Abstract
Objective
A disturbed day-night rhythm is associated with metabolic perturbations that can lead to obesity and type 2 diabetes mellitus (T2DM). In skeletal muscle, a reduced oxidative capacity is also associated with the development of T2DM. However, whether oxidative capacity in skeletal muscle displays a day-night rhythm in humans has so far not been investigated.
Methods
Lean, healthy subjects were enrolled in a standardized living protocol with regular meals, physical activity and sleep to reflect our everyday lifestyle. Mitochondrial oxidative capacity was examined in skeletal muscle biopsies taken at five time points within a 24-hour period.
Results
Core-body temperature was lower during the early night, confirming a normal day-night rhythm. Skeletal muscle oxidative capacity demonstrated a robust day-night rhythm, with a significant time effect in ADP-stimulated respiration (state 3 MO, state 3 MOG and state 3 MOGS, p < 0.05). Respiration was lowest at 1 PM and highest at 11 PM (state 3 MOGS: 80.6 ± 4.0 vs. 95.8 ± 4.7 pmol/mg/s). Interestingly, the fluctuation in mitochondrial function was also observed in whole-body energy expenditure, with peak energy expenditure at 11 PM and lowest energy expenditure at 4 AM (p < 0.001). In addition, we demonstrate rhythmicity in mRNA expression of molecular clock genes in human skeletal muscle.
Conclusions
Our results suggest that the biological clock drives robust rhythms in human skeletal muscle oxidative metabolism. It is tempting to speculate that disruption of these rhythms contribute to the deterioration of metabolic health associated with circadian misalignment.
Keywords: Biological rhythm, Mitochondria, Oxidative capacity, Skeletal muscle, Energy metabolism, Molecular clock
Abbreviations: BMAL1, brain and muscle ARNT-like 1; BMI, body mass index; CLOCK, circadian locomotor output cycles kaput; CRY, cryptochrome; FCCP, carbonyl cyanide-4-trifluoromethoxyphenylhydrazone; NADH, reduced nicotinamide adenine dinucleotide; PER, period; RER, respiratory exchange ratio; RT-QPCR, Real-Time Quantitative Polymerase Chain Reaction; T2DM, type 2 diabetes mellitus; TCA cycle, tricarboxylic acid cycle
Graphical abstract
Highlights
-
•
Mitochondrial oxidative capacity in human skeletal muscle follows a day-night rhythm.
-
•
Oxidative capacity peaks in the late evening and is lowest in the early afternoon.
-
•
Energy expenditure follows a day-night rhythm and is highest in the late evening.
-
•
Human muscle exhibits rhythmic gene expression, with a cycling core molecular clock.
1. Introduction
Many metabolic processes are synchronized to day-night cycles by the circadian clock, thereby anticipating changes in metabolic activity associated with feeding or fasting and physical activity or rest [1]. In our modern “24/7” society, however, many individuals do not adhere to the lifestyle imposed upon us by nature. In this respect, epidemiological studies have shown that circadian misalignment – desynchronization between the intrinsic circadian and behavioral cycles, as is typical in shift-work – is associated with obesity, insulin resistance and type 2 diabetes mellitus (T2DM) [2], [3], [4], [5]. Moreover, intervention studies have shown that challenging behavior by controlled circadian misalignment results in metabolic aberrations like decreased glucose tolerance and insulin sensitivity [6], [7], [8].
Circadian rhythms are governed by a central circadian clock, which is situated in the suprachiasmatic nucleus of the hypothalamus and is sensitive to light as the most important time cue (Zeitgeber) [9]. Interestingly, peripheral tissues have their own clocks. These peripheral clocks are synchronized by the central clock, but they can also be influenced by behavior, such as feeding or exercise [10], [11]. The peripheral clock consists of transcriptional-translational feedback loops. The positive loop consists of the heterodimer of the CLOCK (circadian locomotor output cycles kaput) and BMAL1 (brain and muscle ARNT-like 1) proteins. The negative feedback loop is mediated via heterodimers of the proteins PER (Period) and CRY (Cryptochrome), which repress CLOCK/BMAL1-controlled gene expression [9], [12]. Interestingly, in mouse and cell models, several components of the molecular clock have been causally linked to mitochondrial metabolism [13], mitochondrial integrity and density [14], mitochondrial dynamics [15] and metabolic flexibility [16]. So far however, it is unknown whether mitochondrial metabolism also displays a day-night rhythm in human skeletal muscle. Such data would be relevant, since reduced skeletal muscle oxidative capacity is associated with T2DM [17], [18]. It is tempting to speculate that disturbances in the day-night rhythm may affect muscle mitochondrial metabolism and thereby deteriorate metabolic health.
Here, we investigated whether skeletal muscle mitochondrial function displays day-night rhythmicity by taking multiple muscle biopsies from healthy, lean volunteers within a 24 h period, under tightly controlled experimental conditions. We used a research setting that reflects real life conditions, with regular meals, physical activity and a regular sleep/wake cycle. For this reason we use the term “day-night rhythm” instead of “circadian rhythm” [19]. We here show that gene expression in muscle displays rhythmicity, which is specifically evident for the core components of the molecular clock. Furthermore, we are the first to show the presence of a day-night rhythm in human skeletal muscle oxidative capacity.
2. Material and methods
2.1. Participants
Twelve young lean male Caucasian individuals (age ± SD: 22.2 ± 2.3 years, BMI ± SD: 22.4 ± 2.0 kg/m2) participated in this study. The participants did not engage in exercise more than 3 h per week, were non-smokers, had no active diseases and used no medication, verified by a medical questionnaire. Participants were selected for having a regular sleep duration (normally 7–9 h/night), not having done shift work or having traveled across more than one time zone for at least 3 months. A morningness-eveningness questionnaire (MEQ-SA) was used to exclude extreme morning larks or night owls (MEQ-SA score mean ± SD: 50 ± 7). All participants provided written informed consent. The study was approved by the Ethics Committee of the Maastricht University Medical Center, monitored by the Clinical Trial Center Maastricht and conducted in accordance with the principles of the declaration of Helsinki. All measurements were performed between November 2014 and July 2015. The study was registered at clinicaltrials.gov with identifier NCT02261168.
2.2. Pre-study conditions
One week prior to the study, participants were instructed to maintain a standardized lifestyle. This lifestyle included (trying to) sleep every night from 11 PM until 7 AM, eating breakfast, lunch and dinner at regular times (at 9 AM, 2 PM and 7 PM) with no in-between snacks or drinks other than water. In this period subjects refrained from alcohol and caffeine. Participants were instructed not to engage in exercise three days prior to the study. Two days before the study, we provided standardized meals (see below) to ensure standardized caloric and macronutrient intake for all participants. Lifestyle was monitored by accelerometry (activPAL3 physical activity monitor, PAL Technologies, Glasgow, UK) together with food- and sleep-diaries, which were checked at the start of the first study-day.
2.3. Study design
Participants were admitted to the research unit at noon on study day 1 and stayed for 44 h in total, under standardized conditions mimicking a real-life situation. The first study-day was mainly used to standardize and monitor meals, physical activity and bedtime. Meals were provided at fixed times (9 AM, 2 PM and 7 PM). To prevent a sedentary lifestyle, participants went for a 15-minute low-intensity walk accompanied by a researcher, one hour after every meal. Directly hereafter, participants were instructed to stand for 15 minutes before they were allowed to sit again. In-between meals, physical activity and tests, the participants stayed in a respiration chamber; a small room with a bed, toilet, sink, desk, chair, TV and computer. During the first study-day we performed no measurements. At 11 PM, the lights of the respiration chamber were turned off and the participants were instructed to try to sleep. During this night, sleeping metabolic rate was measured by whole-room indirect calorimetry (Omnical, Maastricht Instruments, Maastricht, The Netherlands) [20].
The second study-day, participants were awakened at 6:30 AM. Hereafter, participants swallowed a telemetric pill for measurement of core-body temperature. Next, an intravenous cannula was placed in the forearm for subsequent blood-draws. The first blood-draw was at 8 AM, followed by an indirect calorimetry measurement using a ventilated hood while awake and at rest in supine posture to calculate resting energy expenditure and substrate oxidation. Directly hereafter, the first skeletal muscle biopsy was taken (described below). These measurements (blood draw, ventilated hood measurement and skeletal muscle biopsy) were repeated five times within 24 h: at 8 AM, 1 PM, 6 PM, 11 PM and 4 AM the next day. Additional blood samples were taken 2-hourly (10 AM, 12 PM, 2 PM, 4 PM, 8 PM, 10 PM, 0 AM, 2 AM, 6 AM and 8 AM). The timing of meals and physical activity was similar to study-day 1 and subjects stayed within the respiration chamber in-between measurements. After the 11 PM biopsy, participants went back to the respiration chamber to sleep with lights off. At 4 AM, the participant was awakened and the last measurements were performed, after which the subject was allowed to sleep until 7 AM. After the 8 AM blood draw the study protocol ended.
2.4. Study meals
Two days before the study and during the study participants were provided with standardized meals, according to Dutch and US dietary guidelines. Caloric intake for consumption at home was calculated by multiplying the estimated resting metabolic rate, obtained with the Harris-Benedict formula [21] with an activity factor of 1.5. Participants were provided with optional extra snacks to eat with their meals if they were still hungry, up to an activity factor of 1.7. For the first study-day in the laboratory, energy requirement was calculated by multiplying the estimated resting metabolic rate with an activity factor of 1.35, because of limited physical activity in the research facility. For the second study-day, energy requirement was calculated by multiplying the sleeping metabolic rate of the first study night (measured by whole-room indirect-calorimetry) by 1.5.
During the study days, participants received 3 meals daily. Breakfast accounted for ∼21 energy%, lunch for ∼30 energy% and dinner for ∼49 energy%. Daily macronutrient composition was ∼52 energy% as carbohydrates, ∼31 energy% as fat (∼9% saturated) and ∼14 energy% as protein. No snacks or drinks other than water were provided in-between meals.
2.5. Skeletal muscle biopsies and respirometry
Five skeletal muscle biopsies were obtained from the m. vastus lateralis according to the Bergström method [22] under local anesthesia (1% lidocaine, without epinephrine). Each biopsy was taken from a separate incision at least 2 cm from the previous incision, moving from distal to proximal. The first biopsy was randomly taken from the left or right leg, and each subsequent biopsy was taken from the other leg. Part of the biopsy was immediately placed in ice-cold preservation medium (BIOPS, OROBOROS Instruments, Innsbruck, Austria) and used for measurement of mitochondrial oxidative capacity. For this analysis, intact muscle fibers were permeabilized and measured for oxygen consumption upon several substrates using an Oxygraph (OROBOROS Instruments). During the assay, muscle fibers were supplemented with malate, octanoylcarnitine, ADP, glutamate, succinate and carbonylcyanide p-trifluoromethoxyphenylhydrazone (FCCP) as described previously [23]. The remaining part of the muscle biopsy was immediately frozen in melting isopentane and stored in −80 °C until further analysis.
2.6. Indirect calorimetry
To calculate whole-body energy expenditure, respiratory exchange ratio (RER), glucose- and fat-oxidation, oxygen consumption and carbon dioxide production were measured with an automated respiratory gas analyzer using a ventilated hood system (Omnical; IDEE, Maastricht, the Netherlands). Calculations of energy expenditure and substrate oxidation were made with the assumption of a negligible protein oxidation [24], [25].
2.7. Measurement of core-body temperature
Core-body temperature was measured by a swallowed telemetric pill (CorTemp HT150002; HQ Inc., Palmetto, FL, USA). The telemetric pill measured the core-body temperature every 10 s and sent the information wirelessly to a portable receiver worn outside the body on the stomach. For further analysis, we used temperature averages of every 10 min, after excluding false measurements.
2.8. Gene transcript quantification
RNA was isolated from 50 mg of muscle material by TRIzol lysis (Qiagen, Hilden, Germany). RNA was further purified by the RNeasy kit from Qiagen (Hilden, Germany).
Microarray analysis: RNA integrity was assessed using a Bioanalyzer 2100 (Agilent) and RIN were above 9.0. RNAs were then processed to generate labeled ssDNA using the Whole Transcript cDNA synthesis and amplification and the Terminal Labeling kit as suggested by the manufacturer (Affymetrix). Samples were hybridized to HuGene 2.0 arrays and native CEL files imported into GeneSpring (v13.1.1, Agilent) after quality controls using the Affymetrix Expression Console. The RMA16 algorithm was used for summarization and data was normalized to the median of all samples. Normalized intensity values were used to generate a self-organized map (distance metric: Euclidean; number of iterations: 500). Protein-encoding genes from specific clusters were searched against the GO Biological Process, KEGG and Reactome databases using the Metascape tool [26]. Microarray data have been deposited in NCBIs Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible through GEO accession number GSE79934.
RT-QPCR: RNA integrity was assessed using a Bioanalyzer (Agilent Technologies, Santa Clara, USA) and yield was measured using a NanoDrop spectrophotometer (Thermo Fisher Scientific, Waltham, USA). The high-capacity RNA-to-cDNA kit from Applied Biosystems (Foster City, USA) was used for transcribing 0.5 μg RNA to cDNA. Transcript abundance was determined using a 7900HT Fast Real-Time PCR System (Applied Biosystems, Waltham, USA). To minimize the variability in reference gene normalization, the geometric mean of three reference genes (RPL26, GUSB and CYPB), which were individually stably expressed in time, was used. This geometric mean was used as the internal reference for comparative gene expression analysis in the remainder of the study [27].
2.9. Protein analysis
Western blot analyses were performed in RIPA-lysates of human muscle tissue. Protein concentration was determined using the Bio-Rad RC/DC kit (Bio-Rad Laboratories, Veenendaal, The Netherlands). Equal amounts of protein were loaded on 12% TGX gels (Bio-Rad Laboratories) or 4–12% Bolt gradient gels (Novex, Thermo Fisher Scientific, Bleiswijk, The Netherlands). Proteins were transferred to nitrocellulose with the Trans-Blot Turbo transfer system (Bio-Rad Laboratories). Primary antibodies: a cocktail of mouse monoclonal antibodies directed against human OXPHOS (dilution 1:10,000; ab110411, Abcam, Cambridge, UK), two mitochondrial markers directed against TOMM20 (dilution 1:10,000; ab186734; Abcam), porin/VDAC (dilution 1:10,000; sc-8828, Santa Cruz Biotechnology, Dallas, Texas), SR-actin (dilution 1:5,000; A-2172; Sigma Aldrich, Zwijndrecht, The Netherlands), PGC-1 (dilution 1:10,000, 516,557, Calbiochem), FIS-1 (dilution 1:1000, sc-98900, Santa Cruz Biotechnology, Dallas, Texas), PINK-1 (dilution 1:2000, sc-33796, Santa Cruz Biotechnology, Dallas, Texas) and OPA-1 (dilution 1:2500, 612,606, Becton Dickinson). The specific proteins were detected using secondary antibodies conjugated with IRDye680 or IRDye800, and were quantified with the CLx Odyssey Near Infrared Imager (Li-COR, Westburg, Leusden, The Netherlands).
2.10. Statistics
Data are presented as mean ± SEM (standard error of the mean) unless indicated otherwise. Statistical analyses were performed with the use of IBM Statistical Package for Social Sciences for MAC, version 23 (SPSS, Inc.). The effect of time on outcome variables was analyzed by repeated measures ANOVA. If the assumption of sphericity was violated (Mauchly's test), we applied Greenhouse-Geisser's correction. In case repeated measures ANOVA revealed significant effects for the oxidative capacity and indirect calorimetry analyses, Bonferroni adjusted post-hoc analyses were applied to look at significant differences between specific time-points. Statistical significance was defined as a p-value < 0.05. In addition, if repeated measures ANOVA resulted in a significant effect of time for targets of mRNA and protein expression and oxidative capacity states, we tested for rhythmicity using the JTK_CYCLE package in R 3.2.1 with Windows 8 [28]. For this analysis, values were normalized to subject mean prior to analysis.
3. Results and discussion
3.1. Participants adhered to a controlled normal lifestyle and displayed a normal day-night rhythm in core-body temperature
To investigate whether mitochondrial oxidative capacity displays a day-night rhythm, we performed an observational study in which twelve young lean male volunteers were admitted to our metabolic research unit for two days under controlled conditions, mimicking our normal daily life. The study protocol is graphically depicted in Figure 1. Prior to the study days, subjects were instructed to adhere to a standardized lifestyle, including fixed sleeping times, which was monitored by accelerometry. During the second study-day we recorded core-body temperature to examine the presence of a normal day-night rhythm (Supplemental Fig. 1). Indeed, all subjects showed a characteristic day–night pattern in core-body temperature with highest temperature at the end of the day and a decrease in temperature after midnight [29], indicating that chronotypes were similar across participants.
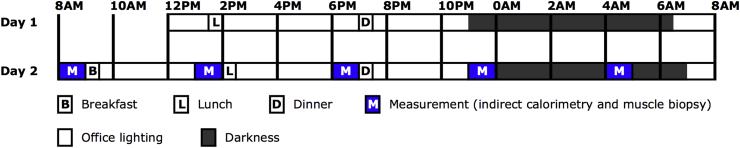
Figure 1.
Study design. Participants stayed in the research facility for 44 h starting at noon on day 1. During the second study-day we performed all measurements. B, breakfast; L, lunch; D, dinner; M, measurement (indirect calorimetry and muscle biopsy).
3.2. Human skeletal muscle displays oscillations in RNA of metabolic and core molecular clock genes
During the second study day, we performed five consecutive skeletal muscle biopsies. To prevent meal interference, the biopsies were taken immediately before the three meals (8 AM, 1 PM, 6 PM), before bedtime (11 PM) and one time during the night (4 AM) after waking up the participant.
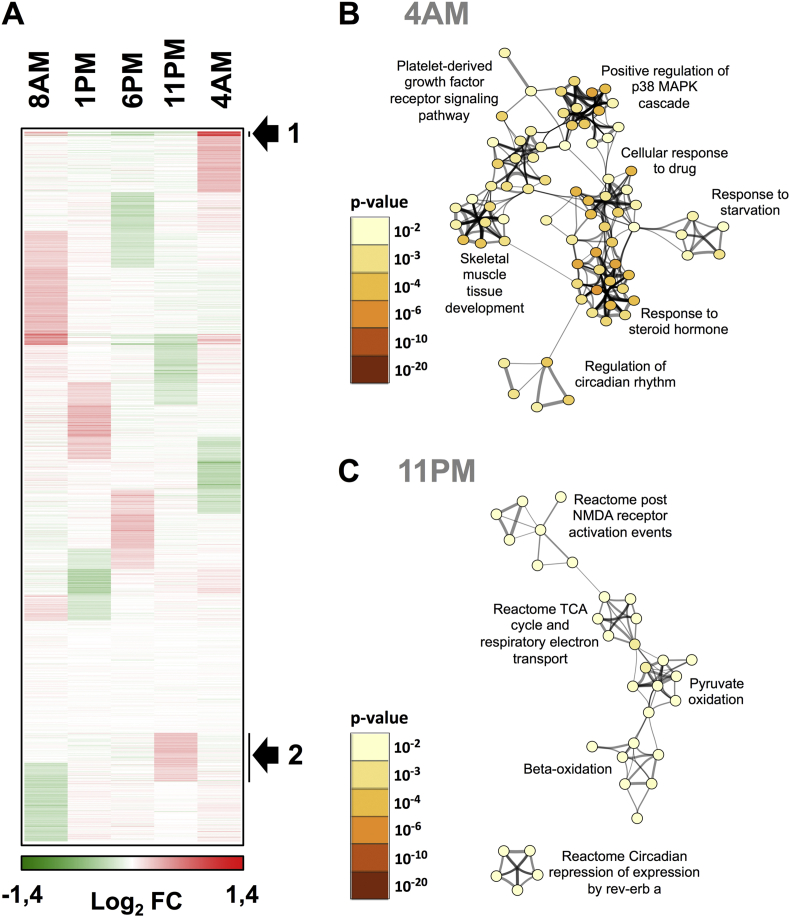
To examine if skeletal muscle displays rhythmicity in gene expression, we analyzed RNA using DNA microarrays in subsequent biopsies from a single donor with a representative rhythm in mitochondrial oxidative capacity. This analysis revealed that 14.5% of transcripts (4094 out of 28,168) displayed a cyclic expression peaking at different times, 75% of them corresponding to protein-encoding transcripts (Figure 2A). Term enrichment against Gene Ontology, KEGG and Reactome databases indicated that gene clusters displaying a maximal expression in the late evening or during the night (11 PM, 4 AM) are notably related to the core clock machinery (4 AM, Figure 2B) including REV-ERB-α, REV-ERB-β, PER3 and DBP and to the tricarboxylic acid (TCA) cycle/respiratory electron transport chain (11 PM, Figure 2C) such as components of NADH dehydrogenase complexes (NDUFA4, NDUFA8) and of ATP synthase (ATP5F1, ATP5G3, APTP5A1, ATP5L). Collectively, these analyses suggested the existence of a functional core clock machinery and oscillations in mitochondrial metabolism in human skeletal muscle.
Figure 2.
Time-dependent expression of transcripts in human skeletal muscle. (A) Time-dependent gene expression was assessed by microarray analysis of RNA extracted from a single donor (N = 1). Protein-encoding transcript expression patterns were visualized as a self-organizing map (Euclidean distance metrics). The color scale indicates upregulated genes (red) and downregulated genes (green) relative to the global median signal of each array. The arrow indicates clusters of genes showing the most prominent peak of expression at the indicated time point (4 AM, cluster 1 and 11 PM, cluster 2). (B and C) Gene annotation enrichment analysis. The genes extracted from cluster 1 and 2 were searched using multiple databases (GeneOntology Biological Processes, KEGG pathways and Reactome) and statistically enriched terms were determined using the Metascape tool. The most significantly enriched terms are indicated for cluster 1 (GO biological processes, panel B, peak expression 4 AM) and cluster 2 (Reactome, panel C, peak expression 11 PM). Statistically significant terms were hierarchically clustered and converted into a network. Each term is represented by a circle node, of which the size is proportional to the number of genes in the term. The color of the node indicates the statistical significance of the term belonging to the cluster (see color scale). The most significant term characterizing each cluster is indicated. Similarities between terms are indicated by connecting lines.
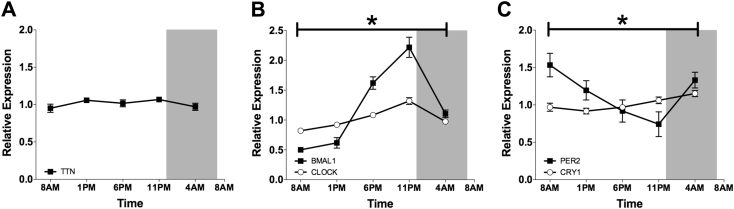
Expression of core clock genes was validated in all participants by RT-QPCR assays. TTN mRNA expression, measured as an arrhythmic muscle filament gene, did not vary throughout the 24-hour period (p = 0.331, Figure 3A). CLOCK mRNA showed significant variation over time (ANOVA p < 0.001), but rhythmicity was of rather low amplitude (JTK_CYCLE p < 0.001; Figure 3B), consistent with previous observations in several mouse tissues [30]. BMAL1 mRNA exhibited a robust sinusoidal rhythm, with highest expression around midnight, and lowest expression in the morning and early afternoon (ANOVA p < 0.001, JTK_CYCLE p < 0.001; Figure 3B). Of the negative feedback loop, PER2 was most abundantly expressed in the early morning (p = 0.019, Figure 3C). CRY1 was most highly expressed in the night, however the amplitude was rather low (p = 0.006, Figure 3C). Both were confirmed to be rhythmic by JTK_CYCLE (p < 0.001).
Figure 3.
Rhythmicity of core molecular clock genes in human skeletal muscle. Diurnal mRNA expression of the arrhythmic muscle filament gene TTN (A), the core clock genes CLOCK and BMAL1 (B), PER2 and CRY1 (C), measured by RT-QPCR. Data are normalized to the geometric mean of 3 housekeeping genes and presented as mean ± SEM. *p < 0.05 for the effect of time in all depicted genes.
Together, our data show robust rhythmicity of the molecular clock in human skeletal muscle, which is consistent with the demonstration of an oscillating molecular clock in other human cells and tissues like leukocytes [31], follicle cells [32] and adipose tissue [33] and in primary cultures of human muscle [34]. In addition, micro-array analysis in sequential biopsies obtained from a single donor revealed cyclic expression of several clusters of genes, including genes involved in important mitochondrial pathways. A previous study investigating the molecular clock in human adipose tissue biopsies demonstrated peak-expression of BMAL1 at the very end of the light-phase, similar to our current results in human skeletal muscle [33]. Interestingly, that study examined adipose tissue gene expression under a standardized circadian rhythm protocol with subjects in the resting condition and with hourly nutritional drinks, thereby limiting the influence of behavior on the core molecular clock expression. The similarity in results suggests that the molecular clock patterns we observe in skeletal muscle may indeed be dominated by circadian rhythmicity. This is further underscored by findings in animals, as in mouse skeletal muscle [35] and mouse liver [13], [15], BMAL1 mRNA is highest at the end of the dark-phase and beginning of the light phase. Since mice are nocturnal animals, this corresponds with a highest BMAL1 expression at the end of the active phase and beginning of the rest phase, similar to our findings in human skeletal muscle.
3.3. Human skeletal muscle oxidative capacity displays a day-night rhythm
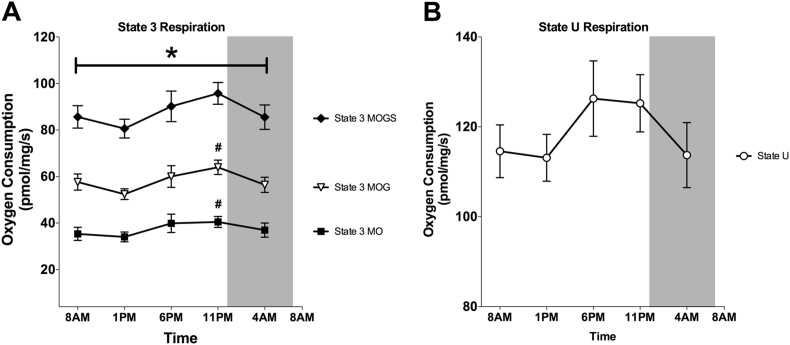
To investigate whether the rhythmicity in mitochondrial and molecular clock genes was also reflected in a day-night rhythm of muscle mitochondrial oxidative capacity, we performed high-resolution respirometry in freshly isolated permeabilized muscle fibers. Skeletal muscle oxidative capacity demonstrated a clear day-night rhythm, with a significant time effect in ADP-stimulated (state 3) respiration fueled by different substrates (state 3 MO, state 3 MOG and state 3 MOGS p = 0.042; 0.016; 0.042 respectively, Figure 4A). Mitochondrial state 3 respiration was lowest at 1 PM, and highest at 11 PM (state 3 MOGS: 80.6 ± 14.0 vs. 95.8 ± 16.3 pmol/mg/sec, mean ± SD). Differences between these time-points were statistically significant for state 3 MO (p = 0.032) and state 3 MOG (p = 0.027), although it just did not reach significance for state 3 MOGS (p = 0.132) after Bonferroni adjustment for multiple testing. Furthermore, JTK_CYCLE analysis of mitochondrial respiration confirmed rhythmicity for state 3 MO (p = 0.007), 3 MOG (p = 0.039) and 3 MOGS (0.041). Maximally uncoupled respiration, reflecting the maximal capacity of the electron transport system, demonstrated similar oscillations although the effect of time did not reach significance (p = 0.121, Figure 4B). It is interesting to note that the difference in mitochondrial state 3 respiration between 1 PM and 11 PM was on average 20%, which is in the same range as the difference in mitochondrial function between patients with T2DM and BMI-matched controls [17], [36], indicating that the magnitude of the day-night rhythm in mitochondrial function is of physiological relevance. Previous studies in animal models of metabolic diseases do show disrupted circadian rhythmicity in the molecular clock of metabolic tissues and in plasma metabolites [37], [38], and it is therefore tempting to speculate that day-night rhythmicity in muscle mitochondrial metabolism is altered in T2DM and pre-diabetic patients. Such disturbances in circadian rhythmicity of mitochondrial function may contribute to reduced metabolic flexibility that is observed in metabolically compromised individuals and thereby contribute to the development of metabolic diseases, consistent with findings that circadian misalignment is associated with obesity and T2DM [2], [3], [4]. If this is indeed the case, restoring circadian rhythmicity may comprise a potential new basis for lifestyle advice and provide treatment options for T2DM.
Figure 4.
Mitochondrial oxidative capacity in human skeletal muscle displays a day-night rhythm. (A) ADP-stimulated respiration of permeabilized muscle fibers upon a lipid substrate (state 3 MO); fueled by complex I-linked substrates (state 3 MOG) and upon parallel electron input into complex I and II (state 3 MOGS). (B) Maximally uncoupled respiration upon FCCP (State U). M, malate; O, octanoylcarnitine; G, glutamate; S, succinate. Data represents oxygen consumption per mg wet weight per second and is depicted as mean ± SEM. *p < 0.05 for the effect of time in all states. #p < 0.05 vs 1 PM for Bonferroni-adjusted post-hoc analysis.
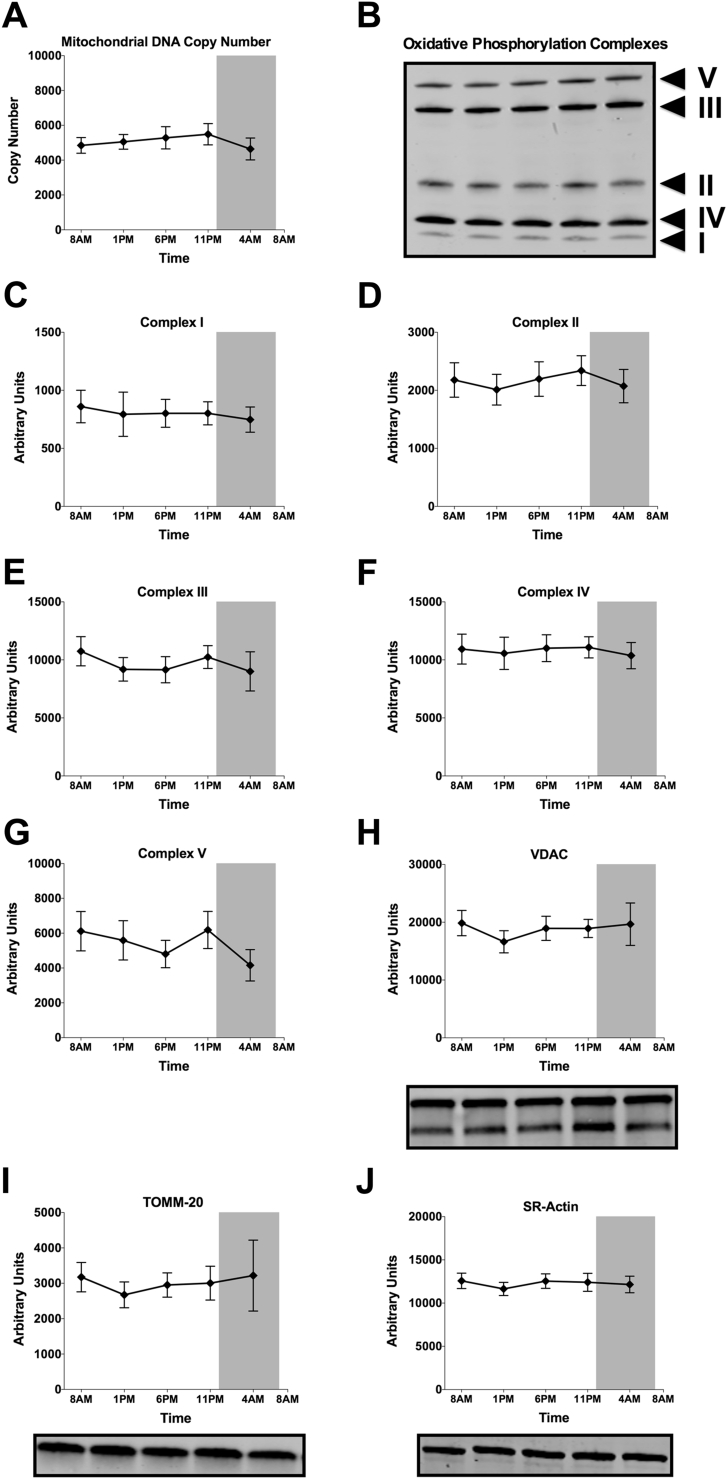
How mitochondrial function can show circadian rhythmicity can not be completely revealed by the current study in humans. However, to investigate if the rhythm in mitochondrial respiration is caused by intrinsic variation in mitochondrial oxidative capacity, rather than by variation in mitochondrial density, we measured several markers of mitochondrial content. Mitochondrial-DNA copy number – the ratio between the DNA copy number of a mitochondrial (ND1) and a nuclear gene (LPL) – did not reveal a day-night rhythm (p = 0.437, Figure 5A). Also, western blot analysis of structural subunits of the five mitochondrial respiratory chain complexes did not reveal a statistically significant effect of time (Figure 5B–G). In addition, the mitochondrial outer membrane proteins VDAC and TOMM-20 did not display day-night rhythmicity (Figure 5H–I). The non-mitochondrial protein SR-Actin was also stable throughout the day (Figure 5J). Together, these results suggest that in skeletal muscle, intrinsic mitochondrial function (rather than mitochondrial content) displays circadian rhythmicity.
Figure 5.
Mitochondrial content and mitochondrial marker proteins do not show rhythmicity. (A) Measurement of mitochondrial DNA copy number (DNA copy numbers of the mitochondrial-encoded gene ND1 divided over the nuclear encoded gene LPL). (B) Representative western blot depicting the oxidative phosphorylation complexes of all time-points (C–G) Protein levels of specific subunits of complex I–V of the mitochondrial electron transport chain, measured by western blotting. (H–J) Protein levels of other mitochondrial (VDAC, TOMM-20) and non-mitochondrial (SR-Actin) proteins measured by western blotting. Representative western blot images are displayed below their respective graphs. Data expressed as mean ± SEM. p > 0.05 for the effect of time in all panels.
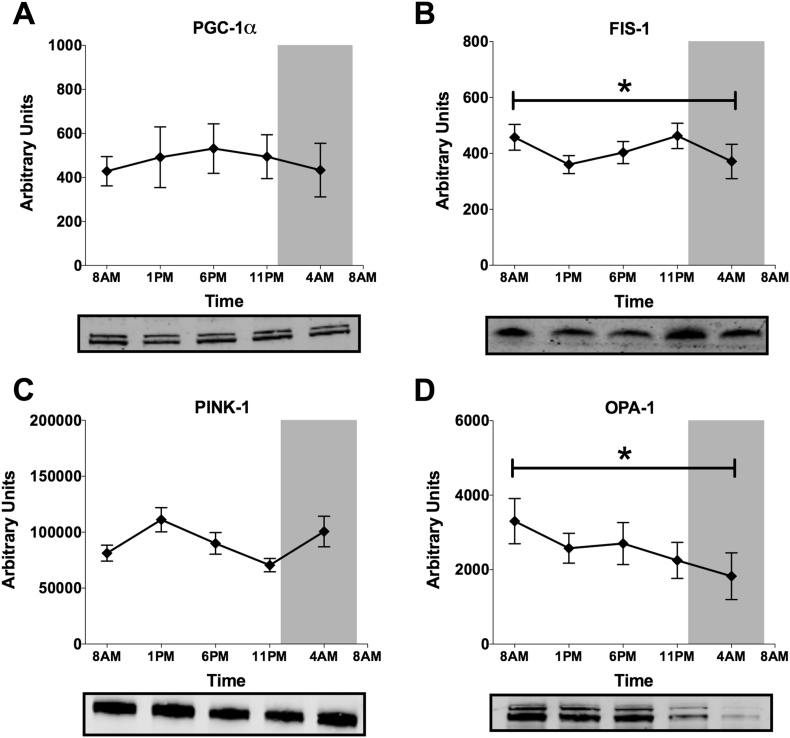
We next investigated whether mitochondrial dynamics or mitochondrial biogenesis could play a role in the circadian rhythmicity of intrinsic mitochondrial function. Mitochondrial fission and fusion occur as a result of changes in energy demand, influence substrate metabolism and are controlled by the circadian clock in mouse liver [15], [39]. Here, we quantified proteins involved in mitochondrial biogenesis and mitochondrial dynamics in skeletal muscle biopsies. PGC-1α, the master regulator of mitochondrial biogenesis, did not show a significant difference over time (p = 0.901, Figure 6A). Interestingly however, FIS1, a marker of mitochondrial fission, displayed significant time-differences, with a pattern matching the rhythm in mitochondrial oxidative capacity (p = 0.016, Figure 6B), although JTK_CYCLE analysis did not confirm significant rhythmicity. PINK-1, a marker of mitophagy, displayed a rhythm opposing FIS-1, although this time-effect just did not reach statistical significance (p = 0.061, Figure 6C). Noteworthy, in murine liver FIS-1 and PINK-1 24 h expression patterns are also opposed [15]. Furthermore, the mitochondrial fusion marker OPA-1 showed a significant decrease over time, across all time points (p = 0.045, Figure 6D, without significant rhythmicity in JTK_CYCLE analysis). These results suggest that mitochondrial dynamics may be involved in governing mitochondrial capacity around the clock, although mechanistic studies in preclinical models are needed to further investigate underlying mechanisms.
Figure 6.
Mitochondrial dynamics rather than mitochondrial biogenesis display a significant time-effect. Protein levels of mitochondrial biogenesis regulator PGC-1α (A), mitochondrial fission protein FIS-1 (B), mitophagy protein PINK-1 (C) and mitochondrial fusion protein OPA-1 (D). Representative western blot images are displayed below their respective graphs. Data presented as mean ± SEM. *p < 0.05 for the effect of time.
The physiological relevance of the rhythmicity in oxidative capacity cannot be deduced from the current study. It is, however, interesting to note that aerobic performance also peaks later in the day [40], [41]. Our results could therefore form a physiological explanation for these findings. An earlier study found that chronotype influences the time-point of peak performance [41]. In our study, we only included participants with an average chronotype. Future studies will be needed to investigate whether early and late chronotypes have a different time-point of peak mitochondrial function.
3.4. Whole-body energy expenditure peaks before midnight
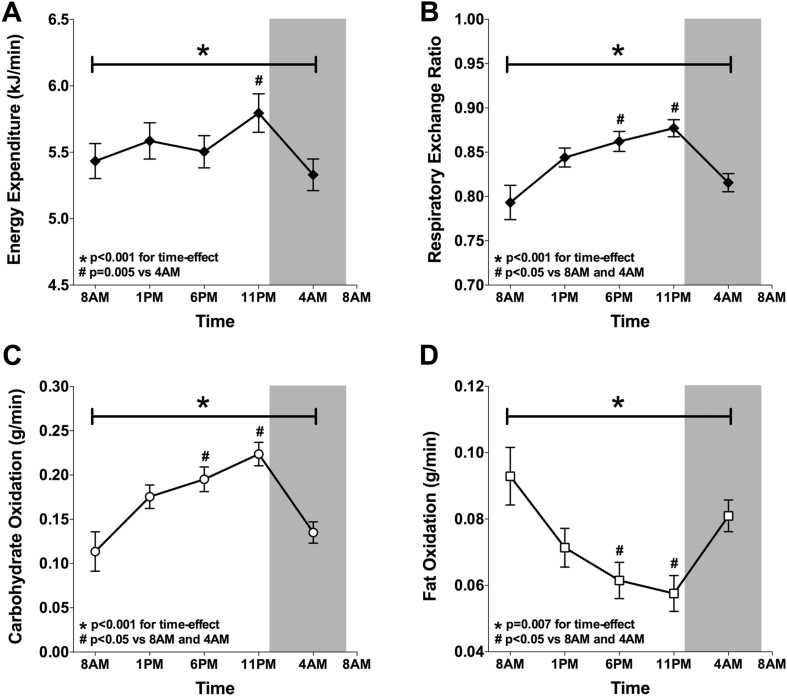
We next examined whether day-night rhythmicity in muscle mitochondrial function was also associated with rhythmicity in whole-body measures of energy metabolism. To this end, we performed indirect calorimetry analyses to determine resting energy expenditure and substrate oxidation at the same time points of the muscle biopsies. Interestingly, similar to mitochondrial respiratory capacity, resting energy expenditure showed a significant time effect, with highest energy expenditure at 11 PM (p < 0.001 for the time-effect; p = 0.005 for Bonferroni adjusted post-hoc analysis 11 PM vs 4 AM; Figure 7A). It is well known that lowest levels of energy expenditure can be measured when subjects are asleep, denoted as sleeping metabolic rate. Even though the participants were awakened before measurements, resting energy expenditure was still lowest at 4 AM. This clearly illustrates day-night rhythmicity in energy expenditure, independent of sleep. RER, carbohydrate oxidation and fat oxidation displayed variation with feeding and fasting periods, with a significantly lower carbohydrate oxidation and RER, and a higher fat oxidation in the fasted state (8 AM and 4 AM) and a higher carbohydrate oxidation and RER, and a lower fat oxidation in the fed state (1 PM, 6 PM and 11 PM) (p < 0.001 for the time-effect in RER and carbohydrate oxidation and p = 0.007 for the time-effect in fat oxidation, Figure 7B–D). Bonferroni-adjusted post-hoc analyses confirmed that RER, carbohydrate- and fat-oxidation were significantly different at 6 PM and 11 PM, when compared with 8 AM and 4 AM (p < 0.05; Figure 7A–D).
Figure 7.
Whole-body resting energy expenditure peaks at the same time as skeletal muscle oxidative capacity whereas substrate oxidation exhibits a clear feeding and fasting pattern. Whole-body resting energy expenditure (A), respiratory exchange ratio (B), carbohydrate oxidation (C) and fat oxidation (D) during the second study-day, calculated from oxygen consumption and carbon-dioxide production measured by indirect calorimetry. Data presented as mean ± SEM. *p < 0.01 for the effect of time. #p < 0.05 for Bonferroni-adjusted post-hoc analysis.
3.5. Plasma metabolites peak according to meals
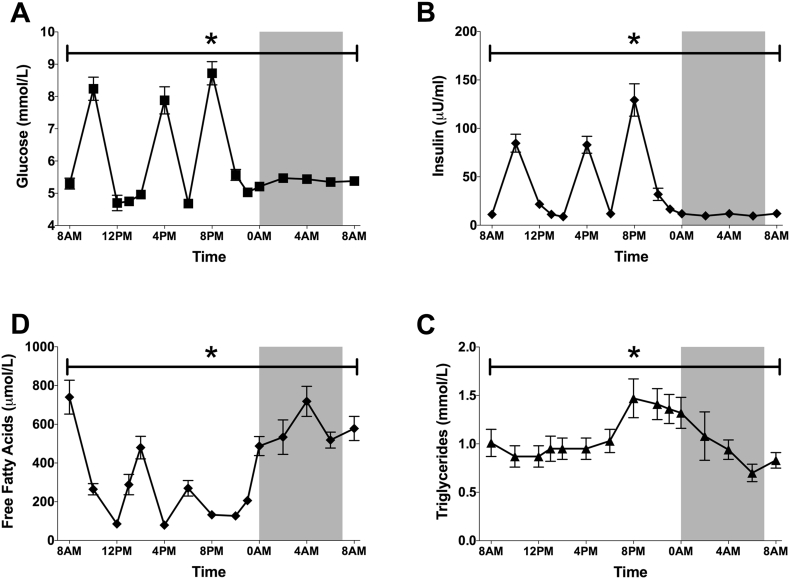
To examine the diurnal pattern in circulating substrates, we took 15 blood samples throughout the second study-day to assess glucose, insulin, free fatty acids and triglycerides (Figure 8A–D). As expected, plasma glucose and insulin displayed marked peaks after the meals, with an overall maximum after the dinner, being the most energy-dense meal. Furthermore, plasma glucose increased slightly during the late evening and night, possibly reflecting an increase in endogenous glucose production together with decreased carbohydrate oxidation. Free fatty acids reflected the fasting-feeding pattern with increased free fatty acids before the meals and during the night when fat oxidation is highest and decreased free fatty acids after the meals. Plasma triglycerides displayed a rise throughout the waking hours with a peak after dinner, being the meal with the highest energy density. During the night, plasma triglycerides decreased again.
Figure 8.
Plasma metabolites and insulin display marked variations over 24 h, mostly associated with feeding-fasting. Plasma levels of glucose (A), insulin (B), free fatty acids (C) and triglycerides (D) throughout the second-study day. Data depicted as mean ± SEM. *p < 0.001 for the effect of time.
3.6. Circadian vs day-night rhythmicity
In this study, we aimed to examine changes in skeletal muscle mitochondrial function and whole-body metabolism throughout 24 h with a standardized protocol designed to mimic our daily lifestyle. To this end, our protocol included a normal sleep–wake cycle, normal meals with a regular calorie distribution and (limited) physical activity. As a result, our protocol does not allow us to evaluate the influence of the endogenous circadian clock per se. However, to control the influence of meals and physical activity, we performed our measurements under controlled conditions with meals and light activity bouts at least 3–4 h before the measurements. Such a protocol therefore reflects day-night, and not circadian rhythmicity, and allows extrapolation of the results to a normal daily lifestyle. The demonstration of a day-night rhythm in mitochondrial function in lean volunteers allows future studies to investigate if such rhythmicity is disturbed in volunteers with compromised metabolic health, such as T2DM. If rhythmicity of mitochondrial metabolism is indeed disturbed in subjects with compromised metabolic health, this may open therapeutic strategies to restore circadian rhythmicity, possibly via REV-ERB-α agonists or nutritional compounds as resveratrol, as they have previously been proven to be successful in mouse studies [45], [46]. Furthermore, it may open a new field of research directed towards the timing of interventions that boost mitochondrial function, and ultimately these studies may reveal how circadian disruption can lead to metabolic disturbances, as has been demonstrated in both human and animal studies [6], [7], [42], [43], [44].
3.7. Conclusion
In conclusion, we here demonstrate the presence of a profound day-night rhythm in human skeletal muscle mitochondrial oxidative capacity. Peak oxidative capacity and highest resting energy expenditure coincide at the end of the day, thereby partly matching the time phase of peak physical performance as described in literature [40], [41]. In addition, we found significant variations over time in proteins involved in mitochondrial dynamics, possibly linking the circadian clock with mitochondrial metabolism. Future investigations should increase the focus on the regulatory mechanisms underlying the current observations. Moreover, it should be examined if disturbances of the rhythm in human skeletal muscle oxidative capacity play a pivotal role in the development of the adverse metabolic consequences of circadian misalignment.
Acknowledgments
We greatly acknowledge all research volunteers for participating in the study. We thank N. Warmke, K. Roumans and R. Stadt (Maastricht University Medical Center) for assistance during the experiments and E. Moonen-Kornips, G. Schaart and J. Bons (Maastricht University Medical Center) for assistance with the biochemical analyses. We thank B. Derudas and C. Gheeraert (Univ Lille) for running the microarray analysis. The technical support of P. Schoffelen (Maastricht University Medical Center) is highly appreciated. We acknowledge the support from the Netherlands Cardiovascular Research Initiative: an initiative with support of the Dutch Heart Foundation (CVON2014-02 ENERGISE). This work is partly financed by the Netherlands Organisation for Scientific Research (TOP 40-00812-98-14047 to P.S.). J.H. is supported by the NUTRIM NWO Graduate Programme which is financially supported by the Netherlands Organization for Scientific Research (022.003.011). F.S. was supported in part by National Institutes of Health grants R01 HL118601 and R01 DK099512. H.D. was supported by grants from the French Foundation for the Study of Diabetes (FFRD), European Foundation for the Study of Diabetes (EFSD) and by the FP7 consortium Eurhythdia. B.S. is a member of the Institut Universitaire de France.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.06.012.
Conflict of interest
None declared.
Appendix A. Supplementary data
The following is the supplementary data related to this article:
References
- 1.Bailey S.M., Udoh U.S., Young M.E. Circadian regulation of metabolism. Journal of Endocrinology. 2014 Aug;222(2):R75–R96. doi: 10.1530/JOE-14-0200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gan Y., Yang C., Tong X., Sun H., Cong Y., Yin X. Shift work and diabetes mellitus: a meta-analysis of observational studies. Occupational and Environmental Medicine. 2015 Jan;72(1):72–78. doi: 10.1136/oemed-2014-102150. [DOI] [PubMed] [Google Scholar]
- 3.Buchvold H.V., Pallesen S., Oyane N.M., Bjorvatn B. Associations between night work and BMI, alcohol, smoking, caffeine and exercise–a cross-sectional study. BMC Public Health. 2015;15(1):1112. doi: 10.1186/s12889-015-2470-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Karlsson B., Knutsson A., Lindahl B. Is there an association between shift work and having a metabolic syndrome? Results from a population based study of 27,485 people. Occupational and Environmental Medicine. 2001 Nov;58(11):747–752. doi: 10.1136/oem.58.11.747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruger M., Scheer F.A. Effects of circadian disruption on the cardiometabolic system. Reviews in Endocrine and Metabolic Disorders. 2009 Dec;10(4):245–260. doi: 10.1007/s11154-009-9122-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Scheer F.A., Hilton M.F., Mantzoros C.S., Shea S.A. Adverse metabolic and cardiovascular consequences of circadian misalignment. Proceedings of the National Academy of Sciences of the United States of America. 2009 Mar 17;106(11):4453–4458. doi: 10.1073/pnas.0808180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Morris C.J., Yang J.N., Garcia J.I., Myers S., Bozzi I., Wang W. Endogenous circadian system and circadian misalignment impact glucose tolerance via separate mechanisms in humans. Proceedings of the National Academy of Sciences of the United States of America. 2015 Apr 28;112(17):E2225–E2234. doi: 10.1073/pnas.1418955112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leproult R., Holmback U., Van Cauter E. Circadian misalignment augments markers of insulin resistance and inflammation, independently of sleep loss. Diabetes. 2014 Jun;63(6):1860–1869. doi: 10.2337/db13-1546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mohawk J.A., Green C.B., Takahashi J.S. Central and peripheral circadian clocks in mammals. Annual Review of Neuroscience. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wolff G., Esser K.A. Scheduled exercise phase shifts the circadian clock in skeletal muscle. Medicine and Science in Sports and Exercise. 2012 Sep;44(9):1663–1670. doi: 10.1249/MSS.0b013e318255cf4c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stokkan K.A., Yamazaki S., Tei H., Sakaki Y., Menaker M. Entrainment of the circadian clock in the liver by feeding. Science. 2001 Jan 19;291(5503):490–493. doi: 10.1126/science.291.5503.490. [DOI] [PubMed] [Google Scholar]
- 12.Kumar Jha P., Challet E., Kalsbeek A. Circadian rhythms in glucose and lipid metabolism in nocturnal and diurnal mammals. Molecular and Cellular Endocrinology. 2015 Dec 15;418(Pt 1):74–88. doi: 10.1016/j.mce.2015.01.024. [DOI] [PubMed] [Google Scholar]
- 13.Peek C.B., Affinati A.H., Ramsey K.M., Kuo H.Y., Yu W., Sena L.A. Circadian clock NAD+ cycle drives mitochondrial oxidative metabolism in mice. Science. 2013 Nov 1;342(6158):1243417. doi: 10.1126/science.1243417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andrews J.L., Zhang X., McCarthy J.J., McDearmon E.L., Hornberger T.A., Russell B. CLOCK and BMAL1 regulate MyoD and are necessary for maintenance of skeletal muscle phenotype and function. Proceedings of the National Academy of Sciences of the United States of America. 2010 Nov 2;107(44):19090–19095. doi: 10.1073/pnas.1014523107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobi D., Liu S., Burkewitz K., Kory N., Knudsen N.H., Alexander R.K. Hepatic Bmal1 regulates rhythmic mitochondrial dynamics and promotes metabolic fitness. Cell Metabolism. 2015 Oct 6;22(4):709–720. doi: 10.1016/j.cmet.2015.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Neufeld-Cohen A., Robles M.S., Aviram R., Manella G., Adamovich Y., Ladeuix B. Circadian control of oscillations in mitochondrial rate-limiting enzymes and nutrient utilization by PERIOD proteins. Proceedings of the National Academy of Sciences of the United States of America. 2016 Feb 9 doi: 10.1073/pnas.1519650113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Phielix E., Schrauwen-Hinderling V.B., Mensink M., Lenaers E., Meex R., Hoeks J. Lower intrinsic ADP-stimulated mitochondrial respiration underlies in vivo mitochondrial dysfunction in muscle of male type 2 diabetic patients. Diabetes. 2008 Nov;57(11):2943–2949. doi: 10.2337/db08-0391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bakkman L., Fernstrom M., Loogna P., Rooyackers O., Brandt L., Lagerros Y.T. Reduced respiratory capacity in muscle mitochondria of obese subjects. Obesity Facts. 2010 Dec;3(6):371–375. doi: 10.1159/000320167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Scheer F., Shea S.A. Fundamentals of the circadian system. In: SRS, editor. Basics of sleep guide [Internet] Vol. 2. Sleep Research Society; 2011. pp. 199–210. [Google Scholar]
- 20.Schoffelen P.F., Westerterp K.R., Saris W.H., Ten Hoor F. A dual-respiration chamber system with automated calibration. Journal of Applied Physiology (1985) 1997 Dec;83(6):2064–2072. doi: 10.1152/jappl.1997.83.6.2064. [DOI] [PubMed] [Google Scholar]
- 21.Harris J.A., Benedict F.G. A biometric study of human basal metabolism. Proceedings of the National Academy of Sciences of the United States of America. 1918 Dec;4(12):370–373. doi: 10.1073/pnas.4.12.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bergstrom J., Hermansen L., Hultman E., Saltin B. Diet, muscle glycogen and physical performance. Acta physiologica Scandinavica. 1967 Oct-Nov;71(2):140–150. doi: 10.1111/j.1748-1716.1967.tb03720.x. [DOI] [PubMed] [Google Scholar]
- 23.Hoeks J., van Herpen N.A., Mensink M., Moonen-Kornips E., van Beurden D., Hesselink M.K. Prolonged fasting identifies skeletal muscle mitochondrial dysfunction as consequence rather than cause of human insulin resistance. Diabetes. 2010 Sep;59(9):2117–2125. doi: 10.2337/db10-0519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Weir J.B. New methods for calculating metabolic rate with special reference to protein metabolism. Journal of Physiology. 1949 Aug;109(1–2):1–9. doi: 10.1113/jphysiol.1949.sp004363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Peronnet F., Massicotte D. Table of nonprotein respiratory quotient: an update. Canadian Journal of Sport Sciences. 1991 Mar;16(1):23–29. [PubMed] [Google Scholar]
- 26.Tripathi S., Pohl M.O., Zhou Y., Rodriguez-Frandsen A., Wang G., Stein D.A. Meta- and orthogonal integration of influenza “OMICs” data defines a role for UBR4 in virus budding. Cell Host Microbe. 2015 Dec 9;18(6):723–735. doi: 10.1016/j.chom.2015.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biology. 2007;8(2):R19. doi: 10.1186/gb-2007-8-2-r19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hughes M.E., Hogenesch J.B., Kornacker K. JTK_CYCLE: an efficient nonparametric algorithm for detecting rhythmic components in genome-scale data sets. Journal of Biological Rhythms. 2010 Oct;25(5):372–380. doi: 10.1177/0748730410379711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Krauchi K. The human sleep-wake cycle reconsidered from a thermoregulatory point of view. Physiology & Behavior. 2007 Feb 28;90(2–3):236–245. doi: 10.1016/j.physbeh.2006.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Bonaconsa M., Malpeli G., Montaruli A., Carandente F., Grassi-Zucconi G., Bentivoglio M. Differential modulation of clock gene expression in the suprachiasmatic nucleus, liver and heart of aged mice. Experimental Gerontology. 2014 Jul;55:70–79. doi: 10.1016/j.exger.2014.03.011. [DOI] [PubMed] [Google Scholar]
- 31.Archer S.N., Viola A.U., Kyriakopoulou V., von Schantz M., Dijk D.J. Inter-individual differences in habitual sleep timing and entrained phase of endogenous circadian rhythms of BMAL1, PER2 and PER3 mRNA in human leukocytes. Sleep. 2008 May;31(5):608–617. doi: 10.1093/sleep/31.5.608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akashi M., Soma H., Yamamoto T., Tsugitomi A., Yamashita S., Yamamoto T. Noninvasive method for assessing the human circadian clock using hair follicle cells. Proceedings of the National Academy of Sciences of the United States of America. 2010 Aug 31;107(35):15643–15648. doi: 10.1073/pnas.1003878107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Otway D.T., Mantele S., Bretschneider S., Wright J., Trayhurn P., Skene D.J. Rhythmic diurnal gene expression in human adipose tissue from individuals who are lean, overweight, and type 2 diabetic. Diabetes. 2011 May;60(5):1577–1581. doi: 10.2337/db10-1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Perrin L., Loizides-Mangold U., Skarupelova S., Pulimeno P., Chanon S., Robert M. Human skeletal myotubes display a cell-autonomous circadian clock implicated in basal myokine secretion. Molecular Metabolism. 2015 Nov;4(11):834–845. doi: 10.1016/j.molmet.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dyar K.A., Ciciliot S., Wright L.E., Bienso R.S., Tagliazucchi G.M., Patel V.R. Muscle insulin sensitivity and glucose metabolism are controlled by the intrinsic muscle clock. Molecular Metabolism. 2014 Feb;3(1):29–41. doi: 10.1016/j.molmet.2013.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Phielix E., Meex R., Moonen-Kornips E., Hesselink M.K., Schrauwen P. Exercise training increases mitochondrial content and ex vivo mitochondrial function similarly in patients with type 2 diabetes and in control individuals. Diabetologia. 2010 Aug;53(8):1714–1721. doi: 10.1007/s00125-010-1764-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kudo T., Akiyama M., Kuriyama K., Sudo M., Moriya T., Shibata S. Night-time restricted feeding normalises clock genes and Pai-1 gene expression in the db/db mouse liver. Diabetologia. 2004 Aug;47(8):1425–1436. doi: 10.1007/s00125-004-1461-0. [DOI] [PubMed] [Google Scholar]
- 38.Kohsaka A., Laposky A.D., Ramsey K.M., Estrada C., Joshu C., Kobayashi Y. High-fat diet disrupts behavioral and molecular circadian rhythms in mice. Cell Metabolism. 2007 Nov;6(5):414–421. doi: 10.1016/j.cmet.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 39.Dahlmans D., Houzelle A., Schrauwen P., Hoeks J. Mitochondrial dynamics, quality control and miRNA regulation in skeletal muscle: implications for obesity and related metabolic disease. Clinical Science (London) 2016 Jun 1;130(11):843–852. doi: 10.1042/CS20150780. [DOI] [PubMed] [Google Scholar]
- 40.Conroy R.T., O'Brien M. Proceedings: diurnal variation in athletic performance. Journal of Physiology. 1974 Jan;236(1):51P. [PubMed] [Google Scholar]
- 41.Facer-Childs E., Brandstaetter R. The impact of circadian phenotype and time since awakening on diurnal performance in athletes. Current Biology. 2015 Feb 16;25(4):518–522. doi: 10.1016/j.cub.2014.12.036. [DOI] [PubMed] [Google Scholar]
- 42.Arble D.M., Bass J., Laposky A.D., Vitaterna M.H., Turek F.W. Circadian timing of food intake contributes to weight gain. Obesity. 2009 Nov;17(11):2100–2102. doi: 10.1038/oby.2009.264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bandin C., Scheer F.A., Luque A.J., Avila-Gandia V., Zamora S., Madrid J.A. Meal timing affects glucose tolerance, substrate oxidation and circadian-related variables: a randomized, crossover trial. International Journal of Obesity (London) 2015 May;39(5):828–833. doi: 10.1038/ijo.2014.182. [DOI] [PubMed] [Google Scholar]
- 44.Salgado-Delgado R.C., Saderi N., Basualdo Mdel C., Guerrero-Vargas N.N., Escobar C., Buijs R.M. Shift work or food intake during the rest phase promotes metabolic disruption and desynchrony of liver genes in male rats. PloS One. 2013;8(4):e60052. doi: 10.1371/journal.pone.0060052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Woldt E., Sebti Y., Solt L.A., Duhem C., Lancel S., Eeckhoute J. Rev-erb-alpha modulates skeletal muscle oxidative capacity by regulating mitochondrial biogenesis and autophagy. Nature Medicine. 2013 Aug;19(8):1039–1046. doi: 10.1038/nm.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J., Zhou B., Yan M., Huang R., Wang Y., He Z. CLOCK and BMAL1 regulate muscle insulin sensitivity via SIRT1 in male mice. Endocrinology. 2016 Jun;157(6):2259–2269. doi: 10.1210/en.2015-2027. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.