Abstract
In recent years, Salmonella enterica serovar Enteritidis has been a primary cause of human salmonellosis in many countries. The major objective of this study was to investigate genetic diversity among Salmonella Enteritidis strains from different origins (food and human) by Enterobacterial Repetitive Intergenic Consensus (ERIC) -PCR, as well as to assess their plasmid profiling and antimicrobial resistance. A total of 30 Salmonella Enteritidis isolates, 15 from food samples (chicken, lamb, beef and duck meats) and 15 from clinical samples were collected in Tehran. Identification of isolates as Salmonella was confirmed by using conventional standard biochemical and serological tests. Multiplex-PCR was used for serotyping of isolates to identify Salmonella Enteritidis. Antimicrobial susceptibility testing to 16 agents founds drug resistance patterns among Salmonella Enteritidis isolates. No resistance was observed to cephalexin, ceftriaxone, ceftazidime and cefotaxime, ciprofloxacin, imipenem or meropenem, chloramphenicol and gentamicin. The highest resistance (96.7%) was observed to nitrofurantoin. Seven plasmid profiles (P1–P7) were detected, and a 68-kb plasmid was found in all isolates. Two different primers; ERIC and (GTG)5 were used for genotyping, which each produced four profiles. The majority of clinical and food isolates fell into two separate common types (CTs) with a similar percentage of 95% by ERIC-PCR. Using primer (GTG)5, 29 isolates incorporated in three CTs with 70% of isolates showing a single banding pattern. Limited genetic diversity among human and food isolates of Salmonella Enteritidis may indicate that contaminated foods were possibly the source of human salmonellosis. These results confirmed that ERIC-PCR genotyping has limited discriminatory power for Salmonella Enteritidis of different origin.
Keywords: Molecular characterization, plasmid profiling, resistance, Salmonella Enteritidis, serotyping
Introduction
Gastroenteritis is the most common manifestation of human Salmonella infections, especially the infection caused by Salmonella Typhimurium and Salmonella Enteritidis serotypes. Epidemiological studies in different countries have shown an increase in Salmonella Enteritidis infections during recent years. According to the WHO, since 1990, Salmonella Enteritidis has been considered the most common cause of gastroenteritis worldwide [1], whereas previously, Salmonella Typhimurium had been the main cause of Salmonella gastroenteritis [2].
In most cases, Salmonella gastroenteritis is self-limiting and usually heals without the need for antibiotic therapy [1], but it may progress to systemic diseases such as bacteraemia [3], meningitis [4] and endocarditis [5], especially in patients with an immunodeficiency [6] or neoplastic disease that are taking antibiotics and immune suppressor drugs. Pregnant women and the elderly are more sensitive to Salmonella infections. For effective epidemiological surveillance and control of Salmonella species, we need accurate subtyping information of strains to identify potential sources of infection. Many traditional ways of typing, such as biochemical and serological characterizations, are tedious and expensive with limited epidemiological value. Moreover, these methods do not have an adequate discriminatory capacity for strains with identical serotype or biochemical characteristics [7]. Different molecular methods have been used by researchers for genotyping enteric bacteria, among which Enterobacterial Repetitive Intergenic Consensus (ERIC) -PCR is a powerful method for DNA fingerprinting. In this method, repeated oligonucleotides are used as a starter of DNA synthesis when there is no need for information on the target DNA sequence, which makes it a powerful method with general applications. This method is faster, simpler and more economical than other genomic typing methods [8].
In recent years, Salmonella Enteritidis has become a major public health problem, and the main cause of both human and animal salmonellosis in many countries. To analyse this problem, the objectives of this study were to determine the extent of genetic variation and clonality among food and clinical strains of Salmonella Enteritidis by examination of susceptibility to common antibiotics, plasmid analysis and ERIC-PCR patterns. In addition, we investigate the prevalence of Salmonella Enteritidis in children’s stool samples.
Materials and methods
Bacterial isolates
A total of 30 Salmonella Enteritidis isolates, including 15 isolates from humans and 15 from food, were used during this study. The human isolates were collected from 1950 patients with diarrhoea admitted to the Children’s Medical Centre Hospital in Tehran, from April to November 2014. All human isolates were recovered from stool of patients suffering from diarrhoea, except for one isolate which was obtained from the blood of a diarrheal patient. This patient was admitted to the hospital with acute gastroenteritis and Salmonella serogroup D was obtained from blood after 3 weeks of initial isolation from stool. Diarrhoeal cases were defined as patients with more than three non-bloody episodes of diarrhoea per day. The food isolates were taken from different food sources, including chicken, lamb, beef and duck meats were obtained from the Microbiology Department of the Veterinary Faculty, Tehran University during the same period of time. Standard microbiological methods were used for identification of Salmonella isolates [9]. The serogroup characterization of isolates was performed via slide-agglutination test with polyvalent and monospecific antisera against somatic and flagella antigens in Phase I and II according to the manufacturer’s instructions (Difco, Franklin Lakes, NJ, USA). The identity of Salmonella Enteritidis isolates was confirmed by a multiplex-PCR as described previously [10].
Antimicrobial susceptibility testing
The disk-diffusion method was used to determine the susceptibility of strains as instructed by the CLSI guidelines [11]. The antimicrobial agents were as follows: amoxicillin (20 μg), gentamicin (10 μg), chloramphenicol (30 μg), cotrimoxazole (25 μg), nalidixic acid (30 μg), ciprofloxacin (5 μg), cefotaxime (30 μg), ceftazidime (30 μg), ceftriaxone (30 μg), cephalexin (30 μg), streptomycin (10 μg), tetracycline (30 μg), imipenem (10 μg), meropenem (10 μg), nitrofurantoin (30 μg), colistin sulphate (25 μg) (Mast Diagnostics, Mast Group Ltd, Merseyside, UK). The strain organism used for quality control was Escherichia coli ATCC 25922.
Plasmid profiling
Plasmids were extracted using a Qiagen Mini Plasmid Kit (Qiagen Mississauga, Ont, Canada). A 0.8% agarose gel was used for plasmid horizontal electrophoresis; moreover, standard E. coli AC11 strain with 68 kb, 7.2 kb and 1.9 kb plasmids was used as the control. The molecular weight of plasmids was determined by Seqaid П software (version 3.5, Kansas State University, Manhattan, KS, USA).
ERIC-PCR and (GTG)5 amplification
One colony of each bacterial strain was purified using a commercial kit (Bioneer, Seoul, Korea) and used as DNA template. Primer pairs for ERIC-PCR amplification were as follows: ERIC-1R (5'-ATGTAAGCTCCTGGGGATTCAC-3') and ERIC-2 (5'-AAGTAAGTGACTGGGGTGAGCG-3') .The (GTG)5 amplification was performed using primers described previously [12]. Amplification was performed in a reaction mixture containing hot start Taqplus Master Mix 12.5 μL, 20 pmol of each primer, 3 μL of DNA, 2.5 μL of PCR buffer and 5 μL ultra-pure water (final volume 25 μL). ERIC-PCR amplification was performed using a programme comprising one denaturation step at 95°C for 5 min; four cycles of 94°C for 4 min, 53°C for 4 min, 72°C for 4 min; 30 cycles of 94°C for 30 s, 53°C for 1 min, 72°C for 1 min; and one cycle at 72°C for 7 min.
The (GTG)5 amplification included one denaturation step at 95°C for 5 min; four cycles of 94°C for 4 min, 50°C for 4 min, 72°C for 4 min; 29 cycles of 94°C for 30 s, 50°C for 1 min, 72°C for 1 min; and one cycle at 72°C for 7 min. Gel images were analysed using Gel compar II version 4.0 (Applied Maths, Sint-Martens-Latem, Belgium). The dendrogram was generated according to the unweighted pair group method with arithmetic (UPGMA) (Fig. 1).
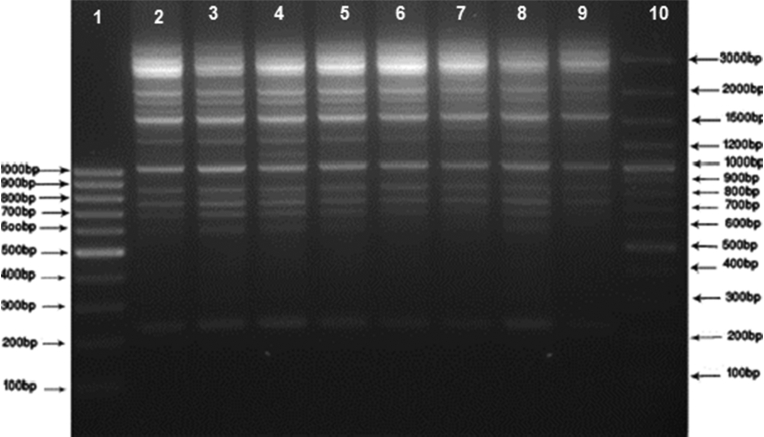
Fig. 1.
DNA fingerprint patterns of Salmonella Enteritidis strains from human and food sources by ERIC-PCR. Lane 1: M1 (SM041) 100 bp DNA Ladder; Lane 10: M2 (SM0321) 100 bp DNA Ladder Plus; Lanes 2–9: clinical and food samples.
Results
In general, 26 different Salmonella serovars were recovered from children (14 boys and 12 girls). The distribution of Salmonella serovars in stool samples was 14 (54%) serogroup D, 6 (23%) Salmonella paratyphi C, 2 (8%) S. paratyphi B, 3 (11%) Salmonella arizonae and 1 (4%) S. paratyphi A, respectively. All isolates belonging to serogroup D were identified as Salmonella Enteritidis by Multiplex PCR.
Antibiotic resistance
In the evaluation of 16 antibiotics, seven antibiotic-resistance patterns were observed among clinical and food isolates. The most common anti-biotype was AB1 (43.2%), which was indicative of resistance to nitrofurantoin. The second anti-biotype was AB2 (40.5%) which characterized a nitrofurantoin-nalidixic acid resistance phenotype. We did not observe any resistance to cephalexin, ceftriaxone, ceftazidime and cefotaxime, ciprofloxacin, imipenem or meropenem, chloramphenicol and gentamicin (Table 1).
Table 1.
The antibiotic resistance pattern of clinical and food isolates
| Resistance pattern | Antibiotics |
% resistance | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CTX | A | T | IN | TS | S | C | CRO | GM | CO | CAZ | CIP | CFX | NA | MEM | IMI | ||
| AB 1 | S | S | S | R | S | S | S | S | S | S | S | S | S | S | S | S | 43.2% |
| AB 2 | S | S | S | R | S | S | S | S | S | S | S | S | S | R | S | S | 40.5% |
| AB 3 | S | R | R | R | R | S | S | S | S | S | S | S | S | R | S | S | 3.3% |
| AB 4 | S | S | R | R | R | R | S | S | S | S | S | S | S | R | S | S | 3.3% |
| AB 5 | S | S | R | R | S | R | S | S | S | S | S | S | S | S | S | S | 3.3% |
| AB 6 | S | S | S | S | S | R | S | S | S | S | S | S | S | S | S | S | 3.3% |
| AB 7 | S | S | S | R | S | R | S | S | S | S | S | S | S | S | S | S | 3.3% |
Abbreviations for antibiotics are as follows: CTX, cefotaxime; A, amoxicillin; T, tetracycline; IN, nitrofurantoin; TS trimethoprim-sulfamethoxazole; S, streptomycin; C, chloramphenicol; CRO, ceftriaxone; GM, gentamicin; Co, colistin sulphate; CAZ, ceftazidime; CIP, ciprofloxacin; CFX, cephalexin; NA, nalidixic acid; MEM, meropenem; IMI, imipenem.
Plasmid profiles
Plasmid profiling of 30 Salmonella Enteritidis isolates revealed seven different profiles (P1–P7) with one to three bands ranging from 1500 bp to 68 000 bp. Two plasmid profiles (P2 and P7) were common among clinical and food isolates; in addition, 7 (47%) clinical isolates and 12 (80%) food isolates were P7. A large plasmid (68 kb) was common in all profiles. The second most common pattern was P2 (16.7 %), which included two other plasmids (1.96 kb and 2.6 kb) as well as 68 kb (Table 2).
Table 2.
Plasmid profiles and antibiotic resistance pattern of Salmonella Enteritidis isolated from human and food origin
| Plasmid profile | Plasmid content | Resistance profile |
|---|---|---|
| P1 | 68000- 6300- 4900 | AB2 (IN-NA) |
| P2 | 68000- 2600- 1960 | AB1 (IN), AB3 (A-T-IN-TS-NA), AB2 (IN-NA) |
| P3 | 68000-2600- 2150 | AB4 (T-IN-TS-S-NA) |
| P4 | 68000- 1500- 2150 | AB5 (T-IN-S) |
| P5 | 68000- 3300 | AB2 (IN-NA) |
| P6 | 68000- 2900 | AB1 (IN) |
| P7 | 68000 | AB1, AB2, AB3 (A-T-IN-TS-NA), AB6 (S), AB7 (IN-S) |
Abbreviations for antibiotics are as follows: A, amoxicillin; T, tetracycline; IN, nitrofurantoin; TS trimethoprim-sulfamethoxazole; S, streptomycin; NA, nalidixic acid.
ERIC-PCR
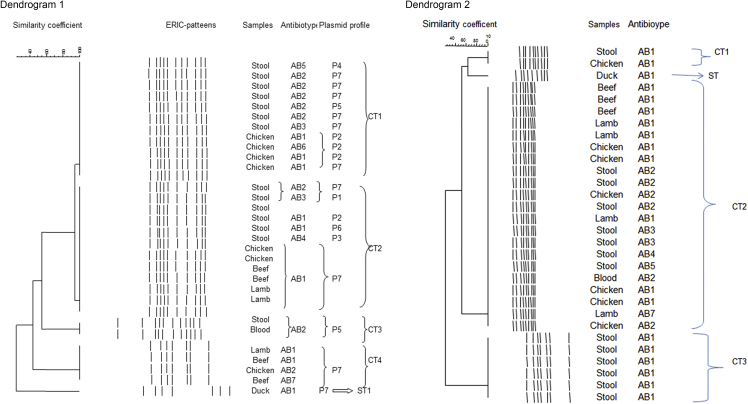
Genotyping of the 30 isolates showed five different banding patterns, with two major common types (CT1 and CT2) comprising 76.6% of total isolates, each of which consisted of both clinical and food isolates. The CT3 with two members only included clinical isolates with the single blood isolate in this category. Four isolates in CT4 were all taken from food samples. One single type (ST1) was also detected, which corresponded to a food isolate. The isolates of duck meat origin were exclusively different from the rest of the samples with single type (ST). In addition, this strain was isolated from north of Iran and both (GTG)5 and ERIC-PCR successfully separated the strain as a single type (Fig. 2; dendrogram 1).
Fig. 2.
Dendograms showing the similarity of Salmonella strains isolated from human and animal food sources as determined by the ERIC-PCR fingerprint analysis (dendrogram 1) and the (GTG)5-PCR fingerprint analysis (dendrogram 2) using the Gelcompar II version 4.0 software program.
(GTG)5-PCR genotyping
This method grouped 21 isolates of different origins (70%) in a single cluster (CT2); two other common types (CT1 and CT3) and one single type were also detected. This genotyping method showed more genetic similarity between human, and food isolates (Fig. 2; dendrogram 2).
Discrimination index
The discriminatory power of ERIC-PCR and (GTG)5-PCR was assessed by Simpson’s diversity index, as presented by Hunter and Gaston [13].
Discussion
Salmonella species have been defined as major food-borne pathogens in humans worldwide. The predominant serotypes have changed over time and are now various in different geographic locations [14]. In this study, the prevalence of Salmonella Enteritidis was 54%, which showed the dominance of this serovar in Iran. Recent reports from Brazil, Poland, Malaysia, China and Greece have shown the same serovar as the dominant serovar with the frequencies ranging from 34% to 86%, which reveals a coincident rise of this serovar around the world [15], [16], [17], [18], [19].
In our study, all isolates were susceptible to cephalexin, ceftriaxone, ceftazidime, cefotaxime and ciprofloxacin. These results indicate that cephalosporins as well as fluoroquinolones are still appropriate drugs that can be used against invasive and systemic infection caused by Salmonella. Furthermore, all isolates were susceptible to monobactams (imipenem or meropenem), which shows that they can be considered as alternative drugs for clinical cases where more resistance to extended-spectrum antimicrobials is encountered. This result is due to limited prescription of carbapenems in Iran. However, their toxic and more harmful nature against microflora should not be ignored. A previous report from Iran showed 100% sensitivity to amikacin, ceftriaxone, xanofloxacin, levofloxacin, norfloxacain and imipenem [20]. In our study, the most frequently antibiotic resistance was observed in nitrofurantoin (100%) and nalidixic acid (73%) among isolates of human origin. The isolates of food origin also showed their highest level of resistance to nitrofurantoin (86.7%) and nalidixic acid (13.3%), but they were significantly different (p <0.005) from clinical isolates. The observed level of resistance to these two agents among human and food isolates was higher than that reported by previous studies in Iran [21]. The data obtained in the present study indicated 73% resistance to this antimicrobial agent. The increased resistance to nalidixic acid among Salmonella Enteritidis isolates was also previously reported in other countries, including some in Europe [22]. This increase might be due to point mutations in DNA gyrase genes (gyrA) or to activation of efflux pumps among this bacterial genus [23]. Morshed and Peighambari showed that 77% of clinical isolates and 24% of food isolates were resistant to nalidixic acid, which is consistent with our results, which showed 73% resistance among clinical isolates and 13.3% among food isolates [20]. Among clinical isolates, 73.3% were multiple drug resistance phenotypes and were resistant to at least three or more antibiotic families, while among food isolates, only 20% had multiple resistance patterns. Generally, in this study, antibiotic resistance rate was much higher than in food isolates. It seems that improper and indiscriminate use of antibacterial drugs in both human and veterinary medicine without careful evaluation of bacterial sensitivity, has led to a widespread resistance to antimicrobial drugs [24]. Horizontal transfer and clonal spread of resistance genes may occur among food-producing animals and humans [25].
Plasmid profiling has been used successfully for the typing of Salmonella Enteritidis strains, but it has some limitations. Many Salmonella Enteritidis strains carry serotype-specific virulence plasmid with different sizes (50–100 kb) [26]. In our study, all isolates successfully typed by plasmid profiling, and one single large plasmid (68 kb) was common in all the profiles, which can be proposed as a serotype-specific virulence plasmid. Morshed and Peighambari have reported six plasmid profiles (A–F) between 49 Salmonella Enteritidis isolates of human and animal origin in Iran. In their study, 98% of isolates had the 68-kb plasmid and 2% had no plasmid [20]. Liebana et al. also reported a single 57-kb plasmid among 56% of their Salmonella Enteritidis isolated from the UK. He showed that the discriminatory power of this method was more than pulsed-field gel electrophoresis (PFGE) and less than ribotyping [27]. Extrachromosomal DNA was considered as an unstable genetic marker. Consequently, strains that show identical chromosomal features may have different plasmid patterns [28]. In addition, the same plasmid patterns can be observed among isolates with different chromosomal characteristics [29].
ERIC-PCR is a powerful DNA-based typing method for fingerprinting. ERIC-PCR analysis is useful for studying the epidemiology of Salmonella Enteritidis [12]. Using ERIC primers, between 30 strains of Salmonella Enteritidis isolates four main clusters had 60% similarity. The majority of isolates (23; 76%) fell in a major cluster (A) with two common types (95% similarity), while the remaining seven strains were classified into two distinct clusters with one single type. Both clinical and food isolates existed in the clusters, which showed different plasmid profiles and antimicrobial resistance properties. The most common type was CT2 (40%), which contained six human isolates and six food isolates, respectively. In this type, food isolates had P7 plasmid patterns and resistance phenotypes (AB1), but the human isolates had different plasmid patterns and resistance phenotypes. This indicates that human isolates had more diverse types. The CT3 had two clinical members with identical resistance phenotypes (AB2) and plasmid patterns (P5); however, they had different origins (stool and blood). The two isolates belonged to one patient, but were obtained on two occasions. The result shows the importance of the treatment and control of Salmonella infections in high-risk individuals, especially for children, which can lead to systematic infections.
Food isolates of duck meat origin were exclusively different from the rest of the samples with single type (ST). This strain was isolated from the north of Iran and both (GTG)5 and ERIC-PCR successfully separated the strain as a single type. The (GTG)5 analysis indicated CT2 as the most common type (70%) with 21 clinical and food isolates falling in this group. The diversity index of population was obtained to be 0.48 and 0.70 by (GTG)5 and ERIC-PCR analysis, respectively, which indicates the higher discriminatory power of the ERIC-PCR method. Using ERIC-PCR, Suh and Song showed a high genetic homogeneity among 22 Salmonella Enteritidis isolates of human and chicken origin in South Korea between 2001 and 2002 [12]. Furthermore, several investigators have shown good discriminatory power and typing capacity of ERIC-PCR for Salmonella Enteritidis isolates from human and food origin [30], [31]. In the study of Campioni et al. performed with 128 Salmonella Enteritidis isolates of human and food origin over a 24-year period in Brazil, ERIC-PCR and PFGE exhibited a high genetic similarity, PFGE being considered the reference standard method for the typing of Salmonella Enteritidis strains [32]. Ammari et al. have shown that ERIC-PCR exhibited a higher discriminatory power than PFGE [33].
In conclusion, Salmonella Enteritidis was phenotypically and genotypically homogeneous and was clonally dispersed among food and human populations in our study. These clones may continue to exist over a considerable period of time in Tehran and spread in different time occasions. This supports the notion that infected animals and humans are important sources of contamination in the environment and the food chain. Using combined genotyping techniques and greater number of isolates makes it more possible to trace the origin of infections in epidemiological investigations.
Acknowledgements
This work was supported by a grant (No. 29218) provided by the Vice-Chancellor for Research of Tehran University of Medical Sciences (Tehran, IR Iran).
Conflict of interest
There were no conflicts of interest.
References
- 1.Chaitram J.M., Jevitt L.A., Tenover F.C., WHO Antimicrobial Resistance Group The World Health Organization's External Quality Assurance System Proficiency Testing Program has improved the accuracy of antimicrobial susceptibility testing and reporting among participating laboratories using NCCLS methods. J Clin Microbiol. 2003;41:2372–2377. doi: 10.1128/JCM.41.6.2372-2377.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.van Pelt W., de Wit M.A., Wannet W.J., Ligtvoet E.J., Widdowson M.A., van Duynhoven Y.T. Laboratory surveillance of bacterial gastroenteric pathogens in The Netherlands, 1991–2001. Epidemiol Infect. 2003;130:431–441. [PMC free article] [PubMed] [Google Scholar]
- 3.Castonguay-Vanier J., Davong V., Bouthasavong L., Sengdetka D., Simmalavong M., Seupsavith A. Evaluation of a simple blood culture amplification and antigen detection method for diagnosis of Salmonella enterica serovar typhi bacteremia. J Clin Microbiol. 2013;51:142–148. doi: 10.1128/JCM.02360-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fomda B.A., Charoo B.A., Bhat J.A., Reyaz N., Maroof P., Naik M.I. Recurrent meningitis due to Salmonella enteritidis: a case report from Kashmir India. Indian J Med Microbiol. 2012;30:474–476. doi: 10.4103/0255-0857.103776. [DOI] [PubMed] [Google Scholar]
- 5.Hibbert B., Costiniuk C., Hibbert R., Joseph P., Alanazi H., Simard T. Cardiovascular complications of Salmonella enteritidis infection. Can J Cardiol. 2010;26:323–325. doi: 10.1016/s0828-282x(10)70444-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ayelo Navarro A., Geronimo Pardo M., Torres Lamberti V., Mateo Cerdan C.M., Jimenez Vizuete J.M., Peyro Garcia R. Salmonella enteritidis bacteraemia as clinical onset of acquired immune deficiency syndrome. Rev Esp Anestesiol Reanim. 2013;60:103–105. doi: 10.1016/j.redar.2012.04.005. [DOI] [PubMed] [Google Scholar]
- 7.Bakeri S.A., Yasin R.M., Koh Y.T., Puthucheary S.D., Thong K.L. Genetic diversity of human isolates of Salmonella enterica serovar Enteritidis in Malaysia. J Appl Microbiol. 2003;95:773–780. doi: 10.1046/j.1365-2672.2003.02033.x. [DOI] [PubMed] [Google Scholar]
- 8.Eriksson J., Lofstrom C., Aspan A., Gunnarsson A., Karlsson I., Borch E. Comparison of genotyping methods by application to Salmonella Livingstone strains associated with an outbreak of human salmonellosis. Int J Food Microbiol. 2005;25:93–103. doi: 10.1016/j.ijfoodmicro.2005.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Washington Winner J.R., Allen S., Janda W., Koneman E., Procop G., P S. 6th edn. Lippincott Williams and Wilkins Press; Philadelphia: 2001. Color Atlas and Textbook of Diagnostic Microbiology. [Google Scholar]
- 10.Firoozeh F., Zahraei-Salehi T., Shahcheraghi F., Karimi V., Aslani M.M. Characterization of class I integrons among Salmonella enterica serovar Enteritidis isolated from humans and poultry. FEMS Immunol Med Microbiol. 2012;64:237–243. doi: 10.1111/j.1574-695X.2011.00883.x. [DOI] [PubMed] [Google Scholar]
- 11.CLSI . Clinical and Laboratory Standards Institute; Wayne, PA: 2008. National Committee for Clinical Laboratory Standards. Performance standards for antimicrobial susceptibility testing: fifteenth informational supplement. M100- S 15. [Google Scholar]
- 12.Suh D.K., Song J.C. Analysis of Salmonella enterica serotype Enteritidis isolated from human and chickens by repetitive sequence-PCR fingerprinting, antibiotic resistance and profiles. J Vet SCi. 2006;7:37–41. doi: 10.4142/jvs.2006.7.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hunter P.R., Gaston M.A. Numerical index of the discriminatory ability of typing systems: an application of Simpson's index of diversity. J Clin Microbiol. 1988;26:2465–2466. doi: 10.1128/jcm.26.11.2465-2466.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barco L., Barrucci F., Olsen J.E., Ricci A. Salmonella source attribution based on microbial subtyping. Int J Food Microbiol. 2013;15(163):193–203. doi: 10.1016/j.ijfoodmicro.2013.03.005. [DOI] [PubMed] [Google Scholar]
- 15.Sadkowska-Todys M., Czarkowski M.P. Salmonellosis in Poland in 2012. Przegl Epidemiol. 2014;68:243–248. [PubMed] [Google Scholar]
- 16.Fernandes S.A., Tavechio A.T., Ghilardi A.C., Dias A.M., Almeida I.A., Melo L.C. Salmonella serovars isolated from humans in Sao Paulo State, Brazil, 1996–2003. Rev Esp Anestesiol Reanim. 2006;48:179–184. doi: 10.1590/s0036-46652006000400001. [DOI] [PubMed] [Google Scholar]
- 17.Lee W.S., Puthucheary S.D., Boey C.C. Non-typhoid Salmonella gastroenteritis. J Paediatr Child Health. 1998;34:387–390. doi: 10.1046/j.1440-1754.1998.00247.x. [DOI] [PubMed] [Google Scholar]
- 18.Spiliopoulou I., Zografou S., Goula A., Dimitracopoulos G., Christofidou M. Molecular epidemiology and antibiotic resistance patterns of Salmonella enterica from southwestern Greece. Chemotherapy. 2007;53:392–396. doi: 10.1159/000109768. [DOI] [PubMed] [Google Scholar]
- 19.Li Y., Xie X., Xu X., Wang X., Chang H., Wang C. Nontyphoidal salmonella infection in children with acute gastroenteritis: prevalence, serotypes, and antimicrobial resistance in Shanghai, China. Foodborne Pathog Dis. 2014;11:200–206. doi: 10.1089/fpd.2013.1629. [DOI] [PubMed] [Google Scholar]
- 20.Morshed R., Peighambari S.M. Drug resistance, plasmid profile and random amplified polymorphic DNA analysis of Iranian isolates of Salmonella enteritidis. New Microbiol. 2010;33:47–56. [PubMed] [Google Scholar]
- 21.Ranjbar R., Giammanco G.M., Aleo A., Plano M.R., Naghoni A., Owlia P. Characterization of the first extended-spectrum β-lactamase-producing nontyphoidal Salmonella strains isolated in Tehran, Iran. Foodborne Pathogens Dis. 2010;7:91–95. doi: 10.1089/fpd.2009.0382. [DOI] [PubMed] [Google Scholar]
- 22.Choi S.H., Woo J.H., Lee J.E., Park S.J., Choo E.J., Kwak Y.G. Increasing incidence of quinolone resistance in human non-typhoid Salmonella enterica isolates in Korea and mechanisms involved in quinolone resistance. J Antimicrob Chemother. 2005;56:1111–1114. doi: 10.1093/jac/dki369. [DOI] [PubMed] [Google Scholar]
- 23.Meakins S., Fisher I.S., Berghold C., Gerner-Smidt P., Tschape H., Cormican M. Antimicrobial drug resistance in human nontyphoidal Salmonella isolates in Europe 2000–2004: a report from the Enter-net International Surveillance Network. Microb Drug Resist. 2008;14:31–35. doi: 10.1089/mdr.2008.0777. [DOI] [PubMed] [Google Scholar]
- 24.Gatto A.J., Peters T.M., Green J., Fisher I.S., Gill O.N., O'Brien S.J. Distribution of molecular subtypes within Salmonella enterica serotype Enteritidis phage type 4 and S. Typhimurium definitive phage type 104 in nine European countries, 2000-2004: results of an international multi-centre study. Epidemiol Infect. 2006;134:729–736. doi: 10.1017/S0950268805005820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hawkey P. Molecular epidemiology of clinically significant antibiotic resistance genes. Br J Pharmacol. 2008;153(S1):S406–S413. doi: 10.1038/sj.bjp.0707632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Angulo F.J., Johnson K.R., Tauxe R.V., Cohen M.L. Origins and consequences of antimicrobial-resistant nontyphoidal Salmonella: implications for the use of fluoroquinolones in food animals. Microb Drug Resist. 2000;6:77–83. doi: 10.1089/mdr.2000.6.77. [DOI] [PubMed] [Google Scholar]
- 27.Liebana E., Garcia-Migura L., Breslin M.F., Davies R.H., Woodward M.J. Diversity of strains of Salmonella enterica serotype enteritidis from English poultry farms assessed by multiple genetic fingerprinting. J Clin Microbiol. 2001;39:154–161. doi: 10.1128/JCM.39.1.154-161.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yu C.Y., Chou S.J., Yeh C.M., Chao M.R., Huang K.C., Chang Y.F. Prevalence and characterization of multidrug-resistant (type ACSSuT) Salmonella enterica serovar Typhimurium strains in isolates from four gosling farms and a hatchery farm. J Clin Microbiol. 2008;46:522–526. doi: 10.1128/JCM.00709-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chiu C.H., Su L.H., Chu C.H., Wang M.H., Yeh C.M., Weill F.X. Detection of multidrug-resistant Salmonella enterica serovar typhimurium phage types DT102, DT104, and U302 by multiplex PCR. J Clin Microbiol. 2006;44:2354–2358. doi: 10.1128/JCM.00171-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Albufera U., Bhugaloo-Vial P., Issack M.I., Jaufeerally-Fakim Y. Molecular characterization of Salmonella isolates by REP-PCR and RAPD analysis. Infection, Genetics and Evolution: Infect Genet Evol. 2009;9:322–327. doi: 10.1016/j.meegid.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 31.Chmielewski R., Wieliczko A., Kuczkowski M., Mazurkiewicz M., Ugorski M. Comparison of ITS Profiling, REP- and ERIC-PCR of Salmonella Enteritidis Isolates from Poland. J Vet Med B. 2002;49:163–168. doi: 10.1046/j.1439-0450.2002.00544.x. [DOI] [PubMed] [Google Scholar]
- 32.Campioni F., Bergamini A.M.M., Falcão J.P. Genetic diversity, virulence genes and antimicrobial resistance of Salmonella Enteritidis isolated from food and humans over a 24-year period in Brazil. Food Microbiol. 2012;32:254–264. doi: 10.1016/j.fm.2012.06.008. [DOI] [PubMed] [Google Scholar]
- 33.Ammari S., Laglaoui A., En-nanei L., Bertrand S., Wildemauwe C., Abid M. Characterization of Salmonella enteritidis isolated from foods and patients in northern Morocco. J Infect Dev Ctries. 2009;3:695–703. doi: 10.3855/jidc.617. [DOI] [PubMed] [Google Scholar]