence of positive selection. In our storm-petrels, we found strong evidence of positive selection at the PBS codons of both of the expressed loci that we assayed. Further tests of our proposed link between random mating and divergent MHC loci will require locus-specific data from more species, but the continued expansion of high throughput sequencing methods might only increase the extent to which future MHC studies sequence amplicons from primers that simultaneously amplify an unknown number of MHC loci.
An additional set of studies of MHC-disassortative mating have examined one locus (see literature review in Supporting Information). This does not generally bear on our model and hypothesis, but a single-locus study of Galápagos sea lions does make an important related argument (Lenz et al. 2013). Galápagos sea lions resemble our storm-petrels in that individual MHC genotypes frequently comprise divergent alleles despite random assortment of mates, because the most common alleles in the population are relatively more diverged from each other (Lenz et al. 2013). However, the two datasets differ in the number of MHC Class II B genes – one for sea lions, and two for storm-petrels – and this has important consequences for whether the system should remain favorably disposed to random mating in the future. If negative frequency-dependent selection by pathogens is a driver of skewed allele frequencies (Ejsmond et al. 2010), there will be changes over time in which alleles are most common. In a single-locus system such as sea lions – or in a two-locus system without monophyly of the two sets of alleles – frequency-dependent selection by pathogens could increase the frequency of alleles that happen to be more similar to each other. In that case, random mating will no longer consistently produce genotypes containing diverged alleles. Thus, allele frequency changes via pathogen-mediated selection can drive the population into a situation in which the consistent production of heterozygous offspring could only occur by disassortative mating. In contrast, a two-locus system with monophyly does not face that constraint. In our case study of storm-petrels, the current difference between the most common allele of each gene (Ocle-DAB1*004 versus Ocle-DAB2*0050) is 15 amino acids. Even if the allele frequencies within each gene shift drastically, the new pairing of the most common allele from each would still differ by at least 8 – and as much as 22 – amino acids. If a species has two genes where a diverged set of alleles cluster by locus, this might ensure that the transiently most common alleles at the two genes will always be quite different from each other, even as allele frequencies change over time. Thus, disassortative mating might remain unnecessary in such a system, whereas it would become needed as a way to achieve functional heterozygosity in systems with a single locus or with two locus with intermingled alleles.
Shifts in allele frequencies are of course expected under pathogen-mediated frequency-dependent selection. Although little is known about pathogen pressures in our storm-petrel population, the frequency of the most common allele of each gene (0.431 and 0.569) is higher than what Ejsmond et al. (Ejsmond et al. 2010) found under simulations of heterozygote advantage alone, and instead is more consistent with a current or historical role for selection by particular pathogens. Leach's storm-petrels have been shown to harbor feather lice and mites (Fitzpatrick & Threlfall 1977) and a hepatazoon (Merino et al. 2014). Other pathogens may be likely given the presence of a high-frequency allele at each gene, and investigation of pathogens may shed light on the likelihood of these allele frequencies shifting radically in the future. Evolutionary response to pathogen-mediated selection pressures may also depend on two other features of MHC: copy number variation and linkage disequilibrium. We found some evidence for CNV in 14% of individuals at one of our two genes; CNV in some systems is an important contributor to individual MHC diversity (de Groot et al. 2015; Eimes et al. 2011; Lighten et al. 2014), and the number of alleles within an individual can be tied to disease resistance (Kurtz et al. 2004; Oliver et al. 2009). We also found evidence of linkage disequilibrium between the two Class II B genes. Multi-locus MHC haplotypes have been shown to correlate with disease resistance in some systems (Hill et al. 1991), but in storm-petrels we do not currently know whether linkage disequilibrium is due to physical linkage, multi-locus selection, or drift (though the latter is unlikely given gene flow in the Atlantic basin (Bicknell et al. 2012) and a local population size of approximately 15,000 pairs).
In Leach's storm-petrels, the divergence of MHC alleles at the two genes fosters MHC-divergent genotypes even without disassortative mating, but there appears to be some scope for further increases in offspring MHC divergence by actively seeking MHC-disassortative mates (Figure 5). In other words, why would birds in this population not choose mates based on disassortative MHC, above and beyond the gains afforded by the diverged duplicated genes? One possibility is that MHC-disassortative mating in storm-petrels might push offspring MHC diversity into the realm of “too different”, given data from some studies showing auto-immune costs of too much MHC diversity (Kalbe et al. 2009; Wegner et al. 2003). A second possibility is that further gains in offspring MHC diversity might be somewhat beneficial but that other aspects of mate choice override this effect. If storm-petrels did not search for MHC-disassortative mates, they might be better able to target other important traits such as parental care (Mauck et al. 2011; Ricklefs et al. 1986) or nest burrow quality (Fricke et al. in press). Broadly speaking, removing one dimension from the mate-optimization equation would give more flexibility to find a mate with a good combination of the other key traits.
The evolution of genes after duplication can unfold in different ways, ranging from neofunctionalization to redundancy to loss of function (Dittmar & Liberles 2010). Based on data from Leach's storm-petrels, we argue that the divergence of allele sequences between duplicated MHC genes can reduce the benefits of actively searching for an MHC-dissimilar mate, in which case the evolutionary history of duplicated genes is shaping the adaptive landscape of sexual selection.
Supplementary Material
Acknowledgments
We thank Shelby Sullivan, Marko Murray, Damon Gannon, and Doug Hinerfeld for important help in the lab or field, and Asant Agyare and Frank Hailer for discussions about MHC. We thank three anonymous reviewers for helpful suggestions, and we thank Tobias Lenz for power analysis scripts. This project was supported by Bates College, the Arthur Vining Davis Foundations, and an Institutional Development Award (P20GM0103423) from the National Institute of General Medical Sciences of the National Institutes of Health. Computational support was provided by the McMaster Service Lab and Repository. AGM holds a Cisco Research Chair in Bioinformatics, supported by Cisco Systems Canada, Inc. This is contribution # 257 from the Bowdoin Scientific Station on Kent Island.
Footnotes
Data Accessibility: DNA sequences: GenBank accessions KP090151-KP090162 and KU232398-KU232400
MHC amino acid sequences, MHC genotypes, microsatellite genotypes, Excel macro, and 4D scripts: Dryad doi:10.5061/dryad.j413n
Author Contributions: DCD designed the study. Field work and lab work were conducted by DCD, ABG, MEG, EM, AGM, and RAM. DCD and AGM analyzed data. RAM coded the models. DCD and ABG drafted the paper, and all authors helped with revisions.
References
- Agbali M, Reichard M, Bryjová A, Bryja J, Smith C. Mate choice for nonadditive genetic benefits correlate with mhc dissimilarity in the rose bitterling (Rhodeus ocellatus) Evolution. 2010;64:1683–1696. doi: 10.1111/j.1558-5646.2010.00961.x. [DOI] [PubMed] [Google Scholar]
- Anmarkrud JA, Johnsen A, Bachmann L, Lifjeld JT. Ancestral polymorphism in exon 2 of bluethroat (Luscinia svecica) MHC class II B genes. Journal of Evolutionary Biology. 2010;23:1206–1217. doi: 10.1111/j.1420-9101.2010.01999.x. [DOI] [PubMed] [Google Scholar]
- Balakrishnan CN, Ekblom R, Völker M, et al. Gene duplication and fragmentation in the zebra finch major histocompatibility complex. BMC Biology. 2010;8 doi: 10.1186/1741-7007-8-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernatchez L, Landry C. MHC studies in nonmodel vertebrates: What have we learned about natural selection in 15 years? Journal of Evolutionary Biology. 2003;16:363–377. doi: 10.1046/j.1420-9101.2003.00531.x. [DOI] [PubMed] [Google Scholar]
- Bichet C, Penn DJ, Moodley Y, et al. Females tend to prefer genetically similar mates in an island population of house sparrows. BMC Evolutionary Biology. 2014;14 doi: 10.1186/1471-2148-14-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bicknell AWJ, Knight ME, Bilton D, et al. Population genetic structure and long-distance dispersal among seabird populations: Implications for colony persistence. Molecular Ecology. 2012;21:2863–2876. doi: 10.1111/j.1365-294X.2012.05558.x. [DOI] [PubMed] [Google Scholar]
- Boehm T, Zufall F. MHC peptides and the sensory evaluation of genotype. Trends in Neurosciences. 2006;29:100–107. doi: 10.1016/j.tins.2005.11.006. [DOI] [PubMed] [Google Scholar]
- Bollmer JL, Dunn PO, Whittingham LA, Wimpee C. Extensive MHC class II B gene duplication in a passerine, the common yellowthroat (Geothlypis trichas) Journal of Heredity. 2010;101:448–460. doi: 10.1093/jhered/esq018. [DOI] [PubMed] [Google Scholar]
- Brown JH, Jardetzky TS, Gorga JC, et al. Three-dimensional structure of the human class II histocompatibility antigen HLA-DR1. Nature. 1993;364:33–39. doi: 10.1038/364033a0. [DOI] [PubMed] [Google Scholar]
- Burri R, Salamin N, Studer RA, Roulin A, Fumagalli L. Adaptive divergence of ancient gene duplicates in the avian MHC class II β. Molecular Biology and Evolution. 2010;27:2360–2374. doi: 10.1093/molbev/msq120. [DOI] [PubMed] [Google Scholar]
- de Groot NG, Blokhuis JH, Otting N, Doxiadis GGM, Bontrop RE. Co-evolution of the MHC class I and KIR gene families in rhesus macaques: Ancestry and plasticity. Immunological Reviews. 2015;267:228–245. doi: 10.1111/imr.12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dearborn DC, Gager AB, Gilmour ME, et al. Non-neutral evolution and reciprocal monophyly of two expressed Mhc class II B genes in Leach's storm-petrel. Immunogenetics. 2015;67:111–123. doi: 10.1007/s00251-014-0813-2. [DOI] [PubMed] [Google Scholar]
- Dittmar K, Liberles DA. Evolution after gene duplication. Wiley-Blackwell; Hoboken, NJ: 2010. [Google Scholar]
- Edwards SV, Hedrick PW. Evolution and ecology of MHC molecules: From genomics to sexual selection. Trends in Ecology and Evolution. 1998;13:305–311. doi: 10.1016/s0169-5347(98)01416-5. [DOI] [PubMed] [Google Scholar]
- Eimes JA, Bollmer JL, Whittingham LA, et al. Rapid loss of MHC class II variation in a bottlenecked population is explained by drift and loss of copy number variation. Journal of Evolutionary Biology. 2011;24:1847–1856. doi: 10.1111/j.1420-9101.2011.02311.x. [DOI] [PubMed] [Google Scholar]
- Eizaguirre C, Yeates SE, Lenz TL, Kalbe M, Milinski M. MHC-based mate choice combines good genes and maintenance of MHC polymorphism. Molecular Ecology. 2009;18:3316–3329. doi: 10.1111/j.1365-294X.2009.04243.x. [DOI] [PubMed] [Google Scholar]
- Ejsmond MJ, Babik W, Radwan J. MHC allele frequency distributions under parasite-driven selection: a simulation model. BMC Evolutionary Biology. 2010;10 doi: 10.1186/1471-2148-10-332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekblom R, Saether SA, Grahn M, et al. Major histocompatibility complex variation and mate choice in a lekking bird, the great snipe (Gallinago media) Molecular Ecology. 2004;13:3821–3828. doi: 10.1111/j.1365-294X.2004.02361.x. [DOI] [PubMed] [Google Scholar]
- Fair J, Paul E, Jones J. Guidelines to the Use of Wild Birds in Research. 3rd. Ornithological Council; Washington, DC: 2010. [Google Scholar]
- Fawcett JA, Innan H. Neutral and non-neutral evolution of duplicated genes with gene conversion. Genes. 2011;2:191–209. doi: 10.3390/genes2010191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Felsenstein J. PHYLIP (Phylogeny Inference Package) Department of Genome Sciences, University of Washington; Seattle: 2005. Distributed by the author. [Google Scholar]
- Fitzpatrick C, Threlfall W. The ectoparasites of three species of seabirds from Newfoundland, Canada. Canadian Journal of Zoology. 1977;55:1205–1209. doi: 10.1139/z77-158. [DOI] [PubMed] [Google Scholar]
- Forsberg LA, Dannewitz J, Petersson E, Grahn M. Influence of genetic dissimilarity in the reproductive success and mate choice of brown trout - Females fishing for optimal MHC dissimilarity. Journal of Evolutionary Biology. 2007;20:1859–1869. doi: 10.1111/j.1420-9101.2007.01380.x. [DOI] [PubMed] [Google Scholar]
- Fricke EC, Blizzard KM, Gannon DP, Mauck RA. Model of burrow selection predicts pattern of burrow switching by Leach's Storm-petrels. Journal of Field Ornithology. (in press) in press. [Google Scholar]
- Fridolfsson A-K, Ellegren H. A simple and universal method for molecular sexing of non-ratite birds. Journal of Avian Biology. 1999;30:116–121. [Google Scholar]
- Gasparini C, Congiu L, Pilastro A. Major histocompatibility complex similarity and sexual selection: Different does not always mean attractive. Molecular Ecology. 2015;24:4286–4295. doi: 10.1111/mec.13222. [DOI] [PubMed] [Google Scholar]
- Goudet J. FSTAT, a program to estimate and test gene diversities and fixation indices. 2002 http://www2.unil.ch/popgen/softwares/fstat.htm.
- Grubb TC., Jr Olfactory navigation to the nesting burrow in Leach's petrel (Oceanodroma leucorrhoa) Animal Behaviour. 1974;22:192–202. doi: 10.1016/s0003-3472(74)80069-2. [DOI] [PubMed] [Google Scholar]
- Gu X, Nei M. Locus specificity of polymorphic alleles and evolution by a birth-and death process in mammalian MHC genes. Molecular Biology and Evolution. 1999;16:147–156. doi: 10.1093/oxfordjournals.molbev.a026097. [DOI] [PubMed] [Google Scholar]
- Haussmann MF, Mauck RA. Telomeres and longevity: testing an evolutionary hypothesis. Molecular Biology and Evolution. 2008;25:220–228. doi: 10.1093/molbev/msm244. [DOI] [PubMed] [Google Scholar]
- Hill AVS, Allsopp CEM, Kwiatkowski D, et al. Common West African HLA antigens are associated with protection from severe malaria. Nature. 1991;352:595–600. doi: 10.1038/352595a0. [DOI] [PubMed] [Google Scholar]
- Huchard E, Albrecht C, Schliehe-Diecks S, et al. Large-scale MHC class II genotyping of a wild lemur population by next generation sequencing. Immunogenetics. 2012;64:895–913. doi: 10.1007/s00251-012-0649-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huchard E, Baniel A, Schliehe-Diecks S, Kappeler PM. MHC-disassortative mate choice and inbreeding avoidance in a solitary primate. Molecular Ecology. 2013;22:4071–4086. doi: 10.1111/mec.12349. [DOI] [PubMed] [Google Scholar]
- Huntington CE, Butler RG, Mauck RA. Leach's Storm-petrel (Oceanodroma leucorhoa) In: Poole A, Gill F, editors. The Birds of North America, no 233. The Academy of Natural Sciences, Philadelphia, and The American Ornithologists' Union; Washington, DC: 1996. [Google Scholar]
- Jeffreys AJ, Kauppi L, Neumann R. Intensely punctate meiotic recombination in the class II region of the major histocompatibility complex. Nature Genetics. 2001;29:217–222. doi: 10.1038/ng1001-217. [DOI] [PubMed] [Google Scholar]
- Juola FA, Dearborn DC. Sequence-based evidence for major histocompatibility complex-disassortative mating in a colonial seabird. Proceedings of the Royal Society B: Biological Sciences. 2012;279:153–162. doi: 10.1098/rspb.2011.0562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalbe M, Eizaguirre C, Dankert I, et al. Lifetime reproductive success is maximized with optimal major histocompatibility complex diversity. Proceedings of the Royal Society B: Biological Sciences. 2009;276:925–934. doi: 10.1098/rspb.2008.1466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalinowski ST, Taper ML, Marshall TC. Revising how the computer program CERVUS accommodates genotyping error increases success in paternity assignment. Molecular Ecology. 2007;16:1099–1106. doi: 10.1111/j.1365-294X.2007.03089.x. [DOI] [PubMed] [Google Scholar]
- Kamiya T, O'Dwyer K, Westerdahl H, Senior A, Nakagawa S. A quantitative review of MHC-based mating preference: The role of diversity and dissimilarity. Molecular Ecology. 2014;23:5151–5163. doi: 10.1111/mec.12934. [DOI] [PubMed] [Google Scholar]
- Knafler GJ, Clark JA, Boersma PD, Bouzat JL. MHC diversity and mate choice in the magellanic penguin, Spheniscus magellanicus. Journal of Heredity. 2012;103:759–768. doi: 10.1093/jhered/ess054. [DOI] [PubMed] [Google Scholar]
- Kurtz J, Kalbe M, Aeschlimann PB, et al. Major histocompatibility complex diversity influences parasite resistance and innate immunity in sticklebacks. Proceedings of the Royal Society of London Series B-Biological Sciences. 2004;271:197–204. doi: 10.1098/rspb.2003.2567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemons PR, Marshall TC, McCloskey SE, et al. A likelihood-based approach for assessment of extra-pair paternity and conspecific brood parasitism in natural populations. Molecular Ecology Resources. 2015;15:107–116. doi: 10.1111/1755-0998.12287. [DOI] [PubMed] [Google Scholar]
- Lenz TL, Mueller B, Trillmich F, Wolf JB. Divergent allele advantage at MHC-DRB through direct and maternal genotypic effects and its consequences for allele pool composition and mating. Proceedings Biological sciences / The Royal Society. 2013;280:20130714. doi: 10.1098/rspb.2013.0714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighten J, van Oosterhout C, Paterson IG, McMullan M, Bentzen P. Ultra-deep Illumina sequencing accurately identifies MHC class IIb alleles and provides evidence for copy number variation in the guppy (Poecilia reticulata) Molecular Ecology Resources. 2014;14:753–767. doi: 10.1111/1755-0998.12225. [DOI] [PubMed] [Google Scholar]
- Mauck RA, Huntington CE, Doherty PF. Experience versus effort: what explains dynamic heterogeneity with respect to age? Oikos. 2012;121:1379–1390. [Google Scholar]
- Mauck RA, Waite TA, Parker PG. Monogamy in Leach's storm-petrel: DNA-fingerprinting evidence. Auk. 1995;112:473–482. [Google Scholar]
- Mauck RA, Zangmeister JL, Cerchiara JC, Huntington CE, Haussmann MF. Male-biased reproductive effort in a long-lived seabird. Evolutionary Ecology Research. 2011;13:19–33. [Google Scholar]
- Merino S, Martínez J, Masello JF, Bedolla Y, Quillfeldt P. First molecular characterization of a hepatozoon species (Apicomplexa: Hepatozoidae) infecting birds and description of a new species infecting storm petrels (Aves: Hydrobatidae) Journal of Parasitology. 2014;100:338–343. doi: 10.1645/13-325.1. [DOI] [PubMed] [Google Scholar]
- Milinski M. The major histocompatibility complex, sexual selection, and mate choice. Annual Review of Ecology, Evolution, and Systematics. 2006:159–186. [Google Scholar]
- Nei M, Rooney AP. Concerted and birth-and-death evolution of multigene families. Annual Review of Genetics. 2005;39:121–152. doi: 10.1146/annurev.genet.39.073003.112240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nevitt GA, Haberman K. Behavioral attraction of Leach's storm-petrels (Oceanodroma leucorhoa) to dimethyl sulfide. Journal of Experimental Biology. 2003;206:1497–1501. doi: 10.1242/jeb.00287. [DOI] [PubMed] [Google Scholar]
- Oliver MK, Telfer S, Piertney SB. Major histocompatibility complex (MHC) heterozygote superiority to natural multi-parasite infections in the water vole (Arvicola terrestris) Proceedings of the Royal Society B: Biological Sciences. 2009;276:1119–1128. doi: 10.1098/rspb.2008.1525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penn DJ, Damjanovich K, Potts WK. MHC heterozygosity confers a selective advantage against multiple-strain infections. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:11260–11264. doi: 10.1073/pnas.162006499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Queller DC, Goodnight KF. Estimating relatedness using genetic markers. Evolution. 1989;43:258–275. doi: 10.1111/j.1558-5646.1989.tb04226.x. [DOI] [PubMed] [Google Scholar]
- Ricklefs RE, Roby DD, Williams JB. Daily energy expenditure by adult Leach's storm-petrels during the nesting cycle. Physiological Zoology. 1986;59:649–660. [Google Scholar]
- Salzburger W, Ewing GB, Von Haeseler A. The performance of phylogenetic algorithms in estimating haplotype genealogies with migration. Molecular Ecology. 2011;20:1952–1963. doi: 10.1111/j.1365-294X.2011.05066.x. [DOI] [PubMed] [Google Scholar]
- Sandberg M, Eriksson L, Jonsson J, Sjöström M, Wold S. New chemical descriptors relevant for the design of biologically active peptides. A multivariate characterization of 87 amino acids. Journal of Medicinal Chemistry. 1998;41:2481–2491. doi: 10.1021/jm9700575. [DOI] [PubMed] [Google Scholar]
- Schwensow N, Eberle M, Sommer S. Compatibility counts: MHC-associated mate choice in a wild promiscuous primate. Proceedings of the Royal Society B: Biological Sciences. 2008;275:555–564. doi: 10.1098/rspb.2007.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepil I, Lachish S, Hinks AE, Sheldon BC. Mhc supertypes confer both qualitative and quantitative resistance to avian malaria infections in a wild bird population. Proceedings of the Royal Society B: Biological Sciences. 2013;280 doi: 10.1098/rspb.2013.0134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepil I, Radersma R, Santure AW, et al. No evidence for MHC class I-based disassortative mating in a wild population of great tits. Journal of Evolutionary Biology. 2015;28:642–654. doi: 10.1111/jeb.12600. [DOI] [PubMed] [Google Scholar]
- Sin YW, Annavi G, Newman C, et al. MHC class II-assortative mate choice in European badgers (Meles meles) Molecular Ecology. 2015;24:3138–3150. doi: 10.1111/mec.13217. [DOI] [PubMed] [Google Scholar]
- Spurgin LG, Richardson DS. How pathogens drive genetic diversity: MHC, mechanisms and misunderstandings. Proceedings of the Royal Society B: Biological Sciences. 2010;277:979–988. doi: 10.1098/rspb.2009.2084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strand T, Wang B, Meyer-Lucht Y, Höglund J. Evolutionary history of black grouse major histocompatibility complex class IIB genes revealed through single locus sequence-based genotyping. BMC Genetics. 2013;14 doi: 10.1186/1471-2156-14-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strandh M, Lannefors M, Bonadonna F, Westerdahl H. Characterization of MHC class I and II genes in a subantarctic seabird, the blue petrel, Halobaena caerulea (Procellariiformes) Immunogenetics. 2011;63:653–666. doi: 10.1007/s00251-011-0534-8. [DOI] [PubMed] [Google Scholar]
- Strandh M, Westerdahl H, Pontarp M, et al. Major histocompatibility complex class II compatibility, but not class I, predicts mate choice in a bird with highly developed olfaction. Proceedings of the Royal Society B: Biological Sciences. 2012;279:4457–4463. doi: 10.1098/rspb.2012.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: Molecular evolutionary genetics analysis version 6.0. Molecular Biology and Evolution. 2013;30:2725–2729. doi: 10.1093/molbev/mst197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Oosterhout C, Hutchinson WF, Wills DPM, Shipley P. MICRO-CHECKER: software for identifying and correcting genotyping errors in microsatellite data. Molecular Ecology Notes. 2004;4:535–538. [Google Scholar]
- Wegner KM, Kalbe M, Kurtz J, Reusch TBH, Milinski M. Parasite selection for immunogenetic optimality. Science. 2003;301:1343. doi: 10.1126/science.1088293. [DOI] [PubMed] [Google Scholar]
- Westerdahl H. No evidence of an MHC-based female mating preference in great reed warblers. Molecular Ecology. 2004;13:2465–2470. doi: 10.1111/j.1365-294X.2004.02238.x. [DOI] [PubMed] [Google Scholar]
- Worley K, Gillingham M, Jensen P, et al. Single locus typing of MHC class I and class II B loci in a population of red jungle fowl. Immunogenetics. 2008;60:233–247. doi: 10.1007/s00251-008-0288-0. [DOI] [PubMed] [Google Scholar]
- Zelano B, Edwards SV. An Mhc component to kin recognition and mate choice in birds: predictions, progress, and prospects. American Naturalist. 2002;160:S225–S237. doi: 10.1086/342897. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.