Abstract
Combining Lipinski’s rule with the docking and steered molecular dynamics simulations and using the PubChem data base of about 1.4 million compounds, we have obtained DNA dyes Hoechst 34580 and Hoechst 33342 as top-leads for the Alzheimer’s disease. The binding properties of these ligands to amyloid beta (Aβ) fibril were thoroughly studied by in silico and in vitro experiments. Hoechst 34580 and Hoechst 33342 prefer to locate near hydrophobic regions with binding affinity mainly governed by the van der Waals interaction. By the Thioflavin T assay, it was found that the inhibition constant IC50 ≈ 0.86 and 0.68 μM for Hoechst 34580 and Hoechst 33342, respectively. This result qualitatively agrees with the binding free energy estimated using the molecular mechanic-Poisson Boltzmann surface area method and all-atom simulations with the AMBER-f99SB-ILDN force field and water model TIP3P. In addition, DNA dyes have the high capability to cross the blood brain barrier. Thus, both in silico and in vitro experiments have shown that Hoechst 34580 and 33342 are good candidates for treating the Alzheimer’s disease by inhibiting Aβ formation.
Electronic supplementary material
The online version of this article (doi:10.1007/s10822-016-9932-1) contains supplementary material, which is available to authorized users.
Keywords: Alzheimer’s disease, DNA dyes, Hoechst 34580, Hoechst 33342, Drug design, Amyloid beta fibril
Introduction
Alzheimer’s disease (AD) is one of the most common forms of dementia [1]. Clinically it is defined as a progressive decline in memory, language and other cognitive functions. AD is the sixth-leading cause of death in the United States and total payments for patients with AD and other dementias are estimated at $226 billion in 2015 [2] posing huge burden to the society. Despite intense research during many decades, the problem of finding efficient drugs for AD remains challenging. Available drugs which are acetylcholinesterase inhibitors and N-methyl-D-aspartate (NMDA) receptor antagonists can treat some symptoms but not cure the disease.
In order to design potential drugs for a given disease one has to know the corresponding target but such a target for AD remains largely uncertain as the cause of AD has not been disclosed yet [3]. There are about twenty hypotheses concerning AD mechanisms [4], but recent experimental evidences strongly support the amyloid cascade hypothesis [5] positing that AD is associated with progressive intra-cerebral accumulation of beta amyloid (Aβ) peptides [6]. In addition, oligomers are presumably more toxic that mature fibrils [7, 8]. Because Aβ peptides are generated by the proteolytic cleavage of amyloid precursor protein (APP) by β- and γ-secretases, AD can be cured by either blocking activity of these secretases or preventing Aβ aggregation. In the latter case Aβ oligomers or fibrils become the drug target. Following this strategy a lot of potential Aβ inhibitors have been identified including short peptides [9], nutraceuticals [10–13], polyamines [14, 15], metal chelators [16], derivatives of vitamin K3 [17], RNA aptamers [18], osmolytes [19], and other compounds [15, 20, 21].
In the present paper, we have carried out the multi-step screening of Aβ aggregation inhibitors from data basic PubChem [22] (http://pubchem.ncbi.nlm.nih.gov) using the Lipinski’s rule [23, 24] in combination with the molecular docking and steered molecular dynamic (SMD) simulations. From predicted top-leads for Aβ40 and Aβ42 fibrils we succeeded to purchase DNA dyes Hoechst 34580 and Hoechst 33342 for in vitro experiment. Using the Thioflavin T (ThT) assay the inhibition constant IC50 was found to be equal 0.86 and 0.68 μM for Hoechst 34580 and Hoechst 33342, respectively. This result is consistent with our estimation of the absolute binding free energy by the molecular mechanic Poisson–Boltzmann surface area (MM-PBSA) [25] method which is more accurate than the docking method. In addition, the QSAR analysis revealed that both DNA dyes are capable to easily cross the blood brain barrier (BBB) implying that they are good candidates for AD treatment.
Materials and methods
Data base of ligands and receptors
Screening of drug candidates has been performed using about 1.4 million compounds from Collaborative Drug Discovery in PubChem [22] (see http://pubchem.ncbi.nlm.nih.gov). Concerning the target (receptor) we chose the structural model of Aβ40 and Aβ42 fibrils. For Aβ40 fibril, the model of truncated fragment Aβ9-40 which is available in the Protein Data Bank with PDB ID: 2LMN [26] with the 8 first disordered residues neglected. This structure has two layers each of which contains 6 strands numbered as A-F and G-L (Figure S1 in Supporting Information (SI)). A full molecular structural model for Aβ40 fibril is the three-fold-symmetric (PDB ID: 2M4J [27]) containing 9 chains (Figure S1). For Aβ42 fibril, the structure of truncated fragment Aβ17-42 (PDB ID: 2BEG_ENREF_25 [28]), which is a model built basically on mutagenesis and H/D exchange experiments, and the solid-state NMR structures of Aβ11-42 (PDB ID 2MXU [29]) were employed (Figure S1).
Lipinski’s rule
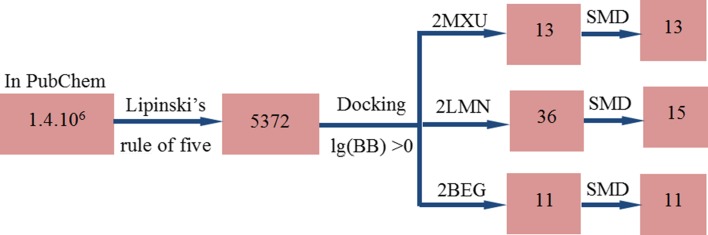
First, ligands were virtually screened by Lipinski’s rule of five [23]. It sets for drug-like properties [23, 24] as molecular weight from 0 to 500 Da, xlogP from 0 to 5, the number of donor hydrogen bonds is from 0 to 5, and the number of acceptor hydrogen bonds is from 0 to 10. The application of Lipinski’s rule reduced the whole set of about 1.4 million compounds to 5372 compounds (Fig. 1).
Fig. 1.
Multi-step screening procedure. From 1.4 million compounds we keep only 5372 compounds satisfying the Lipinski’s rule for drug-like ligands. The further screening by docking method and requirement that drug candidates should have the binding energy ΔEbind < −9.0 kcal/mol and lg(BB) > 0 give the set of 27 ligands for 2MXU, and binding energy ΔEbind < −10.0 kcal/mol and lg(BB) > 0 give the set of 36 ligands for 2LMN. Imposing that candidates should have ΔEbind < −8 kcal/mol and lg(BB) > 0 we obtained 11 ligands for 2BEG, respectively. Applying the SMD method to the set of 36 ligands we obtained 15 top leads for 2LMN and 13 top leads for 2MXU, while this method was just used for re-ranking 11 top leads for 2BEG
Docking method
Autodock Tool 1.5.4 [30] was used to prepare PDBQT file for docking ligands to targets 2LMN, 2BEG, 2MXU, and 2M4J. The docking simulation was performed using the Autodock Vina version 1.1 [31]. For global search, the exhaustiveness was set to 400 which is enough for obtaining reasonable results. Twenty binding modes have been generated starting from random configurations of ligand which had fully flexible torsion degrees of freedom. Because the binding site of Aβ fibrils was not a priori known to cover the whole fibril the boxes of grid dimensions 4.7 × 5.3 × 7.4 nm (2LMN), 2.9 × 4.6 × 25 nm (2BEG), 5.3 × 4.2 × 6.5 nm (2MXU) and 9.2 × 9.2 × 3.5 nm (2M4J) were chosen. The dynamics of receptor was neglected. The lowest binding energy ΔEbind obtained in the best docking mode was chosen as a scoring function for selecting top ligands. Here we selected only those ligands which have ΔEbind less than −10.0, −8.0 and −9.0 kcal/mol for targets 2LMN, 2BEG, and 2MXU, respectively.
Molecular dynamic (MD) simulation
To estimate the binding free energy by the MM-PBSA method, the molecular dynamics simulation was carried out with the AMBER-f99SB-ILDN force field [32] and water model TIP3P [33]. The rationale for our choice of AMBER-f99SB-ILDN is that this force field, as shown by previous work [9, 34–36], provided reasonable results on binding affinity of small molecules to amyloid fibrils. The GAFF force field [37] was used for parameterization of ligands Hoechst 33342 and Hoechst 34580. Restrained electrostatic potential [38] (RESP) point charges were assigned to ligand atoms by the Antechamber package [39] based on electrostatic potential (ESP) calculated by Gaussian09 package [40] at the B3LYP/6-31G* level. Names, types atoms, masses and charges of atoms used in the simulation for Hoechst 33342 and Hoechst 34580 are listed in Table S1 and S2 in SI.
The fibril-ligand complex was placed in the 9.3 × 9.3 × 9.3 (2LMN), 6.8 × 6.8 × 6.8 (2BEG), 7.6 × 7.6 × 7.6 (2MXU) and 11.1 × 11.1 × 11.1 nm3 (2M4J) cubic boxes containing about 78,500, 31,400, 41,448 and 136,200 water molecules with 1 nm distance between the box and solute. The van der Waals (vdW) forces were calculated with a cutoff of 1.4 nm, while the long-range electrostatic interaction was computed by the particle-mesh Ewald summation method [41]. Equations of motion were iterated by a leapfrog algorithm [42] with a time step 2 fs. The overall charge of the systems was set to zero by adding 12, 5, 8 and 27 Na+ ions to 2LMN, 2BEG, 2MXU and 2M4J, respectively. After minimization by the steepest descent method, the position-restrained MD simulations were performed for 500 ps to let water molecules to move into the active site. The equilibration was reached by coupling with temperature and pressure. Constant temperature 300 K was kept using Langevin dynamics with the collision frequency of 2.0 ps−1. The Berendsen barostat [43] was used to maintain the pressure at 1 atm and 300 K with the pressure relaxation time of 1.0 ps.
Steered molecular dynamics
The steered molecular dynamics (SMD) method was developed to study mechanical unfolding of biomolecules [44, 45] and ligand unbinding from receptor along a given direction [46]. Recently, it has been shown that this method is as accurate as the MM-PBSA method but computationally much less demanding [47, 48]. Because the predictive power of the docking method is limited the SMD method was employed to refine docking results as a next step in the multi-step screening procedure. Overall, an spring with spring constant k is attached to a dummy atom at one end and to the first heavy atom of ligand in the pulling direction at the another end. Moving along the pulling direction with a constant loading rate v the dummy atom experiences elastic force F = k(∆x − vt), where ∆x is the displacement of pulled atom from the starting position. We have chosen the spring constant k = 600 kJ/(mol nm2) which is a typical value for cantilever used in AFM experiment [49]. As in our previous works [48, 50, 51], the loading speed was set equal v = 5 nm/ns. This choice of parameters k and v was proved as reasonable for pulling experiment [51]. All Cα-atoms of receptor were restrained to keep the receptor almost at the same place but still maximally maintain its flexibility.
We determined possible pathways of ligands by using CAVER 3.0 [52], Pymol plugin, and chose the easiest path for ligand to exit from receptor as the pulling direction [50]. After equilibration, to completely pull the ligand out of the binding site, 500 ps SMD runs were carried out in NPT ensemble. To obtain reliable results five independent trajectories were performed with different random seeds. In the SMD method the maximum force Fmax in the force-extension/time profile was chosen as a score for binding affinity, i.e. the larger is Fmax, the stronger is the ligand binding.
MM-PBSA method
The MM-PBSA method [25] was used to estimate the binding free energy ΔGbind of DNA dyes to targets 12Aβ9-40 (2LMN), 9Aβ1-40 (2M4J), 5Aβ17-42 (2BEG), and 8Aβ11-42 (2MXU). More details on this method may be found elsewhere [53, 54]. Typically, ΔGbind is given by the following expression:
| 1 |
where ∆Eelec and ∆EvdW are contributions from electrostatic and vdW interactions, respectively. ∆Gsur and ∆GPB are nonpolar and polar solvation energies. The entropic contribution T∆S was estimated using the normal mode approximation. Snapshots collected in equilibrium and Eq. (1) were used to compute ΔGbind.
Blood brain barrier
One of the most important requirements for AD drug candidates is that they should be able to cross the blood brain barrier (BBB) [55] which is created by the brain capillary endothelium. The logarithm base 10 of the ratio of the ligands concentration in the brain to that in the blood, log(BB), is a measure of capability of a given ligand to pass BBB. This quantity is estimated by the QSAR (quantitative structure–activity relationship) approach [56]. The sever preADMET [57, 58] (see http://preadmet.bmdrc.org/) was used to calculate log (BB).
Measures used in data analysis
The backbone root mean square deviation (RMSD), computed using the Gromacs 5.1 package, was used to measure the deviation of structures of the receptor from its initial configuration. A hydrogen bond (HB) was formed provided the distance between donor D and acceptor A is less than 3.5 Å, the H-A distance is less than 2.7 Å and the D-H-A angle is larger than 135 degrees.
Aβ preparation
Aβ was prepared as described in previous literatures [10]. Briefly, Aβ42 peptide (Biopeptide, San Diego, CA) was dissolved in 50 % acetonitrile, divided into aliquots, lyophilized overnight, and stored at −80 °C. Before experiments, Aβ42 was dissolved in hexafluoroisopropanol (HFIP) in 1 mg/mL. The sample was mixed vigorously using a vortex for 5 s and then sonicated for 5 min. After quiescent for 1 h, the HFIP was evaporated in vacuum and Aβ peptides were dissolved by anhydrous dimethyl sulfoxide (DMSO) in 60 mg/mL and then diluted in 10 mM phosphate buffer, pH 7.4. The final Aβ concentration was 50 μM in ThT assays and 25 μM in fluorescence titration.
Compounds preparation
As seen below, Hoechst 34580 and Hoechst 33342 are among the top leads revealed by molecular simulation. They were purchased from Sigma-Aldrich (St. Louis, MD) and were used without purification. Compounds were dissolved respectively in DMSO at 10 mM as stocks. In ThT assays, the desired concentrations of the compounds were serial diluted in DMSO. In fluorescence titration assay, 10 mM compounds were diluted with 10 mM phosphate buffer (pH 7.4) to 5 mM and proceed for titration.
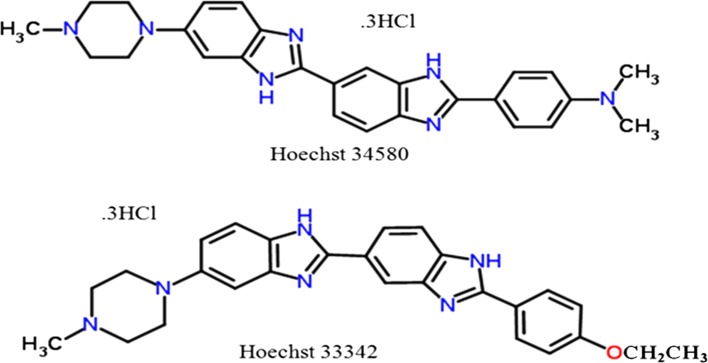
Structures of Hoechst 34580 and Hoechst 33342
In this study, Hoechst 34580 and Hoechst 33342 which are commercially available cell-permeable fluorescent dye for staining DNA and nuclei (Fig. 2). They are identical except the last fragment NCH3CH3 for Hoechst 34580 and OCH2CH3 for Hoechst 33342.
Fig. 2.
2D structures of Hoechst 34580 and Hoechst 33342
ThT assay
Aβ42 fibrillization was measured using a Thioflavin T (ThT) assay. The compounds, from 1.22 μM to 10 mM, were prepared in DMSO and 0.4 μl of each was added to 384 well black plate. Each concentration was prepared in independent triplicates and a solvent control was included. Aβ42 solution at 50 μM in 39.6 μl was prepared with addition of 5 μM ThT. The samples were incubated at 37 °C with agitation for 1 min every hour. ThT fluorescence was monitored using an ELISA microplate reader SpectraMax M5 (Molecular Devices, Sunnyvale, CA) at an excitation wavelength of 442 nm and an emission wavelength of 485 nm. Measurements from independent triplicate trials were averaged and the standard deviations were calculated.
Results and discussion
Simulation results
Docking results
After the first virtual screening step by Lipinski’s rule, the number of compounds is reduced to 5372 (Fig. 1). The Autodock Vina [31] method was then applied to dock this set to targets 2LMN, 2BEG, and 2MXU. We did not perform the similar docking simulation for the whole ligand set to 2M4J because both 2M4J and 2MXU are solid state NMR structures. However, the docking of DNA dyes was carried for the target 2M4J as well. The binding energies ΔEbind, obtained in the best docking modes for 5327 ligands, vary from −0.6 to −11.4 (2LMN), −1.2 to −8.8 (2BEG), and −1.4 to −11.9 kcal/mol (2MXU) (Figure S2 in SI). There are 96 compounds that have the binding energy lower than −10 kcal/mol for 2LMN, 55 compounds have ΔEbind < −8.0 kcal/mol for 2BEG and 57 compounds have ΔEbind < −9.0 kcal/mol for 2MXU. Locations of these compounds in fibrils are presented in Figure S3 in SI. As expected Aβ do not have well defined binding sites because ligands locate either inside or between two layers.
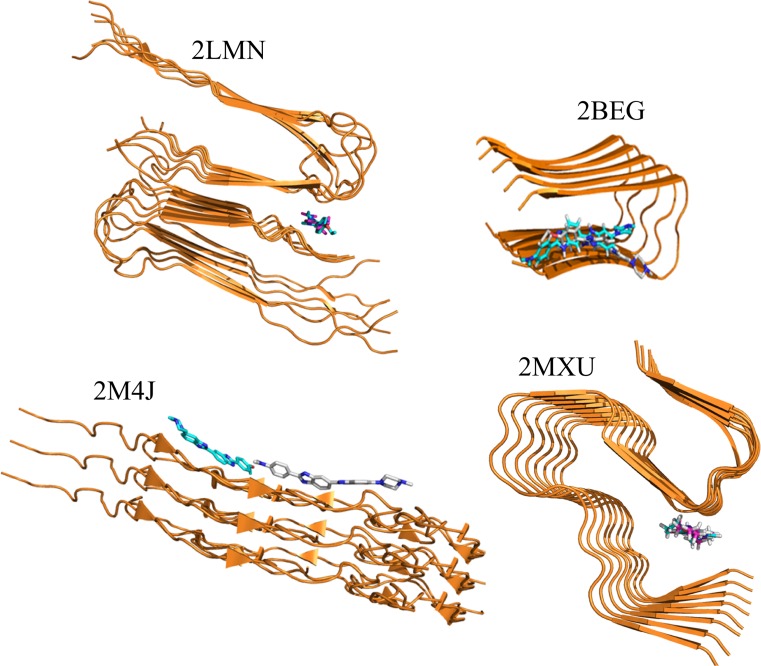
Because we succeeded to purchase DNA dyes Hoechst 34580 (CID: 448202) and Hoechst 33342 (CID: 1464) for in vitro experiment, we consider them in more detail. Their binding poses in 2LM, 2BEG, 2M4J and 2MXU are shown in Fig. 3. Except full length fibril 9Aβ1-40 (2M4J), derived from a human patient, Hoechst 34580 and Hoechst 33342 have nearly the same binding positions in the remaining targets. The DNA dyes are located between two layers near the turn region of 2LMN, inside 2MXU, partially inside 2BEG and outside 2M4J fibrils (Fig. 3). Moreover, for a given target their binding energies are very close to each other (Tables S3–S5). This also holds for 2M4J, where two ligands bind to different places but ΔEbind = −7.27 and −7.15 kcal/mol for Hoechst 34580 and Hoechst 33342, respectively. These results, as shown below, are consistent with the fact that they also have close rupture forces obtained by MD simulations and close binding free energies. The minor difference in their binding affinity is presumably due to high structural similarity (Fig. 2).
Fig. 3.
Binding poses of dyes Hoechst 34580 and Hoechst 33342 in 2LM, 2BEG, 2M4J and 2MXU. The structures were obtained by the docking method
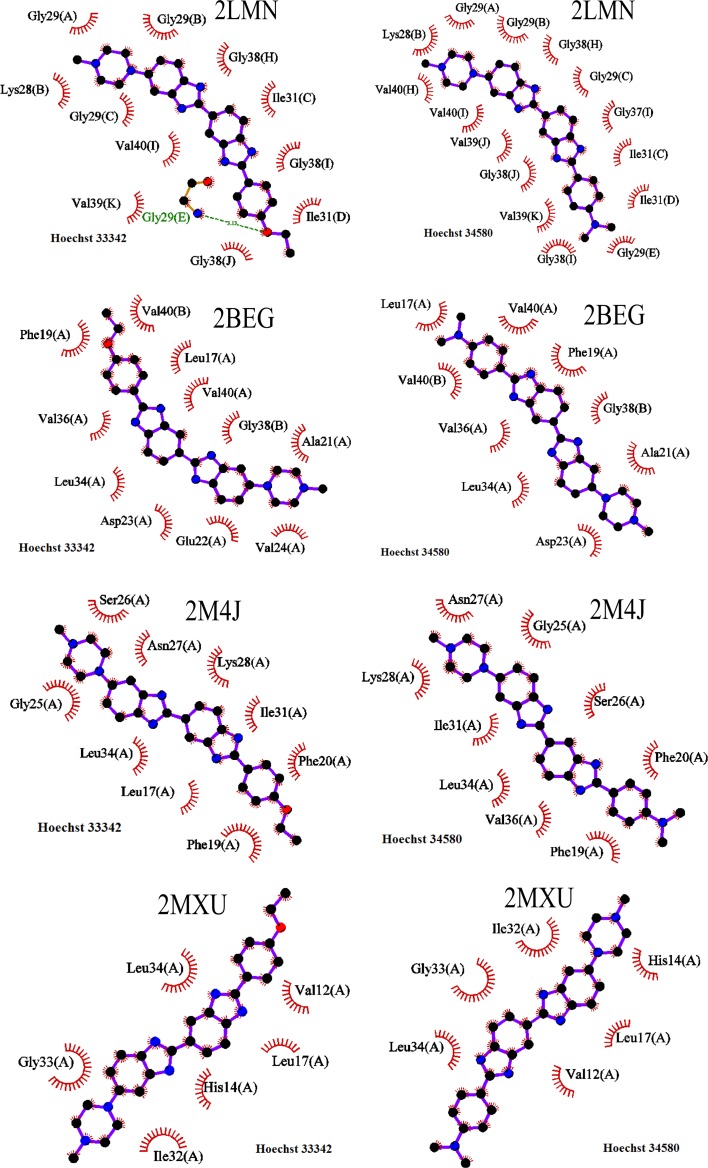
In docking, Hoechst 33342 forms one hydrogen bond (HB) with 2LMN, but none HB was found for 2BEG, 2MXU and 2M4J, while Hoechst 34580 does not have hydrogen binding with four targets (Fig. 4). Because the binding affinity of these compounds is high the poor HB networks indicate that the number HBs alone is not sufficient enough to describe the binding affinity of these complexes.
Fig. 4.
A HBs (green dashed line) and side chain non-bonded contacts (represented by an arc with spokes radiating towards the ligand atoms they contact) between four fibrils and DNA dyes Hoechst 34580 and Hoechst 33342. The plot was prepared using LigPlot + version 1.4.4 [57]
For 2LMN, Hoechst 33342 has 11 side chain (SC) contacts with residues Val39(K), Gly38(J), Ile31(D), Gly38(I), Val40(I), Gly29(C), Ile31(C), Gly38(H), Lys28(B), Gly29(B) and Gly29(A), whereas Hoechst 34580 forms 15 SC contacts with Val39(K), Gly29(E), Ile31(D), Gly38(I), Ile31(C), Gly37(I), Gly29(C), Gly38(H), Gly29(B), Gly29(A), Val40(H), Lys28(B), Val40(I), Val39(J) and Gly38(J) (Fig. 4). Here letters in parentheses refer to chains shown in Figure S1 in SI. In 2BEG, Hoechst 33342 has 11 SC contacts with residues Leu17(A), Gly38(B), Ala21(A), Asp23(A), Val24(A), Leu34(A), Glu22(A), Val36(A), Val40(A), Phe19(A), Val40(B) and Hoechst 34580 forms 9 contacts with residues Leu34(A), Asp23(A), Ala21(A), Val36(A), Phe19(A), Gly38(B), Val40(B), Val40(A), Leu17(A) (Fig. 4). Both DNA dyes have 9 SC contacts with 2M4J including the one with the charged residue Lys28(A) (Fig. 4). Because both ligands at the same place in 2MXU, they have 6 SC contacts with residues Val12(A), Leu17(A), His14(A), Gly33(A), Ile32(A), Leu34(A).
It should be noted that Hoechst 34580 and Hoechst 33342 prefer to stay next to hydrophobic residues of four targets (Fig. 4). In 2LMN and 2M4J they have only one contact with the positively charged residue Lys28 leading to the dominant role of the vdW interaction over the electrostatic interaction in stabilization of fibril-ligand complexes (see below).
Blood brain barrier
Using PreADME, we have calculated log(BB) for ligands revealed by the docking method as the top hits. Choosing only those ligands which have log(BB) > 0 one can further reduce the set to 36 ligands for 2LMN, 11 ligands for 2BEG and 13 ligands for 2MXU (Fig. 1). Hoechst 34580 and Hoechst 33342 are capable to easily cross BBB having log(BB) = 0.73 and 0.67, respectively.
Steered molecular dynamics
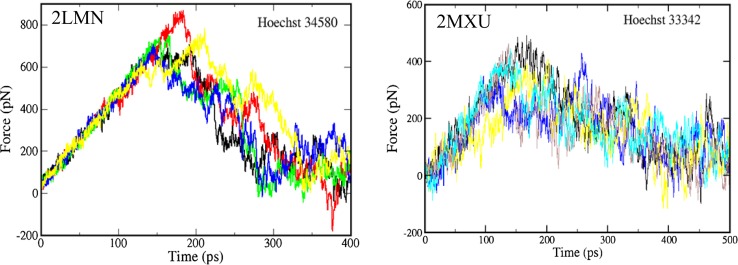
Using the Caver 3.0 [52] one can obtain several possible pulling directions but the easiest pathway with the lowest rupture force Fmax [50] was chosen. Two representative optimal directions are shown in Figure S4 for ligands inside receptor 2MXU and between layers of 2LMN. For each ligand five independent SMD runs were performed and the results were averaged over all trajectories. Typical force–time curves are presented in Fig. 5 showing the sensibility of rupture force on SMD runs.
Fig. 5.
Force-time profiles obtained by the SMD method in five independent trajectories for 2LMN-Hoechst 34580 and 2MXU- Hoechst 33342 complexes
For receptor 2MXU, the SMD method was applied to study the binding affinity of 13 top leads including two DNA dyes. The SMD and docking results are shown in Table S3 together with ligand structures. The ranking of binding affinities based on docking energies is different from that predicted by SMD. Hoechst 34580 is champion in SMD but it is fourth in docking. SMD predicts that among 13 top hits compound CID 447767 is the weakest binder having the lowest rupture force. Consistent with the docking results, Hoechst 34580 and Hoechst 33342 have nearly the same rupture force Fmax (Table S3).
For receptor 2LMN, keeping only those ligands which have Fmax exceeding 470 pN the set of 36 ligands was reduced to 15 compounds as the top leads for AD including Hoechst 34580 and Hoechst 33342 (Table S4). As expected the ranking by docking binding energy is different from the SMD one. Compound CID 5327177 which is strongest in docking becomes seventh in SMD, while CID 6083166 twelfth in docking is first by SMD. Having applied the SMD method to 11 top leads bound with 2BEG we obtained the results shown in Table S5. In SMD Hoechst 34580 and Hoechst 33342 are at positions 10 and 7, respectively.
As evident from Tables S3-S5, in addition to Hoechst 34580 and Hoechst 33342, compound CID 447767 tightly binds to three models of Aβ40 and Aβ42 fibril. Thus, we predict that 3 compounds can interfere with both Aβ40 and Aβ42 aggregation.
MM-PBSA results
To make a direct comparison with experiments, we used the MM-PBSA method to compute ΔGbind of Hoechst 34580 and Hoechst 33342 using Eq. (1). The conformations obtained in the best docking mode (Fig. 3) were used as starting conformations for all-atom MD simulation. For each fibril-ligand complex we performed four 100-150 ns MD runs starting from the same initial conformation but different random seed numbers. From the time dependence of Cα root mean square displacement (RMSD) of Aβ fibril, it is evident that all complexes reach equilibrium after about 40–100 ns (Figures S5 and S6 in SI). Snapshots stored every 20 ps in equilibrium were used to estimate the binding free energy given by Eq. (1).
For all studied targets, the vdW interaction dominates over the electrostatic interaction in directing ligand binding to Aβ because both ΔGelec and ΔGvdW are negative but the absolute value of ΔGvdW is larger than ΔGelec (Table 1). The reason behind this is that, as mentioned above, the DNA dyes locate rather close to hydrophobic residues than to the charged ones (Fig. 4). Due to geometrical similarity of Hoechst 34580 and Hoechst 33342 the entropic contributions are nearly the same for all complexes (Table 1). For a given target including 2BEG, 2M4J and 2MXU the binding free energies of two ligands are the same within error bars. The situation is different for 2LMN where Hoechst 34580 shows binding affinity higher than that of Hoechst 33342. The departure of this target from others is presumably caused by the fact that the DNA dyes are positioned next to the charged residue Lys28(B) (Fig. 4) in 2LMN but it is not the case for other targets where the ligands are surrounded by non-charged residues. This is also supported by the pronounced difference in electrostatic contributions of Hoechst 34580 and Hoechst 33342 to the binding propensity to 2LMN (Table 1).
Table 1.
Binding free energy (kcal/mol), obtained by MM-PBSA method, for Hoechst 34580 and Hoechst 33342 using the AMBER-f99SB-ILDN force field
| Receptor | Complex | ||||||
|---|---|---|---|---|---|---|---|
| 2LMN | Hoechst 34580 | −14.94 | −61.94 | 31.56 | −7.47 | −25.87 | −26.93 ± 1.89 |
| Hoechst 33342 | −8.28 | −58.66 | 32.47 | −5.60 | −23.30 | −16.77 ± 2.43 | |
| 2M4J | Hoechst 34580 | −10.66 | −41.19 | 23.26 | −4.56 | −23.07 | −10.07 ± 6.24 |
| Hoechst 33342 | −8.00 | −36.33 | 14.19 | −5.76 | −21.81 | −14.08 ± 2.27 | |
| 2BEG | Hoechst 34580 | −3.89 | −49.78 | 15.51 | −7.11 | −21.61 | −23.66 ± 4.47 |
| Hoechst 33342 | −10.92 | −51.31 | 22.68 | −5.43 | −22.91 | −22.08 ± 5.73 | |
| 2MXU | Hoechst 34580 | −4.65 | −65.32 | 29.06 | −5.89 | −23.21 | −23.59 ± 1.12 |
| Hoechst 33342 | −4.51 | −52.23 | 21.43 | −8.38 | −23.11 | −20.57 ± 4.37 |
Results were averaged over 4 MD trajectories
The difference in binding free energies of two DNA dyes to 2M4J was also seen (Table 1) due to contact with positively charged residue Lys28, but it is not as pronounced as in 2LMN because the ligands are positioned outside fibril (Fig. 3). Finally, Hoechst 34580 and Hoechst 33342 show the lowest binding affinity to 2M4J (Table 1) presumably because they are not located inside fibril. However, this is valid for a single fibril. There is also a possibility that DNA dyes interfere with the association between fibrils before acting at the individual fibril level. Then the interaction with 2M4J may get enhanced because they can be considered as located inside fibrils. This issue calls for further investigation.
In agreement with the docking simulations, SMD and experimental results (see below), within the error bars ΔGbind of both dyes are the same for Aβ42 fibril. Having ΔGbind < −10 kcal/mol for all targets, Hoechst 34580 and Hoechst 33342 are expected to block the Aβ40 and Aβ42 aggregation.
In order to shed more light on the binding mechanisms, we divided Hoechst 34580 and Hoechst 33342 into five blocks (Figure S7 in SI). The first four blocks are similar and the difference is in blocks 5 with the last atoms 55–62 for Hoechst 33342 and 55–63 for Hoechst 34580.
The contributions from 5 blocks to the vdW interactions depend on targets but for a given target they are similar for two dyes (Tables S6–S9 and Figure S8). The contribution of block 5 is less important than other blocks because it has the least number of atoms (8 and 9 atoms for Hoechst 33342 and Hoechst 34580, respectively). Block 4 contributes to the vdW interactions less than blocks 1–3 and this holds for all fibril models (Tables S6–S9).
Although contributions of individual blocks to the electrostatic interactions are highly diverse, for all targets the difference between Hoechst 34580 and Hoechst 33342 is most pronounced for block 5 (Tables S6–S9 and Figure S9). In 2LMN the Coulomb interaction between block 5 and Hoechst 33342 is 2.78 kcal/mol but it is −24.49 kcal/mol for Hoechst 34580 (Table S6). Due to the proximity with block 5 the difference between two ligands in electrostatic interactions of block 4 is more than blocks 1–3.
Atoms 26, 29, 30, 37, 42 and 43 play a crucial role in electrostatic interaction of Hoechst 34580 with 2LMN having ΔEelec < −30 kcal/mol (Figure S9), while for Hoechst 33342 atoms 24, 26, 29, 30, 38, 42, 43 and 56 are vital. In 2BEG atoms 26, 29, 30, 37, 42 and 43 make a major contribution for Hoechst 33342, whereas atoms 24, 26, 29, 30, 38, 42 and 43 of Hoechst 34580 are dominating. Atoms 29, 30, 42 and 43 drive the electrostatic interaction of two dyes with all four targets including (Figure S9).
Block 3 of Hoechst 33342 is superior in Coulomb interaction with 2LMN, 2M4J and 2MXU while for 2BEG block 2 is the most important (Tables S6–S9). For Hoechst 34580 block 2 tightly binds to 2LMN and 2M4J, but for 2BEG and 2MXU block 1 is dominating.
The contributions of individual blocks to the total interaction (vdW and electrostatic) are shown in Tables S6–S9 and Figure S10. For Hoechst 33342 block 3 is dominating for targets 2LMN, 2M4J and 2MXU but in 2BEG block 2 is the most prominent. Block 5 of Hoechst 34580 is the most important for 2LMN and 2M4J, while block 1 and 3 have the highest binding propensity to 2BEG and 2MXU.
Upon binding of Hoechst 33342 and Hoechst 34580 the solvent accessible surface area (SASA) of all complexes shrinks leading to the negative value of ΔGsur (Table 1). The impact of the dyes depends on targets that Hoechst 34580 changes SASA of 2LMN and 2BEG to a larger extent than Hoechst 33342, while the opposite effect occurs in 2M4J and 2MXU.
Experimental results
Inhibition of Aβ42 aggregation by Hoechst 34580 and Hoechst 33342
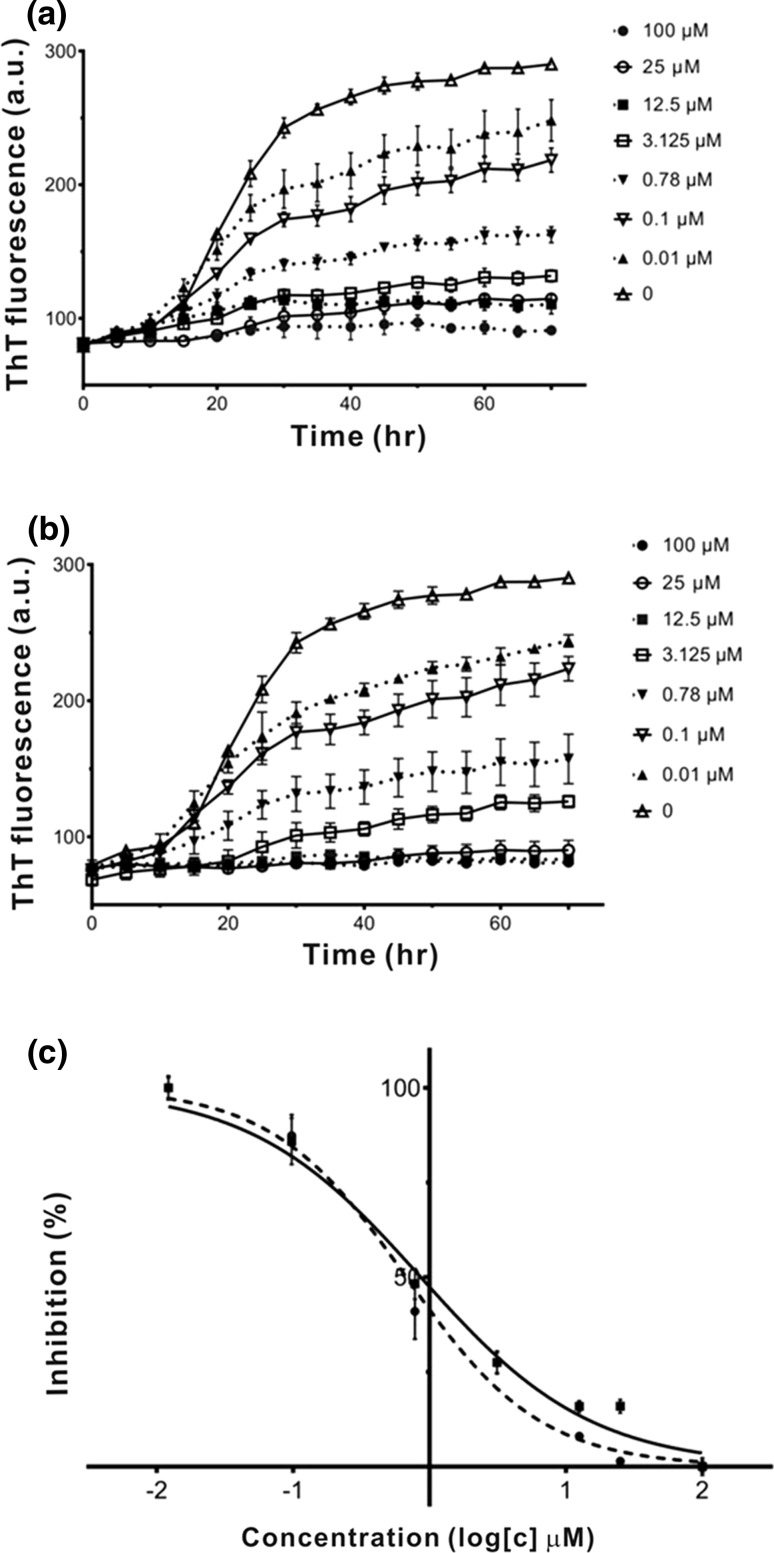
To evaluate the inhibitory effect of these compounds against Aβ42 aggregation, a Thioflavin T (ThT) assay was performed to monitor Aβ fibril formation while respectively co-incubating with various concentrations of the compounds (Fig. 6a, b). When ThT bound to cross β-sheets of fibrils, its emission fluorescence intensity could be measured which indicates the quantity of relative fibril formation. 50 μM Aβ42 solutions co-incubated with 100, 25, 12.5, 3.125, 0.78, and 0.1, 0.01 μM Hoechst 34580 or Hoechst 33342 at 37 °C for 70 h. We found that these compounds could inhibit the aggregation of Aβ42 in a dose-dependent manner. And then we examined the half-maximal concentration (IC50) required (Fig. 6c) to compare the potency of these three compounds. The IC50 was obtained by measuring the concentration of Hoechst 34580 and Hoechst 33342 respectively while maintaining the Aβ42 concentration which gave 0.86 ± 0.05 μM for Hoechst 34580 and 0.68 ± 0.05 μM for Hoechst 33342.
Fig. 6.
Fibrillization kinetics of Aβ42 incubated with and without Hoechst 34580 or Hoechst 33342. a Various concentrations (0.01–100 μM) of Hoechst 34580 was incubated with 50 μM Aβ42 at 37 °C for 70 h; b Various concentrations (0.01–100 μM) of Hoechst 33342 was incubated with 50 μM Aβ42 at 37 °C for 70 h; c Variation in ThT fluorescence intensity as a function of Hoechst 34580 (solid line) and Hoechst 33342 (dashed line). The data were subtracted to background from compound alone. Data were analyzed using GraphPad Prism to obtain IC50 values using log (inhibitor) versus normalized response-variable slope. Dose–response curves showed fractional binding of 5 μM ThT to 50 μM Aβ42 fibrils in the presence of Hoechst 34580 or Hoechst 33342, respectively
To compare the experimental results with the simulation results one cannot use the binding free energy obtained for 2BEG (5Aβ17-42) because this structure is based on H/D exchange and therefore, is not an experimentally observed structure. However, in vitro results may be compared with the results obtained for the solid state NMR structure 8Aβ11-42 (2MXU). With the equation ΔGbind = RT ln(IC50), where gas constant , T = 300 K and inhibition constant IC50 is measured in mol, a binding constant of 1 nM corresponds to ΔGbind ≈ −12.8 kcal/mol. A change in IC50 of one order of magnitude results in a change in the binding free energy of 1.4 kcal/mol. Therefore, the calculated values of ΔGbind for 2MXU (Table 1) imply that IC50 of both DNA dyes could be much less than 1 pM. They are also too far away from the experimentally measured value. The reason behind the discrepancy between theory and experiment is that it is very hard to match the calculated absolute binding free energy with experiments as it depends not only on force fields [53] but also on theoretical methods [59]. However, theoretically estimated binding free energies are presumably useful for ranking binding affinities [59]. This is also evident from our results that, in agreement with experiments, within the error bars Hoechst 34580 and Hoechst 33342 have the same binding free energy (Table 1). Therefore, our theoretical results on ΔGbind are useful for prediction of binding affinity ranking rather than for a direct comparison with experimentally measured inhibition constants.
Conclusion
Using the multi-step virtual screening we have predicted several compounds as potential drugs for AD. The ability of Hoechst 34580 and Hoechst 33342 in blocking Aβ aggregation was confirmed also by in vitro experiments. These compounds are located next to hydrophobic residues of Aβ peptides. The vdW interaction is dominating over the electrostatic interaction in binding propensity. The QSAR analysis showed that Hoechst 34580 and Hoechst 33342 can easily cross BBB having log(BB) greater than 0.5. Because these DNA dyes are known to be not cytotoxic they are recommended for further in vivo studies.
Future directions
In collaboration with experimentalists, our future work will be focused on in vivo study of the impact of DNA dyes Hoechst 34580 and Hoechst 33342 on Aβ aggregation. We plan also to search for new potential inhibitors from other large databases.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Atomic names, types, masses and charges used in the MD simulation of Hoechst 33342 and Hoechst 34580 by AMBER-f99SB-ILDN force field are listed in Table S1 and S2. Binding affinities and ligand ranking obtained by docking and SMD simulations are shown in Table S3–S5. Contributions of individual blocks to the interaction energy of DNA dyes with Aβ fibrils are presented in Tables S6-S9. Figure S1 shows the PDB structures of fibril 12Aβ9–40 (PDB ID 2LMN), 5Aβ17–42 (PDB ID 2BEG), 9Aβ1–40 (PDB ID 2M4J), and 8Aβ11–42 (PDB ID 2MXU). Figure S2 depicts the population of binding energies of 5372 ligands to 2MXU, 2LMN and 2BEG. Figure S3 show binding positions of top hits obtained by docking simulation for 2LMN, 2BEG and 2MXU. Figures S4 and S5 showed the time dependence of RMSD for four targets in complex with two DNA. Figures S6–S9 depict per-atom distribution of vdW and electrostatic interactions of 5 blocks of Hoechst 34580 and Hoechst 33342 with four fibril models. (PDF 1414 kb)
Acknowledgments
We thank P.D.Q. Huy for useful discussions. This work was supported by Vietnam National Foundation for Science and Technology Development (NAFOSTED) under grant number 106-YS.02-2013.01 and Department of Science and Technology at Ho Chi Minh City, Vietnam. The experimental work was supported by Ministry of Science and Technology (MOST), Taiwan under grant number MOST 103-2113-M-001-015. CKH was supported by Grant MOST 104-2112-M-001-002. MSL was supported by the Polish NCN grant 2015/19/B/ST4/02721. Allocation of CPU time at the supercomputer center TASK in Gdansk (Poland) is highly appreciated.
Contributor Information
Chin-Kun Hu, Email: huck@phys.sinica.edu.tw.
Yun-Ru Chen, Email: yrchen@gate.sinica.edu.tw.
Mai Suan Li, Email: masli@ifpan.edu.pl.
References
- 1.Wilson RS, Segawa E, Boyle PA, Anagnos SE, Hizel LP, Bennett DA. The natural history of cognitive decline in Alzheimer’s disease. Psychol Aging. 2012;27(4):1008–1017. doi: 10.1037/a0029857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alzheimer’s A. 2015 Alzheimer’s disease facts and figures. Alzheimer’s Dement J Alzheimer’s Assoc. 2015;11(3):332. doi: 10.1016/j.jalz.2015.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Nasica-Labouze J, Nguyen PH, Sterpone F, Berthoumieu O, Buchete N-V, Coté S, De Simone A, Doig AJ, Faller P, Garcia A. Amyloid β protein and Alzheimer’s disease: when computer simulations complement experimental studies. Chem Rev. 2015;115(9):3518–3563. doi: 10.1021/cr500638n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Herrup K. The case for rejecting the amyloid cascade hypothesis. Nat Neurosci. 2015;18(6):794–799. doi: 10.1038/nn.4017. [DOI] [PubMed] [Google Scholar]
- 5.Hardy J, Selkoe DJ. The amyloid hypothesis of Alzheimer’s disease: progress and problems on the road to therapeutics. Science. 2002;297(5580):353–356. doi: 10.1126/science.1072994. [DOI] [PubMed] [Google Scholar]
- 6.Aguzzi A, O’Connor T. Protein aggregation diseases: pathogenicity and therapeutic perspectives. Nat Rev Drug Discov. 2010;9(3):237–248. doi: 10.1038/nrd3050. [DOI] [PubMed] [Google Scholar]
- 7.Bernstein SL, Dupuis NF, Lazo ND, Wyttenbach T, Condron MM, Bitan G, Teplow DB, Shea J-E, Ruotolo BT, Robinson CV. Amyloid-β protein oligomerization and the importance of tetramers and dodecamers in the aetiology of Alzheimer’s disease. Nat Chem. 2009;1(4):326–331. doi: 10.1038/nchem.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Walsh DM, Klyubin I, Fadeeva JV, Cullen WK, Anwyl R, Wolfe MS, Rowan MJ, Selkoe DJ. Naturally secreted oligomers of amyloid β protein potently inhibit hippocampal long-term potentiation in vivo. Nature. 2002;416(6880):535–539. doi: 10.1038/416535a. [DOI] [PubMed] [Google Scholar]
- 9.Viet MH, Siposova K, Bednarikova Z, Antosova A, Nguyen TT, Gazova Z, Li MS. In silico and in vitro study of binding affinity of tripeptides to amyloid β fibrils: implications for Alzheimer’s disease. J Phys Chem B. 2015;119(16):5145–5155. doi: 10.1021/acs.jpcb.5b00006. [DOI] [PubMed] [Google Scholar]
- 10.Viet MH, Chen C-Y, Hu C-K, Chen Y-R, Li MS. Discovery of dihydrochalcone as potential lead for Alzheimer’s disease: in silico and in vitro study. PLoS ONE. 2013;8(11):e79151. doi: 10.1371/journal.pone.0079151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ngo ST, Li MS. Top-leads from natural products for treatment of Alzheimer’s disease: docking and molecular dynamics study. Mol Simul. 2013;39(4):279–291. doi: 10.1080/08927022.2012.718769. [DOI] [Google Scholar]
- 12.Oken BS, Storzbach DM, Kaye JA. The efficacy of Ginkgo biloba on cognitive function in Alzheimer disease. Arch Neurol. 1998;55(11):1409–1415. doi: 10.1001/archneur.55.11.1409. [DOI] [PubMed] [Google Scholar]
- 13.Yang F, Lim GP, Begum AN, Ubeda OJ, Simmons MR, Ambegaokar SS, Chen PP, Kayed R, Glabe CG, Frautschy SA. Curcumin inhibits formation of amyloid β oligomers and fibrils, binds plaques, and reduces amyloid in vivo. J Biol Chem. 2005;280(7):5892–5901. doi: 10.1074/jbc.M404751200. [DOI] [PubMed] [Google Scholar]
- 14.Yatin SM, Yatin M, Varadarajan S, Ain KB, Butterfield DA. Role of spermine in amyloid β-peptide-associated free radical-induced neurotoxicity. J Neurosci Res. 2001;63(5):395–401. doi: 10.1002/1097-4547(20010301)63:5<395::AID-JNR1034>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 15.Yamin G, Ono K, Inayathullah M, Teplow DB. Amyloid β-protein assembly as a therapeutic target of Alzheimer’s disease. Curr Pharm Des. 2008;14(30):3231–3246. doi: 10.2174/138161208786404137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bush AI. Metal complexing agents as therapies for Alzheimer’s disease. Neurobiol Aging. 2002;23(6):1031–1038. doi: 10.1016/S0197-4580(02)00120-3. [DOI] [PubMed] [Google Scholar]
- 17.Huy PDQ, Yu Y-C, Ngo ST, Van Thao T, Chen C-P, Li MS, Chen Y-C. In silico and in vitro characterization of anti-amyloidogenic activity of vitamin K3 analogues for Alzheimer’s disease. Biochim et Biophys Acta Gen Subj. 2013;4:2960–2969. doi: 10.1016/j.bbagen.2012.12.026. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi T, Tada K, Mihara H. RNA aptamers selected against amyloid β-peptide (Aβ) inhibit the aggregation of Aβ. Mol BioSyst. 2009;5(9):986–991. doi: 10.1039/b903391b. [DOI] [PubMed] [Google Scholar]
- 19.Nitz M, Fenili D, Darabie AA, Wu L, Cousins JE, McLaurin J. Modulation of amyloid-β aggregation and toxicity by inosose stereoisomers. FEBS J. 2008;275(8):1663–1674. doi: 10.1111/j.1742-4658.2008.06321.x. [DOI] [PubMed] [Google Scholar]
- 20.Cummings JL. Alzheimer’s disease. N Engl J Med. 2004;351(1):56–67. doi: 10.1056/NEJMra040223. [DOI] [PubMed] [Google Scholar]
- 21.Hawkes CA, Ng V, McLaurin J. Small molecule inhibitors of Aβ-aggregation and neurotoxicity. Drug Dev Res. 2009;70(2):111–124. doi: 10.1002/ddr.20290. [DOI] [Google Scholar]
- 22.Bolton EE, Wang Y, Thiessen PA, Bryant SH. PubChem: integrated platform of small molecules and biological activities. Annu Rep Comput Chem. 2008;4:217–241. doi: 10.1016/S1574-1400(08)00012-1. [DOI] [Google Scholar]
- 23.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev. 2012;64:4–17. doi: 10.1016/j.addr.2012.09.019. [DOI] [PubMed] [Google Scholar]
- 24.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings1. Adv Drug Deliv Rev. 2001;46(1–3):3–26. doi: 10.1016/S0169-409X(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 25.Kollman PA, Massova I, Reyes C, Kuhn B, Huo S, Chong L, Lee M, Lee T, Duan Y, Wang W. Calculating structures and free energies of complex molecules: combining molecular mechanics and continuum models. Acc Chem Res. 2000;33(12):889–897. doi: 10.1021/ar000033j. [DOI] [PubMed] [Google Scholar]
- 26.Petkova AT, Yau W-M, Tycko R. Experimental constraints on quaternary structure in Alzheimer’s β-amyloid fibrils. Biochemistry. 2006;45(2):498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lu JX, Qiang W, Yau WM, Schwieters CD, Meredith SC, Tycko R. Molecular structure of beta-amyloid fibrils in Alzheimer’s disease brain tissue. Cell. 2013;154(6):1257–1268. doi: 10.1016/j.cell.2013.08.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lührs T, Ritter C, Adrian M, Riek-Loher D, Bohrmann B, Döbeli H, Schubert D, Riek R. 3D structure of Alzheimer’s amyloid-β (1–42) fibrils. Proc Natl Acad Sci USA. 2005;102(48):17342–17347. doi: 10.1073/pnas.0506723102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xiao Y, Ma B. Abeta(1-42) fibril structure illuminates self-recognition and replication of amyloid in Alzheimer’s disease. Nat Struct Mol Biol. 2015;22(6):499–505. doi: 10.1038/nsmb.2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sanner MF. Python: a programming language for software integration and development. J Mol Graph Model. 1999;17(1):57–61. [PubMed] [Google Scholar]
- 31.Trott O, Olson AJ. AutoDock Vina: improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J Comput Chem. 2010;31(2):455–461. doi: 10.1002/jcc.21334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lindorff-Larsen K, Piana S, Palmo K, Maragakis P, Klepeis JL, Dror RO, Shaw DE. Improved side-chain torsion potentials for the Amber ff99SB protein force field. Proteins Struct Funct Bioinform. 2010;78(8):1950–1958. doi: 10.1002/prot.22711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. Comparison of simple potential functions for simulating liquid water. J Chem Phys. 1983;79(2):926–935. doi: 10.1063/1.445869. [DOI] [Google Scholar]
- 34.Zhang T, Xu W, Mu Y, Derreumaux P. Atomic and dynamic insights into the beneficial effect of the 1, 4-naphthoquinon-2-yl-l-tryptophan Inhibitor on Alzheimer’s Aβ1–42 dimer in terms of aggregation and toxicity. ACS Chem Neurosci. 2013;5(2):148–159. doi: 10.1021/cn400197x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huy PDQ, Li MS. Binding of fullerenes to amyloid beta fibrils: size matters. Phys Chem Chem Phys. 2014;16(37):20030–20040. doi: 10.1039/C4CP02348J. [DOI] [PubMed] [Google Scholar]
- 36.Van Vuong Q, Bednarikova Z, Antosova A, Huy PDQ, Siposova K, Tuan NA, Li MS, Gazova Z. Inhibition of insulin amyloid fibrillization by glyco-acridines: an in vitro and in silico study. MedChemComm. 2015;6(5):810–822. doi: 10.1039/C5MD00004A. [DOI] [Google Scholar]
- 37.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 38.Bayly CI, Cieplak P, Cornell W, Kollman PA. A well-behaved electrostatic potential based method using charge restraints for deriving atomic charges: the RESP model. J Phys Chem. 1993;97(40):10269–10280. doi: 10.1021/j100142a004. [DOI] [Google Scholar]
- 39.Wang J, Wang W, Kollman PA, Case DA. Antechamber: an accessory software package for molecular mechanical calculations. J Am Chem Soc. 2001;222:U403. [Google Scholar]
- 40.Frisch M, Trucks G, Schlegel H, Scuseria G, Robb M, Cheeseman J, Scalmani G, Barone V, Mennucci B, Petersson G. Gaussian 09 (Revision-A. 01) Wallingford: Gaussian Inc; 2009. [Google Scholar]
- 41.Darden T, York D, Pedersen L. Particle mesh Ewald: an N· log (N) method for Ewald sums in large systems. J Chem Phys. 1993;98(12):10089–10092. doi: 10.1063/1.464397. [DOI] [Google Scholar]
- 42.Hockney R, Goel S, Eastwood J. Quiet high-resolution computer models of a plasma. J Comput Phys. 1974;14(2):148–158. doi: 10.1016/0021-9991(74)90010-2. [DOI] [Google Scholar]
- 43.Berendsen HJ, Jv Postma, van Gunsteren WF, DiNola A, Haak J. Molecular dynamics with coupling to an external bath. J Chem Phys. 1984;81(8):3684–3690. doi: 10.1063/1.448118. [DOI] [Google Scholar]
- 44.Isralewitz B, Gao M, Schulten K. Steered molecular dynamics and mechanical functions of proteins. Curr Opin Struct Biol. 2001;11(2):224–230. doi: 10.1016/S0959-440X(00)00194-9. [DOI] [PubMed] [Google Scholar]
- 45.Kumar S, Li MS. Biomolecules under mechanical force. Phys Rep. 2010;486(1):1–74. doi: 10.1016/j.physrep.2009.11.001. [DOI] [Google Scholar]
- 46.Grubmüller H, Heymann B, Tavan P. Ligand binding: molecular mechanics calculation of the streptavidin-biotin rupture force. Science. 1996;271(5251):997–999. doi: 10.1126/science.271.5251.997. [DOI] [PubMed] [Google Scholar]
- 47.Suan Li M, Khanh Mai B. Steered molecular dynamics-a promising tool for drug design. Curr Bioinform. 2012;7(4):342–351. doi: 10.2174/157489312803901009. [DOI] [Google Scholar]
- 48.Mai BK, Li MS. Neuraminidase inhibitor R-125489–a promising drug for treating influenza virus: steered molecular dynamics approach. Biochem Biophys Res Commun. 2011;410(3):688–691. doi: 10.1016/j.bbrc.2011.06.057. [DOI] [PubMed] [Google Scholar]
- 49.Gibson CT, Carnally S, Roberts CJ. Attachment of carbon nanotubes to atomic force microscope probes. Ultramicroscopy. 2007;107(10):1118–1122. doi: 10.1016/j.ultramic.2007.02.045. [DOI] [PubMed] [Google Scholar]
- 50.Mai BK, Viet MH, Li MS. Top leads for swine influenza A/H1N1 virus revealed by steered molecular dynamics approach. J Chem Inf Model. 2010;50(12):2236–2247. doi: 10.1021/ci100346s. [DOI] [PubMed] [Google Scholar]
- 51.Vuong QV, Nguyen TT, Li MS. A new method for navigating optimal direction for pulling ligand from binding pocket: application to ranking binding affinity by steered molecular dynamics. J Chem Inf Model. 2015;55(12):2731–2738. doi: 10.1021/acs.jcim.5b00386. [DOI] [PubMed] [Google Scholar]
- 52.Chovancova E, Pavelka A, Benes P, Strnad O, Brezovsky J, Kozlikova B, Gora A, Sustr V, Klvana M, Medek P. CAVER 3.0: a tool for the analysis of transport pathways in dynamic protein structures. PLoS Comput Biol. 2012;8(10):e1002708. doi: 10.1371/journal.pcbi.1002708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nguyen TT, Mai BK, Li MS. Study of Tamiflu sensitivity to variants of A/H5N1 virus using different force fields. J Chem Inf Model. 2011;51(9):2266–2276. doi: 10.1021/ci2000743. [DOI] [PubMed] [Google Scholar]
- 54.Ngo ST, Li MS. Curcumin binds to Aβ1–40 peptides and fibrils stronger than ibuprofen and naproxen. J Phys Chem B. 2012;116(34):10165–11017. doi: 10.1021/jp302506a. [DOI] [PubMed] [Google Scholar]
- 55.Garg P, Verma J. In silico prediction of blood brain barrier permeability: an artificial neural network model. J Chem Inf Model. 2006;46(1):289–297. doi: 10.1021/ci050303i. [DOI] [PubMed] [Google Scholar]
- 56.Rose K, Hall LH, Kier LB. Modeling blood-brain barrier partitioning using the electrotopological state. J Chem Inf Comput Sci. 2002;42(3):651–666. doi: 10.1021/ci010127n. [DOI] [PubMed] [Google Scholar]
- 57.Clark DE. Rapid calculation of polar molecular surface area and its application to the prediction of transport phenomena. 1. Prediction of intestinal absorption. J Pharm Sci. 1999;88(8):807–814. doi: 10.1021/js9804011. [DOI] [PubMed] [Google Scholar]
- 58.Clark DE. Rapid calculation of polar molecular surface area and its application to the prediction of transport phenomena. 2. Prediction of blood–brain barrier penetration. J Pharm Sci. 1999;88(8):815–821. doi: 10.1021/js980402t. [DOI] [PubMed] [Google Scholar]
- 59.Hou T, Wang J, Li Y, Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model. 2010;51(1):69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Atomic names, types, masses and charges used in the MD simulation of Hoechst 33342 and Hoechst 34580 by AMBER-f99SB-ILDN force field are listed in Table S1 and S2. Binding affinities and ligand ranking obtained by docking and SMD simulations are shown in Table S3–S5. Contributions of individual blocks to the interaction energy of DNA dyes with Aβ fibrils are presented in Tables S6-S9. Figure S1 shows the PDB structures of fibril 12Aβ9–40 (PDB ID 2LMN), 5Aβ17–42 (PDB ID 2BEG), 9Aβ1–40 (PDB ID 2M4J), and 8Aβ11–42 (PDB ID 2MXU). Figure S2 depicts the population of binding energies of 5372 ligands to 2MXU, 2LMN and 2BEG. Figure S3 show binding positions of top hits obtained by docking simulation for 2LMN, 2BEG and 2MXU. Figures S4 and S5 showed the time dependence of RMSD for four targets in complex with two DNA. Figures S6–S9 depict per-atom distribution of vdW and electrostatic interactions of 5 blocks of Hoechst 34580 and Hoechst 33342 with four fibril models. (PDF 1414 kb)