Abstract
Dendritic cells (DCs) are capable of processing and presenting exogenous antigens using MHC class I molecules. This pathway is called antigen cross-presentation and plays an important role in the stimulation of naïve CD8+ T cells for infectious and tumor immunity. Our previous studies in DC2.4 cells and bone marrow-derived DCs revealed that exogenously added ovalbumin (OVA) is processed through endoplasmic reticulum (ER)-associated degradation (ERAD) for cross-presentation. In this study, we aimed to further confirm these results by purification of the subcellular compartment in which exogenous antigens undergo ERAD from homogenates of DC2.4 cells pretreated with biotinylated OVA (bOVA). bOVA-containing vesicles were purified using streptavidin (SA)-magnetic beads from cell homogenates and were found to contain ER chaperones and ERAD components together with proteins for antigen presentation. In purified microsomes, bOVA was retained in membranous fractions and degraded by the ubiquitin proteasome system in presence reticulocyte lysates and ATP. These results strongly suggested that DCs processed and degraded exogenous antigens through ERAD for cross-presentation in this purified subcellular compartment.
Keywords: Cell biology, Immunology
1. Introduction
Major histocompatibility (MHC) class I molecules are generally expressed on the cell surface with short peptides generated from endogenous antigens by the ubiquitin-proteasome system and function as immunological self markers [1]. This mechanism of antigen processing and presentation in single cells is called direct presentation [1]. MHC class I molecules associated with non-self antigens, such as specific proteins from cancer cells or infectious agents, serve as non-self markers [1, 2, 3]. Cytotoxic T lymphocytes (CTLs), which destroy these non-self cells by recognizing non-self markers, are derived from naive CD8+ T cells after appropriate stimulation by antigen presenting cells (APCs) harboring both with specific antigen-MHC class I complexes and costimulatory molecules [4, 5]. In APCs, antigens from non-self cells can be internalized and presented on MHC class I molecules [6, 7, 8]. This process is a unique phenomenon among several subsets of dendritic cells (DCs) [9, 10, 11, 12, 13], and is called cross-presentation [6, 7, 8]. Cross-presentation, is a critical mechanism mediating immune responses against non-self cells [14, 15, 16, 17, 18, 19, 20, 21, 22] and parasite-infected cells [23] and can induce peripheral tolerance for self-antigens [8].
In recent years, accumulating evidence has shown that generation of antigenic peptides for cross-presentation is dependent on the ubiquitin-proteasome system [24, 25, 26, 27] and transporter-associated with antigen presentation (TAP) [28], which is required for direct presentation through intracellular pathways [7, 8]. Several groups have described the molecular mechanisms of cross-presentation through intracellular pathways [24, 25, 26, 27, 29]. Some of the above analyses have suggested that cross-presentation occurs through specialized intracellular compartments, such as the endoplasmic reticulum (ER)-phagosome [24, 29] and ERgosome [25, 27]. After internalization of an antigen, only DCs are capable of processing and presenting exogenous antigens on MHC class I molecules [24, 29, 30, 31, 32]. The superior ability of DCs to participate in cross-presentation is largely attributed to their antigen-processing capacity by the ubiquitin-proteasome system [32]. Therefore, in order to avoid lysosomal degradation of exogenous antigens, DCs express low levels of lysosomal proteases [33] with protease inhibitors [33] by harboring phagosomes with a high phagosomal pH [34, 35]. Retained antigens are then retrotransported into the cytosol [36], possibly mediated by Sec61 translocon [28, 38], and are processed through the ubiquitin-proteasome system [28, 38]. Peptides transported into the ER by TAP can then be loaded onto MHC class I molecules [24, 29].
Misfolded and damaged proteins are harmful to cells and are therefore destined for rapid degradation [39, 40, 41]. In the cytoplasm, unfolded proteins are degraded by the ubiquitin-proteasome system. In the ER, proteins with defects in folding, assembly, and glycosylation are recognized and retrograde-transported to the cytoplasm by the ER-associated degradation (ERAD) system, followed by degradation through the ubiquitin-proteasome system [42, 43, 44]. In both pathways, the generated antigenic peptides undergo direct presentation on MHC class I molecules [45, 46, 47, 48]. As shown previously, retrotransport and degradation of endogenous antigens in intracellular pathways occur via a system similar to that of ERAD [28, 38]. However, it is still unclear how exogenous antigens are recognized by the ERAD system.
In this study, we investigated the mechanisms for processing of exogenous antigens by intracellular degradation systems using purified exogenous antigen-containing vesicles and in vitro reconstruction of ERAD for cross-presentation. Our data provide important mechanistic insights into the recognition of exogenous antigens by ERAD.
2. Material and methods
2.1. Cell culture
DC2.4, a DC line [49], was provided by Dr. K. L. Rock (Dana-Farber Cancer Institute, Boston, MA, USA). Cells were cultured in RPMI-1640 (Sigma, St. Louis, MO, USA) supplemented with 2 mM l-glutamine, 1 mM sodium pyruvate, 0.1 mM nonessential amino acids, 100 U/mL penicillin-streptomycin, 55 mM 2-mercaptoethanol, 10 mM HEPES (pH 7.5), and 10% fetal calf serum (FCS) at 37 °C in 5% CO2 unless otherwise indicated. Polymyxin B (50 mg/mL) was added to all cell cultures.
2.2. Antibodies and reagents
The antibodies used in this study were as follows: anti-BiP (rabbit; MBL), anti-calreticulin (for immunoprecipitation: rabbit antibodies from Affinity BioReagents, Golden, CO, USA; for western blotting: mouse antibodies from Stressgen, Victoria, British Columbia, Canada), anti-caveolin 1 (mouse; BD Biosciences, San Diego, CA, USA), anti-CHIP (for western blotting: rabbit antibodies as a gift from Dr. K. Tanaka, Tokyo Metropolitan Institute of Medical Science, Tokyo, Japan; and chicken antibodies as a gift from Mr. S. Seki, MBL, Ina, Japan), anti-Flag (mouse; Sigma), anti-GM-130 (mouse; BD Biosciences), anti-Hsp70 (mouse; Stressgen), anti-KDEL (mouse; Stressgen), anti-LAMP-1 (rat; BD Biosciences), anti-H–2 Kb (mouse; Serotec), anti-multi-ubiquitin (mouse; MBL), anti-ovalbumin (OVA; rabbit; Polysciences, Warrington, PA, USA), anti-protein disulfide isomerase (PDI; rabbit; Stressgen), anti-proteasome 20S subunit alpha 5 (rabbit; Affinity Bio Reagents), anti-Rab5 (mouse; BD Biosciences), anti-Sec61α (rabbit; Upstate Cell Signaling Solutions, New York, NY, USA), anti-TAP1 (goat; Santa Cruz Biotechnology, Santa Cruz, CA, USA), anti-TAP2 (goat; Santa Cruz Biotechnology), anti-Tapasin (rabbit; Stressgen), and anti-VCP (for western blotting: rabbit antibodies from BD Biosciences; for immunoprecipitation: goat antibodies from Santa Cruz Biotechnology) antibodies. As secondary antibodies, streptavidin (SA)-peroxidase conjugate (SA-HRP; Vector Laboratories, Burlingame, CA, USA), goat anti-rabbit IgG peroxidase conjugate (Zymed), goat anti-mouse IgG peroxidase conjugate (Zymed), goat anti-Rat IgG peroxidase conjugate (Zymed), and bovine anti-goat IgG peroxidase conjugate (Santa Cruz Biotechnology) were used. OVA were biotinylated (bOVA) using a FluoReporter Biotin-XX protein labeling kit (Molecular Probes, Eugene, OR, USA). On average, bOVA contained 2 mol biotin per 1 mol OVA. Flag-tagged ubiquitin, MG132, lactacystine, and chloroquine were purchased from Sigma. Reticulocyte lysates (RLs) were purchased from Promega (Madison, WI, USA). Gels were stained using a SilverQuest silver staining kit (Invitrogen, Carlsbad, CA, USA). SA-magnetic beads were purchased from Novagen.
2.3. Preparation of microsome fractions
DC2.4 cells were incubated with bOVA (250 μg/mL) for 4 h, washed twice in phosphate-buffered saline (PBS), resuspended in homogenization medium (0.25 M sucrose, 1 mM EDTA, 10 mM HEPES-NaOH [pH 7.4]), and then disrupted by 10 strokes with a Dounce homogenizer. Unbroken cells and nuclei were removed by centrifugation at 2,000 × g for 10 min twice. When indicated, 2.5 mg/mL bOVA was added to control cell homogenates. The post nuclear supernatant was pelleted at 100,000 × g for 45 min, and pellets were resuspended in homogenization medium. Aliquots were incubated with or without 100 μg/mL trypsin (Sigma) in the presence or absence of 1% Triton X–100 for 30 min at 37 °C.
2.4. Discharge of bOVA from microsomes and degradation of bOVA
Microsomes from DC2.4 cells that had been incubated with bOVA were incubated with or without a 50% volume of RL, ATP (3 mM), and the indicated inhibitors. After incubation at 37 °C for 2 h, bOVA was recovered with SA-magnetic beads and resolved by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) followed by western blotting with SA-HRP.
2.5. Immunoprecipitation
Microsomes from DC2.4 cells that had been incubated with bOVA were incubated with or without a 50% volume of RL, ATP (3 mM), and the indicated inhibitors. After incubation at 37 °C for 2 h, supernatants were collected by 100,000 × g for 45 min. Samples were pre-cleared with protein G sepharose (Amersham Pharmacia Biotech) and incubated with anti-HSP70 antibodies for precipitation by Protein G. Precipitated samples were analyzed by SDS-PAGE and western blotting.
2.6. In vitro ubiquitination of bOVA in vesicles
Microsomes from DC2.4 cells that had been incubated with bOVA were incubated with or without a 50% volume of RL and 0.2 pM Flag-tagged ubiquitin. After incubation at 37 °C for 2 h, bOVA was recovered with SA-magnetic beads and resolved by SDS-PAGE followed by western blotting.
2.7. Purification of microsomes in which bOVA was undergoing ERAD
Microsomes from DC2.4 cells that had been incubated with bOVA at 37 °C for 2 h were incubated with or without SA-magnetic beads (Novagen) at room temperature for 30 min. Bead-bound vesicles were thus purified and used for further experiments. Isolated vesicles from cells incubated with or without bOVA were solubilized in TNE (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.5 M EDTA, 1% Nonidet P-40). Portions of solubilized proteins were resolved on 7.5–15% SDS-PAGE and transferred to Immobilon P membranes for western blotting. Protein bands were visualized by chemiluminescence.
3. Results
3.1. Degradation of exogenously added bOVA in microsomes
We previously found that exogenously added bOVA was accumulated in membranous compartments and processed through ERAD for cross-presentation. Therefore, in this study, we first analyzed the in vitro reconstitution of the ERAD system using exogenously added antigens, microsomes, and RLs. Microsomes were prepared from DC2.4 cells pretreated with bOVA. bOVA that was recovered in the microsomal fraction was resistant to trypsin but could be degraded after addition of TX100, indicating that bOVA was accumulated in membranous compartments as BiP (Fig. 1A). The bOVA found in microsomes was degraded in the presence of the RL, and addition of ATP accelerated degradation, indicating that this processing mechanism was energy dependent (Fig. 1B, C). Moreover, degradation occurred as a function of chase time dependent on RL and was blocked by the proteasome inhibitors MG132 and lactacystine but not by the lysosome inhibitor chloroquine (Fig. 1D, E). In contrast, when bOVA was added after fractionation, the protein was stable and was not degraded, regardless of the addition of RL (Fig. 1D, E). After incubation at 37 °C for 1 h, about 40% of bOVA was still associated with microsomes; however, significant amounts of bOVA were released from microsomes, transported outside of microsomes, and recovered in supernatants in the presence of RL (Fig. 1F). Released bOVA were coimmunoprecipitated with cytosolic molecular chaperone, Hsp70 (Fig. 1G). These results indicated that exogenously added bOVA accumulated in membranous compartments and was then transported outside of microsomes, unfolded and degraded by proteasomes in an energy-dependent manner. Therefore, exogenously added bOVA was processed through an ERAD-like protein degradation mechanism.
Fig. 1.
In vitro reconstitution of retrotransport and degradation using OVA in microsomes (A), Microsomes with (+) or without (−) prior addition of bOVA were treated with (+) or without (−) trypsin and Triton X–100 (TX100). Proteins (2 μg) were resolved on 7.5–15% SDS-PAGE and subjected to western blotting with the indicated antibodies. (B), Microsomes with (+) or without (−) prior addition of bOVA were treated with (+) or without (−) RL and ATP for 1 h. Proteins (2 μg) were subjected to western blotting with SA-HRP. Asterisks in the right indicate non-specific bands with SA-HRP. Equivalent results were attained at least three independent assays. (C), Quantification of the results shown in (B). Bars, mean ± S.D. (error bars) of three independents experiments. Student's test was used to compare (bOVA + RL − ATP −) sample with (bOVA + RL + ATP −) sample, (bOVA + RL − ATP +) sample, and (bOVA + RL + ATP +). **, ρ < 0.01, *,ρ < 0.1, NS, not significant, n = 3. (D), Microsomes with prior addition of bOVA were treated with RL, ATP, and the indicated inhibitors: MG132 (MG: 10 μM), lactacystine (LC: 2 μM), and chloroquine (CQ: 10 μM). Microsomes without prior addition of bOVA were incubated with 2.5 μg of bOVA after fractionation (after) and were treated with RL and ATP. Proteins (2 μg) were subjected to western blotting with SA-HRP. (E), Quantification of the results shown in (D). Bars, mean ± S.D. (error bars) of three independents experiments. Student's test was used to compare (Before, RL + reagent −) sample with (Before RL + reagent +) samples and (After, RL − reagent −) sample with (After, RL + reagent −) sample. *, ρ < 0.01, NS, not significant, n = 3. (F), Microsomes with (+) or without (−) prior addition of bOVA were treated with (+) or without (−) RL in presence of ATP and MG132 (10 μM) for 1 h. Samples were then separated into supernatants and pellets by centrifugation at 100,000 × g for 45 min. Proteins (2 μg) were subjected to western blotting with SA-HRP. Asterisks in the right indicate non-specific bands with SA-HRP. To show the band of bOVA in lane 4, exposure time of this figure is longer than Fig. 1B, results in enhancements of non-specific bands with SA-HRP. Equivalent results were attained at least three independent assays. (G), Microsomes with (bOVA before +) or without (bOVA before −) prior addition of bOVA were treated with RL and ATP in presence or absence of and MG132 (10 μM) for 1 h. Samples without prior addition of bOVA (bOVA before −) were added with 1.0 mg/mL bOVA (bOVA after +) or the same volume of PBS (bOVA after −). Supernatants were subjected to immunoprecipitation (IP) with anti-Hsp70. After SDS-PAGE, blotting was performed with SA-HRP and anti-HSP70. Equivalent results were attained at least three independent assays. Full versions of all sliced images are provided in the Supplementary Figures.
3.2. In vitro ubiquitination of exogenously added bOVA in microsomes
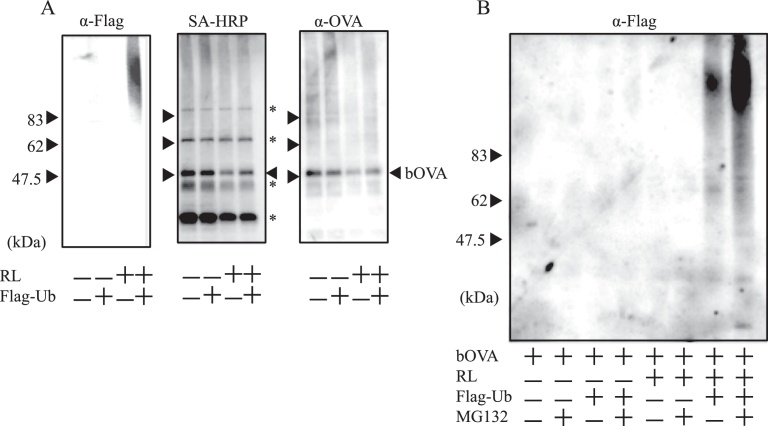
Next, we analyzed in vitro ubiquitination of bOVA in microsomes using Flag-tagged ubiquitin. Microsomes with bOVA were incubated with or without RLs and Flag-tagged ubiquitin. After incubation at 37 °C for 2 h, bOVA was recovered with SA-magnetic beads and resolved by SDS-PAGE followed by western blotting. The majority of bOVA in microsomes showed the same molecular mass as intact bOVA (Fig. 2A). In addition, significant amounts of bOVA were polyubiquitinated, as shown by western blotting with anti-Flag antibodies in the presence of RLs and Flag-Ub (Fig. 2A). The amounts of polyubiquitinated bOVA shown in smear bands increased after the addition of MG132 in the presence of RLs and Flag-Ub (Fig. 2B). These results also suggested that bOVA underwent ERAD-like protein degradation; accumulated in microsomes, retro-transported out of microsomes, ubiqutinated, and then degraded by proteasomes.
Fig. 2.
In vitro reconstitution of ubiquitination using OVA in microsomes (A), Microsomes with prior addition of bOVA were treated with (+) or without (−) RL and Flag-tagged ubiquitin (Flag-Ub) for 1 h and were then solubilized with TNE (20 mM Tris-HCl [pH 7.4], 150 mM NaCl, 0.5 M EDTA, 1% Nonidet P-40). bOVA was purified with SA-magnetic beads and subjected to western blotting with the indicated antibodies. Asterisks in the right indicate non-specific bands with SA-HRP. Equivalent results were attained at least three independent assays. (B), Microsomes with prior addition of bOVA were treated with (+) or without (−) RL, Flag-Ub, and MG132 for 1 h and solubilized using TNE. bOVA was purified with SA-magnetic beads and subjected to western blotting with SA-Flag. Equivalent results were attained at least three independent assays. Full versions of all sliced images are provided in the Supplementary Figures.
3.3. Purification of microsomes containing bOVA
Because exogenously added antigens appeared to be processed through an ERAD-like mechanism, we hypothesized that bOVA in microsomes would undergo translocation dependent on association with membranes via tranlocons, such as Sec61 (Fig. 3A). Because membrane-associated bOVA specifically binds with SA, we purified microsomes containing bOVA using SA-magnetic beads. Microsomes from DC2.4 cells that had been incubated with bOVA were incubated with or without SA-magnetic beads. After incubation at room temperature for 30 min, SA-bound vesicles were isolated using magnetic beads. Microsomes recovered using SA-magnetic beads were resolved by SDS-PAGE followed by silver staining (Fig. 3B, D, F) and western blotting with SA-HRP (Fig. 3C). bOVA was detected in purified vesicles in the presence of exogenously added bOVA and SA-magnetic beads (Fig. 3C). As shown in Fig. 3B, nonspecific proteins appeared to bind with SA-magnetic beads with or without bOVA. In addition to these proteins, we found several unique proteins that appeared only in the presence of exogenously added bOVA and SA-magnetic beads. Addition of free bOVA before incubation with SA-magnetic beads to compete with SA inhibited the purification of both specific and nonspecific proteins (Fig. 3D), indicating that the purification of these proteins was dependent on membrane-associated bOVA. We further confirmed this competition with bOVA by western blotting with SA-HRP (Fig. 3E). Treatment with trypsin before addition of SA-magnetic beads also prevented the purification of microsomes (Fig. 3F), indicating that microsome purification depended on the presence of membrane-associated proteins. The results of bOVA purification following trypsin treatment were also confirmed by western blotting with SA-HRP (Fig. 3G). Collectively, our results showed that purification of microsomes was dependent on membrane-associated bOVA (Fig. 3A).
Fig. 3.
Purification of microsomes with bOVA undergoing ERAD (A), Schematic model of purification of microsomes with bOVA undergoing ERAD. bOVA is associated with the membrane through the Sec61 translocon and targeted with SA-magnetic beads. (B), Microsomes with (+) or without (−) prior addition of bOVA were purified with (+) or without (−) SA-magnetic beads. Proteins (2 μg) or corresponding volumes of purified proteins were resolved on 7.5–15% SDS-PAGE, and silver staining was used to visualize protein bands. Triangles on the right side indicate nonspecific proteins binding to the SA-magnetic beads. Triangles with asterisks indicate unique proteins found only in the presence of exogenously added bOVA and SA-magnetic beads. Arrow indicates bOVA. P.N., post nuclear fraction. Asterisks in the right indicate non-specific bands with SA-HRP. Equivalent results were attained at least three independent assays. (C), Western blotting results of the samples shown in (B) using SA-HRP. Equivalent results were attained at least three independent assays. (D), Microsomes with prior addition of bOVA were purified with (+) or without (−) SA-magnetic beads after addition of 2.5 mg/mL bOVA (+) or the same volume of PBS (−). Proteins (2 μg) or corresponding volumes of purified proteins were resolved on 7.5–15% SDS-PAGE, and silver staining was used to visualize protein bands. Equivalent results were attained at least three independent assays. (E), Western blotting results of the samples shown in (D) using SA-HRP. Equivalent results were attained at least three independent assays. (F), Microsomes with prior addition of bOVA were purified with SA-magnetic beads. Microsomes were treated with (+) or without (−) trypsin and TX-100 before purification (left two lanes) or after purification (right two lanes). Proteins (2 μg) or corresponding volumes of purified proteins were resolved on 7.5–15% SDS-PAGE, and silver staining was used to visualize protein bands. Triangles on the right side indicate nonspecific proteins binding to the SA-magnetic beads. Triangles with asterisks indicate unique proteins found only in the presence of exogenously added bOVA and SA-magnetic beads. Equivalent results were attained at least three independent assays. (G), Western blotting results of the samples shown in (F) using SA-HRP. Equivalent results were attained at least three independent assays. Full versions of all sliced images are provided in the Supplementary Figures.
3.4. In vitro ubiquitination of bOVA by purified vesicles
The bOVA in purified microsomes was degraded in the presence of RLs in a time-dependent manner, and this degradation event was blocked by MG132 and lactacystine but not by chloroquine (Fig. 4A, B) as un-purified microsomes (Fig. 1D). Therefore, we next analyzed the in vitro ubiquitination of bOVA in purified microsomes using Flag-Ub to confirm the accomplishment our purification. After incubation at 37 °C for 2 h with Flag-Ub, bOVA was polyubiquitinated in purified microsomes in the presence of RLs and Flag-Ub (Fig. 4C). The amounts of polyubiquitinated bOVA were augmented in the presence of MG132 (Fig. 4D). These results suggested that purified microsomes contained ERAD machinery proteins.
Fig. 4.
In vitro reconstitution of degradation and ubiquitination using OVA in purified microsomes (A), Purified microsomes were treated with RL, ATP, and the indicated inhibitors. Proteins (2 μg) were subjected to western blotting with SA-HRP. Equivalent results were attained at least three independent assays. (B), Quantification of the results shown in (A). Student's test was used to compare (Before, RL + reagent −) sample with (RL + reagent +) samples. *, ρ < 0.01, NS, not significant, n = 3. (C), Purified microsomes with (+) or without (−) prior addition of bOVA were treated with (+) or without (−) RL and Flag-Ub for 1 h and were solubilized using TNE. bOVA was purified with SA-magnetic beads and subjected to western blotting with the indicated antibodies. Asterisks in the right indicate non-specific bands with SA-HRP. Equivalent results were attained at least three independent assays. (D), Purified microsomes were treated with (+) or without (−) RL, Flag-Ub, and MG132 for 1 h and were then solubilized using TNE. bOVA was purified with SA-magnetic beads and subjected to western blotting with SA-Flag. Asterisks in the right indicate non-specific bands with SA-HRP. Equivalent results were attained at least three independent assays. Full versions of all sliced images are provided in the Supplementary Figures.
3.5. Proteins associated with purified microsomes
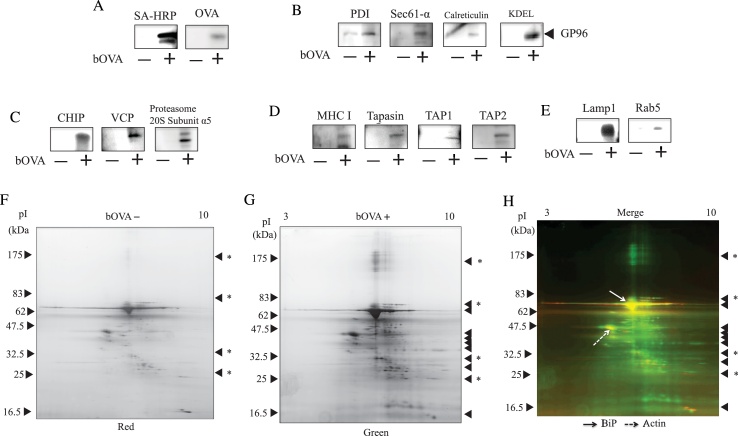
To confirm that purified microsomes were not part of the conventional ER but were specialized compartments, we used western blotting with specific antibodies to detect the presence of ER-associated proteins. bOVA was only detected in bOVA-positive purified microsomes (Fig. 5A). Four ER resident proteins, i.e., PDI, Sec61-α, calreticulin, and GP96, were specifically associated with purified microsomes (Fig. 5B). Molecules for antigen processing and presentation, such as CHIP, VCP, proteasomes, MHC class I H–2 Kb, TAP1, TAP2, and Tapasin, were also associated with purified microsomes (Fig. 5C, D). Copurification of the endosome/lysosome-specific proteins Rab5 and LAMP1 (Fig. 5E) suggest that purified microsomes were not conventional ER vesicles but were specialized compartments including both ERAD proteins and endosome/lysosome proteins. In contrast, proteins found in the caveosome and Golgi apparatus, such as caveolin1 and GM-130, were not detected by specific antibodies in purified microsomes (data not shown). To identify proteins in purified microsomes, we employed two-dimensional difference gel electrophoresis (DIGE). We separated proteins from purified microsomes with bOVA and control microsomes without bOVA on two-dimensional gels and then performed silver staining of the separated proteins. From these data, we searched for proteins that were only present in purified microsomes (Fig. 5F–H). As shown in Fig. 5H, we found many protein spots unique to bOVA-containing purified microsomes (Fig. 5A–E) and two large proteins spots representing nonspecific proteins bound to SA-magnetic beads. These latter two proteins were found to be actin and BiP (data not shown). The unique proteins are now being identified and examined in our laboratory.
Fig. 5.
Proteins in purified microsomes. (A–E), Proteins (2 μg) from purified microsomes with (+) or without (−) prior addition of bOVA were subjected to western blotting with the indicated antibodies. Equivalent results were attained at least three independent assays. (F), Proteins (10 μg) from purified microsomes without (−) prior addition of bOVA were resolved by two-dimensional DIGE. Silver staining was used to visualize protein spots. Triangles on the right side indicate nonspecific proteins binding to the SA-magnetic beads. Equivalent results were attained at least three independent assays. (G), Proteins (10 μg) from purified microsomes with (+) prior addition of bOVA were resolved by two-dimensional DIGE. Silver staining was used to visualize protein spots. Triangles on the right side indicate nonspecific proteins binding to the SA-magnetic beads. Triangles with asterisks indicate unique proteins found only in the presence of exogenously added bOVA and SA-magnetic beads. Equivalent results were attained at least three independent assays. (H), The silver stained image of purified microsomes with (+) prior addition of bOVA was pseudo-colored green (Fig. 5F), and that of control microsomes without (−) prior addition of bOVA was pseudo-colored red (Fig. 5G). Triangles on the right side indicate nonspecific proteins binding to the SA-magnetic beads. Triangles with asterisks indicate unique proteins found only in the presence of exogenously added bOVA and SA-magnetic beads. The two images were merged. A black arrow indicates BiP, and a dotted arrow indicates actin. Full versions of all sliced images are provided in the Supplementary Figures.
4. Discussion
Processing and presentation of exogenous antigens on MHC class I molecules is called antigen cross-presentation. In cross-presentation through the intracellular pathway, exogenous antigens are incorporated by endocytosis, accumulated in ER-like compartments, and retrotransported through the Sec61 complex to the cytosol where they are polyubiquitinated and degraded by the ubiquitin-proteasome system [28, 38]. The latter half of this machinery, i.e., retrotransport and ubiquitin-dependent degradation of exogenous antigens, employs the protein quality control system in the ER, called ERAD. These latter steps require driving forces provided by hydrolyses ATP, retrotransport, polyubiquitination, and Ubiquitin-dependent degradation by the proteasome. However, no previous studies had examined whether exogenous antigens accumulated in conventional ER vesicles or in specialized microsomes for cross-presentation and how exogenous antigens are recognized as substrates for ERAD.
In this report, we purified microsomes containing exogenously added OVA and reconstituted the ERAD-dependent degradation of exogenously added OVA in purified microsomes in vitro. Exogenously added OVA accumulated in microsomal fractions and were degraded in a time-dependent manner in the presence of RLs and ATP. This degradation of OVA was inhibited by proteasome inhibitors but not by a lysosome inhibitor, indicating that bOVA was processed by proteasomes. Requirements of ATP for degradation of OVA also support our hypothesis; exogenously added OVA were processed by proteasomes. We also found that some portion of exogenous OVA was retrotransported to outside of microsomes and associated with Hsp70 in the presence of RLs. The amounts of bOVA associated with Hsp70 were augmented by addition of the proteasome inhibitor suggesting that bOVA was unfolded after retro-translocation to cytosol before degradation by proteasomes.
We also reconstituted ubiquitination of exogenous OVA in the presence of RLs. These ubiquitination was only detected by anti-Flag antibody, but not by neither SA-HRP nor anti-OVA antibody. Since considerable amounts of bOVA incorporated into microsomes was ubiqutinated before fractionation, we could not distinguish these two kinds of bOVA, one was ubiqutinated before fractionation and another was ubiqutinated after addition of Flag-Ub both by SA-HRP and anti-OVA antibody. Addition of MG132 enhanced smear bands detected by anti-Flag antibody indicating that those smear bands were results of in vitro ubiqutination by Flag-Ub.
After retrotransport to the cytosol, OVA was ubiquitinated and then degraded by proteasomes in the presence of RLs and ATP, demonstrating that the latter half of the cross-presentation pathway was reconstituted and that the exogenously incorporated OVA could be recognized as a substrate for ERAD. Since Hsp70 facilitates ERAD substrate selection and targeting [37], coprecipitation of exogenous OVA added before fractionation with Hsp70 also supports our hypothesis. However, when OVA were added after fractionation, it did not localize to the microsomes and was not degraded efficiently. Since conventional ERAD substrates are misfolded proteins that have failed to fold properly in the ER, exogenous OVA added before fractionation should be unfolded after endocytosis. In addition to the above results, four lines of evidence also support the unfolding of incorporated OVA: 1) the bOVA used in this study was soluble and showed equivalent cross-presentation efficiency with normal nonbiotinylated OVA [38]; 2) exogenous bOVA added before solubilization of cells but after solubilization was associated with several ER resident molecular chaperones [38]; 3) OVA, a member of the albumin family of proteins, has a highly conserved globular structure and is soluble in water [50]; 4) serum albumin has a molecular chaperone-like role, associating with extracellular unfolded proteins and inhibiting their aggregation [51]. Therefore, exogenous OVA were unfolded after endocytosis in the microsome fraction, allowing it to be recognized as a substrate for the ERAD machinery; and 5) since bOVA neither degraded nor associated with Hsp70 when OVA was added after fractionation, the only addition of RLs on native OVA never influence upon the folding of this protein.
We and other groups previously reported that exogenous antigens are localized in both the ER and endosomes in DCs [28, 38, 52]. However, these studies did not clarify the cellular fractions in which exogenous antigens undergo ERAD. Therefore, in the current study, we purified microsomes in which exogenous antigens were transported across membranes. We hypothesized that exogenous antigens associated with the membrane via translocons, such as Sec61. Using bOVA as an exogenous antigen, we purified microsomes with SA-magnetic beads. Our results identified several unique proteins that were purified in the presence of both bOVA and SA. Competition with free bOVA and digestion of membranous bOVA with trypsin inhibited the purification of these proteins, indicating that SA targeted membrane-associated bOVA, consistent with our hypothesis. From these purified microsomes, we reconstructed the proteasome-dependent ubiquitination and degradation of bOVA in vitro, supporting the hypothesis that ERAD occurred to promote cross-presentation of exogenous antigens in specialized compartments.
The purified microsomes contained both ER resident proteins and endosome/lysosome-specific proteins, suggesting that purified microsomes showed specific features of both the ER and endosomes/lysosomes. In contrast, proteins from other membranous fractions, such as caveosomes and the Golgi apparatus, were not purified, indicating that non-specific membranous fractions were not included in these purified microsomes. In addition to these membranous proteins, cytosolic proteins involved in antigen processing were purified in the isolated microsomes. Those purified proteins included a set of machinery for cross-presentation, retrotransport, ubiquitination, and processing of extracellular antigens and transport and loading of antigenic peptides on MHC class I molecules, indicating that ERAD and antigenic peptide loading occurred in these purified microsomes and suggesting that purified microsomes were specific for cross-presentation. In addition, the purified microsomes contained more than 100 unique proteins, which are now being experimentally examined in our laboratory. However as shown in Fig. 5H, BiP non-specifically bounds with SA-magnetic beads, indicating a small amounts of ER derived vesicle purified non-specifically by our purification methods. But the amounts of those non-specific proteins, such as PDI, BiP, and Calreticulin, are low enough compared with their positive controls. Essentially these non-specifically purified vesicles showed neither ubiquitination nor degradation against extra added bOVA after purification (data not shown).
DCs have the capacity for cross-presentation in order to efficiently destroy non-self cells, such as cancer cells or virus-infected cells. Although this process requires rapid degradation of endogenous antigens, properly folded endogenous antigens will not degraded promptly; the half-life of properly folded viral antigens is sufficiently long to escape direct presentation. As shown previously and in this report, endogenously added OVA was associated with ER resident molecular chaperones and degraded by ERAD, indicating that OVA was unfolded in DCs [38]. In Herpes simplex virus (HSV)-infected cells, proteins containing disulfide bonds from can be digested by gamma-interferon-inducible lysosomal thiolreductase (GILT), and unfolding of HSV proteins has been shown to be critical for cross-presentation [53]. Because unfolded proteins in cells are harmful, cells rapidly recognize and degrade unfolded proteins through the ubiquitin-proteasome system, and the process of unfolding of extracellular antigens plays an essential role in this rapid cross-presentation. Although DCs are known to use the ERAD system for rapid cross-presentation, the process through which proteins are unfolded has still not been clarified. In this report, we purified microsomes in which endogenous antigens were degraded through the ERAD system. The proteins included in these purified microsomes resembled proteins described in proteomic analyses of ER-phagosomes [24, 29, 54] or ERgosomes [25, 27]. In these compartments, the ER fuses with the plasma membrane, providing the required membrane and protein machinery for cross-presentation [24, 25, 26, 27, 29, 53, 55, 56, 57]. In addition, our data suggested that the ER might function in the recognition of incorporated antigens. Most proteins in endosomes are unfolded as the endosome matures; our data suggested that some mature endosomes in DCs fuse with the ER. In mature endosome exogenous antigens are unfolded, and after fusion with ER ER-resident molecular chaperones recognize the unfolded proteins for effective EARD. Proteomic analyses of the above-purified microsomes are needed in the future in order to test this hypothesis.
Declarations
Author contribution statement
Jun Imai: Conceived and designed the experiments; Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data; Wrote the paper.
Mayu Otani: Performed the experiments.
Takahiro Sakai: Performed the experiments; Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Shinichi Hatta: Analyzed and interpreted the data; Contributed reagents, materials, analysis tools or data.
Funding statement
This work was supported by the Tkasaki University of Health and Welfare.
Competing interest statement
The authors declare no conflict of interest.
Additional information
No additional information is available for this paper.
Appendix A. Supplementary data
References
- 1.Janeway C., Travers P., Walport M., Shlomchik M. 5th edn. Garland Press; New York: 2001. Immunobiology: The Immune System in Health and Disease. [Google Scholar]
- 2.Yewdell J., Anton L.C., Bacik I., Schubert U., Snyder H.L., Bennink J.R. Generating MHC class I ligands from viral gene products. Immunol. Rev. 1999;172:97–108. doi: 10.1111/j.1600-065x.1999.tb01359.x. [DOI] [PubMed] [Google Scholar]
- 3.Huang A.Y., Golumbek P., Ahmadzadeh M., Jaffee E., Pardoll D., Levitsky H. Role of bone marrow-derived cells in presenting MHC class I-restricted tumor antigens. Science. 1994;264:961–965. doi: 10.1126/science.7513904. [DOI] [PubMed] [Google Scholar]
- 4.Banchereau J., Steinman R.M. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 5.Bevan M.J., Minor H. Antigens introduced on H-2 different stimulating cells cross-react at the cytotoxic T cell level during in vivo priming. J. Immunol. 1976;117:2233–2238. [PubMed] [Google Scholar]
- 6.Carbone F.R., Kurts C., Bennett S.R., Miller J.F., Heath W.R. Cross-presentation: a general mechanism for CTL immunity and tolerance. Immunol. Today. 1998;19:368–373. doi: 10.1016/s0167-5699(98)01301-2. [DOI] [PubMed] [Google Scholar]
- 7.Nair-Gupta P., Blander J.M. An updated view of the intracellular mechanisms regulating cross-presentation. Front. Immunol. 2013;4:401. doi: 10.3389/fimmu.2013.00401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Joffre O.P., Segura E., Savina A., Amigorena S. Cross-presentation by dendritic cells. Nat. Rev. Immunol. 2012;12:557–569. doi: 10.1038/nri3254. [DOI] [PubMed] [Google Scholar]
- 9.Bedoui S., Whitney P.G., Waithman J., Eidsmo L., Wakim L., Caminschi I., Allan R.S., Wojtasiak M., Shortman K., Carbone F.R., Brooks A.G., Heath W.R. Cross-presentation of viral and self antigens by skin-derived CD103+ dendritic cells. Nat. Immunol. 2009;10:488–495. doi: 10.1038/ni.1724. [DOI] [PubMed] [Google Scholar]
- 10.Segura E., Villadangos J.A. Antigen presentation by dendritic cells in vivo. Curr. Opin. Immunol. 2009;21:105–110. doi: 10.1016/j.coi.2009.03.011. [DOI] [PubMed] [Google Scholar]
- 11.Cheong C., Matos I., Choi J.H., Dandamudi D.B., Shrestha E., Longhi M.P., Jeffrey K.L., Anthony R.M., Kluger C., Nchinda G., Koh H., Rodriguez A., Idoyaga J., Pack M., Velinzon K., Park C.G., Steinman R.M. Microbial stimulation fully differentiates monocytes to DC-SIGN/CD209(+) dendritic cells for immune T cell areas. Cell. 2010;143:416–429. doi: 10.1016/j.cell.2010.09.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henri S., Poulin L.F., Tamoutounour S., Ardouin L., Guilliams M., de Bevis B., Devilard E., Viret C. CD207+ CD103+ dermal dendritic cells cross-present keratinocyte-derived antigens irrespective of the presence of Langerhans cells. J. Exp. Med. 2010;207:189–206. doi: 10.1084/jem.20091964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shortman K., Heath W.R. The CD8+ dendritic cell subset. Immunol. Rev. 2010;234:18–31. doi: 10.1111/j.0105-2896.2009.00870.x. [DOI] [PubMed] [Google Scholar]
- 14.Schulz O., Diebold S.S., Chen M., Näslund T.I., Nolte M.A., Alexopoulou L., Azuma Y.T., Flavell R.A., Liljeström P., Reis e Sousa C. Toll-like receptor 3 promotes cross-priming to virus-infected cells. Nature. 2005;433:887–892. doi: 10.1038/nature03326. [DOI] [PubMed] [Google Scholar]
- 15.Gabrilovich D.I., Patterson S., Harvey J.J., Woods G.M., Elsley W., Knight S.C. Murine retrovirus induces defects in the function of dendritic cells at early stages of infection. Cell Immunol. 1994;158:167–181. doi: 10.1006/cimm.1994.1265. [DOI] [PubMed] [Google Scholar]
- 16.Fugier-Vivier I., Servet-Delprat C., Rivailler P., Rissoan M.C., Liu Y.J., Rabourdin-Combe C. Measles virus suppresses cell-mediated immunity by interfering with the survival and functions of dendritic and T cells. J. Exp. Med. 1997;186:813–823. doi: 10.1084/jem.186.6.813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Engelmayer J., Larsson M., Subklewe M., Chahroudi A., Cox W.I., Steinman R.M., Bhardwaj N. Vaccinia virus inhibits the maturation of human dendritic cells: a novel mechanism of immune evasion. J. Immunol. 1999;163:6762–6768. [PubMed] [Google Scholar]
- 18.Salio M., Cella M., Suter M., Lanzavecchia A. Inhibition of dendritic cell maturation by herpes simplex virus. Eur. J. Immunol. 1999;29:3245–3253. doi: 10.1002/(SICI)1521-4141(199910)29:10<3245::AID-IMMU3245>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 19.Sigal L.J., Crotty S., Andino R., Rock K.L. Cytotoxic T-cell immunity to virus- infected non-haematopoietic cells requires presentation of exogenous antigen. Nature. 1999;398:77–80. doi: 10.1038/18038. [DOI] [PubMed] [Google Scholar]
- 20.Ignatius R., Marovich M., Mehlhop E., Villamide L., Mahnke K., Cox W.I., Isdell F., Frankel S.S., Mascola J.R., Steinman R.M., Pope M. Canarypox virus-induced maturation of dendritic cells is mediated by apoptotic cell death and tumor necrosis factor alpha secretion. J. Virol. 2000;74:11329–11338. doi: 10.1128/jvi.74.23.11329-11338.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Servet-Delprat C., Vidalain P.O., Bausinger H., Manié S., Le Deist F., Azocar O., Hanau D., Fischer A., Rabourdin-Combe C. Measles virus induces abnormal differentiation of CD40 ligand-activated human dendritic cells. J. Immunol. 2000;164:1753–1760. doi: 10.4049/jimmunol.164.4.1753. [DOI] [PubMed] [Google Scholar]
- 22.Sevilla N., Kunz S., Holz A., Lewick H., Homann D., Yamada H., Campbell K.P., de La Torre J.C., Oldstone M.B. Immunosuppression and resultant viral persistence by specific viral targeting of dendritic cells. J. Exp. Med. 2000;192:1249–1260. doi: 10.1084/jem.192.9.1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.John B., Harris T.H., Tait E.D., Wilson E.H., Gregg B., Ng I.G., Mrass P., Roos D.S., Dzierszinski F., Weninger W., Hunter C.A. Dynamic imaging of CD8 (+) T cells and dendritic cells during infection with Toxoplasma gondii. PLoS Pathog. 2009;5 doi: 10.1371/journal.ppat.1000505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Heath W.R., Carbone F.R. Cross-presentation in viral immunity and self-tolerance. Nat. Rev. Immunol. 2001;1:126–134. doi: 10.1038/35100512. [DOI] [PubMed] [Google Scholar]
- 25.Kovacsovics-Bankowski M., Rock K.L. A phagosome-to-cytosol pathway for exogenous antigens presented on MHC class I molecules. Science. 1995;267:243–246. doi: 10.1126/science.7809629. [DOI] [PubMed] [Google Scholar]
- 26.Guermonprez P., Saveanu L., Kleijmeer M., Davoust J., VanEndert P., Amigorena S. ER-phagosome fusion defines an MHC class I cross-presentation compartment in dendritic cells. Nature. 2003;425:397–402. doi: 10.1038/nature01911. [DOI] [PubMed] [Google Scholar]
- 27.Houde M., Bertholet S., Gagnon E., Brunet S., Goyette G., Laplante A., Princiotta M.F., Thibault P., Sacks D., Desjardins M. Phagosomes are competent organelles for antigen cross-presentation. Nature. 2003;425:402–406. doi: 10.1038/nature01912. [DOI] [PubMed] [Google Scholar]
- 28.Huang A.Y., Bruce A.T., Pardoll D.M., Levitsky H.I. In vivo cross-priming of MHC class I-restricted antigens requires the TAP transporter. Immunity. 1996;4:349–355. doi: 10.1016/s1074-7613(00)80248-4. [DOI] [PubMed] [Google Scholar]
- 29.Ackerman A.L., Giodini A., Cresswell P. A role for the endoplasmic reticulum protein retrotranslocation machinery during crosspresentation by dendritic cells. Immunity. 2006;25:607–617. doi: 10.1016/j.immuni.2006.08.017. [DOI] [PubMed] [Google Scholar]
- 30.Rock K.L., Gamble S., Rothstein L. Presentation of exogenous antigen with class I major histocompatibility complex molecules. Science. 1990;249:918–921. doi: 10.1126/science.2392683. [DOI] [PubMed] [Google Scholar]
- 31.Melief C.J.M. Regulation of cytotoxic T lymphocyte responses by dendritic cells: peaceful coexistence of cross-priming and direct priming? Eur. J. Immunol. 2003;33:2645–2654. doi: 10.1002/eji.200324341. [DOI] [PubMed] [Google Scholar]
- 32.Heath W.R., Carbone F.R. Cross-presentation dendritic cells, tolerance and immunity. Annu. Rev. Immunol. 2001;20:47–64. doi: 10.1146/annurev.immunol.19.1.47. [DOI] [PubMed] [Google Scholar]
- 33.Delamarre L., Pack M., Chang H., Mellman I., Trombetta E.S. Differential lysosomal proteolysis in antigen-presenting cells determines antigen fate. Science. 2005;307:1630–1634. doi: 10.1126/science.1108003. [DOI] [PubMed] [Google Scholar]
- 34.Savina A., Jancic C., Hugues S., Guermonprez P., Vargas P., Moura I.C., Lennon-Duménil A.M., Seabra M.C., Raposo G., Amigorena S. NOX2 controls phagosomal pH to regulate antigen processing during crosspresentation by dendritic cells. Cell. 2006;126:205–218. doi: 10.1016/j.cell.2006.05.035. [DOI] [PubMed] [Google Scholar]
- 35.Claus V., Jahraus A., Tjelle T., Berg T., Kirschke H., Faulstich H., Griffiths G. Lysosomal enzyme trafficking between phagosomes endosomes, and lysosomes in J774 macrophages. Enrichment of cathepsin H in early endosomes. J. Biol. Chem. 1998;273:9842–9851. doi: 10.1074/jbc.273.16.9842. [DOI] [PubMed] [Google Scholar]
- 36.Lin M.L., Zhan Y., Proietto A.I., Prato S., Wu L., Heath W.R., Villadangos J.A., Lew A.M. Selective suicide of cross-presenting CD8+ dendritic cells by cytochrome c injection shows functional heterogeneity within this subset. Proc. Natl. Acad. Sci. USA. 2008;105:3029–3034. doi: 10.1073/pnas.0712394105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Vembar S.S., Brodsky J.L. One step at a time: endoplasmic reticulum-associated degradation. Nat. Rev. Mol. Cell Biol. 2008;9:944–957. doi: 10.1038/nrm2546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Imai J., Hasegawa H., Maruya M., Koyasu S., Yahara I. Exogenous antigens are processed through the endoplasmic reticulum-associated degradation (ERAD) in cross-presentation by dendritic cells. Int. Immunol. 2005;17:45–53. doi: 10.1093/intimm/dxh184. [DOI] [PubMed] [Google Scholar]
- 39.Schild H., Rammensee H.G. Perfect use of imperfection. Nature. 2000;404:709–710. doi: 10.1038/35008165. [DOI] [PubMed] [Google Scholar]
- 40.Sherman M.Y., Goldberg A.L. Cellular defenses against unfolded proteins: a cell biologist thinks about neurodegenerative diseases. Neuron. 2001;29:15–32. doi: 10.1016/s0896-6273(01)00177-5. [DOI] [PubMed] [Google Scholar]
- 41.Schirmbeck R., Bohm W., Reimann J. Stress protein (hsp73)-mediated TAP-independent processing of endogenous, truncated SV40 large T antigen for Db- restricted peptide presentation. Eur. J. Immunol. 1997;27:2016–2023. doi: 10.1002/eji.1830270828. [DOI] [PubMed] [Google Scholar]
- 42.Wiertz E.J., Tortorella D., Bogyo M., Yu J., Mothes W., Jones T.R., Rapoport T.A., Ploegh H.L. Sec61-mediated transfer of a membrane protein from the endoplasmic reticulum to the proteasome for destruction. Nature. 1996;384:432–438. doi: 10.1038/384432a0. [DOI] [PubMed] [Google Scholar]
- 43.Hampton R.Y. ER-associated degradation in protein quality control and cellular regulation. Curr. Opin. Cell Biol. 2002;14:476–482. doi: 10.1016/s0955-0674(02)00358-7. [DOI] [PubMed] [Google Scholar]
- 44.Tsai B., Ye Y., Rapoport T.A. Retro-translocation of proteins from the endoplasmic reticulum into the cytosol. Nat. Rev. Mol. Cell Biol. 2002;3:246–255. doi: 10.1038/nrm780. [DOI] [PubMed] [Google Scholar]
- 45.Schubert U., Anton L.C., Gibbs J., Norbury C.C., Yewdell J.W., Bennink J.R. Rapid degradation of a large fraction of newly synthesized proteins by proteasomes. Nature. 2000;404:770–774. doi: 10.1038/35008096. [DOI] [PubMed] [Google Scholar]
- 46.Reits E.A., Vos J.C., Gromme M., Neefjes J. The major substrates for TA-P in vivo are derived from newly synthesized proteins. Nature. 2000;404:774–778. doi: 10.1038/35008103. [DOI] [PubMed] [Google Scholar]
- 47.Lelouard H., Gatti E., Cappello F., Gresser O., Camossetto V., Pierre P. Transient aggregation of ubiquitinated proteins during dendritic cell maturation. Nature. 2002;417:177–182. doi: 10.1038/417177a. [DOI] [PubMed] [Google Scholar]
- 48.Lelouard H., Ferrand V., Marguet D., Bania J., Camosseto V., David A., Gatti E., Pierre E.P. Dendritic cell aggresome-like induced structures are dedicated areas for ubiquitination and storage of newly synthesized defective proteins. J. Cell Biol. 2004;164:667–675. doi: 10.1083/jcb.200312073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shen Z., Reznikoff G., Dranoff G., Rock K.L. Cloned dendritic cells can present exogenous antigens on both MHC class I and class II molecules. J. Immunol. 1997;158:2723–2730. [PubMed] [Google Scholar]
- 50.Sugio S., Kashima A., Mochizuki S., Noda M., Kobayashi K. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. 1999;12:439–446. doi: 10.1093/protein/12.6.439. [DOI] [PubMed] [Google Scholar]
- 51.Finn T.E., Nunez A.C., Sunde M., Easterbrook-Smith S.B. Serum albumin prevents protein aggregation and amyloid formation and retains chaperone-like activity in the presence of physiological ligands. J. Biol. Chem. 2012;287:21530–21540. doi: 10.1074/jbc.M112.372961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Matheoud D., Moradin N., Bellemare-Pelletier A., Shio M.T., Hong W.J., Olivier M., Gagnon E., Desjardins M., Descoteaux A. Leishmania evades host immunity by inhibiting antigen cross- presentation through direct cleavage of the SNARE VAMP8. Cell Host Microbe. 2013;14:15–25. doi: 10.1016/j.chom.2013.06.003. [DOI] [PubMed] [Google Scholar]
- 53.Singh R., Cresswell P. Defective cross-presentation of viral antigens in GILT-free mice. Science. 2010;328:1394–1398. doi: 10.1126/science.1189176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ackerman A.L., Kyritsis C., Tampe R., Cresswell P. Early phagosomes in dendritic cells form a cellular compartment sufficient for cross presentation of exogenous antigens. Proc. Natl. Acad. Sci. USA. 2003;100:12889–12894. doi: 10.1073/pnas.1735556100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Burgdorf S., Scholz C., Kautz A., Tampe R., Kurts C. Spatial and mechanistic separation of cross- presentation and endogenous antigen presentation. Nat. Immunol. 2008;9:558–566. doi: 10.1038/ni.1601. [DOI] [PubMed] [Google Scholar]
- 56.Gagnon E., Duclos S., Rondeau C., Chevet E., Cameron P.H., Steele-Mortimer O., Paiement J., Bergeron J.J., Desjardins M. Endoplasmic reticulum-mediated phagocytosis is a mechanism of entry into macrophages. Cell. 2002;110:119–131. doi: 10.1016/s0092-8674(02)00797-3. [DOI] [PubMed] [Google Scholar]
- 57.Müller-Taubenberger A., Lupas A.N., Li H., Ecke M., Simmeth E., Gerisch G. Calreticulin and calnexin in the endoplasmic reticulum are important for phagocytosis. EMBO J. 2001;20:6772–6782. doi: 10.1093/emboj/20.23.6772. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.