Abstract
Phenotypic plasticity is a key life history strategy used by many plants and animals living in heterogeneous environments. A multitude of studies have investigated the costs and limits of plasticity, as well as the conditions under which it evolves. Much less well understood are the molecular genetic mechanisms that enable an organism to sense its environment and respond in a plastic manner. The pea aphid wing polyphenism is a compelling laboratory model to study these mechanisms. In this polyphenism, environmental stressors like high density cause asexual, viviparous adult female aphids to change the development of their embryos from wingless to winged morphs. The life history tradeoffs between the two morphs have been intensively studied, but the molecular mechanisms underlying this process remain largely unknown. We therefore performed a genome-wide study of the maternal transcriptome at two time points with and without a crowding stress to discover the maternal molecular changes that lead to the development of winged versus wingless offspring. We observed significant transcriptional changes in genes associated with odorant binding, neurotransmitter transport, hormonal activity, and chromatin remodeling in the maternal transcriptome. We also found that titers of serotonin, dopamine, and octopamine were higher in solitary compared to crowded aphids. We use these results to posit a model for how maternal signals inform a developing embryo to be winged or wingless. Our findings add significant insights into the identity of the molecular mechanisms that underlie environmentally induced morph determination and suggest a possible role for biogenic amine regulation in polyphenisms generally.
Keywords: wing polyphenism, dopamine, serotonin, octopamine, crowding, transcriptional profiling
Introduction
Phenotypic plasticity is a life history strategy used by a variety of species that live in heterogeneous environments. This strategy is particularly widespread in insects, and has likely aided their enormous evolutionary success in surviving in a range of environments (Simpson, et al. 2011). More specifically, insects often display an extreme form of phenotypic plasticity called polyphenism, in which two or more distinct morphs can develop from the same genotype. Examples include different developmental stages of indirect developing insects, seasonal forms of some lepidopterans, and alternative adult morphs of ant castes (Mayr 1963).
Considerable study has addressed the factors that promote the evolution of plasticity versus genetic adaptation (Moran 1992b; Gavrilets and Scheiner 1993; Sultan and Spencer 2002) as well as the potential costs and limits of plasticity (e.g., DeWitt, et al. 1998; Murren, et al. 2015). These studies have greatly advanced our understanding of the ecological and evolutionary role of plasticity. Far less understood, however, are the molecular mechanisms that underlie phenotypic plasticity. Investigations to date have revealed that a variety of mechanisms are involved in alternative morph determination and functioning, such as changes in endocrine factors (Nijhout 1999, 2003), biogenic amines (Anstey, et al. 2009), epigenetic marks (Kucharski, et al. 2008; Simola, et al. 2016), and gene expression (Kijimoto, et al. 2012). Despite these excellent reports, the mechanisms that control polyphenic development are only known for a handful of taxa. Further study is necessary to advance an integrated understanding of how an organism’s developmental program can be environmentally induced to produce discrete, alternative phenotypes. Once this understanding is achieved, it will be possible to more clearly address important evolutionary questions about plasticity, such as why there are costs to plasticity, how plasticity evolves, and whether there are general mechanisms underlying plasticity across taxa (West-Eberhard 2003; Auld, et al. 2010). More broadly, determining how environmental responses occur at the molecular level will allow predictions to be made about how organisms will respond to future environmental alterations, such as those shaped by climate change.
Here we address the fascinating problem of how developmental processes are influenced by environmental cues to cause phenotypic diversity. We focus on the wing polyphenism of aphids, in which dramatically different winged and wingless morphs are produced depending on environmental conditions. Aphids have long been a premier model for investigating the causes and consequences of polyphenism (Dixon 1973; Hardie J. 1985; Moran 1992a). In the wing polyphenism, wingless asexual females produce genetically identical wingless and winged daughters. The wingless females maximize reproduction by investing more energy and resources into the production of offspring, while the winged females preferentially invest their resources into the machinery and fuel for flight (Dixon and Howard 1986). Wingless females are found under environmental conditions of low stress while winged females are produced in response to adverse living conditions such as high aphid densities, poor nutrition, and the presence of predators (Muller, et al. 2001).
The molecular mechanisms underlying the induction of winged versus wingless aphids remain unknown. Several transcriptional studies have identified gene expression differences between the winged and wingless morphs of nymphs or adults, but these studies targeted time points well after embryonic morph determination (Ghanim, et al. 2006; Brisson, et al. 2007; Brisson, et al. 2010; Yang, et al. 2014). Only one differential display study (Ishikawa, et al. 2012) has assayed gene expression changes during morph determination, identifying a handful of individual genes (Uba, McrNaca, and wingless), but not strongly implicating any candidate functional pathways.
Our objective was to perform the first genome-wide transcriptional profiling study on pea aphids (Acyrthosiphon pisum) during wing determination to identify the key molecular pathways involved in this process. Wing morph determination in this species occurs during embryogenesis (Sutherland 1969), which occurs in the ovary. We used crowding as a wing-inducing stimulus in this study. We hypothesized that crowded pea aphid females sense their high-density environmental state and, in response, set off a wave of transcriptional changes that ultimately determine the morphology of the embryos growing within them. Females continue to produce winged offspring for at least 24 hours after their crowding experience (Sutherland 1969), so we also anticipated that this signaling continues even after exposure to the wing-inducing cue has ended. Hence, we examined the maternal transcriptome for gene expression differences when female aphids were experiencing a crowded or uncrowded environment, as well as a later time when they were producing winged or wingless offspring. Our results contribute significant insights into the molecular genetic basis of how an organism relays environmental signals to its developing embryos, and advance the pea aphid as a model for understanding the molecular mechanisms that trigger alternative morph determination.
Material and Methods
Insect rearing and experimental setup
The pea aphid (Acyrthosiphon pisum) line used in this study was ROC-1, collected from Rochester, New York, in 2008. Aphids were reared in cages on fava (Vicia fabae) seedlings. Stocks of the ROC-1 clone were reared in an incubator at long day conditions (16L: 8D,18°C) and relative humidity of 30+/− 5% on fava seedlings. Under these conditions, aphids reproduce asexually. Prior to the start of all the experiments, females were maintained at low density (three per plant) on fava seedlings for three generations to eliminate cross generational effects on offspring morph determination (Sutherland 1969). The females used for the below experiments were adults that had begun larvipositing within the previous 24 hours, approximately three days after the final molt.
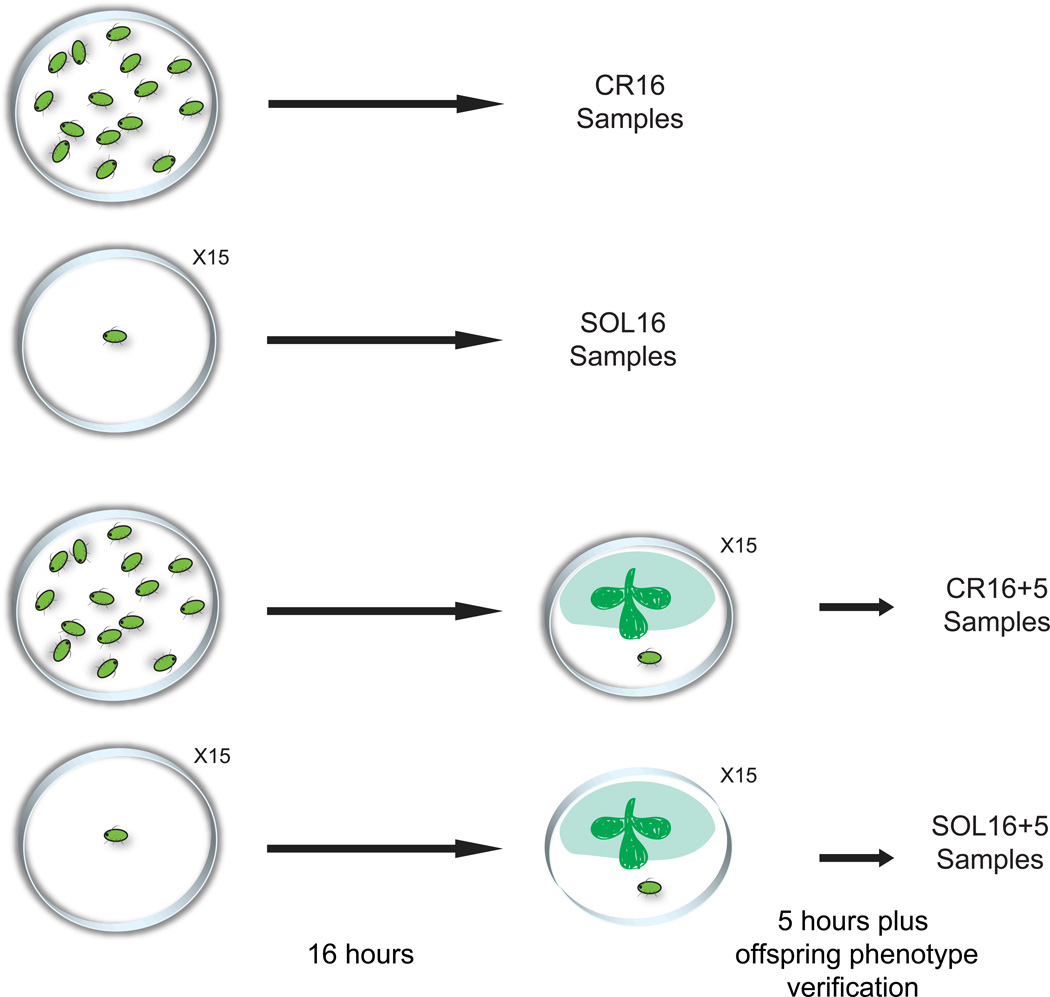
The wing-inducing crowding treatment was applied by placing 15 adult females together in a small (10mm) Petri dish without food. Starvation is necessary because it causes aphids to be mobile, and aphids produce winged offspring when they move around and come in contact with one another (Sutherland 1969). An equal number of adult females were placed individually in Petri dishes without food. This was the solitary treatment. Adult females were subjected to the crowding or solitary treatments for 16 hours. Four sets of crowded and solitary females (15 aphids each) were collected immediately after these 16-hour treatments. Females were dissected to remove and discard ovaries with their developing embryos, and the remaining maternal tissue was stored in TRI Reagent (Molecular Research Centre, Inc.) at −80°C. These are the ‘CR16’ and ‘SOL16’ samples.
Additional sets of 16-hour crowded and solitary females were transferred to Petri dishes, one per plate, with Medicago arborea leaves, and allowed to produce nymphs for five hours. After this five hours of feeding, females were dissected to remove and discard developing ovaries with their embryos, and the remaining maternal tissue was stored separately in TRI Reagent (Molecular Research Centre, Inc.) at −80°C. The nymphs produced in the five hours were allowed to develop, phenotyped as winged or wingless, and the percentage of winged offspring per adult was determined. Only adults producing 100% wingless offspring among the solitary treated aphids were selected and grouped as ‘SOL16+5’ samples. Females that experienced the crowding cue and produced more than 80% winged offspring were selected and grouped as ‘CR16+5’ samples. The complete set of experiments was repeated five times until four replicates of SOL16+5 and CR16+5 samples (each with 15 aphid carcasses) were collected. Figure 1 illustrates this sampling. Note that this experiment allowed us to utilize samples that were verified for their offspring phenotypes, but a potential complication is that crowded aphids were moved to a solitary environment (one per plate) in order to phenotype their offspring. In contrast, solitary aphids stayed in a solitary environment. The crowded to solitary transfer may have induced changes in gene expression that were not induced in the solitary to solitary transfer. We do not expect this to have had a large effect of gene expression, especially given that pea aphids are not social aphids.
Figure 1. Aphid sample collection for RNA-Seq.
As detailed in the Methods, two sets of aphids were used in this study. First, aphids were subjected to a solitary or crowded environment for 16 hours to cause them to produce mainly winged or wingless offspring, respectively. Second, after 16 hours of solitary or crowded environments, some aphids were returned, individually, to a leaf to feed and deposit offspring for five hours. Only females producing greater than 80% winged offspring (CR16+5) from the crowding treatment or 0% winged offspring (SOL16+5) from the solitary treatment were used for transcriptional profiling. For all samples, only aphid carcasses were processed for RNA extraction; ovaries plus embryos were discarded. Four biological replicates were used for RNA-Seq experiments.
RNA isolation, cDNA synthesis and Illumina library preparation
RNA was isolated from pooled maternal tissue (n=15 for each biological replicate) stored in TRI reagent using the RNeasy Kit (Qiagen, CA) according to manufacturer’s instructions. RNA was checked for purity and integrity using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). cDNA libraries were sequenced on the Illumina HiSeq2500 system. Four biological replicates per condition (CR16, SOL16, CR16+5, SOL16+5; total of 16 libraries) were sequenced in four lanes.
Gene expression analyses
Reads were mapped against the pea aphid reference transcriptome v.2.1 using Arraystar v5 (DNASTAR). If 97% of the bases in a read matched, it was aligned to the reference transcriptome. Reads aligning to more than one transcript equally were excluded. Genes with low read counts (less than an average of 10 total counts per sample) were excluded from analysis. The read count data were exported to DESeq2v1.6 (Love, et al. 2014) to identify differentially expressed genes using the negative binomial distribution from normalized read count data. Pairwise comparisons were made between CR16 and SOL16, CR16+5 and SOL16+5, and CR16/SOL16 and CR16+5/SOL16+5. A gene was considered significantly differentially expressed if its corresponding FDR corrected P value (Benjamini and Hochberg 1995) was ≤0.05 and it had a fold change equal or greater than 1.7. Enrichment analysis for GO terms using Blast2GO v.2.6.0 was performed (Conesa, et al. 2005) with all expressed genes as the reference set and the differentially expressed genes as the test set. GO terms with FDR corrected P values ≤0.1 were considered enriched and the “reduce to most specific” function was used to produce tables of enriched GO terms. Genes associated with biogenic amine pathways, as identified using BLASTx searches, were clustered using GENE-E v. 3.0.204. Reads have been submitted to NCBI’s Sequence Read Archive under accession number SRP056026.
Validation of RNA-seq data with real time quantitative PCR
Quantitative real time PCR (qRT-PCR) was used to verify the RNA-Seq results. 11 differentially expressed transcripts were selected for validation based on their putative biological roles. One µg of RNA from the samples used to prepare the RNA-Seq library was reverse transcribed using random hexamer primers (Invitrogen) and Superscript II reverse transcriptase (Life Technologies, Inc.). Reactions were performed on a Real-Time PCR system 7500 (Applied Biosystems) using SYBR green PCR master mix (Applied Biosystems). Primers were designed with Primer3plus (Untergasser, et al. 2007) using the associated gene sequence (Table S1). Primer specificity was verified prior to qRT-PCR using disassociation curve analysis. Three potential reference genes [ACYPI008480 (mediator of RNA polymerase II transcription subunit 27), ACYPI084777 (uncharacterized transcript) and ACYPI009769 (glyceraldehyde-3-phosphate dehydrogenase)] were tested for expression stability across all samples using Normfinder v0.953 (Andersen, et al. 2004). ACYPI008480 was selected as the reference gene for qRT-PCR because of its relative expression stability across samples. Each biological replicate was measured with three technical replicates and fold change was calculated using the 2−ΔΔCT method (Livak and Schmittgen 2001). The correlation between the expression values obtained by RNA-Seq and qRT-PCR was calculated using Pearson’s correlation coefficient.
Whole body biogenic amine titer determination
Three samples of 10 aphids were directly removed from food to comprise the zero hour control. The remaining aphids were placed into ten groups of crowded or solitary conditions for 24 hours. Crowded groups were 10 aphids in a single 10mm Petri dish without a food source, while solitary aphids consisted of 10 aphids in separate Petri dishes, also without food. Aphid whole bodies were collected, weighed, and flash frozen in liquid nitrogen. They were homogenized and suspended in 100µl of acidified water (0.1% formic acid), followed by the addition of 100µl of acidified cold methanol. After centrifugation, the supernatant was dried in a speed vacuum and resuspended in acidified water. LC-MS/MS analysis was carried out in multiple reaction mode, in the positive ion mode, using the Agilent LC1200 HPLC system (Agilent, USA) integrated with a mass spectrometer (Q Trap 4000, AB Sciex). The source conditions include IP = 5500V, temperature = 500°C, GS1 = 50, GS2 = 20, and curtain gas = 30. The sample was separated using reverse phase chromatography employing a C18 column (Thermo) and a gradient method using mobile phase A (water containing 0.1% formic acid) and mobile phase B (acetonitrile containing 0.1% formic acid). External calibration was carried out for the individual compounds from 0.01pg/µl to 500pg/µl with r=0.9995 for each compound. Data were analyzed using Analyst 1.4.2. Statistical significance of the differences in the biogenic amine levels between solitary and crowded aphids was calculated using Mann-Whitney U-tests.
Results
Aphids respond as a group to crowding, but individuals exhibit variability in their polyphenic response
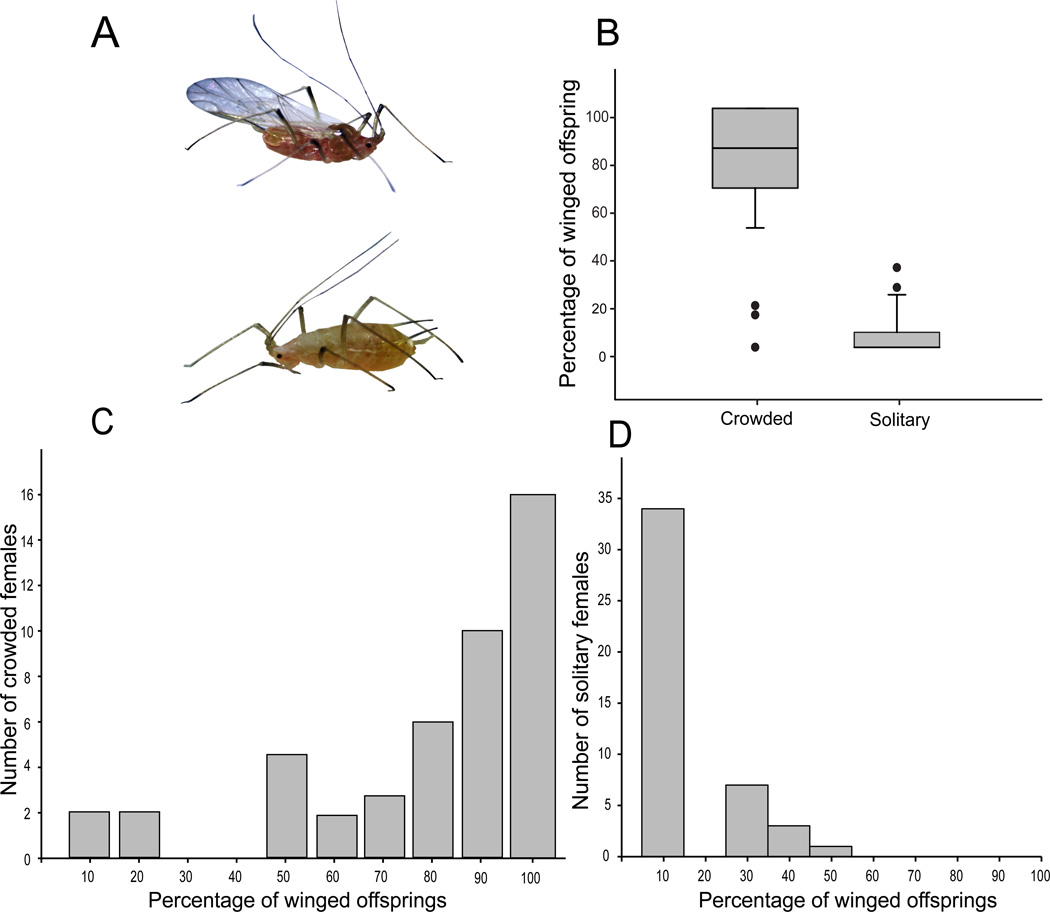
Figure 2 shows the percentage of winged and wingless (A) offspring produced for the five hours after 16 hours of crowding or solitary treatments (B), with data collected from 45 aphid mothers for each treatment, all of the same genotype. The median number of winged offspring produced from solitary aphids was 0%, while the median for the crowded aphids was 83%. We also re-plotted these same data on an aphid-by-aphid basis to more explicitly visualize the polyphenic response exhibited by individual aphids. We observed a large variability in the percentage of winged offspring produced by individual crowded females, ranging from 0% to 100% (Figure 2C). Far less variability was found among the solitary aphids (Figure 2D). This difference in variability was significant (Levene’s test, P = 6E−5).
Figure 2. Aphids respond to a crowding stimulus by producing a greater percentage of winged offspring.
A) The winged and wingless asexual female aphids exhibit dramatic phenotypic differences. B) The percentage of winged offspring produced in the five hours after 16 hours of a solitary or crowding treatment are significantly different (Mann-Whitney U test P = <0.001). Data shown are the phenotypes of the offspring from 45 aphids, with the crowded aphids treated in groups of 15. Boxes represent the interquartile range and the line the median value of each group. Black circles represent outliers. C) and D) show the production of winged offspring from individual females after 16 hours of crowding (C) or solitary treatment (D). The data used in (B) are the same data used in (C) and (D).
To determine whether females continued to produce winged offspring after this initial five hour period, in a separate experiment we treated aphids with a solitary or crowded treatment for 16 hours and then collected their offspring for the next 24 hours. The results are quantitatively identical (Supplemental Figure 1), with no differences in the percentage of winged offspring produced for the five hours after a 16 hour treatment versus 24 hours after a 16 hour treatment (Mann-Whitney U tests, solitary comparison P = 0.19, crowded comparison P = 0.57).
Clustering identifies global patterns in gene expression changes
To identify the maternal transcriptional changes that mediate the pea aphid’s polyphenic response, we performed RNA-Seq profiling on two sets of treated aphids (Figure 1). The first set was aphids that had been subjected to a crowded or solitary environment for 16 hours, called ‘CR16’ and ‘SOL16’, respectively. The second set of aphids had received the crowding or solitary treatment for 16 hours, after which they fed and larviposited for five hours. For this latter set of aphids, we only used females producing greater than 80% winged offspring (our ‘CR16+5’ samples) or 0% winged offspring (our ‘SOL16+5’ samples) for expression profiling. RNA was isolated from the maternal tissues only; the ovaries containing the embryos were removed.
Sequencing of these maternal transcriptome samples yielded more than one billion, 57 to 100-mer single end high quality reads from a total of 16 samples (four replicates each CR16, SOL16, CR16+5, SOL16+5). The average number of raw reads per sample was 97 million. The average mapping percentage of the raw reads to the reference transcripts was 71%. The pairwise correlations between the replicates were very high (r > 0.94) indicating high reproducibility between the replicates.
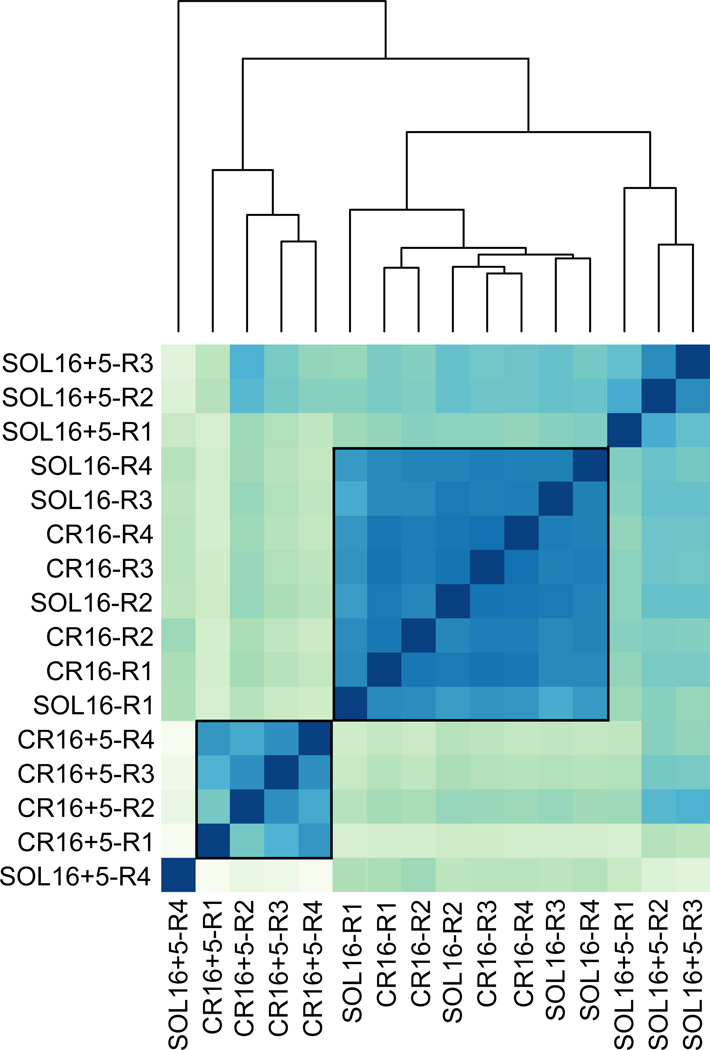
Euclidian distances between samples were calculated using DESeq2v1.6 to examine general patterns of response to the different treatments. Clustering resulted in three, not the expected four, groups of data (Figure 3). The first cluster was composed of the CR16 and SOL16 samples, with no apparent systemic differences between treatments. Second, the CR16+5 samples clustered together. And finally, the SOL16+5 aphids produced a cluster that was more similar to the CR16 and SOL16 samples (except for one replicate which did not group with the others). Overall, the females that were verified as actively producing winged offspring (CR16+5 samples), had the most divergent transcriptomes compared to the other samples (see tree diagram at the top of Figure 3).
Figure 3. Hierarchical clustering of all experiments.
A heat map of the clustered sample distances indicates that CR16 and SOL16 females form a distinct group, as do the CR16+5 females (enclosed in boxes). The SOL16+5 females are the least unified. All four replicates from each experiment are shown (R1-R4). The dendrogram illustrates Euclidian sample-to-sample distances calculated from rlog transformed count data. Darker colors indicate higher correlations among experiments.
Several comparisons were performed to identify differentially expressed genes (see below). 11 of the differentially expressed genes were further analyzed by qRT-PCR to validate the RNA-Seq results (Table S1). We observed a high correlation between the qRT-PCR results and the RNA-Seq expression values (Pearson correlation coefficient, r = 0.95, P <0.001) with the RNA-Seq data (Supplemental Figure 2).
A small number of transcripts differ between CR16 and SOL16 females
The clustering results indicated that the transcriptional profiles of aphids that have been crowded or kept solitary for 16 hours were similar. Indeed, of the 16,438 expressed genes in these samples, only 16 genes were significantly differentially expressed between the CR16 and SOL16 samples. All 16 genes were found at higher levels in the CR16 samples (Table S2). No significantly enriched GO terms were identified.
Large transcriptional changes accompany the production of winged versus wingless offspring
We also compared the transcriptomes of aphids that either were or were not producing winged offspring, as verified by allowing them to lay offspring for five hours after subjecting them to a crowded or solitary environment for 16 hours. A large number of differentially expressed genes were observed in these CR16+5 versus SOL16+5 females (all four replicates for each treatment were used for the differential expression analysis). Of the 16,308 expressed genes in the two sets of samples, 3,515 genes were differentially expressed. 1,509 genes had higher expression in the CR16+5 samples relative to the SOL16+5 samples, and 2,006 genes had lower expression (Tables S3 & S4). Significantly enriched Gene Ontology (GO) categories for the genes with higher expression included functions related to odorant binding, antioxidant activity, hormone binding, translation initiation activity, NADH dehydrogenase (ubiquinone) activity, and peroxiredoxin activity, among others (Table S5). For the genes with lower expression, enriched GO terms included central nervous system development, DNA binding, G-protein coupled receptor activity, chromatin modification, regulation of behavior, ecdysone receptor mediated signaling pathway, synapse organization, chemosensory behavior, and neurotransmitter transport (Table S6). GO terms of particular interest are presented in Table 1 and will be considered in the Discussion.
Table 1.
Selected enriched GO terms and related genes differentially expressed between CR16+5 and SOL16+5 samples
| Locus | Gene | RNAseq P value* |
Log2 fold change** |
|---|---|---|---|
| GO:0005549 | Odorant binding | ||
| ACYPI084264-RA | AP-3 complex subunit sigma-2 | 4.71E-02 | 1.9X up |
| ACYPI31683-RA | Odorant binding protein 10 | 2.34E-04 | 2.6X up |
| ACYPI001753-RA | Odorant binding protein 3 | 7.33E-03 | 2X up |
| ACYPI006495-RA | Odorant binding protein 6 | 1.67E-02 | 2.1X up |
| ACYPI000334-RA | Odorant binding protein 9 | 1.67E-02 | 2.5X up |
| ACYPI003223-RA | Odorant binding protein 1 | 1.04E-02 | 2X up |
| ACYPI006147-RA | Odorant binding protein 2 | 3.79E-02 | 1.7X up |
| ACYPI083147-RA | Odorant binding protein 3 | 2.05E-03 | 2.5X up |
| ACYPI008889-RA | Odorant binding protein 4 | 1.99E-04 | 2.8X up |
| ACYPI003731-RA | Odorant binding protein 8 | 4.52E-02 | 1.7X up |
| GO:0006836 | Neurotransmitter transport | ||
| ACYPI008660-RA | Dopamine transporter | 1.41E-03 | 2.7X down |
| ACYPI080057-RA | Dopamine transporter | 6.59E-08 | 2.4X down |
| ACYPI006527-RA | GABA receptor | 8.38E-13 | 2.9X down |
| ACYPI002046-RA | GABA-gated chloride channel subunit | 1.67E-06 | 2.2X down |
| ACYPI063378-RA | Neurexin | 1.54E-04 | 1.4X down |
| ACYPI005021-RA | Synaptosomal-associated protein 25 | 1.65E-05 | 2Xdown |
| ACYPI005967-RA | Syntaxin 1a | 1.61E-04 | 1.4X down |
| ACYPI008123-RA | Syntrophin | 9.67E-04 | 1.4X down |
| ACYPI005299-RA | Vesicular acetylcholine transporter | 3.58E-13 | 2.7X down |
| ACYPI43841-RA | Voltage-dependent calcium channel type a subunit alpha-1 |
1.36E-05 | 1.9Xdown |
| ACYPI005426-RA | Voltage-dependent calcium channel type a subunit alpha-1 |
1.79E-06 | 1.8X down |
| GO:0005179 | Hormonal activity | ||
| ACYPI005281-RA | Bursicon | 1.40E-02 | 1.9X up |
| ACYPI062598-RA | CCAP | 3.50E-03 | 2.2X up |
| ACYPI002403-RA | Diuretic hormone class2 | 2.61E-02 | 1.8X up |
| ACYPI42083-RA | Eclosion hormone | 3.80E-04 | 2.6X up |
| ACYPI085492-RA | Eclosion hormone | 1.24E-04 | 2.9X up |
| ACYPI007510-RA | Prothoracicostatic peptide precursor (PTSP) | 1.38E-02 | 1.9X up |
| ACYPI009416-RA | TPA glycoprotein hormone beta 5 | 4.11E-02 | 1.8X up |
| GO:0035076 | Ecdysone receptor-mediated signaling pathway | ||
| ACYPI001312-RA | Chromatin-remodeling complex atpase chain iswi | 4.65E-02 | 3.7X down |
| ACYPI004047-RA | Chromatin-Remodeling complex atpase chain iswi | 5.87E-04 | 2.5X down |
| ACYPI001692-RA | Ecdysone receptor | 8.68E-07 | 3.3X down |
| ACYPI005934-RA | Ultraspiracle protein | 2.04E-03 | 1.1X down |
| GO:0006338 | Chromatin remodeling | ||
| ACYPI001312-RA | Chromatin-remodeling complex atpase chain iswi | 4.65E-02 | 3.7X down |
| ACYPI004047-RA | Chromatin-remodeling complex atpase chain iswi | 5.87E-04 | 2.5X down |
| ACYPI56610-RA | Chromodomain-helicase-dna-binding protein mi-2 homolog |
1.11E-04 | 2.6X down |
| ACYPI008000-RA | Histone-arginine methyltransferase carmer-like (CARM-1) |
2.21E-03 | 2.2X down |
| ACYPI005751-RA | Protein hira homolog | 6.48E-03 | 2.3X down |
| ACYPI001480-RA | Protein polybromo-1 | 3.73E-02 | 1.8X down |
| ACYPI008028-RA | Swi snf-related matrix-associated actin-dependent regulator |
2.51E-05 | 9.0X down |
FDR corrected
Fold change compared to SOL16+5 samples (calculated using DESeq2 using read count data)
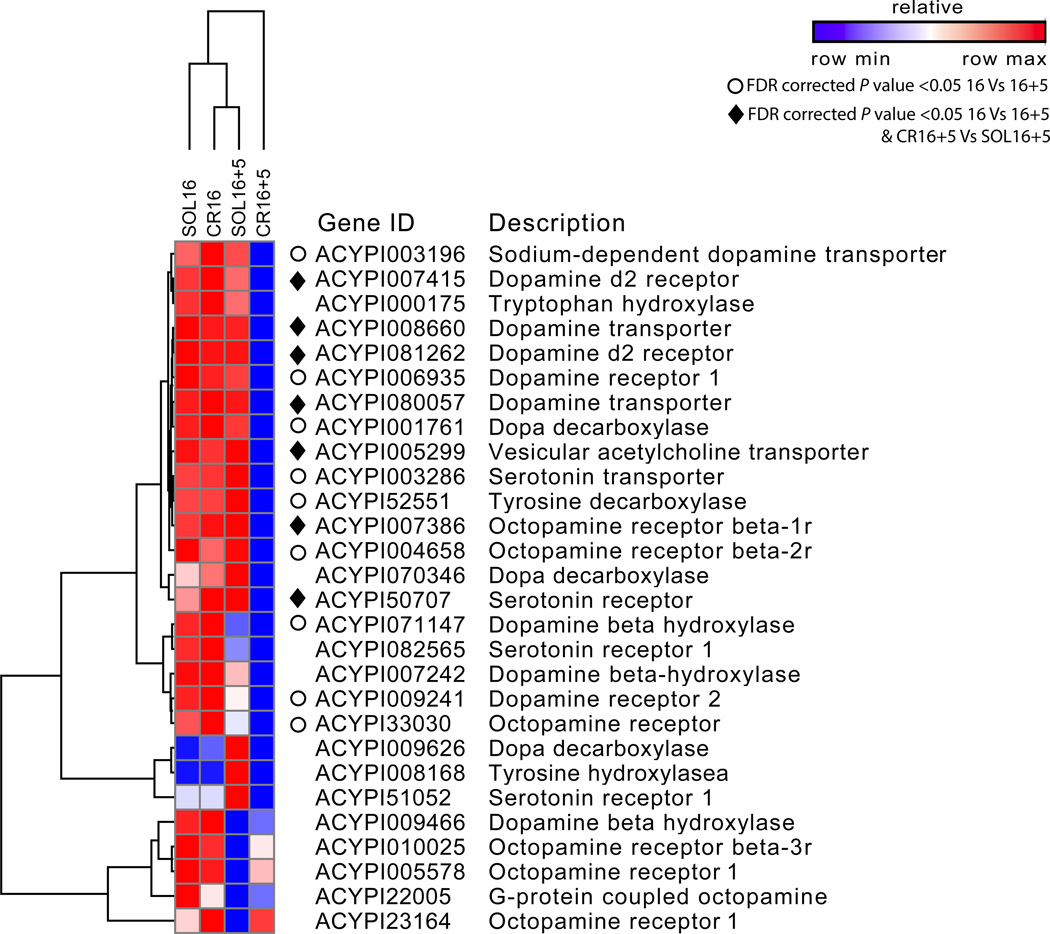
As just mentioned, GO terms associated with the nervous system were enriched in the genes with lower expression in the CR16+5 samples. Closer inspection of the genes with these GO terms revealed genes associated with dopamine, such as two dopamine transporters (ACYPI008660, ACYPI080057). Dopamine, as well as serotonin and octopamine, are biogenic amines known to play roles in regulating aspects of the locust phase polyphenism (Wang and Kang 2014). We were therefore interested in investigating whether they were potentially involved in controlling the pea aphid wing polyphenism. We examined the expression levels of genes associated with the synthesis, transport, and reception of these three compounds from our RNA-Seq data. We clustered their expression patterns across all four conditions using simultaneous hierarchical clustering of genes and experiments. The results were quite striking (Figure 4): the wing-producing, CR16+5 sample consistently showed the lowest expression values for the majority of these genes, indicating that these neurotransmitters are differentially regulated between wing-producing and wingless-producing females (CR16+5 and SOL16+5, respectively). Additionally, approximately half of these genes exhibited higher expression levels in the 16 hour (CR16 and SOL16) relative to the 16+5 (CR16+5 and SOL16+5) hour time point. Recall that aphids in the 16 hour time point are starved, while aphids in the 16+5 hour time point are on food. These latter results indicate that these biogenic amines are an important component of the stress response to starvation in aphids, as has been observed in other systems (Hirashima, et al. 1993; Neckameyer and Weinstein 2005; Mayack and Naug 2015).
Figure 4. Induced aphids have lower expression levels of genes associated with dopamine, serotonin, and octopamine synthesis and signaling pathways.
The expression levels of genes associated with the production and signaling of biogenic amines were clustered via GENE-E using average FPKM expression values for each of the four experiments. Clustering is by experiment and by gene using Pearson correlation coefficients. Expression values are relative to one another within each gene, with red the highest and blue the lowest. ♦ indicates genes that are significantly differentially expressed between the starved 16 hour time point and the fed 16+5 hour time point (16hr samples vs. 16+5hr samples) as well as between the crowded and solitary samples at the 16+5 hour time point (Cr16+5 Vs Sol16+5 samples). ○ indicates genes with significant differences in expression only between the starved 16 hour time point and the 16+5 hour time point comparison (16hr samples vs. 16+5hr samples).
Dopamine, serotonin, and octopamine titers differ between crowded and solitary aphids
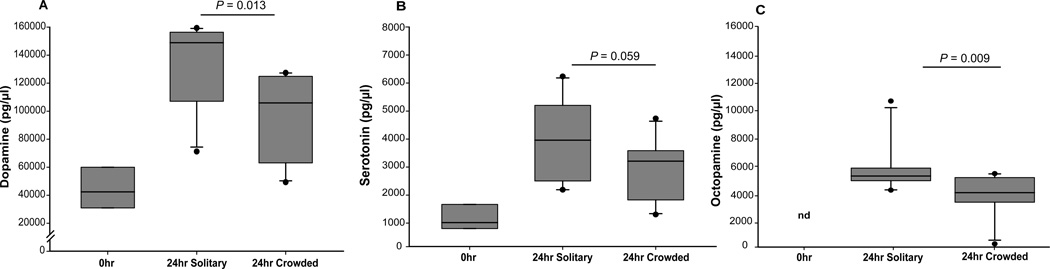
To follow up on these biogenic amine gene expression results, we used LC-MS/MS to quantify whole body titers of serotonin, dopamine, and octopamine. We used three sets of samples: 1) aphids that had been directly removed from leaves (a pre-treatment, 0-hour), 2) aphids that had been exposed to a 24 hour solitary treatment, and 3) aphids that had experienced a 24 hour crowding treatment. Aphids treated with 24 hours of crowding produce a high proportion of winged offspring, comparable to aphids subjected to 16 hours of crowding (Supplemental Figure 3). For all three biogenic amines, the 0-hour treatment had the lowest values, suggesting that these biogenic amine titers are increased following stress in pea aphids (Figure 5). The solitary treatment exhibited significantly higher levels than crowded for both dopamine and octopamine titers, and a near-significantly higher level for serotonin (Mann-Whitney U test, n = 10, P = 0.013 for dopamine, and P = 0.009 for octopamine, P = 0.059 for serotonin). Note that this experiment was likely affected by the same variability of individual aphid polyphenic response noted above in the 16 hour crowded and solitary treatments (Figure 2C & D). We thus would expect larger titer level differences between aphids that had been verified for whether they were producing winged or wingless offspring, which we did not do for this experiment. It is also likely that the additional eight hours of treatment (24 hours versus 16 hours) resulted in a later progression into the wing induction response.
Figure 5. Whole body titers of dopamine, serotonin, and octopamine differ between crowded and solitary treatments.
0hr = aphids taken directly from plants, n = 3 sets of 10 aphids each; 24hr Solitary = solitary aphids for 24 hours, n = 10 sets of 10 aphids each; 24hr Crowded = crowded aphids for 24 hours, n = 10 sets of 10 aphids each. Dots are outliers. The 0hr aphid levels of each biogenic amine were highly significantly different than the S and C treatments for dopamine and serotonin, while octopamine was not detected (“nd”) in the 0hr aphids. P values associated with the differences between the solitary and crowded treatments (Mann-Whitney U test) are noted.
A large transcriptional response occurs when aphids are released from starvation
We also examined how the maternal transcriptome changes after aphids were transferred from starvation onto leaves. To do this, we compared the CR16 and SOL16 females, combined as a single data set with eight replicates, to the CR16+5 and SOL16+5 females, also combined. Of the 17,954 expressed genes, 4,121 were at higher levels when the aphids were feeding at the 16+5 hour time point compared to the starved 16 hour time point, while 3,962 were at lower levels (Tables S7 & S8). Thus, a remarkably large percentage (45%) of the expressed genes were significantly differentially expressed in this comparison, indicating that these treatments represented drastic changes for the aphids; conditions that required massive gene expression responses. Genes with higher expression in the feeding aphids had enriched GO terms related to protein synthesis and metabolic processes (Table S9), while genes with lower expression in the feeding aphids encompassed a diverse array of terms (Table S10). This large transcriptional response is consistent with myriad previous gene expression examinations of organisms undergoing a starvation stress (e.g., Harbison, et al. 2005; Rion and Kawecki 2007; Moskalev, et al. 2015; Price, et al. 2015), including a recent study of the starvation response in the soybean aphid, Aphis glycines (Enders, et al. 2015).
Discussion
Here we use the pea aphid wing polyphenism system to investigate the molecular mechanisms underlying phenotypic plasticity. Our genome-wide RNA-Seq analysis was aimed at identifying the transcriptional response of adult, asexual female aphids as they experience a crowding stress in their environment and then differentially produced winged or wingless offspring. We envision that wing induction is a multi-step process, with females sensing their environment, turning that environmental stimulus into a molecular signal, and then sending that signal to their embryos. The differentially expressed genes that we identified here could be involved in any of these processes, or downstream of these processes. We discuss our results within this context below.
Surprisingly, we found that aphids exposed to a crowded or solitary environment for 16 hours showed very few gene expression differences (only 16 significantly differentially expressed genes identified between the CR16 and SOL16 females, Table S2; also see Fig. 3). One of these 16 genes is ACYPI010137, an arylsulfatase. This gene had over two-fold higher expression in the CR16 relative to SOL16 aphids. Interestingly, an arylsulfatase called eud-1 in Pristionchus pacificus nematodes is the developmental switch that controls a mouth morphology feeding polyphenism (Ragsdale, et al. 2013). In these worms, eud-1 is expressed in neurons and is speculated to act upon a steroid hormone to control the alternative morphologies. Like the aphid wing polyphenism, this feeding polyphenism is modulated by the environmental cue of crowding (Serobyan, et al. 2013). Very few genes controlling polyphenic switches are known, so it is quite remarkable that our gene expression analysis uncovered an arylsulfatase (one of five paralogs in the pea aphid genome with putative orthology to eud-1) as one of only a handful of differentially expressed genes. Further experiments will address whether this gene acts as a developmental switch in the pea aphid wing polyphenism.
The small number of genes discovered with this analysis indicates that the initial stages of the polyphenic response are the result of modest changes in gene expression. Alternatively, it is possible that we are underestimating the gene expression changes due to a biological property of the polyphenism that we uncovered in our experiments. Our crowding cue was sufficient to produce a high production of winged offspring by a group of aphids (more than 80% winged offspring, Figure 2B), but we found that individual aphids were highly variable in terms of the percentage of winged offspring they produced, ranging from 17 to 100% (Figure 2C). So even though we treated the aphids equally, individual aphids may not have each received the same level of wing-inducing signal. Tactile stimulation is likely the predominant mode of how aphids are induced (Sutherland 1969), and individual aphids may have received different amounts of stimulation during the 16 hours of crowding. This differential cue reception despite equal treatment may be part of a ‘stochastic polyphenism’ strategy in which once induced, aphids produce a stochastic output of winged or wingless morphs. This type of bet-hedging strategy is thought to be used in cases where environmental predictability is variable (Walker 1986). Regardless of the cause, the variance in the proportion of winged offspring production by individual aphids likely led to variance in transcriptional profiles within treatments because each of our biological replicates included 15 aphids. This could have resulted in a low number of significantly differentially expressed genes between aphids that were crowded or solitary for 16 hours.
In contrast, we identified a large number of genes (3,515) significantly differentially expressed between the females that were verified as producing predominantly winged or only wingless offspring, the CR16+5 and SOL16+5 comparison. For this experiment, we removed the confounding individual variation in polyphenic response by selecting only females producing 0% (CR16+5) or over 80% winged offspring (SOL16+5). This likely increased the transcriptional similarities among the 15 aphids within each biological replicate and led to the discovery of the differentially expressed genes. Based on the transcriptional profiling results of these aphids, we propose a molecular model that illustrates the events following the crowding cue that leads to the development of winged versus wingless offspring. This model, as yet untested and meant as a guide for future research efforts, is illustrated in Figure 6 and described below.
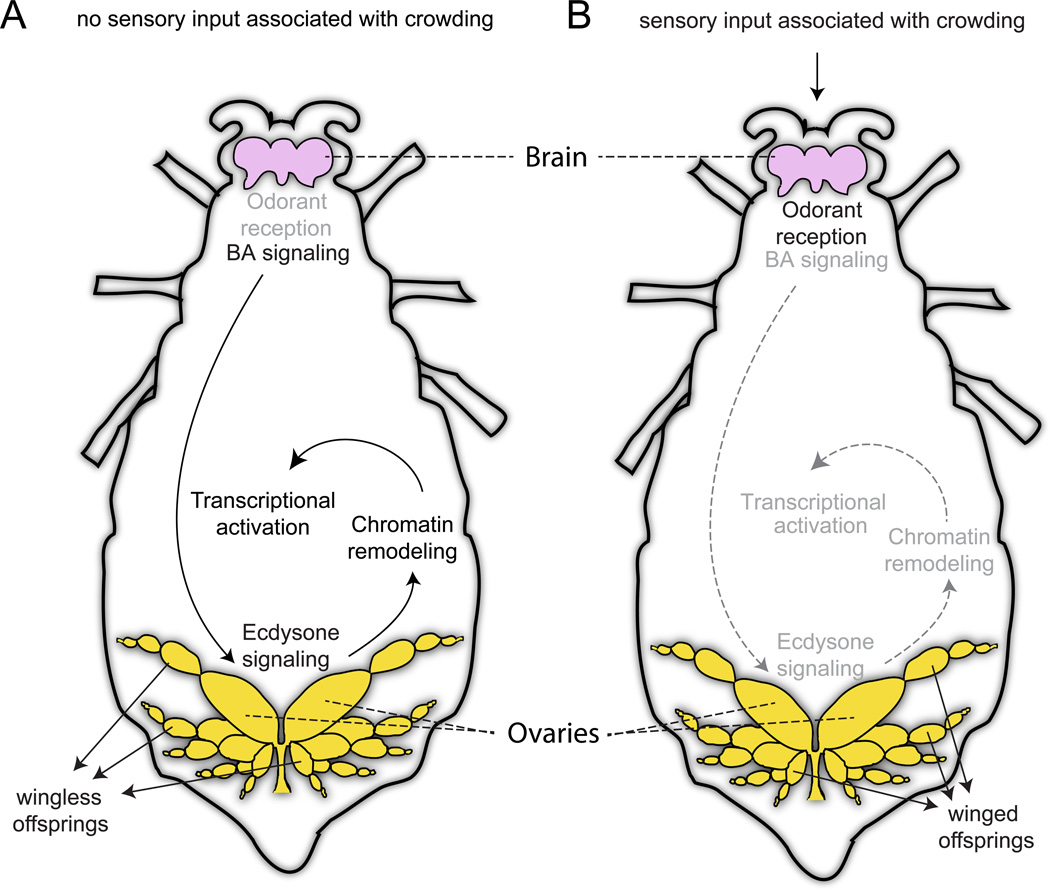
Figure 6. A proposed model for how the maternal crowding stress leads to the development of winged offspring in pea aphids.
A: Uninduced, solitary female aphid. B: Induced, crowded female aphid. Pathways highlighted in black are activated and those in grey are inactivated. In uninduced females, environmental cues do not activate OBP expression. The brain produces biogenic amines that instruct the ovaries to produce ecdysone. Ecdysone signaling causes transcriptional activation via chromatin remodeling. The outcome of these events is the production of wingless offspring. In contrast, in crowded females, unknown chemical cues activate OBP signaling. This ultimately results in downregulation of ecdysone signaling. Without ecdysone-induced transcriptional changes, the default developmental pathway of winged offspring (Ishikawa, et al. 2008) production is followed. This model was constructed based on the gene expression results; components of the model await future testing.
The first component of our model involves sensing the differential environments. Our model posits that transcriptional changes following crowding are likely initiated, at least in part, by odorant perception and associated changes in the brain. In particular, we observed a striking difference in odorant binding protein (OBP) gene expression between the CR16+5 and SOL16+5 females, with nine of the 15 OBP genes in the pea aphid genome (Zhou, et al. 2010) expressed at higher levels in CR16+5 females (Table 1). OBPs are water soluble, extracellular proteins abundant in the sensillum lymph of antennae and other nonsensory organs that carry hydrophobic odorant molecules to odorant receptors (Leal 2013). Insects perceive stressful, crowded conditions either chemically through pheromones (Blum 1985) or mechanically when they come in contact with each other (Simpson, et al. 2001). Early reports on the aphid wing polyphenism emphasized the importance of tactile stimulation in inducing winged offspring production (Sutherland 1969), while more recent reports support a possible role for chemical detection of density in aphids: higher alarm pheromone production is observed by aphids reared in groups (Verheggen, et al. 2009) and an unidentified ‘spacing pheromone’ is released from crowded aphids that can change their behavior (Pettersson, et al. 1995). Our observation of OBP differential expression indicates that in addition to a tactile component, the crowding response might be modulated chemically, at least in part, especially because some of the differentially expressed OBPs are known to be expressed in the aphid’s antennae (De Biasio, et al. 2014).
Our model next posits that neurotransmitters change in response to different environmental stimuli (crowding versus solitary treatments). We observed neural-related genes at higher expression in the wingless producing aphids (SOL16+5 samples). For example, the most significantly enriched GO term in the data set was ‘neuron differentiation’, with a highly significant P value (4.3E−11) and 77 genes in the test set (Table S6). An examination of the genes associated with these neural-related GOs revealed a probable role for GABA and acetylcholine signaling, such as the GABA receptor (ACYPI006527) and the choline transporter (ACYPI009916). We also noted differentially expressed genes associated with biogenic amines that act as neurotransmitters, such as dopamine, serotonin, and octopamine (Figure 3 and Table S6). We followed up on this result by measuring whole body titers of these compounds and found that they differed significantly between crowded and solitary aphids, despite the variation in winged or wingless production by individual aphids we noted above (Figure 4). Thus, these three biogenic amines differed with the induced or uninduced state of the aphids at both the transcriptional and whole body titer levels, indicating a possible functional role in offspring phenotype determination. Future studies will need to reveal whether this relationship is correlative or causative.
Biogenic amines are small molecules synthesized from amino acids within neurons of the central nervous system. They are an integral part of the neuroendocrine system (Roeder 1994). Biogenic amine signaling is of particular interest with respect to polyphenisms, since it plays a causal role in the phase polyphenism induced by crowding in locusts. In the desert locust, Schistocerca gregaria, and the migratory locust, Locusta migratoria, changes in either dopamine or serotonin levels can cause the behavioral differences between the solitary and gregarious morphs, with higher dopamine causing the solitary morph after isolation and lower dopamine causing the gregarious morph after crowding (Anstey, et al. 2009; Ma, et al. 2011; Alessi, et al. 2014). Further, biogenic amine signaling is activated in several insects in response to stressful environmental cues (Iba, et al. 1995; Hirashima, et al. 2000; Chen, et al. 2008; Wada-Katsumata, et al. 2011). Together, these studies raise the possibility that polyphenic insects may have built upon this stress response mechanism to influence alternative developmental pathways. Our results, combined with these previous findings, suggest that the transmission of environmental signals via biogenic amines is a possible factor in density-induced polyphenisms, and may be a common mechanism underlying insect polyphenisms generally.
For our model, we next posit that biogenic amines control ecdysteroid release from the ovaries (ovaries are the main site of ecdysteroid production in adult females (Nijhout 1994)). Biogenic amines likely exert their polyphenic effects via regulating hormones such as juvenile hormone and ecdysteroids (Brown and Nestler 1985; Evans 1985; Shimada-Niwa and Niwa 2014). These hormones, when deployed, are capable of inducing large-scale changes in gene expression in a range of tissues (Li and White 2003; Davis and Li 2013; Zou, et al. 2013). We observed that the GO categories of ‘hormonal activity’ and ‘ecdysone receptor-mediated signaling pathway’ were both overrepresented in the CR16+5 to SOL16+5 comparison, and the former GO category was dominated by ecdysone-associated genes such as bursicon (ACYPI005281), eclosion hormone (ACYPI42083, ACYPI085492) and prothoracicostatic peptide precursor (ACYPI007510) (Table 1). We therefore hypothesize that ecdysone signaling is important in regulating the production of winged versus wingless offspring and that biogenic amine signaling in the brain is upstream of these ecdysone-signaling differences.
Finally, our model posits that the epigenetic process of chromatin remodeling responds to changes in hormone signaling. This is based on our observation of an enriched ‘chromatin modification’ GO term associated with the genes with higher expression in SOL16+5 females (Table S6). Interestingly, ecdysone signaling can control chromatin remodeling (Ables and Drummond-Barbosa 2010). Upon association with ecdysone, the ecdysone receptor binds to cis-regulatory DNA elements and recruits cofactors such as chromatin-modifying proteins and transcription factors to activate or repress transcription (Sedkov, et al. 2003; Privalsky 2004). We observed genes associated with this process significantly differentially expressed between CR16+5 and SOL16+5 samples (Table 1).
Epigenetic reprogramming, such as chromatin remodeling and DNA methylation, is likely critical for environmentally triggered phenotypic plasticity (Moczek and Snell-Rood 2008; Simola, et al. 2013; Simola, et al. 2016). We have already mentioned a possible role for chromatin remodeling in the pea aphid wing polyphenism. DNA methylation’s role in plasticity has received considerable attention in recent years since it was discovered to be involved in the honeybee caste polyphenism (Kucharski, et al. 2008). The pea aphid genome is methylated and contains all the genes necessary for DNA methylation (Walsh, et al. 2010), but neither the maintenance nor de novo DNA methyltransferases (Dnmt1, Dnmt3) were significantly differentially expressed in our experiments. Thus we do not anticipate major DNA methylation differences between winged and wingless producing females. This lack of evidence for a role of DNA methylation is in accordance with recent studies in a wasp (Wang, et al. 2013) and in ants (Libbrecht, et al. 2016) that suggest that DNA methylation patterns are relatively invariable among phenotypes. Our data do, however, confirm the previous observation that crowding increases the expression of the transcript ACYPI007944, a methylation enzyme (Dnmt2, Table S3)(Walsh, et al. 2010). This enzyme has been implicated in t-RNA methylation in Drosophila, especially under stress, which protects it from endonucleolytic cleavage (Schaefer, et al. 2010). Further research is required to understand the exact function of this enzyme in aphids.
Conclusion
Here we have shown for the first time that genome-wide transcriptional profiling of pea aphid females in response to wing-inducing cues reveals changes in olfactory perception, biogenic amines, hormone linked pathways, and chromatin modifications. Thus far, most aspects of our model have not been tested. However, our goal here was to identify potentially functionally important associations between gene expression changes and the production of different offspring phenotypes, which now provide an abundance of information for future experiments. Renewed interest in phenotypic plasticity has recently invigorated investigations into the molecular basis of insect polyphenisms. Our results provide evidence for an emerging theme in this important area of study: that the regulation of alternative developmental programs is multilayered, involving major shifts in hormonal signaling, biogenic amine levels, and epigenetic factors. Our results also provide unique and significant insights into the mechanisms underlying wing polyphenism in pea aphids specifically.
Supplementary Material
The percentage of winged offspring produced in the 24 hours after 16 hours of a solitary or crowding treatment are significantly different (Mann-Whitney U test P =<0.001). Data shown are the phenotypes of the offspring from 45 aphids, with the crowded aphids treated in groups of 15. Boxes represent the interquartile range and the line the median value of each group. Black circles represent outliers.
The percentage of winged offspring produced in the 24 hours after 24 hours of a solitary or crowding treatment are significantly different (Mann-Whitney U test P =<0.001). Data shown are the phenotypes of the offspring from 30 aphids, with the crowded aphids treated in groups of 10. Boxes represent the interquartile range and the line the median value of each group. Black circles represent outliers.
Acknowledgments
We gratefully acknowledge the technical assistance of Kathleen Williams, and thank Ryan D. Bickel for valuable discussions. We are also thankful for the comments from three reviewers that improved the manuscript. This research was supported by award R00ES017367 from the National Institute of Environmental Health Sciences and R01GM116867 from the National Institute of General Medical Sciences to J.A.B.
Footnotes
Author contributions
NNV designed and performed experiments, analyzed data and wrote the manuscript with input from all authors. NM helped with standardizing BA titer protocol and analyzing the data. JAB helped to design the research, obtained funding and helped in writing the manuscript. All authors read and approved the final manuscript.
Data Accessibility
RNA-Seq reads have been submitted to NCBI’s Sequence Read Archive under accession number SRP056026.
References Cited
- Ables ET, Drummond-Barbosa D. The steroid hormone ecdysone functions with intrinsic chromatin remodeling factors to control female germline stem cells in Drosophila. Cell Stem Cell. 2010;7:581–592. doi: 10.1016/j.stem.2010.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alessi AM, O’Connor V, Aonuma H, Newland PL. Dopaminergic modulation of phase reversal in desert locusts. Frontiers in Behavioral Neuroscience. 2014:8. doi: 10.3389/fnbeh.2014.00371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- Andersen CL, Jensen JL, Orntoft TF. Normalization of real-time quantitative reverse transcription-PCR data: A model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Research. 2004;64:5245–5250. doi: 10.1158/0008-5472.CAN-04-0496. [DOI] [PubMed] [Google Scholar]
- Anstey ML, Rogers SM, Ott SR, Burrows M, Simpson SJ. Serotonin mediates behavioral gregarization underlying swarm formation in desert locusts. Science. 2009;323:627–630. doi: 10.1126/science.1165939. [DOI] [PubMed] [Google Scholar]
- Auld JR, Agrawal AA, Relyea RA. Re-evaluating the costs and limits of adaptive phenotypic plasticity. Proceedings of the Royal Society B-Biological Sciences. 2010;277:503–511. doi: 10.1098/rspb.2009.1355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badenhorst P, Xiao H, Cherbas L, Kwon SY, Voas M, Rebay I, Cherbas P, Wu C. The Drosophila nucleosome remodeling factor NURF is required for ecdysteroid signaling and metamorphosis. Genes & Development. 2005;19:2540–2545. doi: 10.1101/gad.1342605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate - a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society Series B-Methodological. 1995;57:289–300. [Google Scholar]
- Blum MS. Alarm pheromones. In: Kerkut GA, Gilbert LI, editors. Comprehensive Insect physiology, Biochemistry and Pharmacology. Pergamon Press; 1985. pp. 193–224. [Google Scholar]
- Brisson JA, Davis GK, Stern DL. Common genome-wide patterns of transcript accumulation underlying the wing polyphenism and polymorphism in the pea aphid (Acyrthosiphon pisum) Evolution & Development. 2007;9:338–346. doi: 10.1111/j.1525-142X.2007.00170.x. [DOI] [PubMed] [Google Scholar]
- Brisson JA, Ishikawa A, Miura T. Wing development genes of the pea aphid and differential gene expression between winged and unwinged morphs. Insect Molecular Biology. 2010;19:63–73. doi: 10.1111/j.1365-2583.2009.00935.x. [DOI] [PubMed] [Google Scholar]
- Brown CS, Nestler C. Catecholamines and indolalkylamines. In: Kerkut GA, Gilbert LI, editors. Comprehensive Insect Physiology, Biochemistry and Pharmacology. Oxford: Pergamon; 1985. pp. 435–497. [Google Scholar]
- Cakouros D, Daish TJ, Mills K, Kumar S. An arginine-histone methyltransferase, CARMER, coordinates ecdysone-mediated apoptosis in drosophila cells. Journal of Biological Chemistry. 2004;279:18467–18471. doi: 10.1074/jbc.M400972200. [DOI] [PubMed] [Google Scholar]
- Chen YL, Hung YS, Yang EC. Biogenic amine levels change in the brains of stressed honeybees. Archives of Insect Biochemistry and Physiology. 2008;68:241–250. doi: 10.1002/arch.20259. [DOI] [PubMed] [Google Scholar]
- Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M. Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Davis MB, Li T. Genomic analysis of the ecdysone steroid signal at metamorphosis onset using and mutants. Genes Genomics. 2013;35:21–46. doi: 10.1007/s13258-013-0061-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Biasio F, Riviello L, Bruno D, Grimaldi A, Congiu T, Sun YF, Falabella P. Expression pattern analysis of odorant-binding proteins in the pea aphid Acyrthosiphon pisum. Insect Science. 2014 doi: 10.1111/1744-7917.12118. [DOI] [PubMed] [Google Scholar]
- DeWitt TJ, Sih A, Wilson DS. Costs and limits of phenotypic plasticity. Trends in Ecology & Evolution. 1998;13:77–81. doi: 10.1016/s0169-5347(97)01274-3. [DOI] [PubMed] [Google Scholar]
- Dixon AFG. Biology of aphids. London: Edward Arnold Ltd.; 1973. [Google Scholar]
- Dixon AFG, Howard MT. Dispersal in aphids, a problem in resource allocation. In: Danthanarayana W, editor. Insect Flight: Dispersal and Migration. Berlin: Springer-Verlag; 1986. pp. 145–151. [Google Scholar]
- Enders LS, Bickel RD, Brisson JA, Heng-Moss TM, Siegfried BD, Zera AJ, Miller NJ. Abiotic and biotic stressors causing equivalent mortality induce highly variable transcriptional responses in the soybean aphid. G3: Genes Genomes Genetics. 2015;5:261–270. doi: 10.1534/g3.114.015149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans PD. Octopamine. In: Kerkut GA, LIGilbert LI, editors. Comprehensive insect physiology, biochemistry and pharmacology. New York: Pergamon Press; 1985. pp. 499–538. [Google Scholar]
- Gavrilets S, Scheiner SM. The genetics of phenotypic plasticity. VI. Theoretical predictions for directional selection. Journal of Evolutionary Biology. 1993;6:49–68. [Google Scholar]
- Ghanim M, Dombrovsky A, Raccah B, Sherman A. A microarray approach identifies ANT, OS-D and takeout-like genes as differentially regulated in alate and apterous morphs of the green peach aphid Myzus persicae (Sulzer) Insect Biochemistry and Molecular Biology. 2006;36:857–868. doi: 10.1016/j.ibmb.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Harbison ST, Chang S, Kamdar KP, Mackay TFC. Quantitative genomics of starvation stress resistance in Drosophila. Genome Biology. 2005;6:R36. doi: 10.1186/gb-2005-6-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardie JLAD. In: Endocrine control of polymorphisms and polyphenism. Kerkut GLIGA, editor. New York: Pergamon; 1985. pp. 441–490. [Google Scholar]
- Hassa PO, Covic M, Bedford MT, Hottiger MO. Protein arginine methyltransferase 1 coactivates NF-kappaB-dependent gene expression synergistically with CARM1 and PARP1. Journal of Molecular Biology. 2008;377:668–678. doi: 10.1016/j.jmb.2008.01.044. [DOI] [PubMed] [Google Scholar]
- Hirashima A, Nagano T, Eto M. Stress-induced changes in the biogenic-amine levels and larval growth of Tribolium Castaneum Herbst . Bioscience Biotechnology and Biochemistry. 1993;57:2085–2089. [Google Scholar]
- Hirashima A, Sukhanova M, Rauschenbach I. Biogenic amines in Drosophila virilis under stress conditions. Biosci Biotechnol Biochem. 2000;64:2625–2630. doi: 10.1271/bbb.64.2625. [DOI] [PubMed] [Google Scholar]
- Iba M, Nagao T, Urano A. Effects of population density on growth, behavior and levels of biogenic amines in the cricket, Gryllus bimaculatus. Zool. Science. 1995;12:695–702. [Google Scholar]
- Ishikawa A, Hongo S, Miura T. Morphological and histological examination of polyphenic wing formation in the pea aphid Acyrthosiphon pisum (Hemiptera, Hexapoda) Zoomorphology. 2008;127:121–133. [Google Scholar]
- Ishikawa A, Ishikawa Y, Okada Y, Miyazaki S, Miyakawa H, Koshikawa S, Brisson JA, Miura T. Screening of upregulated genes induced by high density in the vetch aphid Megoura crassicauda. Journal of Experimental Zoology Part a-Ecological Genetics and Physiology. 2012;317A:194–203. doi: 10.1002/jez.1713. [DOI] [PubMed] [Google Scholar]
- Kijimoto T, Moczek AP, Andrews J. Diversification of doublesex function underlies morph-, sex-, and species-specific development of beetle horns. Proceedings of the National Academy of Sciences of the United States of America. 2012;109:20526–20531. doi: 10.1073/pnas.1118589109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kucharski R, Maleszka J, Foret S, Maleszka R. Nutritional control of reproductive status in honeybees via DNA methylation. Science. 2008;319:1827–1830. doi: 10.1126/science.1153069. [DOI] [PubMed] [Google Scholar]
- Leal WS. Odorant reception in insects: Roles of receptors, binding proteins, and degrading enzymes. Annual Review of Entomology, Vol 58. 2013;58:373–391. doi: 10.1146/annurev-ento-120811-153635. [DOI] [PubMed] [Google Scholar]
- Li TR, White KP. Tissue-specific gene expression and ecdysone-regulated genomic networks in Drosophila. Dev Cell. 2003;5:59–72. doi: 10.1016/s1534-5807(03)00192-8. [DOI] [PubMed] [Google Scholar]
- Libbrecht R, Oxley PR, Keller L, Kronauer DJ. Robust DNA methylation in the clonal raider ant brain. Current Biology. 2016 doi: 10.1016/j.cub.2015.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(T)(−Delta Delta C) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma ZY, Guo W, Guo XJ, Wang XH, Kang L. Modulation of behavioral phase changes of the migratory locust by the catecholamine metabolic pathway. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:3882–3887. doi: 10.1073/pnas.1015098108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayack C, Naug D. Starving honeybees lose self-control. Biol Lett. 2015;11:20140820. doi: 10.1098/rsbl.2014.0820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr E. Animal species and evolution. Cambridge: Harvard University Press; 1963. [Google Scholar]
- Moczek AP, Snell-Rood EC. The basis of bee-ing different: the role of gene silencing in plasticity. Evolution & Development. 2008;10:511–513. doi: 10.1111/j.1525-142X.2008.00264.x. [DOI] [PubMed] [Google Scholar]
- Moran NA. The evolution of aphid life-cycles. Annual Review of Entomology. 1992a;37:321–348. [Google Scholar]
- Moran NA. The evolutionary maintenance of alternative phenotypes Am. Nat. 1992b;139:971–989. [Google Scholar]
- Moskalev A, Zhikrivetskaya S, Krasnov G, Shaposhnikov M, Proshkina E, Borisoglebsky D, Danilov A, Peregudova D, Sharapova I, Dobrovolskaya E, et al. A comparison of the transcriptome of Drosophila melanogaster in response to entomopathogenic fungus, ionizing radiation, starvation and cold shock. BMC Genomics. 2015;16(Suppl 13):S8. doi: 10.1186/1471-2164-16-S13-S8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller CB, Williams IS, Hardie J. The role of nutrition, crowding and interspecific interactions in the development of winged aphids. Ecological Entomology. 2001;26:330–340. [Google Scholar]
- Murren CJ, Auld JR, Callahan H, Ghalambor CK, Handelsman CA, Heskel MA, Kingsolver JG, Maclean HJ, Masel J, Maughan H, et al. Constraints on the evolution of phenotypic plasticity: limits and costs of phenotype and plasticity. Heredity (Edinb) 2015;115:293–301. doi: 10.1038/hdy.2015.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckameyer WS, Weinstein JS. Stress affects dopaminergic signaling pathways in Drosophila melanogaster. Stress. 2005;8:117–131. doi: 10.1080/10253890500147381. [DOI] [PubMed] [Google Scholar]
- Nijhout HF. Control mechanisms of polyphenic development in insects - In polyphenic development, environmental factors alter same aspects of development in an orderly and predictable way. Bioscience. 1999;49:181–192. [Google Scholar]
- Nijhout HF. Development and evolution of adaptive polyphenisms. Evolution & Development. 2003;5:9–18. doi: 10.1046/j.1525-142x.2003.03003.x. [DOI] [PubMed] [Google Scholar]
- Nijhout HF. Inect Hormones. Princeton, NJ: Princeton University Press; 1994. [Google Scholar]
- Pettersson J, Quiroz A, Stephansson D, Niemeyer HM. Odor communication of Rhopalosiphum padi on grasses. Entomologia Experimentalis Et Applicata. 1995;76:325–328. [Google Scholar]
- Price DP, Schilkey FD, Ulanov A, Hansen IA. Small mosquitoes, large implications: crowding and starvation affects gene expression and nutrient accumulation in Aedes aegypti. Parasites & Vectors. 2015;8:252. doi: 10.1186/s13071-015-0863-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Privalsky ML. The role of corepressors in transcriptional regulation by nuclear hormone receptors. Annual Review of Physiology. 2004;66:315–360. doi: 10.1146/annurev.physiol.66.032802.155556. [DOI] [PubMed] [Google Scholar]
- Ragsdale Erik J, Müller Manuela R, Rödelsperger C, Sommer Ralf J. A developmental switch coupled to the evolution of plasticity acts through a sulfatase. Cell. 2013;155:922–933. doi: 10.1016/j.cell.2013.09.054. [DOI] [PubMed] [Google Scholar]
- Rion S, Kawecki TJ. Evolutionary biology of starvation resistance: what we have learned from Drosophila. J Evol Biol. 2007;20:1655–1664. doi: 10.1111/j.1420-9101.2007.01405.x. [DOI] [PubMed] [Google Scholar]
- Roeder T. Biogenic-amines and their receptors in insects. Comparative Biochemistry and Physiology C-Pharmacology Toxicology & Endocrinology. 1994;107:1–12. [Google Scholar]
- Schaefer M, Pollex T, Hanna K, Tuorto F, Meusburger M, Helm M, Lyko F. RNA methylation by Dnmt2 protects transfer RNAs against stress-induced cleavage. Genes Dev. 2010;24:1590–1595. doi: 10.1101/gad.586710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedkov Y, Cho E, Petruk S, Cherbas L, Smith ST, Jones RS, Cherbas P, Canaani E, Jaynes JB, Mazo A. Methylation at lysine 4 of histone H3 in ecdysone-dependent development of Drosophila. Nature. 2003;426:78–83. doi: 10.1038/nature02080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serobyan V, Ragsdale EJ, Muller MR, Sommer RJ. Feeding plasticity in the nematode Pristionchus pacificus is influenced by sex and social context and is linked to developmental speed. Evolution & Development. 2013;15:161–170. doi: 10.1111/ede.12030. [DOI] [PubMed] [Google Scholar]
- Shimada-Niwa Y, Niwa R. Serotonergic neurons respond to nutrients and regulate the timing of steroid hormone biosynthesis in Drosophila. Nature Communications. 2014;5:5778. doi: 10.1038/ncomms6778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simola DF, Graham RJ, Brady CM, Enzmann BL, Desplan C, Ray A, Zwiebel LJ, Bonasio R, Reinberg D, Liebig J, et al. Epigenetic (re)programming of caste-specific behavior in the ant Camponotus floridanus. Science. 2016;351:aac6633. doi: 10.1126/science.aac6633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simola DF, Ye C, Mutti NS, Dolezal K, Bonasio R, Liebig J, Reinberg D, Berger SL. A chromatin link to caste identity in the carpenter ant Camponotus floridanus. Genome Research. 2013;23:486–496. doi: 10.1101/gr.148361.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson SJ, Despland E, Hagele BF, Dodgson T. Gregarious behavior in desert locusts is evoked by touching their back legs. Proceedings of the National Academy of Sciences of the United States of America. 2001;98:3895–3897. doi: 10.1073/pnas.071527998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simpson SJ, Sword GA, Lo N. Polyphenism in insects. Current Biology. 2011;21:R738–R749. doi: 10.1016/j.cub.2011.06.006. [DOI] [PubMed] [Google Scholar]
- Sultan SE, Spencer HG. Metapopulation structure favors plasticity over local adaptation. American Naturalist. 2002;160:271–283. doi: 10.1086/341015. [DOI] [PubMed] [Google Scholar]
- Sutherland ORW. The role of crowding in the production of winged forms by two strains of the pea aphid, Acyrthosiphon pisum. Journal of Insect Physiology. 1969;15:1385–1410. [Google Scholar]
- Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM. Primer3Plus, an enhanced web interface to Primer3. Nucleic Acids Research. 2007;35:71–74. doi: 10.1093/nar/gkm306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verheggen FJ, Haubruge E, De Moraes CM, Mescher MC. Social enviroment influences aphid production of alarm pheromone. Behavioral Ecology. 2009;20:283–288. [Google Scholar]
- Wada-Katsumata A, Yamaoka R, Aonuma H. Social interactions influence dopamine and octopamine homeostasis in the brain of the ant Formica japonica. Journal of Experimental Biology. 2011;214:1707–1713. doi: 10.1242/jeb.051565. [DOI] [PubMed] [Google Scholar]
- Walker TJ. Stochastic polyphenism: coping with uncertainty. Florida Entomolgist. 1986;69:46–62. [Google Scholar]
- Walsh TK, Brisson JA, Robertson HM, Gordon K, Jaubert-Possamai S, Tagu D, Edwards OR. A functional DNA methylation system in the pea aphid, Acyrthosiphon pisum. Insect Molecular Biology. 2010;19:215–228. doi: 10.1111/j.1365-2583.2009.00974.x. [DOI] [PubMed] [Google Scholar]
- Wang X, Kang L. Molecular mechanisms of phase change in locusts. Annual Review of Entomology. 2014;59:225–244. doi: 10.1146/annurev-ento-011613-162019. [DOI] [PubMed] [Google Scholar]
- Wang X, Wheeler D, Avery A, Rago A, Choi JH, Colbourne JK, Clark AG, Werren JH. Function and evolution of DNA methylation in Nasonia vitripennis. PLoS Genet. 2013;9:e1003872. doi: 10.1371/journal.pgen.1003872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- West-Eberhard MJ. Developmental plasticity and evolution. New York: Oxford University Press; 2003. [Google Scholar]
- Yang X, Liu X, Xu X, Li Z, Li Y, Song D, Yu T, Zhu F, Zhang Q, Zhou X. Gene Expression Profiling in Winged and Wingless Cotton Aphids, Aphis gossypii (Hemiptera: Aphididae) International Journal of Biological Sciences. 2014;10:257–267. doi: 10.7150/ijbs.7629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou JJ, Vieira FG, He XL, Smadja C, Liu R, Rozas J, Field LM. Genome annotation and comparative analyses of the odorant-binding proteins and chemosensory proteins in the pea aphid Acyrthosiphon pisum. Insect Molecular Biology. 2010;19:113–122. doi: 10.1111/j.1365-2583.2009.00919.x. [DOI] [PubMed] [Google Scholar]
- Zou Z, Saha TT, Roy S, Shin SW, Backman TW, Girke T, White KP, Raikhel AS. Juvenile hormone and its receptor, methoprene-tolerant, control the dynamics of mosquito gene expression. Proceedings of the National Academy of Sciences of the United States of America. 2013;110:E2173–E2181. doi: 10.1073/pnas.1305293110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
The percentage of winged offspring produced in the 24 hours after 16 hours of a solitary or crowding treatment are significantly different (Mann-Whitney U test P =<0.001). Data shown are the phenotypes of the offspring from 45 aphids, with the crowded aphids treated in groups of 15. Boxes represent the interquartile range and the line the median value of each group. Black circles represent outliers.
The percentage of winged offspring produced in the 24 hours after 24 hours of a solitary or crowding treatment are significantly different (Mann-Whitney U test P =<0.001). Data shown are the phenotypes of the offspring from 30 aphids, with the crowded aphids treated in groups of 10. Boxes represent the interquartile range and the line the median value of each group. Black circles represent outliers.