Abstract
Cold-inducible RNA-binding protein (CIRP) and RNA-binding motif protein 3 (RBM3) are two evolutionarily conserved RNA-binding proteins that are transcriptionally upregulated in response to low temperature. Featuring an RNA-recognition motif (RRM) and an arginine–glycine-rich (RGG) domain, these proteins display many similarities and specific disparities in the regulation of numerous molecular and cellular events. The resistance to serum withdrawal, endoplasmic reticulum stress, or other harsh conditions conferred by RBM3 has led to its reputation as a survival gene. Once CIRP protein is released from cells, it appears to bolster inflammation, contributing to poor prognosis in septic patients. A variety of human tumor specimens have been analyzed for CIRP and RBM3 expression. Surprisingly, RBM3 expression was primarily found to be positively associated with the survival of chemotherapy-treated patients, while CIRP expression was inversely linked to patient survival. In this comprehensive review, we summarize the evolutionary conservation of CIRP and RBM3 across species as well as their molecular interactions, cellular functions, and roles in diverse physiological and pathological processes, including circadian rhythm, inflammation, neural plasticity, stem cell properties, and cancer development.
Keywords: Transcription, Translation, hnRNP, MicroRNA, Neuroscience, Apoptosis, Stress granule
Introduction
Decreased body temperature is a key feature of seasonal hibernation, which is an entrained state of slowed metabolism that widely exists in amphibians and mammals to endure food austerity [1–3]. As a direct cellular consequence of decreased body temperature, global protein synthesis is repressed, thereby switching the cellular program from cell growth to cell preservation. In contrast to the general decrease in protein synthesis, the production of a small group of proteins, including cold-inducible RNA-binding protein (CIRP, alternative abbreviation: CIRBP; synonymous: heterogeneous ribonucleoprotein A18, hnRNP A18) and RNA-binding motif protein 3 (RBM3), increases in hibernating animals [4, 5].
In clinical practice, therapeutic hypothermia (32–34 °C) has been proved a potent tool to alleviate neurological deficits in infants with hypoxic-ischemic encephalopathy [6], and in adults with acute brain injuries [7]. Whereas much deeper hypothermia is used during cardiac and transplant surgery [8], CIRP and RBM3 protein syntheses peak in a range of mild to moderate temperatures (32–34 °C) [9]. Because clinical hypothermia is associated with various life-threatening side effects [10], CIRP and RBM3 are promising research candidates for new therapies.
Apart from their functions under hypothermia, various studies have indicated that CIRP and RBM3 also have important functions in cell protection under general endogenous and environmental stresses at normal temperatures [11]. Here, we provide a comprehensive and systematic overview of biological functions mediated by CIRP and RBM3, within and outside the context of hypothermia by systematically considering almost all papers published on CIRP and RBM3 thus far.
Evolution and protein structure
Whereas the gene coding for human CIRP is localized on chromosome 19p13.3 [12], the gene coding for human RBM3, the homolog of CIRP, has been mapped to the short arm of the X-chromosome at Xp11.23 [13].
Both CIRP and RBM3 belong to a group of stress-responsive proteins that share high amino acid sequence similarity in the N-terminal RNA-binding domain (Fig. 1; Table 1). They both own one conserved RNA-recognition motif (RRM), containing two ribonucleoprotein domains (RNPs), RNP1 and RNP2, which are located at the N-terminal protein end. The consensus sequences of RNP1 and RNP2 are (K/R)G(F/Y)(G/A)FVX(FY) and (L/I)(F/Y)(V/I)(G/K)(G/N)L, respectively [14]. Notably, both RNP1 and RNP2 from CIRP and RBM3 show moderate sequence and function similarity to parts of cold shock proteins (CSPs), with the evolutionary conserved sequences (K/S) G(F/K/Y)G(F/L)IXX and (L/I/V)(F/Q)(V/A/L)HX(STR), respectively [15]. Prokaryotic CSPs exert important functions in response to drastic drops in temperature, e.g., from 37 to 10 °C [14, 15], indicating common features as well as differences between CIRP/RBM3 and CSPs.
Fig. 1.
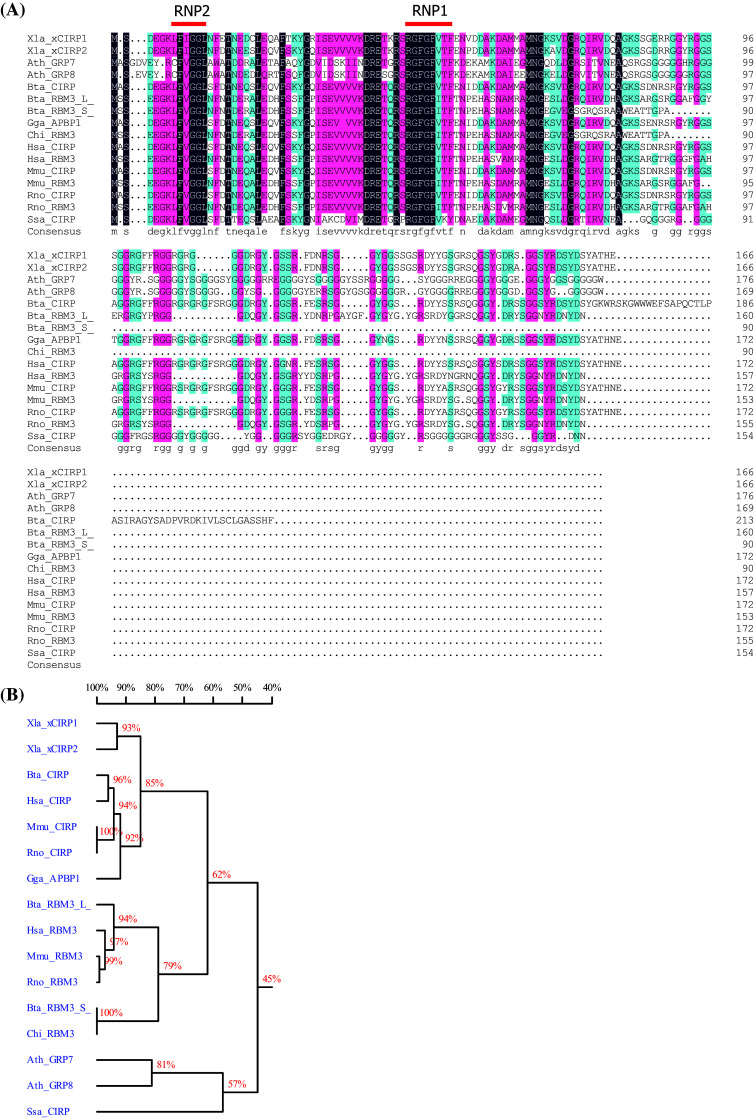
Protein alignment (a) and homology tree (b) of CIRP, RBM3, and their plant homologues in different species. Xla Xenopus laevis, Ath Arabidopsis thaliana, Bta Bos Taurus, Gga Gallus gallus, Chi Capra hircus, Hsa Homo sapiens, Mmu Mus musculus, Rno Rattus norvegicus, SSa Salmo salar, L long full-length RBM3, S short truncated RBM3
Table 1.
Proteins used for alignment in Fig. 1a
| Species | Name abbreviation | Reference ID in UniProtKB | Full name | |
|---|---|---|---|---|
| Plant | Arabidopsis thaliana | Ath_GRP7 | Q03250 | Glycine-rich RNA-binding protein 7 |
| Arabidopsis thaliana | Ath_GRP8 | Q03251 | Glycine-rich RNA-binding protein 8 | |
| Fish | Salmo salar | Ssa_CIRP | B5DGC5 | Cold-inducible RNA-binding protein |
| Amphibian | Xenopus laevis | Xla_xCIRP1 | O93235 | Cold-inducible RNA-binding protein A |
| Xenopus laevis | Xla_xCIRP2 | Q9DED4 | Cold-inducible RNA-binding protein B | |
| Bird | Gallus gallus | Gga_CIRP | Q45KQ2 | Aggrecan promoter binding protein (CIRP homologue) |
| Mammal | Bos Taurus | Bta_CIRP | Q3SZN4 | Cold inducible RNA binding protein |
| Bos Taurus | Bta_RBM3_L | F6RBQ9 | Uncharacterized protein | |
| Bos Taurus | Bta_RBM3_S | Q3ZBA4 | RNA binding motif (RNP1, RRM) protein 3 | |
| Capra hircus | Chi_RBM3 | W8E7I1 | RNA-binding protein 3 | |
| Mus musculus | Mmu_CIRP | P60824 | Cold-inducible RNA-binding protein | |
| Mus musculus | Mmu_RBM3 | O89086 | RNA-binding protein 3 | |
| Rattus norvegicus | Rno_CIRP | P60825 | Cold-inducible RNA-binding protein | |
| Rattus norvegicus | Rno_RBM3 | Q925G0 | RNA-binding protein 3 | |
| Homo sapiens | Hsa_CIRP | Q14011 | Cold-inducible RNA-binding protein | |
| Homo sapiens | Hsa_RBM3 | P98179 | RNA-binding protein 3 |
The C-terminal part of both CIRP and RBM3 contains a less conserved arginine–glycine-rich (RGG) domain, for which reason CIRP and RBM3 belong to the large family of glycine-rich proteins (GRP). In addition, because they are featured with an RRM, they belong to the subfamily Class IVa of GRPs [14, 16]. The evolution of this Class IVa GRP subfamily is highly conserved across vertebrates and higher plants [14] (Fig. 1) with respect to their primary amino acid sequences as well as to their protein functions. For example, in Arabidopsis, the CIRP/RBM3 homologue AtGRP7 is indispensable in cold adaption and drought/osmotic stress responses [17–19]. AtGRP7 also regulates a number of post-transcriptional and translational events [20–23], functions as a circadian oscillator [24], and is involved in pathogen defense [25, 26]. In poikilothermic animals, such as fish, CIRP homologues are also elevated upon environmental osmotic or severe cold stresses (8 °C) but may not change at normal ambient temperatures (20–25 °C) [27–29]. As detailed in the following paragraph, the well-studied amphibian and mammalian CIRP and RBM3 possess biological functions highly similar to AtGRP7 and fish CIRP, implying their preservation of biological activities.
Temporal and spatial distribution
Both CIRP and RBM3 are key factors during early development. In amphibians, which are widely used as models for developmental studies, Xenopus laevis homologue xCIRP-1 is transiently expressed in the developing kidney and brain [30] and is required for embryonic kidney formation [31]. The expression of CIRP homologue AxRBP in the Mexican axolotl starts at gastrula stage day 10–12, peaks at neurula stage day 15 particularly in the neural plate and neural fold, and declines afterward to low levels when hatching [32]. In mammals, RBM3 level peaks during the early postnatal period and then decreases to very low levels in youth and adulthood in most regions of the brain, except for areas where proliferation remains active, such as the subventricular zone (SVZ) and the hippocampal subgranular zone (SGZ) [33, 34], indicating a pivotal role of RBM3 in the maintenance of stemness and proliferation in neural stem/progenitor cells. Regardless to this dynamic temporal expression of CIRP and RBM3 with high abundance in early developmental stages and low in mature organisms, many mature cells maintain their ability to overexpress CIRP and RBM3 in response to stressful conditions, such as cold, see below for details.
The spatial distribution of CIRP and RBM3 in major organs varies between species. In human, RBM3 expression is low or absent in thyroid and heart, whereas CIRP is abundant in these organs [35, 36]. In hibernating animals, RBM3 is upregulated in muscle, liver, and heart tissues of black bears [37, 38], as well as in brain, heart, and liver tissues of squirrels at late torpor [4]. In contrast, CIRP fails to be stimulated in muscle and liver tissues in rats with chronic intermittent cold exposure, but is induced in brain and heart [39]. Within the same tissue, their spatial patterns can be cell type-specific: in mammalian testis, CIRP is predominantly in germ cells [40], whereas RBM3 is mainly in Sertoli cells [41].
At subcellular level, the spatial distribution of CIRP and RBM3 is expected to be mainly in the nucleus, because both proteins are featured with an RGG domain which is a nuclear localization signal and associated with nucleocytoplasmic shuttling. In fact, CIRP and RBM3 are predominantly found in the nucleus [42], where they regulate gene transcription or bind to mRNA for post-transcriptional regulation. In addition, under physiological or stressful conditions, CIRP and RBM3 shuttle between nucleus and cytoplasm [43]. However, there are at least three important exceptions, indicating that the subcellular localization of RBM3 and CIRP is subjected to developmental stage and cell type. First, during the first week after birth, RBM3 resides in the nucleus, then shifts during the second postnatal week toward a more cytoplasmic localization. In sections of adult brain, RBM3 is in general very weekly expressed, and the balance of RBM3 subcellular localization also appears to be highly dependent on cell type [33]. Second, frog xCIRP2 has been found to serve as a major cytoplasmic protein in oocytes [44]. Third, in contrast to strong expression of CIRP in the nucleus of spermatocytes in both mice and humans, round spermatids at stages I–III of mice show CIRP expression in cytoplasm but not in the nucleus, suggesting an additional function of CIRP in the cytoplasm of haploid cells [40].
Interestingly, a nucleocytoplasmic shuttling signal (RG4) has been identified within the RGG domain of frog xCIRP2, which promotes accumulation of xCIRP2 in the cytoplasm once methylated by arginine methyltransferase xPRMT1 [45]. In mammals, methylation of arginine residues in the RGG domain of CIRP due to cytoplasmic stress and endoplasmic reticulum (ER) stress causes CIRP accumulation in cytoplasmic stress granules independent of the major mediator of stress granule formation TIA-1 [46]. For RBM3, a distinct pool of RBM3 proteins can shuttle to the ER upon ER stress where RBM3 regulates the activity of ER membrane protein PERK. The majority of RBM3 proteins, however, remain in the nucleus [47]. The absence of a single arginine residue in the RGG domain of RBM3 promotes the localization of RBM3 in dendrites of neurons rather than in nuclei [48]. Collectively, these studies reveal a critical role of the arginine residue in the RGG domain of CIRP and RBM3 in nucleocytoplasmic shuttling.
Stress-regulated expression
Hypothermia and hyperthermia: CIRP and RBM3 are stress-responsive genes, and their expression is induced by a variety of stressful conditions, including cold stress, which is the first identified condition that increases CIRP and RBM3 expression [35, 49]. In the very early beginnings of CIRP and RBM3 research, both proteins were discovered with high expression in mammalian testis, an organ located outside the body to maintain temperatures slightly less than core body temperature to ensure efficient spermatogenesis [41]. Expression of CIRP and RBM3 decreases under experimental heat stress or cryptorchidism, with a more rapid response of CIRP (6 h) to hypothermia than RBM3 (12 h) [40, 41].
In mammalian cells, the expression levels of both CIRP and RBM3 reach their peaks upon mild to moderate hypothermia (28–34 °C) and drop significantly upon deep hypothermia (15–25 °C) [9, 42, 49]. In contrast, hyperthermia (39–42 °C) causes substantial decreases in CIRP and RBM3 in cultured cells in vitro [35, 49], which is consistent with their change under pathological and experimental conditions in vivo [40, 41]. Notably, RBM3 induction is extremely sensitive to temperature change at least in neural cells, even a 1 °C drop from 37 to 36 °C is sufficient [50]. These facts indicate that CIRP and RBM3 respond to temperature change within a small range in a subtle manner.
In cultured tissues or cells, CIRP also increases more rapidly than RBM3 upon moderate hypothermia at early stage, but declines faster during subsequent rewarming process, indicating a faster dynamic change of CIRP in cold response [9, 51]. When cells are exposed to hypothermia, CIRP is activated within 3 h and reaches maximal expression at 12 h, but drops by 50 % within 8 h during rewarming. In contrast, RBM3 is induced after 3 h and peaks at around 24 h, and its expression level remains unchanged until 8 h after rewarming [11, 35, 51]. However, the dose–response kinetics of CIRP and RBM3 are dependent on the biological systems studied [9, 35, 42, 51].
Hypoxia: Under natural circumstances and when breathing air containing 21 % oxygen at sea level, the oxygen concentrations in the different tissues of the body are considerably heterogeneous [52]. Reduced oxygen tension (hypoxia) compared with physiological tension occurs during diverse acute and chronic injuries or diseases, including cancer [53]. Experimentally, both mild (8 %) and severe (1 %) hypoxia can induce CIRP and RBM3 expression to a comparable level by a mechanism that involves neither hypoxia-inducible factor (HIF) nor mitochondria in vitro [54]. In contrast, severe hypoxia mimicking ischemia in an in vitro model applying cultured neural stem cells (NSCs) suppresses CIRP expression in parallel with cell cycle arrest [55]. Since hydrogen peroxide treatment can inhibit the hypothermia-induced expression of CIRP [56, 57] and a mild level of reactive oxygen species (ROS) is beneficial, whereas a high level is toxic for NSC proliferation, it has been hypothesized that mild hypoxia resulting in mild elevation of ROS increases CIRP expression, whereas severe hypoxia/ischemia inducing an overload of ROS suppresses CIRP expression [55]. Exposing pregnant mice in late gestation to severe systemic hypoxia causes overexpression of HIF-dependent genes and downregulation of RBM3 expression in the placenta and developing brain [58]. In summary, oxygen-regulated expression of CIRP and RBM3 is dose-dependent and subjected to cell vulnerability, and other factors involved in development or pathological changes, such as hypoxic-ischemia, carcinogenesis, and inflammation.
Radiation: Independent of the first characterization of CIRP from Nishiyama et al. in 1997, Sheikh et al. identified the UV light-induced heterogeneous nuclear ribonucleoprotein A18 (hnRNP A18), which was soon proved to be a CIRP homologue in hamster playing a role in DNA damage repair [59]. Similarly, ionizing radiation can stimulate a number of hnRNPs, including CIRP, which promotes the repair of radiation-induced DNA damage [60]. Furthermore, spaceflight increases CIRP and RBM3 expression [61, 62], which may result from irradiation in space.
Miscellaneous: Toxins and drugs can also promote CIRP and RBM3 induction. For instance, the neurotoxin domoic acid elevates CIRP and RBM3 mRNA expression in mouse brain [63]. In fish, CIRP mRNA expression is upregulated upon lipopolysaccharide (LPS) treatment [64]. In addition, as shown in mouse leukemic cell line, RBM3 forms together with other RNA-binding proteins an RNA–protein complex with the first 60 nucleotides of the 3′-UTR of cyclooxygenase-2 (COX-2) mRNA [65]. Growth factors, such as insulin-like growth factor-1 (IGF-1) and fibroblast growth factor 21 (FGF21), can induce CIRP and RBM3 expressions, respectively [50, 66]. Melatonin, a well-studied hormone with both endogenous and exogenous sources, may augment the induction of RBM3 upon mild hypothermia in young neurons but not in mature neurons [50]. Of note, very recently, RBM3 has been shown to be suppressed by metformin and the AMP analog AICAR, probably via the inference of cell metabolism and the activation of AMPK [67]. These observations indicate that stress is not always an inducer, but can be an inhibitor of cold-inducible proteins as well.
Regulation and functions of CIRP
Molecular regulation of CIRP
The precise mechanism by which hypothermia and other stresses modulate the transcription and translation of cold-inducible proteins is poorly understood, although several models have been suggested involving various regulatory levels. At the transcriptional level, a core promoter and an alternative promoter have been identified in the mouse CIRP gene, and both promoters are activated upon mild hypothermia [68]. Furthermore, alternative splicing is one important route in response to cold stress. Hamsters, which are non-hibernating animals, express a long CIRP transcript in their hearts. This transcript has an extra insert containing a stop codon inside the open reading frame (ORF), which probably leads to a truncated translational product and aberrant function. In contrast, hibernating animals predominantly express the short isoform with a complete ORF. Artificial hypothermia can partially promote a shift from the long isoform to the short functional isoform [5]. In mouse fibroblasts, the 5′-UTR and full-length ORF of CIRP are present in two transcripts that are generated under hypothermic conditions, whereas the isoform under euthermic conditions lacks the 5′-UTR and the code for the initial methionine [69]. Cold stress upregulates the level and stability but not translational efficiency of the longer CIRP transcript, which contains a putative internal ribosome entry site (IRES) in the 5′-UTR [69]. In addition, transcription factors may contribute to the modulation of cold-inducible gene transcription. At 32 °C, a greater number of the transcription factor Sp1 are recruited to the mild-cold responsive element (MCRE) in the 5′-flanking region of the CIRP gene than at 37 °C, leading to increased CIRP expression [70]. Overall, CIRP expression levels are altered in response to stress by versatile mechanisms, suggesting a wide range of adaptation to various external and internal challenges.
Molecular and cellular activities of CIRP
Regulation of post-transcriptional and translational events
As other RNA-binding proteins, CIRP has the capacity to bind RNAs, and to modulate them at the post-transcriptional level [71, 72]. In general, such post-transcriptional interactions by RNA-binding proteins involve binding to the target regions within the 3′-UTR, which spans the nucleotide sequence between the stop codon and poly(A) tail [73]. Upon UV irradiation, CIRP binds to the 3′-UTR of two stress-responsive transcripts, replication protein A (RPA) and thioredoxin (TRX), thereby stabilizing the bound mRNA and promoting their translation [74, 75]. Both CIRP RRM domain and RGG domain are required for the maximal binding activity to TRX mRNA [75]. These domains bridge 5′- and 3′-UTRs of the TRX transcript via eIF4G, a key component of the translational machinery to enhance TRX translation [75]. A CIRP-binding motif found in RPA and TRX 3′-UTRs also exists in the 3′-UTR of ataxia telangiectasia mutated- and Rad3-related (ATR) mRNA, a key regulator of the DNA damage response [76]. Thus, CIRP-mediated repair of UV-induced DNA damage involves at least partially ATR [76]. In rat ventricular myocytes, Cirp ablation upregulates the protein levels of potassium channels by post-transcriptional modulation of their α-subunits without changing their transcriptional activity [77].
In addition to the 3′-UTR, the poly(A) tail is an important regulatory element for CIRP-mediated post-transcriptional modulation. Actually, CIRP is enriched in poly(A) sites and controls alternative polyadenylation of a variety of genes, including circadian genes [78]. Moreover, in the regulation of TRX mRNA, the poly(A) tail can strengthen the binding of CIRP to the TRX 3′-UTR and enhance its stability [74]. Furthermore, CIRP putatively associates with the spliceosome [79].
Another RNA-binding protein, human antigen R (HuR), which is known to bind to AU-rich elements of 3′-UTR [80], can strengthen CIRP-mediated regulation. In African-clawed frog, the CIRP homolog xCIRP2 interacts with ElrA (an HuR homologue), and both xCIRP2 and ElrA stabilize mRNA in a cooperative manner [81]. Co-regulation of cyclin E1 mRNA stability by HuR and CIRP has also been discovered in mammalian cancer cells [82].
The function of CIRP in protein translation is unclear. CIRP can associate with ribosomes [44]. The RGG domain of CIRP tethers 3′-UTR and suppresses translation [46, 83]. In addition, there is some evidence to show that CIRP may inhibit gene transcription and translation by targeting regulatory elements inside genes. For example, APBP-1, the chicken homologue of CIRP, binds to a cis-element of the aggrecan gene and represses its expression [84]. In contrast, overexpression of CIRP at 37 °C in an engineered CHO cell line improves the production of recombinant interferon gamma protein [85].
Signaling pathways
As a regulatory protein, CIRP is involved in complex signaling pathways relating to diverse cellular physiological processes, such as cell growth, senescence, and apoptosis.
Stemness: The Wnt/β-catenin pathway is one of the main pathways controlling self-renewal of stem and progenitor cells [86]. The frog xCIRP is a target of xTcf-3, a key mediator in the Wnt/β-catenin pathway [87]. The endogenous inhibitor of β-catenin, GSK-3β kinase, upregulates CIRP transcription, phosphorylates CIRP protein and promotes its cytosolic translocation [75, 76, 88]. Furthermore, CIRP maintains the expression of adhesion molecules, including β-catenin, and is required for embryonic cell movement during development [89].
Cell cycle: Hypothermia is known to slow cell proliferation and to cause cell cycle arrest. In contrast to an early study, which has shown an inhibitory role of CIRP in cell growth upon hypothermia [49], a more recent series of mechanistic investigations have revealed that CIRP positively modulates the cell cycle at different stages. CIRP interacts with HuR and upregulates its expression. In cooperation with CIRP, elevated HuR further increases cyclin E1, a key positive regulator for G1/S transition, and promotes mitosis [82, 90]. Furthermore, CIRP accelerates G0/G1 and G1/S transitions by inhibiting the phosphorylation of cyclin D1 and p27 via the kinase Dyrk1b/Mirk [91]. In addition, CIRP appears to promote cell cycle progression from S phase to G2/M phase [92]. Hence, the current understanding is that CIRP facilitates cell proliferation.
Apoptosis: Apoptosis is induced by numerous exogenous and endogenous signals, involves various signaling pathways, and occurs in a broad range of diseases [93]. RNA-binding proteins are largely known as a family of proteins that modulate apoptosis [94]. Many studies have revealed that CIRP mediates, at least partially, the hypothermic protection of cells from apoptosis. Specifically, CIRP suppresses apoptosis in neural stem cells [95] and cortical neurons probably through mitochondrial pathways [96], which might mediate the protective effect of therapeutic hypothermia [97]. The inhibition of p53, Fas, and caspase-3 pathways also contributes to CIRP-mediated anti-apoptotic effects [57, 98, 99].
Senescence: CIRP activates the ERK1/2 pathway by increasing the phosphorylation of ERK1/2, which promotes cell division and bypasses replicative senescence [92, 100, 101]. The activation of ERK1/2 by CIRP contributes to tumor growth in pituitary corticotroph adenoma [102]. In addition, CIRP has been found at the telomere of HeLa cells [103]. A recent study has unraveled a novel role of CIRP in the maintenance of telomerase activity in both normothermic and hypothermic conditions [104]. These studies collectively support a role of CIRP in anti-senescence.
Biological functions and diseases
Brain disorders
Therapeutic hypothermia can not only efficiently reduce primary injury and prevent secondary injury in acute ischemia [7] and spinal cord injury (SCI) [105], but also delay the progression of chronic neurodegenerative diseases [106]. In vitro, the two cold-inducible proteins CIRP and RBM3 both function against apoptosis in cultured primary neurons or neuron-like PC12 cells [34, 96, 107].
The role of CIRP in brain ischemic injury is controversial. CIRP mRNA levels measured by Northern blot were found to decrease 3–6 h after transient ischemia in rat hippocampus but remained unchanged in the cerebral cortex during a 48 h observation period [56]. However, in the same ischemic model, real-time RT-PCR showed a gradual increase in CIRP mRNA by approximately fivefold until 24 h after cerebral ischemia in rat cortex [108]. In contrast to ischemia, hypothermia considerably induced CIRP expression by approximately 30-fold until 24 h, and the combination of hypothermia and ischemia did not further enhance the CIRP level from 30-fold [108].
An elevated level of ROS is one important detrimental factor in the induction of oxidative stress during ischemia–reperfusion injury in the brain [109]. In PC12 cells, CIRP expression has been observed to be downregulated upon H2O2 treatment, which produces ROS [56]. When CIRP is induced endogenously or overexpressed artificially, H2O2-induced apoptosis in cultured neural cells is dramatically inhibited, indicating a neuroprotective role of CIRP [57, 92]. In contrast to this beneficial intracellular action of CIRP, release of CIRP into the blood system is associated with the activation of detrimental immune responses. Zhou et al. reported that the secretion of CIRP from microglia after cerebral ischemia with subsequent CIRP-mediated TNF-α expression leads to neuroinflammation and causes neuronal damage both in vivo and in vitro [110]. An investigation of alcohol-induced brain inflammation has also demonstrated that extracellular CIRP mediates neuroinflammation by upregulating TNF-α and IL-1β [111]. To summarize, CIRP exerts opposing functions during brain ischemia–reperfusion injury. On the one hand, as long as CIRP remains intracellularly localized, it protects neurons from apoptosis; on the other hand, once CIRP is released, e.g., from microglia, it mediates devastating neuroinflammation at the cellular level.
Circadian rhythm
CIRP and RBM3 show high homology with the two glycine-rich RNA-binding proteins, AtGRP7, and AtGRP8, in Arabidopsis thaliana (Fig. 1). Both of these proteins are pivotal components in a circadian-regulated feedback loop [24, 112]. Similarly, CIRP expression is diurnally regulated in the suprachiasmatic nucleus (SCN) and cerebral cortex of mice [113]. The expression level of CIRP peaks at 6 pm and reaches the bottom at 3 am, altering in a light-dependent manner; the fluctuation occurs only in juvenile and adult mice but not in neonates [113]. Similar to mammals, light signal has been observed to induce CIRP expression in amphibian brain [114]. In 2012, CIRP was identified as a regulator of circadian oscillator genes, including the CLOCK gene, in a post-transcriptional pattern in mammals [71]. Further experiments showed that CIRP is upregulated during the sleep phase [115].
In mammals, the central clock in the SCN systematically synchronizes the body temperature cycles to environmental light–dark cycles [116] and to peripheral clocks (e.g., the liver and pancreas) [117, 118]. A recent study has shown that CIRP contributes to temperature-sensitive oscillation in murine hepatocytes, linking the subtle fluctuation in mammalian body temperature and circadian rhythm in peripheral tissues [118]. Furthermore, as the liver and pancreas are essential metabolic organs, nutrition necessarily influences the peripheral clock in these organs [119]. Ketogenic diets and fasting, which disrupt peripheral clocks, induce CIRP expression in the liver [120]. Meal timing greatly enhances the circadian expression of CIRP in pancreatic adenocarcinoma, pointing to a link between circadian rhythm and cancer therapy involving CIRP [121]. Therefore, CIRP is believed to be a component of mammalian circadian oscillation, which not only is regulated by body temperature and responds to changes in the environment, such as light in a subtle manner, but also controls the expression of downstream circadian genes.
Immune response
Conserved from plant to mammals, pattern recognition receptors (PRRs) are primitive key components to identify pathogen-associated molecular patterns (PAMP) and damage-associated molecular patterns (DAMP) [122, 123]. In 2007, the plant cold-inducible protein, AtGRP7, was first discovered to be involved in plant immunity [25]. AtGRP7 significantly enhances PAMP-triggered immunity by binding to the transcripts and proteins of two PRRs, FLS2 and EFR [124]. It is believed that the mammalian cold-inducible proteins are also involved in innate immune response. In 2013, CIRP was identified as a novel inflammatory mediator released from the heart and liver into the circulatory system during hemorrhagic shock and sepsis [125]. Secreted CIRP acts as a DAMP by binding to TLR4–MD2 complex (one class of mammalian PRRs), triggering inflammatory response by stimulating TNFα and HMGB1 secretion [125]. Conversely, CIRP expression can be impaired by TNFα or TGFβ [126], suggesting a negative feedback loop. Similar to the dual role of CIRP in the brain as discussed above, a recent report has shown that hypothermia-induced CIRP expression in the liver protects hepatocytes by reducing ROS production [127], while anti-CIRP antibody treatment, which neutralizes secreted CIRP in the serum, significantly decreases the inflammatory response and protects the liver from ischemic-reperfusion injury [128]. Other anti-CIRP therapies are also considered to treat inflammation-related disorders, such as abdominal aortic aneurysm based on animal experiments [129]. Today, the quantitative measurement of CIRP levels in peripheral blood using an ELISA kit appears achievable, opening the possibility to investigate CIRP as a new diagnostic marker for sepsis [130]. Furthermore, CIRP deficiency accelerates inflammation phase and wound healing process [131]. Overall, extracellular CIRP induces cell damage by inducing inflammatory responses. However, in the late stage of inflammation, the damaged cells are eliminated by inflammation, and regenerated cells can substitute the dysfunctional ones [132]. It implies that CIRP-mediated immune response may also have favorable aspects.
Cancer
Deduced from the above-summarized features, CIRP and RBM3 are both involved in cell cycle regulation and cell proliferation and are both present in proliferating and malignant cells. Therefore, they are considered as proto-oncogenes, promoting cancer cell proliferation, and transformation in vitro [36, 133, 134] and are differentially expressed in a variety of different cancers compared with normal tissues (Table 2). Despite these common features, their roles in clinical cancer development seem to be opposite. RBM3 expression always correlates with good prognosis and reduced risk of disease progression and recurrence, whereas CIRP seems to be an indicator of poor prognosis (Table 2). Accordingly, high CIRP level is associated with poor chemosensitivity of cancer cells [135, 136].
Table 2.
Roles of CIRP and RBM3 in cancer
| Cancer type | CIRP or RBM3 studied | Proposed mechanisms | Prognosis associated with high expression | References | |||
|---|---|---|---|---|---|---|---|
| Breast cancer | Both | RBM3 | – | Good | [181] | ||
| CIRP | Increase cyclin E1 | Poor | [82, 142] | ||||
| Epithelial ovarian cancer | RBM3 | Inhibit MCM3, Chk1 and Chk2 | Good | [171, 182] | |||
| Endometrial carcinoma | CIRP | – | Unclear | [141] | |||
| Prostate cancer | RBM3 | Involve ERG and PTEN; enhance chemo-sensitivity; regulate CD44 splicing | Good and unclear | [135, 163, 183–185] | |||
| Testicular non-seminomatous germ cell cancer | RBM3 | – | Good | [187] | |||
| Urothelial bladder cancer | RBM3 | – | Good or not determined | [188, 189] | |||
| Oropharyngeal squamous cell carcinoma | Both | RBM3 | – | Not determined | [193] | ||
| CIRP | Induce TLR4-related inflammation | Poor | [140] | ||||
| Esophageal and gastric adenocarcinoma | RBM3 | – | Good | [194] | |||
| Liver cancer | CIRP | Increase ROS, IL-1β and IL-6; suppress p53 | Poor | [99, 137] | |||
| Colorectal cancer | Both | RBM3 | Suppress GSK3β activity and enhance β-catenin signaling | Good | [169, 190–192] | ||
| CIRP | Induce TNF-α and IL-23 | Poor | [138] | ||||
| Melanoma | RBM3 | Inhibit MCM3 | Good | [195–197] | |||
| Astrocytoma | RBM3 | – | Unclear | [198] | |||
| Pituitary adenoma | CIRP | Induce cyclin D1 and decrease p27 via Erk1/2 signaling | Poor | [102, 139] | |||
CIRP is differentially expressed in many cancer types [101]. In hepatocellular carcinoma (HCC), CIRP has been proposed to promote carcinogenesis by controlling ROS accumulation and cancer stem/progenitor cell expansion, and risk of HCC recurrence is positively correlated with Cirp expression in liver [137]. In colorectal tumors, CIRP links tumorigenesis and chronic inflammation by stimulating cytokines, including TNF-α and IL-23 [138]. Thus, inhibiting CIRP could be of therapeutic value at least in liver cancer treatment [99].
In pituitary adenoma, high CIRP expression correlates with proliferating invasive and recurrent tumor, probably via ERK1/2 signaling pathway [102, 139]. In oral squamous cell carcinoma, CIRP is co-expressed with TLR4 and is associated with a short survival rate [140]. In endometrial carcinoma and some endometrial hyperplasia, CIRP expression is absent or markedly decreased compared with normal endometrium, indicating a role of CIRP in normal proliferative events [141]. In addition, CIRP is thought to be involved in the progression of ductal carcinoma into invasive breast cancer presumably by increasing the expression of CyclinE1, an important cell cycle regulator, thereby promoting proliferation and tumor progression in breast cancer transformation [82, 90, 142].
In summary, increasing numbers of clinical studies have demonstrated that CIRP is linked to poor clinical outcome in cancer. Among a variety of possible mechanisms, we would like to highlight the following consideration based on the available above-described data. Major inflammatory pathways are involved in carcinogenesis [143]. The extracellular action of CIRP in immune responses has been linked to detrimental consequences [125]. Albeit no data have yet been published regarding similar extracellular action in cancer, it is hypothesized that extracellular CIRP signaling in cancers may contribute to tumor progression and worse outcome through cytokine activation, as supported by the works from Sakurai et al. [137, 138].
Reproduction
As reported above, two decades ago CIRP was found in the germ cells of mammalian testis [40]. The working temperature of testis is physiologically lower than the core body temperature, which is believed to favor spermatogenesis as well as CIRP expression. Several mechanisms have been proposed to explain the temperature-sensitive function of CIRP in spermatogenesis and testicular injury protection. Post-transcriptionally, CIRP binds and stabilizes mRNAs relating to male infertility in testis [72]. Experimentally increased scrotal temperature causes in testis and epididymis a reduced expression of CIRP and an increase in germ cell apoptosis [144]. In addition, CIRP reduces cryptorchidism-induced testicular damage by suppressing pro-apoptotic p53 and Fas [98]. Upon testicular torsion/detorsion, CIRP prevents testicular damages probably by decreasing oxidative stress and apoptosis in germ cells [145]. Furthermore, CIRP regulates key pathways or components in the cell cycle that affect spermatogenic functions. A comprehensive study has revealed that CIRP promotes the proliferation of undifferentiated spermatogonia by interacting with Dryk1b/Mirk [91]. In addition, when CIRP is downregulated, p44/p42, p38, and SAPK/JNK MAPK pathways are activated in germ cells and impair spermatogenesis [146]. In turtle, CIRP is involved in sex determination in a temperature-dependent manner [147] but with unclear results in the American alligator [148].
Oocytes and embryo cryopreservation are of great importance in reproductive medicine [149]. To freeze an egg or embryo, a novel flash-freeze process called vitrification has replaced the traditional slow-cooling method with more benefits [149]. During vitrification of oocytes, some genes, including CIRP, change their expression profiles and are believed to be involved in the protection against cold stress or crystallization [150, 151]. However, the involvement of CIRP in embryo cryopreservation is controversial. One study has demonstrated that CIRP is elevated in post-warming pro-nuclear stage embryos with higher expression levels in 8-cell stage embryos in the vitrification group [152]. A second group reported that no significant difference in CIRP expression was observed between vitrification and slow-freezing in 8-cell stage embryos [153]. CIRP is also associated with the enhancement of developmental competence of vitrified-warmed oocytes [66]. In addition, high CIRP expression level has been found in cryopreservation of reproductive organs, such as calf testis and sheep ovary [154, 155].
Regulation and functions of RBM3
Molecular regulation of RBM3
There is little information on the mechanisms regulating RBM3 expression. In alternative splicing, a long RBM3 isoform, annotated only in mouse, is more abundant in sleep deprived mice as compared with the commonly known RBM3 transcript in mammals and CIRP, which are both under-expressed during sleep deprivation, suggesting that RBM3 isoforms may play different roles in circadian oscillation in mice [156]. In mammalian neurons, an RBM3 splicing variant lacking a single arginine shows a higher dendritic localization, compared with the isoform having this specific arginine, which may be due to arginine methylation [48] (see paragraph ‘spatial distribution’).
In 2001, Chappell SA et al. discovered an IRES in the 5′-leader sequence of RBM3 [157, 158], which has been applied in viral studies or as a biotechnological tool until very recently [159–161]. However, the presumed RBM3 IRES was unmasked as a cloning artifact in which a cDNA for the Thoc1 gene on chromosome 18 had recombined with an Rbm3 cDNA [162].
Molecular and cellular activities of RBM3
Regulation of post-transcriptional and translational events
RBM3 can bind to and alter the translation of mRNA, as shown for COX-2, IL-8, and vascular endothelial growth factor (VEGF) in macrophages or cancer cells [65, 133], presumably in a cell type-specific manner [36] and involving the interaction with HuR [133]. In addition, RBM3 associates with the spliceosome and is involved in splicing [79]. In prostate cancer cells, RBM3 represses the variant v8–v10 of CD44 mRNA to inhibit stemness and tumorigenesis but enhances the standard spliced CD44 transcript [163]. In addition, RBM3 can modulate alternative polyadenylation in the same way as CIRP [78, 164].
RBM3 is known to modulate the translational process in several ways. In general, RBM3 enhances global protein translation [48, 165]. The underlying mechanisms of this enhancement include (1) binding to 60S ribosomal subunits in an RNA-independent manner; (2) increasing the formation of active polysomes; (3) dephosphorylating eukaryotic initiation factor 2 alpha (eIF2α); and (4) facilitating the phosphorylation of eukaryotic initiation factor 4E (eIF4E) [48, 165]. However, whether RBM3 can regulate translation via microRNAs remains controversial (see below). Moreover, our recent study revealed many ribosomal proteins associating with RBM3 [47], supporting its role in promoting translation. Whether this involvement of RBM3 in translational processes distinguishes fundamentally RBM3 from CIRP is unclear yet as CIRP has not been well-studied in translational processes.
Regulation of microRNA biogenesis
MicoRNAs (miRNAs) are small non-coding RNA molecules with approximately 21 nucleotides. These molecules are widely found in different organisms and regulate numerous developmental and cellular processes post-transcriptionally. Most miRNAs are generated in vivo from the canonical pathway: transcribed primary precursors (pri-miRNAs) are processed by the Drosha/DGCR8 complex into 70-nucleotide precursors (pre-miRNAs) in the nucleus; pre-miRNAs are further exported into cytoplasm and processed by Dicer complex to generate mature miRNA duplexes. One strand of the duplex is incorporated into RISC to guide the degradation of target mRNA [166]. RBM3 is considered to alter miRNA levels, thereby contributing to global protein translation, e.g., under hypothermia [165]. RBM3 binds to 70-nucleotide precursors and facilitates their processing by Dicer complex [167]. RBM3 appears to positively modulate the majority of miRNAs, and negatively modulate only a minor group of miRNAs. However, the finding that RBM3 upregulates most miRNAs is contradictory to the fact that RBM3 increases overall translation. In particular, mature miR-125b expression decreases when RBM3 is overexpressed, as shown by Dresios et al., but the opposite was demonstrated by Pilotte et al., making the RBM3-mediated modulation of specific miRNAs controversial [165, 167]. In addition, a recent study has demonstrated that reduced RBM3 level promotes the expression of a small subset of temperature-sensitive miRNAs, which targets immune genes and prevents pathological hyperthermia [168]. Therefore, RBM3 is believed to execute a regulatory function in miRNA expression, although its exact role remains largely unclear.
Signaling pathways
Stemness: Similar to CIRP, RBM3 is involved in the Wnt/β-catenin signaling pathway as well. In colorectal cancer cells, RBM3 induces stemness through a mechanism involving suppression of GSK3β kinase activity, thereby enhancing β-catenin signaling [169].
Cell cycle: Similar to CIRP, recent studies confirmed a positive role of RBM3 in promoting cell cycle progression. RBM3 modulates cell cycle in G2/M transition [133, 170], instead of G0/G1 and G1/S transitions for CIRP [91]. Specifically, knockdown of RBM3 in tumor cells increases caspase-mediated apoptosis coupled with nuclear cyclin B1, and phosphorylated Cdc25c, Chk1, and Chk2 kinases, implying that under conditions of RBM3 downregulation, cells undergo mitotic catastrophe [133]. Mouse embryonic fibroblasts from RBM3-deficient mice show markedly increased number of G2-phase cells [170], confirming that RBM3 is essential for cells to progress through mitosis. This may explain why tumors with high RBM3 expression show increased sensitivity to chemotherapy and, thus, are associated with better prognosis as compared with RBM3 low or even negative tumors [171].
Apoptosis: RBM3 inhibits staurosporine-induced apoptosis in neuron-like PC12 cells by repressing PARP cleavage [34]. The induction of Bcl-2 and suppression of caspase expression may also be involved in RBM3-mediated survival [47, 172].
ER stress: In the presence of ER stress, unfolded proteins accumulate in the ER lumen and activate unfolded protein response (UPR) to rescue cells. If ER stress exists continuously, UPR initiates the apoptotic program. PERK-eIF2α-CHOP signaling is one of the three main branches of UPR, and it plays the most important role in UPR-induced apoptosis [173]. Under sustained ER stress, RBM3 represses the phosphorylation of PERK and eIF2α, which leads to a decrease in CHOP expression and rescue cells from UPR-induced apoptosis [47]. Notably, although RBM3 is induced by hypothermia, hypothermia itself can activate UPR without inducing apoptosis [42]. Upon ischemia-induced prolonged ER stress, hypothermia has a protective effect by suppressing UPR to prevent apoptosis [174], in accordance with Zhu et al.’s report [47].
Biological functions and diseases
Brain disorders
Very recently, Peretti et al. revealed an important neuroprotective role of RBM3 in Alzheimer’s and prion disease models [175]. Notably, RBM3, but not CIRP, is significantly induced by hypothermia in mouse, preventing neuronal loss and restoring synapse reassembly [175]. Although the underlying mechanism is unknown, one hypothesis may involve the suppression of eIF2α kinase PERK, which has been shown to be a potential therapeutic target in Alzheimer’s disease-related deficits of synapse plasticity [176]. The activity of PERK has recently been linked to RBM3 [47]. However, the details of this mechanism remain to be elucidated.
In response to acute injuries to brain or spinal cord, RBM3 instantly changes its temporal and spatial distributions similar to CIRP. After spinal cord injury (SCI) in a rat model, the number of RBM3-positive cells increases with varying temporal dynamics reported [177, 178]. In one report, RBM3 expression peaked at 1 day after SCI [178], while in the other, RBM3 significantly increased 1 day post-SCI but did not reach the maximal expression level until 5 days post-SCI [177]. The spatial expression of RBM3 in these two reports is also inconsistent. Zhao et al. reported that mostly primary neurons and only a few astrocytes were positive for RBM3 expression under normal conditions, and that RBM3 was induced in both neurons and astrocytes [178]. In the other study, RBM3 is present in both neurons and astrocytes in the sham group, and only astrocytic RBM3 could respond to SCI-induced stress [177]. This discrepancy may result from the instable surgical conditions, although both reports generally support the hypothesis that RBM3 is inducible upon SCI and may exert important pathophysiological functions.
Until now, studies have supported the notion that RBM3 is a general neuroprotective effector, while CIRP can either protect neuronal cells or induce massive neuronal death by mediating neuroinflammation once released.
Circadian rhythm
RBM3 has been studied less frequently in circadian rhythm than CIRP. As described above, RBM3 modulates circadian oscillation by controlling alternative polyadenylation in cooperation with CIRP [78]. In patients suffering from neurological diseases with dysregulation of the sleep–wake cycle, such as bipolar disorders and cluster headache, RBM3 was the most significantly altered gene in peripheral lymphoblasts [179]. In mice, two alternatively spliced RBM3 isoforms may play different roles in sleep [156].
Immune response
In RBM3 knockout mice, no obvious change in cytokine expression has been found in a model of DNA-mediated innate immune response compared with wild-type [170]. Very recently, reduced RBM3 levels have been found to associate with reduced expression of some immune genes, such as IL-6 and TLR2, by modulating a subset of thermos-miRNAs [168]. To the best of our knowledge, no data published thus far suggest that RBM3 might be actively released as it has been demonstrated for CIRP [125].
Cancer
A large variety of immunohistochemical studies, including many tumor types, have shown consistently that loss of RBM3 expression is associated with clinically more aggressive tumors and an independent factor of poor prognosis.
Breast cancer is the leading cancer type in women, and RBM3 is overexpressed in this cancer [180], with a direct correlation of RBM3 expression level and improved clinical outcome [181]. In female genital organs, RBM3 correlates with favorable cisplatin sensitivity and good prognosis in epithelial ovarian cancer (EOC) [182], presumably via a mechanism by which RBM3 suppresses the poor prognostic markers MCM3, Chk1, and Chk2, which are all involved in DNA integrity and cell cycle [171].
In males, prostate cancer is one of the most common cancer types. Very similar to breast cancer and EOC, a high level of RBM3, as an independent biomarker in prostate cancer, predicts a low risk of disease progression and recurrence [183]. Interestingly, high RBM3 expression is found in poorly differentiated prostate tumor [184], whereas experimental downregulation of RBM3 in prostate cancer cells attenuates cell survival and enhances chemosensitivity in vitro [135]. These observations are consistent with the role of RBM3 in promoting cell proliferation and survival but cannot explain the favorable prognosis, indicating the involvement of other mechanisms. One study suggests that the activation of ERG and the depletion of PTEN may contribute to RBM3-mediated good prognosis in radically operated prostate cancer [185]. Another study indicates that RBM3 attenuates the stemness and tumorigenesis of prostate cancer cells by inhibiting CD44 variant splicing [163], although this finding is in contrast to findings in colorectal cancer [169]. Specific cancer types likely differentially affect RBM3 signaling as is known for other proteins, such as estrogen receptors [186]. Moreover, low RBM3 in testicular non-seminomatous germ cell cancer correlates with high risk of treatment failure [187]. Reduced RBM3 levels in urothelial bladder cancer are associated with tumor progression and poor prognosis [188], while high RBM3 expression correlates with lower stage tumors and decreased risk of lymphovascular invasion [189].
Colorectal cancer is the most common type of cancer of the digestive system, and a high level of RBM3 expression is associated with improved prognosis [190], whereas a loss of RBM3 expression is associated with poor prognosis and right-sided localization [191]. Therefore, RBM3 has been proposed as a potential prognostic biomarker, especially in young patients [192].
In the upper digestive system, RBM3 is downregulated in HPV-negative oropharyngeal squamous cell carcinoma compared with normal oral mucosa [193]. In esophageal and gastric adenocarcinoma, high nuclear expression of RBM3 is beneficial, and it correlates with intestinal metaplasia-associated tumors and predicts low risk of recurrence and death independently [194].
Furthermore, low RBM3 expression in other cancer types is associated with poor survival as well. RBM3 is expressed in malignant melanoma [195], and low expression of RBM3 is associated with tumor progression and poor prognosis [196]. High MCM3 expression is observed with a reduced RBM3 level similar to epithelial ovarian cancer [197]. The only exception published thus far is astrocytoma, where higher expression of RBM3 is associated with a higher grade and may promote astrocytic carcinogenesis [198].
A pivotal feature of most tumors is hypoxia [199]. The hypoxic cancer stem cell niche provides a microenvironment for the maintenance of immature cancer cells [200], and moderates hypoxia triggers the induction of RBM3 [54]. In fact, experiments in colorectal cancers have shown that RBM3 regulates Wnt/β-catenin signaling to induce the stemness of cancer cells [169], in contrast to findings in prostate cancer cells [163]. However, the clinical data gathered on RBM3 in various tumor types (Table 2) with RBM3 as a marker of better outcome point toward a more complex network of RBM3 interaction than considered so far. Various specific cell types likely affect RBM3 baseline expression and functions differentially; the cell environment also has an important impact on RBM3.
In conclusion, whereas both CIRP and RBM3 show common characteristics of proto-oncogenes at the cellular level, their roles in the clinical tumor setting are diverse, with CIRP as a marker of poor prognosis and RBM3 as a marker of good prognosis.
Reproduction
Distinct from CIRP, RBM3 is predominantly expressed in Sertoli cells of mammalian testis, rather than in germ cells [41]. Sertoli cells are “nurse” cells to nourish germ cells in the process of spermatogenesis, indicating that RBM3 may only play a supportive role, which is different from a direct role of CIRP. In male rats, the expression level of RBM3 in the sexually dimorphic nuclei of the preoptic area (SDN-POA) neurons is almost twofold as that in female rats, although it declines by 50 % upon NMDA receptor inactivation. In contrast, the expression level of RBM3 in females is unaffected by an NMDA receptor inhibitor, indicating a dose-dependent mechanism related to X-chromosome inactivation [201].
Similar to CIRP, RBM3 is also involved in the vitrification of oocytes [202] and embryos [152, 153], as well as in the cryopreservation of genital organs [154], indicating overlapping activities of RBM3 and CIRP in cold-mediated cell and tissue protection.
Other functions
As RBM3 is involved in a variety of transcriptional and translational events, it is not surprising that it also exerts functions in viral infection. Together with hnRNP A2, RBM3 can interact with vaccinia viral proteins and contribute to the late transcription of virus [203, 204]. More details remain to be determined for RBM3-mediated viral infection and a potential relation to immune response.
In a model of muscle atrophy, rats with hindlimb suspension show higher expression levels of RBM3 in soleus muscles [205]. In particular, RBM3 is involved in the regulation of skeletal muscle size and the prevention of muscle loss, indicating a novel vital function of RBM3 in muscle disease [205]. A subsequent study has further revealed that RBM3 inhibits both necrosis and apoptosis in muscle myoblasts, consistent with a general cytoprotective function of RBM3 [172]. Moreover, RBM3 is suggested to mediate hypothermia-induced overexpression of bone protein alkaline phosphatase and osteocalcin [206]. These studies open new avenues to the understanding of multiple functions of RBM3.
Conclusion and outlook
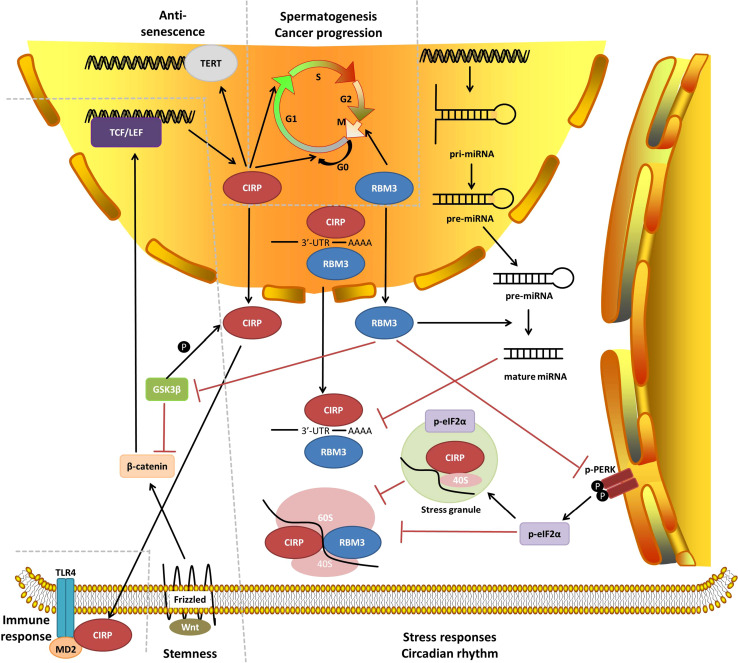
In this review, we comprehensively summarized the biological activities of CIRP and RBM3 by elucidating the upstream and downstream molecular and cellular aspects, and highlighting their relationships to various physiological and pathological processes in vivo (Fig. 2).
Fig. 2.
Molecular network of CIRP and RBM3 functions. Key molecular and cellular functions are briefly illustrated, and their relationships to physiological and pathological functions are indicated
When studying the various research papers on CIRP and RBM3, a particularly striking observation was the variety of diverse cellular functions in which CIRP and RBM3 are involved. In contrast, the upstream mechanisms regulating CIRP and RBM3 appear to be rather similar, including cellular stressors, such as hypothermia and hypoxia. CIRP and RBM3 are unique cellular tools that are present in many cell types that can be activated by various cellular stressors and that are applied by the cells depending on the specific cellular context, namely, the specific presence of other stress response molecules.
Despite the many similarities between both CIRP and RBM3 proteins, particularly regarding evolutionary conservation, sequence homology, expression, and inducibility, their biological functions are distinct. Whereas both proteins are generally upregulated in cancer tissues compared with normal tissue, RBM3 has been identified unanimously as a biomarker for favorable outcome; CIRP for poor outcome. One reason might be the fact that RBM3 facilitates mitosis and increases chemo sensitivity of tumor cells. A second reason for this disparity in cancer progression might be related to the ability of secretion. CIRP has been identified as an important mediator upon severe inflammation or ischemia with specific detrimental functions aggravating cell damage, whereas RBM3 has not been identified extracellularly thus far.
Future research on CIRP and RBM3 in mammals will benefit from reviewing the findings made so far in plants on their homologues AtGRP7 and AtGRP8. And a key for therapeutic implications of CIRP and RBM3 in the future is the ability to translate the current knowledge on the effects of CIRP and RBM3 into specific therapeutic approaches that target either the two proteins directly or the signaling pathways in which they are involved.
Acknowledgments
This work is supported by the Swiss National Science Foundation (SNSF, 31003A_163305). The authors declare no conflicts of interest.
References
- 1.Carey HV, Andrews MT, Martin SL. Mammalian hibernation: cellular and molecular responses to depressed metabolism and low temperature. Physiol Rev. 2003;83(4):1153–1181. doi: 10.1152/physrev.00008.2003. [DOI] [PubMed] [Google Scholar]
- 2.Milsom WK, Jackson DC. Hibernation and gas exchange. Compr Physiol. 2011;1(1):397–420. doi: 10.1002/cphy.c090018. [DOI] [PubMed] [Google Scholar]
- 3.Storey KB. Out cold: biochemical regulation of mammalian hibernation—a mini-review. Gerontology. 2010;56(2):220–230. doi: 10.1159/000228829. [DOI] [PubMed] [Google Scholar]
- 4.Williams DR, Epperson LE, Li W, Hughes MA, Taylor R, Rogers J, Martin SL, Cossins AR, Gracey AY. Seasonally hibernating phenotype assessed through transcript screening. Physiol Genomics. 2005;24(1):13–22. doi: 10.1152/physiolgenomics.00301.2004. [DOI] [PubMed] [Google Scholar]
- 5.Sano Y, Shiina T, Naitou K, Nakamori H, Shimizu Y. Hibernation-specific alternative splicing of the mRNA encoding cold-inducible RNA-binding protein in the hearts of hamsters. Biochem Biophys Res Commun. 2015;462(4):322–325. doi: 10.1016/j.bbrc.2015.04.135. [DOI] [PubMed] [Google Scholar]
- 6.Committee on F, Newborn. Papile LA, Baley JE, Benitz W, Cummings J, Carlo WA, Eichenwald E, Kumar P, Polin RA, Tan RC, Wang KS. Hypothermia and neonatal encephalopathy. Pediatrics. 2014;133(6):1146–1150. doi: 10.1542/peds.2014-0899. [DOI] [PubMed] [Google Scholar]
- 7.Yenari MA, Han HS. Neuroprotective mechanisms of hypothermia in brain ischaemia. Nat Rev Neurosci. 2012;13(4):267–278. doi: 10.1038/nrn3174. [DOI] [PubMed] [Google Scholar]
- 8.Lampe JW, Becker LB. State of the art in therapeutic hypothermia. Annu Rev Med. 2011;62:79–93. doi: 10.1146/annurev-med-052009-150512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tong G, Endersfelder S, Rosenthal LM, Wollersheim S, Sauer IM, Buhrer C, Berger F, Schmitt KR. Effects of moderate and deep hypothermia on RNA-binding proteins RBM3 and CIRP expressions in murine hippocampal brain slices. Brain Res. 2013;1504:74–84. doi: 10.1016/j.brainres.2013.01.041. [DOI] [PubMed] [Google Scholar]
- 10.Choi HA, Badjatia N, Mayer SA. Hypothermia for acute brain injury—mechanisms and practical aspects. Nat Rev Neurol. 2012;8(4):214–222. doi: 10.1038/nrneurol.2012.21. [DOI] [PubMed] [Google Scholar]
- 11.Lleonart ME. A new generation of proto-oncogenes: cold-inducible RNA binding proteins. Biochim Biophys Acta. 2010;1805(1):43–52. doi: 10.1016/j.bbcan.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 12.Nishiyama H, Higashitsuji H, Yokoi H, Itoh K, Danno S, Matsuda T, Fujita J. Cloning and characterization of human CIRP (cold-inducible RNA-binding protein) cDNA and chromosomal assignment of the gene. Gene. 1997;204(1–2):115–120. doi: 10.1016/S0378-1119(97)00530-1. [DOI] [PubMed] [Google Scholar]
- 13.Derry JM, Kerns JA, Francke U. RBM3, a novel human gene in Xp11.23 with a putative RNA-binding domain. Hum Mol Genet. 1995;4(12):2307–2311. doi: 10.1093/hmg/4.12.2307. [DOI] [PubMed] [Google Scholar]
- 14.Ciuzan O, Hancock J, Pamfil D, Wilson I, Ladomery M. The evolutionarily conserved multifunctional glycine-rich RNA-binding proteins play key roles in development and stress adaptation. Physiol Plant. 2015;153(1):1–11. doi: 10.1111/ppl.12286. [DOI] [PubMed] [Google Scholar]
- 15.Horn G, Hofweber R, Kremer W, Kalbitzer HR. Structure and function of bacterial cold shock proteins. Cell Mol Life Sci CMLS. 2007;64(12):1457–1470. doi: 10.1007/s00018-007-6388-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mangeon A, Junqueira RM, Sachetto-Martins G. Functional diversity of the plant glycine-rich proteins superfamily. Plant Signal Behav. 2010;5(2):99–104. doi: 10.4161/psb.5.2.10336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kim JS, Park SJ, Kwak KJ, Kim YO, Kim JY, Song J, Jang B, Jung CH, Kang H. Cold shock domain proteins and glycine-rich RNA-binding proteins from Arabidopsis thaliana can promote the cold adaptation process in Escherichia coli . Nucleic Acids Res. 2007;35(2):506–516. doi: 10.1093/nar/gkl1076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cao S, Jiang L, Song S, Jing R, Xu G. AtGRP7 is involved in the regulation of abscisic acid and stress responses in Arabidopsis . Cell Mol Biol Lett. 2006;11(4):526–535. doi: 10.2478/s11658-006-0042-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yang DH, Kwak KJ, Kim MK, Park SJ, Yang KY, Kang H. Expression of Arabidopsis glycine-rich RNA-binding protein AtGRP2 or AtGRP7 improves grain yield of rice (Oryza sativa) under drought stress conditions. Plant Sci. 2014;214:106–112. doi: 10.1016/j.plantsci.2013.10.006. [DOI] [PubMed] [Google Scholar]
- 20.Streitner C, Hennig L, Korneli C, Staiger D. Global transcript profiling of transgenic plants constitutively overexpressing the RNA-binding protein AtGRP7. BMC Plant Biol. 2010;10:221. doi: 10.1186/1471-2229-10-221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Streitner C, Koster T, Simpson CG, Shaw P, Danisman S, Brown JW, Staiger D. An hnRNP-like RNA-binding protein affects alternative splicing by in vivo interaction with transcripts in Arabidopsis thaliana . Nucleic Acids Res. 2012;40(22):11240–11255. doi: 10.1093/nar/gks873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Koster T, Meyer K, Weinholdt C, Smith LM, Lummer M, Speth C, Grosse I, Weigel D, Staiger D. Regulation of pri-miRNA processing by the hnRNP-like protein AtGRP7 in Arabidopsis . Nucleic Acids Res. 2014;42(15):9925–9936. doi: 10.1093/nar/gku716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lohr B, Streitner C, Steffen A, Lange T, Staiger D. A glycine-rich RNA-binding protein affects gibberellin biosynthesis in Arabidopsis . Mol Biol Rep. 2014;41(1):439–445. doi: 10.1007/s11033-013-2878-7. [DOI] [PubMed] [Google Scholar]
- 24.Heintzen C, Nater M, Apel K, Staiger D. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana . Proc Natl Acad Sci USA. 1997;94(16):8515–8520. doi: 10.1073/pnas.94.16.8515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Fu ZQ, Guo M, Jeong BR, Tian F, Elthon TE, Cerny RL, Staiger D, Alfano JR. A type III effector ADP-ribosylates RNA-binding proteins and quells plant immunity. Nature. 2007;447(7142):284–288. doi: 10.1038/nature05737. [DOI] [PubMed] [Google Scholar]
- 26.Lee HJ, Kim JS, Yoo SJ, Kang EY, Han SH, Yang KY, Kim YC, McSpadden Gardener B, Kang H. Different roles of glycine-rich RNA-binding protein7 in plant defense against Pectobacterium carotovorum, Botrytis cinerea, and tobacco mosaic viruses. Plant Physiol Biochem. 2012;60:46–52. doi: 10.1016/j.plaphy.2012.07.020. [DOI] [PubMed] [Google Scholar]
- 27.Pan F, Zarate J, Choudhury A, Rupprecht R, Bradley TM. Osmotic stress of salmon stimulates upregulation of a cold inducible RNA binding protein (CIRP) similar to that of mammals and amphibians. Biochimie. 2004;86(7):451–461. doi: 10.1016/j.biochi.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 28.Hsu CY, Chiu YC. Ambient temperature influences aging in an annual fish (Nothobranchius rachovii) Aging Cell. 2009;8(6):726–737. doi: 10.1111/j.1474-9726.2009.00525.x. [DOI] [PubMed] [Google Scholar]
- 29.Verleih M, Borchel A, Krasnov A, Rebl A, Korytar T, Kuhn C, Goldammer T. Impact of thermal stress on kidney-specific gene expression in farmed regional and imported rainbow trout. Mar Biotechnol. 2015;17(5):576–592. doi: 10.1007/s10126-015-9640-1. [DOI] [PubMed] [Google Scholar]
- 30.Uochi T, Asashima M. XCIRP (Xenopus homolog of cold-inducible RNA-binding protein) is expressed transiently in developing pronephros and neural tissue. Gene. 1998;211(2):245–250. doi: 10.1016/S0378-1119(98)00102-4. [DOI] [PubMed] [Google Scholar]
- 31.Peng Y, Kok KH, Xu RH, Kwok KH, Tay D, Fung PC, Kung HF, Lin MC. Maternal cold inducible RNA binding protein is required for embryonic kidney formation in Xenopus laevis . FEBS Lett. 2000;482(1–2):37–43. doi: 10.1016/S0014-5793(00)02019-6. [DOI] [PubMed] [Google Scholar]
- 32.Bhatia R, Dube DK, Gaur A, Robertson DR, Lemanski SL, McLean MD, Lemanski LF. Expression of axolotl RNA-binding protein during development of the Mexican axolotl. Cell Tissue Res. 1999;297(2):283–290. doi: 10.1007/s004410051356. [DOI] [PubMed] [Google Scholar]
- 33.Pilotte J, Cunningham BA, Edelman GM, Vanderklish PW. Developmentally regulated expression of the cold-inducible RNA-binding motif protein 3 in euthermic rat brain. Brain Res. 2009;1258:12–24. doi: 10.1016/j.brainres.2008.12.050. [DOI] [PubMed] [Google Scholar]
- 34.Chip S, Zelmer A, Ogunshola OO, Felderhoff-Mueser U, Nitsch C, Buhrer C, Wellmann S. The RNA-binding protein RBM3 is involved in hypothermia induced neuroprotection. Neurobiol Dis. 2011;43(2):388–396. doi: 10.1016/j.nbd.2011.04.010. [DOI] [PubMed] [Google Scholar]
- 35.Danno S, Nishiyama H, Higashitsuji H, Yokoi H, Xue JH, Itoh K, Matsuda T, Fujita J. Increased transcript level of RBM3, a member of the glycine-rich RNA-binding protein family, in human cells in response to cold stress. Biochem Biophys Res Commun. 1997;236(3):804–807. doi: 10.1006/bbrc.1997.7059. [DOI] [PubMed] [Google Scholar]
- 36.Wellmann S, Truss M, Bruder E, Tornillo L, Zelmer A, Seeger K, Buhrer C. The RNA-binding protein RBM3 is required for cell proliferation and protects against serum deprivation-induced cell death. Pediatr Res. 2010;67(1):35–41. doi: 10.1203/PDR.0b013e3181c13326. [DOI] [PubMed] [Google Scholar]
- 37.Fedorov VB, Goropashnaya AV, Toien O, Stewart NC, Chang C, Wang H, Yan J, Showe LC, Showe MK, Barnes BM. Modulation of gene expression in heart and liver of hibernating black bears (Ursus americanus) BMC Genom. 2011;12:171. doi: 10.1186/1471-2164-12-171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fedorov VB, Goropashnaya AV, Toien O, Stewart NC, Gracey AY, Chang C, Qin S, Pertea G, Quackenbush J, Showe LC, Showe MK, Boyer BB, Barnes BM. Elevated expression of protein biosynthesis genes in liver and muscle of hibernating black bears (Ursus americanus) Physiol Genomics. 2009;37(2):108–118. doi: 10.1152/physiolgenomics.90398.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang X, Che H, Zhang W, Wang J, Ke T, Cao R, Meng S, Li D, Weiming O, Chen J, Luo W. Effects of mild chronic intermittent cold exposure on rat organs. Int J Biol Sci. 2015;11(10):1171–1180. doi: 10.7150/ijbs.12161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nishiyama H, Danno S, Kaneko Y, Itoh K, Yokoi H, Fukumoto M, Okuno H, Millan JL, Matsuda T, Yoshida O, Fujita J. Decreased expression of cold-inducible RNA-binding protein (CIRP) in male germ cells at elevated temperature. Am J Pathol. 1998;152(1):289–296. [PMC free article] [PubMed] [Google Scholar]
- 41.Danno S, Itoh K, Matsuda T, Fujita J. Decreased expression of mouse Rbm3, a cold-shock protein, in Sertoli cells of cryptorchid testis. Am J Pathol. 2000;156(5):1685–1692. doi: 10.1016/S0002-9440(10)65039-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Rzechorzek NM, Connick P, Patani R, Selvaraj BT, Chandran S. Hypothermic preconditioning of human cortical neurons requires proteostatic priming. EBioMedicine. 2015;2(6):528–535. doi: 10.1016/j.ebiom.2015.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Thandapani P, O’Connor TR, Bailey TL, Richard S. Defining the RGG/RG motif. Mol Cell. 2013;50(5):613–623. doi: 10.1016/j.molcel.2013.05.021. [DOI] [PubMed] [Google Scholar]
- 44.Matsumoto K, Aoki K, Dohmae N, Takio K, Tsujimoto M. CIRP2, a major cytoplasmic RNA-binding protein in Xenopus oocytes. Nucleic Acids Res. 2000;28(23):4689–4697. doi: 10.1093/nar/28.23.4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aoki K, Ishii Y, Matsumoto K, Tsujimoto M. Methylation of Xenopus CIRP2 regulates its arginine- and glycine-rich region-mediated nucleocytoplasmic distribution. Nucleic Acids Res. 2002;30(23):5182–5192. doi: 10.1093/nar/gkf638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.De Leeuw F, Zhang T, Wauquier C, Huez G, Kruys V, Gueydan C. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp Cell Res. 2007;313(20):4130–4144. doi: 10.1016/j.yexcr.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 47.Zhu X, Zelmer A, Kapfhammer JP, Wellmann S. Cold-inducible RBM3 inhibits PERK phosphorylation through cooperation with NF90 to protect cells from endoplasmic reticulum stress. FASEB J. 2015 doi: 10.1096/fj.15-274639. [DOI] [PubMed] [Google Scholar]
- 48.Smart F, Aschrafi A, Atkins A, Owens GC, Pilotte J, Cunningham BA, Vanderklish PW. Two isoforms of the cold-inducible mRNA-binding protein RBM3 localize to dendrites and promote translation. J Neurochem. 2007;101(5):1367–1379. doi: 10.1111/j.1471-4159.2007.04521.x. [DOI] [PubMed] [Google Scholar]
- 49.Nishiyama H, Itoh K, Kaneko Y, Kishishita M, Yoshida O, Fujita J. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J Cell Biol. 1997;137(4):899–908. doi: 10.1083/jcb.137.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jackson TC, Manole MD, Kotermanski SE, Jackson EK, Clark RS, Kochanek PM. Cold stress protein RBM3 responds to temperature change in an ultra-sensitive manner in young neurons. Neuroscience. 2015;305:268–278. doi: 10.1016/j.neuroscience.2015.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Neutelings T, Lambert CA, Nusgens BV, Colige AC. Effects of mild cold shock (25 degrees C) followed by warming up at 37 degrees C on the cellular stress response. PLoS One. 2013;8(7):e69687. doi: 10.1371/journal.pone.0069687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sharp FR, Bernaudin M. HIF1 and oxygen sensing in the brain. Nat Rev Neurosci. 2004;5(6):437–448. doi: 10.1038/nrn1408. [DOI] [PubMed] [Google Scholar]
- 53.Harris AL. Hypoxia—a key regulatory factor in tumour growth. Nat Rev Cancer. 2002;2(1):38–47. doi: 10.1038/nrc704. [DOI] [PubMed] [Google Scholar]
- 54.Wellmann S, Buhrer C, Moderegger E, Zelmer A, Kirschner R, Koehne P, Fujita J, Seeger K. Oxygen-regulated expression of the RNA-binding proteins RBM3 and CIRP by a HIF-1-independent mechanism. J Cell Sci. 2004;117(Pt 9):1785–1794. doi: 10.1242/jcs.01026. [DOI] [PubMed] [Google Scholar]
- 55.Zhang Q, Wang YZ, Zhang W, Chen X, Wang J, Chen J, Luo W. Involvement of cold inducible RNA-binding protein in severe hypoxia-induced growth arrest of neural stem cells in vitro. Mol Neurobiol. 2016 doi: 10.1007/s12035-016-9761-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xue JH, Nonoguchi K, Fukumoto M, Sato T, Nishiyama H, Higashitsuji H, Itoh K, Fujita J. Effects of ischemia and H2O2 on the cold stress protein CIRP expression in rat neuronal cells. Free Radic Biol Med. 1999;27(11–12):1238–1244. doi: 10.1016/S0891-5849(99)00158-6. [DOI] [PubMed] [Google Scholar]
- 57.Li S, Zhang Z, Xue J, Liu A, Zhang H. Cold-inducible RNA binding protein inhibits H(2)O(2)-induced apoptosis in rat cortical neurons. Brain Res. 2012;1441:47–52. doi: 10.1016/j.brainres.2011.12.053. [DOI] [PubMed] [Google Scholar]
- 58.Trollmann R, Rehrauer H, Schneider C, Krischke G, Huemmler N, Keller S, Rascher W, Gassmann M. Late-gestational systemic hypoxia leads to a similar early gene response in mouse placenta and developing brain. Am J Physiol Regul Integr Comp Physiol. 2010;299(6):R1489–R1499. doi: 10.1152/ajpregu.00697.2009. [DOI] [PubMed] [Google Scholar]
- 59.Sheikh MS, Carrier F, Papathanasiou MA, Hollander MC, Zhan Q, Yu K, Fornace AJ., Jr Identification of several human homologs of hamster DNA damage-inducible transcripts. Cloning and characterization of a novel UV-inducible cDNA that codes for a putative RNA-binding protein. J Biol Chem. 1997;272(42):26720–26726. doi: 10.1074/jbc.272.42.26720. [DOI] [PubMed] [Google Scholar]
- 60.Haley B, Paunesku T, Protic M, Woloschak GE. Response of heterogeneous ribonuclear proteins (hnRNP) to ionising radiation and their involvement in DNA damage repair. Int J Radiat Biol. 2009;85(8):643–655. doi: 10.1080/09553000903009548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Baba T, Nishimura M, Kuwahara Y, Ueda N, Naitoh S, Kume M, Yamamoto Y, Fujita J, Funae Y, Fukumoto M. Analysis of gene and protein expression of cytochrome P450 and stress-associated molecules in rat liver after spaceflight. Pathol Int. 2008;58(9):589–595. doi: 10.1111/j.1440-1827.2008.02275.x. [DOI] [PubMed] [Google Scholar]
- 62.Lebsack TW, Fa V, Woods CC, Gruener R, Manziello AM, Pecaut MJ, Gridley DS, Stodieck LS, Ferguson VL, Deluca D. Microarray analysis of spaceflown murine thymus tissue reveals changes in gene expression regulating stress and glucocorticoid receptors. J Cell Biochem. 2010;110(2):372–381. doi: 10.1002/jcb.22547. [DOI] [PubMed] [Google Scholar]
- 63.Ryan JC, Morey JS, Ramsdell JS, Van Dolah FM. Acute phase gene expression in mice exposed to the marine neurotoxin domoic acid. Neuroscience. 2005;136(4):1121–1132. doi: 10.1016/j.neuroscience.2005.08.047. [DOI] [PubMed] [Google Scholar]
- 64.Prieto-Alamo MJ, Abril N, Osuna-Jimenez I, Pueyo C. Solea senegalensis genes responding to lipopolysaccharide and copper sulphate challenges: large-scale identification by suppression subtractive hybridization and absolute quantification of transcriptional profiles by real-time RT-PCR. Aquat Toxicol. 2009;91(4):312–319. doi: 10.1016/j.aquatox.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 65.Cok SJ, Acton SJ, Sexton AE, Morrison AR. Identification of RNA-binding proteins in RAW 264.7 cells that recognize a lipopolysaccharide-responsive element in the 3-untranslated region of the murine cyclooxygenase-2 mRNA. J Biol Chem. 2004;279(9):8196–8205. doi: 10.1074/jbc.M308475200. [DOI] [PubMed] [Google Scholar]
- 66.Pan Y, Cui Y, He H, Baloch AR, Fan J, Xu G, He J, Yang K, Li G, Yu S. Developmental competence of mature yak vitrified-warmed oocytes is enhanced by IGF-I via modulation of CIRP during in vitro maturation. Cryobiology. 2015 doi: 10.1016/j.cryobiol.2015.10.150. [DOI] [PubMed] [Google Scholar]
- 67.Laustriat D, Gide J, Barrault L, Chautard E, Benoit C, Auboeuf D, Boland A, Battail C, Artiguenave F, Deleuze JF, Benit P, Rustin P, Franc S, Charpentier G, Furling D, Bassez G, Nissan X, Martinat C, Peschanski M, Baghdoyan S. In vitro and in vivo modulation of alternative splicing by the biguanide metformin. Mol Ther Nucleic Acids. 2015;4:e262. doi: 10.1038/mtna.2015.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Al-Fageeh MB, Smales CM. Alternative promoters regulate cold inducible RNA-binding (CIRP) gene expression and enhance transgene expression in mammalian cells. Mol Biotechnol. 2013;54(2):238–249. doi: 10.1007/s12033-013-9649-5. [DOI] [PubMed] [Google Scholar]
- 69.Al-Fageeh MB, Smales CM. Cold-inducible RNA binding protein (CIRP) expression is modulated by alternative mRNAs. RNA. 2009;15(6):1164–1176. doi: 10.1261/rna.1179109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Sumitomo Y, Higashitsuji H, Higashitsuji H, Liu Y, Fujita T, Sakurai T, Candeias MM, Itoh K, Chiba T, Fujita J. Identification of a novel enhancer that binds Sp1 and contributes to induction of cold-inducible RNA-binding protein (cirp) expression in mammalian cells. BMC Biotechnol. 2012;12:72. doi: 10.1186/1472-6750-12-72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Morf J, Rey G, Schneider K, Stratmann M, Fujita J, Naef F, Schibler U. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science. 2012;338(6105):379–383. doi: 10.1126/science.1217726. [DOI] [PubMed] [Google Scholar]
- 72.Xia Z, Zheng X, Zheng H, Liu X, Yang Z, Wang X. Cold-inducible RNA-binding protein (CIRP) regulates target mRNA stabilization in the mouse testis. FEBS Lett. 2012;586(19):3299–3308. doi: 10.1016/j.febslet.2012.07.004. [DOI] [PubMed] [Google Scholar]
- 73.Schwerk J, Savan R. Translating the untranslated region. J Immunol. 2015;195(7):2963–2971. doi: 10.4049/jimmunol.1500756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yang C, Carrier F. The UV-inducible RNA-binding protein A18 (A18 hnRNP) plays a protective role in the genotoxic stress response. J Biol Chem. 2001;276(50):47277–47284. doi: 10.1074/jbc.M105396200. [DOI] [PubMed] [Google Scholar]
- 75.Yang R, Weber DJ, Carrier F. Post-transcriptional regulation of thioredoxin by the stress inducible heterogenous ribonucleoprotein A18. Nucleic Acids Res. 2006;34(4):1224–1236. doi: 10.1093/nar/gkj519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yang R, Zhan M, Nalabothula NR, Yang Q, Indig FE, Carrier F. Functional significance for a heterogenous ribonucleoprotein A18 signature RNA motif in the 3′-untranslated region of ataxia telangiectasia mutated and Rad3-related (ATR) transcript. J Biol Chem. 2010;285(12):8887–8893. doi: 10.1074/jbc.M109.013128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li J, Xie D, Huang J, Lv F, Shi D, Liu Y, Lin L, Geng L, Wu Y, Liang D, Chen YH. Cold-inducible RNA-binding protein regulates cardiac repolarization by targeting transient outward potassium channels. Circ Res. 2015;116(10):1655–1659. doi: 10.1161/CIRCRESAHA.116.306287. [DOI] [PubMed] [Google Scholar]
- 78.Liu Y, Hu W, Murakawa Y, Yin J, Wang G, Landthaler M, Yan J. Cold-induced RNA-binding proteins regulate circadian gene expression by controlling alternative polyadenylation. Sci Rep. 2013;3:2054. doi: 10.1038/srep02054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barbosa-Morais NL, Carmo-Fonseca M, Aparicio S. Systematic genome-wide annotation of spliceosomal proteins reveals differential gene family expansion. Genome Res. 2006;16(1):66–77. doi: 10.1101/gr.3936206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci CMLS. 2001;58(2):266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Aoki K, Matsumoto K, Tsujimoto M. Xenopus cold-inducible RNA-binding protein 2 interacts with ElrA, the Xenopus homolog of HuR, and inhibits deadenylation of specific mRNAs. J Biol Chem. 2003;278(48):48491–48497. doi: 10.1074/jbc.M308328200. [DOI] [PubMed] [Google Scholar]
- 82.Guo X, Wu Y, Hartley RS. Cold-inducible RNA-binding protein contributes to human antigen R and cyclin E1 deregulation in breast cancer. Mol Carcinog. 2010;49(2):130–140. doi: 10.1002/mc.20582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Goncalves Kde A, Bressan GC, Saito A, Morello LG, Zanchin NI, Kobarg J. Evidence for the association of the human regulatory protein Ki-1/57 with the translational machinery. FEBS Lett. 2011;585(16):2556–2560. doi: 10.1016/j.febslet.2011.07.010. [DOI] [PubMed] [Google Scholar]
- 84.Pirok EW, 3rd, Domowicz MS, Henry J, Wang Y, Santore M, Mueller MM, Schwartz NB. APBP-1, a DNA/RNA-binding protein, interacts with the chick aggrecan regulatory region. J Biol Chem. 2005;280(42):35606–35616. doi: 10.1074/jbc.M505380200. [DOI] [PubMed] [Google Scholar]
- 85.Tan HK, Lee MM, Yap MG, Wang DI. Overexpression of cold-inducible RNA-binding protein increases interferon-gamma production in Chinese-hamster ovary cells. Biotechnol Appl Biochem. 2008;49(Pt 4):247–257. doi: 10.1042/BA20070032. [DOI] [PubMed] [Google Scholar]
- 86.Angers S, Moon RT. Proximal events in Wnt signal transduction. Nat Rev Mol Cell Biol. 2009;10(7):468–477. doi: 10.1038/nrn2674. [DOI] [PubMed] [Google Scholar]
- 87.van Venrooy S, Fichtner D, Kunz M, Wedlich D, Gradl D. Cold-inducible RNA binding protein (CIRP), a novel XTcf-3 specific target gene regulates neural development in Xenopus. BMC Dev Biol. 2008;8:77. doi: 10.1186/1471-213X-8-77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Liu GP, Zhang Y, Yao XQ, Zhang CE, Fang J, Wang Q, Wang JZ. Activation of glycogen synthase kinase-3 inhibits protein phosphatase-2A and the underlying mechanisms. Neurobiol Aging. 2008;29(9):1348–1358. doi: 10.1016/j.neurobiolaging.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 89.Peng Y, Yang PH, Tanner JA, Huang JD, Li M, Lee HF, Xu RH, Kung HF, Lin MC. Cold-inducible RNA binding protein is required for the expression of adhesion molecules and embryonic cell movement in Xenopus laevis . Biochem Biophys Res Commun. 2006;344(1):416–424. doi: 10.1016/j.bbrc.2006.03.086. [DOI] [PubMed] [Google Scholar]
- 90.Wu Y, Guo X, Brandt Y, Hathaway HJ, Hartley RS. Three-dimensional collagen represses cyclin E1 via beta1 integrin in invasive breast cancer cells. Breast Cancer Res Treat. 2011;127(2):397–406. doi: 10.1007/s10549-010-1013-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Masuda T, Itoh K, Higashitsuji H, Higashitsuji H, Nakazawa N, Sakurai T, Liu Y, Tokuchi H, Fujita T, Zhao Y, Nishiyama H, Tanaka T, Fukumoto M, Ikawa M, Okabe M, Fujita J. Cold-inducible RNA-binding protein (Cirp) interacts with Dyrk1b/Mirk and promotes proliferation of immature male germ cells in mice. Proc Natl Acad Sci USA. 2012;109(27):10885–10890. doi: 10.1073/pnas.1121524109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Liu J, Xue J, Zhang H, Li S, Liu Y, Xu D, Zou M, Zhang Z, Diao J. Cloning, expression, and purification of cold inducible RNA-binding protein and its neuroprotective mechanism of action. Brain Res. 2015;1597:189–195. doi: 10.1016/j.brainres.2014.11.061. [DOI] [PubMed] [Google Scholar]
- 93.Reed JC. Apoptosis-based therapies. Nat Rev Drug Discov. 2002;1(2):111–121. doi: 10.1038/nrd726. [DOI] [PubMed] [Google Scholar]
- 94.Sutherland LC, Rintala-Maki ND, White RD, Morin CD. RNA binding motif (RBM) proteins: a novel family of apoptosis modulators? J Cell Biochem. 2005;94(1):5–24. doi: 10.1002/jcb.20204. [DOI] [PubMed] [Google Scholar]
- 95.Saito K, Fukuda N, Matsumoto T, Iribe Y, Tsunemi A, Kazama T, Yoshida-Noro C, Hayashi N. Moderate low temperature preserves the stemness of neural stem cells and suppresses apoptosis of the cells via activation of the cold-inducible RNA binding protein. Brain Res. 2010;1358:20–29. doi: 10.1016/j.brainres.2010.08.048. [DOI] [PubMed] [Google Scholar]
- 96.Zhang HT, Xue JH, Zhang ZW, Kong HB, Liu AJ, Li SC, Xu DG. Cold-inducible RNA-binding protein inhibits neuron apoptosis through the suppression of mitochondrial apoptosis. Brain Res. 2015;1622:474–483. doi: 10.1016/j.brainres.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 97.Wu L, Sun HL, Gao Y, Hui KL, Xu MM, Zhong H, Duan ML. Therapeutic hypothermia enhances cold-inducible RNA-binding protein expression and inhibits mitochondrial apoptosis in a rat model of cardiac arrest. Mol Neurobiol. 2016 doi: 10.1007/s12035-016-9813-6. [DOI] [PubMed] [Google Scholar]
- 98.Zhou KW, Zheng XM, Yang ZW, Zhang L, Chen HD. Overexpression of CIRP may reduce testicular damage induced by cryptorchidism. Clin Invest Med. 2009;32(2):E103–E111. doi: 10.25011/cim.v32i2.6027. [DOI] [PubMed] [Google Scholar]
- 99.Lee HN, Ahn SM, Jang HH. Cold-inducible RNA-binding protein, CIRP, inhibits DNA damage-induced apoptosis by regulating p53. Biochem Biophys Res Commun. 2015;464(3):916–921. doi: 10.1016/j.bbrc.2015.07.066. [DOI] [PubMed] [Google Scholar]
- 100.Sakurai T, Itoh K, Higashitsuji H, Nonoguchi K, Liu Y, Watanabe H, Nakano T, Fukumoto M, Chiba T, Fujita J. Cirp protects against tumor necrosis factor-alpha-induced apoptosis via activation of extracellular signal-regulated kinase. Biochim Biophys Acta. 2006;1763(3):290–295. doi: 10.1016/j.bbamcr.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 101.Artero-Castro A, Callejas FB, Castellvi J, Kondoh H, Carnero A, Fernandez-Marcos PJ, Serrano M, Ramon y Cajal S, Lleonart ME. Cold-inducible RNA-binding protein bypasses replicative senescence in primary cells through extracellular signal-regulated kinase 1 and 2 activation. Mol Cell Biol. 2009;29(7):1855–1868. doi: 10.1128/MCB.01386-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Jian F, Chen Y, Ning G, Fu W, Tang H, Chen X, Zhao Y, Zheng L, Pan S, Wang W, Bian L, Sun Q. Cold inducible RNA binding protein upregulation in pituitary corticotroph adenoma induces corticotroph cell proliferation via Erk signaling pathway. Oncotarget. 2016 doi: 10.18632/oncotarget.7037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Dejardin J, Kingston RE. Purification of proteins associated with specific genomic loci. Cell. 2009;136(1):175–186. doi: 10.1016/j.cell.2008.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Zhang Y, Wu Y, Mao P, Li F, Han X, Zhang Y, Jiang S, Chen Y, Huang J, Liu D, Zhao Y, Ma W, Songyang Z. Cold-inducible RNA-binding protein CIRP/hnRNP A18 regulates telomerase activity in a temperature-dependent manner. Nucleic Acids Res. 2016;44(2):761–775. doi: 10.1093/nar/gkv1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Alkabie S, Boileau AJ. The role of therapeutic hypothermia after traumatic spinal cord injury-a systematic review. World Neurosurg. 2015 doi: 10.1016/j.wneu.2015.09.079. [DOI] [PubMed] [Google Scholar]
- 106.Salerian AJ, Saleri NG. Cooling core body temperature may slow down neurodegeneration. CNS Spectr. 2008;13(3):227–229. doi: 10.1017/S1092852900028479. [DOI] [PubMed] [Google Scholar]
- 107.Kita H, Carmichael J, Swartz J, Muro S, Wyttenbach A, Matsubara K, Rubinsztein DC, Kato K. Modulation of polyglutamine-induced cell death by genes identified by expression profiling. Hum Mol Genet. 2002;11(19):2279–2287. doi: 10.1093/hmg/11.19.2279. [DOI] [PubMed] [Google Scholar]
- 108.Liu A, Zhang Z, Li A, Xue J. Effects of hypothermia and cerebral ischemia on cold-inducible RNA-binding protein mRNA expression in rat brain. Brain Res. 2010;1347:104–110. doi: 10.1016/j.brainres.2010.05.029. [DOI] [PubMed] [Google Scholar]
- 109.Sanderson TH, Reynolds CA, Kumar R, Przyklenk K, Huttemann M. Molecular mechanisms of ischemia-reperfusion injury in brain: pivotal role of the mitochondrial membrane potential in reactive oxygen species generation. Mol Neurobiol. 2013;47(1):9–23. doi: 10.1007/s12035-012-8344-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zhou M, Yang WL, Ji Y, Qiang X, Wang P. Cold-inducible RNA-binding protein mediates neuroinflammation in cerebral ischemia. Biochim Biophys Acta. 1840;7:2253–2261. doi: 10.1016/j.bbagen.2014.02.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rajayer SR, Jacob A, Yang WL, Zhou M, Chaung W, Wang P. Cold-inducible RNA-binding protein is an important mediator of alcohol-induced brain inflammation. PLoS One. 2013;8(11):e79430. doi: 10.1371/journal.pone.0079430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Schmal C, Reimann P, Staiger D. A circadian clock-regulated toggle switch explains AtGRP7 and AtGRP8 oscillations in Arabidopsis thaliana . PLoS Comput Biol. 2013;9(3):e1002986. doi: 10.1371/journal.pcbi.1002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Nishiyama H, Xue JH, Sato T, Fukuyama H, Mizuno N, Houtani T, Sugimoto T, Fujita J. Diurnal change of the cold-inducible RNA-binding protein (Cirp) expression in mouse brain. Biochem Biophys Res Commun. 1998;245(2):534–538. doi: 10.1006/bbrc.1998.8482. [DOI] [PubMed] [Google Scholar]
- 114.Sugimoto K, Jiang H. Cold stress and light signals induce the expression of cold-inducible RNA binding protein (cirp) in the brain and eye of the Japanese treefrog (Hyla japonica) Comp Biochem Physiol A Mol Integr Physiol. 2008;151(4):628–636. doi: 10.1016/j.cbpa.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 115.Bellesi M, de Vivo L, Tononi G, Cirelli C. Effects of sleep and wake on astrocytes: clues from molecular and ultrastructural studies. BMC Biol. 2015;13:66. doi: 10.1186/s12915-015-0176-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Morf J, Schibler U. Body temperature cycles: gatekeepers of circadian clocks. Cell Cycle. 2013;12(4):539–540. doi: 10.4161/cc.23670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Mohawk JA, Green CB, Takahashi JS. Central and peripheral circadian clocks in mammals. Annu Rev Neurosci. 2012;35:445–462. doi: 10.1146/annurev-neuro-060909-153128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gerber A, Saini C, Curie T, Emmenegger Y, Rando G, Gosselin P, Gotic I, Gos P, Franken P, Schibler U. The systemic control of circadian gene expression. Diabetes Obes Metab. 2015;17(Suppl 1):23–32. doi: 10.1111/dom.12512. [DOI] [PubMed] [Google Scholar]
- 119.Froy O. The relationship between nutrition and circadian rhythms in mammals. Front Neuroendocrinol. 2007;28(2–3):61–71. doi: 10.1016/j.yfrne.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 120.Oishi K, Yamamoto S, Uchida D, Doi R (2013) Ketogenic diet and fasting induce the expression of cold-inducible RNA-binding protein with time-dependent hypothermia in the mouse liver. FEBS Open Biol 3:192–195. doi:10.1016/j.fob.2013.03.0050 [DOI] [PMC free article] [PubMed]
- 121.Li XM, Delaunay F, Dulong S, Claustrat B, Zampera S, Fujii Y, Teboul M, Beau J, Levi F. Cancer inhibition through circadian reprogramming of tumor transcriptome with meal timing. Cancer Res. 2010;70(8):3351–3360. doi: 10.1158/0008-5472.CAN-09-4235. [DOI] [PubMed] [Google Scholar]
- 122.Dodds PN, Rathjen JP. Plant immunity: towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11(8):539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 123.Rajamuthiah R, Mylonakis E. Effector triggered immunity. Virulence. 2014;5(7):697–702. doi: 10.4161/viru.29091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Nicaise V, Joe A, Jeong BR, Korneli C, Boutrot F, Westedt I, Staiger D, Alfano JR, Zipfel C. Pseudomonas HopU1 modulates plant immune receptor levels by blocking the interaction of their mRNAs with GRP7. EMBO J. 2013;32(5):701–712. doi: 10.1038/emboj.2013.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Qiang X, Yang WL, Wu R, Zhou M, Jacob A, Dong W, Kuncewitch M, Ji Y, Yang H, Wang H, Fujita J, Nicastro J, Coppa GF, Tracey KJ, Wang P. Cold-inducible RNA-binding protein (CIRP) triggers inflammatory responses in hemorrhagic shock and sepsis. Nat Med. 2013;19(11):1489–1495. doi: 10.1038/nm.3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Lopez M, Meier D, Muller A, Franken P, Fujita J, Fontana A. Tumor necrosis factor and transforming growth factor beta regulate clock genes by controlling the expression of the cold inducible RNA-binding protein (CIRBP) J Biol Chem. 2014;289(5):2736–2744. doi: 10.1074/jbc.M113.508200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sakurai T, Kudo M, Watanabe T, Itoh K, Higashitsuji H, Arizumi T, Inoue T, Hagiwara S, Ueshima K, Nishida N, Fukumoto M, Fujita J. Hypothermia protects against fulminant hepatitis in mice by reducing reactive oxygen species production. Dig Dis. 2013;31(5–6):440–446. doi: 10.1159/000355242. [DOI] [PubMed] [Google Scholar]
- 128.Godwin A, Yang WL, Sharma A, Khader A, Wang Z, Zhang F, Nicastro J, Coppa GF, Wang P. Blocking cold-inducible RNA-binding protein protects liver from ischemia-reperfusion injury. Shock. 2015;43(1):24–30. doi: 10.1097/SHK.0000000000000251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Li G, Yang L, Yuan H, Liu Y, He Y, Wu X, Jin X. Cold-inducible RNA-binding protein plays a central role in the pathogenesis of abdominal aortic aneurysm in a murine experimental model. Surgery. 2016 doi: 10.1016/j.surg.2016.01.007. [DOI] [PubMed] [Google Scholar]
- 130.Zhou Y, Dong H, Zhong Y, Huang J, Lv J, Li J. The cold-inducible RNA-binding protein (CIRP) level in peripheral blood predicts sepsis outcome. PLoS One. 2015;10(9):e0137721. doi: 10.1371/journal.pone.0137721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Idrovo JP, Jacob A, Yang WL, Wang Z, Yen HT, Nicastro J, Coppa GF, Wang P. A deficiency in cold-inducible RNA-binding protein accelerates the inflammation phase and improves wound healing. Int J Mol Med. 2016;37(2):423–428. doi: 10.3892/ijmm.2016.2451. [DOI] [PubMed] [Google Scholar]
- 132.Kizil C, Kyritsis N, Brand M. Effects of inflammation on stem cells: together they strive? EMBO Rep. 2015;16(4):416–426. doi: 10.15252/embr.201439702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Sureban SM, Ramalingam S, Natarajan G, May R, Subramaniam D, Bishnupuri KS, Morrison AR, Dieckgraefe BK, Brackett DJ, Postier RG, Houchen CW, Anant S. Translation regulatory factor RBM3 is a proto-oncogene that prevents mitotic catastrophe. Oncogene. 2008;27(33):4544–4556. doi: 10.1038/onc.2008.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Tang C, Wang Y, Lan D, Feng X, Zhu X, Nie P, Yue H. Analysis of gene expression profiles reveals the regulatory network of cold-inducible RNA-binding protein mediating the growth of BHK-21 cells. Cell Biol Int. 2015;39(6):678–689. doi: 10.1002/cbin.10438. [DOI] [PubMed] [Google Scholar]
- 135.Zeng Y, Kulkarni P, Inoue T, Getzenberg RH. Down-regulating cold shock protein genes impairs cancer cell survival and enhances chemosensitivity. J Cell Biochem. 2009;107(1):179–188. doi: 10.1002/jcb.22114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.He H, Altomare D, Ozer U, Xu H, Creek K, Chen H, Xu P. Cancer cell-selective killing polymer/copper combination. Biomaterials science. 2015 doi: 10.1039/c5bm00325c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Sakurai T, Yada N, Watanabe T, Arizumi T, Hagiwara S, Ueshima K, Nishida N, Fujita J, Kudo M. Cold-inducible RNA-binding protein promotes the development of liver cancer. Cancer Sci. 2015;106(4):352–358. doi: 10.1111/cas.12611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Sakurai T, Kashida H, Watanabe T, Hagiwara S, Mizushima T, Iijima H, Nishida N, Higashitsuji H, Fujita J, Kudo M. Stress response protein cirp links inflammation and tumorigenesis in colitis-associated cancer. Cancer Res. 2014;74(21):6119–6128. doi: 10.1158/0008-5472.CAN-14-0471. [DOI] [PubMed] [Google Scholar]
- 139.Wang M, Zhang H, Heng X, Pang Q, Sun A. Expression of cold-inducible RNA-binding protein (CIRP) in pituitary adenoma and its relationships with tumor recurrence. Med Sci Monit. 2015;21:1256–1260. doi: 10.12659/MSM.893128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ren WH, Zhang LM, Liu HQ, Gao L, Chen C, Qiang C, Wang XL, Liu CY, Li SM, Huang C, Qi H, Zhi KQ. Protein overexpression of CIRP and TLR4 in oral squamous cell carcinoma: an immunohistochemical and clinical correlation analysis. Med Oncol. 2014;31(8):120. doi: 10.1007/s12032-014-0120-7. [DOI] [PubMed] [Google Scholar]
- 141.Hamid AA, Mandai M, Fujita J, Nanbu K, Kariya M, Kusakari T, Fukuhara K, Fujii S. Expression of cold-inducible RNA-binding protein in the normal endometrium, endometrial hyperplasia, and endometrial carcinoma. Int J Gynecol Pathol. 2003;22(3):240–247. doi: 10.1097/01.PGP.0000070851.25718.EC. [DOI] [PubMed] [Google Scholar]
- 142.Mange A, Lacombe J, Bascoul-Mollevi C, Jarlier M, Lamy PJ, Rouanet P, Maudelonde T, Solassol J. Serum autoantibody signature of ductal carcinoma in situ progression to invasive breast cancer. Clin Cancer Res. 2012;18(7):1992–2000. doi: 10.1158/1078-0432.CCR-11-2527. [DOI] [PubMed] [Google Scholar]
- 143.Elinav E, Nowarski R, Thaiss CA, Hu B, Jin C, Flavell RA. Inflammation-induced cancer: crosstalk between tumours, immune cells and microorganisms. Nat Rev Cancer. 2013;13(11):759–771. doi: 10.1038/nrc3611. [DOI] [PubMed] [Google Scholar]
- 144.Banks S, King SA, Irvine DS, Saunders PT. Impact of a mild scrotal heat stress on DNA integrity in murine spermatozoa. Reproduction. 2005;129(4):505–514. doi: 10.1530/rep.1.00531. [DOI] [PubMed] [Google Scholar]
- 145.Xia Z, Jiang K, Liu T, Zheng H, Liu X, Zheng X. The protective effect of cold-inducible RNA-binding protein (CIRP) on testicular torsion/detorsion: an experimental study in mice. J Pediatr Surg. 2013;48(10):2140–2147. doi: 10.1016/j.jpedsurg.2013.02.065. [DOI] [PubMed] [Google Scholar]
- 146.Xia ZP, Zheng XM, Zheng H, Liu XJ, Liu GY, Wang XH. Downregulation of cold-inducible RNA-binding protein activates mitogen-activated protein kinases and impairs spermatogenic function in mouse testes. Asian J Androl. 2012;14(6):884–889. doi: 10.1038/aja.2012.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Schroeder AL, Metzger KJ, Miller A, Rhen T. A novel candidate gene for temperature-dependent sex determination in the common snapping turtle. Genetics. 2016 doi: 10.1534/genetics.115.182840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Kohno S, Katsu Y, Urushitani H, Ohta Y, Iguchi T, Guillette LJ., Jr Potential contributions of heat shock proteins to temperature-dependent sex determination in the American alligator. Sex Devel Genet Mol Biol Evolut Endocrinol Embryol Pathol Sex Determ Differ. 2010;4(1–2):73–87. doi: 10.1159/000260374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Edgar DH, Gook DA. A critical appraisal of cryopreservation (slow cooling versus vitrification) of human oocytes and embryos. Hum Reprod Update. 2012;18(5):536–554. doi: 10.1093/humupd/dms016. [DOI] [PubMed] [Google Scholar]
- 150.Wen Y, Zhao S, Chao L, Yu H, Song C, Shen Y, Chen H, Deng X. The protective role of antifreeze protein 3 on the structure and function of mature mouse oocytes in vitrification. Cryobiology. 2014;69(3):394–401. doi: 10.1016/j.cryobiol.2014.09.006. [DOI] [PubMed] [Google Scholar]
- 151.Jo JW, Lee JR, Jee BC, Suh CS, Kim SH. Exposing mouse oocytes to necrostatin 1 during in vitro maturation improves maturation, survival after vitrification, mitochondrial preservation, and developmental competence. Reprod Sci. 2015;22(5):615–625. doi: 10.1177/1933719114556482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Boonkusol D, Gal AB, Bodo S, Gorhony B, Kitiyanant Y, Dinnyes A. Gene expression profiles and in vitro development following vitrification of pronuclear and 8-cell stage mouse embryos. Mol Reprod Dev. 2006;73(6):700–708. doi: 10.1002/mrd.20450. [DOI] [PubMed] [Google Scholar]
- 153.Shin MR, Choi HW, Kim MK, Lee SH, Lee HS, Lim CK. In vitro development and gene expression of frozen-thawed 8-cell stage mouse embryos following slow freezing or vitrification. Clin Exp Reprod Med. 2011;38(4):203–209. doi: 10.5653/cerm.2011.38.4.203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Devi L, Makala H, Pothana L, Nirmalkar K, Goel S. Comparative efficacies of six different media for cryopreservation of immature buffalo (Bubalus bubalis) calf testis. Reprod Fertil Dev. 2014 doi: 10.1071/RD14171. [DOI] [PubMed] [Google Scholar]
- 155.Du T, Chao L, Zhao S, Chi L, Li D, Shen Y, Shi Q, Deng X. Successful cryopreservation of whole sheep ovary by using DMSO-free cryoprotectant. J Assist Reprod Genet. 2015;32(8):1267–1275. doi: 10.1007/s10815-015-0513-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wang H, Liu Y, Briesemann M, Yan J. Computational analysis of gene regulation in animal sleep deprivation. Physiol Genomics. 2010;42(3):427–436. doi: 10.1152/physiolgenomics.00205.2009. [DOI] [PubMed] [Google Scholar]
- 157.Chappell SA, Mauro VP. The internal ribosome entry site (IRES) contained within the RNA-binding motif protein 3 (Rbm3) mRNA is composed of functionally distinct elements. J Biol Chem. 2003;278(36):33793–33800. doi: 10.1074/jbc.M303495200. [DOI] [PubMed] [Google Scholar]
- 158.Chappell SA, Owens GC, Mauro VP. A 5′ leader of Rbm3, a cold stress-induced mRNA, mediates internal initiation of translation with increased efficiency under conditions of mild hypothermia. J Biol Chem. 2001;276(40):36917–36922. doi: 10.1074/jbc.M106008200. [DOI] [PubMed] [Google Scholar]
- 159.Fux C, Langer D, Kelm JM, Weber W, Fussenegger M. New-generation multicistronic expression platform: pTRIDENT vectors containing size-optimized IRES elements enable homing endonuclease-based cistron swapping into lentiviral expression vectors. Biotechnol Bioeng. 2004;86(2):174–187. doi: 10.1002/bit.20028. [DOI] [PubMed] [Google Scholar]
- 160.Wang Z, Wu L, Cheng X, Liu S, Li B, Li H, Kang F, Wang J, Xia H, Ping C, Nassal M, Sun D. Replication-competent infectious hepatitis B virus vectors carrying substantially sized transgenes by redesigned viral polymerase translation. PLoS One. 2013;8(4):e60306. doi: 10.1371/journal.pone.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Cheng X, Gao XC, Wang JP, Yang XY, Wang Y, Li BS, Kang FB, Li HJ, Nan YM, Sun DX. Tricistronic hepatitis C virus subgenomic replicon expressing double transgenes. World J Gastroenterol. 2014;20(48):18284–18295. doi: 10.3748/wjg.v20.i48.18284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Baranick BT, Lemp NA, Nagashima J, Hiraoka K, Kasahara N, Logg CR. Splicing mediates the activity of four putative cellular internal ribosome entry sites. Proc Natl Acad Sci USA. 2008;105(12):4733–4738. doi: 10.1073/pnas.0710650105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Zeng Y, Wodzenski D, Gao D, Shiraishi T, Terada N, Li Y, Vander Griend DJ, Luo J, Kong C, Getzenberg RH, Kulkarni P. Stress-response protein RBM3 attenuates the stem-like properties of prostate cancer cells by interfering with CD44 variant splicing. Cancer Res. 2013;73(13):4123–4133. doi: 10.1158/0008-5472.CAN-12-1343. [DOI] [PubMed] [Google Scholar]
- 164.Hu W, Liu Y, Yan J. Microarray meta-analysis of RNA-binding protein functions in alternative polyadenylation. PLoS One. 2014;9(3):e90774. doi: 10.1371/journal.pone.0090774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Dresios J, Aschrafi A, Owens GC, Vanderklish PW, Edelman GM, Mauro VP. Cold stress-induced protein Rbm3 binds 60S ribosomal subunits, alters microRNA levels, and enhances global protein synthesis. Proc Natl Acad Sci USA. 2005;102(6):1865–1870. doi: 10.1073/pnas.0409764102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Krol J, Loedige I, Filipowicz W. The widespread regulation of microRNA biogenesis, function and decay. Nat Rev Genet. 2010;11(9):597–610. doi: 10.1038/nrg2843. [DOI] [PubMed] [Google Scholar]
- 167.Pilotte J, Dupont-Versteegden EE, Vanderklish PW. Widespread regulation of miRNA biogenesis at the Dicer step by the cold-inducible RNA-binding protein, RBM3. PLoS One. 2011;6(12):e28446. doi: 10.1371/journal.pone.0028446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Wong JJ, Au AY, Gao D, Pinello N, Kwok CT, Thoeng A, Lau KA, Gordon JE, Schmitz U, Feng Y, Nguyen TV, Middleton R, Bailey CG, Holst J, Rasko JE, Ritchie W. RBM3 regulates temperature sensitive miR-142-5p and miR-143 (thermomiRs), which target immune genes and control fever. Nucleic Acids Res. 2016;44(6):2888–2897. doi: 10.1093/nar/gkw041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Venugopal A, Subramaniam D, Balmaceda J, Roy B, Dixon DA, Umar S, Weir SJ, Anant S. RNA binding protein RBM3 increases beta-catenin signaling to increase stem cell characteristics in colorectal cancer cells. Mol Carcinog. 2015 doi: 10.1002/mc.22404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Matsuda A, Ogawa M, Yanai H, Naka D, Goto A, Ao T, Tanno Y, Takeda K, Watanabe Y, Honda K, Taniguchi T. Generation of mice deficient in RNA-binding motif protein 3 (RBM3) and characterization of its role in innate immune responses and cell growth. Biochem Biophys Res Commun. 2011;411(1):7–13. doi: 10.1016/j.bbrc.2011.06.038. [DOI] [PubMed] [Google Scholar]
- 171.Ehlen A, Nodin B, Rexhepaj E, Brandstedt J, Uhlen M, Alvarado-Kristensson M, Ponten F, Brennan DJ, Jirstrom K. RBM3-regulated genes promote DNA integrity and affect clinical outcome in epithelial ovarian cancer. Transl Oncol. 2011;4(4):212–221. doi: 10.1593/tlo.11106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Ferry AL, Vanderklish PW, Dupont-Versteegden EE. Enhanced survival of skeletal muscle myoblasts in response to overexpression of cold shock protein RBM3. Am J Physiol Cell Physiol. 2011;301(2):C392–C402. doi: 10.1152/ajpcell.00098.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Lin JH, Li H, Yasumura D, Cohen HR, Zhang C, Panning B, Shokat KM, Lavail MM, Walter P. IRE1 signaling affects cell fate during the unfolded protein response. Science. 2007;318(5852):944–949. doi: 10.1126/science.1146361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Poone GK, Hasseldam H, Munkholm N, Rasmussen RS, Gronberg NV, Johansen FF. The hypothermic influence on CHOP and Ero1-alpha in an endoplasmic reticulum stress model of cerebral ischemia. Brain Sci. 2015;5(2):178–187. doi: 10.3390/brainsci5020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Peretti D, Bastide A, Radford H, Verity N, Molloy C, Martin MG, Moreno JA, Steinert JR, Smith T, Dinsdale D, Willis AE, Mallucci GR. RBM3 mediates structural plasticity and protective effects of cooling in neurodegeneration. Nature. 2015;518(7538):236–239. doi: 10.1038/nature14142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 176.Ma T, Trinh MA, Wexler AJ, Bourbon C, Gatti E, Pierre P, Cavener DR, Klann E. Suppression of eIF2alpha kinases alleviates Alzheimer’s disease-related plasticity and memory deficits. Nat Neurosci. 2013;16(9):1299–1305. doi: 10.1038/nn.3486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Cui Z, Zhang J, Bao G, Xu G, Sun Y, Wang L, Chen J, Jin H, Liu J, Yang L, Feng G, Li W. Spatiotemporal profile and essential role of RBM3 expression after spinal cord injury in adult rats. J Mol Neurosci. 2014;54(2):252–263. doi: 10.1007/s12031-014-0282-y. [DOI] [PubMed] [Google Scholar]
- 178.Zhao W, Xu D, Cai G, Zhu X, Qian M, Liu W, Cui Z. Spatiotemporal pattern of RNA-binding motif protein 3 expression after spinal cord injury in rats. Cell Mol Neurobiol. 2014;34(4):491–499. doi: 10.1007/s10571-014-0033-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Costa M, Squassina A, Piras IS, Pisanu C, Congiu D, Niola P, Angius A, Chillotti C, Ardau R, Severino G, Stochino E, Deidda A, Persico AM, Alda M, Del Zompo M. Preliminary transcriptome analysis in lymphoblasts from cluster headache and bipolar disorder patients implicates dysregulation of circadian and serotonergic genes. J Mol Neurosci. 2015;56(3):688–695. doi: 10.1007/s12031-015-0567-9. [DOI] [PubMed] [Google Scholar]
- 180.Martinez-Arribas F, Agudo D, Pollan M, Gomez-Esquer F, Diaz-Gil G, Lucas R, Schneider J. Positive correlation between the expression of X-chromosome RBM genes (RBMX, RBM3, RBM10) and the proapoptotic Bax gene in human breast cancer. J Cell Biochem. 2006;97(6):1275–1282. doi: 10.1002/jcb.20725. [DOI] [PubMed] [Google Scholar]
- 181.Jogi A, Brennan DJ, Ryden L, Magnusson K, Ferno M, Stal O, Borgquist S, Uhlen M, Landberg G, Pahlman S, Ponten F, Jirstrom K (2009) Nuclear expression of the RNA-binding protein RBM3 is associated with an improved clinical outcome in breast cancer. Mod Pathol 22(12):1564–1574. doi:10.1038/modpathol.2009.124 [DOI] [PubMed]
- 182.Ehlen A, Brennan DJ, Nodin B, O’Connor DP, Eberhard J, Alvarado-Kristensson M, Jeffrey IB, Manjer J, Brandstedt J, Uhlen M, Ponten F, Jirstrom K. Expression of the RNA-binding protein RBM3 is associated with a favourable prognosis and cisplatin sensitivity in epithelial ovarian cancer. J Transl Med. 2010;8:78. doi: 10.1186/1479-5876-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Jonsson L, Gaber A, Ulmert D, Uhlen M, Bjartell A, Jirstrom K. High RBM3 expression in prostate cancer independently predicts a reduced risk of biochemical recurrence and disease progression. Diagn Pathol. 2011;6:91. doi: 10.1186/1746-1596-6-91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Shaikhibrahim Z, Lindstrot A, Ochsenfahrt J, Fuchs K, Wernert N. Epigenetics-related genes in prostate cancer: expression profile in prostate cancer tissues, androgen-sensitive and -insensitive cell lines. Int J Mol Med. 2013;31(1):21–25. doi: 10.3892/ijmm.2012.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Grupp K, Wilking J, Prien K, Hube-Magg C, Sirma H, Simon R, Steurer S, Budaus L, Haese A, Izbicki J, Sauter G, Minner S, Schlomm T, Tsourlakis MC. High RNA-binding motif protein 3 expression is an independent prognostic marker in operated prostate cancer and tightly linked to ERG activation and PTEN deletions. Eur J Cancer. 2014;50(4):852–861. doi: 10.1016/j.ejca.2013.12.003. [DOI] [PubMed] [Google Scholar]
- 186.Thomas C, Gustafsson JA. The different roles of ER subtypes in cancer biology and therapy. Nat Rev Cancer. 2011;11(8):597–608. doi: 10.1038/nrc3093. [DOI] [PubMed] [Google Scholar]
- 187.Olofsson SE, Nodin B, Gaber A, Eberhard J, Uhlen M, Jirstrom K, Jerkeman M. Low RBM3 protein expression correlates with clinical stage, prognostic classification and increased risk of treatment failure in testicular non-seminomatous germ cell cancer. PLoS One. 2015;10(3):e0121300. doi: 10.1371/journal.pone.0121300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Boman K, Segersten U, Ahlgren G, Eberhard J, Uhlen M, Jirstrom K, Malmstrom PU. Decreased expression of RNA-binding motif protein 3 correlates with tumour progression and poor prognosis in urothelial bladder cancer. BMC Urol. 2013;13:17. doi: 10.1186/1471-2490-13-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Florianova L, Xu B, Traboulsi S, Elmansi H, Tanguay S, Aprikian A, Kassouf W, Brimo F. Evaluation of RNA-binding motif protein 3 expression in urothelial carcinoma of the bladder: an immunohistochemical study. World J Surg Oncol. 2015;13:317. doi: 10.1186/s12957-015-0730-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Hjelm B, Brennan DJ, Zendehrokh N, Eberhard J, Nodin B, Gaber A, Ponten F, Johannesson H, Smaragdi K, Frantz C, Hober S, Johnson LB, Pahlman S, Jirstrom K, Uhlen M. High nuclear RBM3 expression is associated with an improved prognosis in colorectal cancer. Proteom Clin Appl. 2011;5(11–12):624–635. doi: 10.1002/prca.201100020. [DOI] [PubMed] [Google Scholar]
- 191.Melling N, Simon R, Mirlacher M, Izbicki JR, Stahl P, Terracciano LM, Bokemeyer C, Sauter G, Marx AH (2015) Loss of RNA-binding motif protein 3 expression is associated with right-sided localization and poor prognosis in colorectal cancer. Histopathology. doi:10.1111/his.12726 [DOI] [PubMed]
- 192.Wang MJ, Ping J, Li Y, Adell G, Arbman G, Nodin B, Meng WJ, Zhang H, Yu YY, Wang C, Yang L, Zhou ZG, Sun XF. The prognostic factors and multiple biomarkers in young patients with colorectal cancer. Sci Rep. 2015;5:10645. doi: 10.1038/srep10645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Martinez I, Wang J, Hobson KF, Ferris RL, Khan SA. Identification of differentially expressed genes in HPV-positive and HPV-negative oropharyngeal squamous cell carcinomas. Eur J Cancer. 2007;43(2):415–432. doi: 10.1016/j.ejca.2006.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Jonsson L, Hedner C, Gaber A, Korkocic D, Nodin B, Uhlen M, Eberhard J, Jirstrom K. High expression of RNA-binding motif protein 3 in esophageal and gastric adenocarcinoma correlates with intestinal metaplasia-associated tumours and independently predicts a reduced risk of recurrence and death. Biomark Res. 2014;2:11. doi: 10.1186/2050-7771-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Baldi A, Battista T, De Luca A, Santini D, Rossiello L, Baldi F, Natali PG, Lombardi D, Picardo M, Felsani A, Paggi MG. Identification of genes down-regulated during melanoma progression: a cDNA array study. Exp Dermatol. 2003;12(2):213–218. doi: 10.1034/j.1600-0625.2003.00026.x. [DOI] [PubMed] [Google Scholar]
- 196.Jonsson L, Bergman J, Nodin B, Manjer J, Ponten F, Uhlen M, Jirstrom K. Low RBM3 protein expression correlates with tumour progression and poor prognosis in malignant melanoma: an analysis of 215 cases from the Malmo Diet and Cancer study. J Transl Med. 2011;9:114. doi: 10.1186/1479-5876-9-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197.Nodin B, Fridberg M, Jonsson L, Bergman J, Uhlen M, Jirstrom K. High MCM3 expression is an independent biomarker of poor prognosis and correlates with reduced RBM3 expression in a prospective cohort of malignant melanoma. Diagn Pathol. 2012;7:82. doi: 10.1186/1746-1596-7-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198.Zhang HT, Zhang ZW, Xue JH, Kong HB, Liu AJ, Li SC, Liu YX, Xu DG. Differential expression of the RNA-binding motif protein 3 in human astrocytoma. Chin Med J. 2013;126(10):1948–1952. [PubMed] [Google Scholar]
- 199.Wilson WR, Hay MP. Targeting hypoxia in cancer therapy. Nat Rev Cancer. 2011;11(6):393–410. doi: 10.1038/nrc3064. [DOI] [PubMed] [Google Scholar]
- 200.Kise K, Kinugasa-Katayama Y, Takakura N. Tumor microenvironment for cancer stem cells. Adv Drug Deliv Rev. 2015 doi: 10.1016/j.addr.2015.08.005. [DOI] [PubMed] [Google Scholar]
- 201.Hsu HK, Shao PL, Tsai KL, Shih HC, Lee TY, Hsu C. Gene regulation by NMDA receptor activation in the SDN-POA neurons of male rats during sexual development. J Mol Endocrinol. 2005;34(2):433–445. doi: 10.1677/jme.1.01601. [DOI] [PubMed] [Google Scholar]
- 202.Jo JW, Jee BC, Suh CS, Kim SH. The beneficial effects of antifreeze proteins in the vitrification of immature mouse oocytes. PLoS One. 2012;7(5):e37043. doi: 10.1371/journal.pone.0037043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Wright CF, Oswald BW, Dellis S. Vaccinia virus late transcription is activated in vitro by cellular heterogeneous nuclear ribonucleoproteins. J Biol Chem. 2001;276(44):40680–40686. doi: 10.1074/jbc.M102399200. [DOI] [PubMed] [Google Scholar]
- 204.Dellis S, Strickland KC, McCrary WJ, Patel A, Stocum E, Wright CF. Protein interactions among the vaccinia virus late transcription factors. Virology. 2004;329(2):328–336. doi: 10.1016/j.virol.2004.08.017. [DOI] [PubMed] [Google Scholar]
- 205.Dupont-Versteegden EE, Nagarajan R, Beggs ML, Bearden ED, Simpson PM, Peterson CA. Identification of cold-shock protein RBM3 as a possible regulator of skeletal muscle size through expression profiling. Am J Physiol Regul Integr Comp Physiol. 2008;295(4):R1263–R1273. doi: 10.1152/ajpregu.90455.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206.Aisha MD, Nor-Ashikin MN, Sharaniza AB, Nawawi HM, Kapitonova MY, Froemming GR. Short-term moderate hypothermia stimulates alkaline phosphatase activity and osteocalcin expression in osteoblasts by upregulating Runx2 and osterix in vitro. Exp Cell Res. 2014;326(1):46–56. doi: 10.1016/j.yexcr.2014.06.003. [DOI] [PubMed] [Google Scholar]