Abstract
Probiotic supplements are beneficial for animal health and rumen function; and lipopolysaccharides (LPS) from gram negative bacteria have been associated with inflammatory diseases. In this study the transcriptional profile in whole blood collected from probiotics-treated cows was investigated in response to stimulation with lipopolysaccharides (LPS) in vitro. Microarray experiment was performed between LPS-treated and control samples using the Agilent one-color bovine v2 bovine (v2) 4x44K array slides. Global gene expression analysis identified 13,658 differentially expressed genes (fold change cutoff ≥ 2, P < 0.05), 3816 upregulated genes and 9842 downregulated genes in blood in response to LPS. Treatment with LPS resulted in increased expression of TLR4 (Fold change (FC) = 3.16) and transcription factor NFkB (FC = 5.4) and decreased the expression of genes including TLR1 (FC = − 2.54), TLR3 (FC = − 2.43), TLR10 (FC = − 3.88), NOD2 (FC = − 2.4), NOD1 (FC = − 2.45) and pro-inflammatory cytokine IL1B (− 3.27). The regulation of the genes involved in inflammation signaling pathway suggests that probiotics may stimulate the innate immune response of animal against parasitic and bacterial infections. We have provided a detailed description of the experimental design, microarray experiment and normalization and analysis of data which have been deposited into NCBI Gene Expression Omnibus (GEO): GSE75240.
| Specifications | |
|---|---|
| Organism/cell line/tissue | Bos taurus/peripheral blood collected from probiotics treated Holstein-Friesian cows for 60 days |
| Sex | Female |
| Array type | Bovine v2 44k array (Agilent GPL11649) |
| Data format | Raw (Txt) and normalized averages (Txt) |
| Experimental factors | Gene expression analysis in lipopolysaccahride (LPS) vs control |
| Experimental features | Lactating Holstein-Friesian cows were given a recommended oral dose of a commercial probiotics supplement for 60 days. Whole blood collected from the probiotics-treated animals was exposed to lipopolysaccharides (LPS). |
| Consent | N/A |
| Sample source location | Greensboro, USA |
1. Direct link to deposited data
Microarray data from this study are available at [provide URL below]: http://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE75240.
2. Experimental design, materials and methods
2.1. Experimental design
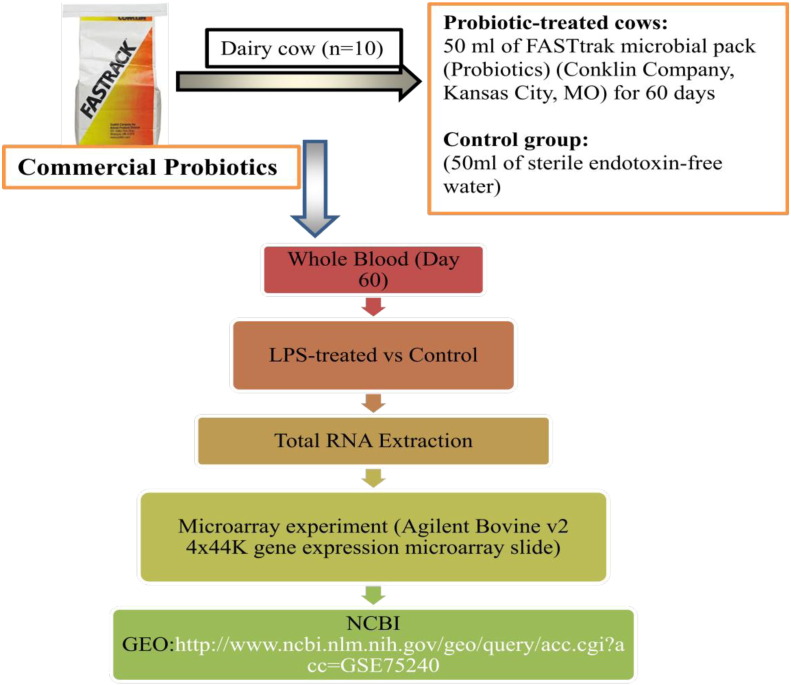
Lactating Holstein-Friesian cows (n = 10) were supplemented with a daily oral dose (50Ml) of the commercial probiotics FASTtrak microbial pack (Conklin Company, Kansas City, MO, USA) for 60 days. Peripheral blood samples were collected aseptically from Probiotics-treated animals into vaccutainer tubes containing anticoagulant ACD. The in vitro effect of lipopolysaccharide (LPS), endotoxin treatment was evaluated using blood samples collected from probiotics-treated animals at (Day 60). Whole blood (5 ml) samples with 1 × 107 viable cells/ml were pretreated with 100 ng/ml of lipopolysaccharide [1] (LPS, Sigma-A) and untreated sample served as control for 30 min using incubation conditions previously described by [2]. After treatment, samples were centrifuged at 10,000 rpm for 15 min and blood cell pellets were used for RNA isolation (Fig. 1).
Fig. 1.
Microarray analysis in lipopolysaccharide treated blood from cows orally administered with probiotics for 60 days. The DNA microarray experiment was performed using the Agilent Bovine v2 4x44K gene expression microarray slides. Raw and normalized average data from the microarray analysis are available at NCBI GEO repository under the accession number: GSE75240.
2.2. RNA isolation and cRNA synthesis
Total RNA from blood cell pellet was isolated using the ZR whole blood RNA miniprep kit (ZYMO RESEARCH, Irvine, CA) following the manufacturer's recommendations. The RNA procedure included differential lysis of red and white blood cells, and an on-column DNase digestion. Total RNA was quantified using a NanoDrop-1000 spectrophotometer (Thermo Fisher Scientific, Waltham, MA), and RNA integrity number (RIN) and quality was determined with the Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). Samples with high RNA integrity (RIN > 7) were stored at -80 °C until used for microarray analysis. A pooled RNA sample was generated by taking an equal concentration of RNA from experimental animals in each group (LPS vs control). Total RNA (500 ng, RIN > 7) was used to prepare Cyanine-3 (Cy3) labeled cRNA using the One-Color Low RNA Input Linear Amplification PLUS kit (Agilent Technologies, Santa Clara, CA, USA) according to the manufacturer's instructions. Sample purification was done with the RNAeasy column purification (QIAGEN, Valencia, CA, USA). Dye incorporation and cRNA yield (Table 1) were checked with the NanoDrop ND-1000 Spectrophotometer (Thermo-Scientific, Waltham, MA).
Table 1.
Total RNA integrity number (RIN), cRNA yield, and specific activity of Cy3 dye in LPS-treated and control samples.
| Sample name | Sample source | RIN | cRNA yield (μg) | Specific activity (pmol Cy3 μg cRNA) |
|---|---|---|---|---|
| LPS-treated | Whole blood | 8.7 | 1.65 | 12.94 |
| Control | Whole blood | 9.0 | 1.69 | 13.13 |
2.3. Microarray hybridization, scanning and data analysis
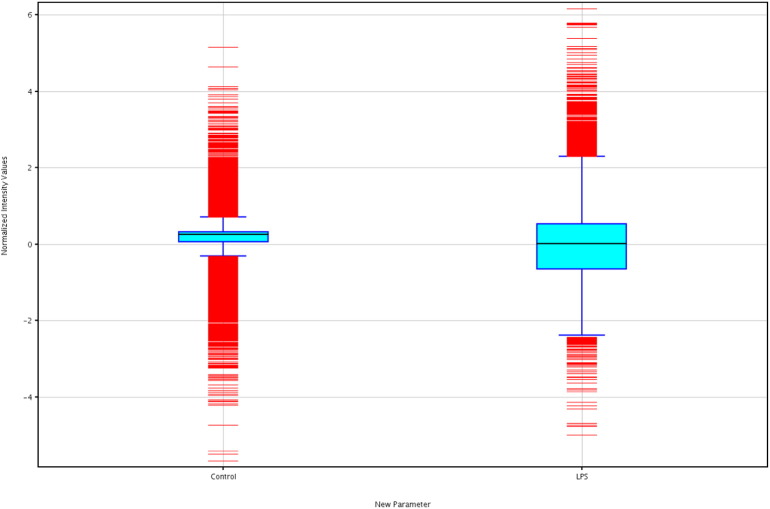
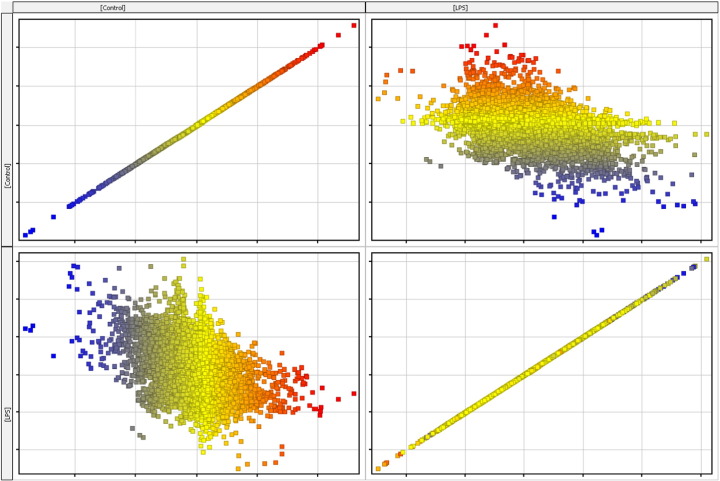
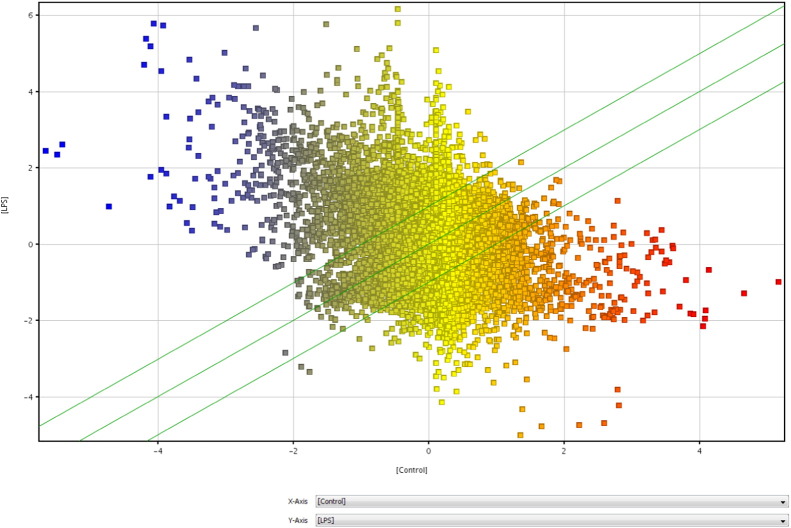
Hybridization was done on the Agilent Bovine v2 4x44K gene expression microarray slide (Agilent Technologies, Santa Clara CA, USA) for 17 h. Slides were scanned immediately after washing on the Agilent DNA Microarray Scanner (G2505B) using one color scan default settings for 4x44k array slides. Signal intensity data were extracted with the Agilent Feature Extraction Software 10.10.1.1 (Agilent) using default parameters (GE1-v5_95_Feb07) according to the manufacturer's protocol. Signal intensity data were extracted with the Agilent Feature Extraction Software (v 9.5.1). Data normalization and statistical analysis were performed using GeneSpring software 13.0 (Agilent Technologies, Santa Clara CA, USA). Boxwhisker, matrix, and scatter plots displaying the log ratio distribution of microarray data after normalization is shown in Fig. 2, Fig. 3, and Fig. 4 respectively. The raw data are available from the NCBI GEO repository, under the accession number GSE75240.
Fig. 2.
Boxwhisker plot of normalized data in LPS and control samples.
Fig. 3.
Matrix plot normalized data in LPS and control samples.
Fig. 4.
Scatter plot normalized data in LPS and control samples.
In conclusion probiotics supplementation had an effect on the response to LPS exposure in bovine whole blood. Treatment with LPS resulted in 13,658 differentially expressed genes (fold change cutoff ≥ 2, P < 0.05), 3816 upregulated genes and 9842 downregulated genes. Exposure to LPS had specific effects on Toll-like receptors genes expression including TLR4, TLR2 and TLR7. Thus, probiotic supplementation may modulate the response to gram negative bacteria.
Conflict of interest
The authors declare no conflict of interests.
Acknowledgement
The authors wish to acknowledge the support of Mr. Corey Burgess. This study was funded by the United States Department of Agriculture (USDA) project NC.X-271-5-13-120-1.
References
- 1.Worku M., Morris A. Binding of different forms of lipopolysaccharide and gene expression in bovine blood neutrophils. J. Dairy Sci. 2009;92(7):3185–3193. doi: 10.3168/jds.2008-1905. [DOI] [PubMed] [Google Scholar]
- 2.Adjei-Fremah S., Jackai L.E., Worku M. Analysis of phenolic content and antioxidant properties of selected cowpea varieties tested in bovine peripheral blood. Am. J. Anim. Vet. Sci. 2015;10(4):235–245. [Google Scholar]