Abstract
Objective
In rodents and humans, besides brown adipose tissue (BAT), islands of thermogenic adipocytes, termed “brite” (brown-in-white) or beige adipocytes, emerge within white adipose tissue (WAT) after cold exposure or β3-adrenoceptor stimulation, which may protect from obesity and associated diseases. microRNAs are novel modulators of adipose tissue development and function. The purpose of this work was to characterize the role of microRNAs in the control of brite adipocyte formation.
Methods/Results
Using human multipotent adipose derived stem cells, we identified miR-125b-5p as downregulated upon brite adipocyte formation. In humans and rodents, miR-125b-5p expression was lower in BAT than in WAT. In vitro, overexpression and knockdown of miR-125b-5p decreased and increased mitochondrial biogenesis, respectively. In vivo, miR-125b-5p levels were downregulated in subcutaneous WAT and interscapular BAT upon β3-adrenergic receptor stimulation. Injections of an miR-125b-5p mimic and LNA inhibitor directly into WAT inhibited and increased β3-adrenoceptor-mediated induction of UCP1, respectively, and mitochondrial brite adipocyte marker expression and mitochondriogenesis.
Conclusion
Collectively, our results demonstrate that miR-125b-5p plays an important role in the repression of brite adipocyte function by modulating oxygen consumption and mitochondrial gene expression.
Keywords: miR-125b-5p, White adipocyte, Brite adipocyte, Mitochondriogenesis
Highlights
-
•
miR-125b-5p levels negatively correlate with UCP1 expression in rodent and human.
-
•
miR125b levels in white adipose tissue are positively correlated with BMI.
-
•
miR-125b-5p modulates oxygen consumption.
-
•
Mitochondriogenesis is controlled by miR-125b-5p.
-
•
In vivo modulation of miR-125b-5p controls brown and brite adipocyte formation.
1. Introduction
A chronic imbalance between energy intake and energy expenditure leads to variation in body weight through modulation of adipose tissue mass. Today, such an imbalance is mostly manifested as obesity, which has become a worldwide socio-economic burden and constitutes a substantial risk factor for hypertension, type 2 diabetes (T2D), cardiovascular diseases, and certain cancers [1], [2], [3]. The adipose organ can be divided into two distinct types of adipose tissues, white and brown. White adipose tissue (WAT) is specialized in the storage and release of fatty acids while brown adipose tissue (BAT) dissipates energy in the form of heat by uncoupling mitochondrial respiratory chain from ATP synthesis [4], [5]. BAT is predominantly composed of brown adipocytes that are characterized by a high mitochondrial content and the expression of uncoupling protein 1 (UCP1), resulting in an exceptionally high capacity of lipid and glucose utilization [6], [7], [8], [9]. In contrast to earlier contention, it is now well accepted that brown adipocytes are present and active in human adults [10], [11], [12], [13]. An inverse relationship between BAT activity and fat content and a decline with increasing age have been reported [10], [14], [15]. Interestingly, also WAT contains thermogenic (i.e., UCP1-positive) fat cells, called “brown-in-white” (“brite”), “beige” or inducible brown adipocytes. These cells are formed upon chronic stimulation with cold, PPARγ ligands, β3-adrenergic agonists, or various other molecules [16], [17], [18], [19]. The precise origin of brite adipocytes remains controversial, as it has been reported that brite adipocytes appearing in subcutaneous WAT (scWAT) upon cold exposure can originate either from trans-differentiation of white adipocytes [20], [21], [22], [23] or from de novo-differentiation of precursors [24]. As BAT constitutes a target to combat obesity and associated diseases [25], [26], [27], [28], identification of cellular and molecular mechanisms involved in the conversion and activation of white adipocytes into brite adipocytes may lead to the development of novel therapeutic tools. Several compounds have already been reported to play key roles [18]. Micro-RNAs (miRNAs) are small non-coding RNAs of approximately 23 nucleotides, which regulate gene expression through RNA interference [29], [30], [31]. miRNAs have recently been recognized as a novel class of modulators in adipose tissue development and function as tissue specific inactivation of Dicer resulted in a dramatic decrease of white fat mass as well as lipodystrophy [32], [33], [34]. Indeed, particular miRNAs have been shown to be involved in human white adipocyte differentiation, lipid metabolism, diabetes and obesity [35], [36], [37], [38], [39], [40]. A few reports demonstrated that miRNAs might be involved in the switch between mature white and brite adipocytes [41], [42]. The miR-193b-365 cluster was described as a novel regulator between closely related brown and myogenic lineages, and miR-196a has been introduced as the first non-coding RNA that acts as key regulator of brite adipocyte development in mouse [38], [43], [44], [45]. Recently, it has been reported that miR-34a acts as an inhibitor of brite and brown adipocyte formation through modulation of FGF21 and SIRT1 function [46]. miR-26 has been reported to be the first in-depth characterized miRNA with an impact on human brite adipogenesis and the ability to improve insulin sensitivity [39], [47]. However, the involvement of miRNAs in the process of conversion (trans-differentiation) of white into brite/brown adipocytes remains to be elucidated.
Herein, we have analyzed the role of miR-125b-5p in the browning/britening of white adipocytes in human and murine cell models and tissues. Altogether, our results show that miR-125b-5p plays an important role in the modulation of brite and brown adipocyte function by targeting mitochondriogenesis and oxygen consumption.
2. Materials and methods
2.1. Reagents
Cell culture media and buffers were purchased from Lonza, foetal bovine serum, insulin and trypsin from Invitrogen, and other reagents from Sigma–Aldrich Chimie.
2.2. Animals
Experiments were conducted in accordance with the French regulation for the care and use of research animals and were approved by local experimentation committees (Nice University and Ciepal Azur: protocol NCE-2013-166). Procedures comply with the ARRIVE guidelines. Animals were maintained under constant temperature near thermoneutrality (28 ± 2 °C) and on 12:12-hour light–dark cycles, with ad libitum access to standard chow diet and water. 10 week-old male C57Bl/6J mice were from Janvier Labs. Thermogenesis activation consisted in an intra-peritoneal injection of the β-adrenergic receptor agonist, CL316,243 (1 mg/kg/day in saline solution, NaCl 0.9%) for 7 days. Control mice were injected with vehicle only.
For miRNA mimic injection, mice were anesthetized with a xylasin/ketamine mixture. miRNA and control mimics were delivered using in vivo-jetPEI® reagent (Ozyme) according to manufacturer's recommendation. In vivo-jetPEI® does not induce any significant inflammatory response. The intra-subcutaneous WAT administration was carried out after a longitudinal incision in the skin at the inguinal area or at the interscapular area for BAT injection as previously described [48]. Mice received five injections per inguinal fat pad and were sacrificed for analysis 2 weeks after injections. Each injection contained 40 ng of miR-125b-5p mimic in 10 μl saline solution. Mice were stitched and reanimated under warm light. Control mice were injected with vehicle (NaCl 0.9%) or mimic control. β-adrenergic receptor stimulation was carried out during the last week (CL316,243, 1 mg/kg/day in saline solution). Control mice were injected with vehicle only.
For Locked Nucleic Acid (LNA™) miR-inhibitor injection (miRCURY LNA™ microRNA Inhibitors, Exiqon), mice received five injections per inguinal fat pad and were sacrificed for analysis 2 weeks after injections. Each injection contained 200 ng LNA inhibitor miR-125b-5p in 10 μl saline solution. β-adrenergic receptor stimulation was carried out during the last week (CL316,243, 0.1 mg/kg/day in saline solution). Control mice groups were injected with vehicle or miRNA inhibitor control.
2.3. Subjects
2.3.1. Human BAT and WAT samples
The study protocol was reviewed and approved by the ethics committee of the Hospital District of Southwestern Finland, and subjects provided written, informed consent following the committee's instructions. The study was conducted according to the principles of the Declaration of Helsinki. All potential subjects were screened for metabolic status, and only those with normal glucose tolerance and normal cardiovascular status (as assessed on the basis of electrocardiograms and measured blood pressure) were included. The age range of the subjects was 23–49 years. We studied a group of 6 healthy volunteers (2 men and 4 women). BAT was sampled from positive FDG-PET scan sites in supraclavicular localization, and subcutaneous WAT was derived via the same incision.
2.3.2. Human adipose sample correlated to the BMI
Primary subcutaneous adipose tissue samples were obtained from patients (n = 35, 30 female, 5 male, age 18–54 years) with a wide range of body mass index (BMI 18.7–70.4 kg m−2, mean 33.8+/− 12.9 kg m−2) who underwent abdominal surgery for nonmalignant diseases or plastic surgery for mammary reduction. The study was approved by the ethical committee of Ulm University (122/99), and all patients gave written, informed consent. Snap-frozen samples were homogenized in TriReagent (Zymo Research, Irvine, USA) and total RNA was subsequently isolated using the Direct-zol RNA MiniPrep kit (Zymo Research). miRNA was reverse transcribed and analyzed with the miScript miRNA PCR Assay System using specific primer assays (Qiagen, Hilden, Germany). Small nucleolar RNA 68 (sno68) was used as reference. cDNA was transcribed using SuperScript II (Thermo Fischer, Waltham, USA) and UCP1 expression was analyzed by qRT-PCR (Roche, Mannheim, Germany).
2.4. Cell culture and stromal vascular fraction preparation
2.4.1. hMADS cells culture
The establishment and characterization of hMADS cells have been described previously [49], [50]. Cells were seeded at a density of 5000 cells/cm2 in Dulbecco's Modified Eagle's Medium (DMEM) supplemented with 10% FBS, 15 mM Hepes, 2.5 ng/ml hFGF2, 60 mg/ml penicillin, and 50 mg/ml streptomycin. hFGF2 was removed when cells reached confluence. Cells were induced to differentiate at day 2 post confluence (designated as day 0) in DMEM/Ham's F12 (1:1) media supplemented with 10 μg/ml transferrin, 10 nM insulin, 0.2 nM triiodothyronine, 1 μM dexamethasone and 500 μM isobutyl-methylxanthine.
Media were changed every other day and cells used at day 18. miRNA mimic (miRIDIAN from Dharmacon), miRNA hairpin inhibitor (miRIDIAN microRNA Hairpin Inhibitors) or LNA inhibitor (miRCURY LNA™ microRNA Inhibitors Exiqon) transfections were performed at day 14 as described previously [39].
2.4.2. Mouse adipose tissue SVF preparation and culture
Animals were euthanized by cervical dislocation to isolate stromal vascular fractions (SVF) as described previously [51], [52], [53].
2.4.3. Human adipose tissue SVF preparation and culture
Abdominal subcutaneous human adipose tissue was collected from healthy patients as res nullus from surgeries (non-pathologic abdominoplasty) for SVF isolation according to the procedure described for mice (see above). SVF cells were plated and maintained in DMEM containing 10% FCS until confluence. Differentiation of primary cultures in white and brite adipocytes was performed according to the protocol described for hMADS cells previously [54], [55].
2.5. Isolation and analysis of RNA
These procedures followed MIQE standard recommendations and were conducted as described previously [56]. The oligonucleotide sequences, designed using Primer Express software, are shown in Supplementary Table 1.
For miRNA analysis, 10 ng of the total RNA were reverse transcribed using Universal cDNA synthesis kit (Exiqon, Denmark). qPCR and data evaluation were performed as described previously [39], [57]. miR-125b-5p oligonucleotide sequence target used for RT-qPCR analysis is: UCCCUGAGACCCUAACUUGUGA. U6 snRNA and 5S rRNA were used as endogenous controls and were purchased from Exiqon.
2.6. mRNA microarray and data analysis
The human oligonucleotide probe set (MWG) consisted of 29,550 50-mer DNA oligonucleotides, enabling the detection of 15,539 distinct RefSeq mRNAs as well as non-RefSeq-annotated transcripts (provided by the manufacturer). Lyophilized oligonucleotides were dissolved in 3× SSC (prepared from 20× stock consisting of 3 M NaCl and 300 mM trisodium citrate, pH 7)/1.5 M betaine (20 μl/well) and spotted in a 4 × 12 print tip group pattern on epoxy-coated glass slides (Nexterion). To analyze the effects of miR-125b-5p on the transcriptome, 4 biological replicates were performed in which hMADS-3 cells were transfected at day 14 of differentiation with mimic miR-125b-5p or mimic miR-ctr. Total RNA was isolated at day 14 and day 16. Detailed protocols for sample labeling and hybridization were performed as described in [39]. Further, the raw data was background corrected, normalized by global mean and dye–swap pairs using ArrayNorm [58], and then exported as text-file containing log2 transformed ratios of gene expression. Filtering for present values and differential expression was conducted with Genesis software [59]. All experimental parameters and raw and processed data files have been deposited in NCBI's Gene Expression Omnibus [60] and are accessible through GEO Series accession number GSE80816 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE80816). Only features that were associated with a RefSeq transcript and detected in 3 or 4 biological replicates were considered for further analysis. miRNA-mRNA interactions were predicted using 10 different prediction tools as described in [39].
2.7. Mitochondrial DNA quantification
Genomic DNA was extracted from cell culture using a DNA extraction kit (Macherey–Nagel EURL, France). 2 ng of the total DNA were used for qPCR analysis, and data evaluation was performed as described previously [39], [57]. The mitochondrial DNA copy number was calculated from the ratio of the DNA of the NADH dehydrogenase gene, a mitochondrial gene, to that of lipoprotein lipase gene, a single copy nuclear gene. The oligonucleotide sequences, designed using Primer Express software, are shown in Supplementary Table 1.
2.8. Histological analysis
Histological analysis was performed as previously described [61].
For immunohistology, tissue sections were incubated in boiling citrate buffer (10 mM, pH 6.0) for 6 min. Cooled sections were rinsed and then permeabilized in PBS with 0.2% triton X-100 at room temperature for 20 min. Sections were saturated in the same buffer containing 3% BSA for 30 min, incubated with anti-perilipin antibody (#RDI-PROGP29, Research Diagnostic Inc., Flanders, USA) for 1 h, and TRITC-coupled anti-guinea pig antibody for 45 min. Nuclear staining was performed with DAPI. UCP1 immunohistochemistry was performed following manufacturer's instructions (LSAB + system-HRP, Dako, Les Ulis, France) and using goat anti-UCP1 (clone C-17, Santa Cruz, Tebu-bio, Le Perray-en-Yvelines, France).
2.9. Measurement of oxygen consumption
For respiration analysis, hMADS cells were seeded in 24 multi-well plates and differentiated as previously described [62]. At day 14 of differentiation, hMADS cells were transfected with mimic miR-125b-5p. Oxygen consumption rate (OCR) of 18 day-old differentiated cells was determined using an XF24 Extracellular Flux Analyzer (Seahorse Bioscience). Isoproterenol (100 nM) was used to characterize β-adrenergic inducible respiration. Uncoupled and maximal OCRs were determined using oligomycin (1.2 μM) and FCCP (1 μM), respectively. Rotenone (1 μM) and Antimycin A (1 μM) were used to inhibit Complex I- and Complex III-dependent respiration, respectively. Parameters were measured for each well as previously described [63].
2.10. Western blot analysis
Proteins were extracted from cells or tissues as previously described [61]. For mitochondrial complex quantification, equal amounts of cellular proteins were separated by electrophoresis using gradient gels (10–20%) and blotted onto nitrocellulose membranes. Following blocking, membranes were incubated overnight with a 1:1000 dilution of total OXPHOS western blot antibody cocktail (#MS601, Mitosciences) before 1 h incubation in 1:10,000 diluted HRP-conjugated anti-mouse immunoglobulins (#7076S, Cell Signaling Technology). Chemiluminescence obtained after adding Clarity ECL western blotting substrate (BioRad, France) was detected using a ChemiDoc MP Imaging System and quantified with Image Lab 5.0 software (Bio-Rad, France).
For other analyses, 40 μg of proteins were blotted using SDS-PAGE basic protocol (10 or 15%). Primary antibody incubation was performed overnight at 4 °C (anti-UCP1, Calbiochem #662045, dilution 1:750; anti-perilipin, Interchim #BP5015, dilution 1:5000; anti-CS, Abcam #ab96600, dilution 1:10,000; anti-β-tubulin, Sigma #T5201, dilution 1:2000; anti-DRP1, Cell Signaling #5391, dilution 1:1000; anti-phosphoDRP1 (ser616), Cell Signaling #4494, dilution 1:1000; anti-LC3, Nanotools #5F10, dilution 1:1000; anti-ATG5, Nanotools #7C6, 1:2000; anti-p62, BD #610833, 1:2000). Primary antibodies were detected with HRP-conjugated anti-rabbit or anti-mouse immunoglobulins (Promega) or anti-guinea pig immunoglobulins (Santa Cruz). Detection was performed using Immobilon Western Chemiluminescent HRP Substrate (Merk-Millipore). OD band intensities were evaluated using PCBas Software.
2.11. Mass spectrometry analysis
Proteins were extracted from brite hMADS cells transfected before conversion with miR-ctr or miR-125b-5p mimics, using lysis buffer (10 mM hepes pH 7, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, and 0.25 M sucrose) containing protease inhibitors (Complete, Roche Diagnostics). Samples were cleared of nucleus by centrifugation (5 min, 800 g), nuclei were spin down, and the mitochondrial-enriched fraction was obtained after centrifugation of the supernatant at 8000 g for 5 min. Samples were resuspended in 50 μl of H2O and stored at −80 °C. Samples were digested by trypsin and concentrated and purified using ZipTip® Pipette Tips (Millipore). Proteomic analysis was performed using the LTQ-Orbitrap (ThermoFisher) as previously described [64]. Spectral accuracy was measured with Mascot version 2.2.0 software (Matrix Science, London, UK) following parameters previously published [65].
2.12. Statistical analyses
Data are expressed as mean values ± SEM and were analyzed using InStat software (GraphPad Software, CA, USA). Data were analyzed by Student's t-test or one-way ANOVA followed by a Student-Newman-Keuls post-test. Differences were considered statistically significant when p < 0.05.
3. Results
3.1. miR-125b-5p levels are downregulated in human and mice brite/brown adipocytes in vitro
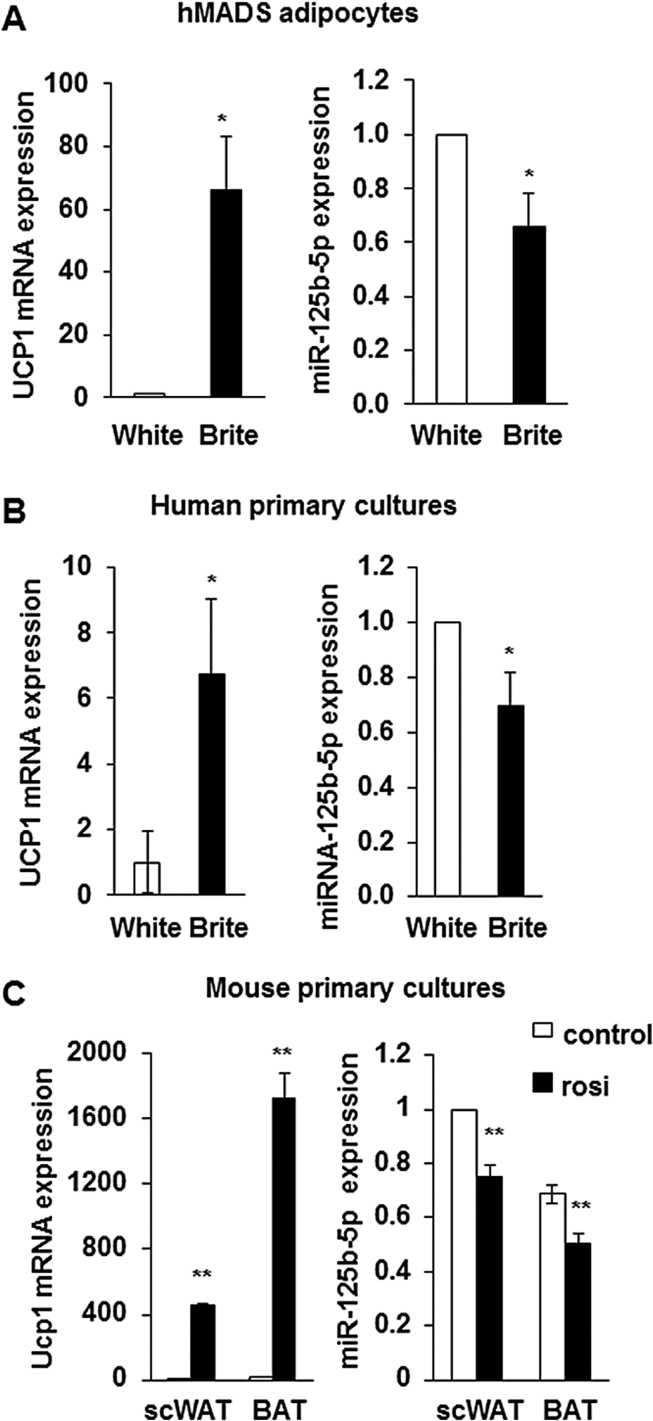
We identified miR-125b-5p as down-regulated during the conversion of hMADS white adipocytes into brite adipocytes. hMADS cells were induced to differentiate into white adipocytes until day 14, followed by cultivation in the absence (white) or the presence (brite) of 100 nM rosiglitazone (a potent PPARγ agonist) for 4 days corresponding to 18 days of differentiation. As expected, UCP1 mRNA levels were high upon rosiglitazone treatment, reflecting the formation of brite adipocytes (Figure 1A). Concomitantly, miR-125b-5p levels decreased by about 40% in brite adipocytes (Figure 1A). These observations were not specific to hMADS cells since similar data were obtained with stromal vascular fraction (SVF) derived white and brite adipocytes isolated from human subcutaneous adipose tissue [55]. UCP1 mRNA expression was confined to brite adipocytes, whereas miR-125b-5p levels were lower in brite adipocytes compared to white adipocytes (Figure 1B). We then aimed at exploring whether this inverse relationship might also exist in murine adipocytes. For that purpose, SVF cells were isolated from murine scWAT and BAT and were induced to differentiate either into white or brite/brown adipocytes. As expected, Ucp1 mRNA was highly abundant in BAT-derived brown adipocytes and scWAT-derived brite adipocytes and almost undetectable in unstimulated (control) adipocytes (Figure 1C) whereas miR-125b-5p levels were significantly lower in brown/brite adipocytes compared to control adipocytes.
Figure 1.
miR-125b-5p levels in human and mice cell models. UCP1 mRNA and miR-125b-5p levels were evaluated by RT-qPCR in A) hMADS white and brite adipocytes, B) human subcutaneous adipose tissue SVF-derived white and brite adipocytes. C) Cells from SVF of BAT and scWAT were differentiated in the absence (control) or the presence (rosi) of rosiglitazone, and used for mRNA and miRNA level quantification by RT-qPCR. Untreated adipocytes from scWAT were considered as white adipocytes and rosiglitazone treated adipocytes from BAT or scWAT as brown or brite adipocytes, respectively. Histograms represent mean ± SEM of 7 experiments (A), 3 independent experiments (B) and 8 mice per group (C). *, **: p < 0.05. *: white vs. brite; **: rosi vs. control.
3.2. miR-125b-5p levels are correlated with recruitment and activation of brown/brite adipocytes in mice and human in vivo
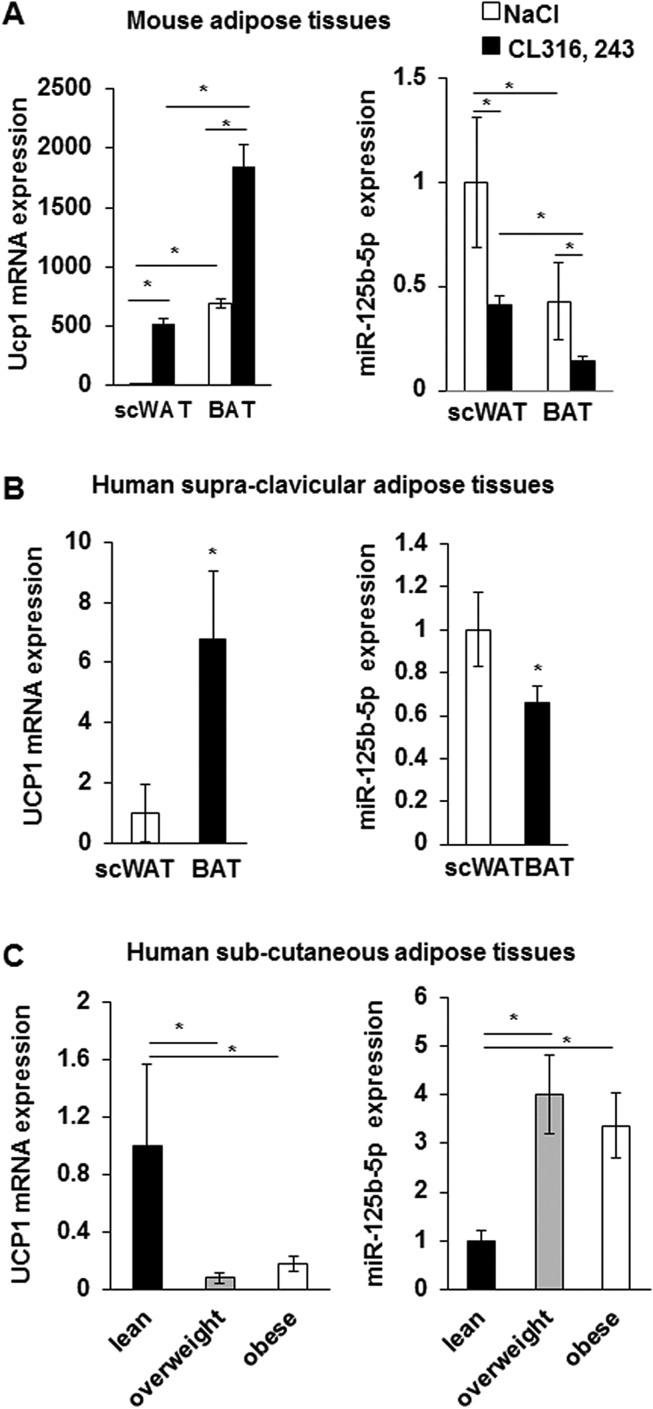
In order to determine whether a correlation exists between miRNA abundance and brown/brite adipocytes activation in vivo, mice maintained at thermoneutrality (28–30 °C) received a β3-adrenergic receptor agonist treatment (CL316,243, 1 mg/kg/day) for 1 week. This stimulation induced a significant increase of Ucp1 mRNA in BAT. Further, Ucp1 was also significantly induced in scWAT corresponding to the recruitment and activation of inducible brite adipocytes (Figure 2A). Notably, miR-125b-5p expression was higher in mice scWAT compared to BAT, and in both tissues, its levels decreased upon CL316,243 stimulation (Figure 2A).
Figure 2.
miR-125b-5p levels in human and mice adipose tissue samples. UCP1 mRNA and miR-125b-5p levels were analyzed by RT-qPCR in A) scWAT and BAT from C57BL/6 mice that received CL316,243 for 1 week, 8 mice per group, B) matched biopsies of scWAT and BAT from 6 healthy adult patients, C) biopsies of abdominal and mammary scWAT from 30 adult patients and correlated to the BMI. *: p < 0.05.
To further test the potential physiological relationship between miR-125-5p and brown adipocyte characteristics, we analyzed WAT and BAT biopsies from healthy volunteers. In line with our in vitro findings on human adipocytes and in vivo findings in mice, UCP1 mRNA was highly expressed in human BAT while miR-125b-5p levels were lower in BAT compared to WAT (Figure 2B). In parallel, we showed that UCP1 mRNA expression was higher in scWAT from lean individuals than in scWAT from overweight or obese patients while miR-125b-5p levels were higher in scWAT from overweight and obese patients (Figure 2C).
Altogether, our data emphasize an inverse relationship between miR-125b-5p and UCP1 mRNA levels, suggesting that miR-125b-5p might play an important role in brown/brite adipocyte formation and/or function.
3.3. Modulation of miR-125b-5p levels does not alter the brite phenotype but affects mitochondriogenesis
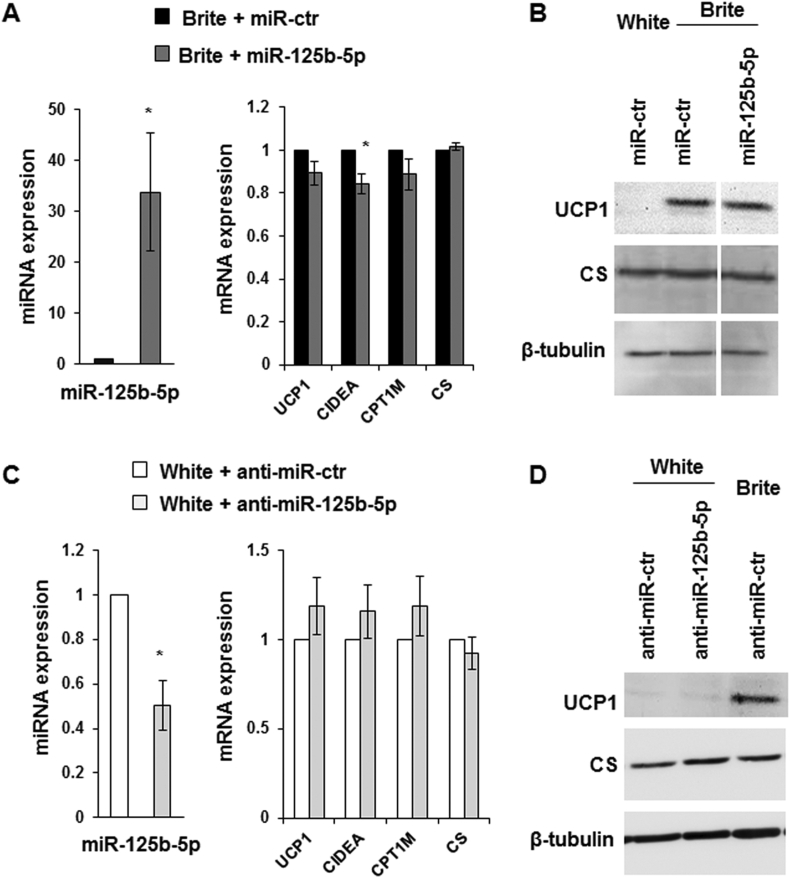
In order to know whether modulation of miR-125b-5p levels affected the human brite adipocyte formation and function, we analyzed a potential inhibitory effect of miR-125b-5p overexpression in hMADS brite adipocytes and a possible promotion of browning due to miR-125b-5p inhibition in hMADS white adipocytes.
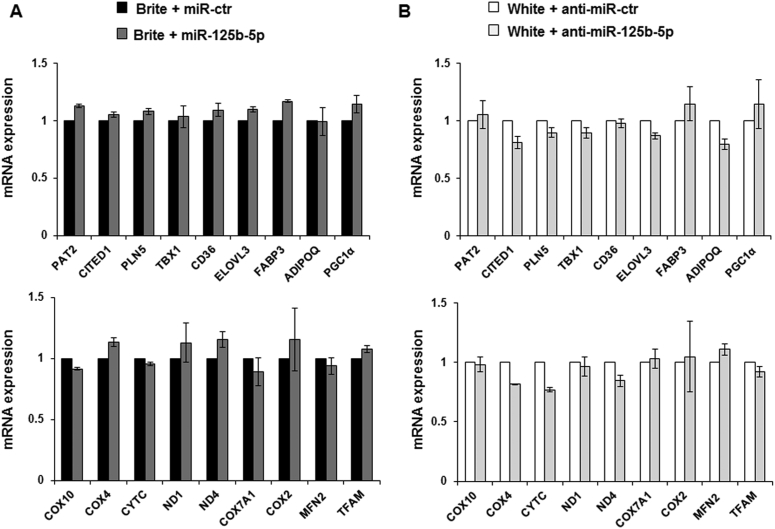
Interestingly, the delivery of an miR-125b-5p mimic (Figure 3A) or miR-125b-5p inhibitor (Figure 3C) did not modify the expression of UCP1 and CS at mRNA (Figure 3A,C) and protein level (Figure 3B,D). Similar results were obtained for genes characteristic of the white (CD36, ADPQ) or brite adipocyte phenotype (CPT1M, PAT2, CITED1, PLN5, TBX1, ELOVL3, FABP3) (Figure 3A,B and Supplementary Figure 1A and B), except a significant slight decrease of CIDEA mRNA levels upon miR-125b-5p mimic transfection (Figure 3A).
Figure 3.
Effects of miR-125b-5p over-expression and inhibition on hMADS brite adipocyte phenotype. Adipocytes transfected with miR-125b-5p mimics (A, B) or miR-125b-5p LNA inhibitor (C, D) or their respective control at day 14 and mRNA, miRNA (A, C) and protein analysis (B, D) were performed at day 18. Histograms display mean ± SEM of 4 experiments, *: p < 0.05, miR-125b-5p mimics or miR-125b-5p LNA inhibitor vs. control.
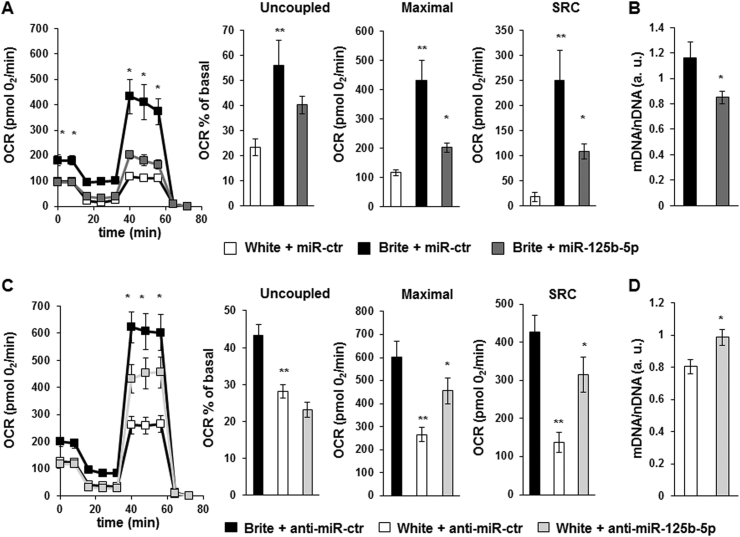
hMADS brite adipocytes displayed a slight increase in mitochondrial DNA content, characteristic of mitochondriogenesis. Overexpression of miR-125b-5p lowered the content of mitochondrial DNA in brite hMADS adipocytes. Interestingly, inhibition of miR-125b-5p increased mitochondrial DNA in hMADS white adipocytes to levels similar to those of brite adipocytes (Figure 4B,D). These modifications in mitochondrial DNA content did not seem to be dependent to TFAM levels as the mRNA of TFAM was not modified in hMADS adipocytes transfected with the mimic or anti-miR for miR-125b-5p (Supplementary Figure 1A and B).
Figure 4.
Effects of miR-125b-5p over-expression and inhibition. hMADS adipocytes transfected with miR-125b-5p mimics (A, B) or miR-125b-5p LNA inhibitor (C, D) or their respective control at day 14 and oxygen consumption measurements (A and C) were performed at day 18. Plots show mitochondrial OCR, and histograms correspond to mitochondrial respiration values. Mitochondrial DNA content was quantified by qPCR upon over-expression or inhibition of miR-125b-5p (B and D), as described in material and methods section. Plots and histograms display mean ± SEM, *, **: p < 0.05, data are from 3 independent experiments (4 replicates per experiment). *: mimic ctr vs. mimic miR-125b-5p; **: white vs. brite.
3.4. Modulation of miR-125b-5p levels affects cellular respiration
In order to determine whether miR-125b-5p levels, in addition to affecting mitochondrial biogenesis, impact mitochondrial function, we analyzed oxygen consumption rates, and we observed that the delivery of an miR-125b-5p mimic led to a significant decrease in basal and maximal oxygen consumption rate and a non-significant decrease of uncoupled respiration (Figure 4A). Inhibition of miR-125b-5p function by delivery of an miR-125b-5p inhibitor led to increased maximal oxygen consumption reaching almost the level of brite adipocytes, whereas the uncoupled respiration rate was not affected (Figure 4C). Further, inhibition of miR-125b-5p did not affect the basal oxygen consumption rate. However, the reserve respiratory capacity quantified by the measure of the spare respiratory capacity (SRC) was inhibited upon overexpression and enhanced upon inhibition of miR-125b-5p (Figure 4A,C), suggesting an important role of this miRNA in the control of mitochondrial respiration.
Using the same approaches, respiratory chain complex proteins were quantified. Analysis of the content of the five oxidative phosphorylation complexes showed that over-expression as well as inhibition of miR-125b-5p affected complexes I and IV, whereas the other complexes were not modified (Supplementary Figure 2). Indeed, over-expression of miR-125b-5p significantly lowered the content of complexes I and IV in brite hMADS adipocytes, and in an opposite manner, transfection with miR-125b-5p inhibitors elevated the levels of these complexes in hMADS white adipocytes (Supplementary Figure 2). These modulations appeared without alterations of the mRNA content of various nuclear (COX4, COX10, COX7A1, CytC) and mitochondrial (ND1, ND4 and COX2) genes encoding respiratory chain complex proteins (Supplementary Figure 1A and B).
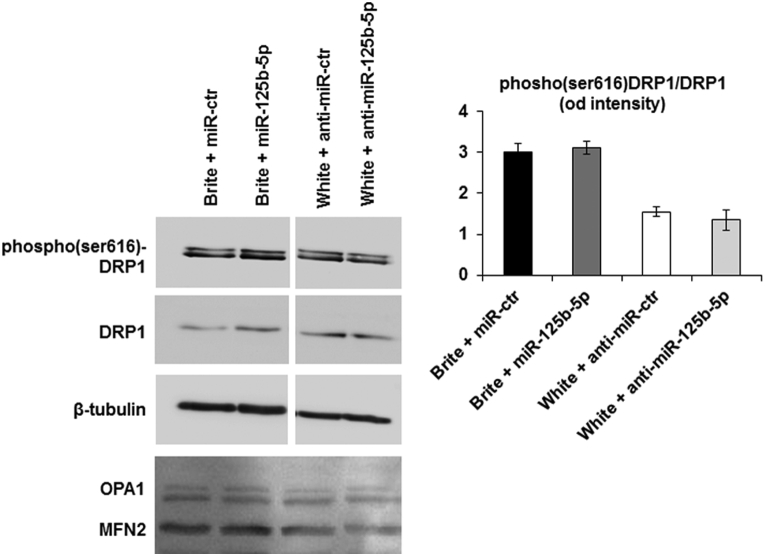
miR-125b-5p seemed to modulate mitochondrial respiratory chain quantities and function, without alteration of mRNA expression of their components. One hypothesis is that miR-125b-5p modifies the dynamics of mitochondria fission and fusion, an important mechanism in brown adipocytes [66] displayed by hMADS cells. Western blot analysis of hMADS adipocytes transfected with mimics and anti-miR-125b-5p did not reveal an alteration of DRP1 ser616 phosphorylation, a key event in fission mechanism. In line with this observation, levels of OPA1 and MFN2, two proteins involved in mitochondria fusion, were not altered (Supplementary Figure 3).
Altogether, these results demonstrated that miR-125b-5p was able to modulate mitochondrial content and function without affecting the general brite adipocyte phenotype.
3.5. Microarray and proteomic approaches
In order to identify a potential mRNA target of miR-125b-5p involved in the white to brite adipocyte conversion, we performed a semi-quantitative proteomic approach (Supplementary Table 2) and global RNA profiling (Supplementary Table 3) using hMADS brite adipocytes transfected with miR-125b-5p mimic or control. The analysis of fractions enriched in mitochondria revealed various modulated proteins (fold <0.8 or >1.25), including proteins from the respiratory chain (i.e. COX5A, ATP synthase subunit) or TCA cycle (i.e. Citrate synthase).
The results from mRNA microarray data have been combined with publicly available data from miRNA target prediction algorithms in order to reveal potential miR-125b-5p targets with a decreased mRNA expression after miR-125b-5p mimic transfection. The top 100 mRNA are listed in Supplementary Table 3. Despite the fact that some of these mRNAs are interesting in the field of adipogenesis, no evident target explaining the defect in mitochondrial activity has been identified.
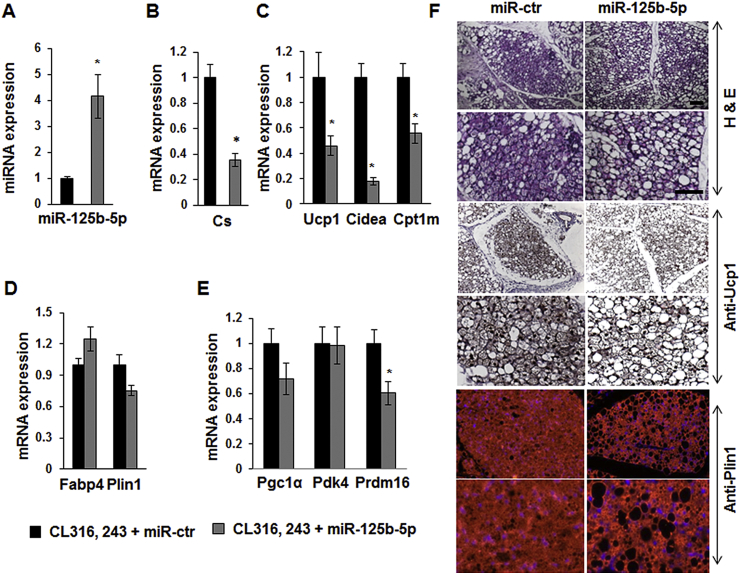
3.6. miR-125b-5p mimic injection in murine scWAT inhibits brite adipocyte formation
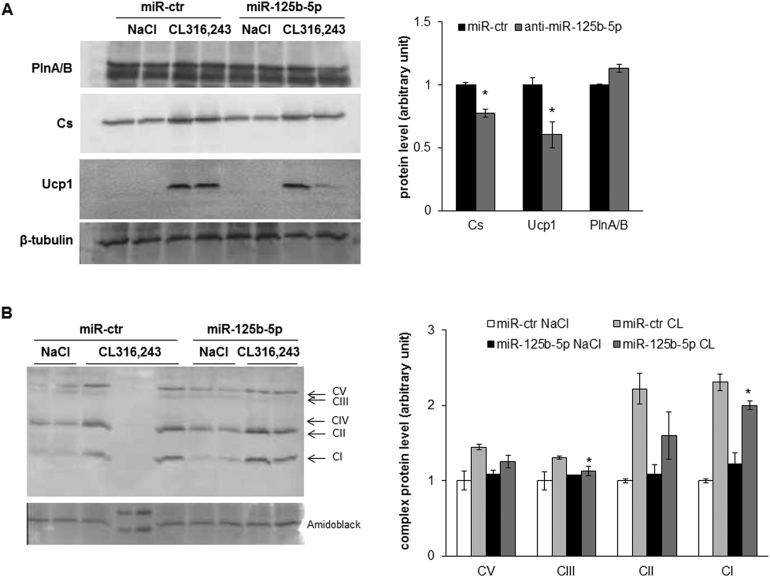
We aimed at investigating in vivo whether miR-125b-5p can affect mitochondrial content and function, and thus brite adipocyte formation and function, in scWAT. For that purpose, we injected miR-125b-5p mimics or control mimics directly into scWAT of C57BL/6 mice for 2 weeks. During the second week, mice received the β3-adrenergic receptor agonist CL316,243 (1 mg/kg/day) or vehicle. Compared to control mimic injections, treatment with miR-125b-5p mimics significantly increased the levels of miR-125b-5p in scWAT (Figure 5A). Molecular analysis of scWAT showed that injection of miR-125b-5p mimic compared to control mimic led to an impairment of the mitochondriogenesis induced by the CL316,243 treatment, as shown by the strong inhibition of citrate synthase (Cs) mRNA expression (Figure 5B). This was linked to a decrease in brite adipocyte markers such as Ucp1, Cpt1m, and Cidea. (Figure 5C). Under these conditions, fatty acid binding protein (Fabp4) and perilipin 1 (Plin1) mRNA levels were not modified, demonstrating that mimic injection did not affected adipocyte content (Figure 5D). Expression of other genes related to brite adipocytes were not affected except a slight yet significant decrease of Prdm16 (Figure 5E). As expected, these alterations at the mRNA level were confirmed at the protein level. Indeed, Ucp1 and Cs protein levels were decreased and PlnA and B (the products of the Plin1 gene) were not affected (Supplementary Figure 5A). In addition, analysis of mitochondrial complex protein levels showed a decrease in all detected complexes, which was significant for complexes I and III (Supplementary Figure 5B).
Figure 5.
Effect of miR-125b-5p mimic injection in scWAT. 10 weeks-old C57BL/6 male mice received miR-125b-5p or control mimic in the subcutaneous WAT and received the β3-adrenergic receptor agonist (CL316,243) for 7 days. miR-125b-5p expression levels were determined in scWAT from mice by RT-qPCR (A). Representative brite adipocytes, mitochondrial (B, C and E), and white adipocyte (D) markers mRNA levels were determined in scWAT by RT-qPCR. Representative histological sections of scWAT (7 μm, paraffin-embedded) HES staining (upper panel), UCP1 (middle panel) and PLIN1 (lower panel) immunostaining are shown (F). *: p < 0.05. Scale bar: 100 μm.
Histological analysis showed an miR-125b-5p-dependent defect in the formation of multilocular lipid droplet-containing adipocytes representative of activated brite/brown adipocytes. The number of unilocular lipid droplet adipocytes in CL316,243 and miR-125b-5p mimic-injected mice was higher compared to control mimic injections (Figure 5F, upper panel). This observation was confirmed by Ucp1 staining, which revealed fewer Ucp1 positive adipocytes upon miR-125b-5p mimic injection (Figure 5F, middle panel). In agreement with the previous observations, perilipin immunostaining showed that activation of β3-adrenergic receptor in the presence of high levels of miR-125b-5p led to less brite adipocytes and to the appearance of adipocytes with large lipid droplets (Figure 5F, lower panel).
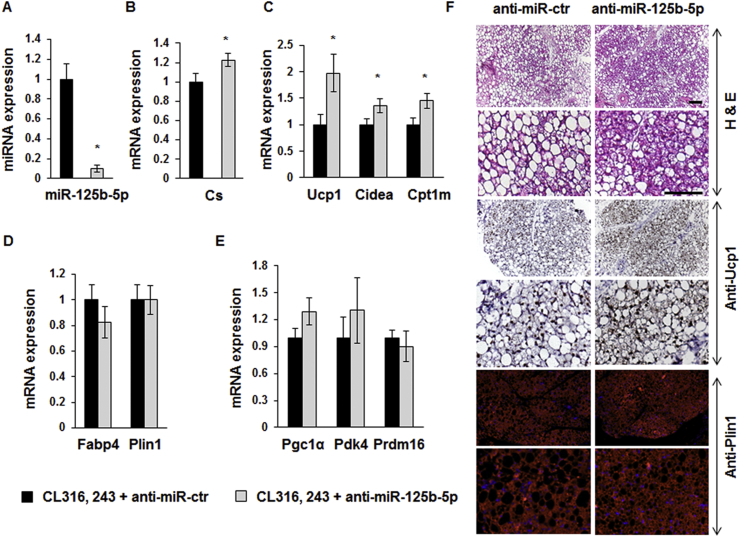
3.7. miR-125b-5p inhibitor injection in murine scWAT promotes brite adipocyte formation
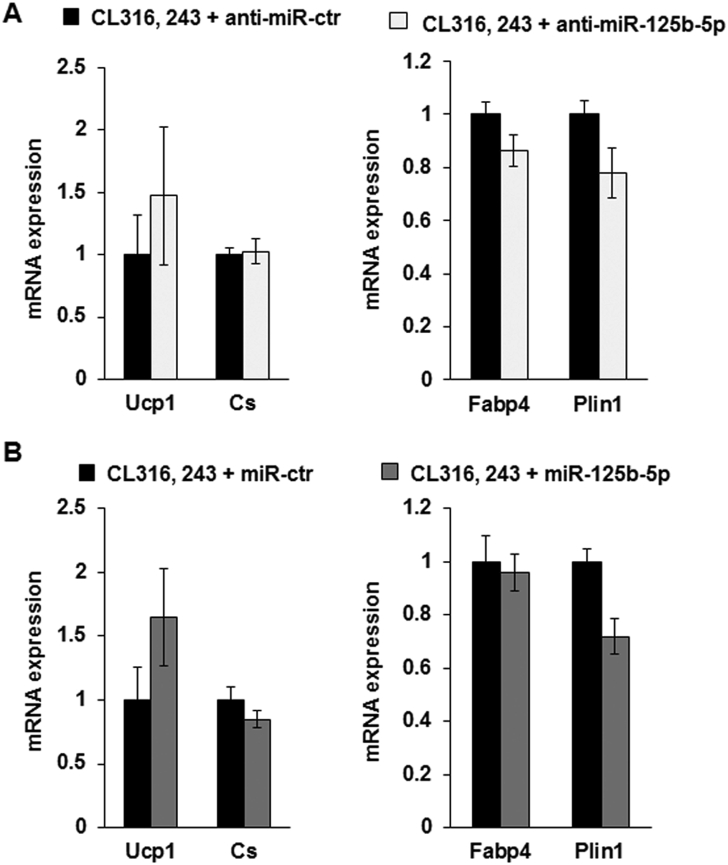
As increased levels of miR-125b-5p negatively affected brown adipocyte formation, we tested whether inhibition of miR-125b-5p expression can enhance brite adipocyte formation in scWAT. miR-125b-5p LNA inhibitors or controls were directly injected into scWAT of C57BL/6 mice for 2 weeks. Compared to control inhibitor injections, treatment with anti-miR-125b-5p inhibitors significantly decreased the levels of miR-125b-5p in scWAT (Figure 6A). As expected, in contrast to the injection of the mimic, injection of the miR-125b-5p inhibitor promoted mitochondrial biogenesis as shown by the significant increase of Cs mRNA (Figure 6B). In agreement with this observation, mitochondrial brite adipocyte markers such as Ucp1, Cpt1m, and Cidea increased in miR-125b-5p LNA inhibitor-injected mice upon CL316,243 treatment (Figure 6C). No variations were observed on Fabp4 and Plin1 mRNA levels (Figure 6D). Further, expression of other brite adipocytes markers was not affected (Figure 6E). Histological analyses showed an increased formation of multilocular lipid droplet-containing adipocytes, representative of activated brite adipocytes, due to miR-125b-5p antagonism (Figure 6F, upper and lower panels). Both Ucp1 and perilipin immunostaining showed that activation of β3-adrenergic receptor in the presence of miR-125b-5p inhibitors led to an increase in the density of brite adipocytes and disappearance of adipocytes with large lipid droplets (Figure 6F, middle and lower panels). Analysis of the non-injected anterior scWAT revealed that brite adipocyte markers such as UCP1 and CS as well as adipogenic markers such as Fabp4 and Plin1 were not affected (Supplementary Figure 4A and B).
Figure 6.
Effect of miR-125b-5p LNA inhibitor injection in scWAT. 10 weeks-old C57BL/6 male mice received miR-125b-5p or control LNA inhibitor in the subcutaneous WAT and received the β3-adrenergic receptor agonist (CL316,243) for 7 days. miR-125b-5p expression levels were determined in scWAT from mice by RT-qPCR (A). Representative brite adipocytes, mitochondrial (B, C, E), and white adipocyte (D) markers mRNA levels were determined in scWAT by RT-qPCR. Representative histological sections of scWAT (7 μm, paraffin-embedded) HES staining (upper panel), UCP1 (middle panel) and PLIN1 (lower panel) immunostaining are shown (F). *: p < 0.05. Scale bar: 100 μm.
4. Discussion
The control of brown/brite adipocyte formation and function represents a promising therapeutic option to combat obesity. Several reports have shown that miRNAs play important roles in adipogenesis and metabolic diseases [40], [42], [67]. The miR-26 family, miR-27, mir-30, miR-34a, miR-106b, miR-133, miR-155, miR-193-365, miR-196 and miR-378 are involved in the control of brown adipocyte formation and function [39], [43], [44], [68], [69], [70], but only a few of them were validated in human so far. Herein, we identified miR-125b-5p as a novel critical regulator of energy-dissipating adipocyte characteristics in mouse and human. This miRNA is perfectly conserved between rodents and human, and known to be involved in carcinoma progression and host immune responses [71]. miR-125b has also been reported as a potential circulating biomarker of rheumatoid arthritis with a predictive potential upon pharmacologic treatments [72]. Importantly, miR-125b-5p promotes murine white adipocyte differentiation probably via SMAD4 inhibition [73]. However, its role in brown/brite adipocyte formation was not known until now. Furthermore, another member of the miR-125 family, miR-125a has been described to be associated with obesity in mouse and human [74]. Our data showed that miR-125b-5p levels are negatively associated with Ucp1 mRNA expression in brite and brown adipocytes derived from mice and humans. Indeed, miR-125b-5p levels were lower in human biopsies of BAT compared to those of WAT and in primary culture-derived brite versus white adipocytes. In agreement with this observation, miR-125b-5p levels were lower in murine BAT compared to scWAT, and its levels dropped under a situation favoring the browning process and BAT activation such as β3-adrenergic agonist treatment. Importantly, miR-125b-5p seems to play an important role in the control of mitochondrial biogenesis and mitochondrial respiration as its overexpression inhibited basal and maximal oxygen consumption as well as mitochondrial DNA content, while antagonism of miR-125b-5p resulted in opposite effects.
In vivo delivery of miR-125b-5p mimics or inhibitors through direct injection in the scWAT depots showed decreased or increased expression of brite associated markers, respectively. These observations support a crucial role of miR-125b-5p in the control of the formation of brite adipocytes in scWAT upon β3-adrenergic receptor activation. Furthermore, histological analysis showed that overexpression of miR-125b-5p negatively affected the formation of brite adipocytes in scWAT, while inhibition of this miRNA did the opposite. Overexpression or inhibition of miR-125b-5p had no effect on the expression of other genes related to adipogenesis (Fabp4, leptin, perilipin), highlighting the involvement of miR-125b-5p in controlling the expression of genes related to brite/brown adipocytes. The in vivo delivery procedure we used has been proven to be efficient as for example in controlling the formation of brite/beige adipocytes by miR-30 [68]. In addition, the analysis of anterior scWAT, representing a non-injected (i.e. control) tissue, confirmed the local efficiency of the treatment and suggested that putative side effects might be non-existent.
In search of potential gene targets of miR-125b-5p, we performed a semi-quantitative proteomic approach and a global mRNA profiling (Supplementary Tables 2 and 3). Most of the candidates identified through the proteomic analysis and RNA profiling with a seed match corresponding to the miR-125b-5p seed sequence were affected only slightly by the miR-125b-5p overexpression. Moreover, even if these targets can be associated with mitochondrial activity, and thus in agreement with the effects of miR-125b-5p obtained in vitro as well as in vivo, none of them were related to a mechanism explaining the effect of miR-125b-5p. Further investigations should allow a better understanding of the role of miR-125b-5p and identification of its targets.
Very recently, miRNAs have been shown to play dual roles, by simultaneously repressing cytoplasmic targets and activating mitochondrial mRNAs [75], highlighting the importance of these regulators in the control of mitochondriogenesis, a hallmark of white to brite adipocyte conversion.
5. Conclusion
Collectively, our data demonstrate that miR-125b-5p levels are associated with brite and brown adipocyte formation and mitochondrial respiration. miR-125b-5p and its responsive target genes represent a potential therapeutic option for the development of new strategies to combat obesity and associated metabolic disorders.
Author contributions
Conceived and designed the experiments: MS and EZA. Performed the experiments: MG, DFP, MK, JCC, VB, RAG and EZA. Analyzed the data: MG, DFP, VB, DL, SH, MK, MS and EZA. Wrote the manuscript: MG and EZA. Provided human AT samples: MW, PFP, UK, DT, TN, MT, PN and KAV. Performed miRNA qPCR analysis in human AT samples: PFP and DT. All authors proofread and approved the manuscript.
Acknowledgments
The authors greatly acknowledge IRCAN animal core facility, IRCAN Cytomed, Bernard Rossi Proteomic and iBV histology platforms. We thank Dr. Baharia Mograbi for helpful discussions. This work was supported by CNRS, EU FP7 project DIABAT (HEALTH-F2-2011-278373), and French Agence Nationale de la Recherche (ANR-10-BLAN-1105 miRBAT). DL is a member of Institut Universitaire de France. DT is supported by grants from the German Research Foundation (DFG TE912/2-1), the Ministry of Science, Research and Arts of Baden-Württemberg (Boehringer Ingelheim University Ulm Biocenter (BIU), 32-7533.-6-10/15/5) and the German Association for Pediatric Endocrinology and Diabetes (DGKED). PFP receives funding from Baden-Württemberg Stiftung, and Martin Wabitsch and Uwe Knippschild receive funding by the Deutsche Forschungsgemeinschaft (SFB 1149/B4).
Footnotes
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.molmet.2016.06.005.
Conflict of interest
The authors declare having no conflict of interest.
Appendix A. Supplementary data
The following are the supplementary data related to this article:
References
- 1.WHO . 2014. Obesity and overweight.http://wwwwhoint/mediacentre/factsheets/fs311/en/ Fact sheet N 311(Updated January 2015) [Google Scholar]
- 2.Dietz W.H., Baur L.A., Hall K., Puhl R.M., Taveras E.M., Uauy R. Management of obesity: improvement of health-care training and systems for prevention and care. Lancet. 2015;385(9986):2521–2533. doi: 10.1016/S0140-6736(14)61748-7. [DOI] [PubMed] [Google Scholar]
- 3.Cawley J., Meyerhoefer C. The medical care costs of obesity: an instrumental variables approach. Journal of Health Economics. 2012;31(1):219–230. doi: 10.1016/j.jhealeco.2011.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Nedergaard J., Golozoubova V., Matthias A., Asadi A., Jacobsson A., Cannon B. UCP1: the only protein able to mediate adaptive non-shivering thermogenesis and metabolic inefficiency. Biochimica et Biophysica Acta. 2001;1504(1):82–106. doi: 10.1016/s0005-2728(00)00247-4. [DOI] [PubMed] [Google Scholar]
- 5.Frontini A., Cinti S. Distribution and development of brown adipocytes in the murine and human adipose organ. Cell Metabolism. 2010;11(4):253–256. doi: 10.1016/j.cmet.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 6.Cannon B., Nedergaard J. Brown adipose tissue: function and physiological significance. Physiological Reviews. 2004;84(1):277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 7.Cinti S., Cigolini M., Sbarbati A., Zancanaro C. Ultrastructure of brown adipocytes mitochondria in cell culture from explants. Journal of Submicroscopic Cytology. 1986;18(3):625–627. [PubMed] [Google Scholar]
- 8.Cinti S., Zancanaro C., Sbarbati A., Cicolini M., Vogel P., Ricquier D. Immunoelectron microscopical identification of the uncoupling protein in brown adipose tissue mitochondria. Biology of the Cell. 1989;67(3):359–362. [PubMed] [Google Scholar]
- 9.Ricquier D., Bouillaud F., Toumelin P., Mory G., Bazin R., Arch J. Expression of uncoupling protein mRNA in thermogenic or weakly thermogenic brown adipose tissue. Evidence for a rapid beta-adrenoreceptor-mediated and transcriptionally regulated step during activation of thermogenesis. Journal of Biological Chemistry. 1986;261(30):13905–13910. [PubMed] [Google Scholar]
- 10.Cypess A.M., Lehman S., Williams G., Tal I., Rodman D., Goldfine A.B. Identification and importance of brown adipose tissue in adult humans. The New England Journal of Medicine. 2009;360(15):1509–1517. doi: 10.1056/NEJMoa0810780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Marken Lichtenbelt W.D., Vanhommerig J.W., Smulders N.M., Drossaerts J.M., Kemerink G.J., Bouvy N.D. Cold-activated brown adipose tissue in healthy men. The New England Journal of Medicine. 2009;360(15):1500–1508. doi: 10.1056/NEJMoa0808718. [DOI] [PubMed] [Google Scholar]
- 12.Virtanen K.A. BAT thermogenesis: linking shivering to exercise. Cell Metabolism. 2014;19(3):352–354. doi: 10.1016/j.cmet.2014.02.013. [DOI] [PubMed] [Google Scholar]
- 13.Virtanen K.A., Lidell M.E., Orava J., Heglind M., Westergren R., Niemi T. Functional brown adipose tissue in healthy adults. The New England Journal of Medicine. 2009;360(15):1518–1525. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 14.Ouellet V., Routhier-Labadie A., Bellemare W., Lakhal-Chaieb L., Turcotte E., Carpentier A.C. Outdoor temperature, age, sex, body mass index, and diabetic status determine the prevalence, mass, and glucose-uptake activity of 18F-FDG-detected BAT in humans. Journal of Clinical Endocrinology and Metabolism. 2011;96(1):192–199. doi: 10.1210/jc.2010-0989. [DOI] [PubMed] [Google Scholar]
- 15.Pfannenberg C., Werner M.K., Ripkens S., Stef I., Deckert A., Schmadl M. Impact of age on the relationships of brown adipose tissue with sex and adiposity in humans. Diabetes. 2010;59(7):1789–1793. doi: 10.2337/db10-0004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petrovic N., Walden T.B., Shabalina I.G., Timmons J.A., Cannon B., Nedergaard J. Chronic peroxisome proliferator-activated receptor gamma (PPARgamma) activation of epididymally derived white adipocyte cultures reveals a population of thermogenically competent, UCP1-containing adipocytes molecularly distinct from classic brown adipocytes. Journal of Biological Chemistry. 2010;285(10):7153–7164. doi: 10.1074/jbc.M109.053942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wu J., Bostrom P., Sparks L.M., Ye L., Choi J.H., Giang A.H. Beige adipocytes are a distinct type of thermogenic fat cell in mouse and human. Cell. 2012;150(2):366–376. doi: 10.1016/j.cell.2012.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sidossis L., Kajimura S. Brown and beige fat in humans: thermogenic adipocytes that control energy and glucose homeostasis. Journal of Clinical Investigation. 2015;125(2):478–486. doi: 10.1172/JCI78362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee P., Werner C.D., Kebebew E., Celi F.S. Functional thermogenic beige adipogenesis is inducible in human neck fat. International Journal of Obesity (London) 2013 doi: 10.1038/ijo.2013.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rosenwald M., Perdikari A., Rulicke T., Wolfrum C. Bi-directional interconversion of brite and white adipocytes. Nature Cell Biology. 2013;15(6):659–667. doi: 10.1038/ncb2740. [DOI] [PubMed] [Google Scholar]
- 21.Cinti S. Transdifferentiation properties of adipocytes in the adipose organ. American Journal of Physiology – Endocrinology and Metabolism. 2009;297(5):E977–E986. doi: 10.1152/ajpendo.00183.2009. [DOI] [PubMed] [Google Scholar]
- 22.Lee Y.H., Petkova A.P., Konkar A.A., Granneman J.G. Cellular origins of cold-induced brown adipocytes in adult mice. FASEB Journal. 2015;29(1):286–299. doi: 10.1096/fj.14-263038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lee Y.H., Petkova A.P., Mottillo E.P., Granneman J.G. In vivo identification of bipotential adipocyte progenitors recruited by beta3-adrenoceptor activation and high-fat feeding. Cell Metabolism. 2012;15(4):480–491. doi: 10.1016/j.cmet.2012.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang Q.A., Tao C., Gupta R.K., Scherer P.E. Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nature Medicine. 2013;19(10):1338–1344. doi: 10.1038/nm.3324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schrauwen P., van Marken Lichtenbelt W.D., Spiegelman B.M. The future of brown adipose tissues in the treatment of type 2 diabetes. Diabetologia. 2015;58(8):1704–1707. doi: 10.1007/s00125-015-3611-y. [DOI] [PubMed] [Google Scholar]
- 26.Yoneshiro T., Aita S., Matsushita M., Kayahara T., Kameya T., Kawai Y. Recruited brown adipose tissue as an antiobesity agent in humans. Journal of Clinical Investigation. 2013;123(8):3404–3408. doi: 10.1172/JCI67803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Yoneshiro T., Saito M. Activation and recruitment of brown adipose tissue as anti-obesity regimens in humans. Annals of Medicine. 2015;47(2):133–141. doi: 10.3109/07853890.2014.911595. [DOI] [PubMed] [Google Scholar]
- 28.Langin D. Recruitment of brown fat and conversion of white into brown adipocytes: strategies to fight the metabolic complications of obesity? Biochimica et Biophysica Acta. 2010;1801(3):372–376. doi: 10.1016/j.bbalip.2009.09.008. [DOI] [PubMed] [Google Scholar]
- 29.Bartel D.P. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Selbach M., Schwanhausser B., Thierfelder N., Fang Z., Khanin R., Rajewsky N. Widespread changes in protein synthesis induced by microRNAs. Nature. 2008;455(7209):58–63. doi: 10.1038/nature07228. [DOI] [PubMed] [Google Scholar]
- 31.Baek D., Villen J., Shin C., Camargo F.D., Gygi S.P., Bartel D.P. The impact of microRNAs on protein output. Nature. 2008;455(7209):64–71. doi: 10.1038/nature07242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Torriani M., Srinivasa S., Fitch K.V., Thomou T., Wong K., Petrow E. Dysfunctional subcutaneous fat with reduced dicer and brown adipose tissue gene expression in HIV-infected patients. Journal of Clinical Endocrinology and Metabolism. 2016;101(3):1225–1234. doi: 10.1210/jc.2015-3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mudhasani R., Puri V., Hoover K., Czech M.P., Imbalzano A.N., Jones S.N. Dicer is required for the formation of white but not brown adipose tissue. Journal of Cellular Physiology. 2011;226(5):1399–1406. doi: 10.1002/jcp.22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mori M.A., Thomou T., Boucher J., Lee K.Y., Lallukka S., Kim J.K. Altered miRNA processing disrupts brown/white adipocyte determination and associates with lipodystrophy. Journal of Clinical Investigation. 2014;124(8):3339–3351. doi: 10.1172/JCI73468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Karbiener M., Fischer C., Nowitsch S., Opriessnig P., Papak C., Ailhaud G. microRNA miR-27b impairs human adipocyte differentiation and targets PPARgamma. Biochemical and Biophysical Research Communications. 2009;390(2):247–251. doi: 10.1016/j.bbrc.2009.09.098. [DOI] [PubMed] [Google Scholar]
- 36.Xie H., Sun L., Lodish H.F. Targeting microRNAs in obesity. Expert Opinion on Therapeutic Targets. 2009;13(10):1227–1238. doi: 10.1517/14728220903190707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kim Y.J., Hwang S.J., Bae Y.C., Jung J.S. MiR-21 regulates adipogenic differentiation through the modulation of TGF-beta signaling in mesenchymal stem cells derived from human adipose tissue. Stem Cells. 2009;27(12):3093–3102. doi: 10.1002/stem.235. [DOI] [PubMed] [Google Scholar]
- 38.Trajkovski M., Lodish H. MicroRNA networks regulate development of brown adipocytes. Trends in Endocrinology & Metabolism. 2013;24(9):442–450. doi: 10.1016/j.tem.2013.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Karbiener M., Pisani D.F., Frontini A., Oberreiter L.M., Lang E., Vegiopoulos A. MicroRNA-26 family is required for human adipogenesis and drives characteristics of brown adipocytes. Stem Cells. 2014;32(6):1578–1590. doi: 10.1002/stem.1603. [DOI] [PubMed] [Google Scholar]
- 40.Peng Y., Yu S., Li H., Xiang H., Peng J., Jiang S. MicroRNAs: emerging roles in adipogenesis and obesity. Cell Signal. 2014;26(9):1888–1896. doi: 10.1016/j.cellsig.2014.05.006. [DOI] [PubMed] [Google Scholar]
- 41.Karbiener M., Scheideler M. MicroRNA functions in brite/brown fat – novel perspectives towards anti-obesity strategies. Computational and Structural Biotechnology Journal. 2014;11(19):101–105. doi: 10.1016/j.csbj.2014.09.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Arner P., Kulyte A. MicroRNA regulatory networks in human adipose tissue and obesity. Nature Reviews Endocrinology. 2015;11(5):276–288. doi: 10.1038/nrendo.2015.25. [DOI] [PubMed] [Google Scholar]
- 43.Mori M., Nakagami H., Rodriguez-Araujo G., Nimura K., Kaneda Y. Essential role for miR-196a in brown adipogenesis of white fat progenitor cells. PLoS Biology. 2012;10(4):e1001314. doi: 10.1371/journal.pbio.1001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sun L., Xie H., Mori M.A., Alexander R., Yuan B., Hattangadi S.M. Mir193b-365 is essential for brown fat differentiation. Nature Cell Biology. 2011;13(8):958–965. doi: 10.1038/ncb2286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Belarbi Y., Mejhert N., Lorente-Cebrian S., Dahlman I., Arner P., Ryden M. MicroRNA-193b controls adiponectin production in human white adipose tissue. Journal of Clinical Endocrinology and Metabolism. 2015 doi: 10.1210/jc.2015-1530. [DOI] [PubMed] [Google Scholar]
- 46.Fu T., Seok S., Choi S., Huang Z., Suino-Powell K., Xu H.E. MicroRNA 34a inhibits beige and brown fat formation in obesity in part by suppressing adipocyte fibroblast growth factor 21 signaling and SIRT1 function. Journal of Molecular Cell Biology. 2014;34(22):4130–4142. doi: 10.1128/MCB.00596-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fu X., Dong B., Tian Y., Lefebvre P., Meng Z., Wang X. MicroRNA-26a regulates insulin sensitivity and metabolism of glucose and lipids. Journal of Clinical Investigation. 2015;125(6):2497–2509. doi: 10.1172/JCI75438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jimenez V., Munoz S., Casana E., Mallol C., Elias I., Jambrina C. In vivo adeno-associated viral vector-mediated genetic engineering of white and brown adipose tissue in adult mice. Diabetes. 2013;62(12):4012–4022. doi: 10.2337/db13-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rodriguez A.M., Elabd C., Delteil F., Astier J., Vernochet C., Saint-Marc P. Adipocyte differentiation of multipotent cells established from human adipose tissue. Biochemical and Biophysical Research Communications. 2004;315(2):255–263. doi: 10.1016/j.bbrc.2004.01.053. [DOI] [PubMed] [Google Scholar]
- 50.Rodriguez A.M., Pisani D., Dechesne C.A., Turc-Carel C., Kurzenne J.Y., Wdziekonski B. Transplantation of a multipotent cell population from human adipose tissue induces dystrophin expression in the immunocompetent mdx mouse. Journal of Experimental Medicine. 2005;201(9):1397–1405. doi: 10.1084/jem.20042224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rodbell M. Metabolism of isolated fat cells. I. Effects of hormones on glucose metabolism and lipolysis. Journal of Biological Chemistry. 1964;239:375–380. [PubMed] [Google Scholar]
- 52.Murholm M., Dixen K., Qvortrup K., Hansen L.H., Amri E.Z., Madsen L. Dynamic regulation of genes involved in mitochondrial DNA replication and transcription during mouse brown fat cell differentiation and recruitment. PLoS One. 2009;4(12):e8458. doi: 10.1371/journal.pone.0008458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pisani D.F., Beranger G.E., Corinus A., Giroud M., Ghandour R.A., Altirriba J. The K+ channel TASK1 modulates beta-adrenergic response in brown adipose tissue through the mineralocorticoid receptor pathway. FASEB Journal. 2016;30(2):909–922. doi: 10.1096/fj.15-277475. [DOI] [PubMed] [Google Scholar]
- 54.Elabd C., Chiellini C., Carmona M., Galitzky J., Cochet O., Petersen R. Human multipotent adipose-derived stem cells differentiate into functional brown adipocytes. Stem Cells. 2009;27(11):2753–2760. doi: 10.1002/stem.200. [DOI] [PubMed] [Google Scholar]
- 55.Barquissau V., Beuzelin D., Pisani D.F., Beranger G.E., Mairal A., Montagner A. White-to-brite conversion in human adipocytes promotes metabolic reprogramming towards fatty acid anabolic and catabolic pathways. Molecular Metabolism. 2016;5(5):352–365. doi: 10.1016/j.molmet.2016.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pisani D.F., Djedaini M., Beranger G.E., Elabd C., Scheideler M., Ailhaud G. Differentiation of human adipose-derived stem cells into “Brite” (brown-in-white) adipocytes. Front Endocrinol (Lausanne) 2011;2:87. doi: 10.3389/fendo.2011.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Karbiener M., Neuhold C., Opriessnig P., Prokesch A., Bogner-Strauss J.G., Scheideler M. MicroRNA-30c promotes human adipocyte differentiation and co-represses PAI-1 and ALK2. RNA Biology. 2011;8(5):850–860. doi: 10.4161/rna.8.5.16153. [DOI] [PubMed] [Google Scholar]
- 58.Pieler R., Sanchez-Cabo F., Hackl H., Thallinger G.G., Trajanoski Z. ArrayNorm: comprehensive normalization and analysis of microarray data. Bioinformatics. 2004;20(12):1971–1973. doi: 10.1093/bioinformatics/bth174. [DOI] [PubMed] [Google Scholar]
- 59.Sturn A., Quackenbush J., Trajanoski Z. Genesis: cluster analysis of microarray data. Bioinformatics. 2002;18(1):207–208. doi: 10.1093/bioinformatics/18.1.207. [DOI] [PubMed] [Google Scholar]
- 60.Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research. 2002;30(1):207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pisani D.F., Ghandour R.A., Beranger G.E., Le Faouder P., Chambard J.C., Giroud M. The omega6-fatty acid, arachidonic acid, regulates the conversion of white to brite adipocyte through a prostaglandin/calcium mediated pathway. Molecular Metabolism. 2014;3(9):834–847. doi: 10.1016/j.molmet.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Loft A., Forss I., Siersbaek M.S., Schmidt S.F., Larsen A.S., Madsen J.G. Browning of human adipocytes requires KLF11 and reprogramming of PPARgamma superenhancers. Genes & Development. 2015;29(1):7–22. doi: 10.1101/gad.250829.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Brand M.D., Nicholls D.G. Assessing mitochondrial dysfunction in cells. Biochemical Journal. 2011;435(2):297–312. doi: 10.1042/BJ20110162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sagot B., Gaysinski M., Mehiri M., Guigonis J.M., Le Rudulier D., Alloing G. Osmotically induced synthesis of the dipeptide N-acetylglutaminylglutamine amide is mediated by a new pathway conserved among bacteria. Proceedings of the National Academy of Sciences of the United States of America. 2010;107(28):12652–12657. doi: 10.1073/pnas.1003063107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bertin S., Samson M., Pons C., Guigonis J.M., Gavelli A., Baque P. Comparative proteomics study reveals that bacterial CpG motifs induce tumor cell autophagy in vitro and in vivo. Molecular & Cellular Proteomics. 2008;7(12):2311–2322. doi: 10.1074/mcp.M800100-MCP200. [DOI] [PubMed] [Google Scholar]
- 66.Wikstrom J.D., Mahdaviani K., Liesa M., Sereda S.B., Si Y., Las G. Hormone-induced mitochondrial fission is utilized by brown adipocytes as an amplification pathway for energy expenditure. EMBO Journal. 2014;33(5):418–436. doi: 10.1002/embj.201385014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Xu S., Chen P., Sun L. Regulatory networks of non-coding RNAs in brown/beige adipogenesis. Bioscience Reports. 2015 doi: 10.1042/BSR20150155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hu F., Wang M., Xiao T., Yin B., He L., Meng W. miR-30 promotes thermogenesis and the development of beige fat by targeting RIP140. Diabetes. 2015;64(6):2056–2068. doi: 10.2337/db14-1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Trajkovski M., Ahmed K., Esau C.C., Stoffel M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nature Cell Biology. 2012;14(12):1330–1335. doi: 10.1038/ncb2612. [DOI] [PubMed] [Google Scholar]
- 70.Pan D., Mao C., Quattrochi B., Friedline R.H., Zhu L.J., Jung D.Y. MicroRNA-378 controls classical brown fat expansion to counteract obesity. Nature Communications. 2014;5:4725. doi: 10.1038/ncomms5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sun Y.M., Lin K.Y., Chen Y.Q. Diverse functions of miR-125 family in different cell contexts. Journal of Hematology & Oncology. 2013;6:6. doi: 10.1186/1756-8722-6-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Duroux-Richard I., Pers Y.M., Fabre S., Ammari M., Baeten D., Cartron G. Circulating miRNA-125b is a potential biomarker predicting response to Rituximab in rheumatoid arthritis. Mediators of Inflammation. 2014;2014:342524. doi: 10.1155/2014/342524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ouyang D., Ye Y., Guo D., Yu X., Chen J., Qi J. MicroRNA-125b-5p inhibits proliferation and promotes adipogenic differentiation in 3T3-L1 preadipocytes. Acta Biochimica et Biophysica Sinica (Shanghai) 2015;47(5):355–361. doi: 10.1093/abbs/gmv024. [DOI] [PubMed] [Google Scholar]
- 74.Diawara M.R., Hue C., Wilder S.P., Venteclef N., Aron-Wisnewsky J., Scott J. Adaptive expression of microRNA-125a in adipose tissue in response to obesity in mice and men. PLoS One. 2014;9(3):e91375. doi: 10.1371/journal.pone.0091375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Zhang X., Zuo X., Yang B., Li Z., Xue Y., Zhou Y. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell. 2014;158(3):607–619. doi: 10.1016/j.cell.2014.05.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.