Abstract
Previously published studies revealed that variation in expression of the DNA-binding protein ARID3a in B lymphocytes from patients with systemic lupus erythematosus (SLE) correlated with levels of disease activity (“Disease activity in systemic lupus erythematosus correlates with expression of the transcription factor AT-rich-interactive domain 3A” (J.M. Ward, K. Rose, C. Montgomery, I. Adrianto, J.A. James, J.T. Merrill et al., 2014) [1]). The data presented here compare DNA methylation patterns from SLE peripheral blood mononuclear cells obtained from samples with high numbers of ARID3a expressing B cells (ARID3aH) versus SLE samples with normal numbers of ARID3a+ B cells (ARID3aN). The methylation data is available at the gene expression omnibus (GEO) repository, “Gene Expression Omnibus: NCBI gene expression and hybridization array data repository” (R. Edgar, M. Domrachev, A.E. Lash, 2002) [2]. Isolated B cells from SLE ARID3aH and ARID3aN B samples were also evaluated via qRT-PCR for Type I interferon (IFN) signature and pathway gene expression levels by qRT-PCR. Similarly, healthy control B cells and B cells stimulated to express ARID3a with the TLR agonist, CpG, were also compared via qRT-PCR. Primers designed to detect 6 IFNa subtype mRNAs were tested in 4 IFNa, Epstein-Barr Virus-transformed B cell lines (“Reduced interferon-alpha production by Epstein-Barr virus transformed B-lymphoblastoid cell lines and lectin-stimulated lymphocytes in congenital dyserythropoietic anemia type I” (S.H. Wickramasinghe, R. Hasan, J. Smythe, 1997) [3]). The data in this article support the publication, “Human effector B lymphocytes express ARID3a and secrete interferon alpha” (J.M. Ward, M.L. Ratliff, M.G. Dozmorov, G. Wiley, J.M. Guthridge, P.M. Gaffney, J.A. James, C.F. Webb, 2016) [4].
Keywords: SLE, B cells, ARID3a
Specifications Table
| Subject area | Immunology |
| More specific subject area | SLE and ARID3a+B cells |
| Type of data | Figure, Tables, link |
| How data was acquired | Electrophoresis and BIOMARK HD |
| Data format | Raw, analyzed |
| Experimental factors | FACS-purified SLE and healthy B lymphocytes (+/- CpG-stimulation) |
| Experimental features | DNA was isolated from ARID3aHand ARID3aNtotal PBMCs; RNA was extracted from LCLs, peripheral blood SLE B cells, and healthy control B cells with or without CpG-stimulation for 24 hours. |
| Data source location | Oklahoma City, OK; USA |
| Data accessibility | Data is available within this article and deposited in NCBI׳s Gene Expression Omnibushttp://www.ncbi.nlm.nih.gov/geo/ accessiblevia GEO series accession number: GEO:GSE84965 |
Value of the data
-
•
DNA gene methylation data derived from SLE peripheral blood mononuclear cells were subdivided based on levels of ARID3a expression, a transcription factor which correlated with disease activity indices [1], allowing comparison of patient samples with high and low ARID3a levels.
-
•
Data for expression of a subset of IFNa associated genes obtained from SLE samples with high or low ARID3a expression, and from healthy control blood cells or those stimulated to express increased levels of ARID3a, allow comparison of effects of high and low ARID3a expression on gene expression.
-
•
Data provide validation of primer sets useful for studying Type I interferon signature genes.
1. Data
One database link, three tables, and one figure are provided in this article. Methyl-seq data from SLE PBMCs segregated based on high or normal numbers of ARID3a+ B cells was deposited in NCBI׳s GEO database under the following accession number GEO: GSE84965 [2]. Table 1, Table 2 show qRT-PCR data obtained via Biomark HD for Type I IFN pathway genes from RNA derived from SLE B cells subdivided based on ARID3a levels [1], and for healthy control B cells with or without CpG induced ARID3a expression [4]. IFN signature genes are in bold. Primers for RT-PCR and qRT-PCR are given in Table 3. Fig. 1 shows the results of RT-PCR of IFNa in four EBV-transformed lymphoblastoid B cell lines [3].
Table 1.
Upregulated genes in ARID3aH versus ARID3aN SLE B cells.
| Gene | ARID3aH | ARID3aN | P-value |
|---|---|---|---|
| ARID3a | 0.6843 | 0.0692 | 0.0008 |
| BCL2 | 4.6000 | 1.3283 | 0.0081 |
| BCL2L1 | 7.6980 | 0.3608 | 0.0049 |
| EPSTI1 | 1958.1900 | 17.6625 | 0.0034 |
| HERC5 | 17.4250 | 1.9833 | 0.0043 |
| IFI6 | 46.0745 | 3.7483 | 0.0227 |
| IFI27 | 1958.1900 | 17.6625 | 0.0034 |
| IFI44 | 23.6242 | 16.6467 | 0.3387 |
| IFI44L | 130.6440 | 29.8092 | 0.0369 |
| IFIT3 | 33.2118 | 14.8208 | 0.0691 |
| IFNA2 | 49.0570 | 22.9925 | 0.2194 |
| IFNAR1 | 1.0425 | 1.2433 | 0.5319 |
| IFNB1 | 9.3942 | 1.1575 | 0.0004 |
| IRF3 | 2.1863 | 0.1408 | 0.0008 |
| IRF5 | 1.3742 | 0.3783 | 0.0035 |
| IRF7 | 3.5360 | 0.2510 | 0.0006 |
| ISIG15 | 5.6233 | 0.6150 | 0.0118 |
| Ly6E | 21.4388 | 1.5683 | 0.0023 |
| MX1 | 33.6467 | 4.3542 | 0.0009 |
| MYD88 | 4.1236 | 2.2740 | 0.0999 |
| OAS1 | 11.7725 | 0.3267 | 0.0007 |
| OAS2 | 0.8475 | 0.2542 | 0.0847 |
| OAS3 | 6.0257 | 0.2313 | 0.0018 |
| PLSCR1 | 17.5975 | 0.8400 | 0.0526 |
| SIGLEC1 | 144.2100 | 55.8713 | 0.0570 |
| STAT1 | 2.2225 | 1.0158 | 0.0660 |
| TLR 7 | 5.7500 | 2.0808 | 0.0165 |
| TLR9 | 3.8250 | 2.8425 | 0.4706 |
| USP18 | 17.5975 | 0.8400 | 0.0010 |
Table 2.
Upregulated or downregulated genes in CpG-stimulated versus unstimulated healthy control B cells.
| Gene | CpG | Unstimulated | P-value |
|---|---|---|---|
| Upregulated | |||
| EPST1 | 3.14125 | 1.01875 | <0.000001 |
| HERC5 | 6.9225 | 0.9875 | <0.000001 |
| IFI6 | 1.95 | 1.00125 | 0.001894 |
| IFI27 | 4.9 | 1.58 | 0.001146 |
| IFI44 | 1.805 | 1.05125 | 0.003712 |
| IFI44L | 2.2325 | 1.23 | 0.001263 |
| IFIT3 | 3.06375 | 1.19625 | 0.000121 |
| IFNA2 | 3.16625 | 1.07125 | 0.015070 |
| IFNAR1 | 2.12 | 1.0075 | <0.000001 |
| IFNB1 | 3.3125 | 1.325 | 0.017893 |
| IRF3 | 1.94875 | 1.01875 | <0.000001 |
| IRF5 | 1.01375 | 1.04125 | 0.710859 |
| IRF7 | 1.5325 | 0.98625 | <0.000001 |
| ISG15 | 0.7825 | 0.8975 | 0.385249 |
| Ly6E | 1.6825 | 1.015 | 0.000261 |
| MX1 | 2.61125 | 1.0375 | 0.000002 |
| MYD88 | 1.31125 | 0.96625 | 0.000083 |
| OAS1 | 4.3175 | 0.945 | <0.000001 |
| OAS2 | 1.7975 | 0.99 | 0.000954 |
| OAS3 | 2.1375 | 1.0725 | 0.001380 |
| PLSCR1 | 1.5125 | 0.99625 | 0.000332 |
| STAT1 | 1.0025 | 1.01375 | 0.808948 |
| TLR7 | 5.30625 | 1.0375 | 0.000002 |
| TLR9 | 2.28375 | 0.99875 | 0.001518 |
| Downregulated | |||
| BCL2L1 | 0.7475 | 0.9525 | 0.009423 |
IFN signature genes are in bold.
Table 3.
Primer sequences.
| Gene | Primer sequence (5′ to 3′) | Figure |
|---|---|---|
| IFIT1 | CTCCTTGGGTTCGTCTATAAATTG AGTCAGCAGCCAGTCTCAG |
Fig. 1a in [4] |
| HPRT1 | TTGGTCAGGCAGTATAATCC GGGCATATCCTACAACAAAC |
Fig. 1a in [4] |
| GAPDH | GCCGCATCTTCTTTTGCGT GCCCAATACGACCAAATCCGT |
Fig. 1a in [4] |
| CMYC | ACTCTGAGGAGGAACAAGAA TGGAGACGTGGCACCTCTT |
Fig. 1e in [4] |
| ARID3A | AACAAGAAGCTGTGGCGTGA TCATGTATTGGGTCCGCAGG |
Fig. 1c in [4] |
| ACTIN | ATCTGGCACCACACCTTCTACAATGAGCTGCG CGTCATACTCCTGCTTGCTGATCCACATCTGC |
Fig. 1 |
| IFNA | CCTGGCACAAATGAGGAGAA AGCTGCTGGTAAAGTTCAGTATAG |
Fig. 2a in [4] Fig. 1 |
| OAS1 | TACCCTGTGTGTGTGTCCAA AGAGGACTGAGGAAGACAACC |
Fig. 3a, 4d in [4]Table 1 |
| OAS2 | TGGTGAACACCATCTGTGAC CCATCGGAGTTGCCTCTTAA |
|
| OAS3 | AGGACTGGATGGATGTTAGCC ACTTGTGGCTTGGGTTTGAC |
Table 1 |
| ISG15 | CTGAGAGGCAGCGAACTCA GCTCAGGGACACCTGGAA |
Table 1 |
| PLSCR1 | GTTGTCCCTGCTGCCTTCA TGGGTGCCAAGTCTGAATAACA |
Table 2 |
| HERC5 | TTCAGATCACATGTGGAGATTACC GTTCTGTCCCCAGGCAAAA |
Table 1, Table 2 |
| IFI44 | GGCTTTGGTGGGCACTAATA TGCCATCTTTCCCGTCTCTA |
|
| IFIT3 | ACTGGCAATTGCGATGTACC GCTCAATGGCCTGCTTCAAA |
Table 2 |
| LY6E | TGCTCCGACCAGGACAACTA GGCTGTGGCCAAATGTCAC |
Table 1, Table 2 |
| MX1 | ATGCTACTGTGGCCCAGAAA GGCGCACCTTCTCCTCATA |
Table 1, Table 2 |
| USP18 | TGAATGTGGACTTCACCAGGATA GCAGCAGAAGCATCTGGAAA |
Table 1 |
| IFI44L | GCAAAAGTGAAGCAAGTTCACA GAACCTCACTGCAATCATCCA |
Table 1, Table 2 |
| IFI6 | TGCTACCTGCTGCTCTTCA TCAGGGCCTTCCAGAACC |
Table 1 |
| SIGLEC1 | AGGAGGCGTGTTTGTAAGCA TGTGGCTGCATCAGGATCAA |
Fig. 3a in [4] |
| IFI27 | TTGTGGCTACTCTGCAGTCA CCCAGGATGAACTTGGTCAA |
Table 1 |
| EPSTI1 | GCAAGAGCAAGAAAGAGCCAAA CCTTGGAGTCGGTCCAGAAAA |
Table 1, Table 2 |
| IRF3 | ACCAATGGTGGAGGCAGTAC TGGGGCCAACACCATGTTA |
Fig. 3b, 4e in [4] |
| IRF5 | AGATCTACGAGGTCTGCTCCAA CCTCTCCTGCACCAAAAGAGTA |
|
| IRF7 | GGCAGAGCCGTACCTGTCA ACCGTGCGGCCCTTGTA |
|
| TLR7 | TCTTCAACCAGACCTCTACATTCC AGCCCCAAGGAGTTTGGAAA |
Table 1, Table 2 |
| TLR9 | TGCAACTGGCTGTTCCTGAA ACAAGGAAAGGCTGGTGACA |
Table 2 |
| MYD88 | CTGCAGAGCAAGGAATGTGAC TGCTGGGGAACTCTTTCTTCA |
|
| IFNAR1 | AGTGACGCTGTATGTGAGAAAA ACGGGAGAGCAAATAATGCA |
Fig. 3b in [4] Table 2 |
| STAT1 | ATGCTGGCACCAGAACGAA GCTGGCACAATTGGGTTTCAA |
|
| IFNA2 | AGGATTCAGCGGGAACACAA CAATCTCAAACTCTGGTGGTTCAAA |
Table 2 |
| IFNB1 | ATGAGCAGTCTGCACCTGAA GACTGTACTCCTTGGCCTTCA |
Table 2 |
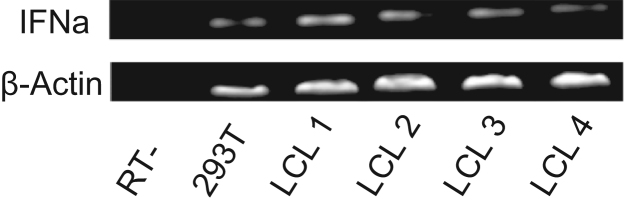
Fig. 1.
EBV-transformed lymphoblastoid B cell lines (LCLs) express IFNa. RT-PCR analysis of IFNa expression in 4 distinct EBV-transformed lymphoblastoid lines was measured in comparison to the positive control cell line, 293T. A no template (NT) negative control is also shown. The housekeeping gene, β-actin, was amplified to demonstrate relative levels of IFNa RNA in each cell line.
2. Experimental design, materials and methods
2.1. Peripheral blood cells and cell lines
Total peripheral blood mononuclear cells (PBMCs) were obtained via Ficoll purification, and were stained for the pan-B cell marker CD20 and intracellular ARID3a prior to analyses by flow cytometry, as previously described [1]. These data allowed subdivision of SLE samples into ARID3a high and ARID3a normal patient samples, such that ARID3aH SLE samples had numbers of ARID3a+ B cells >2 standard deviations above the average numbers of ARID3a+ B cells in healthy controls (>9830 ARID3a+ B cells/ml), versus ARID3aN (<9830 ARID3a+ B cells/ml), as defined previously [1]. B lymphocytes purified by flow cytometric sorting (>97% purity via post-sort analyses) were used immediately for RNA preparation in the case of SLE samples, or in the case of healthy control cells, were grown in complete RPMI media (RPMI 1640, 5×10−5 M β-mercaptoethanol, 100 U/ml penicillin, 100 µg/ml streptomycin, 2 mM glutamine and 1 mM sodium pyruvate) supplemented with 4% heat inactivated fetal bovine serum (FBS), with or without 5 µg/ml Class CpG oligonucleotide for 24 h, as previously described [4]. Epstein-Barr Virus (EBV)-transformed lymphoblastoid B cell lines (LCLs) were generated from 4 SLE patient samples and maintained in complete RPMI media.
2.2. Methyl-seq
To determine if increased expression of ARID3a within SLE patient samples was associated with alterations in DNA methylation, genomic DNA was isolated using standard phenol/Chloroform extraction protocols from total PBMCs obtained from each of two SLE patient samples characterized as ARID3aH and two independent SLE samples characterized as ARID3a low. DNA was fragmented on a Covaris S2 sonicator (Covaris, Woburn, MA) to an average size of ~350 bp in length and methylated DNA was isolated using the MethylMiner Methylated DNA Enrichment Kit (Life Technologies, Carlsbad, CA). Illumina sequencing libraries were prepared from each sample using the Illumina Truseq DNA LT Sample Prep Kit (Illumina, San Diego, CA) by the Genomics Core facility at Oklahoma Medical Research Foundation. Libraries were sequenced on an Illumina Hiseq 2000 instrument with paired-end 100 bp reads. Quality control metrics were assessed with Picard tools v. (https://broadinstitute.github.io/picard/). After sequencing, reads were aligned to the human reference genome hg19 using the aligner BWA-MEM [5] followed by local realignment around problematic indel sequences using the Genome Analysis Tool Kit (GATK) [6]. Genes with statistically significant methylation differences were defined using EpiCenter v. 1-6-1-8 [7]. Methylation differences were tested over promoters of the genes, defined as 2000 bp regions upstream of gene’ transcription start sites. The differentially methylated regions were visualized in the IGV integrative genomics viewer [8]. For visualization in the UCSC Genome Browser BigWig files were created from the final BAM files using a combination of BEDTools [9] and UCSC conversion utilities [10].
2.3. Biomark HD assays
Peripheral blood mononuclear cells were isolated from peripheral blood of 6 SLE patients and 2 healthy individuals, and were analyzed for ARID3a expression as described above by flow cytometry. B lymphocytes were enriched from the remaining PBMCs via negative selection using magnetic beads containing other lineage markers, and the remaining cells were stained with CD20 for fluorescence activated cell sorting (FACS) using a FACSAria II (BD Biosciences). Post-sort analyses revealed >98% CD20+ B lymphocytes. RNA was isolated, quantified and assessed for integrity using Agilent Total RNA Pico chips on the 2100 Bioanalyzer (Agilent Technologies, Boblingen, Germany). The DELTAgene assay designer was used for primer design for optimal performance on the Biomark HD system. Primer specificity was determined via melting curve analysis at 400 nM. cDNA preparation (Fluidigm preamp master mix, PM100-5580), amplification (Fluidigm, DELTAgene assay kit), qRT-PCR and analyses were all performed as previously described [11]. Data in Table 1, Table 2 are normalized to the housekeeping gene Hprt1. A list of primers for the genes assessed is given in Table 3.
2.4. IFNa analyses of EBV lines
For qRT-PCR, RNA was extracted using Tri-Reagent (MRC, Inc.) and chloroform:isoamyl alcohol 24:1 (Sigma), precipitated in isopropanol, and collected via centrifugation. cDNA was synthesized at 37 °C for 1 h with M-MLV reverse transcriptase (Promega) and random primers (Promega), and amplified for 40 cycles at 60 °C for 30 s, 72 °C for 1 min, and 95 °C for 30 s for IFNa (IFNA2, IFNA5, IFNA6, IFNA8, IFNA14, IFNA16) gene expression. Amplified products were electrophoresed through 2% agarose gel.
Acknowledgments
The authors thank V. Roberts and T.D. Templeton for technical assistance. Funding from National Institutes of Health: AIO90343 and AI044215 (CFW), and AR053483, GM103510, AI101934, AI082714, GM104938 (JMG, JAJ), and AR056360, AR063124, and GM110766 (PMG) and Lupus Foundation of America (CFW) supported these studies.
Footnotes
Transparency data associated with this article can be found in the online version at http://dx.doi.org/10.1016/j.dib.2016.08.049.
Contributor Information
Julie M. Ward, Email: julie-ward@ouhsc.edu.
Michelle L. Ratliff, Email: michelle-ratliff@ouhsc.edu.
Mikhail G. Dozmorov, Email: Mikhail.dozmorov@vcuhealth.org.
Graham Wiley, Email: graham-wiley@omrf.org.
Joel M. Guthridge, Email: joel-guthridge@omrf.org.
Patrick M. Gaffney, Email: patrick-gaffney@omrf.org.
Judith A. James, Email: judith-james@omrf.org.
Carol F. Webb, Email: carol-webb@ouhsc.edu.
Transparency document. Supplementary material
Supplementary material
.
References
- 1.Ward J.M., Rose K., Montgomery C., Adrianto I., James J.A., Merrill J.T. Disease activity in systemic lupus erythematosus correlates with expression of the transcription factor AT-rich-interactive domain 3A. Arthritis Rheumatol. 2014;66:3404–3412. doi: 10.1002/art.38857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Edgar R., Domrachev M., Lash A.E. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wickramasinghe S.N., Hasan R., Smythe J. Reduced interferon-alpha production by Epstein-Barr virus transformed B-lymphoblastoid cell lines and lectin-stimulated lymphocytes in congenital dyserythropoietic anaemia type I. Br. J. Haematol. 1997;98:295–298. doi: 10.1046/j.1365-2141.1997.1993016.x. [DOI] [PubMed] [Google Scholar]
- 4.Ward J.M., Ratliff M.L., Dozmorov M.G., Wiley G., Guthridge J.M., Gaffney P.M., James J.A., Webb C.F. Human effector B lymphocytes express ARID3a and secrete interferon alpha. J. Autoimmun. 2016 doi: 10.1016/j.jaut.2016.08.003. NIHMS 809919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A. The Genome analysis toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang W., Umbach D.M., Vincent Jordan N., Abell A.N., Johnson G.L., Li L. Efficiently identifying genome-wide changes with next-generation sequencing data. Nucleic Acids Res. 2011;39:e130. doi: 10.1093/nar/gkr592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Robinson J.T., Thorvaldsdottir H., Winckler W., Guttman M., Lander E.S., Getz G. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kent W.J., Zweig A.S., Barber G., Hinrichs A.S., Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ratliff M.L., Mishra M., Frank M., Guthridge J.M., Webb C.F. The transcription factor ARID3a is important for in vitro differentiation of human hematopoietic progenitors. J. Immunol. 2015;192:614–623. doi: 10.4049/jimmunol.1500355. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material