Abstract
Objective
Parental obesity can induce metabolic phenotypes in offspring independent of the inherited DNA sequence. Here we asked whether such non-genetic acquired metabolic traits can be passed on to a second generation that has never been exposed to obesity, even as germ cells.
Methods
We examined the F1, F2, and F3 a/a offspring derived from F0 matings of obese prediabetic Avy/a sires and lean a/a dams. After F0, only lean a/a mice were used for breeding.
Results
We found that F1 sons of obese founder males exhibited defects in glucose and lipid metabolism, but only upon a post-weaning dietary challenge. F1 males transmitted these defects to their own male progeny (F2) in the absence of the dietary challenge, but the phenotype was largely attenuated by F3. The sperm of F1 males exhibited changes in the abundance of several small RNA species, including the recently reported diet-responsive tRNA-derived fragments.
Conclusions
These data indicate that induced metabolic phenotypes may be propagated for a generation beyond any direct exposure to an inducing factor. This non-genetic inheritance likely occurs via the actions of sperm noncoding RNA.
Keywords: Paternal effects, Epigenetic inheritance, Noncoding RNA, Sperm RNA
Highlights
-
•
Paternal obesity induces latent defects in metabolism in F1 sons.
-
•
Metabolic disease in F1 sons is exposed by short challenge with a Western diet.
-
•
F1 sons transmit their phenotype to F2 grandsons in the absence of dietary challenge.
-
•
F1 sperm exhibit changes to prominent small RNA species.
1. Introduction
Offspring phenotypes can be affected by parental health or parental environmental exposures, independent of variation in the inherited DNA sequence. Examples of such non-genetic transgenerational effects occur with a broad range of stressors, from dietary stress and toxin exposure to psychological stress or trauma (reviewed in [1]). In some instances, induced phenotypes can be observed across multiple generations, but whether such observations represent true inheritance of an acquired trait, or are merely a residuum of the original exposure, is not clear. This distinction has adaptive significance for organisms living in a changing environment, particularly given the potential for environmentally-induced epigenetic states to respond to selection [2], [3].
In those cases in which induced phenotypes have been observed over more than one generation, determination of true inheritance is confounded by alternative scenarios, which can be very difficult to dissect. Persistence of an induced phenotype into the grand-offspring of compromised mothers can generally be attributed to the direct exposure of developing offspring germ cells to the initial stressor. Such a scenario underpins several recent reports of multi-generational programming by parental metabolism. For example, male mice exposed to a poor intrauterine environment can transmit metabolic defects to their own offspring [4], [5], [6]. Another potential confounder is ‘serial’ programming of the induced phenotype: that is, when an induced phenotype in F1 programs defects in F2, then those defects in F2 program F3, et cetera: in this scenario an induced phenotype can theoretically be propagated indefinitely, with no requirement for ‘epigenetic inheritance’. It has been proposed that serial programming resulting from a repeatedly compromised gestational environment may underlie most, if not all, examples of multigenerational maternal programming [7].
It is currently unclear whether an induced metabolic phenotype can be transmitted into successive generations without the continued influence of the stimulus; in other words, whether true, non-genetic inheritance can occur via gametes that were never exposed [8]. True inheritance of parental effects is arguably best studied through the male lineage, where confounding effects of the intrauterine environment can be largely excluded. Many paternal effects have been reported across a broad range of stressors [9]. In rodents, perturbed paternal metabolism can induce metabolic defects in first-generation offspring [5], [10], [11], [12], but whether these effects can be carried into a second unexposed generation is not known. Epidemiological observations suggest it could be the case [13], [14], however inheritance of something acquired is difficult to ascertain in human cohorts: genetic heterogeneity, lifestyle factors, and the necessarily retrospective nature of longitudinal studies are major confounders.
We have addressed the question of inheritance of induced metabolic traits using a congenic rodent model of obesity and pre-diabetes in which the dominant obesogenic allele can be segregated away from the offspring. In this model we find that paternal obesity induces a latent metabolic phenotype in F1 sons that is unmasked by short exposure to a Western-style high-fat diet. By breeding F1 sons maintained on a healthy control diet (those offspring exposed to paternal obesity but metabolically normal), we find that the induced but latent phenotype is inherited into a second, unexposed generation. We also find that this inheritance is associated with changes to the small RNA profile of F1 sperm, despite these sperm developing and maturing in metabolically normal environments.
2. Materials and methods
2.1. Mice and diets
All animals were handled in accordance with good practice as defined by the National Health and Medical Research Council (Australia) Statement on Animal Experimentation, and requirements of state government legislation. The study was approved by the Garvan/St Vincent's Animal Ethics Committee (12/09 and 13/35).
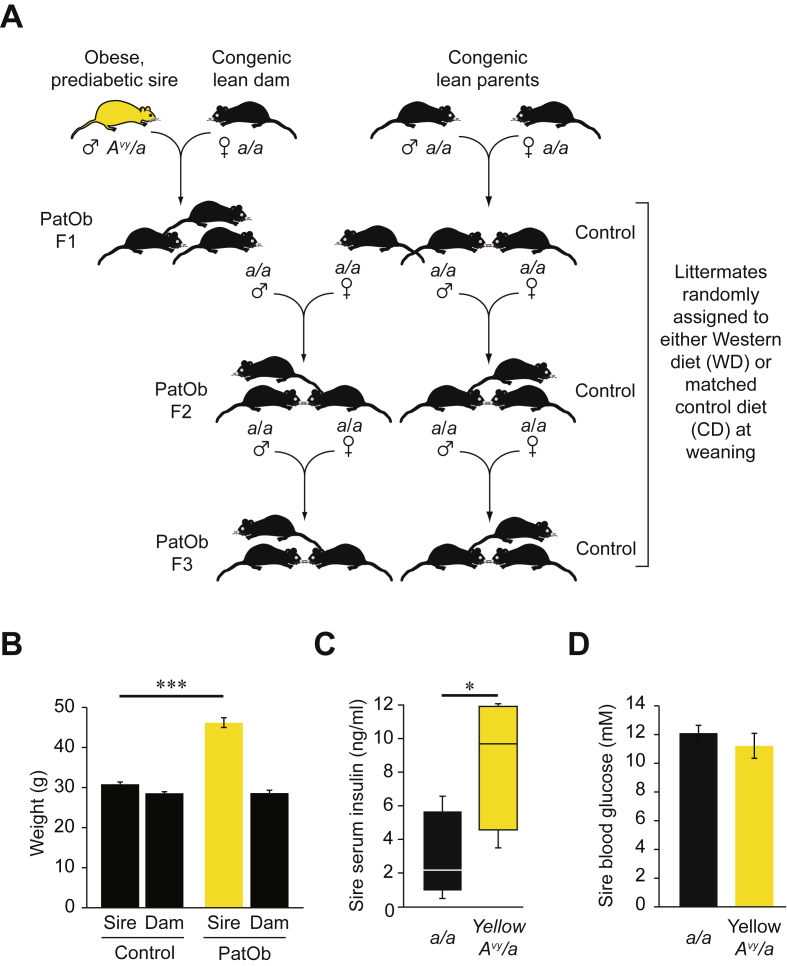
The Avy mice used in this study were descended from an isogenic C57BL/6 colony at Oak Ridge National Laboratories (Oak Ridge, TN, USA). Avy/a mice display a variety of phenotypes from obese yellow through degrees of coat-colour mottling to lean agouti [15]. To avoid any effects of ancestral obesity in our stock Avy colony, we maintain it by breeding only lean agouti Avy/a mice to congenic a/a mice. The breeding strategy and breeding statistics are outlined in Figure 1A and Table T1 respectively.
Figure 1.
Experimental design and metabolic characteristics of obese founder males. (A) Schematic diagram showing breeding strategy. Obese yellow Avy/a males were mated with congenic lean a/a dams to generate PatObF1 offspring. PatObF1 males were mated with Control F1 daughters, generated by mating lean a/a mice, to generate PatObF2. PatObF3 were generated by intercrossing PatObF2 males and females. For simplicity, Avy/a offspring of obese yellow Avy/a sires are not shown. (B) Weights of lean a/a sires and their a/a dams (Control, n = 62) and of obese yellow Avy/a sires and their a/a dams (PatOb, n = 21) measured one week after birth of offspring. (C) Blood glucose of lean a/a and obese yellow Avy/a males at 12 weeks of age (n = 12). (D) Serum insulin of lean a/a and obese yellow Avy/a males at 12 weeks of age (n = 5). Error bars represent SEM; *p < 0.05, ***p < 0.001.
Experimental a/a mice were derived from mating obese yellow Avy/a males to lean a/a females. Control mice were derived from mating of lean a/a males and females. Control and experimental mice were generated contemporaneously with experimental mice the generations. Data from multiple generations of control mice were pooled for each metabolic test as statistical analyses ensured that the metabolic characteristics of each generation were equivalent.
The mice in this study were produced over the course of two years in the same facility. To maximise breeding output (obese yellow Avy/a mice are poor breeders), in general males were allowed to cohabit with females throughout the breeding period. To control for social effects on the females, some F0 animals were time-mated: males were placed in the female's cage overnight and removed early the next morning along with any faeces.
All breeding mice were fed ad libitum on NIH-31 control diet from weaning (5% w/w fat, 13.5 MJ/kg). a/a experimental offspring were randomly assigned at weaning to either SF00-219 high-fat diet designed to mimic a Western fast-food diet (WD, equivalent to Harlan Teklad TD88137; 22% w/w fat (40% digestible energy), 19.4 MJ/kg or matched normal fat control (CD; 6% w/w fat, 16.1 MJ/kg)). Feeds were obtained from Specialty Feeds (Glen Forrest, WA, Australia). Avy/a offspring in F1 were culled at weaning. Experimental a/a mice were culled at 12 weeks of age; metabolic organs were harvested and weighed and blood collected, and sperm were harvested from some animals (see 2.3).
2.2. Metabolic profiling
Glucose tolerance testing was performed on six week old mice; the tester was unaware of the group allocation of the individuals being tested. Mice were fasted for 6 h and given an intraperitoneal injection of 1.5 g glucose/kg weight. Blood glucose levels were measured at 0, 15, 30, 45, 60 and 90 min using an AlphaTrak glucose monitor (Abbott). Animals were excluded from the analysis if their blood glucose did not increase by at least 75% by 15 min post-injection, or if excessive fighting between animals before injection raised the basal glucose measurement more than two standard deviations above the group mean.
Serum insulin levels were measured in 12 week blood samples using the Mouse Insulin ELISA kit (Millipore); the tester was unaware of the group allocation of the samples being tested.
Lipidomic profiling was performed on 12 week liver samples, as described previously [16]; the tester was blinded to the group allocation of samples. Samples more than two standard deviations from the group mean were excluded.
2.3. Sperm isolation and RNA preparation
Mature motile sperm were isolated from the cauda epididymis of 12 week old experimental mice on CD. Caudae were dissected and sliced open, and sperm allowed to swim out into DMEM media on a heating pad. Media was collected after 30 min and either snap frozen in liquid nitrogen or processed immediately. After thawing, samples were centrifuged at 50 g to remove any tissue debris. The supernatant was collected and sperm were centrifuged for 3 min at 2500 g, washed twice in PBS, incubated in somatic cell lysis buffer (0.5% Triton-X, 0.1% SDS) for 5 min, washed again and resuspended in PBS. A proportion of the sperm were examined microscopically for the presence of somatic cells: only sperm samples with no somatic cells visible in 500–1000 sperm visualised were used for RNA analysis. RNA was extracted from pelleted sperm using Trizol; sperm lysis was assisted by the addition of 200 mM β-mercaptoethanol, and repeated passage through a 29 g needle.
2.4. Small RNA-Seq
Barcoded small RNA sequencing libraries were prepared from ∼100 ng sperm total RNA using the NEBNext Small RNA for Illumina kit (NEB). Ligation of the 3′ adapter was allowed to proceed overnight to encourage maximal ligation of 3′O-methylated RNAs. Libraries were sequenced on an Illumina HiSeq 2500 in Rapid Run mode.
Fastq files were trimmed using Cutadapt (v1.6) to lengths of 18–51 nt, with 10% adapter sequence error allowed. Trimmed reads were aligned to genome build mm10 using Bowtie (v1.1.1) with parameters: --sam -n 1 -l 18 -e 70 (one mismatch allowed in an 18 base seed and a maximum sum of quality scores for mismatched bases of 70). Multimappers were randomly assigned among best hits. Mapped reads were annotated against miRNAs (mm10 mirbase20; mirbase.org), tRNAs (UCSC golden path), nested repeats (UCSC golden path), refgene (UCSC golden path) using custom perl scripts; annotation ties were broken by considering (in order) the orientation of the read relative to the genetic element, biotype hierarchy (according to the list above) and the number of bases overlapping the genetic element. For miRNA comparisons we used generalised linear modelling with EdgeR [17], applying an abundance threshold of 0.02% total miRNA (i.e. 200 reads per million mapped miRNA reads).
2.5. FLAG-Ago2 immunoprecipitation
HeLa cells were transfected with a FLAG-Ago2 construct in p3XFLAG-CMV-10 plasmid (Sigma). After 48 h, ∼5 × 106 cells were lysed and the lysate incubated with Anti-Flag M2 antibody or isotype control IgG (Sigma) bound to Dynabeads Protein G (Life Technologies). After washing, 10% of the immunoprecipitation was removed for Western blotting for Flag-Ago2, and the remaining 90% was used for small RNA quantification. For RNA preparation, beads were treated with Proteinase K and removed, and RNA was prepared using Trizol.
tRF5-GluCTC, tRF5-GlyGCC and hsa-miR-21-5p were assayed in HeLa cell total RNA (to confirm tRF expression) and IP RNA (to test for Ago2 enrichment) by Taqman MicroRNA assay (Life Technologies).
2.6. miRNA/tRF target and GO analysis
To generate lists of target genes for miR-10a/b and tRF5-GluCTC, we used TargetScan v4.0, which allows input of a user-defined seed sequence [18]. We used DAVID [19], [20] to determine significantly enriched ontologies. For analysis of miRNA/tRF target expression in oocytes and early embryo, we used data from DBTMEE [21] using an abundance threshold of 3 FKPM.
2.7. Statistical analyses
Weights trajectories were fitted using a linear mixed model [22] and comparisons performed with a Wald test. For metabolic measurements (GTT, serum insulin, liver lipids), groupwise comparisons were performed with one-way ANOVA. Because sample size was not equivalent between groups, we confirmed the homogeneity of variances using Levene's test. Where variances were unequal, Welch's modified F statistic was used to assess significance; equivalent conclusions were obtained using the Welch ANOVA and the standard ANOVA. Post-hoc pairwise comparisons between groups of interest were performed with a one-sided Student's t-test.
3. Results
3.1. Paternal obesity induces a latent predisposition to hepatic insulin resistance in F1 sons
To model paternal obesity, we used C57BL/6 mice carrying the dominant agouti viable yellow (Avy) mutation [23]. On the C57 background, the Avy allele exhibits epigenetic inheritance through the maternal germline [24], and this facet of the strain has given it prominence in the study of mammalian epigenetic inheritance, in particular as an ‘epigenetic biosensor’ [25], [26], [27], [28]. However the use of Avy mice in this experiment is unrelated to inheritance, epigenetic or otherwise, of Avy itself. Here we merely exploit the obese and insulin resistant phenotype of Avy/a mice that display pan-somatic epigenetic activity of the Avy allele (obese yellow mice). We study only their a/a offspring, which do not carry the Avy allele. Yellow Avy/a mice overeat because Avy expression interferes with hypothalamic melanocortin receptor-mediated satiety signalling [23], a pathway linked to genetic obesity in humans [29]. Yellow Avy mice model human obesity in the absence of any dietary intervention: they simply overeat their standard food, become progressively heavier and fatter, and by maturity are obese and insulin resistant [30].
Obese yellow Avy/a heterozygote sires were mated to congenic lean a/a dams to generate a/a offspring that are genetically identical to offspring from a/a matings. In this way, we generated two isogenic groups of F1 offspring (PatOb and Control) differing only in the Avy genotype, and the phenotypes, of their sires (Figure 1A). Obese Avy/a sires were profoundly heavier than congenic lean a/a sires, and also hyperinsulinemic, but not hyperglycaemic (Figure 1B–D). These characteristics model the majority of obese male humans of reproductive age who are insulin resistant but not frankly diabetic [31]. Offspring of each group were randomly assigned at weaning to either control diet (CD) or to a Western-style diet (WD) high in saturated fat and sugar. Breeding statistics and diet composition can be found in Tables T1 and T2, respectively.
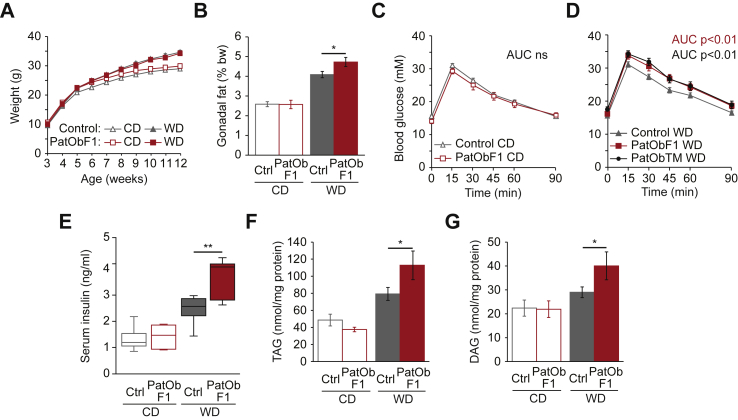
When maintained on either CD or WD, weight trajectories of male a/a offspring of obese sires (PatObF1) were no different to offspring of lean sires (Figure 2A). But while all animals gained weight on the WD, PatObF1 animals gained more fat than controls (Figure 2B), suggesting that paternal obesity confers an increased sensitivity to the adipogenic effects of a high-fat diet. Similarly, glucose tolerance was normal in all F1 mice when maintained on CD (Figure 2C), but after only three weeks of exposure to the WD, PatObF1 males showed a significantly impaired response in an intraperitoneal glucose tolerance test (Figure 2D). The same degree of intolerance was observed whether the offspring were generated from an overnight timed mating or continuous co-housing of sire and dam (Figure 2D), indicating that the programmed defect was transmitted at the time of mating, rendering programming via social effects of the obese sire on the dam or offspring improbable. PatObF1 males also had exacerbated hyperinsulinemia in response to WD (Figure 2E), along with hepatic steatosis: intrahepatic triacylglycerol (TAG), and the insulin resistance driver diacylglycerol (DAG) [32] were significantly elevated in PatObF1 males (Figure 2F,G). Together these data indicate that paternal obesity programmed a latent predisposition to hepatic insulin resistance that was unmasked by short challenge with a Western diet. We observed this programming only in males: for the duration of the experiment, female PatObF1 exhibited normal glucose homeostasis on both CD and WD (Figure S1). Sexual dimorphism is often reported in both maternal and paternal programming [10], [13], [14], [33], so this finding is not unusual, but underscores the importance of examining both genders. The origin of gender discordance in environmental programming, or in non-communicable disease in general, is not currently understood: the actions of sex hormones, differing placental function [34], and autosomal gender-specific epigenetic differences (e.g. [35]) may all be involved.
Figure 2.
Paternal obesity programs latent metabolic defects in F1 male offspring. (A,B) Body weights (A) and relative gonadal fat weights at 12 weeks (B) of Control and PatObF1 offspring fed control diet (CD) or Western diet (WD); CD: Control n = 46, PatOb n = 16; WD: Control n = 43, PatOb n = 19. (C,D) Glucose tolerance tests on six-week old CD offspring (C) and WD offspring (D), including those from time-mated obese sires (PatObTM, n = 16; other animal numbers as in A). (E) Serum insulin in 12-week old offspring; CD: Control n = 9, PatOb n = 5; WD: Control n = 8, PatOb n = 5. (F,G) Hepatic triacylglyceride (TAG, F) and diacylglyceride (DAG, G) levels in 12-week old offspring; CD: Control n = 7, PatOb n = 8; WD: Control n = 8, PatOb n = 8. Error bars represent SEM; *p < 0.05, **p < 0.01.
3.2. The latent metabolic phenotype induced by paternal obesity is transmitted from F1 sons to F2 grandsons
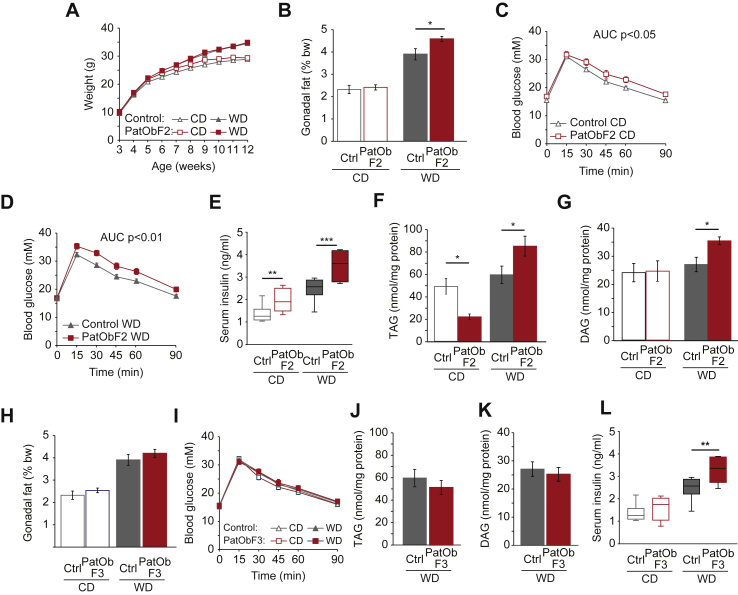
We then asked whether the defects programmed by paternal obesity could be inherited, that is, passed into a second, unexposed, generation. Here the latency of the phenotype in PatObF1 provides a material advantage in distinguishing inheritance from serial programming, since the germ cells and sperm of F1 mice on CD were not exposed to any metabolic dysfunction. When we bred control females with PatObF1 males on CD we found that the resultant PatObF2 males faithfully recapitulated the phenotype of their F1 sires. Again the weight trajectories of PatObF2 grandsons were no different from control (Figure 3A), but PatObF2 had a greater gonadal fat mass on WD (Figure 3B). With the WD challenge, PatObF2 grandsons exhibited the same pathognomonic signs of hepatic insulin resistance displayed by PatObF1 sons: marked glucose intolerance (Figure 3D), hyperinsulinemia (Figure 3E) and elevated intrahepatic TAG and DAG (Figure 3F,G). The only contrast with the phenotype of F1 was that PatObF2 exhibited mild hyperinsulinema and a mild glucose handling impairment even on CD (Figure 3C,E), suggesting that the phenotype in F2 is slightly more overt than in F1. Again, this induced phenotype was not apparent in females (Figure S2). Nor was it apparent in an F3 generation of males: by the third generation, the Western diet challenge made no difference to male offspring fat mass, glucose tolerance or hepatic lipids (Figure 3H–K). The only remaining manifestation of the latent phenotype was hyperinsulinemia in response to the WD (Figure 3L).
Figure 3.
Metabolic programming by paternal obesity is inherited by a second, but not third, generation. (A) Body weights of Control and PatObF2 offspring fed control diet (CD) or Western diet (WD); CD: Control n = 46, PatObF2 n = 17; WD: Control n = 43, PatObF2 n = 17. (B) Relative gonadal fat weights of animals in A. (C,D) Glucose tolerance tests on six week old Control and PatObF2 mice on CD (C) and WD (D); animal numbers as in A. (E) Serum insulin in 12-week old Control and PatObF2 mice; CD: Control n = 9, PatObF2 n = 8, WD: Control n = 8, PatObF2 n = 7. (F,G) Liver triacylglyceride (TAG, F) and diacylglyceride (DAG, G) levels in 12-week old Control and PatObF2 on CD and WD; CD: Control, n = 8, PatObF2 n = 8; WD: Control, n = 8, PatObF2 n = 7. (H) Relative gonadal fat weights in 12-week old Control and PatObF3 F3 mice; CD: Control n = 46, PatObF3 n = 31; WD: Control n = 43, PatObF3 n = 27. (I) Glucose tolerance tests on six week old Control and PatObF3 CD and WD mice; animal numbers as in H. (J, K) Liver triacylglyceride (TAG, J) and diacylglyceride (DAG, K) levels in 12-week old Control and PatObF3 mice on WD: Control, n = 8, PatObF2 n = 7. (L) Serum insulin in 12-week old Control and PatObF3 mice; CD: Control n = 9, PatObF3 n = 5, WD: Control n = 8, PatObF3 n = 7. Error bars represent SEM in all panels; *p < 0.05, **p < 0.01, ***p < 0.001.
Taken together, these data indicate that the latent predisposition to metabolic defects induced by paternal obesity is inherited into a second, unexposed generation, but not a third. The germ cells that gave rise to F2 developed while F1 mice were gestating in a normal intrauterine environment, and matured inside an F1 individual that was not obese and had no overt metabolic disturbance. Thus we have observed true, non-genetic transgenerational inheritance of an acquired metabolic phenotype induced by ancestral obesity.
3.3. Inheritance of the latent metabolic phenotype into F2 is associated with changes in F1 sperm RNA content
The characteristics of the inheritance we observe implicate an epigenetic inheritance system. Small noncoding RNAs direct epigenetic states in the germline [36], [37] and have been implicated in invertebrate models of epigenetic inheritance [38], [39], [40], [41]. Recently two separate studies have shown that the small RNA content of murine sperm is sensitive to dietary perturbations: both a low-protein diet and a high-fat diet were linked to changes in the abundance of specific small RNA species in exposed sperm [12], [42]. It is therefore likely that yellow founders, with their hyperphagia, profound obesity and metabolic abnormalities, also have deranged sperm small RNA profiles. By contrast, PatObF1 mice are not obese, nor do they exhibit any apparent metabolic abnormality, yet despite eating a normal healthy diet they are able to transmit the induced latent phenotype to PatObF2. We therefore asked whether small RNA perturbations may also underlie the inheritance of the induced phenotype by examining the small RNA content of sperm from the ‘normal’ PatObF1 males on control diet. We compared PatObF1 sperm small RNA profiles with those from control-fed control males.
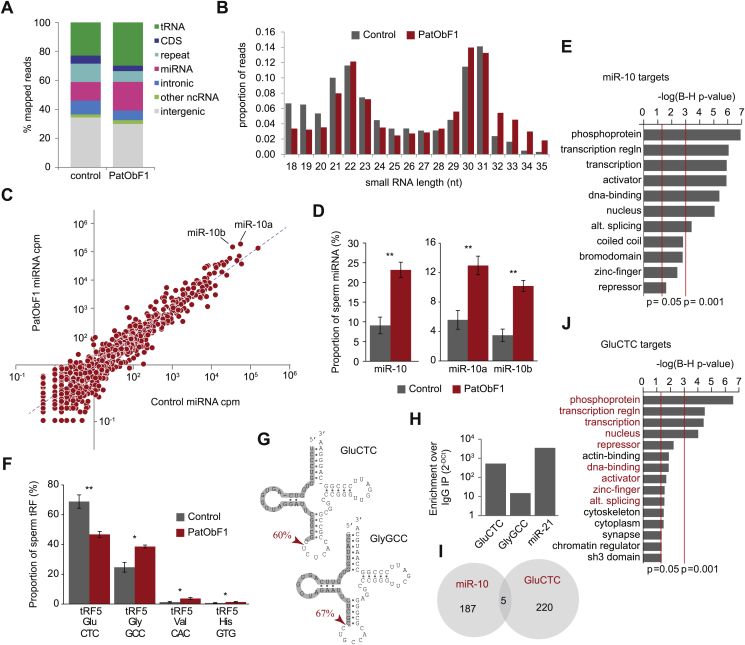
The distribution of small RNA lengths and biotypes between the two groups was very similar (Figure 4A,B), but within two biotypes – miRNA and tRNA-derived fragments (tRFs) – there were major differences in abundance of some species. Using stringent thresholds (only considering miRNA with counts >200 per million, and adjusted p-value of <0.05) we found 24 miRNAs with altered expression in PatObF1 (Figure 4C, Table 1). One sperm miRNA, miR-10, exhibited such dramatic changes that it increased by 2.5-fold to constitute ∼25% of all sperm miRNA in PatObF1 (Figure 4D). Both miR-10 isotypes, −10a and −10b (encoded by different genes) were significantly elevated. Differing only by one central nucleotide outside of the seed sequence, these two miRs target the same mRNAs. These targets were found to be heavily and significantly enriched for functions related to the regulation of transcription (Figure 4E).
Figure 4.
Exposure to paternal obesity alters small RNAs in the sperm of F1 sons. Small RNA biotype (A) and size distribution (B) of all 18-35 nt sRNA reads in Control and PatObF1 sperm. (C) Scatterplot of sperm miRNA abundance from Control (x-axis) versus PatObF1 (y-axis). (D) miR-10 as a fraction of all sperm miRNA in PatObF1 (red) relative to Control (grey); panel at right shows levels of the two miR-10 isoforms **q < 0.01 (E) Gene ontologies significantly enriched in the targets of miR-10. (F) tRFs significantly altered in PatObF1 sperm (red) relative to control sperm (grey) represented as proportion of all sperm tRFs * p < 0.05, **p < 0.01 (G) Clover leaf structures of GluCTC (top) and GlyGCC (bottom); dominant sperm tRF isoform shaded grey and red arrowheads show major cleavage point. (H) Results of Taqman RT-PCR on RNA purified from an Ago2 immunoprecipitation (IP) showing tRF enrichment in the IP over the negative control IgG IP; miR-21 is a positive control.(I) Individual gene targets of miR-10 and tRF5-Glu-CTC show little overlap. (J) Gene ontologies significantly enriched in the targets of tRF5-Glu-CTC; ontologies in common with miR-10 are shown in red.
Table 1.
microRNAs with altered abundance in sperm of PatObF1.a
| microRNA | Average counts per millionb |
Fold change | q-value | ||
|---|---|---|---|---|---|
| Control | PatOb | ||||
| Upregulated | mmu-miR-204 | 717 | 3749 | 5.23 | 0.0002 |
| mmu-miR-182 | 694 | 2298 | 3.31 | 0.0043 | |
| mmu-miR-429-3p | 386 | 1182 | 3.06 | 0.0003 | |
| mmu-miR-10b | 35,008 | 101,932 | 2.91 | 0.0012 | |
| mmu-miR-146b | 566 | 1634 | 2.88 | 0.0192 | |
| mmu-miR-200a | 280 | 793 | 2.83 | 0.0004 | |
| mmu-miR-148a-3p | 7619 | 21,046 | 2.76 | 0.0033 | |
| mmu-miR-200a-3p | 6752 | 18,443 | 2.73 | 0.0033 | |
| mmu-miR-200b-3p | 8257 | 22,186 | 2.69 | 0.0039 | |
| mmu-miR-451a | 632 | 1666 | 2.64 | 0.0257 | |
| mmu-miR-183 | 1226 | 2985 | 2.44 | 0.0088 | |
| mmu-miR-10a | 55,867 | 129,719 | 2.32 | 0.0088 | |
| mmu-let-7e | 628 | 1205 | 1.92 | 0.0267 | |
| mmu-miR-210-3p | 350 | 648 | 1.85 | 0.0266 | |
| mmu-miR-187-3p | 245 | 378 | 1.54 | 0.0438 | |
| Downregulated | mmu-miR-328-3p | 1661 | 582 | 0.35 | 0.0265 |
| mmu-miR-676-3p | 2003 | 699 | 0.35 | 0.0262 | |
| mmu-miR-378a-3p | 5059 | 1740 | 0.34 | 0.0246 | |
| mmu-miR-669c | 903 | 309 | 0.34 | 0.0433 | |
| mmu-miR-92a-3p | 12,593 | 4390 | 0.35 | 0.0341 | |
| mmu-miR-425 | 5340 | 1592 | 0.30 | 0.0405 | |
| mmu-miR-127-3p | 1470 | 360 | 0.24 | 0.0033 | |
| mmu-miR-184-3p | 7487 | 1808 | 0.24 | 0.0103 | |
| mmu-miR-34b-3p | 12,678 | 2073 | 0.16 | 0.0052 | |
Where sequencing abundance >200 reads per million and q < 0.05.
Reads per million mapped miRNA reads.
Around a quarter of all sperm small RNAs were tRFs of two predominant species, tRF5-GluCTC and tRF5-GlyGCC: collectively, sequences of these two isotypes contributed ∼90% of all sperm tRFs. The relative ratios of these tRFs were inverted in PatObF1 sperm relative to control sperm, with an ∼30% reduction in tRF5-GluCTC and a corresponding increase in tRF5-GlyGCC (Figure 4F). Another two tRFs, ValCAC and HisGTG were also significantly elevated in PatObF1 sperm, although these tRFs were much less abundant, constituting less than 5% of all sperm tRFs. Both GluCTC and GlyGCC tRFs displayed precise cleavage at the 5′ side of the anticodon loop of the mature tRNA to generate fragments of predominantly 30 and 31 nucleotides, respectively (Figure 4G). These two tRFs are almost precisely the same tRFs that were recently reported to be perturbed in male sperm by exposure to an altered diet [12], [42].
Mature sperm are transcriptionally and translationally inert, so any function of sperm-borne small RNA is likely to manifest post-fertilisation. The recently reported diet-responsive tRFs induce transcriptional changes in the early embryo when injected into zygotes, but whether this is a direct or indirect transcriptional effect is unknown. Available evidence indicates that tRFs reside almost exclusively in the cytoplasm [43] where they can associate with polyribosomes and affect translation [44], [45], [46]. The majority of miRNAs are also found in the cytoplasm and functionally associate with Ago2. Ago2 is highly expressed in the oocyte and early embryo and is required for the maternal-to-zygotic transition (MZT) [47]. We asked whether the two sperm tRFs can also associate with Ago2, and found that tRF5-GluCTC, and to a lesser extent tRF5-GlyGCC, was enriched in an Ago2 pulldown (Figure 4H). This indicates that at least tRF5-GluCTC binds Ago2, signifying its potential to act in a miRNA-like manner in the zygote. Using a miRNA seed-sequence based algorithm we identified putative tRF5-GluCTC targets; these targets shared very little overlap with those of miR-10 at the gene level (Figure 4I; Table T3), yet remarkably resided in strikingly similar ontologies related to transcriptional regulation (Figure 4J). Using publicly available datasets of murine early embryonic transcription [21] we found that 114/220 miR-10 targets and 77/191 tRF-5-GluCTC targets are expressed in the oocyte and approximately half of these targets are reduced by at least two-fold at the 4-cell stage (Table T4), consistent with a repressive post-transcriptional function of miR-10 and tRF-5-GluCTC.
4. Discussion
We have identified an acquired metabolic phenotype that is induced in two succeeding generations by ancestral paternal obesity. This phenotype occurs independent of the inherited genotype, the gestating environment, the fathers' diet, or continued germ cell exposure to metabolic disturbance. The characteristics of our model – in particular the latency of the induced phenotype – strongly suggest that the transmission we observe represents true inheritance of an acquired metabolic phenotype. There have by now been many reports of intergenerational transmission of induced phenotypes, but whether these represent true environmentally-induced epigenetic inheritance is a matter of current debate [48]. We are aware of only one previous report of a paternally-induced (behavioural) phenotype that was inherited into a second generation without exposure of germ cells to the stressor [49]. Thus, our findings may be viewed as a clear demonstration of the true inheritance of a metabolic phenotype induced by ancestral obesity. Our findings have immediate relevance to understanding the consequences of the human obesity epidemic, and add to the knowledge on potential mechanisms by which the memory of ancestral exposures may be maintained through multiple generations.
In our model, obese founder males had a syndrome of obesity and hyperinsulinemia, but the F1 males that gave rise to the F2 generation did not. When maintained on a regular chow diet, F1 males were effectively indistinguishable from isogenic controls in terms of weight, glucose metabolism, insulin levels, and hepatic lipids; this argues strongly against the serial transmission of the metabolic defects from F1 to F2, and implies epigenetic inheritance. The possibility that the latent phenotype was transmitted by behavioural factors is also unlikely: dams partnered with obese sires were no heavier than dams of control sires (Figure 1B), indicating that they did not acquire the hyperphagic behaviour of their partners, and the programmed phenotype was just as robust when the influence of the sire was restricted to an overnight timed-mating (Figure 2D). The most parsimonious explanation for the transmission of the latent metabolic phenotype from F1 to F2 is transgenerational epigenetic inheritance: some mark or memory of the exposure to paternal obesity passaged via the F1 germline.
While F2 males exhibited a phenotype identical to, if not more pronounced, than F1 males, they did not program any phenotype in their F3 offspring (Figure 3). The only aspect of the induced phenotype that we observed to be maintained into F3 was elevated serum insulin on exposure to the Western diet. In the absence of any glucose handling defect, this could be indicative of subclinical insulin resistance in PatObF3 mice, and indicates that by the third generation after exposure to obesity the programmed phenotype has all but disappeared. These data lend further support to the view that what we observe is true inheritance as opposed to serial programming: if some aspect of the metabolic phenotype observed in F1 is responsible for programming the F2 phenotype, then that phenotype should in turn program F3 – but it does not. The reversion of the phenotype three generations after exposure is reminiscent of very recent findings in Caenorhabditis elegans, where induced RNAi responses are heritable for only a limited number of generations in the absence of selection or further exposure to the inducing agent [50]. Such ‘tunable mechanisms’ of epigenetic inheritance might also be at play with the response to paternal obesity in mammals; reversion could reflect an adaptive mechanism to counter the maintenance of epigenetic responses in rapidly changing environmental conditions [50].
Some insight into potential mechanism is provided by our finding of alterations to multiple prominent small RNA species in the sperm of F1 mice born of obese fathers (Figure 4). The idea that sperm RNA is involved in transgenerational epigenetic inheritance is well established in invertebrate systems. In the fly, small RNAs influence the transgenerational inheritance of maternal immunity to paternally-introduced transposons [41]. In the nematode C. elegans, small noncoding RNAs are implicated in multiple models of transgenerational inheritance [38], [39], [51] including a metabolic starvation-induced response [40]. There is more limited, and somewhat controversial, evidence for small RNA involvement in inheritance of vertebrate phenotypes: injection of sperm RNA into fertilised oocytes has been reported to recapitulate several induced phenotypes, including white tail tipping [52], cardiac hypertrophy [53] and fear [54].
Molecular changes to mouse sperm (including alterations to DNA methylation, miRNAs, and tRFs), have been described in response to environmental factors, including dietary perturbations [4], [6], [11], [12], [42], [54], but in all of these studies sperm were directly exposed to the environmental stimulus, either as developing germ cells or as maturing gametes. In this study we did not examine the directly exposed sperm (i.e. that from the obese founder males), because identification of any agent of inheritance would be confounded by changes induced by direct exposure of the sperm to the overt syndrome of obesity and prediabetes. It is remarkable, however, that we find alterations to the same sperm tRF species as have been recently reported to be sensitive to male diet [12], [42]. The sperm from F1 males in our study were not exposed to obesity or an altered diet, or indeed to any apparent metabolic insult at all, but they still transmitted the induced metabolic phenotype to the F2 generation. Together these data provide compelling evidence implicating tRFs and other small RNAs in the inheritance of acquired metabolic traits in vertebrate species. How these RNA perturbations are maintained in germ cells and gametes for an entire life cycle beyond the inducing stressor is currently a mystery.
Given that mature sperm are transcriptionally and translationally inert, it is likely that sperm small RNAs function post-fertilisation. Recent work in C. elegans found that around half of all small noncoding RNAs in the one-cell embryo are paternally derived [55]. This raises the possibility that sperm-borne small RNAs play a role in very early development prior to, or during, the maternal-to-zygotic transition [56]. Both of the recent studies that identified diet-regulated sperm tRFs invoked a direct transcriptional interference mechanism for sperm-borne tRFs in the early embryo [11], [25]. This is curious as available evidence indicates that tRFs reside almost exclusively in the cytoplasm [43] where they can associate with polyribosomes and affect translation [44], [45], [46]. Our data suggest that tRFs may have a post-transcriptional regulatory function that affects embryonic transcription indirectly. The targets of the two small RNA species in sperm most affected by paternal obesity, tRF5-GluCTC and miR-10, are heavily enriched in transcriptional regulatory functions, implying that changes in their abundance in sperm may perturb transcriptional regulation in the developing embryo post-transcriptionally. That a large (>50%) of these targets are significantly downregulated by the 4-cell stage, when the MZT is complete, implies that early perturbations in these pathways may set in train alterations that lead to a reduced resilience to environmental insults (such as over-nutrition) later in life.
5. Conclusions
In summary, we demonstrate transgenerational inheritance of an acquired metabolic trait in mammals. Our data implicate small RNAs in the mechanism of inheritance. If the phenomenon we observe here translates to humans, our findings suggest that individuals with obese paternal ancestry could avoid the deleterious effects of metabolic programming with dietary intervention, yet still propagate the propensity for metabolic dysfunction to their offspring. These findings might explain, at least in part, the dramatic rise in incidence of the metabolic syndrome observed in the 20th and 21st century.
Author contributions
C.M.S. and J.E.C. conceived and designed the project with input from S.A.E. and D.I.K.M.; S.A.E. and A.A. performed animal husbandry and metabolic analyses, with assistance from P.E.Y., J.E.C. and M.E.B.; J.E.C. performed small RNA profiling; S.P.K. performed Ago2 immunoprecipitation and Taqman assays; P.E.Y., D.T.H, E.G. and J.W.K.H performed bioinformatics analysis; K.G.L. and D.C.H. performed mass spectrometry; C.M.S., J.E.C., M.F., G.H. and D.I.K.M. analysed and interpreted data; C.M.S., J.E.C. and D.I.K.M. wrote the paper, with input from all authors.
Acknowledgements
Raw sequencing data has been deposited in the NCBI Sequencing Read Archive under study accession SRP051542. J.E.C. is supported by an Australian Research Council (ARC) DECRA Fellowship (DE120100723). G.H. is supported by an ARC Future Fellowship (FT110100455). M.A.F. is supported by an National Health and Medical Research Council (NHMRC) Senior Principal Research Fellowship (APP1021168). C.M.S. is supported by an ARC Future Fellowship (FT120100097). The study was also supported by the Victor Chang Cardiac Research Institute, St Vincents Clinic Foundation, and in part by the Mostyn Foundation, ARC DP120100825 and ARC DP130103027.
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.06.008.
Contributor Information
Jennifer E. Cropley, Email: j.cropley@victorchang.edu.au.
Catherine M. Suter, Email: c.suter@victorchang.edu.au.
Competing financial interests
The authors declare no competing financial interests.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.Hanson M.A., Gluckman P.D. Early developmental conditioning of later health and disease: physiology or pathophysiology? Physiological Reviews. 2014;94:1027–1076. doi: 10.1152/physrev.00029.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Cropley J.E., Dang T.H., Martin D.I., Suter C.M. The penetrance of an epigenetic trait in mice is progressively yet reversibly increased by selection and environment. Proceedings of the Royal Society of London Series B: Biological Sciences. 2012:2347–2353. doi: 10.1098/rspb.2011.2646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sollars V., Lu X., Xiao L., Wang X., Garfinkel M.D., Ruden D.M. Evidence for an epigenetic mechanism by which Hsp90 acts as a capacitor for morphological evolution. Nature Genetics. 2003;33:70–74. doi: 10.1038/ng1067. [DOI] [PubMed] [Google Scholar]
- 4.Martinez D., Pentinat T., Ribo S., Daviaud C., Bloks V.W., Cebria J. In utero undernutrition in male mice programs liver lipid metabolism in the second-generation offspring involving altered Lxra DNA methylation. Cell Metabolism. 2014;19:941–951. doi: 10.1016/j.cmet.2014.03.026. [DOI] [PubMed] [Google Scholar]
- 5.Jimenez-Chillaron J.C., Isganaitis E., Charalambous M., Gesta S., Pentinat-Pelegrin T., Faucette R.R. Intergenerational transmission of glucose intolerance and obesity by in utero undernutrition in mice. Diabetes. 2009;58:460–468. doi: 10.2337/db08-0490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Radford E.J., Ito M., Shi H., Corish J.A., Yamazawa K., Isganaitis E. In utero undernourishment perturbs the adult sperm methylome and intergenerational metabolism. Science. 2014 doi: 10.1126/science.1255903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aiken C.E., Ozanne S.E. Transgenerational developmental programming. Human Reproduction Update. 2014;20:63–75. doi: 10.1093/humupd/dmt043. [DOI] [PubMed] [Google Scholar]
- 8.Dias B.G., Ressler K.J. Experimental evidence needed to demonstrate inter- and trans-generational effects of ancestral experiences in mammals. Bioessays. 2014;36:919–923. doi: 10.1002/bies.201400105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Rando O.J. Daddy issues: paternal effects on phenotype. Cell. 2012;151:702–708. doi: 10.1016/j.cell.2012.10.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ng S.-F., Lin R., Laybutt D., Barres R., Owens J., Morris M. Chronic high-fat diet in fathers programs β-cell dysfunction in female rat offspring. Nature. 2010;467:963–969. doi: 10.1038/nature09491. [DOI] [PubMed] [Google Scholar]
- 11.Carone B.R., Fauquier L., Habib N., Shea J.M., Hart C.E., Li R. Paternally induced transgenerational environmental reprogramming of metabolic gene expression in mammals. Cell. 2011;143:1084–1096. doi: 10.1016/j.cell.2010.12.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen Q., Yan M., Cao Z., Li X., Zhang Y., Shi J. Sperm tsRNAs contribute to intergenerational inheritance of an acquired metabolic disorder. Science. 2016;351:397–400. doi: 10.1126/science.aad7977. [DOI] [PubMed] [Google Scholar]
- 13.Pembrey M.E., Bygren L.O., Kaati G., Edvinsson S., Northstone K., Sjostrom M. Sex-specific, male-line transgenerational responses in humans. European Journal of Human Genetics. 2006;14:159–166. doi: 10.1038/sj.ejhg.5201538. [DOI] [PubMed] [Google Scholar]
- 14.Kaati G., Bygren L.O., Edvinsson S. Cardiovascular and diabetes mortality determined by nutrition during parents' and grandparents' slow growth period. European Journal of Human Genetics. 2002;10:682–688. doi: 10.1038/sj.ejhg.5200859. [DOI] [PubMed] [Google Scholar]
- 15.Duhl D.M., Vrieling H., Miller K.A., Wolff G.L., Barsh G.S. Neomorphic agouti mutations in obese yellow mice. Nature Genetics. 1994;8:59–65. doi: 10.1038/ng0994-59. [DOI] [PubMed] [Google Scholar]
- 16.Matthews V.B., Allen T.L., Risis S., Chan M.H., Henstridge D.C., Watson N. Interleukin-6-deficient mice develop hepatic inflammation and systemic insulin resistance. Diabetologia. 2010;53:2431–2441. doi: 10.1007/s00125-010-1865-y. [DOI] [PubMed] [Google Scholar]
- 17.Robinson M.D., McCarthy D.J., Smyth G.K. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26:139–140. doi: 10.1093/bioinformatics/btp616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lewis B.P., Burge C.B., Bartel D.P. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 19.Huang da W., Sherman B.T., Lempicki R.A. Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- 20.Huang da W., Sherman B.T., Lempicki R.A. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Park S.J., Shirahige K., Ohsugi M., Nakai K. DBTMEE: a database of transcriptome in mouse early embryos. Nucleic Acids Research. 2014 doi: 10.1093/nar/gku1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Laird N.M., Ware J.H. Random-effects models for longitudinal data. Biometrics. 1982;38:963–974. [PubMed] [Google Scholar]
- 23.Yen T.T., Gill A.M., Frigeri L.G., Barsh G.S., Wolff G.L. Obesity, diabetes, and neoplasia in yellow A(vy)/- mice: ectopic expression of the agouti gene. FASEB Journal. 1994;8:479–488. doi: 10.1096/fasebj.8.8.8181666. [DOI] [PubMed] [Google Scholar]
- 24.Morgan H.D., Sutherland H.G., Martin D.I., Whitelaw E. Epigenetic inheritance at the agouti locus in the mouse. Nature Genetics. 1999;23:314–318. doi: 10.1038/15490. [DOI] [PubMed] [Google Scholar]
- 25.Waterland R.A., Jirtle R.L. Transposable elements: targets for early nutritional effects on epigenetic gene regulation. Molecular and Cellular Biology. 2003;23:5293–5300. doi: 10.1128/MCB.23.15.5293-5300.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cropley J.E., Suter C.M., Beckman K.B., Martin D.I. Germ-line epigenetic modification of the murine A vy allele by nutritional supplementation. Proceedings of National Academy of Sciences United States of America. 2006;103:17308–17312. doi: 10.1073/pnas.0607090103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dolinoy D.C., Huang D., Jirtle R.L. Maternal nutrient supplementation counteracts bisphenol A-induced DNA hypomethylation in early development. Proceedings of National Academy of Sciences United States of America. 2007;104:13056–13061. doi: 10.1073/pnas.0703739104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dolinoy D.C. The agouti mouse model: an epigenetic biosensor for nutritional and environmental alterations on the fetal epigenome. Nutrition Reviews. 2008;66(Suppl. 1):S7–S11. doi: 10.1111/j.1753-4887.2008.00056.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Farooqi S., O'Rahilly S. Genetics of obesity in humans. Endocrine Reviews. 2006;27:710–718. doi: 10.1210/er.2006-0040. [DOI] [PubMed] [Google Scholar]
- 30.Wolff G.L., Roberts D.W., Mountjoy K.G. Physiological consequences of ectopic agouti gene expression: the yellow obese mouse syndrome. Physiological Genomics. 1999;1:151–163. doi: 10.1152/physiolgenomics.1999.1.3.151. [DOI] [PubMed] [Google Scholar]
- 31.Zimmet P., Alberti K.G., Shaw J. Global and societal implications of the diabetes epidemic. Nature. 2001;414:782–787. doi: 10.1038/414782a. [DOI] [PubMed] [Google Scholar]
- 32.Perry R.J., Samuel V.T., Petersen K.F., Shulman G.I. The role of hepatic lipids in hepatic insulin resistance and type 2 diabetes. Nature. 2014;510:84–91. doi: 10.1038/nature13478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gabory A., Attig L., Junien C. Sexual dimorphism in environmental epigenetic programming. Molecular and Cellular Endocrinology. 2009;304:8–18. doi: 10.1016/j.mce.2009.02.015. [DOI] [PubMed] [Google Scholar]
- 34.Tarrade A., Panchenko P., Junien C., Gabory A. Placental contribution to nutritional programming of health and diseases: epigenetics and sexual dimorphism. Journal of Experimental Biology. 2015;218:50–58. doi: 10.1242/jeb.110320. [DOI] [PubMed] [Google Scholar]
- 35.Reizel Y., Spiro A., Sabag O., Skversky Y., Hecht M., Keshet I. Gender-specific postnatal demethylation and establishment of epigenetic memory. Genes & Development. 2015;29:923–933. doi: 10.1101/gad.259309.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moazed D. Small RNAs in transcriptional gene silencing and genome defence. Nature. 2009;457:413–420. doi: 10.1038/nature07756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stuwe E., Toth K.F., Aravin A.A. Small but sturdy: small RNAs in cellular memory and epigenetics. Genes and Development. 2014;28:423–431. doi: 10.1101/gad.236414.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ashe A., Sapetschnig A., Weick E.M., Mitchell J., Bagijn M.P., Cording A.C. piRNAs can trigger a multigenerational epigenetic memory in the germline of C. elegans. Cell. 2012;150:88–99. doi: 10.1016/j.cell.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buckley B.A., Burkhart K.B., Gu S.G., Spracklin G., Kershner A., Fritz H. A nuclear Argonaute promotes multigenerational epigenetic inheritance and germline immortality. Nature. 2012;489:447–451. doi: 10.1038/nature11352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rechavi O., Houri-Ze'evi L., Anava S., Goh W.S., Kerk S.Y., Hannon G.J. Starvation-induced transgenerational inheritance of small RNAs in C. elegans. Cell. 2014;158:277–287. doi: 10.1016/j.cell.2014.06.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Grentzinger T., Armenise C., Brun C., Mugat B., Serrano V., Pelisson A. piRNA-mediated transgenerational inheritance of an acquired trait. Genome Research. 2012 doi: 10.1101/gr.136614.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sharma U., Conine C.C., Shea J.M., Boskovic A., Derr A.G., Bing X.Y. Biogenesis and function of tRNA fragments during sperm maturation and fertilization in mammals. Science. 2016;351:391–396. doi: 10.1126/science.aad6780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Garcia-Silva M.R., Cabrera-Cabrera F., Guida M.C., Cayota A. Hints of tRNA-derived small RNAs role in RNA silencing mechanisms. Genes (Basel) 2012;3:603–614. doi: 10.3390/genes3040603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kumar P., Anaya J., Mudunuri S.B., Dutta A. Meta-analysis of tRNA derived RNA fragments reveals that they are evolutionarily conserved and associate with AGO proteins to recognize specific RNA targets. BMC Biology. 2014;12:78. doi: 10.1186/s12915-014-0078-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sobala A., Hutvagner G. Small RNAs derived from the 5′ end of tRNA can inhibit protein translation in human cells. RNA Biology. 2013;10:553–563. doi: 10.4161/rna.24285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gebetsberger J., Zywicki M., Kunzi A., Polacek N. 2012 tRNA-derived fragments target the ribosome and function as regulatory non-coding RNA in Haloferax volcanii. Archaea. 2012:260909. doi: 10.1155/2012/260909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lykke-Andersen K., Gilchrist M.J., Grabarek J.B., Das P., Miska E., Zernicka-Goetz M. Maternal Argonaute 2 is essential for early mouse development at the maternal-zygotic transition. Molecular Biology of the Cell. 2008;19:4383–4392. doi: 10.1091/mbc.E08-02-0219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fernandez-Twinn D.S., Constancia M., Ozanne S.E. Intergenerational epigenetic inheritance in models of developmental programming of adult disease. Seminars in Cell & Developmental Biology. 2015;43:85–95. doi: 10.1016/j.semcdb.2015.06.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dias B.G., Ressler K.J. Parental olfactory experience influences behavior and neural structure in subsequent generations. Nature Neuroscience. 2014;17:89–96. doi: 10.1038/nn.3594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Houri-Ze'evi L., Korem Y., Sheftel H., Faigenbloom L., Toker I.A., Dagan Y. A tunable mechanism determines the duration of the transgenerational small RNA inheritance in C. elegans. Cell. 2016;165:88–99. doi: 10.1016/j.cell.2016.02.057. [DOI] [PubMed] [Google Scholar]
- 51.Alcazar R.M., Lin R., Fire A.Z. Transmission dynamics of heritable silencing induced by double-stranded RNA in Caenorhabditis elegans. Genetics. 2008;180:1275–1288. doi: 10.1534/genetics.108.089433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rassoulzadegan M., Grandjean V., Gounon P., Vincent S., Gillot I., Cuzin F. RNA-mediated non-mendelian inheritance of an epigenetic change in the mouse. Nature. 2006;441:469–474. doi: 10.1038/nature04674. [DOI] [PubMed] [Google Scholar]
- 53.Wagner K.D., Wagner N., Ghanbarian H., Grandjean V., Gounon P., Cuzin F. RNA induction and inheritance of epigenetic cardiac hypertrophy in the mouse. Developmental Cell. 2008;14:962–969. doi: 10.1016/j.devcel.2008.03.009. [DOI] [PubMed] [Google Scholar]
- 54.Gapp K., Jawaid A., Sarkies P., Bohacek J., Pelczar P., Prados J. Implication of sperm RNAs in transgenerational inheritance of the effects of early trauma in mice. Nature Neuroscience. 2014;17:667–669. doi: 10.1038/nn.3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Stoeckius M., Grun D., Rajewsky N. Paternal RNA contributions in the Caenorhabditis elegans zygote. EMBO Journal. 2014;33:1740–1750. doi: 10.15252/embj.201488117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Stoeckius M., Grun D., Kirchner M., Ayoub S., Torti F., Piano F. Global characterization of the oocyte-to-embryo transition in Caenorhabditis elegans uncovers a novel mRNA clearance mechanism. EMBO Journal. 2014;33:1751–1766. doi: 10.15252/embj.201488769. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.