Summary
Human genomics is identifying candidate genes for congenital heart disease (CHD), but discovering the underlying mechanisms remains challenging. In a patient with CHD and Heterotaxy (Htx), a disorder of left-right patterning, we previously identified a duplication in Nup188. However, a mechanism to explain how a component of the nuclear pore complex (NPC) could cause Htx/CHD was undefined. Here, we show that knockdown of Nup188 or its binding partner Nup93 leads to a loss of cilia during embryonic development while leaving NPC function largely intact. Multiple data, including the localization of endogenous Nup188/93 at cilia bases supports their direct role at cilia. Super-resolution imaging of Nup188 shows two barrel-like structures with dimensions and organization incompatible with a NPC-like ring, arguing against a proposed “ciliary pore complex.” We suggest that the nanoscale organization and function of nups are context-dependent in a way that is required for the structure of the heart.
Keywords: congenital heart disease, left-right patterning, Xenopus tropicalis, nuclear pore complex, heterotaxy, nucleoporin, super resolution, 3D nanoscopy
eTOC
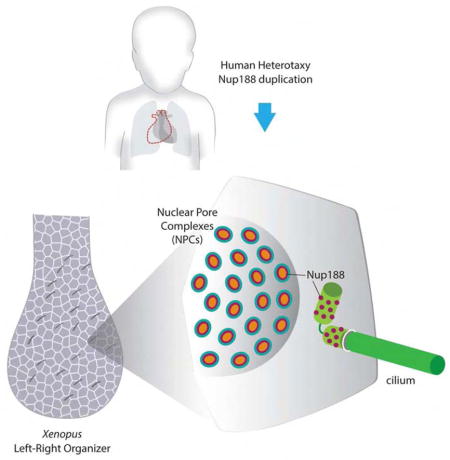
del Viso, Huang et al. show that Nup188 encases the cilium base in a supra-molecular organization distinct from nuclear pore complexes. Depletion of inner ring nups, including Nup188, leads to cilia loss and aberrant heart patterning in the developing Xenopus embryo, possibly explaining Heterotaxy in a patient with Nup188 duplication.
Introduction
Nuclear pore complexes (NPCs) are massive ~100 MD protein assemblies that span the inner and outer nuclear membranes of the nuclear envelope; there, they control the exchange of molecules between the nucleus and cytoplasm. The archetypal 8-fold radially symmetric NPC is built from ~30 individual proteins termed nucleoporins (nups) many of which are found within stable subcomplexes (Figure 1H) (Alber et al., 2007; Amlacher et al., 2011; Cronshaw et al., 2002; Rout et al., 2000). The Nup107–160 “Y” and Nup93 complexes are thought to build concentric outer and inner rings, respectively, of the NPC scaffold (Alber et al., 2007; Amlacher et al., 2011; Hurt and Beck, 2015; Kelley et al., 2015; Vollmer and Antonin, 2014; von Appen et al., 2015). The scaffold provides anchor points for the Nup62 complex, and other unstructured nups rich in Phe-Gly (FG) amino acids that also help form the cytoplasmic filaments and nuclear basket (Alber et al., 2007; Chug et al., 2015; Hurt and Beck, 2015; Stuwe et al., 2015). Together, the ‘FG-nups’ establish a size-selective diffusion barrier while also providing binding sites for rapidly shuttling nuclear transport receptors bound to signal-bearing cargo (Schmidt and Gorlich, 2016).
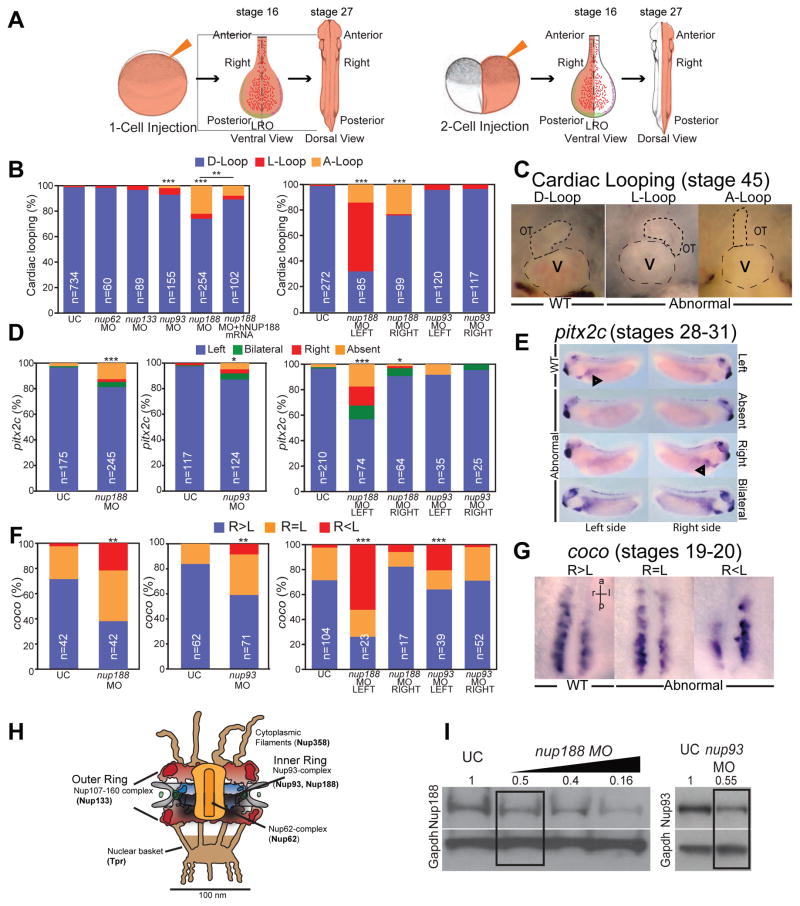
Figure 1. Depletion of inner ring nups alters LR patterning.
(A) 1- (left) and 2-cell (right) injection schematic (not to scale) showing a salmon colored dye as a tracer. When injections are done in one cell at 2-cell stage, embryos are selected where either the left or the right side of the embryo is targeted, as shown in the diagram of dissected LRO (stage 16) or in a later stage 27 embryo. Small red lines in LRO represent cilia.
(B) Percentage of embryos with abnormal cardiac looping (see Figure 1C for key to score cardiac looping) after MO injection at 1-cell stage (left) or 2-cell stage (right). nup188 morphant heart looping defects are rescued by human NUP188 expression (left). n=total number of embryos from 3 independent experiments. Chi-square/Fisher’s Exact Test, *** p<0.0005, ** p<0.005. UC – uninjected controls.
(C) Examples of cardiac looping phenotypes of WT and abnormal Xenopus hearts at stage 45 viewed ventrally with anterior to the top. D-Loop: normal outflow track (OT) loops right (short dashed line), L-Loop: abnormal OT loops left, A-Loop: abnormal OT is unlooped. V, ventricle is indicated by long dashed line.
(D) Percentage of embryos with abnormal pitx2c (see Figure 1E for key to score pitx2c staining) after MO injection at 1-cell stage (left, middle) or 2-cell stage (right). n=total number of embryos from 3 independent experiments. Chi-square/Fisher’s Exact Test, *** p<0.0005, * p<0.05.
(E) Example key of pitx2c expression showing lateral views of embryos (stages 28–31) with dorsal at top. Arrowheads point to pitx2c in the lateral plate mesoderm.
(F) Percentage of embryos with abnormal coco expression (see Figure 1G for key for coco scoring) after MO injection at 1-cell stage (left, middle) or 2-cell stage (right). L, R are left and right side of LRO, respectively. n=total number of embryos from 3 independent experiments. Chi-square/Fisher’s Exact Test, *** p<0.0005, ** p<0.005.
(G) Example key of coco expression patterns in the LRO at stages 19–20. Ventral views with anterior to the top.
(H) Nuclear pore complex (NPC) schematic. Nups referred to in this study are in bold.
(I) Nup188 and Nup93 protein levels determined by Western Blot. Numbers are relative protein levels normalized to Gapdh with triangle reflecting dose of MO. Boxed lane indicates dose used for all experiments.
See also Figures S1 and S2.
While it is likely that most NPCs are biochemically and thus functionally identical, work in multicellular eukaryotes supports the tissue-specific expression of certain nups raising the possibility of biochemically distinct NPCs with cell-type tailored transport properties (Kelley et al., 2015; Ori et al., 2013; Raices and D’Angelo, 2012; Reza et al., 2016). Moreover, changes to the relative levels of nups have been observed during cellular differentiation suggesting critical roles for nups in determining cell fate (D’Angelo et al., 2012; Gomez-Cavazos and Hetzer, 2015; Lupu et al., 2008; Smith et al., 2011). However, we are lacking tools capable of assessing the biochemical composition of individual NPCs or differentiating between the functions of nups at the NPC versus at other subcellular locations. Therefore, identifying the underlying mechanisms of how nups might play cell type or tissue specific roles remains a major challenge.
The concept that nups might have roles outside of nuclear transport was established by work showing that the entire ‘Y’ complex localized to kinetochores during mitosis (Belgareh et al., 2001; Loiodice et al., 2004), which contributes to mitotic progression (Mishra et al., 2010; Orjalo et al., 2006). Consistent with an intimate link between nups and the mitotic machinery, both Nup62 and Nup188 might also localize to centrosomes (Hashizume et al., 2013; Itoh et al., 2013). There is also growing interest in direct roles for nups in regulating gene expression within the nucleoplasm (Buchwalter et al., 2014; Capelson et al., 2010; Jacinto et al., 2015; Kalverda et al., 2010; Liang et al., 2013; Light et al., 2013; Pascual-Garcia et al., 2014). Intriguingly, nups were recently localized at the bases of cilia (Kee et al., 2012). Indeed, many scaffold and FG-nups were localized to the cilium raising the possibility that they would install a diffusion barrier with size-selective properties and structure similar to the NPC. Additionally, the similarity of 1) some “ciliary localization signals” to “nuclear localization signals”, 2) the involvement of the nuclear transport receptor karyopherin/importin β2 in contributing to the localization of some proteins to the cilium, and 3) the presence of Ran-GTP in the axoneme made the concept of a “ciliary pore complex” analogous to the NPC extremely attractive (Dishinger et al., 2010; Kee et al., 2012; Kee and Verhey, 2013; Takao et al., 2014). Other work, however, failed to reproduce the localization of nups at the cilium base (Breslow et al., 2013). Moreover, the cilium base diffusion barrier was unaffected by inhibitors of the NPC barrier suggesting that its molecular composition does not include the FG-nups (Breslow et al., 2013). Therefore, the “ciliary pore complex” model remains controversial. Nonetheless, there are sufficient studies to support the concept that nups may be localized to different subcellular locations; whether these nups might also port their structural and functional organization is a key question, which needs to be addressed using nanoscale imaging approaches like super-resolution microscopy. Such approaches would be able to address, for example, whether nups are capable of forming NPC-like structures at the cilium base.
Cilia have emerged as key organelles with diverse roles including generating extracellular fluid flow, mechanical force sensing, and compartmentalized cell signaling (Eggenschwiler and Anderson, 2007; Ishikawa and Marshall, 2011; Pennekamp et al., 2015). Interestingly, some ciliopathies (human diseases linked to cilia malfunction) lead to abnormal positioning of organs across the left-right (LR) axis resulting in a disorder called heterotaxy (Htx), which can lead to a severe form of congenital heart disease (CHD) (Basu and Brueckner, 2008; Bisgrove and Yost, 2006; Fliegauf et al., 2007; Sutherland and Ware, 2009). During embryonic development, the midline heart tube loops to the right, establishing the cardiac asymmetry that is essential for adult function. This LR asymmetry begins with signaling at the LR Organizer (LRO), which is formed from mesendodermal cells near the end of gastrulation (Basu and Brueckner, 2008; Pennekamp et al., 2015). Motile cilia in the LRO beat to generate leftward extracellular fluid flow sensed by immotile cilia, which leads to a cascade of asymmetric gene expression and, ultimately, proper organ situs (Boskovski et al., 2013; McGrath et al., 2003; Tabin and Vogan, 2003). Remarkably, we recently identified a duplication of Nup188 in a patient with Htx and CHD (Fakhro et al., 2011). A major motivation for this study is to integrate Nup188 into the LR patterning cascade to support the disease relevance for Nup188 in Htx/CHD.
In the following, we demonstrate that depletion of Nup188 and its binding partner Nup93 leads to the loss of cilia in multiple cell types including mammalian cell lines and the LRO of Xenopus. Our data are consistent with a model in which inner ring components of the NPC, but not other nups, directly localize and function at the cilium base. Moreover, by leveraging super-resolution approaches, we demonstrate that the nanoscale distribution of Nup188 at the cilium base is distinct from NPCs supporting a previously undescribed context-dependent organization and function of nucleoporins.
Results
Inner ring nups are required for LR patterning
To explore the mechanism of how Nup188 could impact cardiac looping, we first tested depletion of Nup188 in Xenopus. Xenopus is ideal for these studies as gene dosage can be titrated to test both gain and loss of function; similar strategies are more difficult and expensive to perform in murine models. Additionally, when compared to other aquatic models, Xenopus has cardiac structures including a septated atria and cardiac trabeculations that are more closely related to humans reinforcing its utility for examining LR patterning and cardiac looping (Blum et al., 2009; Warkman and Krieg, 2007). In fact, by injecting morpholino oligos (MOs) or mRNA into one cell of the 2-cell embryo, embryos can be identified in which only the left or right side is targeted (Figure 1A), making it a truly unique tool for investigating LR patterning. With these advantages in mind, we tested whether partial depletion of nup188 using MOs caused cardiac looping defects similar to the Htx disease phenotype. Remarkably, in ~25% of injected embryos (Figure 1B, left panel) (Fakhro et al., 2011) we observed abnormal A- and L-loop cardiac morphologies (Figure 1C). Critically, we confirmed the specificity of our MOs by co-injecting human NUP188 mRNA, which rescued these laterality phenotypes (Figure 1B, left panel).
As Nup188 forms the inner ring complex of the NPC with Nup93, Nup205, Nup155 and Nup53/35 (Figure 1H), we next evaluated whether the heart defects were specific to this nup subcomplex. After depletion of representative components of the outer ring (Nup133), the central transport channel (Nup62) or an additional inner ring nup, Nup93, only knockdown of nup93 significantly altered cardiac looping in ~10% of embryos despite similar, or more severe reductions (~50% to 75%) in nup levels for each nup tested (Figures 1B, left panel, 1I, S1A). These results suggest that the heart looping defects occur after a specific disruption of inner ring nup function, but not other essential nup subcomplexes.
To better understand the mechanism by which the inner ring nups affected heart development, we examined the expression of two additional LR markers at different stages of embryonic development: pitx2c and coco (dand5, cerl2). Pitx2 is a homeodomain containing transcription factor that is expressed in the left lateral plate mesoderm and is essential for laterality of the internal organs (Lin et al., 1999; Yoshioka et al., 1998). Coco, an extracellular nodal antagonist, is upstream of pitx2c and is initially expressed symmetrically across the LR axis but becomes asymmetric as the LR axis is specified (stage 19 in Xenopus) (Schweickert et al., 2010; Vonica and Brivanlou, 2007). In both nup188 and nup93 morphants, the pattern of pitx2c expression was abnormal (see Figure 1E for phenotype key) in ~20% and ~10% of the embryos, respectively; the most prevalent phenotype was an absence of detectable pitx2c expression (Figure 1D, left and middle panels). In addition, depletion of Nup188 and Nup93 led to the loss of coco asymmetry (i.e. its expression on the right and left sides of the embryo were indistinguishable (R=L)) in significantly more morphants compared to control embryos (Figures 1F, left and middle panels and 1G). Combined, these results indicate a global defect in LR patterning at the earliest steps in the LR signaling cascade likely reflecting abnormal signaling at the LRO.
Specific nups are required for cilia
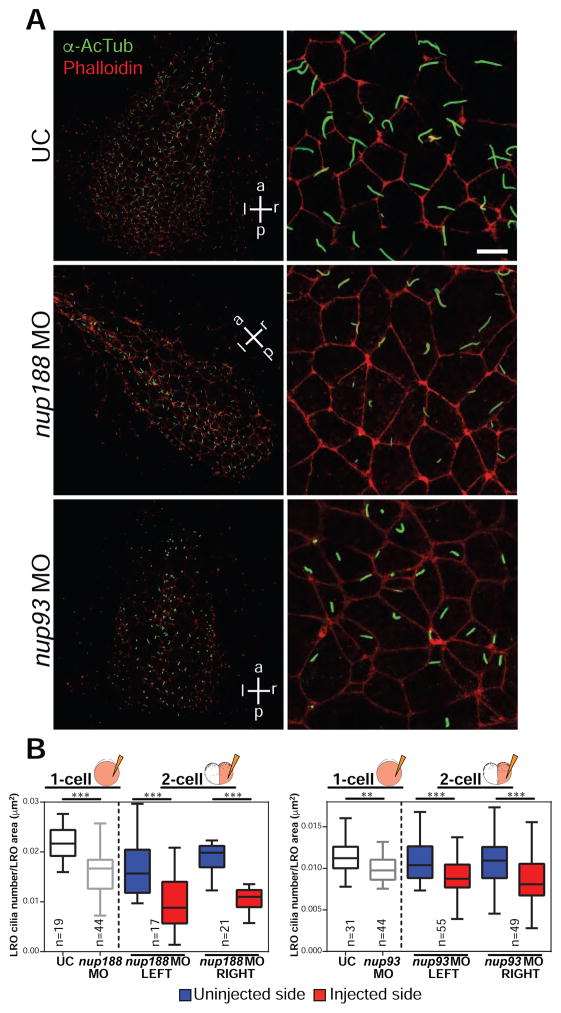
LR asymmetry is established by the beating of cilia in the LRO, which generates leftward extracellular fluid flow sensed by immotile cilia (Basu and Brueckner, 2008; Boskovski et al., 2013; Pennekamp et al., 2015). The equal bilateral coco staining and loss of pitx2c in Nup93/188 knockdown embryos was suggestive of a disruption of this cilia motility or signaling (Schweickert et al., 2010; Vick et al., 2009). In Xenopus, cilia motility is essential only on the left side of the LRO (Vick et al., 2009). Strikingly, by specifically targeting the left-side of the embryo (Figure 1A) ~70% of nup188 morphant embryos showed defects in heart structure, compared to ~20% with a right-sided depletion (Figure 1B, right panel). These more significant left-sided effects were mirrored by aberrant pitx2c and coco staining and cumulatively pointed to cilia dysfunction in the LRO (Figures 1D and 1F, right panels). We thus directly examined cilia in the LRO by immunostaining for acetylated tubulin (AcTub). In the LROs depleted of Nup188, there was a significant ~28% reduction in the density of cilia compared to uninjected controls (Figures 2A and 2B). Similarly, depleting nup188 in half of the LRO reduces the number of cilia specifically in the targeted half (Figure 2B). In the LROs of nup93 morphants, we observed a more modest but still significant ~12% reduction in cilia (Figures 2A and 2B). Finally, to test if this effect on cilia could be generalized to human cells, we depleted Nup188 in retinal pigmented epithelial (RPE) cells using small interfering RNAs (siRNA), which had significantly fewer cilia than wild type cells (~20% less cilia; Figures S1B and S1C, see Figure 4F for degree of knockdown).
Figure 2. nup188 and nup93 depletion decreases cilia in the LRO.
(A) LROs from embryos treated with the indicated MOs (or UC) stained with anti-acetylated α-tubulin (α-AcTub, green) to detect primary cilia, Phalloidin (red) marks the cell borders. p,a,l,r: posterior, anterior, left, right, respectively. Right panels show a magnification of a representative region of the LRO.
(B) Box plots of cilia number per LRO area (μm2) of embryos derived from injections at the 1 or 2-cell stage. Box depicts 25th to 75th percentiles with median marked by horizontal line in box. Whiskers indicate data range from smallest to largest values. n=total number of embryos from 3 independent experiments, T-test, *** p<0.0005, ** p<0.005.
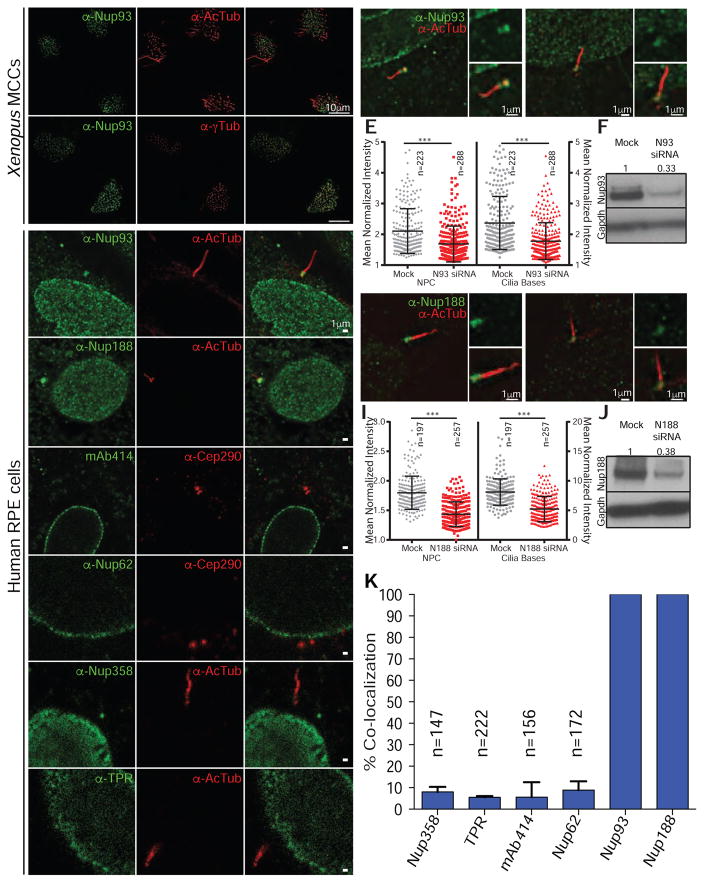
Figure 4. Nup188 and Nup93 specifically localize to the bases of cilia.
(A) Immunofluorescence images of Xenopus epidermal MCCs stained with anti-Nup93 (green) and either anti-AcTub (red, top) or anti-γTub (red, bottom) antibodies with merge.
(B) Immunofluorescence images of human RPE cells stained with the indicated anti-nup antibodies (left) with anti-AcTub, anti-γTub or anti-Cep290 (middle) and merge (right). See Figure S3B for location of cilia-specific epitopes.
(C) Anti-Nup93 antibodies are specific. Fluorescence image (merge of green and red channels) of representative scrambled (mock) siRNA-treated RPE cell stained with anti-Nup93 (green) and anti-AcTub (red) antibodies. Right panels are higher magnification of cilium base showing just the anti-Nup93 signal (green; top) and merge of anti-Nup93 and anti-AcTub (red) images (bottom). Arrowhead points to cilium base. N denotes the nucleus.
(D) Identical to (C) except cells are treated with siRNAs specific to NUP93 (N93).
(E) Plot of normalized fluorescence intensity in arbitrary units (a.u.) of anti-Nup93 signal at cilium base and NPCs of individual cells treated with scrambled (mock) or specific NUP93 (N93) siRNAs. Mean +/− SD indicated. n=total number of cells/cilia from 3 independent experiments, T-test, *** p<0.0005.
(F) Western blot of Nup93 levels after siRNA transfection; numbers are quantification of protein levels relative to Gapdh.
(G–J) Anti-Nup188 antibodies are specific. Panel layout identical to (C–F) except for cells are treated with scrambled (mock) or siRNAs specific to NUP188 (N188).
(K) Plot of percentage of cells in which the anti-nup signal colocalizes with a cilium base marker. Mean and SD are shown. n=total number of cells from 3 independent experiments.
See also Figures S1 and S3.
We next tested whether the loss of cilia in the LROs was observed in other ciliated tissues. In Xenopus, the embryonic epidermis has an array of multiciliated cells (MCCs) whose cilia beat to generate extracellular fluid flow similar to the mammalian tracheal epithelium or the oviduct. Interestingly, in the course of our experiments, we noted that nup93 morphants failed to glide over the surface of petri dishes (Figures S1D–S1F and Movie S1); this gliding behavior is driven by the beating of cilia in the MCCs. When examining cilia in nup93 morphants, we observed a near total loss of cilia in the MCCs of the epidermis that was specific to the side that had received the MO injection (Figure 3A, 100% of embryos n=73). Critically, this cilia loss was rescued by expressing human NUP93 in nup93 morphants (Figure S1G). These results were again specific for the inner ring nups, as we observed a modest reduction of epidermal cilia in nup188 morphants (Figure 3A, 41% of embryos n=70) but no appreciable loss of cilia after depletion of Nup62 or Nup133, despite the severe malformations of Nup133-depleted embryos (Figures 3B and S1A). In comparison, we were struck by the ‘normal’ overall morphology of the nup93/nup188 morphants, suggesting that depletion of the inner ring nups does not result in a global developmental defect that might be expected with a disruption in NPC function. Rather, the partial depletion of Nup93/188 results in a reduction in cilia in multiple tissues suggesting cilia-specific effects.
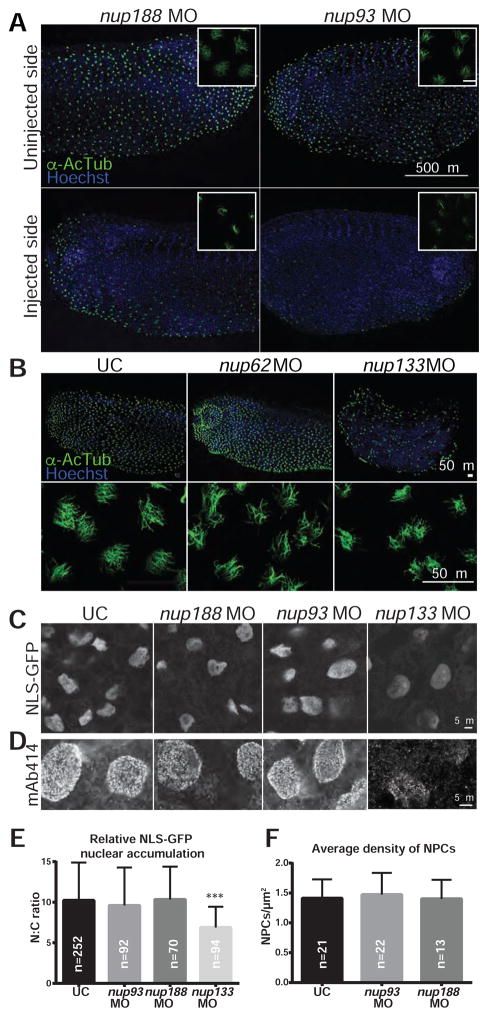
Figure 3. Depletion of Nup93/188 affects cilia but not NPCs.
(A) Fluorescence images of lateral views, with dorsal at the top, of both sides (uninjected and injected) of a single embryo with the cilia of the multiciliated cells (MCCs) labeled with α-AcTub (green). Nuclei stained with Hoechst (blue). Insets (top right) are a magnification of a cluster of MCCs, scale bar: 20 μm. The disruption of cilia was found in 100% (n=73) of nup93 morphants and in 41% (n=70) of nup188 morphants.
(B) Fluorescence images of Xenopus embryos injected with nup62 (n=30) or nup133 (n=33) MOs at the 1-cell stage labeled with anti-AcTub (green). Lateral views of embryos, with dorsal to the top (top panels) and higher magnification of the MCCs at bottom. Nuclei stained with Hoechst (blue). Level of nup depletion in these morphants is shown in Figure S1A.
(C) Embryos were co-injected with mRNA encoding NLS-GFP and nup93, nup188 or nup133 MOs. Animal caps were dissected and allowed to develop MCCs before imaging the distribution of NLS-GFP. Cilia in these explants are shown in Figure S1J.
(D) Staining of NPCs with the mAb414 antibody in animal caps of UC and embryos injected with the indicated MOs.
(E) Graph shows the mean nuclear/cytoplasm (N:C) fluorescence ratios of NLS-GFP with SD. n=total number of nuclei from 3 independent experiments. T-test, *** p<0.0005
(F) Graph shows the mean and SD of the NPC density in animal caps of UCs and embryos injected with the indicated MOs. n= number of nuclei from 3 independent experiments. See also Figures S1 and S2.
NPC function is intact
We considered the possibilities that cilia loss was due to a change in cilia cell-type specification and/or inhibition of nuclear transport. The former idea was tested by examining the expression of foxj1, a critical regulator of motile ciliogenesis (Stubbs et al., 2008), which remained unchanged between wildtype and nup93/nup188 morphants at stages relevant for LRO cilia and MCCs (Figures S1H and S1I). To test the latter hypothesis, we first assessed the localization of a classic nuclear localization signal “NLS”-GFP reporter. To do this, we took advantage of the capacity in Xenopus to explant embryonic stem cells, “the animal cap”, from blastula stage embryos either depleted for Nup93/188 (or not). The animal caps are grown on fibronectin-coated dishes and differentiate into embryonic epidermis complete with functional MCCs without any additional treatment (Werner and Mitchell, 2013). Strikingly, while we observed the specific loss of cilia in nup93/nup188 morphant MCCs (Figure S1J), little change was observed in the steady state distribution of NLS-GFP, which accumulated ~10-fold over the cytoplasm in wildtype and morphant explants (Figures 3C and 3E). These results support the conclusion that the level of Nup93/188 knockdown in the explants has little impact on the karyopherin/importin α/β1 transport pathway, at least at steady state, despite the clear effect on cilia (Figure S1J). Second, to rule out a reduction in NPCs, we evaluated NPC density with the mAb414 antibody, which recognizes at least four FG-nups (Davis and Blobel, 1986; Sukegawa and Blobel, 1993). We were unable to detect significant differences in mAb414 staining between wildtype and nup morphants (Figures 3D and 3F).
We were concerned that the lack of any overt NPC defect in nup93/188 morphants might reflect that our assays were not sensitive enough to pick up changes to nuclear transport. We therefore depleted Nup133, a key component of the Y complex required for NPC assembly. First, at knockdown levels less severe than Nup93/188 (Figure S1A), we observed that the embryos were malformed suggesting a global defect in development upon NPC disruption (Figure 3B). Consistent with this, both the nuclear accumulation of NLS-GFP and mAb414 staining were reduced (Figures 3C–3E). Remarkably, however, despite these obvious defects in nuclear transport, the epidermal cilia were largely unaffected (Figure 3B). Thus, the knockdown of Nup133 reinforces that Nup93 and Nup188 might have a specific role at cilia.
Nup188 overexpression mimics loss of function
Cognizant that our studies began with a Htx/CHD patient with a duplication of the NUP188 locus (Fakhro et al., 2011), we tested whether the overexpression of NUP188 also impacted heart development in Xenopus. As shown in Figure S2A, the overexpression of either Xenopus or human NUP188 recapitulated a Htx-like (cardiac looping) phenotype in ~15% of the embryos. We also detected a loss of cilia in the LRO (Figures S2B and S2C) and in the MCCs of 35% of the embryos (Figure S2D). However, we did not detect an alteration in the mAb414 staining of NPCs or in the nuclear transport of the NLS-GFP reporter using the assays described above (Figures S2E–S2G). From these experiments, we conclude that Nup188 overexpression can lead to a loss of cilia without affecting NPC function, similar to the loss of function scenario. Cumulatively, our results point to a specific role for inner ring nups in disrupting cilia but not NPC function, providing a plausible mechanism for disease pathogenesis in the Htx/CHD patient.
Specific nups localize to cilia bases
Cumulatively, our data suggest that the cardiac looping phenotypes observed specifically when inner ring nups are depleted reflects a direct function for these nups at cilia rather than at NPCs. As there are conflicting reports on the localization of nups at the cilium base (Breslow et al., 2013; Kee et al., 2012), we felt it was critical to re-examine this subject. We began by testing the localization of endogenous Nup93 in the MCCs of the Xenopus epidermis. We observed the accumulation of anti-Nup93 signal in puncta at the cell surface that was specific for cells with cilia. This signal was adjacent to an anti-AcTub stain, which labels the axoneme, and overlapped with gamma tubulin (anti-γTub) (Figure 4A). As gamma tubulin is a major component of the basal bodies, this result is consistent with a specific Nup93 localization at cilia bases (Figure 4A). This distribution was mirrored in human RPE cells where we detected the anti-Nup93 signal below the cilium in virtually all cells (Figures 4B and 4K). A similar immunolocalization was observed using antibodies directed towards native Nup188 (Figures 4B and 4K). To provide an explanation for the disparate reports on the localization of nups at the bases of cilia and to address the propensity for antibodies to non-specifically stain centrosomes, we tested the specificity of the Nup93 and Nup188 antibodies through the use of siRNAs. Critically, we observed a specific reduction of Nup93/188-staining at NPCs and at the cilium base after treatment of cells with Nup93- (Figures 4C–4F) or Nup188-targeted siRNAs (Figures 4G–4J), which was not observed in cells transfected with the scrambled siRNA control (Mock), confirming the specific localization of these two inner ring nups at the cilium base. Finally, we also detected a pool of an overexpressed SNAP-tagged version of Nup188 at centrioles (Figure S3A) (Itoh et al., 2013; Keppler et al., 2003), supporting that both endogenous and overexpressed Nup188 share specific centriolar distributions.
We next closely inspected the distribution of Nup93/188 in reference to established cilia and centriole landmarks (Figure S3B). In most cells, the anti-Nup93 and anti-Nup188 signals showed two distinct peaks of intensity at the cilium base that overlapped with both the mother and daughter centrioles when stained by an anti-γTub antibody (Figures S3C and S3D). Consistent with this, the two peaks flanked an anti-rootletin antibody signal (rootletin connects the two centrioles (Yang et al., 2002); Figures S3B–S3D) and only one peak colocalized with an anti-ninein stain that specifically labels the mother centriole (Bouckson-Castaing et al., 1996) (Figures S3B–S3D). Thus, Nup93 and Nup188 localize to the bases of cilia, where they colocalize with centrioles.
To assess whether cilium base staining could be observed for additional nups, we tested several nup-specific antibodies, including mAb414, and those that recognize Nup62, Nup358 and Tpr (Figure 1H). However, we observed (conservatively) colocalization of cytosolic nup foci with cilium base markers in less than 10% of cells (Figures 4B and 4K), even after increasing our exposure times to saturate the NPC-specific fluorescence. The lack of mAb414 staining was somewhat surprising since Nup62, one of the nups recognized by mAb414, directly interacts with Nup93 at the NPC (Chug et al., 2015; Grandi et al., 1995; Sachdev et al., 2012; Stuwe et al., 2015) and might bind centrosomes during mitosis (Hashizume et al., 2013). However, even when using antibodies specific for Nup62, we failed to detect cilium base signal despite robust NPC fluorescence (Figures 4B and 4K). Thus, several nups that contribute to forming the NPC diffusion barrier and facilitating active nuclear transport are undetectable at the cilium base under our experimental conditions.
Nanoscale spatial organization of nups at cilia
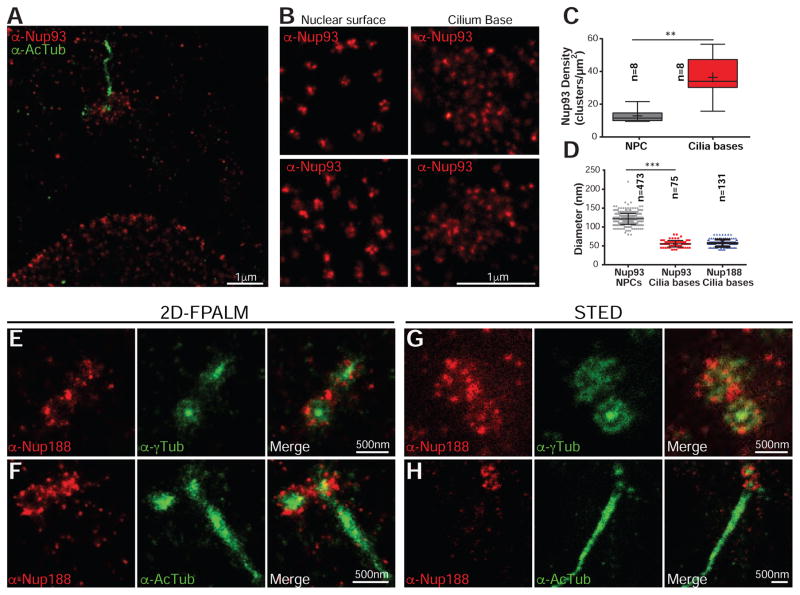
Traditional loss-of-function approaches for inhibiting nup function at different subcellular locations are difficult to interpret because of potential off-target effects of inhibiting nuclear transport. Thus, our approach for investigating the mechanism of nup function at cilia bases was to directly test the hypothesis that nups form a “ciliary pore complex” structurally analogous to the NPC, and thus be capable of organizing into ~100 nm diameter rings at cilia bases (Kee et al., 2012). As such structures would not be visible by conventional light microscopy, we took advantage of our super-resolution 2D-Fluorescence Photoactivation Localization Microscopy (FPALM) super-resolution microscope, which generates images with resolutions of ~30 nm in x,y (Huang et al., 2013). A super-resolution image of an RPE cell stained with anti-Nup93 antibody shows a concentration of anti-Nup93 signal below the cilium (Figure 5A; anti-AcTub staining shown in green). The anti-Nup93 localization ‘spots’ were distributed evenly with a density of ~36/μm2 (Figure 5B, right panels and Figure 5C), whereas the localization clusters were less dense at the nuclear envelope (~12/μm2; Figure 5B, left panels and Figure 5C), consistent with them being NPCs. There were no obvious ring structures at the cilium base, but importantly this was not a limitation of the antibody staining or resolution as we observed rings at the nuclear surface (Figure 5B, left panels). Even when rings could not be conclusively discerned, the diameters of the nuclear envelope clusters were on average ~125 nm, consistent with the dimensions of NPCs (Figure 5D). In contrast, the average diameter of the localizations at cilia bases were ~50 nm (Figure 5D), too small to form an analogous structure. Lastly, to ensure that the clusters of anti-Nup93 localizations represent labeling of multiple copies of Nup93, as opposed to the local repetitive switching of individual fluorophore labeled antibodies, we provide a comparison showing much higher localization numbers at clusters representing NPCs and those at the cilium base compared to background labeling in the same dataset (Figures S4A–S4D).
Figure 5. Nups do not form NPC-like rings at cilia bases.
(A) 2D FPALM super-resolution image of a cell co-stained with anti-Nup93 (red; AF647) and anti-AcTub antibodies (green; Cy3B).
(B) Higher magnification views of the nuclear surface (left panels) and the cilium base (right panels) of two different cells stained with anti-Nup93 (AF647).
(C) Box plot where the density of localization clusters at the nuclear envelope and cilia bases are directly compared. The box depicts the 25th to 75th percentiles with the median marked as horizontal line in box. The whiskers mark the data range from the smallest to the largest value. “+” indicates the mean of the distribution (n=number of cells). T-test, ** p<0.005.
(D) Plot of the diameter (mean +/− SD) of localization clusters at the nuclear envelope and cilia bases. n=number of localization clusters from 3 independent experiments. T-test, *** p<0.0005.
(E) 2D FPALM images of anti-Nup188 (red; AF647) and anti-γTub antibodies (green; Cy3B) with merge on right.
(F) As in (E) except with anti-AcTub co-stain.
(G) STED images of a cell co-stained with anti-Nup188 (red; ATTO647N) and anti-γTub antibodies (green; ATTO594) with merge on right.
(H) As in (G) except with anti-AcTub co-stain.
See also Figure S4.
We acquired similar data for the anti-Nup188 cilium base localizations (Figures 5E and 5F), but a low labeling density of NPCs (likely due to differences in local epitope accessibility) with our specific antibody precluded a direct comparison to anti-Nup188 stained NPCs. Nonetheless, the overall density and distribution of Nup188 immunolocalizations at the cilium base were similar to Nup93 (Figure 5D). Interestingly, in cases where we imaged down the basal body/centriole axis (lower centrioles in Figures 5E and 5F), the anti-Nup188 clusters encircled the anti-γTub or anti-AcTub signal in a ~400 nm diameter ring. Similar results were found using STED microscopy (Figures 5G and 5H), which does not rely on the stochastic blinking of fluorophores and serves to emphasize the specific clustered distribution of the anti-Nup188 signal (see also Figure S4E for a comparison of fluorescence intensities, which are 2–3 fold higher at cilium base). Taken together, the difference in relative ‘size’ and density of the localizations at cilia bases compared to the nuclear envelope support the conclusion that Nup93 and Nup188 are not organized as ~100 nm diameter rings and thus do not contribute to the formation of one, or a series, of NPC-like “ciliary pore complexes.” Interestingly, our data support that Nup188 is found in the pericentriolar material (PCM) surrounding and encapsulating the centriole core.
Nup188 clusters form two barrels at the cilium base
To better understand the spatial organization of the nup clusters around the basal body/centrioles, we took advantage of our recently developed Whole-cell 4Pi Single-Molecule Switching Nanoscopy (W-4PiSMSN) microscope that offers an x-y and z-resolution of ~10–20 nm over an ~1 μm optical section; it is thus ideal for imaging cellular structures at the dimensions of the cilium base (Huang et al., 2016). In order to visualize the underlying ultrastructure using W-4PiSMSN, we focused primarily on imaging Nup188 which provides a dense labeling.
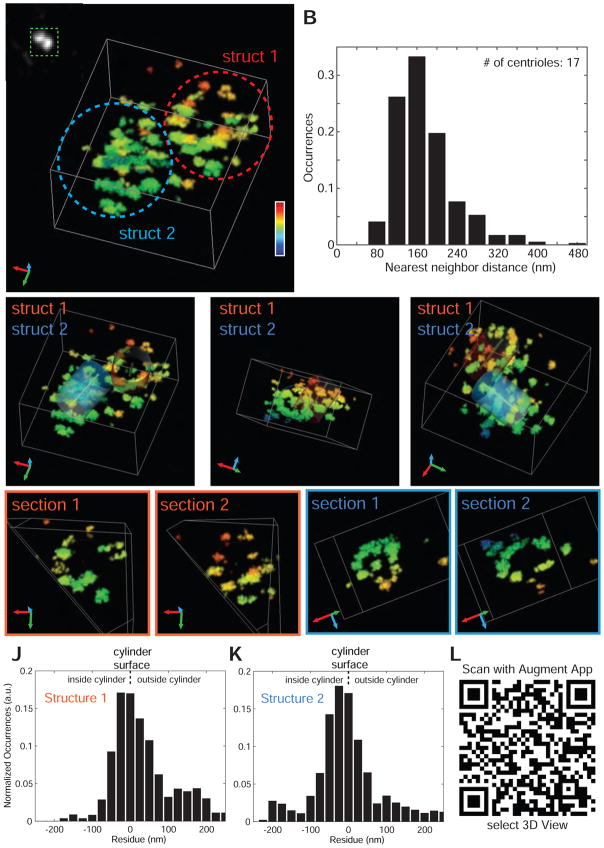
Strikingly, 3D super-resolution images of the anti-Nup188 signal revealed two discrete structures, each composed of dozens of individual localization clusters that could not be resolved by confocal microscopy (Figure 6A, confocal image in inset at lower magnification; Movie S2). The number of localizations in these clusters far exceeded those outside of the structure (i.e. background) confirming that these clusters do not represent the repetitive blinking of individual antibodies but multiple copies of Nup188 (compare Figure S5A left panel [cilia bases] to right panel [background] and see cumulative density plot in Figure S5B). In addition, the reader is encouraged to manipulate the structures using the Augment 3D visualization app (available in Apple App Store or Google Play), which recognizes the QR code in Figure 6L. Interestingly, the two structures were of similar size and shape but were offset to each other (Movie S2) in an arrangement consistent with the basal body and daughter centriole, which are often found perpendicular to each other (Figure S3B). As the structures resembled barrels, we fitted cylinders into each structure (Figures 6C, 6E (rotations), 6L, and Movie S2) where a 436 nm diameter cylinder best fit the data. In Figures 6J and 6K, we have plotted the distance of single molecule localizations (including outliers) both inside and outside of the barrel structures, which shows a tight peak at the cylinder surface and a relatively empty center. Consistent with this, by sectioning along the central axes of both cylinders, a clear opening is visible (Figures 6F–6I) supporting the concept that Nup188 would encircle and encapsulate the centrioles within a barrel-like structure. Five additional structures with fitted cylinder models can be found in Movie S3 (see Figure S6 for corresponding histograms that support the cylinder models). Moreover, we performed a statistical analysis on each of the structures using a bootstrapping method to provide uncertainty values of the cylinder radii estimates, which ranged between 0.1 and 6.7 nm (Table S1).
Figure 6. Nup188 forms two barrel-like structures that encase the centrioles.
(A) Diffraction-limited (inset) and 3D W-4PiSMSN super-resolution image of anti-Nup188 signal at the cilium base. Heat map coloration reflects depth in z as depicted by scale. Individual structures are circled. See also Movie S2.
(B) Nearest neighbor distances from all localization clusters in all structures imaged were calculated and plotted.
(C–E) Rotated views of image in (A) where two cylinders (blue and red) have been fitted into both structures denoted by “1” and “2”. See also Movie S2.
(F–G) and (H–I) Planar sections of each structure cut across the cylinder axis (central point). Dotted lines represent cylinders.
(J–K) Distribution of the distances of individual localizations from W-4PiSMSN data inside (negative values) and outside (positive values) the modeled cylinder surface of Struct 1 and 2.
(L) QR code to load Nup188 W-4PiSMSN structure into the 3D visualization application Augment™. See Experimental Procedures for instructions.
See also Figures S5 and S6.
The distribution of Nup188 around the basal body/centrioles was analogous to components of the PCM. To better understand if the distribution of Nup188 reflects an underlying organization of the PCM, we calculated the nearest neighbor distances of all of the localization clusters from all of our structures (n=17) in 3D (Figure 6B; Figure S5C also shows dispersed cluster distances in background). This analysis revealed a striking organization where the localization clusters showed a peak nearest neighbor distance of 160 nm suggesting that Nup188 either establishes or reflects a structural unit of the PCM and/or centriole.
Discussion
This study provides one of only a handful of examples of the power of emerging super-resolution approaches to reveal new insights into the organization of proteins at the nanoscale (Huang et al., 2016; Xu et al., 2013). The data make a compelling case that a specific subset of nups localize to cilia bases as previously undescribed components of the PCM. Indeed, our data support that Nup188 is capable of organizing into barrel-like structures with ~400 nm diameters that encase the basal body and daughter centriole. Thus, this work marries well with the recent reconceptualization of the PCM (driven by super-resolution studies (Fu and Glover, 2012; Lawo et al., 2012; Mennella et al., 2012; Sonnen et al., 2012)) from a more traditional amorphous organization to one that has more discrete “layers” (Mennella et al., 2014). Indeed, the super-resolution images of the Nup188 clusters are remarkably similar to those of pericentrin (Mennella et al., 2012), which has been suggested to have a 9-fold radial symmetry scaffolded by the centriole itself. While our data do not definitively establish such a 9-fold radial symmetry for Nup188, we think that the remarkably consistent ~160 nm distance between Nup188 clusters (Figure 6B) reflects binding to an underlying architecture; the centriole is the most obvious candidate to template this architecture and provides a likely hypothesis for future testing. Such a hypothesis must also be informed by a comprehensive biochemical analysis of nups at the cilium base, which is a necessity to precisely determine the stoichiometry and nearest neighbor interactions that drive this unique nup organization between the nuclear and cilia compartments.
The different organization of nups at the cilium base versus NPCs suggests a context-dependent function for nups distinct from their role in nuclear transport, but can we functionally decouple these roles? Several lines of evidence support that the loss of cilia in Xenopus tissues reflects a direct role for the inner ring nups at the cilium base, and not at NPCs. First, we observe a distinct specificity in the capacity of the inner ring nups to affect cilia (Figures 1B, 2A, 3A and 3B). What is most striking is that this functional specificity is mirrored by the unique capacity of these nups, and not others, to localize to cilia bases (Figures 4A, 4B and 4K). Second, as nuclear transport is essential in all cells, a disruption of NPCs should lead to devastating consequences for the embryo. Indeed, depletion of the outer ring Y-complex component Nup133 leads to severe embryonic malformations (due to NPC disruption; Figures 3C–3E), yet remarkably, the cilia are only mildly affected (Figure 3B). This suggests that nuclear transport and cilia are uncoupled under these conditions. Thirdly, and consistent with these two processes being uncoupled, the knockdown of both Nup188 and Nup93 leads to a loss of cilia throughout the embryo, without affecting its gross morphology (Figures 2A and 3A). This result is further reflected by the normal accumulation of NLS-GFP in the nucleus and no obvious changes to NPC density in nup188/93 morphants (Figures 3C–3F). Taken in the aggregate, it appears unlikely that a disruption of nuclear transport underlies the cilia-specific effects of depleting the inner ring nups; we conclude instead that we have uncovered a new (and direct) role for these nups at cilia.
The clear difference in the organization of nups at the cilium base versus the NPC support that they function outside of the attractive “ciliary pore complex” model (Kee et al., 2012). Consistent with this idea, the distribution of nups at the cilium base is far from the ciliary transition zone where such a structure would also be presumed to gate the cilium (Figures 5 and S3). In addition, while one could argue that a nup-based diffusion barrier need not be formed from ~100 nm diameter channels (for example, hydrogels of FG-nups generated from recombinant proteins can recapitulate the barrier properties of the NPC (Frey and Gorlich, 2007)), a pre-requisite would be the presence of the critical FG-nups that define the NPC barrier, which we were unable to detect at the cilium base (Figures 4B and 4K). Therefore, we suggest that the cilium base diffusion barrier is likely established by other proteins that would also be consistent with studies indicating it has a larger pore size (7.9–9 nm) (Breslow et al., 2013; Lin et al., 2013) versus that of the NPC (~2.6–5 nm) (Keminer and Peters, 1999; Mohr et al., 2009; Paine et al., 1975).
One could speculate many possible roles for the inner ring nups at the bases of cilia but we favor a structural or scaffold model; that is, Nup188 and Nup93 generate a critical platform on which other proteins assemble. One clear candidate is the mRNA export factor Gle1, which interacts with the inner ring nup, Nup155 (Rayala et al., 2004). Interestingly, Gle1 was recently shown to localize to the cilium base in Zebrafish and in RPE cells (S. Wente, personal communication); Gle1 contributes to inositol signaling, shown previously to be required for cilia motility and LR patterning (Sarmah et al., 2005; Sarmah et al., 2007). Thus, a hypothesis is the disruption of inner ring nups leads to the loss of Gle1 function at cilia bases. Alternatively, as a component of PCM required for cilia, Nup188 might provide a unique molecular signature that triggers basal body versus centrosome maturation. To address these and other hypotheses will again require biochemical approaches capable of defining the interactome of nups at cilia, a key goal for our future work as it will also open the door to generate tools capable of more definitively decoupling the role of nups at NPCs and cilia.
While a molecular mechanism for how nups impact cilia structure/function has yet to be defined, a major motivation for these studies was a desire to elucidate a plausible mechanism for NUP188 in Htx and CHD, which was far from obvious when we identified the NUP188 duplication in our patient (Fakhro et al., 2011). With established technologies to interrogate the human genome, we are beginning to define large numbers of genetic lesions that may cause CHD and congenital malformations more generally (Fakhro et al., 2011; Glessner et al., 2014; Greenway et al., 2009; Zaidi et al., 2013). However, assigning disease causality remains a challenge. In most cases, second alleles are unavailable with the limited cohorts analyzed to date, and the diversity and novelty of the genes identified rarely provides a parsimonious explanation of a pathogenic developmental mechanism that could support disease relevance. Together, these factors confound the disease relevance for a specific candidate gene such as NUP188. As our results demonstrate that Nup188 is essential for cilia, LR patterning, and normal cardiac looping, we have established a basis for assigning NUP188 as a ciliopathy gene. More broadly, elucidating a plausible mechanism for disease pathogenesis, i.e. a mechanism for Nup188 in causing Htx/CHD, is a powerful alternative strategy for defining disease relevance especially in the many cases where only single alleles are available (Fakhro et al., 2011; Glessner et al., 2014; Greenway et al., 2009; Zaidi et al., 2013). In sum, this work provides a compelling case study for how patient driven gene discovery can help uncover unanticipated mechanisms that impact our understanding of LR axis patterning and heart development, and the subcellular organization of protein complexes at the nanoscale.
Experimental procedures
See Supplemental Experimental procedures for more detailed methods.
Xenopus and mammalian cell culture
Xenopus were housed and cared for according to established protocols that were approved by Yale IACUC. Using standard protocols, we injected MOs, fluorescent tracers, and/or mRNAs into 1 or 2-cell Xenopus embryos and assayed gene expression by in situ hybridization and cardiac looping by visual inspection (Boskovski et al., 2013; del Viso and Khokha, 2012; Khokha et al., 2002). Animal caps were cultured in standard frog media on fibronectin coated plates, while Human TERT RPE cells were cultured in DMEM/Ham’s F12 media supplemented with 10% fetal bovine serum (FBS). For siRNA experiments, cells were transfected with siRNAs using Lipofectamine RNAiMAX transfection Reagent (Life Technologies) following the manufacturer’s instructions. Cells were incubated with siRNA mix overnight in media without FBS. The next day cells were transferred to media supplemented with 10% FBS for 36 h and then were starved in media without FBS for an additional 48 h to grow cilia, before fixation.
Immunofluorescence and conventional microscopy
For both Xenopus and cell culture lines, cells were first briefly permeabilized by 0.1% saponin followed by fixation in −20°C Methanol. Primary and secondary antibodies are listed in Supplemental Experimental Procedures. Immunostained cells or embryos were imaged on either a Zeiss Axiovert fitted with optical interference (Apotome) microscopy (Figures 3A, 3B, S1G, and S2D), a Leica TCS SP8 confocal microscope (Figures 2A, 3C, 3D, 4A, 4B, S1J, S2B, S2E and S2F), or a Deltavision (Applied Precision/GE Healthcare) with deconvolution (Figures 4C–4D and 4G–4H and Figure S3). Fluorescent images were processed and analyzed with Fiji/ImageJ (Schindelin et al., 2012). To localize SNAP-Nup188, HeLa cells were transfected with SNAP-Nup188 and then stained with the cell permeable SNAP substrate Oregon Green (New England Biolabs; 1:500) following the manufacturer’s instructions prior to fixation (Keppler et al., 2003).
Statistical Analysis
All experiments were performed a minimum of three times (except Figures S1H, S1I, S2C, and S2D which were done twice) and numbers stated in graphs are the composite of multiple experiments. Statistical significance of cardiac looping, pitx2c and coco abnormalities, as well as the nup93 rescue experiment and the cilia reduction in RPE cells upon Nup188 siRNA, was tested using Chi-squared or Fisher’s Exact test. The significance of the amount of cilia in LROs was evaluated using paired (for injections at 2-cell stage) or unpaired, two-tailed student’s t- tests (Figures 2 and S2C) using GraphPad Prism version 6.00, GraphPad Software, La Jolla California USA, www.graphpad.com. The statistical significance of the intensity reduction after siRNA depletion of NUP93 and NUP188 at the nuclei or base of cilia in RPE cells was tested using the unpaired, two-tailed student’s t-tests (non-parametric) using GraphPad (Figures 4E and 4I). In all Figures, statistical significance was defined as p<0.05.
Super-resolution imaging
Immunolabeled samples were imaged on either a custom built 2D FPALM setup as described previously (Huang et al., 2013; Lin et al., 2015) or a custom built W-4PiSMSN microscope (Huang et al., 2016). For STED based imaging, we performed fixation and immunolabeling but used secondary antibodies conjugated to ATTO647N or ATTO594 dyes. These samples were imaged on a Leica TCS SP8 Gated STED 3x microscope.
Conventional fluorescence image analysis
All fluorescent micrographs were processed and analyzed using Fiji/ImageJ software (Schindelin et al., 2012). In some cases (images presented in Figures 4C, 4D, 4G, 4H, S3B and S3C), images were first deconvolved using the iterative algorithm in softWoRx (version 5.5; Applied Precision). All quantification in Fiji, however, was performed using unprocessed images. To determine relative intensities, we selected a region of interest at the nuclear surface or the base of cilia and measured the mean intensity which in some cases (Figures 4E and 4I) was then normalized to the mean intensity of the background for each individual image. In Figure S3B and S3C, line profiles were generated using the “plot profile” function. To count cilia in LROs, we used the “count particles” function. To evaluate NPC density (Figures 3D and 3F), images were made binary and a Gaussian blur filter was applied. To count NPCs, the “find maxima” function was employed. NPC density was assessed by dividing the maxima count over the area in which it was determined. Further quantification, data plotting and statistical analyses was performed in GraphPad Prism version 6.00, GraphPad Software, La Jolla California USA, www.graphpad.com.
Super-Resolution image analysis
For 2D FPALM image analysis, we measured the density of the Nup93/Nup188 localizations at both the nuclear envelope and bases of cilia using Fiji/ImageJ software by manually thresholding and using the “find maxima” tool. Diameters of clusters were measured using the line profile tool in Fiji/ImageJ software. For cluster intensity measurements in STED (Figure S4E), intensity values are average counts per pixel in the selected square regions (white boxes). For 3D image analysis (W-4PiSMSN), we first de-noised the data and then defined clusters as a group of single-molecule localizations if the number of localizations within the group was above 20. Subsequently, the centers of the clusters were determined by calculating their centroids, and the nearest neighbor distance of a cluster center was calculated based on its minimum Euclidian distance to other centers within a Nup188 structure. To model cylinders in the data, the cylinder’s central axis and diameter were determined by regression. To determine the uncertainty of the cylinder radius estimates, we implemented a bootstrapping method.
3D Visualization with Augmented Reality of Nup188
To visualize and manipulate a 3D image of Nup188, the QR code in Figure 6L can be scanned with the “Augment 3D” visualization platform, which is available for your smartphone or tablet from the App store (iPhone/iPad) or Google Play (Android). 1) Download and launch the “Augment” app. 2) Scan the QR code (Figure 6L). Launch Augment and select Scan. Point your camera at the QR code which will be scanned and the image loaded. Select 3D view in the menu at the bottom of the screen and manipulate the image by dragging with your figure or zoom in and out by pinching or spreading motions with two fingers. NOTE: no personal information is required or stored by the Augment 3D app.
Supplementary Material
Highlights.
Cilia loss is a likely cause of Heterotaxy in a patient with a Nup188 duplication
Disruption of inner ring nups leads to defects in heart structure
Inner ring nups localize to the cilium base and are required for cilia
3D nanoscopy reveals nups encasing centrioles in evenly spaced assemblies
Acknowledgments
We thank the patients and their families, who are the inspiration for this study. The authors wish to thank Michael Slocum and Sarah Kirschner for animal husbandry and Mark Lessard for invaluable help with STED microscopy. We are indebted to the CCMI core for fluorescent microscopy resources. We thank George Takahashi from the Purdue Envision Center for help on data visualization and 3D Augmented Reality. We thank Wolfram Antonin, Chin-Bin Chien, Mary Dasso, Elias Coutavas, Günter Blobel, and Martin Hetzer for reagents including antibodies and plasmids and Susan Wente for sharing unpublished data. FdV was supported by a Brown-Coxe Postdoctoral Fellowship. This work was supported by the NIH (R01 HL124402 to JB, CPL, and MKK and P30 DK45735 to JB) and the Wellcome Trust (095927/A/11/Z) to JB. MKK is a Mallinckrodt Scholar. MC was supported by an NIH training grant (T32GM007223). JB discloses significant financial interest in Bruker Corp. JB and FH disclose significant financial interest in Hamamatsu Photonics.
Footnotes
Author Contributions
FdV, CPL, and MKK conceived, designed, contributed, and analyzed all the data from each of the experiments. FdV performed all the Xenopus experiments. FdV, FH, JM, and YZ performed all super-resolution experiments. FdV, JM, and MC performed all immunolocalization experiments. FdV and NR performed Xenopus gliding assays. FH and JB designed and built custom super-resolution microscopes and analyzed these data with FdV, CPL, and MKK. Manuscript was written by FdV, CPL and MKK with input from all authors.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References and Notes
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, et al. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. doi: 10.1038/nature06405. [DOI] [PubMed] [Google Scholar]
- Amlacher S, Sarges P, Flemming D, van Noort V, Kunze R, Devos DP, Arumugam M, Bork P, Hurt E. Insight into structure and assembly of the nuclear pore complex by utilizing the genome of a eukaryotic thermophile. Cell. 2011;146:277–289. doi: 10.1016/j.cell.2011.06.039. [DOI] [PubMed] [Google Scholar]
- Basu B, Brueckner M. Cilia multifunctional organelles at the center of vertebrate left-right asymmetry. Curr Top Dev Biol. 2008;85:151–174. doi: 10.1016/S0070-2153(08)00806-5. [DOI] [PubMed] [Google Scholar]
- Belgareh N, Rabut G, Bai SW, van Overbeek M, Beaudouin J, Daigle N, Zatsepina OV, Pasteau F, Labas V, Fromont-Racine M, et al. An evolutionarily conserved NPC subcomplex, which redistributes in part to kinetochores in mammalian cells. The Journal of cell biology. 2001;154:1147–1160. doi: 10.1083/jcb.200101081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisgrove BW, Yost HJ. The roles of cilia in developmental disorders and disease. Development. 2006;133:4131–4143. doi: 10.1242/dev.02595. [DOI] [PubMed] [Google Scholar]
- Blum M, Beyer T, Weber T, Vick P, Andre P, Bitzer E, Schweickert A. Xenopus, an ideal model system to study vertebrate left-right asymmetry. Dev Dyn. 2009;238:1215–1225. doi: 10.1002/dvdy.21855. [DOI] [PubMed] [Google Scholar]
- Boskovski MT, Yuan S, Pedersen NB, Goth CK, Makova S, Clausen H, Brueckner M, Khokha MK. The heterotaxy gene GALNT11 glycosylates Notch to orchestrate cilia type and laterality. Nature. 2013;504:456–459. doi: 10.1038/nature12723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bouckson-Castaing V, Moudjou M, Ferguson DJ, Mucklow S, Belkaid Y, Milon G, Crocker PR. Molecular characterisation of ninein, a new coiled-coil protein of the centrosome. Journal of cell science. 1996;109(Pt 1):179–190. doi: 10.1242/jcs.109.1.179. [DOI] [PubMed] [Google Scholar]
- Breslow DK, Koslover EF, Seydel F, Spakowitz AJ, Nachury MV. An in vitro assay for entry into cilia reveals unique properties of the soluble diffusion barrier. The Journal of cell biology. 2013;203:129–147. doi: 10.1083/jcb.201212024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchwalter AL, Liang Y, Hetzer MW. Nup50 is required for cell differentiation and exhibits transcription-dependent dynamics. Mol Biol Cell. 2014;25:2472–2484. doi: 10.1091/mbc.E14-04-0865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capelson M, Liang Y, Schulte R, Mair W, Wagner U, Hetzer MW. Chromatin-bound nuclear pore components regulate gene expression in higher eukaryotes. Cell. 2010;140:372–383. doi: 10.1016/j.cell.2009.12.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chug H, Trakhanov S, Hulsmann BB, Pleiner T, Gorlich D. Crystal structure of the metazoan Nup62*Nup58*Nup54 nucleoporin complex. Science. 2015;350:106–110. doi: 10.1126/science.aac7420. [DOI] [PubMed] [Google Scholar]
- Cronshaw JM, Krutchinsky AN, Zhang W, Chait BT, Matunis MJ. Proteomic analysis of the mammalian nuclear pore complex. The Journal of cell biology. 2002;158:915–927. doi: 10.1083/jcb.200206106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D’Angelo MA, Gomez-Cavazos JS, Mei A, Lackner DH, Hetzer MW. A change in nuclear pore complex composition regulates cell differentiation. Dev Cell. 2012;22:446–458. doi: 10.1016/j.devcel.2011.11.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis LI, Blobel G. Identification and characterization of a nuclear pore complex protein. Cell. 1986;45:699–709. doi: 10.1016/0092-8674(86)90784-1. [DOI] [PubMed] [Google Scholar]
- del Viso F, Khokha M. Generating diploid embryos from Xenopus tropicalis. Methods in molecular biology. 2012;917:33–41. doi: 10.1007/978-1-61779-992-1_3. [DOI] [PubMed] [Google Scholar]
- Dishinger JF, Kee HL, Jenkins PM, Fan S, Hurd TW, Hammond JW, Truong YN, Margolis B, Martens JR, Verhey KJ. Ciliary entry of the kinesin-2 motor KIF17 is regulated by importin-beta2 and RanGTP. Nature cell biology. 2010;12:703–710. doi: 10.1038/ncb2073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eggenschwiler JT, Anderson KV. Cilia and developmental signaling. Annu Rev Cell Dev Biol. 2007;23:345–373. doi: 10.1146/annurev.cellbio.23.090506.123249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhro KA, Choi M, Ware SM, Belmont JW, Towbin JA, Lifton RP, Khokha MK, Brueckner M. Rare copy number variations in congenital heart disease patients identify unique genes in left-right patterning. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2915–2920. doi: 10.1073/pnas.1019645108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nature reviews Molecular cell biology. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- Frey S, Gorlich D. A saturated FG-repeat hydrogel can reproduce the permeability properties of nuclear pore complexes. Cell. 2007;130:512–523. doi: 10.1016/j.cell.2007.06.024. [DOI] [PubMed] [Google Scholar]
- Fu J, Glover DM. Structured illumination of the interface between centriole and peri-centriolar material. Open Biol. 2012;2:120104. doi: 10.1098/rsob.120104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glessner J, Bick AG, Ito K, Homsy J, Rodriguez-Murillo L, Fromer M, Mazaika EJ, Vardarajan B, Italia MJ, Leipzig J, et al. Increased Frequency of De Novo Copy Number Variations in Congenital Heart Disease by Integrative Analysis of SNP Array and Exome Sequence Data. Circulation research. 2014 doi: 10.1161/CIRCRESAHA.115.304458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gomez-Cavazos JS, Hetzer MW. The nucleoporin gp210/Nup210 controls muscle differentiation by regulating nuclear envelope/ER homeostasis. The Journal of cell biology. 2015;208:671–681. doi: 10.1083/jcb.201410047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grandi P, Schlaich N, Tekotte H, Hurt EC. Functional interaction of Nic96p with a core nucleoporin complex consisting of Nsp1p, Nup49p and a novel protein Nup57p. EMBO J. 1995;14:76–87. doi: 10.1002/j.1460-2075.1995.tb06977.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenway SC, Pereira AC, Lin JC, DePalma SR, Israel SJ, Mesquita SM, Ergul E, Conta JH, Korn JM, McCarroll SA, et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nat Genet. 2009;41:931–935. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hashizume C, Moyori A, Kobayashi A, Yamakoshi N, Endo A, Wong RW. Nucleoporin Nup62 maintains centrosome homeostasis. Cell cycle. 2013;12:3804–3816. doi: 10.4161/cc.26671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Hartwich TM, Rivera-Molina FE, Lin Y, Duim WC, Long JJ, Uchil PD, Myers JR, Baird MA, Mothes W, et al. Video-rate nanoscopy using sCMOS camera-specific single-molecule localization algorithms. Nature methods. 2013;10:653–658. doi: 10.1038/nmeth.2488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang F, Sirinakis G, Allgeyer ES, Schroeder LK, Duim WC, Kromann EB, Phan T, Rivera-Molina FE, Myers JR, Irnov I, et al. Ultra-High Resolution 3D Imaging of Whole Cells. Cell. 2016 doi: 10.1016/j.cell.2016.06.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hurt E, Beck M. Towards understanding nuclear pore complex architecture and dynamics in the age of integrative structural analysis. Current opinion in cell biology. 2015;34:31–38. doi: 10.1016/j.ceb.2015.04.009. [DOI] [PubMed] [Google Scholar]
- Ishikawa H, Marshall WF. Ciliogenesis: building the cell’s antenna. Nature reviews Molecular cell biology. 2011;12:222–234. doi: 10.1038/nrm3085. [DOI] [PubMed] [Google Scholar]
- Itoh G, Sugino S, Ikeda M, Mizuguchi M, Kanno S, Amin MA, Iemura K, Yasui A, Hirota T, Tanaka K. Nucleoporin Nup188 is required for chromosome alignment in mitosis. Cancer science. 2013;104:871–879. doi: 10.1111/cas.12159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto FV, Benner C, Hetzer MW. The nucleoporin Nup153 regulates embryonic stem cell pluripotency through gene silencing. Genes Dev. 2015;29:1224–1238. doi: 10.1101/gad.260919.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalverda B, Pickersgill H, Shloma VV, Fornerod M. Nucleoporins directly stimulate expression of developmental and cell-cycle genes inside the nucleoplasm. Cell. 2010;140:360–371. doi: 10.1016/j.cell.2010.01.011. [DOI] [PubMed] [Google Scholar]
- Kee HL, Dishinger JF, Blasius TL, Liu CJ, Margolis B, Verhey KJ. A size-exclusion permeability barrier and nucleoporins characterize a ciliary pore complex that regulates transport into cilia. Nature cell biology. 2012;14:431–437. doi: 10.1038/ncb2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kee HL, Verhey KJ. Molecular connections between nuclear and ciliary import processes. Cilia. 2013;2:11. doi: 10.1186/2046-2530-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelley K, Knockenhauer KE, Kabachinski G, Schwartz TU. Atomic structure of the Y complex of the nuclear pore. Nat Struct Mol Biol. 2015;22:425–431. doi: 10.1038/nsmb.2998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keminer O, Peters R. Permeability of single nuclear pores. Biophys J. 1999;77:217–228. doi: 10.1016/S0006-3495(99)76883-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keppler A, Gendreizig S, Gronemeyer T, Pick H, Vogel H, Johnsson K. A general method for the covalent labeling of fusion proteins with small molecules in vivo. Nat Biotechnol. 2003;21:86–89. doi: 10.1038/nbt765. [DOI] [PubMed] [Google Scholar]
- Khokha MK, Chung C, Bustamante EL, Gaw LW, Trott KA, Yeh J, Lim N, Lin JC, Taverner N, Amaya E, et al. Techniques and probes for the study of Xenopus tropicalis development. Dev Dyn. 2002;225:499–510. doi: 10.1002/dvdy.10184. [DOI] [PubMed] [Google Scholar]
- Lawo S, Hasegan M, Gupta GD, Pelletier L. Subdiffraction imaging of centrosomes reveals higher-order organizational features of pericentriolar material. Nature cell biology. 2012;14:1148–1158. doi: 10.1038/ncb2591. [DOI] [PubMed] [Google Scholar]
- Liang Y, Franks TM, Marchetto MC, Gage FH, Hetzer MW. Dynamic association of NUP98 with the human genome. PLoS Genet. 2013;9:e1003308. doi: 10.1371/journal.pgen.1003308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Light WH, Freaney J, Sood V, Thompson A, D’Urso A, Horvath CM, Brickner JH. A conserved role for human Nup98 in altering chromatin structure and promoting epigenetic transcriptional memory. PLoS Biol. 2013;11:e1001524. doi: 10.1371/journal.pbio.1001524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CR, Kioussi C, O’Connell S, Briata P, Szeto D, Liu F, Izpisua-Belmonte JC, Rosenfeld MG. Pitx2 regulates lung asymmetry, cardiac positioning and pituitary and tooth morphogenesis. Nature. 1999;401:279–282. doi: 10.1038/45803. [DOI] [PubMed] [Google Scholar]
- Lin Y, Long JJ, Huang F, Duim WC, Kirschbaum S, Zhang Y, Schroeder LK, Rebane AA, Velasco MG, Virrueta A, et al. Quantifying and optimizing single-molecule switching nanoscopy at high speeds. PloS one. 2015;10:e0128135. doi: 10.1371/journal.pone.0128135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin YC, Niewiadomski P, Lin B, Nakamura H, Phua SC, Jiao J, Levchenko A, Inoue T, Rohatgi R, Inoue T. Chemically inducible diffusion trap at cilia reveals molecular sieve-like barrier. Nature chemical biology. 2013;9:437–443. doi: 10.1038/nchembio.1252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiodice I, Alves A, Rabut G, Van Overbeek M, Ellenberg J, Sibarita JB, Doye V. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis. Mol Biol Cell. 2004;15:3333–3344. doi: 10.1091/mbc.E03-12-0878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lupu F, Alves A, Anderson K, Doye V, Lacy E. Nuclear pore composition regulates neural stem/progenitor cell differentiation in the mouse embryo. Dev Cell. 2008;14:831–842. doi: 10.1016/j.devcel.2008.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrath J, Somlo S, Makova S, Tian X, Brueckner M. Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell. 2003;114:61–73. doi: 10.1016/s0092-8674(03)00511-7. [DOI] [PubMed] [Google Scholar]
- Mennella V, Agard DA, Huang B, Pelletier L. Amorphous no more: subdiffraction view of the pericentriolar material architecture. Trends Cell Biol. 2014;24:188–197. doi: 10.1016/j.tcb.2013.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mennella V, Keszthelyi B, McDonald KL, Chhun B, Kan F, Rogers GC, Huang B, Agard DA. Subdiffraction-resolution fluorescence microscopy reveals a domain of the centrosome critical for pericentriolar material organization. Nature cell biology. 2012;14:1159–1168. doi: 10.1038/ncb2597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra RK, Chakraborty P, Arnaoutov A, Fontoura BM, Dasso M. The Nup107-160 complex and gamma-TuRC regulate microtubule polymerization at kinetochores. Nature cell biology. 2010;12:164–169. doi: 10.1038/ncb2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohr D, Frey S, Fischer T, Guttler T, Gorlich D. Characterisation of the passive permeability barrier of nuclear pore complexes. EMBO J. 2009;28:2541–2553. doi: 10.1038/emboj.2009.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ori A, Banterle N, Iskar M, Andres-Pons A, Escher C, Khanh Bui H, Sparks L, Solis-Mezarino V, Rinner O, Bork P, et al. Cell type-specific nuclear pores: a case in point for context-dependent stoichiometry of molecular machines. Mol Syst Biol. 2013;9:648. doi: 10.1038/msb.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orjalo AV, Arnaoutov A, Shen Z, Boyarchuk Y, Zeitlin SG, Fontoura B, Briggs S, Dasso M, Forbes DJ. The Nup107-160 nucleoporin complex is required for correct bipolar spindle assembly. Mol Biol Cell. 2006;17:3806–3818. doi: 10.1091/mbc.E05-11-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paine PL, Moore LC, Horowitz SB. Nuclear envelope permeability. Nature. 1975;254:109–114. doi: 10.1038/254109a0. [DOI] [PubMed] [Google Scholar]
- Pascual-Garcia P, Jeong J, Capelson M. Nucleoporin Nup98 associates with Trx/MLL and NSL histone-modifying complexes and regulates Hox gene expression. Cell Rep. 2014;9:433–442. doi: 10.1016/j.celrep.2014.09.002. [DOI] [PubMed] [Google Scholar]
- Pennekamp P, Menchen T, Dworniczak B, Hamada H. Situs inversus and ciliary abnormalities: 20 years later, what is the connection? Cilia. 2015;4:1. doi: 10.1186/s13630-014-0010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raices M, D’Angelo MA. Nuclear pore complex composition: a new regulator of tissue-specific and developmental functions. Nature reviews Molecular cell biology. 2012;13:687–699. doi: 10.1038/nrm3461. [DOI] [PubMed] [Google Scholar]
- Rayala HJ, Kendirgi F, Barry DM, Majerus PW, Wente SR. The mRNA export factor human Gle1 interacts with the nuclear pore complex protein Nup155. Mol Cell Proteomics. 2004;3:145–155. doi: 10.1074/mcp.M300106-MCP200. [DOI] [PubMed] [Google Scholar]
- Reza N, Khokha M, Del Viso F. Nucleoporin gene expression in Xenopus tropicalis embryonic development. Int J Dev Biol. 2016 doi: 10.1387/ijdb.150317nr. In press. [DOI] [PubMed] [Google Scholar]
- Rout MP, Aitchison JD, Suprapto A, Hjertaas K, Zhao Y, Chait BT. The yeast nuclear pore complex: composition, architecture, and transport mechanism. The Journal of cell biology. 2000;148:635–651. doi: 10.1083/jcb.148.4.635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdev R, Sieverding C, Flotenmeyer M, Antonin W. The C-terminal domain of Nup93 is essential for assembly of the structural backbone of nuclear pore complexes. Mol Biol Cell. 2012;23:740–749. doi: 10.1091/mbc.E11-09-0761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmah B, Latimer AJ, Appel B, Wente SR. Inositol polyphosphates regulate zebrafish left-right asymmetry. Dev Cell. 2005;9:133–145. doi: 10.1016/j.devcel.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Sarmah B, Winfrey VP, Olson GE, Appel B, Wente SR. A role for the inositol kinase Ipk1 in ciliary beating and length maintenance. Proc Natl Acad Sci U S A. 2007;104:19843–19848. doi: 10.1073/pnas.0706934104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, et al. Fiji: an open-source platform for biological-image analysis. Nature methods. 2012;9:676–682. doi: 10.1038/nmeth.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmidt HB, Gorlich D. Transport Selectivity of Nuclear Pores, Phase Separation, and Membraneless Organelles. Trends Biochem Sci. 2016;41:46–61. doi: 10.1016/j.tibs.2015.11.001. [DOI] [PubMed] [Google Scholar]
- Schweickert A, Vick P, Getwan M, Weber T, Schneider I, Eberhardt M, Beyer T, Pachur A, Blum M. The nodal inhibitor Coco is a critical target of leftward flow in Xenopus. Current biology: CB. 2010;20:738–743. doi: 10.1016/j.cub.2010.02.061. [DOI] [PubMed] [Google Scholar]
- Smith ER, Zhang XY, Capo-Chichi CD, Chen X, Xu XX. Increased expression of Syne1/nesprin-1 facilitates nuclear envelope structure changes in embryonic stem cell differentiation. Dev Dyn. 2011;240:2245–2255. doi: 10.1002/dvdy.22717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonnen KF, Schermelleh L, Leonhardt H, Nigg EA. 3D-structured illumination microscopy provides novel insight into architecture of human centrosomes. Biol Open. 2012;1:965–976. doi: 10.1242/bio.20122337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stubbs JL, Oishi I, Izpisua Belmonte JC, Kintner C. The forkhead protein Foxj1 specifies node-like cilia in Xenopus and zebrafish embryos. Nat Genet. 2008;40:1454–1460. doi: 10.1038/ng.267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuwe T, Bley CJ, Thierbach K, Petrovic S, Schilbach S, Mayo DJ, Perriches T, Rundlet EJ, Jeon YE, Collins LN, et al. Architecture of the fungal nuclear pore inner ring complex. Science. 2015;350:56–64. doi: 10.1126/science.aac9176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sukegawa J, Blobel G. A nuclear pore complex protein that contains zinc finger motifs, binds DNA, and faces the nucleoplasm. Cell. 1993;72:29–38. doi: 10.1016/0092-8674(93)90047-t. [DOI] [PubMed] [Google Scholar]
- Sutherland MJ, Ware SM. Disorders of left-right asymmetry: heterotaxy and situs inversus. Am J Med Genet C Semin Med Genet. 2009;151C:307–317. doi: 10.1002/ajmg.c.30228. [DOI] [PubMed] [Google Scholar]
- Tabin CJ, Vogan KJ. A two-cilia model for vertebrate left-right axis specification. Genes Dev. 2003;17:1–6. doi: 10.1101/gad.1053803. [DOI] [PubMed] [Google Scholar]
- Takao D, Dishinger JF, Kee HL, Pinskey JM, Allen BL, Verhey KJ. An assay for clogging the ciliary pore complex distinguishes mechanisms of cytosolic and membrane protein entry. Current biology: CB. 2014;24:2288–2294. doi: 10.1016/j.cub.2014.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vick P, Schweickert A, Weber T, Eberhardt M, Mencl S, Shcherbakov D, Beyer T, Blum M. Flow on the right side of the gastrocoel roof plate is dispensable for symmetry breakage in the frog Xenopus laevis. Dev Biol. 2009;331:281–291. doi: 10.1016/j.ydbio.2009.05.547. [DOI] [PubMed] [Google Scholar]
- Vollmer B, Antonin W. The diverse roles of the Nup93/Nic96 complex proteins - structural scaffolds of the nuclear pore complex with additional cellular functions. Biological chemistry. 2014;395:515–528. doi: 10.1515/hsz-2013-0285. [DOI] [PubMed] [Google Scholar]
- von Appen A, Kosinski J, Sparks L, Ori A, DiGuilio AL, Vollmer B, Mackmull MT, Banterle N, Parca L, Kastritis P, et al. In situ structural analysis of the human nuclear pore complex. Nature. 2015;526:140–143. doi: 10.1038/nature15381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonica A, Brivanlou AH. The left-right axis is regulated by the interplay of Coco, Xnr1 and derriere in Xenopus embryos. Dev Biol. 2007;303:281–294. doi: 10.1016/j.ydbio.2006.09.039. [DOI] [PubMed] [Google Scholar]
- Warkman AS, Krieg PA. Xenopus as a model system for vertebrate heart development. Semin Cell Dev Biol. 2007;18:46–53. doi: 10.1016/j.semcdb.2006.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner ME, Mitchell BJ. Using Xenopus skin to study cilia development and function. Methods in enzymology. 2013;525:191–217. doi: 10.1016/B978-0-12-397944-5.00010-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu K, Zhong G, Zhuang X. Actin, spectrin, and associated proteins form a periodic cytoskeletal structure in axons. Science. 2013;339:452–456. doi: 10.1126/science.1232251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J, Liu X, Yue G, Adamian M, Bulgakov O, Li T. Rootletin, a novel coiled-coil protein, is a structural component of the ciliary rootlet. The Journal of cell biology. 2002;159:431–440. doi: 10.1083/jcb.200207153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshioka H, Meno C, Koshiba K, Sugihara M, Itoh H, Ishimaru Y, Inoue T, Ohuchi H, Semina EV, Murray JC, et al. Pitx2, a bicoid-type homeobox gene, is involved in a lefty-signaling pathway in determination of left-right asymmetry. Cell. 1998;94:299–305. doi: 10.1016/s0092-8674(00)81473-7. [DOI] [PubMed] [Google Scholar]
- Zaidi S, Choi M, Wakimoto H, Ma L, Jiang J, Overton JD, Romano-Adesman A, Bjornson RD, Breitbart RE, Brown KK, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220–223. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.